Abstract
GMrepo (data repository for Gut Microbiota) is a database of curated and consistently annotated human gut metagenomes. Its main purpose is to facilitate the reusability and accessibility of the rapidly growing human metagenomic data. This is achieved by consistently annotating the microbial contents of collected samples using state-of-art toolsets and by manual curation of the meta-data of the corresponding human hosts. GMrepo organizes the collected samples according to their associated phenotypes and includes all possible related meta-data such as age, sex, country, body-mass-index (BMI) and recent antibiotics usage. To make relevant information easier to access, GMrepo is equipped with a graphical query builder, enabling users to make customized, complex and biologically relevant queries. For example, to find (1) samples from healthy individuals of 18 to 25 years old with BMIs between 18.5 and 24.9, or (2) projects that are related to colorectal neoplasms, with each containing >100 samples and both patients and healthy controls. Precomputed species/genus relative abundances, prevalence within and across phenotypes, and pairwise co-occurrence information are all available at the website and accessible through programmable interfaces. So far, GMrepo contains 58 903 human gut samples/runs (including 17 618 metagenomes and 41 285 amplicons) from 253 projects concerning 92 phenotypes. GMrepo is freely available at: https://gmrepo.humangut.info.
INTRODUCTION
Increasing evidence has linked gut microbiota to many aspects of human life, including health (1–3), diseases (4–13), development (14–18), responses to drugs and treatments (19–23). In recent years, the number and total volume of human gut metagenomic data (including both 16S and metagenomic sequencing data) have been increasing rapidly (24). Most of the raw sequencing data have been deposited into several general purpose databases, such as NCBI Sequence Read Archive (SRA) (25) (https://www.ncbi.nlm.nih.gov/sra) and European Nucleotide Archive (ENA) (26) (https://www.ebi.ac.uk/ena). A few other databases, including EBI Metagenomics (now MGnify) (24), gcMeta (27), MSE (28) and Qiita (29), have provided processed data and organized them according to the habitats from which the samples were taken. These public resources greatly facilitate data reuse, especially meta-analyses across multiple related studies for the purpose of cross-study validation and discovery of novel causal microbial taxa underlying certain phenotypes (11,12,30).
Despite these existing efforts to deposit, organize and analyze the rapidly growing human metagenomic data, major obstacles to their reusability and accessibility remain, especially incomplete and/or inaccurate phenotype information and/or missing meta-data. Recently, a study reported an initial effort to curate human metagenomic data; however, the data were limited in the number of samples reported (5716 samples collected from 26 projects as of January 2017 when the results were first published), contained metagenomic data from other body sites and could only be accessed using R (31). In addition, so far there have been no systematic efforts to help users filter human gut samples and/or projects with biologically relevant questions. For example, there is no easy way to find fecal samples that were taken from healthy individuals of 18–25 years of age with healthy body mass indexes (BMIs, 18.9–24.9) from any of the existing databases and data sources; also it is very difficult to find all the projects that are related to colorectal neoplasms studies, contain >100 samples and contain both patients and healthy controls.
To address these issues, and more importantly to facilitate the reusability and accessibility of the rapidly growing human metagenomic data, we developed GMrepo as a database of curated human gut metagenomic data (including both 16S and metagenomic sequencing data). The main features of GMrepo include: (i) manually curated phenotype information for each collected run/sample and all possible related meta-data, such as the age, sex, country, body-mass-index (BMI) and even recent antibiotics usage; more meta-data could be included in the future; (ii) consistently annotated microbial contents, including taxonomic assignments of sequencing reads and precommutated species/genus relative abundances using state-of-art toolsets; (iii) collected samples organized according to their associated phenotypes and statistics, including species-prevalence, abundances and co-occurrences; (iv) in addition to the online database, GMrepo also provides programmable access to most of its contents through representational state transfer (REST) application programming interfaces (APIs); (v) more importantly, GMrepo is equipped with powerful and easy-to-use graphical query builders to allow users to make customized, biologically meaningful queries to the collected samples and projects.
CONSTRUCTION AND CONTENTS OF GMrepo
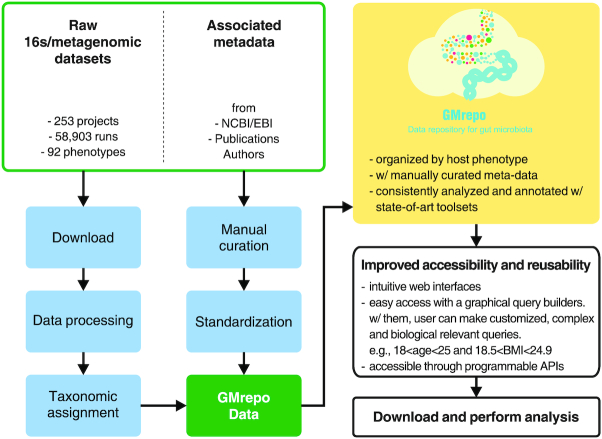
Figure 1 illustrates the overall workflow of GMrepo, while Figure 2 shows the detailed analysis pipeline of the collected human gut metagenomic data. Below is a brief summary of the materials and methods used in this study.
Figure 1.
Overall workflow of GMrepo. Processing steps are indicated in the blue rounded boxes.
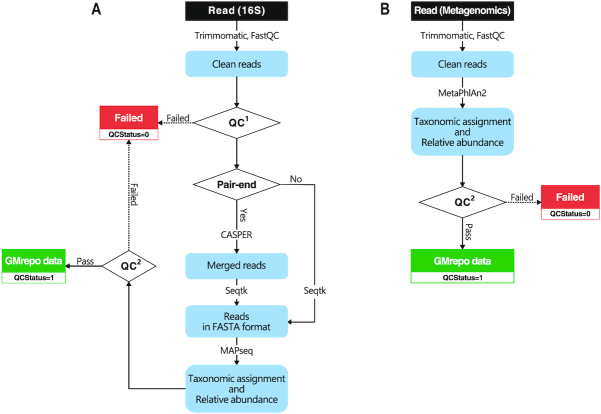
Figure 2.
Schematic representations of the GMrepo metagenomics pipeline for amplicon data (A) and metagenomic data (B). Processing steps are indicated in the blue rounded boxes and tools are marked on the arrows. Input and output files as colored rectangles (black, green, red). Conditional judgments are in trapezoids. QC1: a run will be marked as ‘failed’ (QCStatus = = 0) if less than 20k reads or <50% of reads were retained after trimming; QC2: arun will be marked as ‘failed’ (QCStatus = = 0) if a single taxon accounts for >99.99% of the total abundance.
Data acquisition of sequencing reads and manual curation of meta-data
Raw sequencing reads were downloaded from the EBI ENA (26) (European Nucleotide Archive, https://www.ebi.ac.uk/ena) and NCBI SRA (25) (Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra) databases using command line tools from enaBrowserTools (https://github.com/enasequence/enaBrowserTools) and SRA-Tools (https://ncbi.github.io/sra-tools/) facilitated by Aspera (a high-speed data transfer tool). Related meta-data of the sequencing platforms, corresponding biosamples, experiments, projects and the human hosts from which the fecal samples were taken, were obtained from EBI Metagenomics (now MGnify) (24) and related databases of the NCBI.
Two rounds of manual curation were then performed on the meta-data. For the first round, meta-information, such as phenotypes (health or diseases), age, sex and BMI of the associated samples/runs were extracted using in-house R and Perl scripts and were manually curated and supplemented with the materials obtained from the related publications and/or even from the authors (Figure 1). The extracted meta-data include sequencing related meta-data, including the sequencing platform, type of sequences obtained (i.e. 16S or metagenomic) number of sequences, and human host related meta-data including phenotypes (i.e., diseases or healthy), age, sex, country, BMI and antibiotic usage. For the second round, different curators from the first round reviewed the collected meta-data and made necessary corrections.
Processing of raw sequencing reads
FastQC (version 0.11.8, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to evaluate the overall quality of the downloaded data, followed by the use of Trimmomatic (32) to remove sequencing vectors and low-quality bases. Sequences shorter than two-thirds of the original read length were removed from the subsequent analysis (Figure 2).
For 16S sequences, single-ended sequencing reads were used directly for subsequent analysis, while the pair-ended reads were first merged using Casper (33) before downstream processing. Metagenomic sequences were used directly for subsequent analysis, regardless of whether they were single- or pair-ended.
The processed data were referred as to ‘clean data’. When necessary, Seqtk (https://github.com/lh3/seqtk) was used to convert FASTQ sequences to FASTA format.
Taxonomic assignment to processed sequencing reads and calculation of relative abundances
For 16S sequences, MAPseq version 1.2 (34) was used to analyze the obtained clean data and assign taxonomic classification information to the reads. Reads with a combined score higher than 0.4 at the genus level were used for subsequent analysis, as recommended by the authors of MAPseq. Relative abundances were then calculated at the genus and species levels for each sample/run, with total abundance values of 100%.
For metagenomic sequences, MetaPhlAn2 (35) was used with default parameters for the taxonomic assignments to the sequencing reads and calculating the relative abundances at species and genus levels.
Two-step quality controls
A two-step quality control process was used to ensure the quality of the data (Figure 2). First, amplicon sequencing samples/runs with <20 000 reads were removed from subsequent analysis and were marked as ‘failed QC (QC status = 0)’ in GMrepo. The second step of quality control is for both amplicon sequences and metagenomic sequences. After taxonomy assignment, samples/runs containing only a single taxon, i.e., a species or a genus accounted for more than or equal to 99.99% of the total abundance, will also be marked as ‘failed QC (QC status = 0)’.
Species co-occurrence analysis
Species co-occurrences were performed separately for phenotypes with more than 50 related samples/runs. For each species-species and genus-genus pair of phenotypes, Fisher's exact text (fisher.test() function in R) was used; the four required numbers as input are: the number of samples/runs in which both taxa are found, the numbers of samples in which either taxa are found and the number of samples in which neither of the taxa are found. Taxon pairs with an Odds ratio (OR) value larger than 1 and a P-value < 0.05 are considered to significantly co-occur in a phenotype.
In addition to the presence/absence information, the relative abundances of the co-occurring pairs were used to calculate Person and Spearman correlations in order to further describe the directions of the interactions between the two taxa.
Database construction and web development
All data were loaded into a MySQL database. The frontend (the webpages) of the website was coded using HTML and JavaScript, while the backend was coded using PHP with a Slim framework to support queries to the MySQL database and provide representational state transfer (REST) application programming interfaces (APIs) for programmable access to our data. The AngularJS framework was used to bride the front- and back- ends. D3.js and plotly.js were used for visualizations at the front-end. Various other open-source JavaScript libraries were also used, including jQuery and jQuery QueryBuilder. The website is hosted on an Apache server.
USAGE, UTILITY AND FUTURE DIRECTIONS
Human gut metagenomic data organized according to host phenotypes
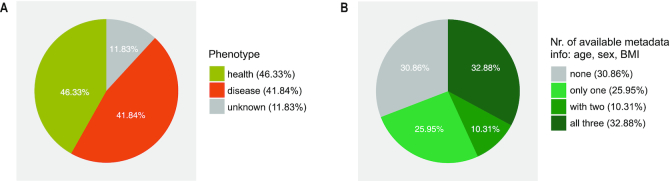
Through multiple rounds of manual curation, we collected meta-data for a total of 58 903 runs of human gut metagenomic data from 253 projects, including 17 618 metagenomes and 41 285 amplicons spanning 92 phenotypes (health and 91 diseases). Figure 3 summarizes statistics of some of the metadata we have collected. For example, we were able to assign explicit phenotype information to most of the collected samples (88.17%, Figure 3A); however, despite our efforts, we were able to obtain only very basic meta-data including age, sex and BMI for only one third of the samples. As shown in Figure 3B, 30.86% of the samples contained none of the basic meta-data, while the rest contained only one or two (25.95% and 10.31%, respectively). These results highlight the challenges in reusing metagenomic data and call for reporting standards of minimal meta-data information or metagenomic samples.
Figure 3.
Statistics of some of the metadata we collected. (A) The unknown phenotype means that the health status of the sample provider is not clearly indicated. For data from the American Gut Project (AGP), we only use diagnoses from medical professionals (doctor, physician assistant). Samples with unknown phenotypes are mainly from AGP. (B) The integrity of the metadata is assessed based on age, sex and BMI.
In addition to the project-run relationships, we organized the collected gut metagenomic data according to their associated host phenotypes. We adopted the MeSH system (36) (Medical Subject Headings, a hierarchically organized controlled vocabulary for biomedical information) to describe and organize these phenotypes. Listed in Table 1 are the top 10 phenotypes included in GMrepo.
Table 1.
Top 10 phenotypes included in GMrepo
| Phenotype | No. of runs | No. of processed runs | No. of valid runs | No. of failed runs | No. of associated species | No. of associated genus |
|---|---|---|---|---|---|---|
| Health | 27 329 | 20 320 | 12 485 | 7835 | 6189 | 1613 |
| Colitis, Ulcerative | 2509 | 2440 | 1175 | 1265 | 4183 | 1285 |
| Irritable Bowel Syndrome | 2092 | 2091 | 954 | 1137 | 3320 | 1064 |
| Infant, Premature | 1443 | 1443 | 1240 | 203 | 260 | 97 |
| Colorectal Neoplasms | 1374 | 1374 | 1256 | 118 | 4596 | 1380 |
| Diarrhea | 1355 | 1354 | 470 | 884 | 2775 | 906 |
| Constipation | 1244 | 1244 | 611 | 633 | 3146 | 1022 |
| Migraine Disorders | 1235 | 1235 | 574 | 661 | 2894 | 964 |
| Lung Diseases | 1228 | 1228 | 592 | 636 | 2817 | 958 |
| Autoimmune Diseases | 1154 | 1154 | 547 | 607 | 2848 | 956 |
No. of runs: all runs with curated meta-data,
No. of processed runs: number of all runs with the sequence data processed; please note all runs will be processed eventually,
No. of valid runs: number of runs whose data passed our QC procedure and the corresponding species/genus relative abundances are available in our database,
No. of failed runs: number of runs whose data DID NOT passed our QC procedure,
No. of associated species: number of species associated with the processed and valid runs.
No. of associated genus: number of genus associated with the processed and valid runs.
For each phenotype, we summarized the total numbers of associated species and genera. For example, in total there are 6189 species (and/or strains) associated with healthy individuals (https://gmrepo.humangut.info/phenotypes/D006262), which were assigned to a total of 1613 genera. However, only 389 (∼6.3% of the total) species, assigned to 91 (∼5.6% of total) genera, were found in more than one sample with a median relative abundance higher than 0.01%. Similar results were found in other phenotypes. These results indicate that most of these taxa were found in only a small number of runs, and/or are presented with limited abundances.
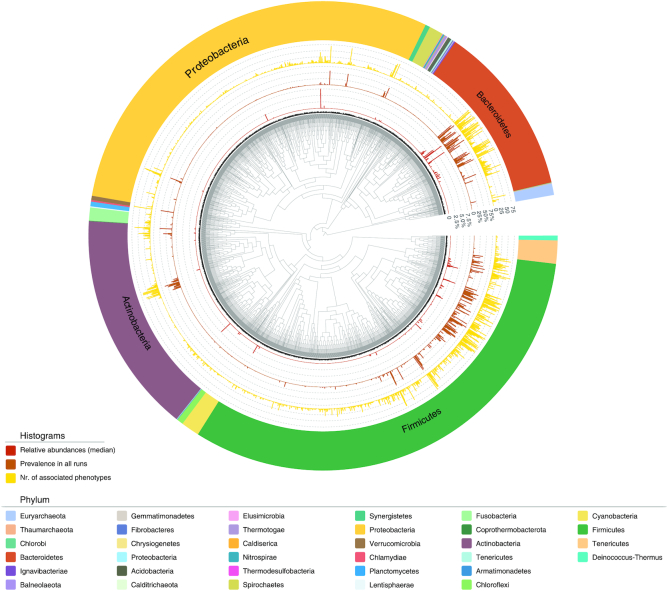
In all of the 28 252 valid runs in our database, we found that in total 6973 species were assigned to 1710 genera. Among these, 2685 species, assigned to 834 genera, were found in more than one sample with a median relative abundance higher than 0.01% within one or more phenotypes (Figure 4, the phylogenetic relationships of these species were obtained from the NCBI taxonomy database (37) and were visualized using Evolview v3 (38)); these numbers are close to recently published results (39). Although the prevalence of most species is low, our results have expanded the known species repertoire of the collective human gut microbiota. Diet, region, and disease are known to affect the abundance and diversity of the human gut microbiota. We believe that the total number of species/strains in the human gut flora will be further increased as more samples are analyzed in the future.
Figure 4.
Phylogenetic tree comprising the 2685 included species, based on NCBI taxonomy. These 2685 species were found in more than one samples with a median relative abundance higher than 0.01% within one or more phenotypes. The three inner layers show the statistics of these species in our database, including the median relative abundance of the species (red) and the species prevalence in the samples (brown) and phenotypes (yellow). The outermost layer shows the corresponding phyla of these species.
Additional links to the NCBI BioProject, NCBI SRA and NCBI MeSH Browser were also provided for each of the projects, runs and phenotypes, in order to facilitate researchers to obtain more information or download raw sequencing data. More external databases will be included in the future.
Species abundance, prevalence and co-occurrence within and across phenotypes
With the availability of precalculated relative abundance information for all valid runs in GMrepo, we allow users to visualize the species/genus abundance distribution in a phenotype of interest as a scatter plot (Supplementary Figure S1); if a user chooses a disease (e.g. Crohn's Disease or colorectal neoplasms), the abundances of the same taxon in healthy controls will also be retrieved and visualized side-by-side with the disease in the scatter plot and boxplot (Figure 5A, B). Visualization and comparison of the taxon abundances across all phenotypes is also supported (Supplementary Figures S2 and S3).
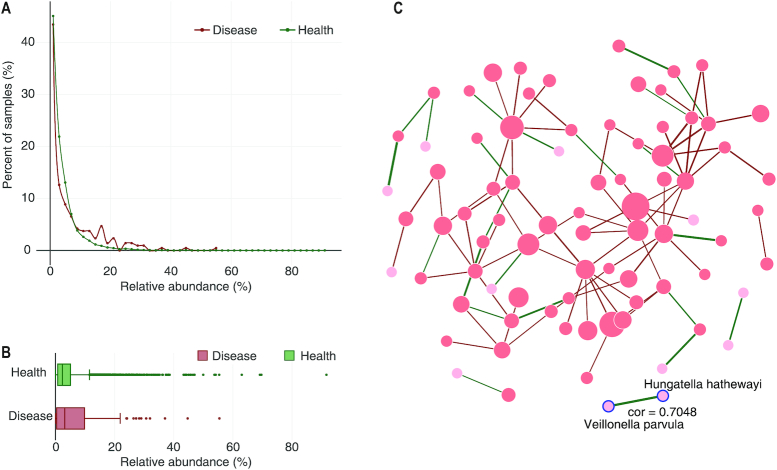
Figure 5.
Details of a species in Crohn's Diseases. Faecalibacterium prausnitzii is chosen to show its distributions (A) and relative abundances (B) in Crohn's Disease. For various disease phenotypes, the relative abundances of the species of interest in healthy controls (green) will also be retrieved and visualized side-by-side with the disease (red). (C) A species co-occurrence network constructed based on the significantly co-occurred pairs for a phenotype (Crohn's Disease). Nodes: species co-occurred with others in samples of this phenotype with sizes proportional to the number of connected nodes in the network. Links: indicate co-occurring relationships between species with widths proportional to the absolute value of the correlation coefficient (Pearson correlation), while the colors indicate positive (green) or negative (red) correlations. Placing a mouse over a node can highlight the node and its direct neighbors and show the names of the node and its direct neighbors.
We also calculated the species/genus prevalence for each species (Supplementary Figure S4). Based on the presence/absence information, we calculated pairwise co-occurrences within each phenotype for all possible species-species and genus-genus pairs. For significantly co-occurred pairs (see the ‘Construction and contents of GMrepo’ for details), we also provided precalculated Person and Spearman correlation coefficient values based on their relative abundances, in order to further describe the directions of the interactions between the two taxa. For example, a significant positive correlation coefficient may indicate the two taxa prefer similar environments and/or are beneficial to each other's’ growth, while a significant negative correlation coefficient may indicate the two taxa prefer different environments and/or are competitive. A co-occurrence network can then be constructed based on significantly co-occurred pairs, as shown in Figure 5C.
Additional links to external databases were also provided for each of the species and genera identified in GMrepo, in order to facilitate researchers in obtaining related information on these taxa. So far we have linked GMrepo to NCBI taxonomy, ENA taxonomy, genome annotations (40), microbe to bacteriophage interactions (41), bacteria to drug interactions (http://www.bugdrug-db.info) and a few others (42). More external databases will be included in the future.
Complex and biologically relevant queries to our data are facilitated by graphical query builders
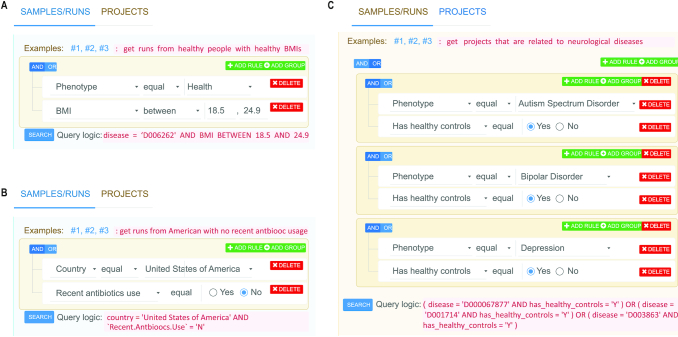
One of the most important features of GMrepo is the collection and manual curation of related meta-data. To further take advantage of this data, we equipped GMrepo with graphical query builders (powered by the jQueryBuilder widget) to allow users to perform complex queries. We provided two query builders and three examples for each. As shown in Figure 6, the query builders are easy to use because of their straightforward and self-explanatory interface. They support complex logic combinations (AND, OR and grouping) that allow users to perform biologically relevant queries. For example, Figure 6A shows how to find runs/samples from healthy individuals with BMIs between 18.5 and 24.9; Figure 6B allows users to find fecal samples of Americans who have not used antibiotics recently; Figure 6C shows how to find projects that are related to neurological diseases (including autism spectrum disorder, bipolar disorder and depression) and each contains healthy controls. More examples can be found at https://gmrepo.humangut.info.
Figure 6.
Graphical selectors and three examples. These selectors support complex logic combinations (AND, OR and grouping) that allow users to perform biologically relevant queries. (A) Shows how to find samples from healthy individuals with BMIs between 18.5 and 24.9; (B) allows users to find fecal samples of Americans who did not recently use antibiotics; (C) shows how to find projects that are related to neurological diseases (e.g. including autism spectrum disorder, bipolar disorder and depression) and each contains healthy controls.
More query builders will be added in the near future to allow users to search for species/genera based on their abundances, prevalence, co-occurrences and differential abundances in different phenotypes.
Future directions
In addition to the continuous collection of new human gut metagenomic data in the coming years, we plan to add new contents to GMrepo, including (but not limited to) viral abundances, functional profiles and metabolic pathway profiles for the collected samples. We also plan to include more utilities, allowing users to perform on-site cross-sample comparisons, differential abundance analysis and mathematical modeling. These will further facilitate the reusability and accessibility of human gut metagenomic data and will contribute to better understanding of the relationships between gut microbiota dysbiosis and human diseases.
CONCLUSIONS
In this study, we introduced GMrepo, an online database of curated, consistently annotated meta-data and human gut metagenomic data. With 58 903 samples/runs collected from 253 projects and 92 phenotypes, GMrepo is one of the largest databases dedicated to human gut metagenomes (including both 16S and metagenomic sequences). We carefully curated meta-data and applied stringent criteria to keep only high quality data. To facilitate reusability and accessibility, we included precomputed species/genus relative abundances, prevalence within and across phenotypes, as well as pairwise co-occurrence information. These data are available at the website and can be accessed through programmable interfaces. To make relevant information easier to access, we equipped GMrepo with a graphical query builder, allowing users to make customized, complex and biologically relevant queries. We will continue developing GMrepo in the near future by including more manually curated human gut metagenomic data, more functional annotated data, and more utilities.
DATA AVAILABILITY
All data are freely accessible to all academic users. This work is licensed under a Creative Commons Attribution-Non-Commercial 3.0 Unported License (CC BY-NC 3.0). In addition to various download functions on many webpages, users can download all data from the ‘Data downloads’ section of the ‘Help’ page. Programmable access through REST APIs is also supported; detailed instructions on using R, Perl and Python to access our data can be found at the ‘Programmable access’ section of the ‘Help’ page or our GitHub page: https://github.com/evolgeniusteam/GMrepoProgrammableAccess.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Key Research and Development Program of China [2018YFC0910502 to W.H.C.]. This work was partly supported by National Key R&D Program of China (2018YFC0910500), National Natural Science Foundation of China (61932008, 61772368, 61572363), Natural Science Foundation of Shanghai (17ZR1445600), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01) and ZJLab.
Conflict of interest statement. None declared.
REFERENCES
- 1. Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.M. et al.. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T. et al.. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I.. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 4. Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., Guo J., Le Chatelier E., Yao J., Wu L. et al.. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014; 513:59–64. [DOI] [PubMed] [Google Scholar]
- 5. Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A., Forslund K., Hildebrand F., Prifti E., Falony G. et al.. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016; 535:376–381. [DOI] [PubMed] [Google Scholar]
- 6. Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D. et al.. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490:55–60. [DOI] [PubMed] [Google Scholar]
- 7. Noguera-Julian M., Rocafort M., Guillen Y., Rivera J., Casadella M., Nowak P., Hildebrand F., Zeller G., Parera M., Bellido R. et al.. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016; 5:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frye R.E., Slattery J., MacFabe D.F., Allen-Vercoe E., Parker W., Rodakis J., Adams J.B., Krajmalnik-Brown R., Bolte E., Kahler S. et al.. Approaches to studying and manipulating the enteric microbiome to improve autism symptoms. Microb. Ecol. Health Dis. 2015; 26:26878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F. et al.. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013; 155:1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J., Zhao F., Wang Y., Chen J., Tao J., Tian G., Wu S., Liu W., Cui Q., Geng B. et al.. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017; 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A., Fleck J.S., Voigt A.Y., Palleja A., Ponnudurai R. et al.. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019; 25:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas A.M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., Beghini F., Manara S., Karcher N., Pozzi C. et al.. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019; 25:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dai D., Wang T., Wu S., Gao N.L., Chen W.H.. Metabolic dependencies underlie interaction patterns of gut microbiota during enteropathogenesis. Front. Microbiol. 2019; 10:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Backhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H. et al.. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015; 17:690–703. [DOI] [PubMed] [Google Scholar]
- 15. Forsgren M., Isolauri E., Salminen S., Rautava S.. Late preterm birth has direct and indirect effects on infant gut microbiota development during the first six months of life. Acta Paediatr. 2017; 106:1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wall R., Ross R.P., Ryan C.A., Hussey S., Murphy B., Fitzgerald G.F., Stanton C.. Role of gut microbiota in early infant development. Clin. Med. Pediatr. 2009; 3:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart C.J., Ajami N.J., O’Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., Ross M.C., Lloyd R.E., Doddapaneni H., Metcalf G.A. et al.. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018; 562:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pronovost G.N., Hsiao E.Y.. Perinatal interactions between the microbiome, immunity, and neurodevelopment. Immunity. 2019; 50:18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N. et al.. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017; 170:548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., Prifti E., Vieira-Silva S., Gudmundsdottir V., Pedersen H.K. et al.. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015; 528:262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C. et al.. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018; 359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., Gajewski T.F.. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018; 359:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillere R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P. et al.. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018; 359:91–97. [DOI] [PubMed] [Google Scholar]
- 24. Mitchell A.L., Scheremetjew M., Denise H., Potter S., Tarkowska A., Qureshi M., Salazar G.A., Pesseat S., Boland M.A., Hunter F.M.I. et al.. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018; 46:D726–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kodama Y., Shumway M., Leinonen R., Database International Nucleotide Sequence. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrison P.W., Alako B., Amid C., Cerdeno-Tarraga A., Cleland I., Holt S., Hussein A., Jayathilaka S., Kay S., Keane T. et al.. The european nucleotide archive in 2018. Nucleic Acids Res. 2019; 47:D84–D88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi W., Qi H., Sun Q., Fan G., Liu S., Wang J., Zhu B., Liu H., Zhao F., Wang X. et al.. gcMeta: a Global Catalogue of Metagenomics platform to support the archiving, standardization and analysis of microbiome data. Nucleic Acids Res. 2019; 47:D637–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su X., Jing G., McDonald D., Wang H., Wang Z., Gonzalez A., Sun Z., Huang S., Navas J., Knight R.J.M.. Identifying and predicting novelty in microbiome studies. MBio. 2018; 9:e02099-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gonzalez A., Navas-Molina J.A., Kosciolek T., McDonald D., Vazquez-Baeza Y., Ackermann G., DeReus J., Janssen S., Swafford A.D., Orchanian S.B. et al.. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods. 2018; 15:796–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J.. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017; 8:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pasolli E., Schiffer L., Manghi P., Renson A., Obenchain V., Truong D.T., Beghini F., Malik F., Ramos M., Dowd J.B. et al.. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods. 2017; 14:1023–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwon S., Lee B., Yoon S.. CASPER: context-aware scheme for paired-end reads from high-throughput amplicon sequencing. BMC Bioinformatics. 2014; 15(Suppl. 9):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matias Rodrigues J.F., Schmidt T.S.B., Tackmann J., von Mering C.. MAPseq: highly efficient k-mer search with confidence estimates, for rRNA sequence analysis. Bioinformatics. 2017; 33:3808–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C.. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012; 9:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lipscomb C.E. Medical Subject Headings (MeSH). Bull. Med. Libr. Assoc. 2000; 88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 37. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subramanian B., Gao S., Lercher M.J., Hu S., Chen W.H.. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019; 47:W270–W275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D.. A new genomic blueprint of the human gut microbiota. Nature. 2019; 568:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mende D.R., Letunic I., Huerta-Cepas J., Li S.S., Forslund K., Sunagawa S., Bork P.. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 2017; 45:D529–D534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao N.L., Zhang C., Zhang Z., Hu S., Lercher M.J., Zhao X.-M., Bork P., Liu Z., Chen W.-H.. MVP: a microbe–phage interaction database. Nucleic Acids Res. 2018; 46:D700–D707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W.H., Lu G., Chen X., Zhao X.M., Bork P.. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017; 45:D940–D944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are freely accessible to all academic users. This work is licensed under a Creative Commons Attribution-Non-Commercial 3.0 Unported License (CC BY-NC 3.0). In addition to various download functions on many webpages, users can download all data from the ‘Data downloads’ section of the ‘Help’ page. Programmable access through REST APIs is also supported; detailed instructions on using R, Perl and Python to access our data can be found at the ‘Programmable access’ section of the ‘Help’ page or our GitHub page: https://github.com/evolgeniusteam/GMrepoProgrammableAccess.