Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that function as diverse endogenous gene regulators at the post-transcriptional level. In the past two decades, as research effort on miRNA identification, function and evolution has soared, so has the demand for miRNA databases. However, the current plant miRNA databases suffer from several typical drawbacks, including a lack of entries for many important species, uneven annotation standards across different species, abundant questionable entries, and limited annotation. To address these issues, we developed a knowledge-based database called Plant miRNA Encyclopedia (PmiREN, http://www.pmiren.com/), which was based on uniform processing of sequenced small RNA libraries using miRDeep-P2, followed by manual curation using newly updated plant miRNA identification criteria, and comprehensive annotation. PmiREN currently contains 16,422 high confidence novel miRNA loci in 88 plant species and 3,966 retrieved from miRBase. For every miRNA entry, information on precursor sequence, precursor secondary structure, expression pattern, clusters and synteny in the genome, potential targets supported by Parallel Analysis of RNA Ends (PARE) sequencing, and references is attached whenever possible. PmiREN is hierarchically accessible and has eight built-in search engines. We believe PmiREN is useful for plant miRNA cataloguing and data mining, therefore a resource for data-driven miRNA research in plants.
INTRODUCTION
In the past two decades, extensive research effort has been devoted to discover non-coding regulatory RNAs and study their functions. In particular, microRNAs (miRNAs), mainly 20–22 nucleotide (nt) small RNAs (sRNAs), are emerging as an important class of endogenous gene regulators acting at the post-transcriptional level (1–5). Mature miRNAs are processed from much longer primary transcripts, called pri-miRNAs, via stem-loop-structured intermediates called pre-miRNAs (1–4,6,7), and are usually incorporated into the RNA-induced silencing complex where they interact with their target transcripts (1–4,6,7). It is well established that miRNAs widely exist in animals and plants, and are crucial for many fundamental biological processes (2,3,8). In plants, miRNAs play significant roles in development and responses to environmental stresses (9–16).
Availability of next generation sequencing (NGS) methods prompted the development of computational tools for miRNA identification and annotation by capturing miRNA biogenesis characteristics such as stem-loop structure and preferential accumulation of sequence reads corresponding to mature and the partially complementary star miRNAs (17–20). Consequently, the numbers of annotated miRNAs have soared. There are several databases, such as miRBase (21), PMRD (plant miRNA database) (22) and PmiRKB (plant miRNA knowledgebase) (23), which are devoted to archiving miRNA information related to plants and have greatly facilitated miRNA studies. Nevertheless, they suffer from common problems such as uneven annotation standards, lack of entries for many important plant species, and insufficient information attachment (24,25).
For instance, miRBase, which is considered the gold standard of miRNA database, hosts only 7025 miRNA entries from 86 plant species, including 33 species of which no more than 20 miRNAs are annotated each. As miRBase is based on community-curation, the uneven annotation standards adopted by different researchers have resulted in a large number of questionable entries (24). In addition, 28 of the 86 species lack reference genomes, and for the remaining species with genome references, only basic information such as sequences, precursor secondary structures, genomic coordinates and support from sRNA-Seq datasets is provided for the miRNA entries. Thus, it is highly desired to establish a plant miRNA database with more uniform and strict annotation standards, greater data integrity, and more comprehensive annotations. A well-curated database for miRNAs would aid functional characterization of individual miRNAs, elucidation of the evolutionary trajectory of miRNAs across lineages, and application of miRNAs in biotechnology (25).
Towards a one stop plant miRNA database, we employed a standardized method based on our own miRDeep-P2 tool (26), the updated version of miRDeep-P (18), and newly updated plant miRNA annotation criteria (25) to systematically annotate 20,388 miRNAs in 88 phylogenetically representative plant species. These entries were comprehensively annotated and accessible in PmiREN (Plant miRNA Encyclopedia). PmiREN also provides an easy-to-use interface to browse, search, and download the data, enabled by eight built-in search engines. Taken together, PmiREN is a comprehensive functional database amenable for data mining and database-driven research and therefore a useful resource for the plant miRNA research community.
MATERIALS AND METHODS
Data sources
The current version of PmiREN (version 1.0) consists of miRNA entries from 88 plants, including 2 chlorophytes, 1 moss, 1 lycophyte, 2 gymnosperms, 1 basal angiosperm, 1 magnoliidae, 18 monocotyledons and 62 eudicotyledons (Supplementary Figure S1 and Table S1). Whole genome references and gene annotations were downloaded from Assembly of NCBI (27) (https://www.ncbi.nlm.nih.gov/assembly/) and/or Phytozome V12.1 (28) (https://phytozome.jgi.doe.gov/pz/portal.html). More details are included in Supplementary Table S1. 1537 sRNA-Seq and 116 PARE-Seq (Parallel Analysis of RNA Ends sequencing) (29,30) datasets were obtained from the NCBI GEO DataSets (31) (https://www.ncbi.nlm.nih.gov/geo/; details in Supplementary Tables S2 and S3, respectively).
Data analysis pipelines
Data pre-processing
The format of sRNA-Seq or PARE-Seq datasets from GEO DataSets is not uniform. Linux version 2.8.2 of SRA toolkit (32) (https://www.ncbi.nlm.nih.gov/sra/docs/toolkitsoft/) was employed to convert original compressed files into Fastq format. Trim Galore (version 0.5.0) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to trim adapter sequences with parameters ‘–length 18 –max_length 28 –small_rna –stringency 3′. To format sRNA-Seq datasets, trimmed Fastq files were further compacted to Fasta format with identical reads collapsed by an in-house Perl script.
Annotation of miRNAs
A standardized process centred on miRDeep-P2 (26) and newly updated plant miRNA criteria (25) was employed to identify miRNAs. For each species, whole genome reference and sRNA-Seq datasets were used as input files while the new plant miRNA criteria were added as a filter (Figure 1A). All miRNA candidates (including those having counterparts in miRBase) retrieved by this process were designated as high confidence entries with three stars. Candidates in miRBase that were not retrieved by the automated pipeline were re-examined and annotated. Candidates with low expression supported by sRNA-Seq datasets (RPM (reads per million) cut-off value < 10) and meeting the structure criteria of miRNA precursors were designated intermediate confidence with two stars. Candidates only meeting structure criteria of miRNA precursors and without any expression support by sRNA-Seq datasets were considered low confidence with one star. Other entries in miRBase were discarded.
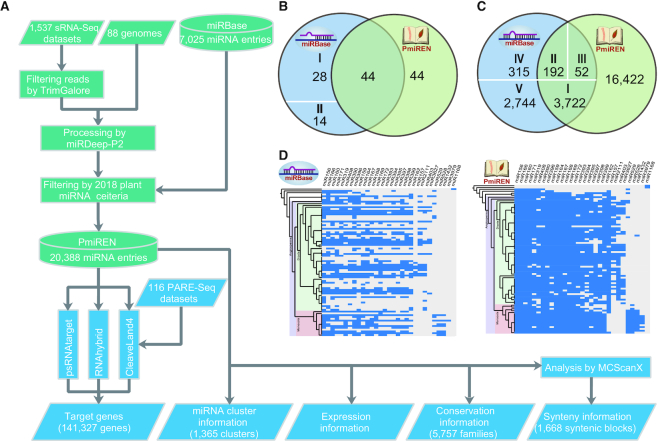
Figure 1.
A data processing workflow of PmiREN and the comparison with miRBase. (A) The data processing workflow and outcomes of PmiREN. (B) The comparison of plant species between PmiREN and miRBase (21). I, species lack of both genome references and sRNA-Seq datasets. II, species with available genome references but without available sRNA-Seq datasets. (C) The comparison of miRNA entries between PmiREN and miRBase (21). I, entries retrieved with identical annotation. II, entries corrected by name or genomic coordinates. III, entries whose mature/star miRNAs re-annotated. IV, entries unable to be mapped to genome references and discarded in PmiREN. V, entries filtered by newly updated miRNA criteria and abandoned in PmiREN. (D) The comparison of miRNA annotation integrity between miRBase (left) (21) and PmiREN (right). 30 most conserved plant miRNA families were selected, and a box is highlighted by blue if at least one member of a given miRNA family was annotated.
Target prediction and validation
Two suites of plant miRNA target prediction methods, psRNATarget (33) and RNAhybrid (34), were used to predict miRNA targets. The mature miRNA sequences and mRNA transcripts of the corresponding species were uploaded to psRNATarget webserver. Newest default parameters of Schema V2 (2017 release) (33), except that the default expectation threshold of 5 was reduced to a more restrict value of 3, was used. RNAhybrid was used to predict energetically plausible miRNA:mRNA duplexes with plant-specific constraints as previously described (34,35). A cut-off value for minimum free energy/minimum duplex energy of 0.70 was used (35). CleaveLand4 (36) was used to process PARE-Seq datasets and only category 0, 1, 2 results were kept to reduce false positives.
Expression analysis
The expression values of mature and star miRNAs were normalized by RPM as previously described (37). For each sRNA-Seq dataset, reads mapped to pri-miRNAs (mismatches not allowed) and localized in genomic positions of mature miRNAs (no more than 2 nt shift allowed) were deemed to correspond to mature miRNAs. The total numbers of these reads were counted to calculate the RPM value for the mature miRNAs. The same method was applied to calculate RPM values for the star miRNAs. In case multiple sRNA-Seq datasets from the same tissue were available, the mean RPM value was used.
Syntenic analysis
The synteny analysis of miRNAs was carried out using MCScanX (38). Firstly, GFF (general feature format) files and Fasta files including all protein-coding genes and pre-miRNAs were generated in species within the same family. Subsequently, DNA sequences of protein-coding genes and pre-miRNAs were combined and used as a query to search against itself using BLASTN (39) with E-value 1e–10. The GFF files and BLAST output files of all genes and pre-miRNAs were imported into MCScanX to scan the collinearity pairs. Circos (40) was used to display the results.
Conservation analysis
The conservation of miRNAs was assigned based on the similarity in mature miRNA sequences. For each annotated miRNA families, distribution of all members among all species was determined. The correspondence between miRNA families and species containing these families were highlighted in the species phylogenetic tree in each miRNA locus information page.
RESULTS
Data content of PmiREN
Data in the current version of PmiREN were based on 1537 sRNA-Seq datasets from 88 plant species phylogenetically ranging from chlorophytes to angiosperms (Figure 1B, Supplementary Table S1 and Figure S1). In parsing these sRNA-Seq datasets using a standard method centred on miRDeep-P2 with newly updated miRNA annotation criteria (Figure 1A) (25,26), 16,422 high confidence novel miRNA loci were discovered in comparison to the current release of miRBase (Release 22.1) (21). In PmiREN, 44 species were overlapped with miRBase. For these species, we updated miRNA annotations with up-to-date genome references and retrieved 3722 entries in miRBase that were considered as high confidence and designated with three stars (Figure 1C). Information associated with 244 entries from miRBase, either family designation, genome coordinate, or mature/star miRNA location, was corrected in PmiREN (Figure 1C). 3059 entries in miRBase were discarded for lack of supporting evidence (Figure 1C). Collectively, PmiREN currently contains 20 388 miRNAs (Figure 1C). As an example, the 30 most conserved plant miRNA families were selected for comparison between miRBase and PmiREN. It is clear that coverage of miRNA annotation was greatly increased in PmiREN (Figure 1D). In addition, PmiREN provides information on conservation of 5757 miRNA families in different species, 1668 synteny blocks, expression pattern across organ types and developmental stages for all species, and target genes (141 327 miRNA-target pairs) based on prediction and validation using PARE-Seq data whenever possible.
Features of PmiREN annotation
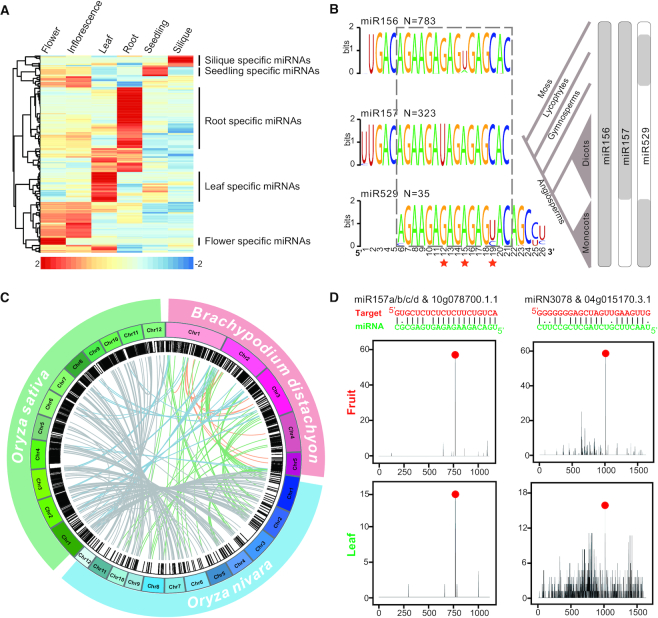
Similar to other databases, PmiREN includes all basic information of a given miRNA, including mature and star miRNA sequences, predicted secondary structure of the pre-miRNA, and genome coordinates. Moreover, miRNA annotation in PmiREN has several unique features. First, since all miRNAs were parsed from sRNA-Seq datasets, miRNA expression information was directly obtained from normalization of these sRNA-Seq datasets. For instance, in the model plant Arabidopsis thaliana, expression patterns of all miRNAs from different tissues and developmental stages can be used to cluster miRNAs that are expressed in specific tissues (Figure 2A). Second, PmiREN entries have corrected and modified information associated with many miRNAs including family assignment, the mature and/or star miRNA sequences, etc., that were erroneously annotated in miRBase. This improvement is helpful to discern the evolutionary relationship of miRNAs from different species. For example, after manual curation of the miR156/157/529 superfamily based on consensus nucleotides and phylogenetic relationship, a pattern became clear that the miR156 subfamily is present in all species while miR157 only exists in dicots and miR529 only appears in monocots (Figure 2B). Third, by incorporating results from collinear analysis, PmiREN contains collinearity information of miRNAs in the same family across the species. This information is useful to detect evolutionary changes such as gain or loss of family members among different species (Figure 2C). Forth, the prediction of miRNA targets in PmiREN was done using two different programs, psRNATarget (33) and RNAhybrid (34), and also validated by PARE-Seq datasets. Currently, PmiREN includes miRNA targets supported by PARE-Seq data in 21 species (Supplementary Table S3). Figure 2D shows two exemplar miRNA-target pairs supported by PARE-Seq data, one pair is between the conserved miR157 and its target, and the other pair between a newly discovered miRN3078 (all new entries in PmiREN are designated with ‘N’ and numbered consecutively) and its target.
Figure 2.
Unique features of PmiREN. (A) An example of miRNA expression patterns. Expression values (RPM) of all miRNAs in Arabidopsis were achieved from 39 sRNA-Seq datasets from different tissues including flower, inflorescence, leaf, root, seeding and silique. The log10 values of RPM means of each miRNA in a specific tissue was employed to make heat map analysis. The hierarchical clustering was generated by Pearson correlation coefficients of these values. Gradient colors from red to blue indicate expression values from high to low. (B) A possible evolutionary model for the dynamic divergence of miR156/157/529 superfamily in land plants. Weblogo (45) on the left side shows the sequence similarity and difference of miR156, miR157 and miR529 while the cartoon on the right side presents their existence in different evolutionary branches. Red Stars on the bottom indicate the variations of superfamily core region. (C) An examples of miRNA syntenic analyses. Syntenic miRNA pairs of 3 species in grass family including Oryza sativa (rice), Brachypodium distachyon and Oryza nivara were displayed by Circos diagram (40). (D) Examples of predicted miRNA targets and their validations by PARE-Seq datasets. Two examples including one conserved miRNA isoform (miR157a/b/c/d) and one non-conserved one (miRN3078) from tomato (Solanum lycopersicum) were displayed. The complimentary between miRNAs and targets was predicted by both RNAhybrid (34) and psRNATarget (33). Validations from two tissues (fruit and leaf) by PARE-Seq data were achieved by CleaveLand4 (36).
Functional sections of PmiREN
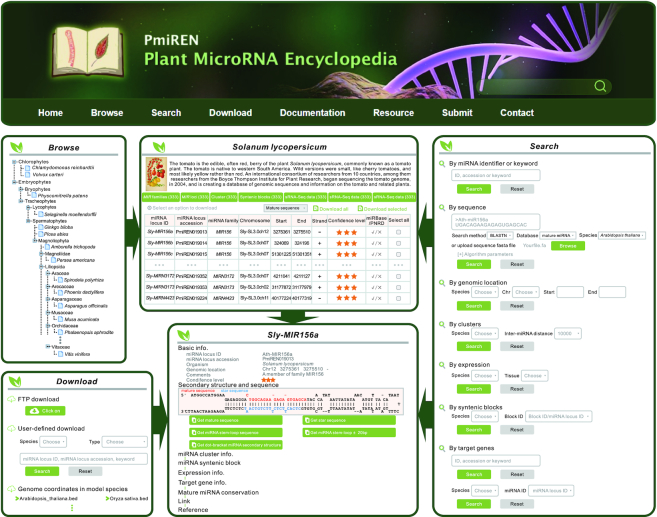
PmiREN provides convenient accesses and eight search engines (Figure 3). Users can browse all data by shortcuts and multiple layers of webpages. All data can be downloaded in bulk and customized manners (Figure 3).
Figure 3.
A schematic view of PmiREN features.
Browse
Basically, users can browse miRNA entries by species shortcuts at home page or through the Browse tag in toolbar. Browse tag will lead users to species list where the species phylogenetic information is included. When clicking the Latin name of selected species, a name card including summary of genome version, number of miRNAs, etc., is displayed. The next level of Browse is the summary of all miRNAs in a specific species where users can visit clusters, syntenic blocks, and sRNA-Seq and PARE-Seq datasets as well. Further, when clicking each miRNA entry, the link will direct users to a detailed information page, consisting of nine sections such as basic genomic information, cluster information, syntenic block, expression pattern, targets, conservation of the miRNA, links to miRBase (21) and/or PNRD (the updated version of PMRD) (22,41), and related references from Pubmed.
Search
Users can search the whole database by eight search engines. Via keyword or miRNA identifier search engine located on webpage banner, the users could search all fields of the whole database, and the result will return a summary of all searched fields and a list of hit miRNAs. The second search engine is by sequence, and a BLAST (39) web interface is deployed where parameters are adjustable. For the convenience of quickly searching miRNA information such as genome coordinates, clusters, expression values, and syntenic blocks, PmiREN provides correspondent search engines respectively. Using cluster search as an example, users can choose species and select the search region. Last, a dual layer search engine was built in to search miRNA targets by which users could directly input target genes or miRNA identifiers in a selected species.
Download
Through the Download tag located in toolbar, all basic information on miRNAs in each species could be downloaded in bulk or item by item. Users can download data by virtue of three ports on the Download page. First, by means of FTP, users can browse all available information and download their needs in bulk. Second, via the port of user-defined Download, users can choose the information they are interested in by either selecting species and data types or searching by an input keyword. Third, a quick access is provided to users so that the genome coordinates of miRNAs in model plants could be handily downloaded.
Documentation
The page of Documentation includes six sections where users can obtain almost all information on PmiREN. Section I is a brief introduction of PmiREN whereas Section II embraces information where we fetched and processed all meta-datasets. Section III is Frequently Asked Questions (FAQ), which serves as a detailed manual of PmiREN, and many shortcuts or useful tips are included as well. Section IV-VI list such information as where we acquired the collection of plant hand drawings, citation, etc.
Resource
At the Resource page, we collected important and classified resources on plant miRNAs including miRNA databases, tools of miRNA annotation (especially for plant miRNA annotation), tools of miRNA target prediction, tools of pri/pre-miRNA secondary structure prediction, and classical references on miRNA annotation. In addition, we provide quick links to these tools and hyperlinks of their corresponding references.
Submit
As an effort to make PmiREN a one stop community resource, we started to accept submission of plant miRNAs with required information via an official email address of Submit@PmiREN.com. All submitted miRNA items will be processed by our standard procedure described in Material and Methods. By this process, the submitted miRNAs will be grouped into categories with different confidence levels.
DISCUSSION
Research effort on plant miRNA identification, function and evolution has dramatically increased in the past two decades. Meanwhile, abundant data of plant genome references and sRNA-Seq data, two prerequisites for accurately annotating miRNAs (25), have accumulated exponentially after NGS methods were introduced. These together laid a solid foundation for plant miRNA annotation and inherently required a comprehensive database to store and organize all knowledge on plant miRNAs. miRBase started as a centralized database for archiving community annotated miRNAs and assigning each entry a unique numerical identifier (21). However, it became increasingly clear that the minimal intervention approach adopted by miRBase leads to compromised annotation due to uneven standards (24), even though the research community has released two successive versions of plant miRNA annotation criteria (25,42). Therefore, we were compelled to use a standardized pipeline centered on miRDeep-P2 (26) in combination with the latest plant miRNA annotation criteria to re-annotate miRNAs for all plant species with genome references and sRNA-Seq datasets available (Figure 1A).
Compared to miRBase and other miRNA databases, not only were more plant species annotated, both the coverage and integrity of miRNA annotation were also improved (Supplementary Table S4). As shown in Figure 1, a large number of novel miRNA entries were added, questionable entries removed, and family assignment corrected (Figure 1B–D). A direct consequence of this improvement is that users can now make more efficient comparative analyses of miRNAs in the broad context of evolution. A fitting example is miR156/157/529 (43,44), which was previously referred to as the miR156 superfamily (Figure 2B). In fact, after curation based on consensus nucleotides in PmiREN, we found that the miR157 clad is actually present in dicots alone while miR529 exists only in monocots after this separation of angiosperms (Figure 2B). Although this finding alone is not sufficient to uncover the exact evolutionary history of this family or the functional implication of this diversification, it does provide justification and foundation for new investigations on this important family.
Additionally, several lines of information on plant miRNAs were provided in PmiREN. By normalizing sRNA-Seq datasets in different tissues or developmental stages, miRNA expression patterns in species with multiple available sRNA-Seq libraries were established (Figure 2A). Analyses of miRNA conservation and synteny cross different species clearly demonstrated evolutionary trajectory of specific miRNA families and birth/death of family members (Figure 2BC). The target collections with validation from PARE-Seq dataset strengthened reliability of predicted miRNA targeting (Figure 2D). Beside examples shown here, it is conceivable similar analyses could be performed, albeit in greater detail and in combination with functional studies, to greatly raise our understanding of plant miRNAs based on abundant information that is already available. At the same time, PmiREN provides a friendly interface to browse and access all data via multi-layered webpages, powerful search engines and download ports (Figure 3). For these reasons, we believe PmiREN is a comprehensive functional database amenable for data mining and database-driven research and therefore a useful resource for plant miRNA research.
As with all databases, PmiREN needs future upgrades to improve. The current version of PmiREN contains annotation for 88 species but there are over 300 plants with reference genome sequences in NCBI Assembly (27), although many of these species still lack sRNA-Seq datasets. Therefore, processing available sRNA-Seq data to annotate the corresponding miRNA candidates is a top priority in future versions. At the same time, we will make efforts to update PmiREN by accepting data from peer databases and individual researchers. We will use the same unified method and standard for annotating and screening the new entries, and then add them into the newer versions of PmiREN. In this way, PmiREN can be a useful database continuingly serving the plant miRNA research community.
DATA AVAILABILITY
PmiREN is freely available at http://www.pmiren.com.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members in Dr Yang and Dr Li's laboratories for their comments and criticisms on this study.
Author contributions: X. Yang, L. Li, Y. Wang, X. Cao, Y. Zhao and J. Wei, designed the project. Z. Guo, Z. Kuang, Y. Tan, C. Cheng, J. Yang, X. Lu, C. Hao and T. Wang performed the computational analysis and constructed the database. Y. Wang, Z. Guo, Z. Kuang, X. Yang and L. Li wrote the manuscript. All authors commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Beijing Academy of Agriculture and Forestry Sciences (BAAFS) [KJCX201907-2 and KJCX20180204 to X.Y.]; National Natural Science Foundation of China (NSFC) [31621001 to L.L.]. Funding for open access charge: Beijing Academy of Agriculture and Forestry Sciences (BAAFS) [KJCX201907-2 and KJCX20180204]; National Natural Science Foundation of China (NSFC) [31621001].
Conflict of interest statement. None declared.
REFERENCES
- 1. Axtell M.J., Westholm J.O., Lai E.J.. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011; 12:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009; 136:669–687. [DOI] [PubMed] [Google Scholar]
- 3. Rogers K., Chen X.. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013; 25:2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones-Rhoades M.W., Bartel D.P., Bartel B.. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006; 57:19–53. [DOI] [PubMed] [Google Scholar]
- 5. Llave C.A.L., Kasschau K.D., Rector M.A., Carrington J.C.. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002; 14:1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel D.P. MicroRNAs genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 7. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 9. Khraiwesh B., Zhu J.K., Zhu J.H.. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta. 2012; 1819:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samad A.F.A., Sajad M., Nazaruddin N., Fauzi L.A., Murad A.M.A., Zainal Z., Ismail I.. MicroRNA and transcription factor: key players in plant regulatory network. Front. Plant Sci. 2017; 8:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sunkar R., Li Y.F., Jagadeeswaran G.. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012; 17:196–203. [DOI] [PubMed] [Google Scholar]
- 12. Sunkar R. MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 2010; 21:805–811. [DOI] [PubMed] [Google Scholar]
- 13. Sunkar R., Zhu J.K.. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004; 16:2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J.W., Czech B., Weigel D.. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009; 138:738–749. [DOI] [PubMed] [Google Scholar]
- 15. Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S.. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009; 138:750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sunkar R., Chinnusamy V., Zhu J., Zhu J.K.. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007; 12:301–309. [DOI] [PubMed] [Google Scholar]
- 17. Friedländer M.R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., Rajewsky N.. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008; 26:407–415. [DOI] [PubMed] [Google Scholar]
- 18. Yang X., Li L.. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011; 27:2614–2615. [DOI] [PubMed] [Google Scholar]
- 19. Lei J., Sun Y.. miR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics. 2014; 30:2837–2839. [DOI] [PubMed] [Google Scholar]
- 20. Paicu C., Mohorianu I., Stocks M.B., Xu P., Coince A., Billmeier M., Dalmay T., Moulton V., Moxon S.. miRCat2: accurate prediction of plant and animal microRNAs from next-generation sequencing datasets. Bioinformatics. 2017; 16:2446–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2018; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z., Yu J., Li D., Zhang Z., Liu F., Zhou X., Wang T., Ling Y., Su Z.. PMRD: plant microRNA database. Nucleic Acids Res. 2009; 38:D806–D813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meng Y., Gou L., Chen D., Mao C., Jin Y., Wu P., Chen M.. PmiRKB: a plant microRNA knowledge base. Nucleic Acids Res. 2010; 39:D181–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor R.S., Tarver J.E., Hiscock S.J., Donoghue P.C.. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014; 19:175–182. [DOI] [PubMed] [Google Scholar]
- 25. Axtell M.J., Meyers B.C.. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell. 2018; 30:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuang Z., Wang Y., Li L., Yang X.. miRDeep-P2: accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics. 2018; 35:2521–2522. [DOI] [PubMed] [Google Scholar]
- 27. Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A. et al.. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2015; 44:D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2011; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. German M.A., Luo S., Schroth G.P., Meyers B.C., Green P.J.. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009; 4:356–362. [DOI] [PubMed] [Google Scholar]
- 30. Zhai J., Arikit S., Simon S.A., Kingham B.F., Meyers B.C.. Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods. 2014; 67:84–90. [DOI] [PubMed] [Google Scholar]
- 31. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al.. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kodama Y., Shumway M., Leinonen R.. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2011; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dai X., Zhuang Z., Zhao P.X.. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018; 46:W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krüger J., Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006; 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alves-Junior L., Niemeier S., Hauenschild A., Rehmsmeier M., Merkle T.. Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic. Acids. Res. 2009; 37:4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Addo-Quaye C., Miller W., Axtell M.J.. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2008; 25:130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang X., Zhang H., Li L.. Global analysis of gene-level microRNA expression in Arabidopsis using deep sequencing data. Genomics. 2011; 98:40–46. [DOI] [PubMed] [Google Scholar]
- 38. Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H. et al.. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J.S., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yi X., Zhang Z., Ling Y., Xu W., Su Z.. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2014; 43:D982–D989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyers B.C., Axtell M.J., Bartel B., Bartel D.P., Baulcombe D., Bowman J.L., Cao X., Carrington J.C., Chen X., Green P.J. et al.. Criteria for annotation of plant microRNAs. Plant Cell. 2008; 20:3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morea E.G., da Silva E.M., Silva G.F., Valente G.T., Rojas C.H., Vincentz M.G., Nogueira F.T.. Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol. 2016; 16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cui J., You C., Chen X.. The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 2017; 35:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crooks G.E., Hon G.C., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PmiREN is freely available at http://www.pmiren.com.