Abstract
Recent advances in genome sequencing and functional genomic profiling have promoted many large-scale quantitative trait locus (QTL) studies, which connect genotypes with tissue/cell type-specific cellular functions from transcriptional to post-translational level. However, no comprehensive resource can perform QTL lookup across multiple molecular phenotypes and investigate the potential cascade effect of functional variants. We developed a versatile resource, named QTLbase, for interpreting the possible molecular functions of genetic variants, as well as their tissue/cell-type specificity. Overall, QTLbase has five key functions: (i) curating and compiling genome-wide QTL summary statistics for 13 human molecular traits from 233 independent studies; (ii) mapping QTL-relevant tissue/cell types to 78 unified terms according to a standard anatomogram; (iii) normalizing variant and trait information uniformly, yielding >170 million significant QTLs; (iv) providing a rich web client that enables phenome- and tissue-wise visualization; and (v) integrating the most comprehensive genomic features and functional predictions to annotate the potential QTL mechanisms. QTLbase provides a one-stop shop for QTL retrieval and comparison across multiple tissues and multiple layers of molecular complexity, and will greatly help researchers interrogate the biological mechanism of causal variants and guide the direction of functional validation. QTLbase is freely available at http://mulinlab.org/qtlbase.
INTRODUCTION
Genome-wide association studies (GWAS) have revealed tens of thousands of genomic loci associated with numerous traits and diseases; however, the underlying causal mechanism often remains poorly understood (1–3). Causal genetic variants alter initial molecular phenotypes, such as chromatin states, and induce a cascade of biological effects, ultimately contributing to the development of traits and diseases (4–6). Exploiting such genotype–phenotype causality would facilitate in-depth understanding of the genetic basis of complex traits. Recent advances in genome sequencing and functional genomic profiling have promoted many large-scale quantitative trait locus (QTL) studies (7), which connect genotypes with tissue/cell type-specific cellular functions (i.e. molecular traits quantified by various high-throughput assays) in different biological stages. For example, QTL mapping has been used to study the genetic determination of chromatin accessibility (caQTL), DNA methylation (mQTL) and histone modification (hQTL) in epigenetic regulation; gene expression (eQTL) and alternative splicing (sQTL) in transcriptional regulation; RNA editing (reQTL) and competing endogenous RNA expression (cerQTL) in post-transcriptional regulation; ribosome occupancy (riboQTL) and protein expression (pQTL) in translational regulation; and cell metabolism (metaQTL) in post-translational regulation (7,8). Here, genomic loci that explain all or a fraction of variation in a given molecular trait are referred to as xQTLs.
For fundamental interpretation of the genetic basis of human complex traits, researchers have in the last decade performed extensive eQTL studies on most human tissues (9) and immune cell types (10) by leveraging gene expression as molecular readouts. As the majority of GWAS trait/disease-associated variants are located in the noncoding genomic regions, the overwhelming increase in eQTL data has greatly facilitated the discovery of novel disease genes and benefits the dissection of variant regulatory mechanisms in a context-dependent manner (11). Several resources, including eQTL Browser (http://eqtl.uchicago.edu), seeQTL (12), ExSNP (13) and ImmuneRegulation (14), have been developed for compiling the eQTL results for certain tissue/cell types. However, gene expression only partially explains trait heritability and is not likely to describe most risk mechanisms (15,16); therefore, there is an urgent need to comprehensively integrate the published xQTL summary statistics for various molecular phenotypes. Although several GWAS databases, such as GRASP (17) and PhenoScanner (18), have begun to provide xQTL summary statistics in recent updates, they only incorporate a small fraction of studies on limited tissue/cell types. In addition, emerging data show that specific types of QTL can be mediated through other fine-scale molecular processes, for example, most eQTLs exert their genetic effect on gene expression through altering open chromatin (19), DNA methylation (20), histone modification (21,22), transcription factor binding (23), chromatin interaction (24) or other post-transcriptional events (25); pQTLs without corresponding cis-eQTLs can exert genetic effects on mRNA decoys (26), alternative splicing (27) or ribosome occupancy (28). Therefore, exploring the causal relationships among the molecular phenotypes at specific risk loci is essential for in-depth understanding of the genetic mechanism of complex traits. Unfortunately, no resource can facilitate cross-QTL investigation of genetic variants and the various molecular phenotypes. Last but not least, translating genetic association with causality from genome-wide QTL signatures requires functional prioritization and follow-up validation; hence, there is a great need for the integration of large-scale tissue/cell type-specific functional genomics data and allele-specific annotations for identifying causal variants (29).
In this work, we manually curated 233 independent QTL studies across 13 human molecular traits and mapped them to 78 human tissue/cell types. We standardized heterogeneous QTL results using a uniform process and compiled 712 unique QTL summary statistics. By designing a highly interactive web function, we constructed a versatile database, QTLbase, which allows users to query, compare and visualize QTLs at tissue-wise, phenome-wise and variant-wise levels. QTLbase also incorporates large-scale tissue/cell type-specific genomic features and functional annotations to interpret the underlying QTL mechanisms. To the best of our knowledge, QTLbase is unique and the most comprehensive resource for exploring the association between genetic variants and the diverse molecular phenotypes in humans. It is free to all and can be accessed at http://mulinlab.org/qtlbase or http://mulinlab.tmu.edu.cn/qtlbase.
MATERIALS AND METHODS
Data curation and processing
We manually curated QTL studies of human molecular phenotypes from the literature by searching PubMed and Google Scholar using QTL-relevant keywords, such as specific xQTL terms, as well as specific descriptions of molecular phenotypes. Both cis- and trans-QTL studies were included, and we incorporated molecular traits, whether studied in normal, treated or diseased tissue/cell types. To create an unbiased collection and to avoid potential data redundancy, we considered QTL data only from published articles that reported genome-wide primary QTL mapping or meta-analysis. Studies involving the reanalysis of existing QTL data or data from specific genomic loci were generally excluded. We also discarded less informative QTL results if variant, trait or P-value information were missing. A single QTL study may involve multiple tissue/cell types [e.g. GTEx (Genotype-Tissue Expression project) (9) and DICE (10)], various molecular phenotypes [e.g. BLUEPRINT (30)], several human populations (31) or different mapping strategies (e.g. genotype-based and allele-specific); we split such QTL results into multiple sets and assigned unique source IDs to distinguish the data. To ensure recognized tissue/cell type naming, we mapped QTL-relevant tissue/cell types to a commonly used anatomogram that has been adopted in the GTEx and the EMBL-EBI Expression Atlas (32). For tissues with numerous QTLs analyzed in finely sorted subregions or cell types, such as whole blood and brain, we recorded both the main tissue together with the sub-tissue/cell type information. Population information was mapped to the five super populations (AFR [African], AMR [Ad Mixed American], EAS [East Asian], EUR [European] and SAS [South Asian]) in the 1000 Genomes project (1 KGP). If QTL mapping was conducted on samples from multiple populations, we designated the population as ‘MIX’.
QTL summary statistics normalization
Variant normalization
As variant information may have been heterogeneous among the collected QTL data, we synchronized the originally recorded dbSNP IDs with those in dbSNP build 151 (33). For variants whose records provide only chromosome position, we first converted them to GRCh37 (Genome Reference Consortium Human Build 37) position using LiftOver (34) and filled in the corresponding dbSNP ID. We identified the effective allele of each QTL from the original publication or related documents, and extracted the reference and alternative alleles from dbSNP build 151.
Molecular trait normalization
Given the complexity of molecular trait description and genomic coordinate recording, we normalized the molecular phenotypes according to different criteria. Briefly, for genes (including long noncoding RNA and small RNA) or transcript phenotypes, such as eQTL and sQTL, we transformed the name or position (if not provided) according to GENCODE Release 30 (GRCh37) (35). For phenotypes measured by microarray, such as most mQTLs, we recorded the probe ID as the trait name and the corresponding position described in the chip manifest file. For non-gene phenotypes measured by next-generation sequencing, the trait name was an abbreviation of the molecular phenotype together with the actual genomic position, for example, a hQTL was recorded as H3K27ac (chr1:1234–5678). If trans-QTL summary statistics were not provided, we defined a QTL as a trans-QTL if the associated trait was far from the variant location (>10 Mb).
Statistical value normalization
Given the diverse QTL mapping strategies, the summary statistics format of the collected QTLs differed substantially. Typically, we expect QTLbase to contain important statistical values, including effective allele, P-value, effect size, standard error and false discovery rate. Effect size was normalized according to applied statistical methods, including β value, t-statistic, chi-squared statistic, odds ratio or logarithm of the odds (LOD) score. Standard error (if not provided) was inferred from the P-value, effect size and sample size using quantile function.
Variant annotation integration
We integrated 20 functional annotations into QTLbase, these annotations can be divided into four major categories according to usage: variant information, functional prediction, functional evidence and disease association. For variant information, we collected variant identity from dbSNP; and variant allele frequency was derived from gnomAD (36) and 1 KGP Phase 3 (37). For functional prediction, we incorporated many frequently used prediction scores, including aggregated conservation scores from CADD (38), aggregated noncoding variant prediction scores from regBase (39), aggregated missense mutation pathogenic scores from dbNSFP (40), aggregated splicing altering prediction scores from dbscSNV (41), aggregated miRNA target altering prediction scores from dbMTS (42), and several function predictions from HaploReg (43), RegulomeDB (44) and InterVar (45). For functional evidence, we integrated large-scale tissue/cell type-specific epigenomic profiling (e.g. histone modifications, transcription factor binding, open chromatin and nascent transcription) from different resources, such as the Roadmap Epigenomics Project (46), Cistrome DB (47) and the FANTOM5 project (48). For disease association, we included ClinVar-reported variants (49), DisGeNET-recorded variants (50) and ICGC somatic mutation information (51) (Supplementary Table S1).
Database design
QTLbase is built on a JAVA-based web framework. The QTL summary statistics and annotation information are stored in MySQL or retrieved by Tabix (52). Several dynamic web pages are implemented by D3.js, jQuery and related JavaScript modules.
RESULTS
Data summary of QTLbase
The literature search initially retrieved hundreds of QTL analysis studies involving human molecular phenotypes. After strict screening and processing, up to August 2019, QTLbase included a total of 233 independent genome-wide QTL studies involving 712 unique QTL summary statistics across 13 molecular phenotypes and 78 tissue/cell types (Supplementary Figures S1 and S2). Among all collected xQTL summary statistics, 66.85% and 15.81% are from eQTL and mQTL, respectively; however, other QTL types, such as sQTL, pQTL, caQTL and hQTL, have recently emerged due to the increasing affordability of omics technologies (Supplementary Figure S1). A large number of xQTLs were from sub-tissue/cell types from whole blood (30.79%) and brain (14.56%) (Supplementary Figure S3) to ensure uniform naming, QTLbase mapped them to fine-scale terms, yielding 30 more tissue/cell types than the GTEx project (Supplementary Figure S3). Most xQTLs were identified in tissue/cell types from European populations (47.43%) or mixed populations (44.8%) (Supplementary Figure S4). Compared with GWAS on common traits/diseases, the majority of QTL studies were performed on limited sample sizes, with only 20 studies using >3000 samples (Supplementary Figure S5). However, xQTL analysis potentially has higher statistical power than GWAS.
Use of QTLbase
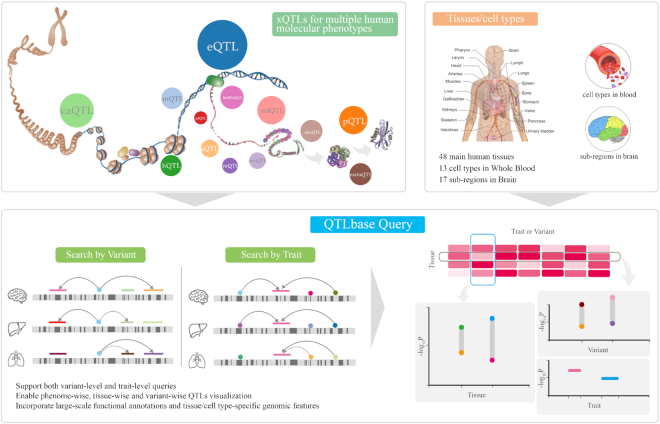
The August 2019 version of QTLbase incorporates 171 524 441 significant associations, namely 159 197 054 cis-QTLs and 12 327 387 trans-QTLs (P-value ≤ 0.05), across numerous genomic loci, molecular traits and tissue/cell types. Such a multidimensional data structure presents challenges in QTL querying and visualization. To facilitate novel discoveries using this high-content repository, we developed a rich web client by incorporating many interactive and user-friendly features. Generally, users can perform both variant- and trait-level queries, and the upcoming web pages enable highly interactive xQTL exploration at phenome-wise, tissue-wise and variant-wise levels (Figure 1).
Figure 1.
The data structure and general function of QTLbase.
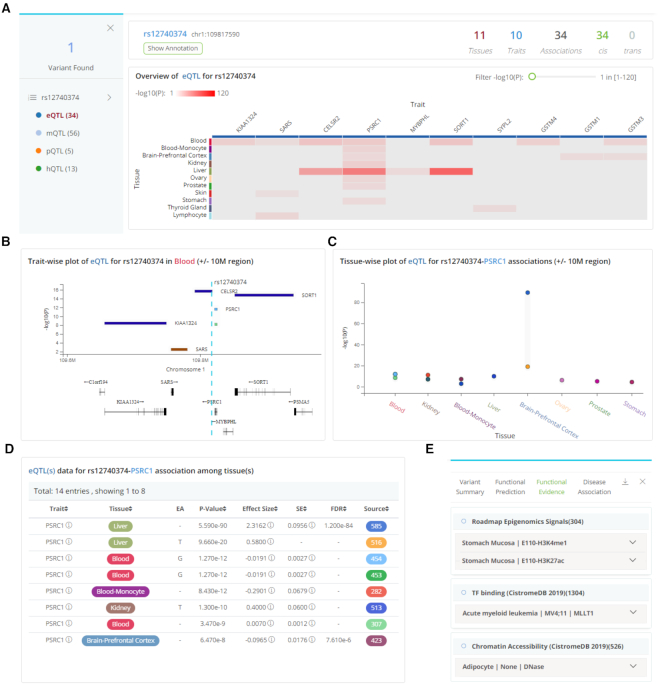
Search by variant
QTLbase accepts variant-level queries by either dbSNP ID or genomic position and displays query results on a dynamic web page. The left panel shows matched cis-xQTL types and the number of associated traits. Users can click on each QTL type to inspect detailed information in the panel on the right (Figure 2A). The top-right panel displays summary information about the selected QTL type, including the query variant, allele information, number of associated tissue/cell types, number of associated molecular traits, total QTL associations, and total cis- and trans-QTLs (Figure 2A). Below this summary information, a zoomable heat map visualizes the distribution of the associated traits across tissue/cell types. Each column represents a QTL-associated trait; each row depicts a separate tissue/cell type. The grid color represents the median P-value of the QTLs on a particular trait and tissue (Figure 2A). Clicking on each tissue row in the heat map opens a phenome-wise plot that displays the genomic distribution of the QTL-associated traits, accompanied by their significance and gene annotations (Figure 2B). Clicking on each trait column in the heat map brings a tissue-wise plot to the front and shows the significance of the QTLs across the related tissue/cell types (Figure 2C). Users can adjust focused region size and decorate the plot using several advanced options. Clicking on each grid in the heat map will highlight the associated QTLs with the specific trait and tissue in both phenome- and tissue-wise plots. The plots are highly interactive and can be synchronized with the bottom summary statistics table, which is searchable and downloadable (Figure 2D). Furthermore, users can check variant functional annotations by clicking the ‘Show Annotation’ button: a floating panel will display extensive annotation information (Figure 2E). Importantly, users can also switch to trans-xQTLs viewer or a tissue-oriented viewer for xQTL comparisons in this result page (Supplementary Figures S6 and S7).
Figure 2.
QTLbase variant-level query results. (A) Major result panels show matched QTL types, basic QTL statistics, and a heat map plot of associated QTLs across traits and tissues for the queried variant. (B) Trait-wise plot of associated QTLs in specific tissue/cell type for queried variant. (C) Tissue-wise plot of associated QTLs on specific trait for queried variant. (D) Summary statistics table of associated QTLs. (E) Floating panel for functional annotations.
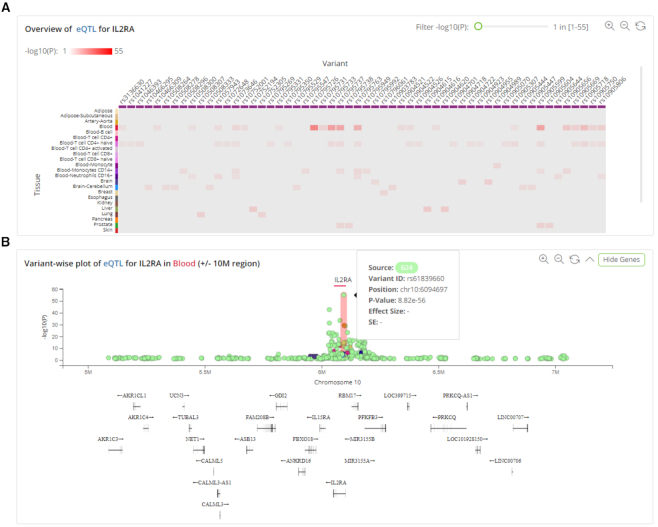
Search by trait
QTLbase also accepts trait-level queries by either trait name or genomic position. The results page layout is similar to that of a variant-level query: the left panel shows the matched QTL types and the top-right panel displays summary information about the query trait and selected QTL type. On this page, the heat map plot shows the distribution of trait-associated variants across tissue/cell type (Figure 3A). Each column represents a trait-associated variant; each row shows a separate tissue/cell type. The grid color represents the median P-value of the QTLs for a specific variant and tissue. Clicking on each tissue row in the heat map opens a variant-wise plot displaying the genomic distribution of the trait-associated variants with their significance and gene annotations (Figure 3B). Most of the other functions on this dynamic page are the same as in the descriptions of variant-level queries.
Figure 3.
QTLbase trait-level query results. (A) Heat map plot of associated QTLs across variants and tissues. (B) Variant-wise plot of associated QTLs in specific tissue for a queried trait.
Variant annotations
QTLbase integrates and compiles 20 annotations (Supplementary Table S1) from four major categories according to the attribute and usage of the collected datasets: variant information, functional prediction, functional evidence and trait association (details in ‘Materials and Methods’ section). Users can activate the annotation panel and download all related annotations by clicking on the corresponding buttons in the query results pages.
Comparison with existing databases
We compared QTLbase with five existing QTL resources for human molecular phenotypes. First, the existing QTL databases collect small numbers of independent QTL studies, and the largest ones, namely PhenoScanner (18) and GRASP (17), contain <30 independent studies. Second, the majority of these databases, such as ImmuneRegulation (14), ExSNP (13) and seeQTL (12), only report eQTL results or involve far fewer other QTL types. Third, the current resources incorporate limited tissue/cell types, and the tissue/cell type descriptions are unclassified. However, QTLbase integrates far more xQTLs for various molecular phenotypes, and includes the most QTL-related tissue/cell types so far, mapping them to standardized terms. Importantly, QTLbase provides a highly interactive web page to facilitate researchers to compare QTLs at tissue-wise, phenome-wise and variant-wise levels, which is largely absent from the current resources. Finally, QTLbase incorporates large-scale functional annotations and tissue/cell type-specific genomic features for in-depth interpretation of xQTLs (see details in Supplementary Table S2). In summary, to the best of our knowledge, QTLbase largely outperforms other databases and establishes the most comprehensive knowledge base for studying the associations between human genetic variants and molecular phenotypes.
Case study
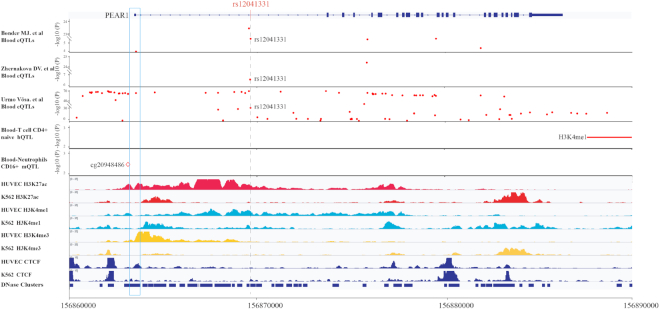
We used a previously reported case to illustrate the validity and usefulness of QTLbase for identifying the potential functional effects of genetic variants. The case involved a platelet reactivity and cardiovascular disease-associated variant, rs12041331, that could reinforce its enhancer activity and affect PEAR1 gene expression during megakaryopoiesis via an allele-specific effect on DNA methylation (53). Specifically, the G allele of rs12041331 ensures fully methylated status and has higher enhancer activity; it was also found that the G allele is highly correlated with a CpG island in the PEAR1 promoter and could reduce CTCF-binding affinity at the promoter through higher methylation, thereby liberating its enhancer activity and increasing PEAR1 expression. QTLbase showed many pieces of evidence that could validate and support the genetic effect and possible regulatory mechanism of rs12041331 in different molecular traits (Figure 4). First, we found that rs12041331 was supported by three blood eQTL studies with large sample sizes (>2000), and it was significantly associated with expression of the PEAR1 gene (54–56). Second, QTLbase showed that rs12041331 is also a hQTL linked to the enhancer marker H3K4me1 in several immune cells. The A allele of rs12041331 was associated with lower H3K4me1 levels around the PEAR1 gene in naïve CD4+ T cells (30), indicating that it could modulate PEAR1 gene expression by affecting histone modification. Notably, we found much mQTL evidence regarding rs12041331; it could alter a specific CpG site (cg20948486) at the PEAR1 promoter in CD16+ neutrophils (30). Finally, QTLbase functional annotations identified that rs12041331 overlapped with many tissue/cell-type specific epigenomic signals, such as histone modifications H3K4me1, H3K4me3 and H3K27ac in widespread cell types; DNase I-hypersensitive sites in immune cells; and transcription factor (e.g. NF-κB, STAT1, SPI1) binding in endothelial and immune cells. Taken together, these QTLbase-derived observations not only are consistent with the investigations of the original study, but also provide additional biological insights into the regulatory mechanism of rs12041331.
Figure 4.
Supporting evidence from QTLbase for the regulatory mechanism of rs12041331. rs12041331 is supported by three independent eQTL studies [Bonder et al. (55), Zhernakova et al. (54) and Urmo Võsa et al. (56)] in blood and is associated with PEAR1 gene expression. rs12041331 is also a hQTL linked to a nearby enhancer marker H3K4me1 in naïve CD4+ T cells [Lu Chen et al. (30)] and has been reported as a mQTL affecting a CpG site (cg20948486) in the PEAR1 promoter (highlighted with a blue box) in CD16+ neutrophils [Lu Chen et al. (30)]. H3K27ac, H3K4me1 and H3K4me3 histone modification ChIP-seq profiles, CTCF ChIP-seq profile and DHS clusters for HUVECs and K562 cells are shown (data from ENCODE).
CONCLUSIONS
In the post-GWAS era, interpreting risk variants that cause the development of complex diseases, and how, is a major, challenging task. Increasing xQTL studies on diverse molecular phenotypes have opened up a new avenue for interrogating mediation paths and cascade effects between genetic factors and complex diseases. By systematically curating and processing xQTL summary statistics, we developed the comprehensive knowledge base QTLbase for biologists and geneticists to query, compare and visualize xQTLs in a highly efficient and interactive manner. To the best of our knowledge, QTLbase is the largest database to integrate various QTL types across numerous tissue/cell types to enable evaluation of the possible molecular functions of genetic variants. We believe that this database will facilitate the QTL retrieval process for researchers and exploration of the underlying mechanisms of the genetic association of complex traits.
Given the rapid development of advanced biotechnologies, new QTL types will continue to be identified at different molecular levels. During the curation and construction of QTLbase, we found that several newer QTL studies had connected genetic variants to more complex molecular phenotypes, such as CRD-QTLs, which correlate genetic effect with cis-regulatory domains representing local chromatin organization (24). Therefore, QTLbase will integrate more novel QTLs in the future and be updated frequently.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31701143, 31871327]; Natural Science Foundation of Tianjin [18JCZDJC34700]. Funding for open access charge: National Natural Science Foundation of China [31701143].
Conflict of interest statement. None declared.
REFERENCES
- 1. Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J.. 10 Years of GWAS Discovery: biology, function, and translation. Am. J. Hum. Genet. 2017; 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tam V., Patel N., Turcotte M., Bosse Y., Pare G., Meyre D.. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019; 20:467–484. [DOI] [PubMed] [Google Scholar]
- 3. Mills M.C., Rahal C.. A scientometric review of genome-wide association studies. Commun. Biol. 2019; 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Albert F.W., Kruglyak L.. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015; 16:197–212. [DOI] [PubMed] [Google Scholar]
- 5. Schaid D.J., Chen W., Larson N.B.. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018; 19:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang D., Yi X., Zhang S., Zheng Z., Wang P., Xuan C., Sham P.C., Wang J., Li M.J.. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res. 2018; 46:W114–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vandiedonck C. Genetic association of molecular traits: a help to identify causative variants in complex diseases. Clin. Genet. 2018; 93:520–532. [DOI] [PubMed] [Google Scholar]
- 8. Li M.J., Yan B., Sham P.C., Wang J.. Exploring the function of genetic variants in the non-coding genomic regions: approaches for identifying human regulatory variants affecting gene expression. Brief. Bioinform. 2015; 16:393–412. [DOI] [PubMed] [Google Scholar]
- 9. GTEx Consortium Genetic effects on gene expression across human tissues. Nature. 2017; 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G. et al.. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018; 175:1701–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li M.J., Li M., Liu Z., Yan B., Pan Z., Huang D., Liang Q., Ying D., Xu F., Yao H. et al.. cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol. 2017; 18:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia K., Shabalin A.A., Huang S., Madar V., Zhou Y.H., Wang W., Zou F., Sun W., Sullivan P.F., Wright F.A.. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012; 28:451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu C.H., Pal L.R., Moult J.. Consensus Genome-Wide expression quantitative trait loci and their relationship with human complex trait disease. OMICS. 2016; 20:400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kalayci S., Selvan M.E., Ramos I., Cotsapas C., Harris E., Kim E.Y., Montgomery R.R., Poland G., Pulendran B., Tsang J.S. et al.. ImmuneRegulation: a web-based tool for identifying human immune regulatory elements. Nucleic Acids Res. 2019; 47:W142–W150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.R., Lareau C., Shoresh N. et al.. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018; 50:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chun S., Casparino A., Patsopoulos N.A., Croteau-Chonka D.C., Raby B.A., De Jager P.L., Sunyaev S.R., Cotsapas C.. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 2017; 49:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X., Gierman H.J., Levy D., Plump A., Dobrin R., Goring H.H., Curran J.E., Johnson M.P., Blangero J., Kim S.K. et al.. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics. 2014; 15:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamat M.A., Blackshaw J.A., Young R., Surendran P., Burgess S., Danesh J., Butterworth A.S., Staley J.R.. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019; doi:10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Degner J.F., Pai A.A., Pique-Regi R., Veyrieras J.B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., Crawford G.E. et al.. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012; 482:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banovich N.E., Lan X., McVicker G., van de Geijn B., Degner J.F., Blischak J.D., Roux J., Pritchard J.K., Gilad Y.. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014; 10:e1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waszak S.M., Delaneau O., Gschwind A.R., Kilpinen H., Raghav S.K., Witwicki R.M., Orioli A., Wiederkehr M., Panousis N.I., Yurovsky A. et al.. Population variation and genetic control of modular chromatin architecture in humans. Cell. 2015; 162:1039–1050. [DOI] [PubMed] [Google Scholar]
- 22. Grubert F., Zaugg J.B., Kasowski M., Ursu O., Spacek D.V., Martin A.R., Greenside P., Srivas R., Phanstiel D.H., Pekowska A. et al.. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell. 2015; 162:1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tehranchi A.K., Myrthil M., Martin T., Hie B.L., Golan D., Fraser H.B.. Pooled ChIP-Seq Links variation in transcription factor binding to complex disease risk. Cell. 2016; 165:730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Delaneau O., Zazhytska M., Borel C., Giannuzzi G., Rey G., Howald C., Kumar S., Ongen H., Popadin K., Marbach D. et al.. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science. 2019; 364:6439. [DOI] [PubMed] [Google Scholar]
- 25. Li M.J., Zhang J., Liang Q., Xuan C., Wu J., Jiang P., Li W., Zhu Y., Wang P., Fernandez D. et al.. Exploring genetic associations with ceRNA regulation in the human genome. Nucleic Acids Res. 2017; 45:5653–5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pai A.A., Cain C.E., Mizrahi-Man O., De Leon S., Lewellen N., Veyrieras J.B., Degner J.F., Gaffney D.J., Pickrell J.K., Stephens M. et al.. The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 2012; 8:e1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K.. RNA splicing is a primary link between genetic variation and disease. Science. 2016; 352:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., Gilad Y.. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015; 347:664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gallagher M.D., Chen-Plotkin A.S.. The Post-GWAS Era: From association to function. Am. J. Hum. Genet. 2018; 102:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martin D., Watt S., Yan Y., Kundu K., Ecker S. et al.. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016; 167:1398–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu L., Candille S.I., Choi Y., Xie D., Jiang L., Li-Pook-Than J., Tang H., Snyder M.. Variation and genetic control of protein abundance in humans. Nature. 2013; 499:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Papatheodorou I., Fonseca N.A., Keays M., Tang Y.A., Barrera E., Bazant W., Burke M., Fullgrabe A., Fuentes A.M., George N. et al.. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018; 46:D246–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F. et al.. The UCSC Genome browser database: update 2006. Nucleic Acids Res. 2006; 34:D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. et al.. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. 2019; 13 August 2019, preprint: not peer reviewed 10.1101/531210. [DOI]
- 37. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H. et al.. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang S., He Y., Liu H., Zhai H., Huang D., Yi X., Dong X., Wang Z., Zhao K., Zhou Y. et al.. regBase: whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants. Nucleic Acids Res. 2019; doi:10.1093/nar/gkz774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X., Wu C., Li C., Boerwinkle E.. dbNSFP v3.0: A One-Stop database of functional predictions and annotations for human nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016; 37:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jian X., Boerwinkle E., Liu X.. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014; 42:13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li C., Swartz M.D., Yu B., Bai Y., Liu X.. dbMTS: a comprehensive database of putative human microRNA target site SNVs and their functional predictions. 2019; 26 May 2019, preprint: not peer reviewed 10.1101/554485. [DOI] [PMC free article] [PubMed]
- 43. Ward L.D., Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016; 44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al.. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Q., Wang K.. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017; 100:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.H., Brown M., Zhang X., Meyer C.A. et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Consortium, F., the, R.P., Clst Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T. et al.. A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. International Cancer Genome, C. Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabe R.R., Bhan M.K., Calvo F., Eerola I. et al.. International network of cancer genome projects. Nature. 2010; 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011; 27:718–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Izzi B., Pistoni M., Cludts K., Akkor P., Lambrechts D., Verfaillie C., Verhamme P., Freson K., Hoylaerts M.F.. Allele-specific DNA methylation reinforces PEAR1 enhancer activity. Blood. 2016; 128:1003–1012. [DOI] [PubMed] [Google Scholar]
- 54. Zhernakova D.V., Deelen P., Vermaat M., van Iterson M., van Galen M., Arindrarto W., van ’t Hof P., Mei H., van Dijk F., Westra H.J. et al.. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017; 49:139–145. [DOI] [PubMed] [Google Scholar]
- 55. Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J. et al.. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017; 49:131–138. [DOI] [PubMed] [Google Scholar]
- 56. Võsa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., Saha A., Kreuzhuber R., Kasela S. et al.. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. 2018; 19 October 2018, preprint: not peer reviewed 10.1101/447367. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.