Abstract
As NLM’s Conserved Domain Database (CDD) enters its 20th year of operations as a publicly available resource, CDD curation staff continues to develop hierarchical classifications of widely distributed protein domain families, and to record conserved sites associated with molecular function, so that they can be mapped onto user queries in support of hypothesis-driven biomolecular research. CDD offers both an archive of pre-computed domain annotations as well as live search services for both single protein or nucleotide queries and larger sets of protein query sequences. CDD staff has continued to characterize protein families via conserved domain architectures and has built up a significant corpus of curated domain architectures in support of naming bacterial proteins in RefSeq. These architecture definitions are available via SPARCLE, the Subfamily Protein Architecture Labeling Engine. CDD can be accessed at https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
CDD CONTENT
At the time of writing, the CDD version v3.17 is the live production version with 52,910 protein- and protein domain-models obtained from Pfam (1), SMART (2), the COGs collection (3), TIGRFAMS (4), the NCBI Protein Clusters collection (5), NCBIfam (6) and CDD’s in-house data curation effort (7). CDD version v3.18 will be released in the winter 2019/2020 and will include Pfam version 32 and a total of 55 434 protein and protein-domain models. For CDD v3.18, the fixed assumed size of the domain model database has again been increased to match the current size of the model collection, resulting in marginally higher E-values reported by RPS-BLAST (8).
The NCBIfam collection in CDD is a set of models derived from HMMs that have been developed for improving the annotation of bacterial genomes. Currently, CDD excludes NCBIfam models that were built to identify proteins involved in antimicrobial resistance, due to their narrow scope.
Table 1 shows the 20 largest classifications for common and functionally diverse domain families that have recently been updated or added to CDD. In total, over 4,700 models curated by the CDD group have been newly published or updated since CDD release v3.16. At the time of the CDD v3.17 release, CDD annotated about 85% of the sequences in the Entrez/protein database (excluding sequences from environmental sampling). CDD also covered about 94% of the protein sequences (longer than 30 residues) derived from protein 3-dimensional structures as provided via MMDB. CDD curation staff monitors structure-derived sequences that do not yet have coverage in CDD for novel protein domain families with wide taxonomic distribution and generates corresponding domain family models de novo.
Table 1.
The largest domain family hierarchies created or updated since CDD release v3.16
| Root | models | Name |
|---|---|---|
| cd14964 | 589 | Seven-transmembrane G protein-coupled receptor superfamily |
| cd00196 | 315 | Beta-grasp ubiquitin-like fold |
| cd01165 | 242 | BTB/POZ domain superfamily |
| cd00083 | 202 | basic Helix Loop Helix (bHLH) domain superfamily |
| cd01391 | 192 | Type 1 periplasmic binding fold superfamily |
| cd06174 | 187 | Major Facilitator Superfamily |
| cd14494 | 172 | Cys-based protein tyrosine phosphatase and dual-specificity phosphatase superfamily |
| cd17912 | 169 | N-terminal helicase domain of the DEAD-box helicase superfamily |
| cd09852 | 137 | PIN (PilT N terminus) domain superfamily |
| cd02208 | 113 | RmlC-like cupin superfamily |
| cd00021 | 97 | B-box-type zinc finger superfamily |
| cd06660 | 96 | Aldo-keto reductase (AKR) superfamily |
| cd14733 | 93 | BACK (BTB and C-terminal Kelch) domain |
| cd08161 | 86 | SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain superfamily |
| cd04433 | 85 | Adenylate forming domain, Class I superfamily |
| cd03873 | 82 | Zinc peptidases M18, M20, M28, and M42 |
| cd00156 | 82 | phosphoacceptor receiver (REC) domain of response regulators/pseudo response regulators |
| cd16961 | 77 | Type I restriction-modification system specificity (S) subunit Target Recognition Domain |
| cd07346 | 75 | Six-transmembrane helical domain of the ATP-binding cassette transporters |
| cd00172 | 74 | SERine Proteinase INhibitors (serpin) family |
| cd00301 | 71 | lipocalin/cytosolic fatty acid-binding protein family |
| cd00048 | 69 | double-stranded RNA binding motif (DSRM) superfamily |
The table lists the root node of each hierarchy, the number of models in the hierarchy (including the root node and intermediate nodes if present), and the name of the protein domain (super)family.
For CDD v3.18, a total of 33 980 site annotations are available on 12 418 out of 16 069 CDD staff-curated domain models. Sequence patterns have been recorded for 3250 of these site annotations, so that pattern matches determine whether a site annotation is being mapped onto a query sequence.
SPARCLE
Protein domain architectures can be defined as a sequential (N- to C-terminal) list of one or more domain footprints annotated on a protein sequence. In CDD, we distinguish between superfamily architectures (where hits to several different models that are redundant or related to each other are treated as the same superfamily hit) and specific or subfamily domain architectures (SDAs), where high-confidence (specific) domain annotation is taken into consideration. The CDART (Conserved Domain Architecture Retrieval Tool) service (9) groups proteins in the Entrez database by common domain superfamily architecture. SPARCLE for ‘Subfamily Protein Architecture Labeling Engine’, on the other hand, groups proteins by SDA, and we have engaged in a curation effort that reviews SDAs that are well-represented in the protein sequence collection and associates them with protein name suggestions and short functional descriptions. To date, CDD curators have assigned names and functional labels to ∼25 000 SDAs, with a focus on SDAs common in bacterial genomes. A publicly accessible Entrez database supports text queries and points to summary information for SDAs as well as links to other databases, most importantly the NCBI protein collection.
CD-Search displays not only domain and feature annotation, but also the name and functional characterization assigned to the corresponding SDA, if available via SPARCLE.
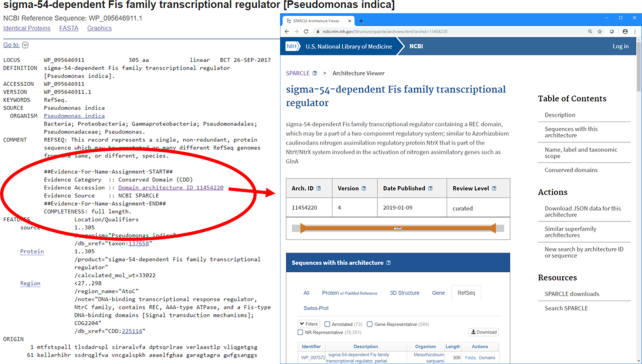
The SPARCLE curation effort is focused on architectures common in bacteria, and supports the automated, evidence-based assignment of names to proteins in RefSeq and the Prokaryotic Genome Annotation Pipeline (PGAP)6. Protein names provided by the curated subset of SPARCLE have relatively low preference in the hierarchy of naming evidence sources but cover a lot of ground and often provide the only suggestion available for a gene product. At this time, about 42 million bacterial RefSeq proteins are named via SPARCLE (out of 126 million total bacterial proteins and 92 million proteins with naming evidence). Figure 1 shows an example of how naming evidence is currently being displayed by the sequence ‘flatfile’ (GenPept format) viewer.
Figure 1.
‘flatfile’ (GenPept format) view of a bacterial protein from the RefSeq collection. Bacterial of proteins with the accession prefix ‘WP’ are now being equipped with evidence for the name assignment (highlighted with a red oval). Evidence accessions are hot-linked to provide more information about the specific annotation rule, in this case a conserved domain architecture curated in SPARCLE. Other evidence types with hotlinks to an annotation rule viewer are Hidden Markov Models (HMMs) and BLAST rules, which have higher precedence than domain architectures and will overrule the name suggested by SPARCLE.
DATA AVAILABILITY
Table 2 lists URLs for services, tools, and data collections provided by CDD. RPS-BLAST is part of NCBI’s BLAST software distribution. Pre-formatted RPS-BLAST search databases are available so that conserved domain searches can be run locally, and the results can be formatted with the rpsbproc utility so that they correspond to reports generated by CD-Search (10) and BATCH CD-Search, including site annotations. A new utility, sparclbl (SparcleLabel) is available via FTP; sparclbl processes results from local RPS-BLAST searches and provides suggestions for protein names based on domain architecture. An in-house version of sparclbl is part of NCBI’s prokaryotic genome annotation pipeline (PGAP) (6).
Table 2.
URLs and other resources associated with the CDD project
| URL | Description |
|---|---|
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation |
| https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | BATCH CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation. Up to 4000 protein queries may be submitted per request |
| https://www.ncbi.nlm.nih.gov/cdd | Entrez interface to CDD |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml | CDD project home page |
| https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi | CDART domain architecture viewer |
| https://ftp.ncbi.nih.gov/pub/mmdb/cdd | CDD FTP site, see README file for content |
| https://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml | Domain hierarchy editor/viewer and protein structure/alignment viewer |
| https://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: https://www.ncbi.nlm.nih.gov/BLAST/download.shtml | RPS-BLAST stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution |
| https://www.ncbi.nlm.nih.gov/sparcle | Entrez interface to SPARCLE (Subfamily Protein Architecture Labeling Engine) |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/rpsbproc | Standalone utility for enriching and formatting RPS-BLAST results |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/SparcleLabel/ | Standalone utility for naming/labeling proteins using SPARCLE |
CDD shares domain models with the InterPro at the European Bioinformatics Institute to supplement sequence annotation with data that are uniquely provided by the CDD curation effort, including protein domain models for very specific subfamilies and the annotation of functional sites. To date, >3100 domain signatures provided by CDD have been integrated by InterPro (11).
FUTURE WORK
The CDD group is investigating whether model-specific word-score thresholds can be applied when building RPS-BLAST search databases and help speed searching while keeping the loss of annotation at a minimum. Instructions for how to use such a search set will be announced via the CDD news page at https://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_news.html, once available.
ACKNOWLEDGEMENTS
We thank the NCBI Information Engineering Branch and the NCBI RefSeq team for continuing support and assistance with software and database development. We are indebted to the authors of Pfam, SMART, COGs, TIGRFAMs, NCBIfam and NCBI’s Protein Clusters database for providing access to their resources and data, and the users of CDD for their acknowledgements and invaluable feedback.
Comments, suggestions, and questions are welcome and should be directed to: info@ncbi.nlm.nih.gov.
FUNDING
Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding for open access charge: Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Conflict of interest statement. None declared.
REFERENCES
- 1. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Letunic I., Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018; 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V.. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001; 29:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haft D.H., Selengut J.D., Richter A.R., Harkins D., Basu M.K., Beck E.. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013; 41:D387–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klimke W., Agarwala R., Badretdin A., Chetvernin S., Ciufo S., Fedorov B., Kiryutin B., O’Neill K., Resch W., Resenchuk S. et al.. The National center for biotechnology information's protein clusters database. Nucleic Acids Res. 2009; 37:D216–D223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R. et al.. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018; 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. et al.. CDD/SPARCLE: functional classification of proteins via subfamily domain architecture. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchler-Bauer A., Panchenko A.R., Shoemaker B.A., Thiessen P.A., Geer L.Y., Bryant S.H.. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002; 30:281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geer L.Y., Domrachev M., Lipman D.J., Bryant S.H.. CDART: protein homology by domain architecture. Genome Res. 2002; 12:1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marchler-Bauer A., Bryant S.H.. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2005; 32:W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.Y., El-Gebali S., Fraser M. et al.. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019; 47:D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table 2 lists URLs for services, tools, and data collections provided by CDD. RPS-BLAST is part of NCBI’s BLAST software distribution. Pre-formatted RPS-BLAST search databases are available so that conserved domain searches can be run locally, and the results can be formatted with the rpsbproc utility so that they correspond to reports generated by CD-Search (10) and BATCH CD-Search, including site annotations. A new utility, sparclbl (SparcleLabel) is available via FTP; sparclbl processes results from local RPS-BLAST searches and provides suggestions for protein names based on domain architecture. An in-house version of sparclbl is part of NCBI’s prokaryotic genome annotation pipeline (PGAP) (6).
Table 2.
URLs and other resources associated with the CDD project
| URL | Description |
|---|---|
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation |
| https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | BATCH CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation. Up to 4000 protein queries may be submitted per request |
| https://www.ncbi.nlm.nih.gov/cdd | Entrez interface to CDD |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml | CDD project home page |
| https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi | CDART domain architecture viewer |
| https://ftp.ncbi.nih.gov/pub/mmdb/cdd | CDD FTP site, see README file for content |
| https://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml | Domain hierarchy editor/viewer and protein structure/alignment viewer |
| https://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: https://www.ncbi.nlm.nih.gov/BLAST/download.shtml | RPS-BLAST stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution |
| https://www.ncbi.nlm.nih.gov/sparcle | Entrez interface to SPARCLE (Subfamily Protein Architecture Labeling Engine) |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/rpsbproc | Standalone utility for enriching and formatting RPS-BLAST results |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/SparcleLabel/ | Standalone utility for naming/labeling proteins using SPARCLE |
CDD shares domain models with the InterPro at the European Bioinformatics Institute to supplement sequence annotation with data that are uniquely provided by the CDD curation effort, including protein domain models for very specific subfamilies and the annotation of functional sites. To date, >3100 domain signatures provided by CDD have been integrated by InterPro (11).