Abstract
ClinVar is a freely available, public archive of human genetic variants and interpretations of their relationships to diseases and other conditions, maintained at the National Institutes of Health (NIH). Submitted interpretations of variants are aggregated and made available on the ClinVar website (https://www.ncbi.nlm.nih.gov/clinvar/), and as downloadable files via FTP and through programmatic tools such as NCBI’s E-utilities. The default view on the ClinVar website, the Variation page, was recently redesigned. The new layout includes several new sections that make it easier to find submitted data as well as summary data such as all diseases and citations reported for the variant. The new design also better represents more complex data such as haplotypes and genotypes, as well as variants that are in ClinVar as part of a haplotype or genotype but have no interpretation for the single variant. ClinVar's variant-centric XML had its production release in April 2019. The ClinVar website and E-utilities both have been updated to support the VCV (variation in ClinVar) accession numbers found in the variant-centric XML file. ClinVar's search engine has been fine-tuned for improved retrieval of search results.
INTRODUCTION
ClinVar (1,2) is a freely available, public archive of human genomic variants and interpretations of their relationships to diseases and other conditions. It is maintained at the National Center for Biotechnology Information (NCBI), within the National Library of Medicine (NLM) at the National Institutes of Health (NIH). Interpretations of variants in ClinVar have been submitted by >1300 organizations, including clinical testing laboratories, research laboratories, locus-specific databases, clinicians, patient registries, expert panels, and other organizations (https://www.ncbi.nlm.nih.gov/clinvar/docs/submitter_list/). Each submitted record (SCV record, or submission to ClinVar) includes a description of the variant or set of variants that was interpreted; the condition for which the variant was interpreted; the interpretation of the clinical significance of the variant; and the submitter's evidence for that interpretation, structured as observations of the variant. ClinVar aggregates submitted data by the variant (VCV records, or variation in ClinVar) and by the variant-condition pair (RCV records, or reference ClinVar record). For each VCV and RCV record, an aggregate interpretation is calculated, indicating whether there is consensus or conflict among submitters on the interpretation. A review status (https://www.ncbi.nlm.nih.gov/clinvar/docs/review_status/) is assigned to each SCV, RCV, and VCV record to convey the level of review that supports the interpretation. The review status is calculated based on three factors: (i) whether a submitter is approved as an expert panel or practice guideline provider; (ii) whether the submitter(s) provided both their assertion criteria (https://www.ncbi.nlm.nih.gov/clinvar/docs/review_status/#ac) and some form of evidence for their interpretation (or a public contact person who could provide that evidence); and (iii) consensus across submitters in the interpretation of the variant. Data in ClinVar may be searched and browsed on the website (https://www.ncbi.nlm.nih.gov/clinvar/) and downloaded from the ftp site (ftp.ncbi.nlm.nih.gov/pub/clinvar/). Programmatic access is also available using NCBI’s E-utilities and Entrez direct (https://www.ncbi.nlm.nih.gov/books/NBK25501/).
In its seventh year of operation, ClinVar holds more than 825 000 submitted records, representing more than half a million variants. ClinVar includes both germline and somatic variants; >4000 variants in ClinVar were observed in somatic tissue. Most variants in ClinVar are small variants, such as single nucleotide variants (SNVs) and short insertions or deletions; however, >16 000 variants in ClinVar are greater than one kilobase (kb) in length, which is an often-used definition of structural variants. These structural variants include copy number gains and losses, as well as more complex structural variants like inversions and translocations. ClinVar is driven by submissions of data, and its scope is limited to variants that have been interpreted for clinical or functional significance, not merely observed. Thus the database is not comprehensive for all variants, or all clinically relevant variants. For example, somatic variants are known to be not well-represented in ClinVar. It is difficult to estimate what percentage of all clinically relevant variants are found in ClinVar, because much of the data is only housed in siloed databases, some of which are proprietary. However, it is likely that most clinically relevant variants are now being identified by clinical genetics testing laboratories. Of the 402 laboratories that participate in the NIH Genetic Testing Registry (3), which are largely from the United States, 42% submit data to ClinVar. While the comprehensiveness of ClinVar is difficult to estimate, the dataset is growing steadily; the number of variants in ClinVar increases by an average of 60% each year.
In recent years, several professional societies have called for sharing interpretations of variants in a public database like ClinVar. Recommendations for data sharing have been published by the American Medical Association (AMA, 2013), the American College of Medical Genetics and Genomics (4); and the National Society of Genetic Counselors (5). ClinVar provides a unique opportunity for clinical testing laboratories to share their interpretations of variants. ClinVar has no requirement for a clinical laboratory to publish the data in a scientific journal; the clinical lab can submit all their data to a single database; and data submitted to ClinVar is made freely available to all users. Within the first two years of ClinVar's operations, 46 laboratories had submitted interpretations from clinical testing. Since then, that number has continued to increase by an average 78% each year, a demonstration of increased commitment to data sharing by clinical genetics testing laboratories.
NEW DISPLAY FOR THE CLINVAR VARIATION PAGES
A variant or set of variants with interpretations is represented by a VCV accession number and is displayed on the Variation page, the default view on the ClinVar website. The Variation page is useful for viewing all available data for a variant, regardless of the disease for which the variant was interpreted. In contrast, an RCV accession number represents the relationship between a variant and the disease for which the variant was interpreted.
On average, ∼8000 users access the ClinVar web site each day. The Variation page in ClinVar was redesigned to improve access to the most important data in a VCV record. Prior to software development, interviews with users were conducted to learn how they interacted with the page and to determine which parts of the VCV record were of primary importance. The results of those interviews informed the updates that were made to the layout of the page. The new, state-of-the-art page design and layout improves usability of the data.
Summary section
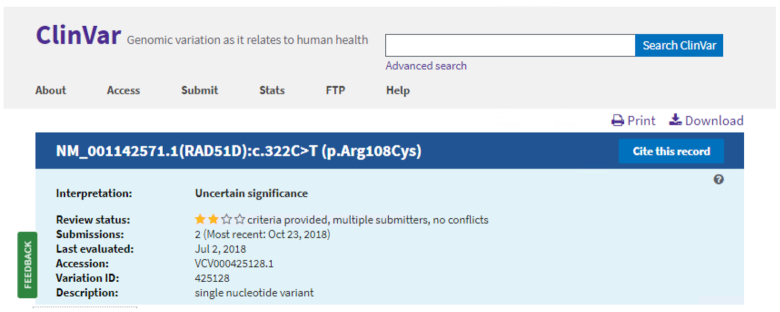
The top section of the Variation page highlights the information of highest value to most users. Beneath the summary, users can view detailed information in three selectable tabs—Variant details, Conditions and Gene(s). The summary section is displayed regardless of the navigation among tabs. As in the previous web design, the new Variation page summary section displays aggregate data including the interpretation, review status, number of submissions, the date the variant was most recently evaluated by a submitter, and the Variation ID. In addition, the summary section displays two key features in the redesign (Figure 1). The first feature is the VCV accession and version number. This accession number uniquely identifies the variant or set of variants and it can be used to search ClinVar on the web site (see Improvements to searching ClinVar, below). The VCV accession number is also reported in the variant-centric XML file, ClinVarVariationRelease (see Production release for the variant-centric XML, below). The second feature is a brief description of the variant. For most variants in ClinVar, the description is the length of the variant and the variant type, e.g. ‘single nucleotide variant’ or ‘3bp deletion’. For variants larger than 1 kb, the length is rounded to one decimal place, e.g. for a copy number loss with a length of 4 419 752 bp, the variant description is ‘4.4 Mb copy number loss’. For combinations of variants interpreted together, the description is either ‘haplotype’ or ‘genotype’.
Figure 1.
The ClinVar Variation page Summary section displays the most important aggregate data on a ClinVar VCV record.
Variant details tab
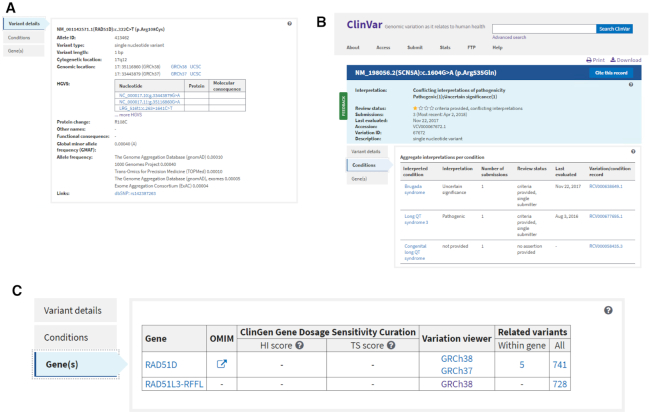
The Variant details tab is shown by default (Figure 2A) in the set of tabs below the summary section; the Conditions and Gene(s) tabs are described below. The Variant details tab corresponds to the Allele(s) section in the previous web display. The major change to this section is a table that displays sets of HGVS (6) expressions on corresponding nucleotide and protein sequences, along with the molecular consequence calculated for the variant on that set of sequences. The table includes HGVS expressions that were submitted and those that are calculated by NCBI. In addition, links to genome browsers (NCBI’s Variation Viewer (7) and the UCSC browser (8)) have been added to this section, next to the genomic location, to make it easier to view the variant in its genomic context.
Figure 2.
The tabs on the ClinVar Variation page. (A) The Variant details tab displays general information about the variant, including submitted and calculated HGVS expressions. (B) The Conditions tab displays a summary of data aggregated by variant and condition. In this example, note that the aggregate interpretation for the variant indicates that there is a conflict. The conflict occurs between interpretations for different conditions, however, there are no conflicts in the interpretations aggregated by variant and condition. (C) The Gene(s) tab displays generation information about the gene, or genes, for the variant. In this example, the variant is located in two overlapping genes so both genes are displayed.
Conditions tab
The Conditions tab is a new feature on the redesigned variant page. This tab provides a summary of each condition that has been reported for a variant. For example, Variation ID 67672 represents NM_198056.2:c.1604G>A which has been reported to ClinVar for Brugada syndrome, Long QT syndrome 3, and Congenital long QT syndrome (Figure 2B). The Conditions tab provides a tabular summary of the data for the variant with each condition, that includes a link to MedGen (9) to learn more about the condition; the interpretation; the number of submissions; the review status; and the date that the variant was last evaluated by submitters for that condition. There is also a link to the RCV record that is specific to each variant-disease pair. The data in the Conditions tab can help provide clarity when there is a conflict in the interpretation at the level of the variant, but no conflict when the interpretations are aggregated by variant-condition pairs, as in Figure 2B.
Gene(s) tab
The Gene(s) tab is a new feature that provides a tabular display of the gene, or genes, affected by the variant (Figure 2C). Most variants in ClinVar are SNVs, which typically are in a single gene; thus the Gene(s) tab most often displays a single gene. However, some SNVs lie in genes that overlap each other; in this case the Gene(s) tab lists all genes for the variant. ClinVar also includes large structural variants that may span one or more genes. For structural variants, the Gene(s) tab lists up to 10 genes affected by the variant; if there are >10 genes, there are links to view the complete list of genes in Variation Viewer and in the ClinGen Dosage Sensitivity Map (https://www.ncbi.nlm.nih.gov/projects/dbvar/clingen/). Each gene is linked to Entrez Gene (10) and OMIM® (11), to expedite access to more information about the gene. Scores for haploinsufficiency and triplosensitivity from the ClinGen Dosage Sensitivity Map are provided with links to curation results, when available. Included also are links to Variation Viewer so that variants can be viewed in a genomic context. Additionally, there are links to find all other variants in ClinVar that are within this gene (typically SNVs and small insertions/deletions) and all other variants in ClinVar that affect this gene (all variants, including structural variants).
Submitted interpretations and evidence
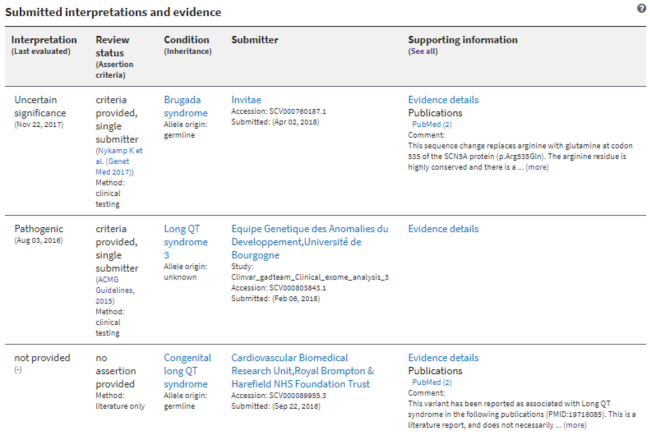
The ‘Submitted interpretations and evidence’ table is a summary of all submitted records, or SCVs, for the variant (Figure 3). Each row represents one submitted record. For each record, the table displays key information such as the submitter's interpretation of the variant; the review status of the submitted record; a link to assertion criteria, when provided; the condition for which the submitter interpreted the variant; the name of the submitter; links to citations; and the submitter's comment on clinical significance, when provided.
Figure 3.
The Submitted interpretations and evidence table displays a summary of data from each submitted record for the variant. The link ‘(See all)’ in the last column header opens a configurable display (see Figure 4).
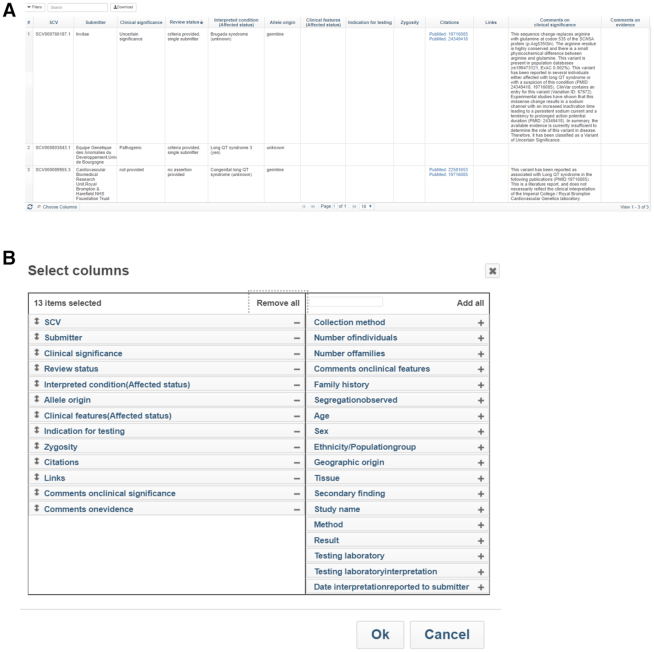
The last column of the ‘Submitted interpretations and evidence’ table includes a link to an ‘Evidence details’ page which displays more details provided by the submitter such as the observations, or evidence, for the submitter's interpretation of the variant (Figure 4A). These details include data such as age, sex, clinical features, zygosity, and ethnicity/population group. The Evidence details page can be configured to add more columns of data using the ‘Choose Columns’ button at the lower left of the table. Additional fields that can be added in this way include ‘Method’ and ‘Result’, which are provided for submissions with statements of functional evidence for variants, and ‘Testing laboratory’ and ‘Testing laboratory interpretation’ which may be provided for submissions from patient registries (Figure 4B).
Figure 4.
The Evidence details page. (A) The table displays details of each observation on each submitted record for the variant. (B) The column selector lets the submitter configure the columns the user wants to view.
Citations table
The bottom of the new variation page displays a table of all citations reported to ClinVar for the variant with links to PubMed, except for citations provided as assertion criteria and citations that are submitted specifically for the disease (not the variant). Users can leverage this information in their search of the literature for a variant. Note that this table is not curated independently of submissions; thus it is not expected to be comprehensive for all variants in ClinVar.
Removed features
A few features were removed from the redesigned pages. The previous web display included a section on the right for Variant frequency in dbGaP. The data in this section was out-of-date; in the future, ClinVar can link to aggregate allele frequencies based on studies in dbGap as reported in dbSNP. The right side of the previous Variation page also included a section called ‘Related information’, comprised of computed links to related data in other Entrez databases. This section was rarely used; some of the links, such as the link to dbSNP, were incorporated into existing sections of the redesigned page.
In the previous Variation page, the section of submitted data was divided into three sections: interpretations based on germline observations of the variant, interpretations based on somatic observations of the variant, and interpretations of pharmacogenomic variants. In the new Variation pages, the submitted data is displayed in a single table. Based on user feedback, this feature may be added back.
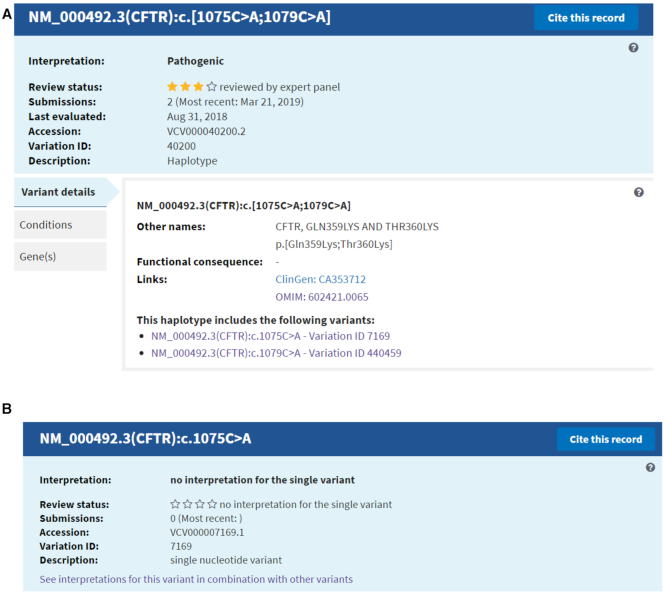
Haplotypes, genotypes and included variants
Some interpretations in ClinVar are for sets of more than one variant, such as haplotypes and genotypes. The redesigned Variation page provides several improvements to the display of these complex variations (Figure 5A). In the Summary section, the ‘Description’ of the variation is either ‘Haplotype’ or ‘Genotype’. The Variant details tab provides a hierarchical representation of the variants that make up each haplotype and genotype, so that the phase of the variants is clear, and includes links to each individual variant page for more details.
Figure 5.
(A) An example of a Variation ID that represents a haplotype, noted in the Description. The Variant details section lists the variants that comprise the haplotype and links to the ClinVar record for each individual variant. (B) The summary section for one of the individual variants in the haplotype. There is no interpretation for the single variant in ClinVar. Note the link ‘See interpretations for this variant in combination with other variants.’ which makes it easier to review the contexts in which this variant has been interpreted.
Another new feature is the display of variants that are only in ClinVar because they are included in an interpreted haplotype or genotype, but for which there is no submitted interpretation for the single variant. These ‘included variants’ are most often short variants like SNVs. However, a haplotype can also be considered an included variant, if ClinVar has a submission for a genotype, in which the variation on one chromosome is a haplotype (e.g. VCV000431012). Included variants display “no interpretation for the single variant" in both the interpretation and the review status fields (Figure 5B). Included variants also have a link in the Summary section to ‘See interpretations for this variant in combination with other variants’, to guide the user quickly to records for haplotypes or genotypes that include the variant.
Future work
The ClinVar team will continue to engage with users to improve ClinVar and its web display. Improvements to usability of the Evidence details page are planned, including making it easier to add columns to the table. User research is also being performed to understand how ClinVar users perceive the review status (represented by stars on web pages) and variants reviewed by expert panels and in practice guidelines, so that these concepts can be made more obvious to users.
UPDATES TO E-UTILITIES
ClinVar continues to support four functions for E-utilities, NCBI’s API for data retrieval: esearch, esummary, efetch, and elink. The redesigned Variation page uses ClinVar's new variation-centric XML as the source of data including the new nine-digit accession numbers, prefixed with VCV. The E-utilities efetch function in ClinVar supports queries for VCV accessions and returns the new XML format. This update allows programmatic retrieval of ClinVar data in XML format that corresponds to data displayed on the Variation page.
The latest VCV record can be retrieved using an efetch query for a VCV accession number (without the version) OR a Variation ID. For example:
VCV accession: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=clinvar&rettype=vcv&id=VCV000014206.
Variation ID: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=clinvar&rettype=variation&id=14206,41472.
The latest XML for a VCV accession.version can be retrieved using an efetch query for the VCV accession and version. For example: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=clinvar&rettype=vcv&id=VCV000014206.1.
The RCV XML can still be retrieved by efetch also. The latest RCV record can be retrieved using an efetch query for an RCV accession number (without the version). For example: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=clinvar&rettype=clinvarset&id=RCV000000606.
The latest XML for an RCV accession.version can be retrieved using an efetch query for the RCV accession and version. For example: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=clinvar&rettype=clinvarset&id=RCV000000606.3.
The comprehensive guide to access via APIs is maintained on the ClinVar web site under the Access tab: https://www.ncbi.nlm.nih.gov/clinvar/docs/maintenance_use/#api.
PRODUCTION RELEASE FOR THE VARIANT-CENTRIC XML
The beta release of ClinVar's variant-centric XML, ClinVarVariationRelease, was described previously (12). This format went into its production release in April 2019. Similar to ClinVarFullRelease (12), the variant-centric XML is generated and archived monthly. Between the monthly releases, files are also generated weekly for users who want updates that are synchronized with the ClinVar website, but the weekly releases are not archived. The monthly releases for ClinVarVariationRelease are found in the following directory: ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/xml/clinvar_variation/ and the weekly releases are in: ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/xml/clinvar_variation/weekly_release/.
Each record in ClinVarVariationRelease includes the VCV accession number for the record. Versioning of the VCV accession numbers started in April 2019 with the first production release of the variant-centric XML file. The VCV version number is incremented when the content of any underlying submitted (SCV) record changes, including addition or deletion of SCV records and updates to SCV records. The version number is not increased when updates are made by NCBI staff, e.g., when rs numbers are added by automated processes or when curators add an HGVS expression or a citation.
Each ClinVarVariationRelease includes only the VCV records that are current at the time of release. VCV records may be removed if all of the underlying SCV records are deleted by the submitter or if the variant has been identified to be a duplicate. Future plans include development of a deletion report, as a supplement to the variant-centric XML file. The deletion report will list VCV records that were once public but are no longer current, so that it is clear that a VCV record is not missing from the release in error. VCV records that have been identified to be redundant are reported in the element ‘ReplacedList’.
The schema for ClinVarVariationRelease is in: ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/xsd_public/clinvar_variation/ The schema is updated as needed.
IMPROVEMENTS TO SEARCHING CLINVAR
VCV accession numbers are now searchable on the ClinVar website. Users can view the latest record for a variation by searching with a VCV accession number, e.g., https://www.ncbi.nlm.nih.gov/clinvar/?term=VCV000633778. Searching with a VCV accession.version number returns the latest record for a specific version of the variation, e.g., https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000042300.1/. Note that searching ClinVar by accession.version is more specific than the same search on NCBI’s general search page: https://www.ncbi.nlm.nih.gov/search/?term=. For example, if you search ClinVar for VCV000042300.1, you get version 1 of VCV000042300; if you search for VCV000042300.1 on the NCBI general search page, you get the most recent version of VCV000042300 (version 3, currently).
Searching by Variation ID remains the same, returning the latest instance of that VCV record. Searching for any other term, like a gene symbol or a disease name, remains unchanged; the results are the latest instance of each VCV record for the search term.
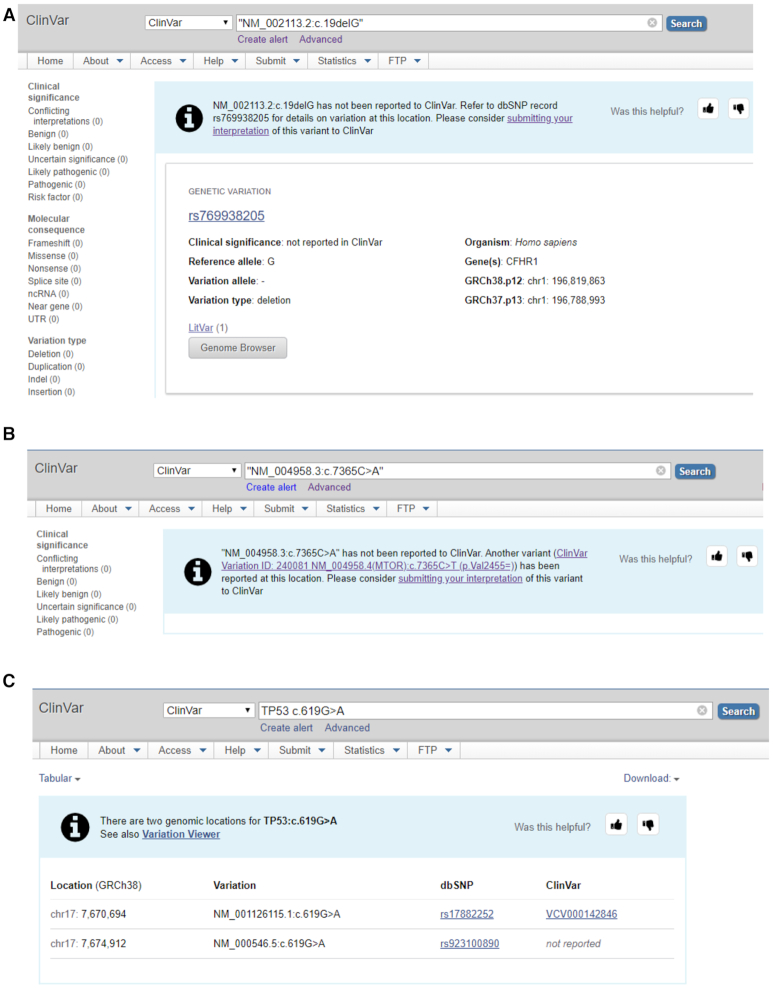
Several improvements to search have been made to reduce the likelihood that a query for a variant has no result in ClinVar. When the user searches for a term in ClinVar and gets no result, it can be confusing. No search result may be the outcome if the variant has not been reported to ClinVar or if there is a problem processing the search term. For example, a valid HGVS expression is an excellent search term because it is unambiguous. However, a user may find no result when searching for an HGVS expression, because the user may have unknowingly entered an invalid HGVS expression or the variant may not exist in the database. When the variant is not in ClinVar but is known to dbSNP, the improved search engine provides results for both ClinVar and dbSNP (9) (Figure 6A). This gives the user confidence that the search term is valid, and there is no data for the variant in ClinVar. The user can then choose to follow the link to dbSNP to learn what is known about the variant, including any available population frequency data.
Figure 6.
(A) Search result for a variant that has not been reported to ClinVar but is found in dbSNP. (B) Search result for a variant that is not in ClinVar, but a different variant at the same location is in the database. (C) Search result for TP53 c.619G>A, an ambiguous query for which there are two possible results.
In some cases, the specific allele is not in ClinVar, but another allele at that location is in the database. In this case, the search results indicate explicitly that the specific variant in the query has not been reported to ClinVar, and notes that there is another variant at the same location, with a link to that record (Figure 6B). This feature helps users identify related variants that affect the same nucleotide and amino acid and may be pathogenic, which is part of the criteria in the ACMG guidelines for the interpretation of sequence variants (13).
A common query in ClinVar is a combination of the gene symbol and the c. portion of an HGVS expression, e.g. TP53 c.619G>A. This type of query is ambiguous, because the c. portion of the HGVS expression refers to a transcript, but there may be more than one transcript for the gene. In the updated search engine, for this type of query, the c. portion is combined with each transcript for the gene, and all valid results on all transcripts in both ClinVar and dbSNP are returned (Figure 6C).
USING SPDI IN SUBMISSION PROCESSING
Validation, identification, and aggregation of variant descriptions are central to processing submissions to ClinVar. The misidentification of variants can lead to duplicate variant records. For example, a submitted variant may be described in HGVS as an insertion, when the accurate HGVS nomenclature (6) is a duplication, which is a more specific kind of insertion. A second example is a variant that can be described at different locations (e.g. in repetitive sequence), either left-shifted or right-shifted on the reference sequence. A third example is a variant described on genomic coordinates on different assemblies (e.g. on GRCh37 and GRCh38). If different descriptions for the same variant are not recognized during submission processing, ClinVar may have redundant records for the variant. ClinVar's submission processing was recently updated to use NCBI’s SPDI notation and Variant Overprecision Correction algorithm (VOCA) (14) to validate the submitted description, which may be either an HGVS expression or chromosomal coordinates with reference and alternate alleles. VOCA uses alignments in NCBI’s Alignment Data Sets (ADS; https://www.ncbi.nlm.nih.gov/variation/services/remapping/) which include alignments between RefSeq transcripts and non-chromosomal genomic sequences (prefixed NM, NR, NG), as well as assembly-associated genomic sequences (NCs, NTs and NWs). Thus variants can be easily remapped from transcripts to multiple assemblies and between different assemblies, including any future assemblies that use the current assembly model (15). SPDI and VOCA also allow ClinVar processing to check whether an HGVS expression and a variant description on chromosomal coordinates are consistent, when a submitter provides both descriptions for a variant. The normalized description of the variant in SPDI notation supports aggregation with previously submitted variants known to the ClinVar database. The adoption of SPDI and VOCA resulted in a 50% decrease in the number of duplicate variant records that must be manually curated and merged by NCBI staff.
One limitation of SPDI is that it does not handle variants with an imprecise location, such as exon deletions and large copy number variants, nor does it handle intronic variants described on a transcript sequence. Some validation is currently performed in ClinVar for these types of variants using in-house software tools; however, this validation is not as robust as SPDI in terms of normalizing variant descriptions. Future work is planned to provide more robust validation of intronic variants and variants with an imprecise location.
SUMMARY
In the last two years, the ClinVar team has focused on improving the display, quality, and access to data in ClinVar. A new, modern design for the ClinVar Variation page provides a more intuitive display of the data. Using SPDI and VOCA in ClinVar's submission processing improves the validation and identification of variants, and decreases the number of redundant variant records in the database. Refinements to the online search engine improve access to the data for clinicians and web users. Improvements to E-Utilities and the production release of the variant-centric XML enable programmatic access via APIs for bioinformaticians in clinical laboratories and research settings to download bulk data. Over the next two years, the focus of the ClinVar team will shift to improving the speed of submission processing, to facilitate rapid sharing of valuable data from submitters.
ACKNOWLEDGEMENTS
The authors thank the ClinVar users who have participated in user experience interviews. We welcome feedback from the community. Please contact us at clinvar@ncbi.nlm.nih.gov with your feedback, suggestions, and testimonials. The authors also thank Marilu Hoeppner for her generosity and diligence in editing this manuscript.
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health
Conflict of interest statement. None declared.
REFERENCES
- 1. Landrum M.J., Kattman B.L.. ClinVar at five years: Delivering on the promise. Hum. Mutat. 2018; 39:1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R.. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014; 42:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C. et al.. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013; 41:D925–D935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ACMG Board of Directors Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017; 19:721–722. [DOI] [PubMed] [Google Scholar]
- 5. National Society of Genetic Counselors (NSGC) 2015; Clinical Data Sharing (Position Statement) [Blog post].
- 6. den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E.. HGVS Recommendations for the description of sequence variants: 2016 Update. Hum. Mutat. 2016; 37:564–569. [DOI] [PubMed] [Google Scholar]
- 7. NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015; 43:D6–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D. et al.. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018; 46:D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016; 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018; 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A.. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019; 47:D1038–D1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015; 17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmes J.B., Moyer E., Phan L., Maglott D., Kattman B.L.. SPDI: data model for variants and applications at NCBI. Bioinformatics. 2019; doi: 10.1093/bioinformatics/btz856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.C., Agarwala R., McLaren W.M., Ritchie G.R. et al.. Modernizing reference genome assemblies. PLoS Biol. 2011; 9:e1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]