Abstract
CancerGeneNet (https://signor.uniroma2.it/CancerGeneNet/) is a resource that links genes that are frequently mutated in cancers to cancer phenotypes. The resource takes advantage of a curation effort aimed at embedding a large fraction of the gene products that are found altered in cancer cells into a network of causal protein relationships. Graph algorithms, in turn, allow to infer likely paths of causal interactions linking cancer associated genes to cancer phenotypes thus offering a rational framework for the design of strategies to revert disease phenotypes. CancerGeneNet bridges two interaction layers by connecting proteins whose activities are affected by cancer drivers to proteins that impact on the ‘hallmarks of cancer’. In addition, CancerGeneNet annotates curated pathways that are relevant to rationalize the pathological consequences of cancer driver mutations in selected common cancers and ‘MiniPathways’ illustrating regulatory circuits that are frequently altered in different cancers.
INTRODUCTION
The ability to sequence a whole genome in a day at a cost that compares favorably with traditional diagnostic approaches has contributed to assemble large collections of cancer genomes that are freely accessible in public repositories (1). The comparison with the genome sequences of the patient non-transformed tissues has, in turn, allowed to define a cancer genomic landscape by identifying genes that are frequently mutated in a specific tumor type (2). Using this information, the COSMIC Cancer Gene Census (CGC) (3) has curated lists of genes that are causally implicated in the onset and progression of different cancers. Additional independent curation efforts such as for instance the ones by the Comparative Toxicogenomics Database (4), by IntOGen (5), by DriverDBv2 (6) or by the UniProtKB consortium (7) and others have aimed at similar goals, albeit with different strategies and using different criteria for filtering experimental support for gene-cancer associations. These sparse undertakings have been collated in a single resource by the DisGeNET project (8) that lists 4145 genes associated to 738 neoplastic diseases. However, although these studies have, in some cases, led to the fine-mapping of underlying causal variations and generated some mechanistic insight, our understanding of the connections between genetic variations and cancer onset and progression remains limited. A seminal paper by Hanahan and Weinberg has provided a widely used conceptual framework (Hallmarks of cancer) to rationalize the complexity of neoplastic diseases (9). The challenge is now that of establishing functional paths between the genes that are frequently mutated in cancers and the ‘Hallmark phenotypes’. This information may assist in the design of therapeutic strategies that revert the cancer phenotypes.
The CGC group has started a curation effort and screened the literature to recover experimental information relating cancer genes to cancer phenotypes (3).
The database CancerGeneNet (https://signor.uniroma2.it/CancerGeneNet/) was conceived to address a similar challenge by using a different unbiased approach aimed at precisely identifying chains of causal relationships explaining the impact of cancer-gene mutations on cancer phenotypes. The resource is based on the annotation of experimental information that permits to embed the cancer genes into the cell network of causal protein relationships. It is based on three curation efforts and a set of graph algorithms to connect gene lists and to find shortest paths between any two nodes on the network.
CancerGeneNet takes advantage of the global causal network curated in SIGNOR (10), a database of curated logic relationships which offers a framework for investigating the information flow between different signaling proteins thereby linking genes to phenotypes.
MATERIALS AND METHODS
Cancer-gene lists
We have taken advantage of two lists of cancer related genes (Supplementary Table S2). The first list is an expert curated list and was downloaded from the CGC web site (v89) (3). The census associates 710 genes to 354 tumor types. The second list was obtained from the file ‘Curated gene-disease associations’ (DisGeNET v 6.0) (8). We selected the records with disease type ‘disease’ and disease semantic type ‘neoplastic process’, resulting in a list of 4145 cancer genes associated to 710 tumor types. While the CGC is a curated list, the DisGeNET resource integrates data from different resources and is much less selective in inclusion criteria. As a result, some tumors are associated to a large number of genes, up to ∼1000. The DisGeNET list, however, can be ranked according to a score that depends on the supporting evidence. In CancerGeneNet the DisGeNET cancer gene lists are truncated to the 50 genes with the highest score.
Curation rules and strategies
Information about logic relationships linking cancer genes, cancer phenotypes and cancer drugs was searched in the scientific literature by standard methods (PubMed and Google searches). The retrieved articles were filtered by expert curators and relevant information annotated in the database according to the SIGNOR curation model.
The cancer phenotypes, forming the end point of the CancerGeneNet graph are shortened versions of the ‘hallmarks of cancers’ (9). The mapping between the hallmarks of cancers and the cancer phenotypes in CancerGeneNet is reported in Supplementary Table S1. We have not considered ‘Avoiding immune destruction’ since this hallmark involves interaction between different cell types and we have added the phenotype ‘differentiation’. The hallmark ‘Dysregulating cellular energetics’ comprise a large number of very different ‘metabolic phenotypes’. Including all these phenotypes in our analysis would complicate the curation effort, increase the burden of the search algorithms and confuse the result page. We limited our effort to the phenotype ‘Glycolysis’ as the modulation of this metabolic pathway is possibly the best studied and aerobic glycolysis is the clearest readout of the Warburg effect.
Finally, we took advantage of a recent paper (11) that provides a list of FDA approved anticancer drugs and their targets to manually annotate 91 FDA drugs and 105 gene targets.
Database structure and portal implementation
The data is presented to the user through a web interface based on HTML5, integrated with JavaScript scripting to provide a more dynamic and responsive user interaction. Most data manipulation and display are handled through PHP (version 7.0.33). R scripting is used to extract further meaningful information. The network viewer is created using SPV (Signaling Pathway Visualizer, v1.0) (12).
Graph algorithms
Identification of the shortest paths between a specific gene and cancer phenotypes was programmatically implemented using the shortest path function of the R igraph (13). This function returns all shortest paths linking any two nodes in an oriented graph, such as the one in SIGNOR. To offer the user the possibility to explore paths that are different from the shortest ones, but that may be of biological relevance, we used the function, all_simple_paths, implemented in the NetworkX module (14) of the Python language. The implemented function returns all the shortest paths linking the query gene to the target phenotype and adds those paths that are one step longer. The resulting path list are ranked for relevance by using a measure of path length that takes into consideration the ‘reliability’ of each edge forming the path, as defined in SIGNOR (Licata et al. under submission to the NAR database issue). This reliability score combines four features supporting the functional relevance of the curated relation: (i) the number of references (PMID) that support the interaction in SIGNOR, (ii) the number of SIGNOR pathways that include the interaction, (iii) the number of occurrences of the relation in the Reactome interactome and (iv) the number of times the target entity is mentioned in the UNIPROT source entity page. The combined reliability score ranges from 0 to 1. We next defined node distance (d) which is linked to reliability (r) by the following simple relation d = 1 – r.
The total path length is the sum of the length of all the edges in the path. We also evaluated the effect of the protein activity on the phenotype where path containing 0 or an even number of inhibitory steps are defined as phenotype activators, otherwise are considered as phenotype inhibitors.
Network of cancer similarity
The network of cancer similarity offers an alternative access to the data in the resource. To draw the network, we have first calculated the Jaccard index by comparing the lists of cancer related genes between any pair of cancers. For the DisGeNET gene lists, we limited the calculation to the 50 genes with higher tumor-association score. Cancers with a Jaccard index above an arbitrarily chosen threshold are joined by an edge in the graph. The network is displayed in the homepage with Cytoscape.js (16), a Javascript library for graph visualization and analysis. Node (cancer) size is mapped to the number of associated genes. Edge width is related to the –log10 of the P-value calculated by the Fisher's exact test. Graphs are rendered with the Euler layout for both the CGC and DisGeNET datasets. To avoid overcrowding of the DisGeNET graph, cancers with three associated genes or less are not shown.
RESULTS
Retrieving experimental information on causal interactions
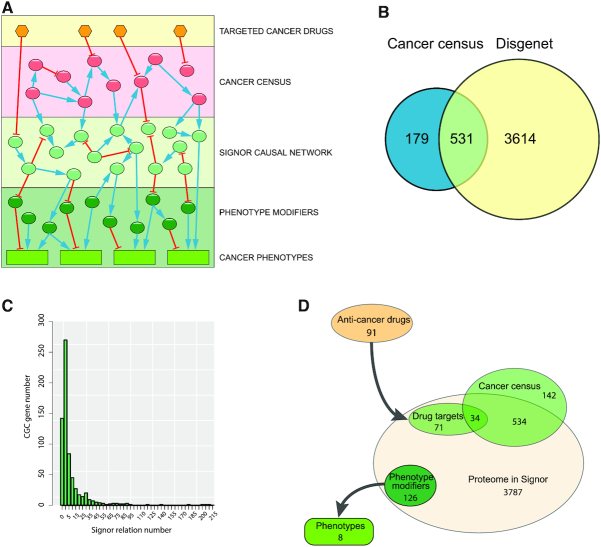
The CancerGeneNet project aims at linking genes implicated in cancer to cancer phenotypes via causal interactions between proteins (Figure 1A). Two lists of cancer related genes were derived from the CGC (3) and from DisGeNET (8) respectively (Supplementary Table S2). The two resources use different inclusion strategies and stringency criteria. As a consequence, the two lists are different in size (710 and 4145 gene products respectively) and only 531 genes are common to both lists (Figure 1B). At the beginning of this project we monitored the fraction of the CGC that has at least one interaction in SIGNOR. It turned out that ∼300 CGC genes (∼40%) were not annotated in the SIGNOR network. Thus, we started a curation campaign searching for published information on the impact that perturbation of a cancer gene had on the activity of other proteins in the cell proteome. At the end of this effort the percentage of cancer genes, which were integrated in the causal network, rose to ∼80%. The number of relationships associated to the genes in the CGC varies from zero (142 genes) to 214 (for AKT1) as shown in Figure 1C
Figure 1.
Database content. (A) schematic representation of the layered information in the CancerGeneNet database. The second layer represents the cancer gene network that was specifically curated for this project. The yellow hexagons in the top layer represent the anticancer drugs that target sensitive signaling proteins. The cancer genes impact on the causal interactome network in SIGNOR (light green circles) that directly affect cancer phenotypes (green rectangles). (B) Venn diagram of the cancer genes annotated in the Cancer Gene Census and in DisGeNET. (C) Histogram representing the distribution of the number of relationships for each cancer gene in the Cancer Gene Census. (D) The diagram illustrates the size of the protein ensembles that are (i) annotated in Signor, (ii) included in the Cancer gene Census, (iii) specific targets of anti-cancer drugs or (iv) have been annotated as phenotype modifiers.
The graph that can be extracted from the curated information in CancerGeneNet is a tripartite signed directed graph linking cancer related gene-products to cancer phenotypes and anticancer drugs (Figure 1A). The interactions annotated for this project have been selected for their relevance to the modulation of cancer pathways and have been curated according to the SIGNOR data model (10) and curation rules, following the ‘Causal Tab’ standard (17). We have first looked for experimental information on causal relationships (i.e. protein A activates protein B) between gene products listed in the CGC and proteins in the human proteome. Causal relations and their signs depend on the genetic context. All the annotations in CancerGeneNet refer to a wild type context. The effect of each activating/inactivating mutation on the information flow in a disease context needs to be evaluated after inferring the effect of the mutation on protein activity.
By this approach we have curated ∼6000 new interactions. However, 142 cancer genes (∼20%) remain uncharacterized and therefore disconnected from the global causal cell interactome (Supplementary Table S3). For most of these genes no robust information could be found about direct logic interactions underlying their link to cancer promotion.
Next, we have identified proteins that are demonstrated to affect cancer phenotypes (say p53 for ‘cell death’), either in overexpression or when inactivated (∼120 proteins). This step has a certain degree of arbitrariness as many genes have been linked to some hallmarks more or less directly. We have selected eight phenotypes that are easily implementable in our data model and can be mapped to the cancer hallmarks (Supplementary Table S1). Finally, we have embedded these two interaction layers into the global human-cell causal-interactome, as annotated in the SIGNOR database.
In a third curation undertaking we have annotated causal interactions between the anti-cancer drugs that are used in therapy and their targeted proteins (Figure 1D) (11).
In addition, cancer pathways and MiniPathways, which are frequently altered in different tumors, have been manually curated, allowing analysis of pathway cross talk and comparison of inferred cancer networks with processes that are frequently altered in cancers. We have defined for each annotated cancer pathway a set of 20–30 proteins that are often mutated or have their expression perturbed in the tumor. A cancer pathway is an ensemble of relationships that link the ‘seed’ entities. A MiniPathway is smaller network (about 10–15 entities) that describes a signal transduction module that is frequently perturbed in different types of cancers.
Database interface
The main motivation for the development of the CancerGeneNet resource was that of offering a database of causal relationships and tools that could help (i) inferring paths linking cancer associated genes to cancer related phenotypes and (ii) identifying functional correlations between genes that are found mutated in cancers by building networks of causal relationships.
The website homepage offers four entry points to search and interrogate the database.
The panel in the top left corner allows to enter any gene (Uniprot ID or gene name), or list of genes, and query the database for the shortest causal path(s) linking the gene of interest and eight phenotypes characterizing cancers. It is possible to search for path linking any gene and the eight hallmarks, separately or simultaneously. The results of the paths searching algorithm, are shown as a graph displaying the interactions connecting the query gene(s) to the phenotype(s) and as a table listing all the shortest paths, and those that are one step longer. Paths are ranked and listed in a table according to a measure of reliability as described in methods and outlined with green or red colors to identify activating and inhibitory paths respectively.
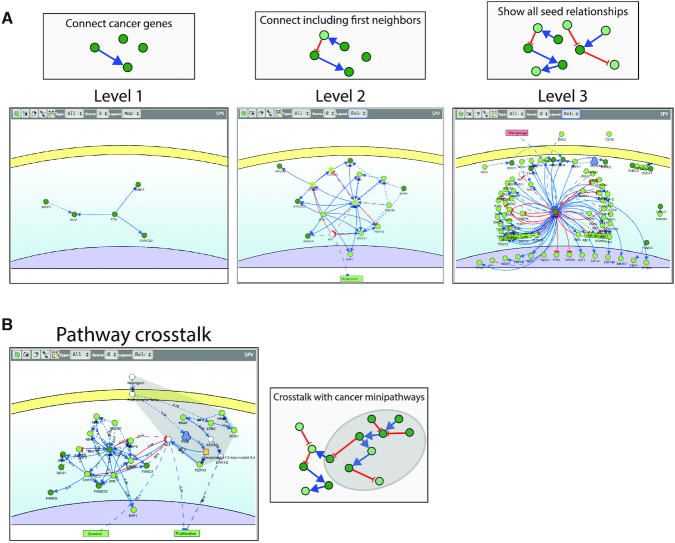
CancerGeneNet also offers a second functionality that assists in addressing the task of finding whether genes in a list, for instance a list of genes associated to a given cancer, are linked by causal relations. This can be achieved by searching a specific cancer either in a drop-down menu or in a network representing cancer similarities, as determined by the calculation of the Jaccard index for each pair of tumors in the cancer gene lists. Both entry points can be used to access either the CGC or the DisGeNET datasets. By selecting a given cancer in the drop-down menu or by clicking the ‘connect driver genes’ in the node pop up window in the similarity network, users can launch a search looking for logic relationships between the cancer associated genes. Three types of graphs are assembled and may be graphically displayed (Figure 2A). In a first display (level 1) only direct connections between the query genes are shown. A second algorithm (level 2) aims at connecting the query cancer genes by including also proteins that, by interacting with two cancer genes, allow to form a bridge between them (first neighbors). Finally, at level 3 it is possible to show all the logical interactions that are annotated to the query genes in the database. A check-box also allows to add to the network the physical interactions that can be retrieved from the mentha database (18). Once any of these graphs is displayed it is possible to edit it by adding or removing nodes or by filtering the graph according to interaction score and type. Gene products that are targeted by FDA approved anti-cancer drugs are outlined with a cyan border. By clicking the node, the list of inhibitors can be visualized via a hyperlink in the popup window.
Figure 2.
Connecting cancer associated genes. A tool based on graph algorithms offers the possibility to find functional connections in list of genes that can be user-defined or obtained from the Cancer Gene Census or DisGeNET. (A) Three types of graphs can be obtained. At level 1 only direct connections between query genes are shown. At level 2, two query genes can be linked by causal interactions with common proteins in the global causal interactome. Finally, at level 3, all the interactions of the query cancer genes are shown. (B) Once one of these graphs are shown it is possible to explore cross talks with a list of curated cancer MiniPathways.
One additional feature allows to count the number of nodes that are in common between the displayed networks and the cancer MiniPathways that are curated in the database and to calculate the statistical significance of the overlap using the same procedure described for the DISNOR database (19). This offers the possibility to infer cancer pathways that are likely perturbed in the tumor specific network that is under scrutiny. Finally, the cross-talk between the displayed graph and cancer MiniPathways can be shown graphically (Figure 2B). The annotated information that is used to generate the graphs can be downloaded in a tab delimited format for local use.
The approach described above is not limited to gene lists annotated in the CGC and in DisGeNET, as any user defined gene list can be entered in the ‘connect proteins’ frame to query the database and to draw networks connecting the genes in the list (Figure 2A).
DISCUSSION
CangerGeneNet is offered to the users to explore mechanisms underlying the cancer-driver nature of selected gene alterations. This is unique in the cancer resource panorama and useful as it can stimulate new hypothesis to be tested experimentally. Since the CancerGeneNet network is directed and signed its analysis also permits to infer whether any chain of causal relationships has the potential to activate or to inactivate the end node of the chain. One may find surprising that the answer to this type of queries is not univocal and depends on the specific path that is considered. In other words, a gene may turn out to act as an onco-gene or an onco-suppressor depending on the specific path that is taken to join the gene to the cancer phenotype. There might be trivial explanations of these findings, some of which are mentioned below. However, we also need to consider that a gene/protein-function depends on the context and reports of genes that may be classified as oncogenes in one context and onco-suppressors in another are becoming more and more common (20–24).
We have made our best effort to ensure high coverage and accuracy in our annotation endeavor. However, we are aware, that important logic relationships might have escaped our attention while some others might have been annotated inaccurately. This can be improved with a continuous curation and revision effort. One additional important caveat is that our ‘naïve approach’ assumes that all proteins in the proteome are expressed in all cells and, as such, it may suggest paths linking proteins that are not expressed in the specific tumor. Proteome expression databases such as the Human Protein Atlas (25) can help filtering paths involving proteins that are not expressed in the biological system of interest.
Finally, our model might not be completely adequate to represent the intricacy of cell signaling. As an example, in our model proteins have ‘one’ function that can be stimulated or inhibited. The model does not allow to consider that a protein has two different functions that can be modulated independently. For instance, a kinase that has two targets where only one of the two activities is modulated by an inhibitor.
The application of graph algorithms to the network underlying CancerGeneNet permits to achieve tasks that are relevant for the inference of the phenotypic impact of altering the expression or activity of any given gene product. In the database front-hand, we have implemented a number of graph algorithms that manipulate the signed directed ‘cancer graph’ to help accomplishing the following tasks: (i) determine the paths linking any given gene to the cancer hallmark-phenotypes, (ii) given a list of apparently unlinked tumor associated genes, as in the CGC or in DisGeNET, connect them via causal relationships, (iii) given any user defined gene list, connect them via causal relationships and link them to the hallmarks of cancer.
The value of CancerGeneNet lies on the unbiased coverage of the cell causal network linking cancer related genes without much prejudice on the causal relationships that are relevant. As such the resource is able to provide novel hypotheses. However, it should be kept in mind that biology is more complex than graph theory and that the resource outputs should be taken as suggestions and filtered critically by expert cancer biologists.
Finally, an unexpected outcome of our curation effort is the finding that for up to 20% of the cancer genes we know little about the molecular mechanisms underlying their tumorigenic function.
Many resources focus on cancer signaling pathways (15,26,27). To our knowledge, however, the manually annotated information-content and the tasks that can be accomplished with CancerGeneNet cannot be matched by any other public resource. CancerGeneNet offers a unique tool that permits to bridge the gap between genomic data and cancer phenotypes.
DATA AVAILABILITY
CancerGeneNet is available at https://signor.uniroma2.it/CancerGeneNet/. The underlying curated information can be downloaded from https://signor.uniroma2.it/.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (AIRC) Project IG 2017 [20322 to G.C.]; EMBL-EBI core funding and the Italian Association for Cancer Research (triennial fellowship Starwood Hotels & Resorts) [18137 to L.P.]. Funding for open access charge: Italian Association for Cancer Research [20322].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hutter C., Zenklusen J.C.. The cancer genome Atlas: creating lasting value beyond its data. Cell. 2018; 173:283–285. [DOI] [PubMed] [Google Scholar]
- 2. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. et al.. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A.. The COSMIC cancer gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018; 18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J.. The comparative toxicogenomics Database: update 2019. Nucleic Acids Res. 2019; 47:D948–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Tamborero D., Schroeder M.P., Jene-Sanz A., Santos A., Lopez-Bigas N.. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods. 2013; 10:1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chung I.-F., Chen C.-Y., Su S.-C., Li C.-Y., Wu K.-J., Wang H.-W., Cheng W.-C.. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res. 2016; 44:D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. UniProt Consortium, T UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018; 46:2699–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., García-García J., Sanz F., Furlong L.I. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanahan D., Weinberg R.A.. Hallmarks of Cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 10. Perfetto L., Briganti L., Calderone A., Perpetuini A.C., Iannuccelli M., Langone F., Licata L., Marinkovic M., Mattioni A., Pavlidou T. et al.. SIGNOR: a database of causal relationships between biological entities. Nucleic Acids Res. 2016; 44:D548–D554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun J., Wei Q., Zhou Y., Wang J., Liu Q., Xu H.. A systematic analysis of FDA-approved anticancer drugs. BMC Syst. Biol. 2017; 11:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calderone A., Cesareni G.. SPV: a JavaScript Signaling Pathway Visualizer. Bioinformatics. 2018; 34:2684–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Csárdi G., N.T.. The igraph software package for complex network research. Inter. J. Comp. Syst. 2006; 1695:1–9. [Google Scholar]
- 14. Hagberg A.A., Schult D.A., Swart P.J.. Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science Conference. 2008; SciPy 2008; 11–16. [Google Scholar]
- 15. Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. et al.. The reactome pathway knowledgebase. Nucleic Acids Res. 2018; 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2015; 32:btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perfetto L., Acencio M.L., Bradley G., Cesareni G., Del Toro N., Fazekas D., Hermjakob H., Korcsmaros T., Kuiper M., Lægreid A. et al.. CausalTAB:the PSI-MITAB2.8updated format for signalling data representation and dissemination. Bioinformatics. 2019; 35:3779–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calderone A., Castagnoli L., Cesareni G.. mentha: a resource for browsing integrated protein-interaction networks. Nat. Methods. 2013; 10:690–691. [DOI] [PubMed] [Google Scholar]
- 19. Lo Surdo P., Calderone A., Iannuccelli M., Licata L., Peluso D., Castagnoli L., Cesareni G., Perfetto L.. DISNOR: a disease network open resource. Nucleic Acids Res. 2018; 46:D527–D534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams T.M., Lisanti M.P.. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am. J. Physiol. Physiol. 2005; 288:C494–C506. [DOI] [PubMed] [Google Scholar]
- 21. Eskelinen E.-L. The dual role of autophagy in cancer. Curr. Opin. Pharmacol. 2011; 11:294–300. [DOI] [PubMed] [Google Scholar]
- 22. Bard-Chapeau E.A., Li S., Ding J., Zhang S.S., Zhu H.H., Princen F., Fang D.D., Han T., Bailly-Maitre B., Poli V. et al.. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 2011; 19:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H., Wu Y., Zhu S., Liang W., Wang Z., Wang Y., Lv T., Yao Y., Yuan D., Song Y.. PTP1B promotes cell proliferation and metastasis through activating src and ERK1/2 in non-small cell lung cancer. Cancer Lett. 2015; 359:218–225. [DOI] [PubMed] [Google Scholar]
- 24. Neel J.-C., Humbert L., Lebrun J.-J.. The dual role of TGFβ in human Cancer: From tumor suppression to cancer metastasis. ISRN Mol. Biol. 2012; 2012:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. et al.. A pathology atlas of the human cancer transcriptome. Science. 2017; 357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 26. Kuperstein I., Bonnet E., Nguyen H.-A., Cohen D., Viara E., Grieco L., Fourquet S., Calzone L., Russo C., Kondratova M. et al.. Atlas of Cancer Signalling Network: a systems biology resource for integrative analysis of cancer data with Google Maps. Oncogenesis. 2015; 4:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CancerGeneNet is available at https://signor.uniroma2.it/CancerGeneNet/. The underlying curated information can be downloaded from https://signor.uniroma2.it/.