Abstract
Small non-coding RNAs have gained substantial attention due to their roles in animal development and human disorders. Among them, microRNAs are special because individual gene sequences are conserved across the animal kingdom. In addition, unique and mechanistically well understood features can clearly distinguish bona fide miRNAs from the myriad other small RNAs generated by cells. However, making this distinction is not a common practice and, thus, not surprisingly, the heterogeneous quality of available miRNA complements has become a major concern in microRNA research. We addressed this by extensively expanding our curated microRNA gene database - MirGeneDB - to 45 organisms, encompassing a wide phylogenetic swath of animal evolution. By consistently annotating and naming 10,899 microRNA genes in these organisms, we show that previous microRNA annotations contained not only many false positives, but surprisingly lacked >2000 bona fide microRNAs. Indeed, curated microRNA complements of closely related organisms are very similar and can be used to reconstruct ancestral miRNA repertoires. MirGeneDB represents a robust platform for microRNA-based research, providing deeper and more significant insights into the biology and evolution of miRNAs as well as biomedical and biomarker research. MirGeneDB is publicly and freely available at http://mirgenedb.org/.
INTRODUCTION
In the last two decades, the small non-coding RNA field has significantly expanded beyond such well known small RNAs as transfer RNAs (tRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNA) (1) to include small interfering RNAs (siRNAs) (2), Piwi-interacting RNAs (piRNAs) (3), and, in particular microRNAs (miRNAs) (4–7). Although both tRNAs (8) and ribosomal RNAs (9) can generate small regulatory RNAs, miRNAs are characterized by a distinctive suite of sequence features, in addition to striking sequence conservation, not seen in other types of small RNAs (10–12). Unfortunately, recognition and utilization of these clear and mechanistically well understood features is not common practice (13–23) and has, for instance, led to extreme overestimations of the human microRNA complement (24–27). Because of the fundamental roles miRNAs play in establishing robustness of gene regulatory networks across Metazoa (28,29), and their importance in development (30), formation of cell identity (31) and numerous human diseases including cancer (32,33), it is imperative that homologous miRNAs in different species are correctly identified, annotated, and named using consistent criteria against the backdrop of numerous other types of coding and non-coding RNA fragments (23,34,35). Further, it is vital that bona fide miRNAs are clearly distinguished from non-miRNAs to avoid spurious conclusions (e.g. (36–38)) concerning the role small RNAs play in human disease.
Nonetheless, these goals are largely ignored for existing databases, such as miRBase (39), which has developed organically through community-wide submissions of published miRNA calls, and miRCarta (40), a repository that aims to provide miRNA candidates from ultra-deep sequencing experiments in human. With respect to miRBase, several research groups have shown that up to two-thirds of the entries are false positives, with many entries being fragments of other classes of small RNAs including tRNAs and snoRNAs, in addition to numerous rRNA fragments (13–23). The interpretation of these non-miRNA fragments as bona fide miRNAs affects our understanding of not only how miRNAs evolve (41), but also incorrectly annotated bona fide miRNAs can lead to erroneous conclusions on miRNA biology (see, e.g. (42,43)). Inconsistencies in nomenclature and changes between miRBase releases have made it challenging to use miRBase throughout the years leading to numerous community efforts to both independently identify changes to miRBase (44–49) and to develop independent (see (50)) and study-specific databases (14,51–55).
To address these concerns, we previously developed a manually curated and open source miRNA gene database, MirGeneDB, which is based on consistent annotation and nomenclature criteria (23). But because it contained only four species, the usefulness for comparative studies was severely limited. Here, we present a major update to our database, MirGeneDB version 2.0 (http://mirgenedb.org), which now contains high-quality annotations of 10 899 bona fide and consistently named miRNAs constituting 1275 miRNA families from 45 species, representing every major metazoan group, including many well-established and emerging invertebrate and vertebrate model organisms (Figure 1).
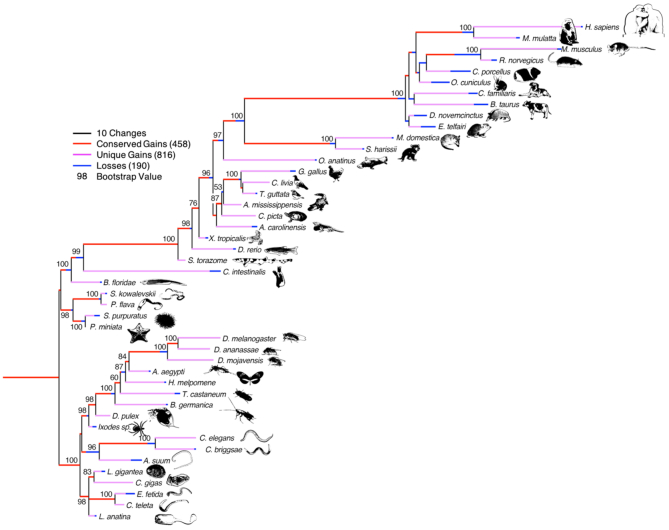
Figure 1.
The evolution of the 1275 microRNA families across the 45 metazoan species currently annotated in MirGeneDB. Conserved gains are shown in red; species-specific gains are shown in pink, and losses are shown in blue; and these gains and losses are mapped onto a generally accepted topology of these species rooted between the deuterostomes and protostomes with branch lengths corresponding to gains and losses, respectively. Note though that this topology is largely recovered when just analyzing the gains and losses of the miRNA families themselves as shown by the bootstrap values indicated at the nodes (Supplementary Methods); the only known nodes not recovered are nodes within the placental mammals and Ecdysozoa, primarily due to losses in rodents and nematodes, respectively.
EXPANSION OF MirGeneDB
For the expansion from version 1.0 to 2.0, we analyzed more than 400 publicly available smallRNA sequencing datasets with at least one representative dataset for each organism, that were automatically downloaded and processed using sRNAbench (56) and miRTrace (57), respectively. This allowed for a consistent and uniform annotation of miRNAomes for each species using MirMiner (11) (see Supplementary Table, ‘file_info’ for files and see Supplementary Information for detailed methods) (23).
The existing MirGeneDB.org miRNA complements for human, mouse, chicken and zebrafish were expanded from our initial effort by 32, 54, 41 and 103 genes to a total of 556, 447, 270 and 390 genes, respectively (Supplementary Table, ‘table’), and annotation-accuracy for human and zebrafish was further improved using available Cap Analysis of Gene Expression (CAGE) data (Supplementary Figure S1, Supplementary Table, ‘CAGE’; Supplementary Information) (58). We further used Dicer-, Drosha- and Exportin 5-knockout data (59), as well as primary cell expression data (58,60–62) to refine human annotations.
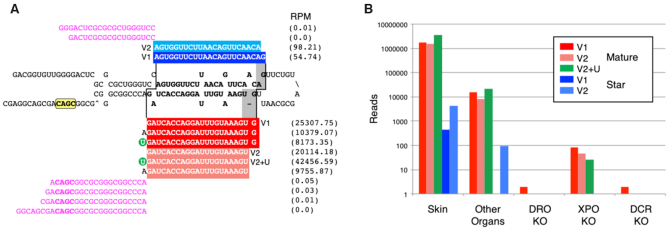
Although since its inception MirGeneDB gave special attention to the precise annotation of both the 5p and 3p arms (and thus allowing for better annotation of miRNA isoforms (63,64)), with a clear distinction made between sequenced reads and predicted reads for each miRNA entry, MirGeneDB 2.0 includes four new features related to the transcription and processing of miRNAs (Figure 2A). First, Group 2 miRNAs (65,66)—those miRNA precursor transcripts that are mono-uridylated at their 3′ end, what we term the 3′ non-templated uridine (3′NTU)—are specifically tabulated, allowing the user to easily discriminate Group 2 from ‘Group 1’ (or canonical) miRNAs. Second, sequence motifs, including the 5′ ‘UG’ motif, the loop ‘UGU/G’ motif, as well as the 3′ CNNC motif (67–69) are bioinformatically identified for every miRNA primary transcript. Third, processing variants, where alternative Drosha/Dicer cuts significantly (>10% of available reads) affect the processed mature seed sequence of the locus (see for example ref. (70)), are added as distinct entries (indicated with the ‘v’ in the name). Some loci, like the Mir-203 gene (Figure 2A) show both mono-uridylation and variant processing such that only one of the two major variants is classified as a Group 2 miRNA. Finally, we also annotate anti-sense loci (‘-as’) for miRNA genes where again significant expression (>10%) of both sense and antisense strands is observed (71).
Figure 2.
The annotation of microRNA sequences and the implementation of transcriptional and processing information for each miRNA gene in MirGeneDB. (A) The structure and read stacks for Hsa-Mir-203. The precursor sequence is shown in bold; mature reads are shown in red and star reads in blue with the reads per million for each major transcript detected shown to the far right. A ‘CNNC’ processing motif (68) is shown in yellow. Also shown are the 5′ and 3′ miRNA offset reads (magenta), which clearly conform to the indicated Drosha cut (staggered line, left) given the reads processed from this locus. The Dicer cut (staggered line, right) results in two primary mature forms (dark vs light red), what we term ‘variants’ (v) that are offset from one another by 1 nucleotide (gray). The 5′ end of variant one starts with the ‘G’ whereas the 5′ end of variant two is moved 1 nucleotide 3′ and starts with the ‘U.’ Each of these two major Dicer products is accompanied by the appropriate star sequence, with variant 1 shown in dark blue and variant 2 in light blue. The mature form of variant 2—but not version 1—is heavily mono-uridylated at its 3′ end (green circle) and is thus a ‘Group 2’ miRNA (59,66). (B) The quantification of Hsa-Mir-203 read across various human-specific data sets. As expected (e.g. (72)) expression in skin is about ∼2 orders of magnitude higher relative to other organs sampled (e.g., brain, liver, stomach, lung, uterus, pancreas, testes, colorectum, small intestine and kidney) and the detection of the mature form is nearly 3 orders of magnitude relative to the star. Consistent with Mir-203 being a bona fide miRNA, expression is nearly abrogated in DROSHA and DICER knock-outs, and greatly diminished in the EXPORTIN-5 knock-out (59).
QUALITY OF MirGeneDB ANNOTATIONS
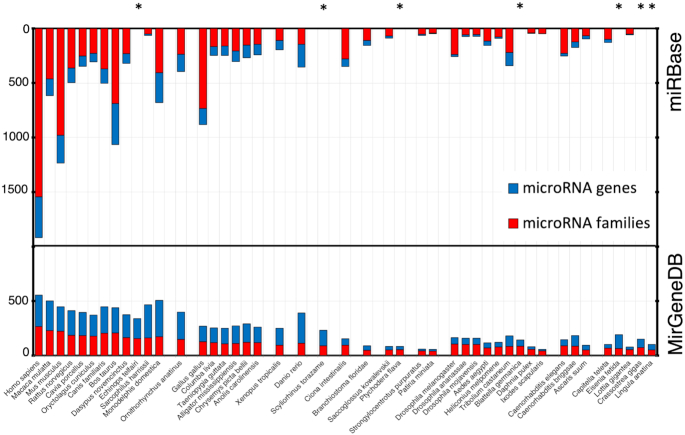
A robust database must be free of both false positive and false negative entries. MiRBase categorizes a subset of their entries as high-confidence miRNAs, which are those that are highly expressed and show clear indications of proper processing, and further has introduced a public voting system to identify more high-quality candidates (73). MirGeneDB takes an alternative approach: rather than allowing for community annotation, the near-complete miRNA repertoire of each taxon is added to MirGeneDB using a consistent and well-defined set of criteria (23,34,74). When comparing MirGeneDB 2.0 and miRBase, the number of miRNAs conforming to the annotation criteria is about three times higher in MirGeneDB than it is in miRBase (2844 for the miRBase ‘high confidence’ set (73)). Further, because the primary requirement for the inclusion of a putative miRNA to miRBase is publication in a peer-reviewed journal, over time, miRBase has become increasingly heterogeneous with respect to the number of miRNAs for closely related species, such as the often studied human and rarely studied macaque (75) (Figure 3). This focus on model systems—in particular human, mouse and chicken—has resulted in miRBase having, on the one hand, a much larger number of annotated sequences for some of the 38 taxa shared with MirGeneDB2.0, accounting for estimated 5631 false positives, and, on the other hand, miRBase lacking 19% of all MirGeneDB2.0 genes, accounting for 2097 false negatives (Supplementary Figure S2, Supplementary Table, ‘overview’). These disparities have obstructed comparative genomic approaches in the miRNA field: for example, missing miRNA families have been misinterpreted as secondary losses, questioning then the fundamental conservation of miRNA families (76). However, very similar miRNA complements in terms of total miRNA genes and miRNA families are observed in closely related groups in MirGeneDB (Figure 3), supporting earlier evolutionary studies arguing for the utility of miRNAs as excellent phylogenetic markers (11,41,57,74) (Figure 1).
Figure 3.
Metazoan miRNA complements are homogeneous between closely related species. Top: miRBase community-report based complements show high heterogeneity in the numbers of families (red) and genes (blue) for closely related species. For instance, in miRBase, human and macaque differ by 1300 genes (Hsa 1917, Mml 617) and 1081 families (Hsa: 1543, Mml: 462). Bottom: MirGeneDBs curated complements are homogeneous for both gene and family numbers (see Supplementary Figure S3 for conserved families, genes in comparison to novel families and genes). For instance, in MirGeneDB, human and macaque differ by 55 genes (Hsa 556, Mml 501) and only one conserved family (Hsa: 206, Mml: 205). Asterisks mark species that are found in MirGeneDB, but not in miRBase.
Thus, while it is inevitable that some cell-type specific or lowly expressed miRNAs are missing from our annotations, MirGeneDB can be considered essentially free of false positives. Further, because MirGeneDB is focused on identification of miRNA genes and families, rather than sequences (23), a bona fide miRNA gene identified in one taxon is identified as such in all, in contrast to miRBase, where the same gene can be identified as generating a high-confidence miRNA sequence in one taxon, but a low-confidence sequence in another (23). Hence, we are confident that there are few (if any) missing miRNA genes that are conserved between two (or more) of the 45 currently included taxa.
IMPROVED WEB INTERFACE OF MirGeneDB
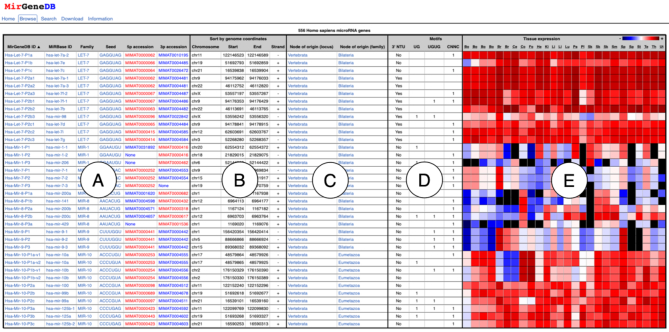
The expanded web-interface of MirGeneDB 2.0 allows browsing (http://mirgenedb.org/browse), searching (http://mirgenedb.org/search) and now also downloading (http://mirgenedb.org/download) of miRNA-complements for each organism, in addition to a general information page about the criteria used for miRNA annotation (http://mirgenedb.org/information), as well as false negatives for each taxon (where known), and links to previous versions of MirGeneDB. On the browse-pages for each organism (e.g. http://mirgenedb.org/browse/hsa), a table is available that includes MirGeneDB IDs and miRBase IDs (if available), family- and seed-assignment and the strandedness of the miRNA (i.e. whether the mature arm is the 5p arm, the 3p arm, or both) (Figure 4, ‘A’); overview information on location in the genome (Figure 4, ‘B’); and the phylogenetic origin of each miRNA locus and family (Figure 4, ‘C’). The new features in MirGeneDB 2.0, including the 3′ NTU’s and sequence motifs (see Figure 2) are also indicated (Figure 4, ‘D’). Finally, a heatmap of the expression of each miRNA for all available tissues is available to orient users on expression patterns (Figure 4, ‘E’).
Figure 4.
Improved web interface of MirGeneDB. For each species in MirGeneDB an overview browse page exists that lists all genes. For each gene the following information is provided and sortable: hyperlinked names (both MirGeneDB ID and miRBase ID linking to MirGeneDB and miRBase, respectively), family- and seed- assignments, and arm preference (A), genomic coordinates (B); inferred phylogenetic origin of both the gene locus and family (C); information on the presence or absence of 3′ NTU’s and sequence motifs (D); and a normalized heatmap for available datasets (E).
From here, gene-pages for each miRNA gene can be opened (e.g. http://mirgenedb.org/show/hsa/Let-7-P1a) that contain names, family and seed, orthologues and paralogues, sequences, such as the mature seeds, structure, and a range of other information, including genomic coordinates with hyperlinks to UCSC or ENSEMBLs genome browsers when available. Further, interactive read-pages are also provided for each gene (e.g., http://mirgenedb.org/static/graph/hsa/results/Hsa-Let-7-P1a.html) that show an overview of read-stacks on the corresponding extended precursor sequence of each gene-page. These pages contain detailed representations of templated and 3′-end non-templated reads for individual datasets for each gene, including reports on miRNA isoforms and downloadable read-mappings, and the information can be used to quantify expression of any miRNA across known data sets (e.g. Figure 2B).
For the miRNA repertoire of each species, the members of all miRNA families, or for each miRNA entry, we provide sub-annotations of the precursor, mature, loop, co-mature, star and seed sequences. In addition, we also provide 30-nucleotide flanking regions on both arms for each miRNA to generate an extended precursor transcript for the discovery of regulatory sequence motifs, and lastly separate annotations of seed sequences. On the search-pages, these annotations can be searched independently, either by sequence using Blast (77), the MirGeneDB name, or, if existing, by the full miRBase name (78). Users can also search specific 7-nt seed sequences, and all searches can be done either for individual species or over the entire database. Finally, on the download-pages, fasta, gff, or bed-files for all miRNA components are downloadable for each species.
MiRNA NOMENCLATURE
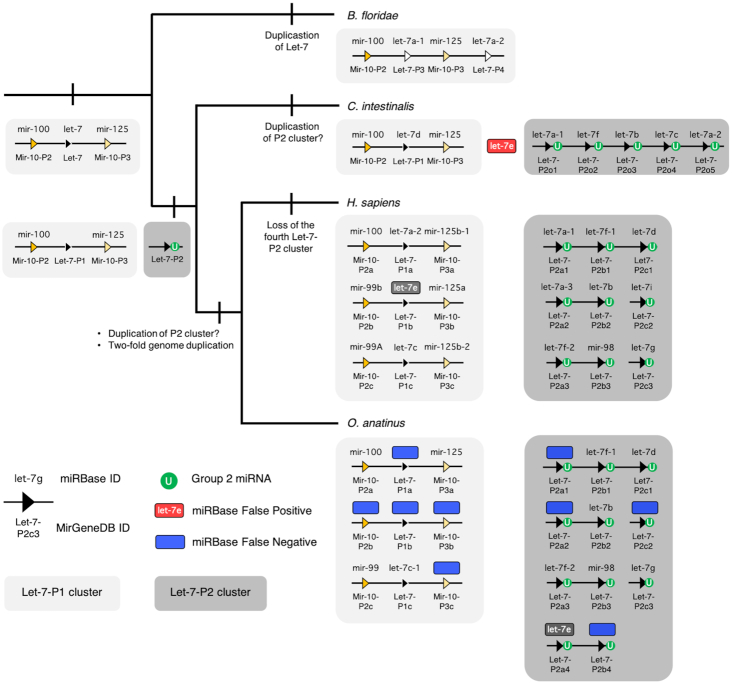
Following Ambros et al. (34), MirGeneDB 2.0 employs an internally consistent nomenclature system where genes of common descent are assigned the same miRNA family name, allowing for the easy recognition of both orthologues in other species, and paralogues within the same species, as described earlier (23). The advantages—and limitations—of the nomenclature system employed by MirGeneDB are exemplified by the LET-7 family of miRNAs (Figure 5). Let-7 is an ancient miRNA gene evolving sometime after the bilaterian split from cnidarians, but before the divergence between protostomes and deuterostomes (79,80), and was (and, in many taxa, still is) syntenically linked to two MIR-10 family members, mir-99/100 and mir-125). However, before the last common ancestor of urochordates and vertebrates (collectively called the Olfactores, (81)), this original gene duplicated, generating two paralogues, one still linked to the two MIR-10 genes (paralogue 1, light gray box), and a second, now located elsewhere in the genome (paralogue 2, dark gray box) (82), that is mono-uridylated at the 3′ end (66). This second paralogue likely duplicated several times before the divergence between urochordates and vertebrates, and then, early in vertebrate evolution, the entire genome duplicated twice. Thus, the last common ancestor of gnathostomes had three clusters of P1 with one Let-7 gene and four clusters of P2, each consisting of 2–3 Let-7 genes (Figure 5). This was followed by breakage of some of the clusters and the loss of the fourth cluster in some taxa, in particular therian mammals. Although both the urochordate and the vertebrates have multiple linked P2 Let-7 genes, none of these genes can be directly orthologized with any of the five P2 genes in urochordates, and thus these five genes are called ‘orphans’ (23) in the urochordate to highlight this fact. However, if new information comes to light that will allow for robust phylogenetic insight, these names would be changed accordingly.
Figure 5.
Nomenclature comparison between MirGeneDB and miRBase for representative chordate Let-7s. Shown is the accepted topology (81) for the three major subgroups of chordates, and for each taxon, a (unscaled) representation of the genomic organization of its Let-7 genes/sequences. MirGeneDB names are shown below each of the loci symbols, and the miRBase sequence names are above. The primitive condition is to possess a single Let-7 gene linked to the two Mir-10 genes (light gray box), as is still found in many bilaterian taxa. In the amphioxus Branchiostoma floridae, this single Let-7 duplicated, and this new paralogue is now positioned at the 3′ end of the cluster. In the Olfactores there is a separate gene duplication event generating another paralogue that is not linked to the original Let-7 cluster in any known urochordate, like Ciona intestinalis, or any vertebrate, including human (H. sapiens) and the platypus (O. anatinus). Further distinguishing this paralogue is that in all Olfactores these Let-7 genes (shown in the dark gray boxes) are Group 2 miRNAs, each with an untemplated mono-uridylated 3′ end (green circles) (see (66)). False negatives (i.e. loci present and transcribed that are present in MirGeneDB, but not in miRBase) are shown in blue. A single false positive (i.e. a sequence present in miRBase—cin-let-7e—but without a corresponding locus in the genome) is shown in red. Note that let-7e also names two sequences derived from two non-orthologous genes in human and platypus—a canonical Group 1 Let-7 (Let-7-P1b) in human, but a Group 2 miRNA (Let-7-P2a4) in platypus. This locus is also present in diapsids (birds and ‘reptiles’), as well as in the teleost fish Danio rerio, but is lost in therian (i.e. placental and marsupial) mammals (see also (82)). Despite the fact that the monophyly of these Group 2 Let-7s in Olfactores appears robust, how the ancestral cluster of the three Let-7-P2s in vertebrates is related to the five linked P2 genes in C. intestinalis remains unknown. Hence, MirGeneDB identifies these genes with this phylogenetic opacity in mind.
The nomenclature system employed by MirGeneDB has several distinct advantages. First, non-orthologous genes are never given the same name. For example, both human and platypus have let-7e sequences, but let-7e in human is derived from the ancestral P1 gene, is linked to MIR-10 genes, and is a Group 1 miRNA; let-7e in platypus is derived from the ancestral P2 gene, is not linked to MIR-10 genes, is mono-uridylated at its 3′ terminus, and maybe most importantly is a gene lost in all therian (i.e. placental and marsupial) mammals (Figure 5).
Second, simply from the name, one can get an accurate picture of the evolutionary history of the gene within the context of a monophyletic miRNA family (23). For example, there are two Let-7 genes in the amphioxus Branchiostoma floridae, a close chordate relative to urochordates and vertebrates, that are amphioxus-specific gene duplicates of the MIR-10 associated Let-7 gene. Although they are called let-7a-1 and let-7a-2, the same names employed by two human miRNAs, they are in fact amphioxus-specific gene duplicates of the MIR-10 associated Let-7 gene. MirGeneDB then necessarily identifies them accordingly, naming them Bfl-Let-7-P3 and Bfl-Let-7-P4 to distinguish these unique paralogues from the two Let-7 paralogues (P1 and P2) of Olfactores (Figure 5).
A third advantage to this system is that misnamed genes will not be orphaned in literature searches or functional studies. For example, one of the 12 human Let-7 paralogue was originally named mir-98 (see Figure 5), and although miRBase lists this gene correctly within the LET-7 family, it is not obvious from the name itself. Notably, in the latest release, applying a novel text mining approach for literature searches, the miRBase authors state that there are only 11 Let-7 family members in human, failing to account for Mir-98 (39). This example clearly highlights the importance of consistent naming and the risks of non-uniform nomenclature systems.
Finally, because MirGeneDB uses this natural classification and nomenclature system, it allows for an accurate reconstruction of ancestral miRNA repertoires—both at the family-level and at the gene-level—that is now provided in MirGeneDB 2.0 for all nodes leading to the 45 terminal taxa considered. This allows users to easily assess both gains and losses of miRNA genes and families through time. Again, with respect to the LET-7 family, it is clear that therians lost two ancestral Let-7 genes, genes that are still retained in platypus (Let-7-P2a4 and -Pb4, see Figure 5) and were present in their last common ancestor.
FUNCTIONAL CLASSIFICATION OF MiRNA-SEEDS
The binding and repression of miRNA targets is primarily mediated by the reverse complementarity between the miRNA seed (nucleotide positions 2–8 of the mature miRNA) and the corresponding target region (83,84). Although highly conserved across vast distances of geologic time, seed sequences can and do change (11,23), expanding the functional repertoire of an ancestral seed sequence. Further, because there are only 16,384 (47) possible seed sequences, sequence space is highly limited necessitating the inevitability of convergence in two evolutionary independent miRNA families. For example, in Caenorhabditis briggsae, there are four LET-7 paralogues (http://mirgenedb.org/browse/cbr?family=LET-7) that all share the seed ‘GAGGUAG’. Interestingly, however, when listing all miRNAs with this seed (http://mirgenedb.org/browse/cbr?seed=GAGGUAG) 8 genes from C. briggsae are listed including four paralogues of the MIR-7594 family. These genes though house the mature sequence - and hence the seed sequence - on the 3p arm, as opposed to the 5p arm as found in LET-7 genes, and thus are a clear case of evolutionary convergence. Nonetheless, because there might be some interesting functional overlap between the Let-7 and Mir-7594 sequences MirGeneDB also now has a ‘seed’ category for each miRNA that summarizes all miRNA entries with the exact same seed sequence (Figure 4, ‘A’). This interface allows the user to find all miRNAs with identical seed within a given species, or among all MirGeneDB organisms in both orthologous and non-orthologous genes. Further, different seeds in similarly named genes allows the user to easily recognize divergence of the seed sequence itself. Finally, a search function is provided that allows the user to search for any known seed sequence.
FUTURE DEVELOPMENTS
The establishment of this carefully curated database of miRNA genes, supplementing existing databases, including miRBase and miRCarta, represents a stable and robust foundation for reproducible miRNA research, in particular studies that rely on cross-species comparisons to explore the roles miRNAs play in development and disease, as well as the evolution of miRNAs and animals themselves. Our long-term goal is to have a wider representation of metazoan species, and for each of these organisms a large number of comparable datasets for a comprehensive set of organs, tissues and cell types.
We hasten to stress that although ∼11 000 genes currently in MirGeneDB have been hand curated, mistakes are inevitable, both in terms of the inclusion of species-specific false positives, missing false negatives, as well as processing errors, mistakes in understanding evolutionary history (possibly resulting in nomenclature errors), and other factors. We would ask the community to alert us to any such errors as only through community-wide collaboration can these inevitable mistakes be eliminated from the database, and MirGeneDB promises to resolve any errors in a timely fashion.
DATA AVAILABILITY
All MirGeneDB data are publicly and freely available under the Creative Commons Zero license. Data are available for bulk download from http://mirgenedb.org/download. All previous versions of MirGeneDB can be found under the Information tab (http://mirgenedb.org/information). Feedback on any aspect of the MirGeneDB database is welcome by email to BastianFromm@gmail.com or Kevin.J.Peterson@dartmouth.edu, or via Twitter (@MirGeneDB).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marc Halushka, and Gianvito Urgese for discussions, and Georgios Magklaras and Sveinung Gundersen for IT support, and two anonymous reviewers for helpful comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Research Area (SFO) program of the Swedish Research Council (VR) through Stockholm University (to B.F., W.K., M.R.F.); South-Eastern Norway Regional Health Authority [2014041, 2018014 to B.F., E.H.]; Russian Science Foundation [18-15-00098 to V.O.]; Dr Mary J. O’Connell and the School of Life Sciences at the University of Nottingham; A.M. has been supported by the Norwegian Research Council, South-Eastern Norway Regional Health Authority and the University of Oslo through the Centre for Molecular Medicine Norway (NCMM), which is part of the Nordic European Molecular Biology Laboratory partnership for Molecular Medicine; K.J.P. has been supported by the National Science Foundation, NASA-Ames and Dartmouth College.
Conflict of interest statement. None declared.
REFERENCES
- 1. Matera A.G., Terns R.M., Terns M.P.. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007; 8:209–220. [DOI] [PubMed] [Google Scholar]
- 2. Hamilton A.J., Baulcombe D.C.. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999; 286:950–952. [DOI] [PubMed] [Google Scholar]
- 3. Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E.. Characterization of the piRNA complex from rat testes. Science. 2006; 313:363–367. [DOI] [PubMed] [Google Scholar]
- 4. Lee R.C., Feinbaum R.L., Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 5. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T.. Identification of novel genes coding for small expressed RNAs. Science. 2001; 294:853–858. [DOI] [PubMed] [Google Scholar]
- 6. Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P.. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294:858–862. [DOI] [PubMed] [Google Scholar]
- 7. Lee R.C., Ambros V.. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001; 294:862–864. [DOI] [PubMed] [Google Scholar]
- 8. Goodarzi H., Liu X., Nguyen H.C.B., Zhang S., Fish L., Tavazoie S.F.. Endogenous tRNA-Derived fragments suppress breast cancer progression via YBX1 Displacement. Cell. 2015; 161:790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chak L.-L., Mohammed J., Lai E.C., Tucker-Kellogg G., Okamura K.. A deeply conserved, noncanonical miRNA hosted by ribosomal DNA. RNA. 2015; 21:375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G.. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403:901–906. [DOI] [PubMed] [Google Scholar]
- 11. Wheeler B.M., Heimberg A.M., Moy V.N., Sperling E.A., Holstein T.W., Heber S., Peterson K.J.. The deep evolution of metazoan microRNAs. Evol. Dev. 2009; 11:50–68. [DOI] [PubMed] [Google Scholar]
- 12. Sempere L.F., Cole C.N., McPeek M.A., Peterson K.J.. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. B. Mol. Dev. Evol. 2006; 306:575–588. [DOI] [PubMed] [Google Scholar]
- 13. Castellano L., Stebbing J.. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res. 2013; 41:3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E. et al.. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010; 24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones-Rhoades M.W. Conservation and divergence in plant microRNAs. Plant Mol. Biol. 2012; 80:3–16. [DOI] [PubMed] [Google Scholar]
- 16. Ludwig N., Becker M., Schumann T., Speer T., Fehlmann T., Keller A., Meese E.. Bias in recent miRBase annotations potentially associated with RNA quality issues. Sci. Rep. 2017; 7:5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langenberger D., Bartschat S., Hertel J., Hoffmann S., Tafer H., Stadler P.F.. MicroRNA or Not MicroRNA? Advances in Bioinformatics and Computational Biology. 2011; Berlin Heidelberg: Springer; 1–9. [Google Scholar]
- 18. Meng Y., Shao C., Wang H., Chen M.. Are all the miRBase-registered microRNAs true? A structure- and expression-based re-examination in plants. RNA Biol. 2012; 9:249–253. [DOI] [PubMed] [Google Scholar]
- 19. Tarver J.E., Donoghue P.C., Peterson K.J.. Do miRNAs have a deep evolutionary history. Bioessays. 2012; 34:857–866. [DOI] [PubMed] [Google Scholar]
- 20. Taylor R.S., Tarver J.E., Hiscock S.J., Donoghue P.C.. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014; 19:175–182. [DOI] [PubMed] [Google Scholar]
- 21. Wang X., Liu X.S.. Systematic curation of miRBase annotation using integrated small RNA High-Throughput sequencing data for C. elegans and Drosophila. Front. Genet. 2011; 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Axtell M.J., Meyers B.C.. Revisiting criteria for plant MicroRNA Annotation in the era of Big Data. Plant Cell. Am. Soc Plant Biol. 2018; 30:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fromm B., Billipp T., Peck L.E., Johansen M., Tarver J.E., King B.L., Newcomb J.M., Sempere L.F., Flatmark K., Hovig E. et al.. A uniform system for the annotation of vertebrate microRNA Genes and the evolution of the human microRNAome. Annu. Rev. Genet. 2015; 49:213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Londin E., Loher P., Telonis A.G., Quann K., Clark P., Jing Y., Hatzimichael E., Kirino Y., Honda S., Lally M. et al.. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1106–E1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jha A., Panzade G., Pandey R., Shankar R.. A legion of potential regulatory sRNAs exists beyond the typical microRNAs microcosm. Nucleic Acids Res. 2015; 43:8713–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng W.-C., Chung I.-F., Tsai C.-F., Huang T.-S., Chen C.-Y., Wang S.-C., Chang T.-Y., Sun H.-J., Chao J.Y.-C., Cheng C.-C. et al.. YM500v2: a small RNA sequencing (smRNA-seq) database for human cancer miRNome research. Nucleic Acids Res. 2015; 43:D862–D867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alles J., Fehlmann T., Fischer U., Backes C., Galata V., Minet M., Hart M., Abu-Halima M., Grässer F.A., Lenhof H.-P. et al.. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019; 47:3353–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ebert M.S., Sharp P.A.. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012; 149:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gosline S.J.C., Gurtan A.M., JnBaptiste C.K., Bosson A., Milani P., Dalin S., Matthews B.J., Yap Y.S., Sharp P.A., Fraenkel E.. Elucidating MicroRNA regulatory networks using transcriptional, Post-transcriptional, and histone modification measurements. Cell Rep. 2016; 14:310–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alberti C., Cochella L.. A framework for understanding the roles of miRNAs in animal development. Development. 2017; 144:2548–2559. [DOI] [PubMed] [Google Scholar]
- 31. Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M.. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005; 433:769–773. [DOI] [PubMed] [Google Scholar]
- 32. Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. et al.. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838. [DOI] [PubMed] [Google Scholar]
- 33. Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M. et al.. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M. et al.. A uniform system for microRNA annotation. RNA. 2003; 9:277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tosar J.P., Rovira C., Cayota A.. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 2018; 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hou X.-S., Han C.-Q., Zhang W.. MiR-1182 inhibited metastasis and proliferation of ovarian cancer by targeting hTERT. Eur. Rev. Med. Pharmacol. Sci. 2018; 22:1622–1628. [DOI] [PubMed] [Google Scholar]
- 37. Zhang D., Xiao Y.-F., Zhang J.-W., Xie R., Hu C.-J., Tang B., Wang S.-M., Wu Y.-Y., Hao N.-B., Yang S.-M.. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015; 360:151–159. [DOI] [PubMed] [Google Scholar]
- 38. Zhou J., Dai W., Song J.. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem. Biophys. Res. Commun. 2016; 470:445–452. [DOI] [PubMed] [Google Scholar]
- 39. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Backes C., Fehlmann T., Kern F., Kehl T., Lenhof H.-P., Meese E., Keller A.. miRCarta: a central repository for collecting miRNA candidates. Nucleic Acids Res. 2018; 46:D160–D167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tarver J.E., Taylor R.S., Puttick M.N., Lloyd G.T., Pett W., Fromm B., Schirrmeister B.E., Pisani D., Peterson K.J., Donoghue P.C.J.. Well-annotated microRNAomes do not evidence pervasive miRNA loss. Genome Biol. Evol. 2018; 10:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Engkvist M.E., Stratford E.W., Lorenz S., Meza-Zepeda L.A., Myklebost O., Munthe E.. Analysis of the miR-34 family functions in breast cancer reveals annotation error of miR-34b. Sci Rep. 2017; 7:9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fromm B., Tosar J.P., Lu Y., Halushka M.K., Witwer K.W.. Human and Cow Have Identical miR-21-5p and miR-30a-5p Sequences, Which Are Likely Unsuited to Study Dietary Uptake from Cow Milk. The Journal of Nutrition. 2018; 148:1506–1507. [DOI] [PubMed] [Google Scholar]
- 44. Van Peer G., Lefever S., Anckaert J., Beckers A., Rihani A., Van Goethem A., Volders P.J., Zeka F., Ongenaert M., Mestdagh P. et al.. miRBase Tracker: keeping track of microRNA annotation changes. Database. 2014; 2014:bau080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong X., Heinicke F., Rayner S.. miRBaseMiner, a tool for investigating miRBase content. RNA Biol. 2019; 16:1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu T.-P., Lee C.-Y., Tsai M.-H., Chiu Y.-C., Hsiao C.K., Lai L.-C., Chuang E.Y.. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One. 2012; 7:e42390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bonnal R.J.P., Rossi R.L., Carpi D., Ranzani V., Abrignani S., Pagani M.. miRiadne: a web tool for consistent integration of miRNA nomenclature. Nucleic Acids Res. 2015; 43:W487–W492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu T., Su N., Liu L., Zhang J., Wang H., Zhang W., Gui J., Yu K., Li J., Le T.D.. miRBaseConverter: an R/Bioconductor package for converting and retrieving miRNA name, accession, sequence and family information in different versions of miRBase. BMC Bioinformatics. BioMed Central. 2018; 19:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haunsberger S.J., Connolly N.M.C., Prehn J.H.M.. miRNAmeConverter: an R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics. 2017; 33:592–593. [DOI] [PubMed] [Google Scholar]
- 50. Budak H., Bulut R., Kantar M., Alptekin B.. MicroRNA nomenclature and the need for a revised naming prescription. Brief. Funct. Genomics. 2016; 15:65–71. [DOI] [PubMed] [Google Scholar]
- 51. Grimson A ( Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008; 455:1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jan C.H., Friedman R.C., Ruby J.G., Bartel D.P.. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011; 469:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P.. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006; 20:3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruby J.G. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006; 127:1193–1207. [DOI] [PubMed] [Google Scholar]
- 55. Ruby J.G. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007; 17:1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aparicio-Puerta E., Lebrón R., Rueda A., Gómez-Martín C., Giannoukakos S., Jaspez D., Medina J.M., Zubkovic A., Jurak I., Fromm B. et al.. sRNAbench and sRNAtoolbox 2019: intuitive fast small RNA profiling and differential expression. Nucleic Acids Res. 2019; 47:W530–W535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kang W., Eldfjell Y., Fromm B., Estivill X., Biryukova I., Friedländer M.R.. miRTrace reveals the organismal origins of microRNA sequencing data. Genome Biol. BioMed Central. 2018; 19:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., Åström G., Babina M., Bertin N., Burroughs A.M. et al.. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017; 35:872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim Y.-K., Kim B., Kim V.N.. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McCall M.N., Kim M.S., Adil M., Patil A.H., Lu Y., Mitchell C.J., Leal-Rojas P., Xu J., Kumar M., Dawson V.L. et al.. Toward the human cellular microRNAome. Genome Res. 2017; 27:1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Juzenas S., Venkatesh G., Hübenthal M., Hoeppner M.P., Du Z.G., Paulsen M., Rosenstiel P., Senger P., Hofmann-Apitius M., Keller A. et al.. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017; 45:9290–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Halushka M.K., Fromm B., Peterson K.J., McCall M.N.. Big strides in cellular MicroRNA Expression. Trends Genet. Elsevier Curr. Trends. 2018; 34:165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Desvignes T., Loher P., Eilbeck K., Ma J., Urgese G., Fromm B., Sydes J., Aparicio-Puerta E., Barrera V., Espín R. et al.. Unification of miRNA and isomiR research: the mirGFF3 format and the mirtop API. Bioinformatics. 2019; doi:10.1093/bioinformatics/btz675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neilsen C.T., Goodall G.J., Bracken C.P.. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012; 28:544–549. [DOI] [PubMed] [Google Scholar]
- 65. Kim B., Ha M., Loeff L., Chang H., Simanshu D.K., Li S., Fareh M., Patel D.J., Joo C., Kim V.N.. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 2015; 34:1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Heo I., Ha M., Lim J., Yoon M.-J., Park J.-E., Kwon S.C., Chang H., Kim V.N.. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012; 151:521–532. [DOI] [PubMed] [Google Scholar]
- 67. Fang W., Bartel D.P.. The menu of features that define primary MicroRNAs and Enable de novo design of MicroRNA Genes. Mol. Cell. 2015; 60:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bartel D.P. Metazoan MicroRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manzano M., Forte E., Raja A.N., Schipma M.J., Gottwein E.. Divergent target recognition by coexpressed 5′-isomiRs of miR-142-3p and selective viral mimicry. RNA. 2015; 21:1606–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tyler D.M., Okamura K., Chung W.-J., Hagen J.W., Berezikov E., Hannon G.J., Lai E.C.. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008; 22:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yi R., Poy M.N., Stoffel M., Fuchs E.. A skin microRNA promotes differentiation by repressing “stemness”. Nature. 2008; 452:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tarver J.E., Sperling E.A., Nailor A., Heimberg A.M., Robinson J.M., King B.L., Pisani D., Donoghue P.C., Peterson K.J.. miRNAs: small genes with big potential in metazoan phylogenetics. Mol. Biol. Evol. 2013; 30:2369–2382. [DOI] [PubMed] [Google Scholar]
- 75. Backes C., Meder B., Hart M., Ludwig N., Leidinger P., Vogel B., Galata V., Roth P., Menegatti J., Grässer F. et al.. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. 2016; 44:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thomson R.C., Plachetzki D.C., Mahler D.L., Moore B.R.. A critical appraisal of the use of microRNA data in phylogenetics. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E3659–E3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang Z., Schwartz S., Wagner L., Miller W.. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000; 7:203–214. [DOI] [PubMed] [Google Scholar]
- 78. Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J.. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006; 34:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pasquinelli A.E. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000; 408:86–89. [DOI] [PubMed] [Google Scholar]
- 80. Pasquinelli A.E., McCoy A., Jiménez E., Salo E., Ruvkun G., Martindale M.Q., Baguna J.. Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution? Evol Dev. Wiley Online Library. 2003; 5:372–378. [DOI] [PubMed] [Google Scholar]
- 81. Delsuc F., Brinkmann H., Chourrout D., Philippe H.. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006; 439:965–968. [DOI] [PubMed] [Google Scholar]
- 82. Hertel J., Bartschat S., Wintsche A., Otto C., Stadler P.F. SBCL . Evolution of the let-7 microRNA Family. RNA Biol. 2012; 9:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002; 30:363–364. [DOI] [PubMed] [Google Scholar]
- 84. Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B.. Prediction of mammalian microRNA targets. Cell. 2003; 115:787–798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MirGeneDB data are publicly and freely available under the Creative Commons Zero license. Data are available for bulk download from http://mirgenedb.org/download. All previous versions of MirGeneDB can be found under the Information tab (http://mirgenedb.org/information). Feedback on any aspect of the MirGeneDB database is welcome by email to BastianFromm@gmail.com or Kevin.J.Peterson@dartmouth.edu, or via Twitter (@MirGeneDB).