Abstract
Alternative polyadenylation (APA) is an important post-transcriptional regulation that recognizes different polyadenylation signals (PASs), resulting in transcripts with different 3′ untranslated regions, thereby influencing a series of biological processes and functions. Recent studies have revealed that some single nucleotide polymorphisms (SNPs) could contribute to tumorigenesis and development through dysregulating APA. However, the associations between SNPs and APA in human cancers remain largely unknown. Here, using genotype and APA data of 9082 samples from The Cancer Genome Atlas (TCGA) and The Cancer 3′UTR Altas (TC3A), we systematically identified SNPs affecting APA events across 32 cancer types and defined them as APA quantitative trait loci (apaQTLs). As a result, a total of 467 942 cis-apaQTLs and 30 721 trans-apaQTLs were identified. By integrating apaQTLs with survival and genome-wide association studies (GWAS) data, we further identified 2154 apaQTLs associated with patient survival time and 151 342 apaQTLs located in GWAS loci. In addition, we designed an online tool to predict the effects of SNPs on PASs by utilizing PAS motif prediction tool. Finally, we developed SNP2APA, a user-friendly and intuitive database (http://gong_lab.hzau.edu.cn/SNP2APA/) for data browsing, searching, and downloading. SNP2APA will significantly improve our understanding of genetic variants and APA in human cancers.
INTRODUCTION
Alternative polyadenylation (APA) is a widespread phenomenon that generates transcript isoforms with different lengths of 3′ untranslated regions (3′UTR) by recognizing different polyadenylation signals (PASs) (1). More than 70% of human genes have multiple polyadenylation sites (2). As a common post-transcriptional modification mechanism, APA events may cause the alteration of important regulatory elements, such as miRNA binding sites and RNA protein binding sites, thus impacting the stability, localization and translation rate of mRNAs (3). APA modulation has been investigated in cells, tissues and different diseases. Previous studies have shown that APA often functions in a tissue- or cell-specific manner (4,5), and several APA dysregulations have been identified in human diseases (6–9), including cancers (10). A significant global 3′UTR shortening has been found in cancer cell lines and tumor samples, compared with normal samples (11). Another study pointed out that shortening or lengthening of 3′UTR might lead to a worse prognosis in some cancers. For example, kidney cancer samples with the shorter isoforms TMCO7 and PLXDC2 were found to have lower survival rates (12). However, research on the APA role and APA regulation in cancer is still at an early stage.
As the most common genetic variant, single nucleotide polymorphisms (SNPs) are major contributors to the differences in human disease susceptibility (13). Genome-wide association studies (GWAS) have identified thousands of SNPs associated with complex traits and diseases. Currently, most studies of the disease/trait-related SNPs remain at statistical level, and the biological mechanism underlying them is still largely unknown (14). Quantitative trait locus (QTL) mapping, such as eQTL and meQTL analysis, is a method used to evaluate the effects of genetic variants on intermediate molecular phenotypes, and has been demonstrated as a powerful tool to decipher the function of SNPs and prioritize genetic variants within GWAS loci (15–19). Recent studies have confirmed the associations between several APA quantitative trait loci (apaQTLs) and cancer. For example, the presence of a SNP in a canonical PAS within TP53 (AATAAA to AATACA) has been found to be highly associated with the processing of the impaired 3′ end of TP53 transcripts and increase the susceptibility to cancers including cutaneous basal cell carcinoma, prostate cancer, glioma and colorectal adenoma (20). However, large-scale genome-wide analyses of apaQTL have rarely been reported, and no database for apaQTLs in cancer is available. Recently, Feng et al. have used Percentage of Distal polyA site Usage Index (PDUI) to quantify APA events for 10,537 tumor samples across TCGA 32 cancer types (21). Therefore, it is feasible to add APA as an additional dimension to the existing cancer genomic analysis.
In this study, by using the genotype and PDUI data, we developed a new computational pipeline to systematically perform apaQTL analyses across 32 cancer types. We further identified apaQTLs associated with patient overall survival time and apaQTLs located in GWAS linkage disequilibrium (LD) regions. The SNP2APA database (http://gong_lab.hzau.edu.cn/SNP2APA/) was constructed for browsing, searching and downloading the apaQTL data.
MATERIALS AND METHODS
Collection and processing of genotype data
We downloaded the genotype data across 32 cancer types from the TCGA data portal (https://portal.gdc.cancer.gov/) (22), which contained 898,620 SNPs called by Affymetrix SNP 6.0 array. We extracted 9082 samples with both genotype data and APA data available (Figure 1A). To increase the power for apaQTL discovery, IMPUTE2 was used to impute autosomal variants of all samples in each cancer type with haplotypes of 1000 Genomes Phase 3 as the reference panel (23,24). After imputation, SNPs of each cancer type were selected in terms of the following criteria (25): (i) imputation confidence score, INFO ≥0.4, (ii) minor allele frequency (MAF) ≥5%, (iii) SNP missing rate <5% for best-guessed genotypes at posterior probability ≥0.9 and (iv) Hardy-Weinberg equilibrium P-value > 1 × 10−6 estimated by Hardy-Weinberg R package (26).
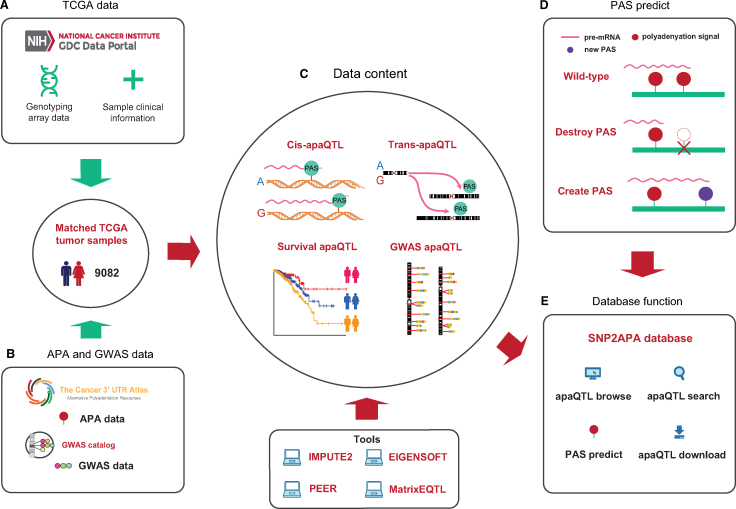
Figure 1.
Simplified schematic showing the workflow of SNP2APA database. (A) Collection of genotype and clinical data. (B) Collection of APA data and GWAS data. (C) Database content in SNP2APA. (D) The online PAS predict tool in SNP2APA. (E) Main functions in SNP2APA.
Collection and processing of data for APA events
To quantify dynamic APA events, we used the PDUI value as the indicator and downloaded them from The TC3A Data Portal (http://tc3a.org/) for 32 cancer types (Figure 1B) (21). PDUI value was a novel, intuitive ratio for quantifying APA events based on RNA-Seq data (12). PDUI was calculated by the number of transcripts with distal polyA site divided by the total number of transcripts with both distal and proximal polyA sites. The greater PDUI represented the more transcripts using the distal polyA site, and vice versa. For example, value 1 indicated that all transcripts of the gene used the distal polyA site, while value 0 indicated that all transcripts of the gene used the proximal polyA site. For each cancer type, APA events were selected as follows: (i) the missing rate of PDUI data <0.1, (ii) the standard deviation of PDUI > 5%. After filtering, an average of 4143 APA events per cancer type were included for the further analyses. To minimize the effects of outliers on the regression scores, the PDUI values of each gene across all samples were transformed into a standard normal based on rank (25).
Obtaining of covariates
To improve the sensitivity in QTL analyses, we collected several known and unknown confounders as covariates for apaQTL analysis (25). We first used the smartpca in the EIGENSTRAT program (27) to perform principal component analysis (PCA) of the genotype data for each cancer type. The top five principal components in genotype data were included as covariates for correcting the ethnicity differences. We additionally used PEER software (28) to analyse the APA data and obtained the first 15 PEER factors as covariates which were used for eliminating the possible batch effects and other confounders. Finally, other common confounders such as gender, age and tumor stage (25,29,30), were also included as covariates for apaQTL analysis.
Identification of cis- and trans-apaQTL using MatrixEQTL
For each cancer type, we evaluated pairwise associations between autosomal SNPs and APA events through linear regression by MatrixEQTL (31), a software for efficient QTL analysis. The SNP locations (hg19) were downloaded from dbSNP database (https://www.ncbi.nlm.nih.gov/projects/SNP) and distal PAS locations were extracted from the APA datasets. The SNPs with false discovery rates (FDRs) <0.05 calculated by MatrixEQTL and the absolute value of correlation coefficient (r) ≥0.3 were defined as apaQTLs (Figure 1C). Of them, we further defined the apaQTLs within 1 Mb from the distal PAS as the cis-apaQTLs (25), while defined the apaQTLs beyond that region or on another chromosome as the trans-apaQTLs.
Identification of survival-associated apaQTLs
To prioritize promising apaQTLs, we further examined the association between apaQTLs and patient survival time. The clinical data including survival time of patient were downloaded from TCGA data portal. For each apaQTL, the samples were divided into three groups by genotypes: homozygous genotype (AA), heterozygous genotype (Aa), and homozygous genotype (aa). Then the log-rank test was performed to examine the differences in survival time, and Kaplan–Meier (KM) curves were plotted for intuitive visualization of the survival time for each group. Finally, apaQTLs with FDR <0.05 were designated as survival-associated apaQTLs.
Identification of GWAS-associated apaQTLs
GWAS has been successfully used for identifying thousands of disease susceptibility loci, but it remains a challenge to pinpoint the causal variants and decipher their underlying mechanisms. To facilitate the interpretation of GWAS results, we integrated apaQTLs with the existing GWAS risk loci to explore trait/disease-associated apaQTLs. We downloaded all the risk tag SNPs identified in GWAS studies from GWAS catalog (http://www.ebi.ac.uk/gwas, accessed September 2018) (32). Then the SNPs in linkage disequilibrium (LD) regions with GWAS tag SNPs were extracted from SNAP (https://personal.broadinstitute.org/plin/snap/ldsearch.php) (33). The parameters were set as follows: (i) SNP dataset: 1000 Genomes, (ii) r2 (the square of the Pearson correlation coefficient of LD) threshold: 0.5, (iii) population panel: CEU (Utah residents with northern and western European ancestry), (iv) distance limit: 500 kb. Finally, we defined apaQTLs that overlapped with these GWAS tag SNPs and LD SNPs as GWAS-associated apaQTLs.
DATABASE CONSTRUCTION AND CONTENT
All results mentioned above were stored into MongoDB database (version 3.4.2) in the form of relation tables. A user-friendly web interface, SNP2APA (http://gong_lab.hzau.edu.cn/SNP2APA/), was constructed to support data browsing, searching, downloading and PAS online prediction (Figure 1D and E), based on Flask (version 1.0.3) framework with Angularjs (version 1.6.1) as the JavaScript library. It was running on Apache2 web server (version 2.4.18). We have tested SNP2APA on various web browsers, including Chrome (recommended), Firefox, Opera, Internet Explorer, Windows Edge and Safari of macOS.
Data summary of SNP2APA
In total, SNP2APA included 9082 tumor samples across 32 cancer types with both genotype data and APA data available for apaQTL analysis. The sample sizes for each cancer type ranged from 36 in cholangiocarcinoma (CHOL) to 1,091 in invasive breast carcinoma (BRCA) with a median of 221 (Table 1). After genotype imputation and quality control, 4 390 660 SNPs on average per each cancer type were included for further analysis, ranging from 2 746 335 for BRCA to 5 143 663 for acute myeloid leukemia (LAML). After filtering APA events by both the rate of missing PDUI value >0.1 and PDUI standard deviation >0.05, we obtained an average of 4143 APA events per cancer type, ranging from 519 for thyroid carcinoma (THCA) to 6978 for stomach adenocarcinoma (STAD).
Table 1.
Summary of apaQTLs in SNP2APA
| Cis | Trans | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cancer type | No. of amples | No. of enotypes | No. of PA events | Pairs | APA events | apaQTLs | Pairs | APA events | apaQTLs |
| ACC | 77 | 3 567 954 | 3114 | 3026 | 135 | 2864 | 1566 | 158 | 1422 |
| BLCA | 408 | 4 190 525 | 3780 | 17 072 | 218 | 16 472 | 883 | 82 | 819 |
| BRCA | 1 091 | 2 746 335 | 5379 | 11 941 | 212 | 11 376 | 501 | 7 | 470 |
| CESC | 299 | 4 291 784 | 3268 | 14 767 | 211 | 14 358 | 773 | 114 | 745 |
| CHOL | 36 | 4 012 152 | 3564 | 1 710 | 54 | 1610 | 1980 | 34 | 1153 |
| COAD | 285 | 4 499 815 | 3356 | 15 797 | 231 | 15 264 | 1341 | 231 | 1280 |
| DLBC | 48 | 4 845 461 | 3658 | 1630 | 67 | 1580 | 2640 | 126 | 2171 |
| ESCA | 184 | 4 457 611 | 4510 | 27 484 | 615 | 26 009 | 665 | 122 | 644 |
| GBM | 150 | 4 556 998 | 5353 | 36 614 | 801 | 34 381 | 575 | 126 | 539 |
| HNSC | 518 | 4 254 665 | 4646 | 19 960 | 254 | 19 162 | 715 | 18 | 655 |
| KICH | 66 | 3 771 774 | 4477 | 3047 | 136 | 3010 | 1477 | 128 | 1313 |
| KIRC | 525 | 4 577 720 | 4906 | 20 978 | 240 | 19 596 | 905 | 25 | 867 |
| KIRP | 290 | 4 895 360 | 4355 | 19 494 | 280 | 18 258 | 2390 | 330 | 2156 |
| LAML | 122 | 5 143 663 | 3754 | 7675 | 159 | 7588 | 517 | 81 | 501 |
| LGG | 515 | 4 634 138 | 5251 | 29 267 | 330 | 27 826 | 1150 | 41 | 1008 |
| LIHC | 369 | 4 158 963 | 3127 | 10 779 | 159 | 10 511 | 842 | 131 | 738 |
| LUAD | 511 | 4 384 429 | 4471 | 19 628 | 241 | 18 763 | 1210 | 23 | 1160 |
| LUSC | 500 | 3 744 419 | 5126 | 21 804 | 296 | 20 915 | 718 | 14 | 673 |
| MESO | 87 | 4 784 882 | 3999 | 9077 | 237 | 8447 | 1082 | 120 | 1019 |
| OV | 291 | 2 963 431 | 6174 | 21 159 | 382 | 19 702 | 285 | 57 | 285 |
| PAAD | 178 | 4 996 008 | 4466 | 20 351 | 462 | 19 177 | 1065 | 178 | 951 |
| PCPG | 178 | 4 721 561 | 3696 | 25 042 | 571 | 23 185 | 1133 | 131 | 1130 |
| PRAD | 494 | 4 828 721 | 4704 | 30 998 | 332 | 29 312 | 1842 | 15 | 1796 |
| SARC | 258 | 4 088 267 | 3910 | 13 158 | 232 | 12 582 | 897 | 320 | 536 |
| SKCM | 103 | 4 854 570 | 4179 | 12 811 | 310 | 11 672 | 1766 | 144 | 1702 |
| STAD | 414 | 4 310 492 | 6978 | 23 045 | 334 | 21 499 | 478 | 97 | 465 |
| TGCT | 150 | 4 825 013 | 4616 | 20 876 | 487 | 19 369 | 1118 | 204 | 1068 |
| THCA | 503 | 4 877 853 | 519 | 2999 | 35 | 2896 | 10 | 9 | 9 |
| THYM | 120 | 4 940 146 | 3773 | 12 939 | 325 | 12 255 | 971 | 117 | 957 |
| UCEC | 176 | 4 950 486 | 2588 | 8903 | 288 | 8788 | 987 | 212 | 920 |
| UCS | 56 | 3 888 385 | 3733 | 2206 | 99 | 1999 | 1185 | 143 | 1112 |
| UVM | 80 | 4 737 552 | 3149 | 8021 | 186 | 7516 | 552 | 66 | 457 |
cis- and trans-apaQTLs in SNP2APA
SNP2APA mainly provided four kinds of datasets: cis- and trans-apaQTLs, survival apaQTLs and GWAS-associated apaQTLs (Figure 2A and B). In the cis-apaQTL analysis, a total of 467 942 cis-apaQTLs across 32 cancer types were identified at the level of FDR < 0.05 and |r| ≥ 0.3, with a median of 14 811 apaQTLs per cancer type, minimum of 1580 in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), and maximum of 34 381 in glioblastoma multiforme (GBM). In the trans-apaQTL analysis, a total of 30 721 trans-apaQTLs across 32 cancer types were identified at P-value < 1 × 10−8 and |r| ≥ 0.3, with a median of 936 apaQTLs per cancer type, minimum of nine in thyroid carcinoma (THCA), and maximum of 2171 in DLBC.
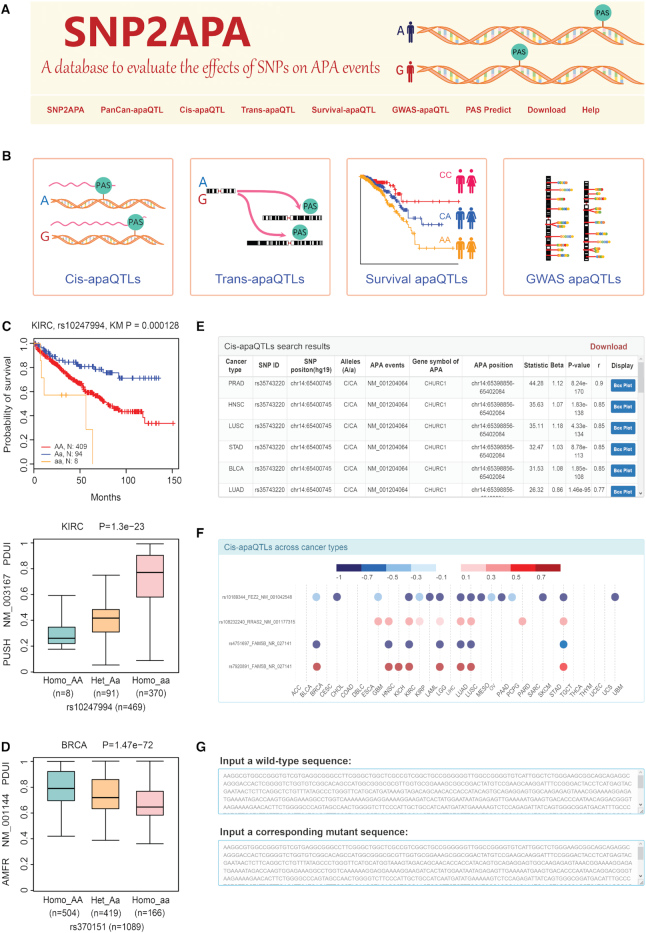
Figure 2.
The interface of SNP2APA database. (A) Browser bar in SNP2APA. (B) Modules of cis- and trans-apaQTL, survival apaQTL and GWAS apaQTL. (C) An example of survival apaQTL. KM-plot indicated that rs10247994 in KIRC was highly association with patient survival time, and box plot indicated that rs10247994 in KIRC was highly associated with PDUI values of the APA event in PUSH gene. (D) An example of GWAS apaQTL. Box plot indicated that GWAS associated apaQTL rs370151 in BRCA was highly associated with PDUI values of the APA event in AMFR. (E) Search results of cis-apaQTL dataset. (F) The heatmap displaying the correlation coefficient (r) of apaQTLs in the ‘Pancan-apaQTL’ page. The label for y-axis contains SNP ID, gene symbol of APA and APA event. (G) The input of online PAS prediction tool.
Survival and GWAS associated apaQTLs
To prioritize promising apaQTLs, we associated apaQTLs with the survival data of patients downloaded from the TCGA portal. A total of 2154 apaQTLs associated with overall survival time across 32 cancer types at FDR < 0.05, were identified and included in SNP2APA. For example, we found that rs10247994 was highly associated with patient overall survival time in kidney renal clear cell carcinoma (KIRC) (Figure 2C). The significant differences in PDUI values among corresponding genotypes of rs10247994 were observed, indicating that this SNP might play an important role in regulating the APA event of PUSH gene in KIRC (Figure 2C).
We further mapped apaQTL results to SNPs in GWAS regions and identified a total of 151 342 apaQTLs overlapping with GWAS LD regions with one or multiple traits. For example, rs2303282, as a risk SNP, was reported to be associated with BRCA (34). In our study, we found that rs370151 was in LD with the rs2303282 (LD r2 = 0.87) and was highly associated with APA event of AMFR gene. AMFR was reported to encode a tumor motor stimulating protein receptor (35). Thus, it could be inferred that rs370151 might play an important role in breast cancer by affecting APA events (Figure 2D).
THE FUNCTION AND USAGE OF SNP2APA DATABASE
SNP2APA provided a user-friendly web interface (http://gong_lab.hzau.edu.cn/SNP2APA/) that enabled users to browse, search, and download four datasets: cis-apaQTLs, trans-apaQTLs, survival-apaQTLs, and GWAS-apaQTLs. In addition, we designed a ‘Pancan-apaQTL’ page for batch search and visualization. A ‘PAS Predict’ page was constructed for online predicting whether a SNP could destroy or create the PAS of APA.
On the homepage, we provided a quick search option for users. After inputting an interested SNP, gene or APA event, users could obtain the corresponding results presented as four dynamic tables containing the information of cis-apaQTLs, trans-apaQTLs, survival-apaQTLs and GWAS-apaQTLs. By querying the cis/trans-apaQTL page, we could obtain a table containing the information of SNP ID, SNP genomic position, SNP alleles, APA events, gene symbol of APA, APA position, beta value (effect size of SNP on PDUI value), r value and P-value of apaQTL (Figure 2E). For each record, a vector diagram of the boxplot was embedded to display the association between SNP genotypes and PDUI values. By querying the survival-apaQTL page, the SNP ID, SNP genomic position, SNP alleles, sample size, log-rank test P-value, and median survival time of different genotypes will be displayed. For each record, a vector diagram of the KM-plot was provided for visualizing the association between SNP genotypes and overall survival time. On the ‘GWAS-apaQTL’ page, the information of the SNP, related APA event, gene symbol of APA and related traits would be available.
On the ‘PanCan-apaQTL’ page, users could submit multiple SNPs or gene symbols of APA events. Then they would obtain two heatmaps displaying the correlation coefficient (r) of cis-apaQTLs and trans-apaQTLs across the cancer types (Figure 2F).
PAS is the most important regulatory element during the regulation of APA events (3). To further explore the impact of SNP on PAS, we developed a web-based tool by utilizing Dragon PolyA Spotter (http://www.cbrc.kaust.edu.sa/dps/Capture.html) (36) and designed the ‘PAS Predict’ page. On this page, users could submit a wild-type sequence and the corresponding mutant sequence to predict the effect of SNP on polyadenylation signals (PAS) so as to determine whether SNP could destroy or create the PAS (Figure 2G).
In SNP2APA, four main datasets for each cancer type are freely available from the ‘Download’ page. The ‘Help’ page provided the basic information on database, pipeline of database construction, result summary, and contact. SNP2APA was open to any feedback with email address provided at the bottom of the ‘Help’ page.
CONCLUSION AND FUTURE DIRECTIONS
We developed SNP2APA as a resource providing comprehensive apaQTLs across 32 cancer types. To the best of our knowledge, this is the first database systematically evaluating the effects of the genetic variants on APA, especially in multiple cancer types with a large sample size. In recent years, increasing studies have suggested that APA is likely to play important roles in cancer. Therefore, it is urgent to add APA as an additional dimension to existing cancer genomic analysis. In this version of TC3A, by using genotype and APA data of 9082 tumor samples, we provided numerous apaQTLs among multiple cancer types and identified abundant apaQTLs associated with patient survival time or located in known GWAS loci. To explore the impact of SNPs on PAS, we also designed an online tool for users to predict functional apaQTLs. The SNP2APA database will greatly facilitate the interpretation of risk SNPs identified in genetic studies. In the future, with the increasing number of RNA-Seq datasets and genotype data from large consortium projects, we will continue to update the SNP2APA database. We believe that our database will be of particular interest to researchers in the field of genetic variants and APA in cancer.
FUNDING
National Natural Science Foundation of China (NSFC) [31970644 to J.G., 31822030 and 31771458 to A.Y.G.]; Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [11041810351 to J.G.]. Funding for open access charge: Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [11041810351].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gruber A.J., Schmidt R., Gruber A.R., Martin G., Ghosh S., Belmadani M., Keller W., Zavolan M.. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016; 26:1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tian B., Manley J.L.. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017; 18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elkon R., Ugalde A.P., Agami R.. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013; 14:496–506. [DOI] [PubMed] [Google Scholar]
- 4. MacDonald C.C. Tissue-specific mechanisms of alternative polyadenylation: Testis, brain, and beyond (2018 update). Wires RNA. 2019; 10:e1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Di Giammartino D.C., Nishida K., Manley J.L.. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011; 43:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang J.W., Yeh H.S., Yong J.. Alternative polyadenylation in human diseases. Endocrinol. Metab. (Seoul). 2017; 32:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bacchetta R., Barzaghi F., Roncarolo M.G.. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Ann. N. Y. Acad. Sci. 2018; 1417:5–22. [DOI] [PubMed] [Google Scholar]
- 8. Bennett C.L., Brunkow M.E., Ramsdell F., O’Briant K.C., Zhu Q., Fuleihan R.L., Shigeoka A.O., Ochs H.D., Chance P.F.. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA → AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001; 53:435–439. [DOI] [PubMed] [Google Scholar]
- 9. Garin I., Edghill E.L., Akerman I., Rubio-Cabezas O., Rica I., Locke J.M., Maestro M.A., Alshaikh A., Bundak R., del Castillo G. et al.. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erson-Bensan A.E., Can T.. Alternative polyadenylation: another foe in cancer. Mol. Cancer Res. 2016; 14:507–517. [DOI] [PubMed] [Google Scholar]
- 11. Xiang Y., Ye Y., Lou Y., Yang Y., Cai C., Zhang Z., Mills T., Chen N.Y., Kim Y., Muge Ozguc F. et al.. Comprehensive characterization of alternative polyadenylation in human cancer. J. Natl. Cancer Inst. 2018; 110:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia Z., Donehower L.A., Cooper T.A., Neilson J.R., Wheeler D.A., Wagner E.J., Li W.. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 2014; 5:5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shastry B.S. SNP alleles in human disease and evolution. J. Hum. Genet. 2002; 47:561–566. [DOI] [PubMed] [Google Scholar]
- 14. Do C., Shearer A., Suzuki M., Terry M.B., Gelernter J., Greally J.M., Tycko B.. Genetic-epigenetic interactions in cis: a major focus in the post-GWAS era. Genome Biol. 2017; 18:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. et al.. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013; 45:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K.C., Hua Y.M., Gueroussov S., Najafabadi H.S., Hughes T.R. et al.. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015; 347:1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takata A., Matsumoto N., Kato T.. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat. Commun. 2017; 8:14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gong J., Mei S.F., Liu C.J., Xiang Y., Ye Y.Q., Zhang Z., Feng J., Liu R.Y., Diao L.X., Guo A.Y. et al.. PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 2018; 46:D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong J., Wan H., Mei S.F., Ruan H., Zhang Z., Liu C.J., Guo A.Y., Diao L.X., Miao X.P., Han L.. Pancan-meQTL: a database to systematically evaluate the effects of genetic variants on methylation in human cancer. Nucleic Acids Res. 2019; 47:D1066–D1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stacey S.N., Sulem P., Jonasdottir A., Masson G., Gudmundsson J., Gudbjartsson D.F., Magnusson O.T., Gudjonsson S.A., Sigurgeirsson B., Thorisdottir K. et al.. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 2011; 43:1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng X., Li L., Wagner E.J., Li W.. TC3A: the cancer 3′ UTR atlas. Nucleic Acids Res. 2018; 46:D1027–D1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howie B.N., Donnelly P., Marchini J.. A flexible and accurate genotype imputation method for the next generation of Genome-Wide association studies. PLoS Genet. 2009; 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., Gabriel S.B. et al.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ardlie K.G., DeLuca D.S., Segre A.V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., Lek M. et al.. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Graffelman J. Exploring diallelic genetic markers: The hardyweinberg package. J. Stat. Softw. 2015; 64:1–23. [Google Scholar]
- 27. Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D.. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006; 38:904–909. [DOI] [PubMed] [Google Scholar]
- 28. Stegle O., Parts L., Piipari M., Winn J., Durbin R.. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012; 7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schulz H., Ruppert A.K., Herms S., Wolf C., Mirza-Schreiber N., Stegle O., Czamara D., Forstner A.J., Sivalingam S., Schoch S. et al.. Genome-wide mapping of genetic determinants influencing DNA methylation and gene expression in human hippocampus. Nat. Commun. 2017; 8:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ongen H., Andersen C.L., Bramsen J.B., Oster B., Rasmussen M.H., Ferreira P.G., Sandoval J., Vidal E., Whiffin N., Planchon A. et al.. Putative cis-regulatory drivers in colorectal cancer. Nature. 2014; 512:87–90. [DOI] [PubMed] [Google Scholar]
- 31. Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012; 28:1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al.. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O’Donnell C.J., de Bakker P.I.W.. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008; 24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michailidou K., Lindstrom S., Dennis J., Beesley J., Hui S., Kar S., Lemacon A., Soucy P., Glubb D., Rostamianfar A. et al.. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017; 551:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang W.G., Raz A., Douglas-Jones A., Mansel R.E.. Expression of autocrine motility factor (AMF) and its receptor, AMFR, in human breast cancer. J. Histochem. Cytochem. 2006; 54:231–241. [DOI] [PubMed] [Google Scholar]
- 36. Kalkatawi M., Rangkuti F., Schramm M., Jankovic B.R., Kamau A., Chowdhary R., Archer J.A.C., Bajic V.B.. Dragon PolyA Spotter: predictor of poly(A) motifs within human genomic DNA sequences. Bioinformatics. 2012; 28:127–129. [DOI] [PMC free article] [PubMed] [Google Scholar]