Abstract
The National Center for Biotechnology Information (NCBI) provides a large suite of online resources for biological information and data, including the GenBank® nucleic acid sequence database and the PubMed database of citations and abstracts published in life science journals. The Entrez system provides search and retrieval operations for most of these data from 35 distinct databases. The E-utilities serve as the programming interface for the Entrez system. Custom implementations of the BLAST program provide sequence-based searching of many specialized datasets. New resources released in the past year include a new PubMed interface, a sequence database search and a gene orthologs page. Additional resources that were updated in the past year include PMC, Bookshelf, My Bibliography, Assembly, RefSeq, viral genomes, the prokaryotic genome annotation pipeline, Genome Workbench, dbSNP, BLAST, Primer-BLAST, IgBLAST and PubChem. All of these resources can be accessed through the NCBI home page at www.ncbi.nlm.nih.gov.
INTRODUCTION
NCBI overview
The National Center for Biotechnology Information (NCBI), a center within the National Library of Medicine at the National Institutes of Health, was created in 1988 to develop information systems for molecular biology. Since that time the amount and variety of data that NCBI maintains has expanded enormously and can be generally grouped into six categories: Literature, Health, Genomes, Genes, Proteins and Chemicals (Table 1). NCBI provides facilities for submitting and downloading data, analysis and visualization software, educational events and materials about NCBI products, and software and services to support an expanding developer community. These services, along with all other data resources, are available through the NCBI home page at www.ncbi.nlm.nih.gov/. In most cases, the data underlying these resources and executables for the software described are available for download at ftp.ncbi.nlm.nih.gov.
Table 1.
The Entrez Databases (as of 4 September 2019)
| Database | Records | Description |
|---|---|---|
| Literature | www.ncbi.nlm.nih.gov/home/literature/ | |
| PubMed | 30 090 705 | scientific and medical abstracts/citations |
| PubMed Central | 5 706 133 | full-text journal articles |
| NLM Catalog | 1 604 689 | index of NLM collections |
| Books | 758 285 | books and reports |
| MeSH | 279 004 | ontology used for PubMed indexing |
| Health | www.ncbi.nlm.nih.gov/home/health/ | |
| ClinVar | 561 351 | human variations of clinical significance |
| dbGaP | 1 357 | genotype/phenotype interaction studies |
| MedGen | 327 671 | medical genetics literature and links |
| GTR | 60 029 | genetic testing registry |
| Genomes | www.ncbi.nlm.nih.gov/home/genomes/ | |
| SNP | 686 600 501 | short genetic variations |
| Nucleotide | 409 361 023 | DNA and RNA sequences |
| Probe | 32 407 891 | sequence-based probes and primers |
| BioSample | 11 474 586 | descriptions of biological source materials |
| SRA | 8 945 479 | high-throughput DNA and RNA sequence read archive |
| dbVar | 5 954 211 | genome structural variation studies |
| Taxonomy | 2 254 489 | taxonomic classification and nomenclature catalog |
| BioProject | 374 697 | biological projects providing data to NCBI |
| Assembly | 462 179 | genome assembly information |
| Genome | 47 065 | genome sequencing projects by organism |
| BioCollections | 8 092 | museum, herbaria, and other biorepository collections |
| Genes | www.ncbi.nlm.nih.gov/home/genes/ | |
| GEO Profiles | 128 414 055 | gene expression and molecular abundance profiles |
| Gene | 25 258 008 | collected information about gene loci |
| GEO DataSets | 3 345 732 | functional genomics studies |
| PopSet | 331 479 | sequence sets from phylogenetic and population studies |
| HomoloGene | 141 268 | homologous gene sets for selected organisms |
| Proteins | www.ncbi.nlm.nih.gov/home/proteins/ | |
| Protein | 771 516 322 | protein sequences |
| Identical Protein Groups | 249 297 183 | protein sequences grouped by identity |
| Protein Clusters | 1 137 329 | sequence similarity-based protein clusters |
| Sparcle | 173 523 | conserved domain architectures |
| Structure | 154 783 | experimentally-determined biomolecular structures |
| Conserved Domains | 57 242 | conserved protein domains |
| Chemicals | www.ncbi.nlm.nih.gov/home/chemicals/ | |
| PubChem Substance | 245 012 057 | deposited substance and chemical information |
| PubChem Compound | 96 257 804 | chemical information with structures, information and links |
| PubChem BioAssay | 1 067 621 | bioactivity screening studies |
| BioSystems | 983 968 | molecular pathways with links to genes, proteins and chemicals |
This article provides a brief overview of the NCBI Entrez system of databases, followed by a summary of resources that were either introduced or significantly updated in the past year. More complete discussions of NCBI resources can be found on the home pages of individual databases, on the NCBI Learn page (www.ncbi.nlm.nih.gov/learn/) or in the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/).
The Entrez system
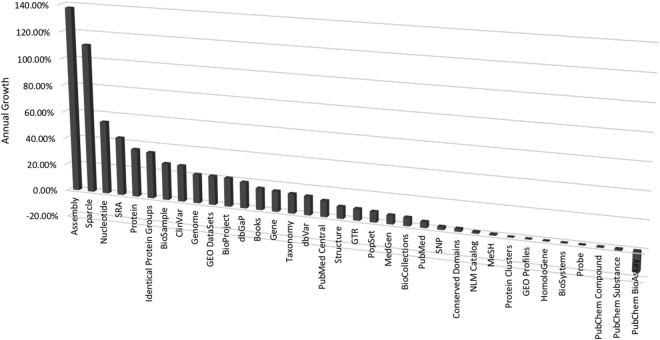
Entrez (1) is an integrated database retrieval system that provides access to a diverse set of 35 databases that together contain 2.7 billion records (Table 1 and Figure 1). Links to the web portal for each of these databases are provided on the Entrez global search page (www.ncbi.nlm.nih.gov/search/). Entrez supports text searching using simple Boolean queries, downloading of data in various formats and linking records between databases based on asserted relationships. The LinkOut service expands the range of links to include external resources, such as organism-specific genome databases. The records retrieved in Entrez can be displayed in many formats and downloaded singly or in batches. An Application Programming Interface for Entrez functions (the E-utilities) is available, and detailed documentation is provided at eutils.ncbi.nlm.nih.gov.
Figure 1.
Annual growth rates of the number of records in each Entrez database as of 4 September 2019.
Data sources and collaborations
NCBI receives data from three sources: direct submissions from researchers, national and international collaborations or agreements with data providers and research consortia, and internal curation efforts. For example, NCBI manages the GenBank database (2) and participates with the EMBL-EBI European Nucleotide Archive (ENA) (3) and the DNA Data Bank of Japan (DDBJ) (4) as a partner in the International Nucleotide Sequence Database Collaboration (INSDC) (5). Details about direct submission processes are available from the NCBI Submit page (www.ncbi.nlm.nih.gov/home/submit.shtml) and from the resource home pages (e.g. the GenBank page, www.ncbi.nlm.nih.gov/genbank/). NCBI staff provide identifiers to submitters for their data usually within 2–5 business days, depending on the destination database and the complexity of the submission. More information about the various collaborations, agreements and curation efforts are also available through the home pages of the individual resources.
RECENT DEVELOPMENTS
Literature updates
PubMed and PubMed labs
An updated version of PubMed, which will eventually replace the current version, is now available on the PubMed Labs platform (www.pubmed.gov/labs). We anticipate that this new version will become the default PubMed interface in early 2020. The older version will continue to run in parallel for a period after the new site is launched. We continuously validate the new interface by prioritizing and aligning features based on user research including usability testing and feedback from users. The updated interface features a mobile-first, responsive layout that offers better support for accessing PubMed content with the increasingly popular small-screen devices such as mobile phones and tablets. The new display of search results will include snippets, highlighted text fragments from the article abstract selected based on their relatedness to the query. These snippets help users decide if an article is useful to them. Additional improvements to the interface make it easier for users to discover related content, such as similar articles, references and associated data.
In addition to the user interface, we continued to improve the PubMed search algorithm. In 2018, we introduced a new relevant search algorithm called Best Match (6), and have further improved this machine learning-based ranking model and have retrained it using more recent user clickthrough data from December 2018 through May 2019. We have also strengthened how PubMed handles synonyms. Synonyms are very important in searching, as they allow articles to be retrieved even when the author and the searcher use different terms for the same concept. We increased the PubMed Labs synonym list to nearly 130 000 using both word co-occurrence and word embedding-based similarity to identify synonym pairs. We also used highly accurate synonym pairs from MeSH and UMLS. Finally, we enhanced PubMed's Computed Authors algorithm that determines which authors with the same name are the same person and which are not. So that the results are as accurate as possible, we now incrementally update these data twice each week and perform full updates twice a year.
PubMed Central (PMC)
In November 2018, PubMed Central (PMC) began aggregating data citations, data availability statements and Supplementary Data in an Associated Data box. PMC displays this box on articles that have one or more of these features. By exposing this content, readers can more readily discover datasets to accelerate discovery and advance health. Since taking this step, PMC has seen a notable increase in the number of daily downloads of Supplemental Files.
In support of the National Library of Medicine's strategic plan (www.nlm.nih.gov/pubs/plan/lrp17/NLM_StrategicReport2017_2027.html), PMC increased efforts over the past year to ‘connect the resources of a digital research enterprise.’ PMC released guidance on the supply of peer review documents (July 2019) and clinical trial information in machine-readable formats (January 2019) and began actively working with data providers to supply these materials and linkages. These efforts aim to increase the transparency of scholarly communication and support large-scale analysis of the scientific literature.
Bookshelf
The NCBI Bookshelf provides free online access to over 7000 books and documents in life science and healthcare from over 150 content providers. In the past year, Bookshelf has simplified the process of finding major types of content—such as monographs, textbooks, systematic reviews, and statistical works, as well as prominent subjects like medical genetics and toxicology—by indexing publication (www.nlm.nih.gov/mesh/pubtypes.html) and resource types (www.ncbi.nlm.nih.gov/books/NBK45615/-search.Resource_Type_RT). Bookshelf has also enhanced resources integrated with other NCBI databases, such as the Genetics Testing Registry (GTR) and PubChem. For instance, users may now view and download PDFs for GeneReviews and LactMed summaries.
My Bibliography
My Bibliography is a component of My NCBI that allows users to create an online collection of published work. Users can either import citations directly from PubMed or add them manually using fielded templates. Accounts linked to NIH’s eRA Commons can associate citations with awards and manage compliance with the NIH Public Access Policy. My Bibliography now has an updated interface that makes it easier to manage very large bibliographies and share an author's body of work. The new version supports searching within the bibliography for keywords, author names and grant numbers to quickly filter the view to only the most relevant citations.
Genome updates
Sequence database search
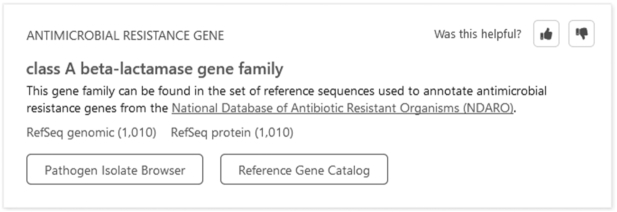
NCBI recently made available a new and improved search experience that interprets plain language for commonly performed categories of sequence searches. Results are presented in new, easy-to-interpret interfaces at the top of standard results pages and highlight the data and relevant tools likely to be of greatest interest to most users (Figure 2). The new search experience is available in several NCBI resources, including Nucleotide, Protein, Gene, Genome, Assembly and the ‘All Databases’ search page. Users receive a consistent result from this new service regardless of the database in which they initiate their search. As-you-type suggestions further facilitate these searches and reflect a subset of the most popular NCBI queries associated with the entered text. Use of these resource-specific suggestions reduces typographical errors that can derail searches and facilitates the entry of hard-to-spell organism names. Selection of a result from the drop-down menu guarantees a result from NCBI’s new search experience; queries that do not use the drop-down menu are processed by the standard Entrez system.
Figure 2.
Featured result box that appears above standard sequence search results in response to the query class A beta lactamase.
The new search functionality addresses well-defined queries that often failed to return results previously, such as organism-gene (e.g. human BRCA1) or organism-assembly (e.g. dog reference genome), and makes it easier to identify useful exemplars from queries that return large numbers of results (e.g. Escherichia coli recA). It offers refined handling of searches for genes in a particular organism, regardless of whether the gene is annotated at the species or subspecies level. The featured result highlights the gene annotated on the organism's reference assembly if available. Search results for a virus, such as HIV-1, include an interactive graphical representation of the viral genome that provides a contextual view of the annotated viral proteins. In turn, the graphics allow access to sequences, publications and analysis tools for the selected protein. The new search also offers greater access to antimicrobial resistance (AMR) protein information at NCBI. Supported queries include AMR alleles, genes or proteins in any of the aforementioned supported databases. In addition, the new service recognizes queries for families (e.g. class A beta-lactamase) of AMR genes and retrieves the best representative DNA sequence referenced from the National Database of Antibiotic Resistant Organisms (NDARO, www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/). It additionally facilitates the identification of curated gene sets included in the Reference Sequence (RefSeq) Targeted Loci Project. This project includes genes that are useful for phylogenetic and evolutionary analysis: ribosomal RNA genes in bacteria, archaea, and fungi, and internal transcribed spacer regions in fungi and oomycetes. In addition, NCBI has enhanced the ClinVar search experience to recognize more genetic variation expressions and thereby reduce the incidence of false negative search results. Details of the ClinVar search updates are provided in the companion article in this issue.
Searching homologous genes
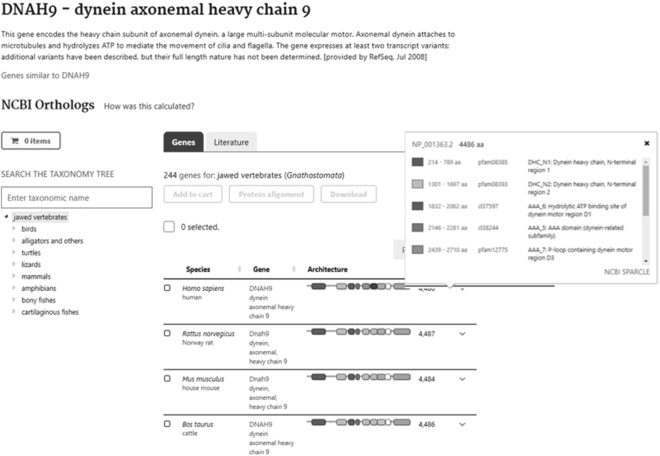
NCBI recently added a new way for users to find evolutionarily related genes within and across organisms represented in the NCBI RefSeq dataset. The goal of this new service is to facilitate comparative genomic research by allowing users to easily access sequence data as well as visualization and analysis tools for homologous gene sets from the increasing number of annotated eukaryotic reference genomes. These gene sets can be found by entering a gene symbol combined with a taxonomic group (e.g. mammals DNAH9) or by selecting the ‘orthologs’ option from the suggest menu (e.g. DNAH9 orthologs) in the search box within NCBI sequence databases (Nucleotide, Protein, Gene, Assembly, Genome) or the ‘All Databases’ search page (Figure 3). Additionally, vertebrate ortholog sets can be accessed through the new search interface for organism-specific genes. The search result provides links to two types of related genes sets. The first is a set of vertebrate orthologous genes calculated by NCBI Gene based on a combination of protein sequence similarity, local synteny information, and manual assertion (www.ncbi.nlm.nih.gov/kis/info/how-are-orthologs-calculated/). The second link provides a set of genes that share protein architecture with the orthologous gene set and includes genes from all metazoans as well as from selected plant, fungal and protist species (www.ncbi.nlm.nih.gov/kis/info/how-are-similar-genes-calculated/). Both pages display genes in rows that can be expanded to reveal more detailed information on transcripts and proteins, as well as links to the NCBI genome browser and InterPro protein families. Gene lists can be searched and filtered based on categories defined in a taxonomy tree. A cart feature allows users to download or align customized datasets.
Figure 3.
Gene ortholog page for DNAH9. The hover box displayed shows conserved domains within the human protein NP_001363.2.
Prokaryotic Genome Annotation Pipeline (PGAP)
The Prokaryotic Genome Annotation Pipeline (PGAP) (7,8) is now publicly available from GitHub to users interested in predicting genes on public or privately owned bacterial and archaeal genomes (github.com/ncbi/pgap). This pipeline is a re-implementation in the Common Workflow Language (CWL) of the version of PGAP currently used by NCBI to annotate RefSeq and some GenBank genomes. Users can execute PGAP on individual computers, a compute farm, or in the cloud. PGAP is packaged in a Docker container with the necessary binaries and cwltool, the CWL reference implementation. Datasets curated at NCBI for prokaryotic annotation, such as proteins representing homology clusters, Hidden Markov Models (HMMs) and other annotation rules are also distributed with the tool. Provided a multiFASTA file and a minimal set of metadata (the species that the assembly represents in particular), PGAP will produce an annotation that conforms to what the internal NCBI pipeline would generate and that can be readily submitted to GenBank through the genome submission portal.
RefSeq
NCBI’s Reference Sequence (RefSeq) project (9) celebrated its 20th anniversary in 2019. The dataset now includes over 206 million sequences from over 93 000 taxa, reflecting 26% and 15% annual growth, respectively. The eukaryote dataset incorporates genomic data from over 960 species. Of these, NCBI has annotated 562 genomes with the NCBI Eukaryotic Genome Annotation Pipeline, including all vertebrates and most other multicellular eukaryotes in RefSeq. This common processing pipeline provides consistent and high-quality annotation to aid cross-species studies, with over 90% of annotations incorporating evidence from RNA-seq data. The RefSeq annotation for the human GRCh38 reference assembly is now being updated every 2–3 months to more rapidly incorporate ongoing improvements based on evidence from PacBio and nanopore RNA sequencing, CAGE, proteomics and additional data.
The prokaryote dataset (7) incorporates data from over 165 000 assemblies representing over 11 000 species, primarily annotated with NCBI’s PGAP. Microbial RefSeq protein records (with the WP_ prefix) now include the curated evidence used to assign protein function and, where possible, gene symbols, publications and Enzyme Commission numbers. The set of evidence used for naming is a hierarchical collection of curated HMM-based and BLAST-based protein families as well as conserved domain architectures (7). Hyperlinks in the Evidence Accession field of the Evidence-For-Name-Assignment comment block in the protein records lead to web pages with more information about the naming evidence, including the thresholds used for defining a match between the evidence and the protein, along with the list of all the prokaryotic proteins that match the evidence and define the protein family (go.usa.gov/xVQu6). As of August 2019, 72% of prokaryotic proteins in RefSeq (90 out of 125 million) were named based on such evidence.
Assembly
Bacterial genome sequencing has become sufficiently inexpensive that it is being used to explore variation within a species and to monitor the spread of pathogens. NCBI has made several changes to accommodate the resulting explosion in the number of genome assemblies for a small number of pathogenic bacteria (over 125 000 genome assemblies from Salmonella enterica alone have been added this year, plus another 58 000 genome assemblies from only three other species). Genome assemblies from projects that generate more than 100 assemblies for a species are tagged as ‘derived from surveillance project’. Such assemblies are filtered out of the results of searches in the Assembly database (www.ncbi.nlm.nih.gov/assembly/) (10) by default; however, an option is provided to clear the filter and view all assemblies matching the query. Users can view ‘derived from surveillance project’ assemblies in the Assembly resource and can download their genome data from the genomes File Transfer Protocol (FTP) site, even though such assemblies are excluded from RefSeq. Also, genomes FTP directories in the taxonomic hierarchy no longer list each assembly when a species has more than 1000 assemblies; however, the assembly_summary.txt file in the species FTP directory can be used to find assemblies of interest along with their FTP path.
NCBI has also made several improvements that facilitate finding and downloading genome data sets of interest. Notably, links have been added between members of a pair of genome assemblies derived from the same diploid individual. New file types have been added to the genomes FTP site, including a file with annotation in Gene Transfer Format (GTF), a ‘genomic gaps’ file providing the locations and types of gaps in a genome assembly, and many new files generated by the NCBI Eukaryotic Genome Annotation Pipeline. These files provide transcript-to-genome alignments for RefSeq sequences in Binary Alignment Map (BAM) format, alignments of same- and cross-species transcripts used as evidence, annotations of Gnomon models, and comparisons to the previous annotation. In addition, a new download API allows users to retrieve genome assembly data files programmatically (api.ncbi.nlm.nih.gov/genome/v0/download). NCBI also now provides an API that can be used to check the size of a genome assembly prior to submission and avoid a potential validation issue that could delay processing the submission (api.ncbi.nlm.nih.gov/genome/v0/expected_genome_size).
Viral genomes
NCBI continues to make it easier to find and use viral genome sequence data. Because viral sequences are submitted over time by multiple groups, they often lack standardized isolate attributes. This creates inconsistencies across otherwise comparable data and necessitates prior knowledge of possible synonyms to use the data effectively. For example, accurate search strategies for viruses isolated from human hosts would require use of several terms including ‘Homo sapiens,’ ‘human,’ ‘male,’ and ‘patient.’ Building upon our experiences with the Virus Variation and Influenza resources (11), we have been experimenting with computational approaches to validate taxonomy and normalize host and isolation source data from viral genomes. These data are available on the NCBI Labs (12) testing platform through an experimental interface built to support both sequence-based and attribute-based searches (www.ncbi.nlm.nih.gov/labs/virus/vssi/). This resource includes data from our Viral Genomes Resource (13) and is available now for use by the public. Collected user feedback will be used to inform future development.
Genome Workbench
NCBI’s Genome Workbench is a desktop GUI software package designed to manipulate and visualize complex molecular biology data, such as sequences, annotation and expression, variation and alignments. In addition to supporting popular bioinformatics data formats (FASTA, GFF3, VCF and BAM), Genome Workbench can also connect to NCBI data sources such as GenBank and RefSeq to retrieve content. Genome Workbench is also fully compatible with user-supplied data not found in public databases, and during processing such data remains within the user's trusted local environment. Genome Workbench makes biological content visible in graphical displays that users can export as high-quality images in PDF and SVG format, making them suitable for use in publications and posters.
Genome Workbench offers seamless integration with NCBI-provided and other popular bioinformatics analysis tools. These include alignment creation algorithms (BLAST) and comparative genomics multiple alignment tools (such as Clustal (14), KAlign (15) and MAFFT (16)) that enable users to build comparative alignments and reconstruct phylogenetic trees. All tools and views run in a fully interactive framework that does not require engagement with heavy duty bioinformatics pipelines or programming experience.
Released in July 2019, Genome Workbench v3.0.0 also offers tools that enable users to prepare genome data for submission to NCBI. Submitters can import genome sequence data from FASTA files and annotations from GFF3 and GTF files and then edit the data in a convenient GenBank Flat File view to prepare the submission. The new submission editing package also runs validation algorithms to help submitters interactively find and fix errors before sending the data to NCBI, supporting a smoother submission experience. The enhanced editing and data exploration capabilities that Genome Workbench now offers extend beyond what was possible in Sequin. Genome Workbench is a cross platform package that works on Windows, MacOS and variants of Linux. Version 3.0.0 is also compatible with the popular Google and Amazon cloud environments.
dbSNP
The Database of Single Nucleotide Polymorphisms (dbSNP) is a repository of human genomic variations, including both common and rare single-nucleotide variations and other small-scale variations, along with frequency data (17). In 2019 dbSNP celebrated its 20th anniversary hallmarked by 2 billion submitted SNP (SS) records and new improvements. dbSNP released new products that provide more precise Reference SNP (RS) clustering and that employ NCBI remapping, the new SPDI variant notation (18), and the Variant Overprecision Correction Algorithm (VOCA) (https://www.ncbi.nlm.nih.gov/variation/notation/). Recent releases based on these new build systems were Build 152 (December 2018) containing 683 million RS records and dbSNP Build 153 (August 2019) containing 695 million RS records, including 552 million with population frequency data. These releases also included updates to RefSNP report pages, FTP downloads, and the E-utilities API (https://www.ncbi.nlm.nih.gov/snp/docs/entrez/refsnp_change/).
BLAST updates
BLAST and Docker
NCBI has made the BLAST+ command-line tools (19) available as part of a Docker container. Wrapping BLAST+ in a container makes it easier to both install and maintain these programs. We have tested the Docker version of BLAST+ extensively on Google Cloud Platform (GCP) and have also staged some of the most popular BLAST databases in a Google bucket. This solution is convenient for users whose compute needs come in bursts, as they can simply start up many machines at a cloud provider in order to finish the task quickly. The Docker version of BLAST+ should run on any machine that has Docker installed. For more information on this resource, see https://github.com/ncbi/blast_plus_docs.
Web BLAST
In 2019 NCBI released an update to the default web BLAST report, which now has prominent controls allowing users to filter results by organism, percent identity, and expect value. The Descriptions, Graphic Summary, Alignments and Taxonomy reports are now presented in four tabs, allowing users to easily switch between these views.
Primer-BLAST
The Primer-BLAST (20) web page now allows users to force Primer-BLAST to ignore certain off-target matches. For example, it may be useful to ignore tissue-specific splice variants and predicted sequences. This feature can give Primer-BLAST more flexibility in the primers it designs, thereby providing users with better results.
IgBLAST
IgBLAST (21) can now detect Ig rearrangements with ultra-long D and N regions such as those found in human anti-HIV antibodies. We have improved the algorithm to determine the V(D)J rearrangement frame as well as the CDR3 end for cases that have base insertions or deletions near the rearrangement junction or CDR3 end boundary. In addition, IgBLAST now uses standard gene locus names such as IGH and TRB instead of traditional names like VH and VB.
Chemical updates
PubChem (22–24) (pubchem.ncbi.nlm.nih.gov) is a public chemical data repository at NCBI. In the past year, PubChem integrated chemical information from more than 70 new data sources. Notably, thanks to a data contribution by the publisher Thieme Chemistry, PubChem added more than 1.2 million links between nearly 700 000 chemicals and approximately 700 000 scientific articles with a focus on synthetic organic chemistry and pharmaceutical substances (go.usa.gov/xEDCA). These links dramatically expand the findability, accessibility, interoperability and reusability (FAIR) of synthesis-related chemical information.
In 2019 PubChem released a new web interface, including a redesigned home page and record summary pages that provide easier and faster access to chemical information (go.usa.gov/xEM4Y; go.usa.gov/xEuP7). As part of this effort, PubChem updated the data model for the PUG-View interface (25), which serves information needed to render interactive web pages and provides programmatic access to chemical annotation content in PubChem (go.usa.gov/xEHXj). The updated home page includes a new unified search interface supporting many query types, including text, chemical structure, molecular formula, and patent and article identifiers. The unified search interface allows users to simultaneously search the three primary PubChem databases (Compound, Substance and BioAssay) as well as other closely associated collections, including aspects of gene/protein targets, patent documents and literature articles. In addition, PubChem introduced the Pathway View page, which provides information about chemicals, proteins, genes and diseases involved in or associated with a given biological pathway. The Pathway View can increase a user's understanding of the biological roles of gene/protein targets and their interaction with chemicals. PubChem also released a periodic table and pages for each element containing authoritative and curated data. These pages are closely integrated with PubChem Compound summary pages and provide a RESTful, machine accessible interface to periodic table information. This was a timely addition considering 2019 was the International Year of the Periodic Table (IYPT) (go.usa.gov/xmUQ9).
FOR FURTHER INFORMATION
The resources described here include documentation, other explanatory materials and references to collaborators and data sources on their respective web sites. An alphabetical list of NCBI resources is available from a link above the category list on the left side of the NCBI home page. The NCBI Help Manual and the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/), both available as links in the common page footer, describe the principal NCBI resources in detail. The NCBI Learn page (www.ncbi.nlm.nih.gov/learn/) provides links to documentation, tutorials, webinars, courses and upcoming conference exhibits. A variety of video tutorials are available on the NCBI YouTube channel that can be accessed through links in the standard NCBI page footer. A user-support staff is available to answer questions at info@ncbi.nlm.nih.gov, and users can view support articles at support.nlm.nih.gov. Updates on NCBI resources and database enhancements are described on the NCBI Insights blog (ncbiinsights.ncbi.nlm.nih.gov), NCBI social media sites (FaceBook, Twitter and LinkedIn) and the several mailing lists and RSS feeds that provide updates on services and databases. Links to these resources are in the NCBI page footer and on NCBI Insights.
ACKNOWLEDGEMENTS
The authors would like to thank all of the NCBI staff who through their dedicated efforts continue to allow NCBI to provide our full collection of services to the community. In particular, the authors would like to thank these individuals for their contributions to the resources described in this work: Ray Anderson III, Alex Astashyn, Azat Badretdin, Rostyslav Bryzgunov, Michael Cervantes, Jessica Chan, Vyacheslav Chetvernin, Jinna Choi, Marie Collins, George Coulouris, Mike DiCuccio, Olga Ermolaeva, Bob Falk, Asta Gindulyte, Dan Haft, Kurtis Haro, Vichet Hem, Wratko Hlavina, Sharmin Hussain, Michael Kholodov, George Khoroshavtsev, Andrew Kim, Evgeny Kireev, Vamsi Kodali, Vladimir Korobtchenko, Sergey Koshelkov, Alex Kotliarov, Chris Lanczycki, Martin Latterner, Carl Leubsdorf, Wenjun Li, Hanguan Liu, Bonnie Maidak, Aron Marchler-Bauer, Patrick Masterson, Peter Meric, Vadim Miller, Eyal Mozes, Kathleen O’Neil, Maxim Osipov, Sergey Petrunin, Ievgeniia Radetska, Greg Schuler, Douglas Slotta, Robert Smith, Guangfeng Song, Grisha Starchenko, Craig Wallin, Mingzhang Yang and Xuan Zhang.
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A.. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996; 266:141–162. [DOI] [PubMed] [Google Scholar]
- 2. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2019; 47:D94–D99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harrison P.W., Alako B., Amid C., Cerdeno-Tarraga A., Cleland I., Holt S., Hussein A., Jayathilaka S., Kay S., Keane T. et al.. The European Nucleotide Archive in 2018. Nucleic Acids Res. 2019; 47:D84–D88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kodama Y., Mashima J., Kosuge T., Ogasawara O.. DDBJ update: the Genomic Expression Archive (GEA) for functional genomics data. Nucleic Acids Res. 2019; 47:D69–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karsch-Mizrachi I., Takagi T., Cochrane G. International Nucleotide Sequence Database Collaboration . The international nucleotide sequence database collaboration. Nucleic Acids Res. 2018; 46:D48–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fiorini N., Canese K., Starchenko G., Kireev E., Kim W., Miller V., Osipov M., Kholodov M., Ismagilov R., Mohan S. et al.. Best Match: New relevance search for PubMed. PLoS Biol. 2018; 16:e2005343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R. et al.. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018; 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J.. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016; 44:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A. et al.. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016; 44:D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schaffer A.A., Brister J.R.. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017; 45:D482–D490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sayers E.W., Agarwala R., Bolton E.E., Brister J.R., Canese K., Clark K., Connor R., Fiorini N., Funk K., Hefferon T. et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019; 47:D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brister J.R., Ako-Adjei D., Bao Y., Blinkova O.. NCBI viral genomes resource. Nucleic Acids Res. 2015; 43:D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson J.D., Higgins D.G., Gibson T.J.. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lassmann T., Sonnhammer E.L.. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005; 6:298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katoh K., Misawa K., Kuma K., Miyata T.. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002; 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smigielski E.M., Sirotkin K., Ward M., Sherry S.T.. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000; 28:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes J.B., Moyer E., Phan L., Maglott D., Kattman B.L.. SPDI: data model for variants and applications at NCBI. 2019; BiorXiv doi:23 March 2019, preprint: not peer reviwed 10.1101/537449. [DOI] [PMC free article] [PubMed]
- 19. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L.. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012; 13:134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye J., Ma N., Madden T.L., Ostell J.M.. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013; 41:W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S. Getting the most out of PubChem for virtual screening. Expert Opin. Drug Discov. 2016; 11:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al.. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim S., Thiessen P.A., Cheng T., Zhang J., Gindulyte A., Bolton E.E.. PUG-View: programmatic access to chemical annotations integrated in PubChem. J. Cheminform. 2019; 11:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]