Abstract
Liquid-liquid phase separation (LLPS) leads to a conversion of homogeneous solution into a dense phase that often resembles liquid droplets, and a dilute phase. An increasing number of investigations have shown that biomolecular condensates formed by LLPS play important roles in both physiology and pathology. It has been suggested the phase behavior of proteins would be not only determined by sequences, but controlled by micro-environmental conditions. Here, we introduce LLPSDB (http://bio-comp.ucas.ac.cn/llpsdb or http://bio-comp.org.cn/llpsdb), a web-accessible database providing comprehensive, carefully curated collection of proteins involved in LLPS as well as corresponding experimental conditions in vitro from published literatures. The current release of LLPSDB incorporates 1182 entries with 273 independent proteins and 2394 specific conditions. The database provides a variety of data including biomolecular information (protein sequence, protein modification, nucleic acid, etc.), specific phase separation information (experimental conditions, phase behavior description, etc.) and comprehensive annotations. To our knowledge, LLPSDB is the first available database designed for LLPS related proteins specifically. It offers plenty of valuable resources for exploring the relationship between protein sequence and phase behavior, and will enhance the development of phase separation prediction methods, which may further provide more insights into a comprehensive understanding of LLPS in cellular function and related diseases.
INTRODUCTION
Liquid-liquid phase separation (LLPS) is a reversible process of a homogenous fluid de-mixing into two distinct liquid phases, one condensed phase and one dilute phase (1). In biological system, a growing number of studies have indicated that LLPS is a critical principle underlying intracellular organization, forming liquid condensates with no membrane enveloped, known as membraneless organelles (MLOs), such as stress granules, P bodies and nucleolus (2,3). Moreover, many biological processes including RNA metabolism, chromatin rearrangement and signal transduction, have been revealed to be regulated by LLPS (4–6). Another reason why LLPS and the formation of biomolecular liquid condensates have drawn much attention is that in some situations the liquid condensates can transform into solid aggregates or amyloid fibers which are deemed to be implicated in a range of incurable neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) (7,8), frontotemporal dementia (FTD) (9) and Alzheimer's disease (AD) (10).
A rapidly increasing number of reports have focused on the phase separation mechanism for a variety of biomolecules in recent years. It has been demonstrated that a number of proteins, such as P granule protein LAF-1 (11,12), RNA helicase DDX4 (13), RNA-binding protein FUS (8,14) and TDP-43 (15), could undergo LLPS both in vivo and in vitro. The liquid condensates formed through LLPS are generally deemed as the result of multivalent weak interactions between multiple folded domains or the multiple interacting motifs in intrinsically disordered regions (IDRs) (16). The low complexity regions (LCRs) that IDRs generally encompass, in which specific amino acids are overrepresented compared to the amino acid proportions in proteome, such as prion-like domains, RGG motifs and PR/GR repeats, are suggested to play critical roles in driving LLPS through electrostatic, hydrophobic, Pi-Pi and cation-Pi interactions with the motifs of themselves or other molecules (17–19). It indicates that, just like how the amino acid sequence of a globular protein determines its folded structure, the sequences of the proteins involved in phase separation play a major role in determining the phase behavior. Meanwhile, RNAs and/or DNAs have been revealed to be able to regulate the phase separation tendency, when mixed with protein, especially with RNA- or DNA-binding proteins (20–22). Additionally, environmental conditions such as temperature, pH, pressure, ionic strength and crowding agent, etc., are also important factors for protein LLPS (23), and the condensates formed in specific condition could be essential for cell processes or adaption (24,25).
A detailed and reliable database of the biomolecular properties and pertinent environmental conditions in the biological phase separation systems can facilitate the comprehensive understanding of what elements determine or control the phase behavior of biomolecules as well as what the biological functions of phase separation systems are in cell. However, resources of this kind are still lacking. The Database UniProt contains the most comprehensive protein information, but has not yet collected the newly reported phase separation data (26). Another database DisProt (27), provides comprehensive information of intrinsically disordered proteins or regions (IDPs or IDRs), and it even provides the ‘liquid–liquid phase separation’ functional annotation for some deposited proteins such as RNA-binding protein FUS (ID no.: DP01102), in the updated 7.0 version, however, the annotations about LLPS are quite limited and lack corresponding details.
Here, we present a carefully curated resource—LLPSDB, which is designed specifically for LLPS related proteins that have been validated by experiments in vitro. All data in LLPSDB are curated manually by carefully mining published literatures and related resources. It provides two aspects of information, one regarding biomolecular properties, including protein sequence, sequence modification, ability of coalescence with nucleic acid, etc., and the other regarding experimental conditions such as protein concentration, ionic concentration, temperature, pH, pressure, crowding agent, detected technique, description of phase behavior and so on. A couple of related databases, such as Uniprot (26), MobiDB (28), DisProt (27), OMIM (29), IDEAL (30), AmyPro (31), FuzzDB (32) and PubMed, are linked from LLPSDB. Additionally, an online ‘blastp’ server and various searching options are offered. The user-friendly web interface (http://bio-comp.ucas.ac.cn/llpsdb or http://bio-comp.org.cn/llpsdb) designed for LLPSDB is easily accessible to scientists with a wide range of expertise.
MATERIALS AND METHODS
Data collection, standardization and curation
All LLPS data in LLPSDB were curated manually from literatures. The literature mining was performed via retrieving in ‘PubMed’ and ‘Web of Science’ by key words: phase separation, phase transition, liquid, protein, de-mixing, assembly, condensate, condensation, coacervate, segregate and segregation. From the total retrieved 3603 papers published up to July 2019, 154 articles were screened out. Only original research articles were collected, and those containing the data of LLPS in vitro were retained. Review papers were excluded to avoid redundancy and confusion. We considered proteins and nucleic acids involved in LLPS as ‘main components’, and constructed entries accordingly. Each entry was identified based on specific protein sequence and nucleic acid type (such as the wild type and mutant of FUS belong to different entry, similarly, component of RNA with 15nt and that with 30nt belong to different entry). Other molecules such as salts, buffer molecules and crowding agents, along with temperature and pH, etc., were considered as experimental conditions. These conditions, in their original units of measurement in the paper, together with the detected phase behavior (a detailed phase diagram or a tag ‘Yes’ or ‘No’ for whether the system phase separates or not) were extracted manually from the screened articles. All the data were checked at least twice. Any incomplete/ambiguous entry was consolidated either by contacting the authors of the article or tracking related references. All the specific information extracted from literatures is listed in the top box of Figure 1.
Figure 1.
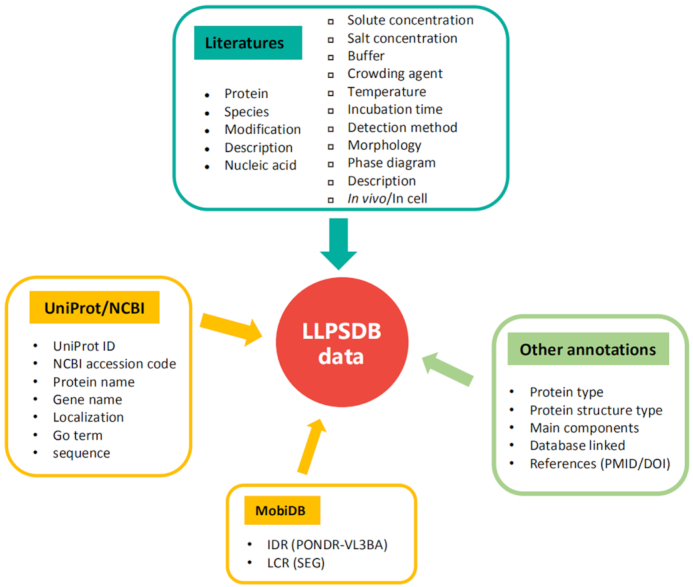
Data structure of LLPSDB. Most of the data are curated from literatures including information of main components (proteins and nucleic acids, as listed in left column of the top box), experimental conditions and phase behavior description (listed in the right column of the top box). Some protein sequences and important annotations of natural protein are from UniProt/NCBI (26) and MobiDB (28) (boxes with yellow frame). Other annotations are added to organize data methodically (box with green frame).
In LLPSDB, some protein annotations (mainly for natural proteins) such as localization, Gene Ontology (GO) term and sequences of some proteins (if not provided by the literature), were obtained from UniProt/NCBI. As IDRs and LCRs in proteins have been demonstrated to be generally critical in LLPS, the sequences of them are presented in visualization. IDRs were identified via searching MobiDB (28) or by PONDR VL3-BA algorithm (33) for those sequences not deposited in MobiDB, and only those segments with no fewer than 15 residues were taken into account (33). LCRs data were also from MobiDB or predicted by SEG algorithm (34) with default parameters. Additionally, in order to organize the data methodically, other information such as protein type (natural or designed), protein structure type (IDR, IDR-fold or Fold) and main components (whether DNAs or RNAs are included) were annotated. Other databases related with the corresponding protein, such as DisProt (27), OMIM (29), IDEAL (30), AmyPro (31), FuzzDB (32), as well as the PMID/DOI of the literature, were also linked from LLPSDB.
It is important to emphasize that LLPSDB focuses on situations where protein alone or with other components (proteins or nucleic acids) was validated to undergo LLPS (or NOT) in vitro. Since in many investigated systems, the mixtures of RNA such as total mRNAs (35,36), instead of specific nucleic acid were added, the sequences of nucleic acid are not presented in LLPSDB. Those systems with only nucleic acids as main component (which means there is no protein) in solution were not included. In addition, systems with segregation of antibodies (such as IgG) and materials designed as drug carrier were also excluded. LLPSDB contains systems where the condensates were observed to flow, fuse, drop, wet and reverse (which were characterized as typical liquid-like droplets), or in which liquid morphology was identified by FRAP (fluorescence recovery after photobleaching), EM (electron microscopy) or other techniques. Systems in which assemblies change morphology from liquid in ripening, such as droplet-to-gel or droplet-to-aggregate were recorded in the database, but those form gels or aggregates directly from homogeneous solution were not deposited.
Database and web site implementation
LLPSDB is implemented as an ASP.net web application with database access. We provide a user-friendly web interface designed by HTML, CSS and JavaScript, with all the data stored and organized through SQL Server and C# programming language.
RESULTS
Datasets classification and statistics
As a novel resource of biomolecular LLPS, LLPSDB currently contains 1182 carefully curated, experimentally validated phase separation systems including 273 independent proteins and 2394 experimental conditions (included 209 phase diagrams). Nine hundred fifty-four entries deposited in LLPSDB are in physiological conditions, namely at temperature within 277–300 K, the concentration of NaCl or KCl within 50–200 mM/L, and pH value in 6.0–8.0. To better curate the deposited data, we classified all the entries by three parallel classification methods. The first classification is based on protein type: the overall 273 proteins are grouped into two subclasses, one for 198 natural proteins and the other for 75 designed proteins, which include 1054 and 191 entries, respectively. The UniProt ID or NCBI accession code of each natural protein is provided, and more annotations from these databases, along with IDRs and LCRs visualization as well as the linkage of related databases, are presented. For the designed proteins, protein name and a short description from literatures are provided. The second classification is based on the main components in the LLPS system, categorizing all entries into three subsets—‘protein(s)’, ‘Protein(s) + RNA’ and ‘Protein(s) + DNA’, depending on whether nucleic acids participate in the coacervation. There are 913 entries where the condensates only contain protein(s), meantime 183 entries containing RNA and 86 entries containing DNA. The third classification is based on the number of main components, by which the entries are grouped into systems including one, two and more main components. We note that, for some systems in which nucleic acids were added as a mixture and the specific nucleic acid sequences were not specified, we consider the added ‘RNA’ or ‘DNA’ as a single component. This classification differentiates simple and complex coacervation (37), where the formation of transient dynamic networks might be formed via distinct interaction patterns due to the different complexity of the systems (such as multiple components, or proteins with different structure type like folded, disordered or mixed). The third classification includes 542 entries with one component, 502 with two components, and 128 with more than two components are deposited. The overview of all subsets according to the three classifications is shown in Table 1.
Table 1.
Overview of the data subsets in LLPSDB
| Classifications | Data type | Number of entry | Proportion (%) |
|---|---|---|---|
| Protein type | Natural protein | 1054 | 85 |
| Designed protein | 191 | 15 | |
| Main components type | Protein(s) | 913 | 77 |
| Protein(s) + RNA | 183 | 16 | |
| Protein(s) + DNA | 86 | 7 | |
| Main components number | One component | 542 | 46 |
| Two components | 502 | 43 | |
| More components | 128 | 11 |
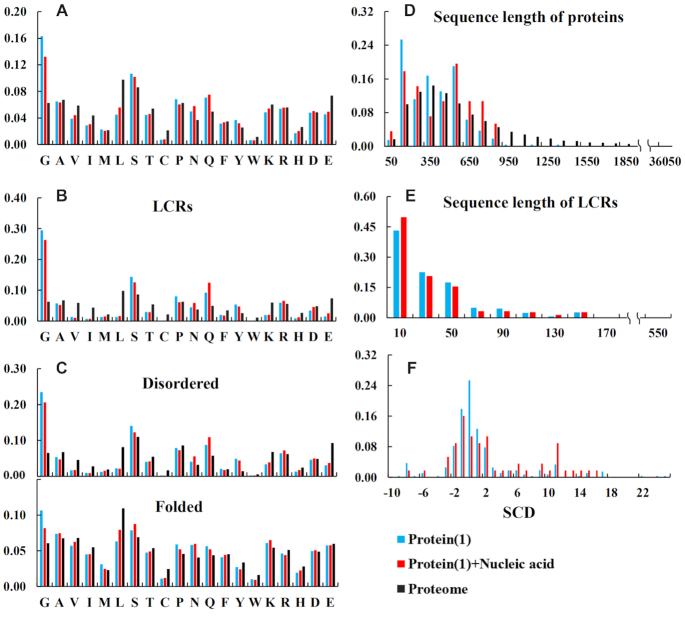
Given that multivalent interactions among IDRs or LCRs are deemed as the main driving forces for protein LLPS (17,18), we analyzed amino acid composition and sequence length distribution of deposited natural proteins with simple coacervation, which means the system includes only one main component, that is an IDR-contained protein which undergoes LLPS in solution by itself (labeled as ‘Protein(1)’ in Figure 2). Considering that many studies have shown that nucleic acids can modulate phase separation tendency of proteins, we also analyzed the subset in which the systems undergo LLPS including one IDR-contained protein and nucleic acid (labeled as ‘Protein(1)+Nucleic acid’ in Figure 2). As mentioned above, IDRs were identified as segments of no shorter than 15 residues obtained by searching MobiDB (28), or by PONDR VL3-BA algorithm (33) if no data are available in MobiDB. We also performed the same analyses on the LCRs, the folded and disordered regions of the proteins in the two subsets ‘Protein(1)’ and ‘Protein(1)+Nucleic acid’. LCRs were predicted by the SEG algorithm (34) with default parameters. The corresponding statistics of the human proteome (from refseq30 in NCBI) was performed as a control.
Figure 2.
Statistical analysis of protein sequence in simple LLPS entries. ‘Protein(1)’ means there is only one IDR-contained protein in the LLPS system, and ‘Protein(1)+Nucleic acid’ means the system also contains DNA or RNA. For the two subsets, the amino acid compositions of the proteins (A), LCRs (B), as well as the disordered (top panel of C) and the folded (bottom panel of C) regions are presented on the left side. On the right side, the sequence length distributions of the proteins and LCRs in the subsets are shown in (D) and (E), respectively; (F) exhibits the distribution of SCD (sequence charge decoration) for the whole protein sequences in the subsets. The statistical analysis from the human proteome is shown as a comparison (black bar), where all sequences were used in (A), (B) and (D), and disordered regions and folded ones were used in the top and the bottom panels of (C) respectively.
The statistical results shown in Figure 2 (also shown on the ‘statistics’ page of LLPSDB web site) indicate that, in both the proteins and their LCRs in the subsets ‘Protein(1)’ and ‘Protein(1)+Nucleic acid’, glycine is the most abundant among the 20 amino acids. In addition, glycine is much more abundant in the two subsets than in the control subset, i.e. the human proteome (Figure 2A and B). It implies that the flexibility of the peptide chain arising from the large fraction of glycine can be favorable for the formation of condensed liquid. Surprisingly, the occupations of arginine and tyrosine, which have attracted much attention in recent studies (38,39), do not exhibit apparent difference from the proteome. Further, the amino acid compositions of disordered regions in the subsets (top panel of Figure 2C) are similar with those of LCRs, while the folded regions seem to have slightly higher abundance of glycine and lower abundance of leucine comparing with the human proteome. The sequence lengths of the proteins in the subsets generally range from 50 to 900 amino acids (Figure 2D), and the distributions are roughly close to those of the human proteome, except the higher fractions at around 150 amino acids and 550 amino acids. LCRs within the both subsets contain generally fewer than 100 amino acids (Figure 2E). Overall, the two subsets ‘Protein(1)’ and ‘Protein(1)+Nucleic acid’ display similar features in the above analysis.
As a recent report suggested that the decoration pattern of charged residues in protein sequence is correlated with its phase behavior (40), we calculated two characterizing parameters κ (41) and SCD (sequence charge decoration) (42) for all proteins (using the whole sequence) in the two subsets. Only 3% of the sequences have rational κ values (with blob up to 9), while most of the SCD values are from –4 to 4 (Figure 2F).
Web interface
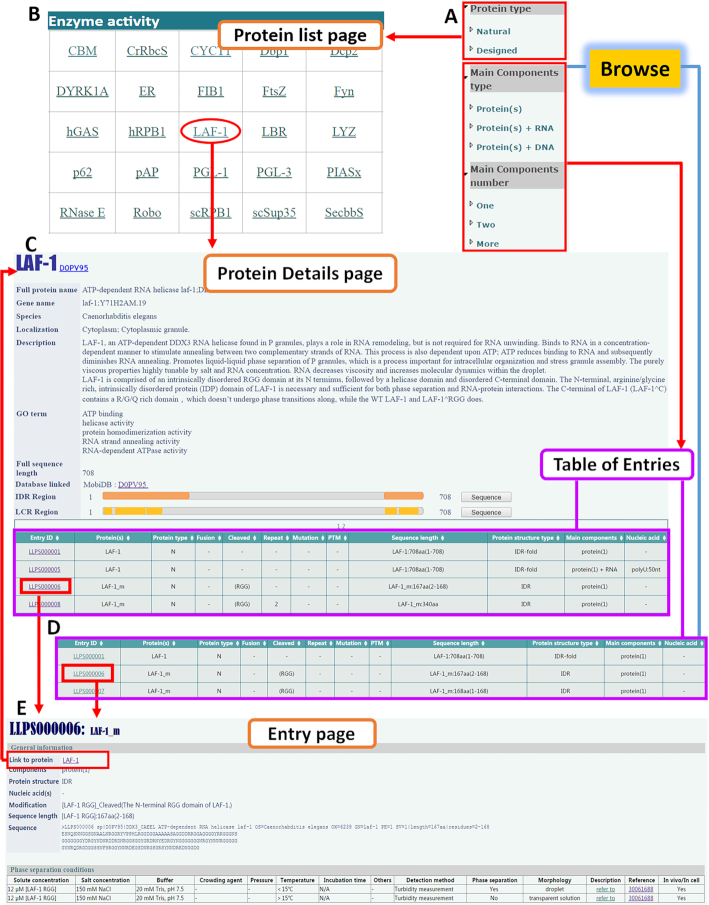
The user-friendly web interface of LLPSDB allows the data to be browsed, searched and downloaded conveniently. The database includes Home, Browse, Search, Submit, Statistics, Download and Help modules. Browse module is the core of LLPSDB. All data deposited in LLPSDB can be classified into different subsets as mentioned in the above section and presented on the left panel on ‘Browse’ page (also shown in Figure 3A). When either ‘natural’ or ‘designed’ protein subset classified by ‘protein type’ is browsed, an extra ‘Protein List page’ will be displayed at first (Figure 3B), which is only presented in this classification. Natural proteins on this page have been divided into different groups according to their molecular biological functions. Upon clicking on the name of specific protein, users can access into a ‘Protein Details page’ (Figure 3C), where the general information of this protein is recorded, followed by a ‘Table of Entries’ which lists all entries that this protein is involved in. The ‘Table of Entries’ displays the brief information of main components in each entry, and can be browsed directly for the other two classifications (Figure 3C and D). More detailed information of each entry is available on the expanded ‘Entry page’ linked by Entry ID (Figure 3E). On the ‘Entry page’, the ‘Protein Details page’ can be linked through the corresponding protein name.
Figure 3.
The Browse module in LLPSDB. (A) Dataset can be browsed by three distinct classifications. (B) An extra ‘Protein List page’ is displayed when browsing natural or designed proteins. (C) The ‘Protein Details page’ consists of two parts: protein information and related ‘Table of Entries’. (D) The ‘Table of Entries’ displays brief information of main components in each entry. (E) The expanded ‘Entry page’ displays the comprehensive information of the entry, including general information for main components and phase separation conditions.
In addition to Browse, the web-accessible LLPSDB provides other easy-to-use modules. Users can search the database separately or combined by ‘Species’, ‘Protein structure type’, ‘Sequence length’, ‘PTM’, ‘Mutation type’, ‘Protein type’, ‘Main components type’ and ‘Main components number’, or by inputting a ‘Keyword’. The ‘Keywords’ can be a protein name, UniProt ID, Entry ID, Localization or GO term. Besides the various options for searching, an online ‘blastp’ server is offered to users for searching entries with specific protein sequence in LLPSDB. Additionally, through Submit module, researchers can submit their own published data to LLPSDB. All datasets in the three classifications are available on the ‘Download’ page. Detailed guidance on the use of LLPSDB could be found on the ‘Help’ page.
DISCUSSION AND FUTURE DEVELOPMENT
LLPSDB is a novel, carefully curated database of proteins undergoing liquid-liquid phase separation in vitro with experimental validation. It not only records the comprehensive annotations of all LLPS related proteins, but also includes the details of corresponding experimental conditions, as well as explicit descriptions of the phase behavior. The database contains both simple condensed systems, such as those with only one single domain protein, and complex systems such as those with components more than two different proteins. The IDRs, LCRs, sequence modifications (including cleaved, mutation, post-translational modification, etc.), combined with the recorded phase separation conditions in LLPSDB, have a huge potential for helping elucidate the relationship between protein sequence, interaction type and phase behavior. In addition, the functional annotations, and the linkage to related databases (such as OMIM (29), IDEAL (30), AmyPro (31), FuzzDB (32)), could help users to discover the biological function mechanism of the LLPS protein in physiology and pathology. Given the plenty resources provided in LLPSDB, we anticipate it will facilitate the achievement of a deeper understanding of biomolecular phase separation.
As all entries in LLPSDB are based on experimental data in vitro, it will not only provide validated testing datasets for the emerging theoretical models (43–45), but may enhance the development of phase separation prediction methods (46–48). LLPSDB will be maintained for a long-term and updated regularly by regular internal updates through scanning newly published literatures and data submissions from the research community.
ACKNOWLEDGEMENTS
We thank Prof. Luhua Lai, Prof. Zhirong Liu, Prof. Jianfeng Pei, Prof. Tiejian Luo, Prof. Jizhong Lou, Prof. Wei Feng, Dr Hao Ruan, Dr Huabin Zhou and Dr Changsheng Zhang for their helpful suggestions. We are grateful to Dr Yi-Hsun Lin for checking and polishing the English language of the manuscript. We also thank the authors of some retrieved publications for their kind replying to our request about LLPS data. This work was supported by University of Chinese Academy of Sciences.
FUNDING
National Natural Science Foundation of China [21633001, 31870718]. Funding for open access charge: National Natural Science Foundation of China [31870718].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dolgin E. What lava lamps and vinaigrette can teach us about cell biology. Nature. 2018; 555:300–302. [DOI] [PubMed] [Google Scholar]
- 2. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L. et al.. Protein phase Separation: A new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P. et al.. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018; 25:833–840. [DOI] [PubMed] [Google Scholar]
- 5. Tatavosian R., Kent S., Brown K., Yao T., Duc H.N., Huynh T.N., Zhen C.Y., Ma B., Wang H., Ren X.. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019; 294:1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang G., Wang Z., Du Z., Zhang H.. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018; 174:1492–1506. [DOI] [PubMed] [Google Scholar]
- 7. Gui X., Luo F., Li Y., Zhou H., Qin Z., Liu Z., Gu J., Xie M., Zhao K., Dai B. et al.. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat. Commun. 2019; 10:2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. et al.. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 9. Mann J.R., Gleixner A.M., Mauna J.C., Gomes E., DeChellis-Marks M.R., Needham P.G., Copley K.E., Hurtle B., Portz B., Pyles N.J. et al.. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019; 102:321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kostylev M.A., Tuttle M.D., Lee S., Klein L.E., Takahashi H., Cox T.O., Gunther E.C., Zilm K.W., Strittmatter S.M.. Liquid and hydrogel phases of PrP(C) linked to conformation shifts and triggered by alzheimer's amyloid-beta oligomers. Mol. Cell. 2018; 72:426–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schuster B.S., Reed E.H., Parthasarathy R., Jahnke C.N., Caldwell R.M., Bermudez J.G., Ramage H., Good M.C., Hammer D.A.. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 2018; 9:2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C., Eckmann C.R., Myong S., Brangwynne C.P.. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D. et al.. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015; 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monahan Z., Ryan V.H., Janke A.M., Burke K.A., Rhoads S.N., Zerze G.H., O’Meally R., Dignon G.L., Conicella A.E., Zheng W. et al.. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017; 36:2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R.G., Shorter J., Bonini N.M.. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell. 2018; 71:703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harmon T.S., Holehouse A.S., Pappu R.V.. Differential solvation of intrinsically disordered linkers drives the formation of spatially organized droplets in ternary systems of linear multivalent proteins. New J. Phys. 2018; 20:045002. [Google Scholar]
- 17. Lin Y.H., Forman-Kay J.D., Chan H.S.. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry. 2018; 57:2499–2508. [DOI] [PubMed] [Google Scholar]
- 18. Martin E.W., Mittag T.. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry. 2018; 57:2478–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franzmann T.M., Alberti S.. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J. Biol. Chem. 2019; 294:7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou H., Song Z., Zhong S., Zuo L., Qi Z., Qu L.J., Lai L.. Mechanism of DNA-induced phase separation for transcriptional repressor VRN1. Angew. Chem. Int. Ed. Engl. 2019; 58:4858–4862. [DOI] [PubMed] [Google Scholar]
- 21. Du M., Chen Z.J.. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018; 361:704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drino A., Schaefer M.R.. RNAs, phase separation, and membrane-less organelles: are post-transcriptional modifications modulating organelle dynamics. Bioessays. 2018; 40:e1800085. [DOI] [PubMed] [Google Scholar]
- 23. Posey A.E., Holehouse A.S., Pappu R.V.. Phase Separation of Intrinsically Disordered Proteins. Methods Enzymol. 2018; 611:1–30. [DOI] [PubMed] [Google Scholar]
- 24. Winter R.H.A., Cinar H., Fetahaj Z., Cinar S., Vernon R.M., Chan H.S.. Temperature, hydrostatic pressure, and osmolyte effects on liquid–liquid phase separation in protein condensates: physical chemistry and biological implications. Chemistry (Easton). 2019; 25:22–1. [DOI] [PubMed] [Google Scholar]
- 25. Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 26. Bateman A., Martin M.J., Orchard S., Magrane M., Alpi E., Bely B., Bingley M., Britto R., Bursteinas B., Busiello G. et al.. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piovesan D., Tabaro F., Micetic I., Necci M., Quaglia F., Oldfield C.J., Aspromonte M.C., Davey N.E., Davidovic R., Dosztanyi Z. et al.. DisProt 7.0: a major update of the database of disordered proteins. Nucleic Acids Res. 2017; 45:D219–D227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piovesan D., Tabaro F., Paladin L., Necci M., Micetic I., Camilloni C., Davey N., Dosztanyi Z., Meszaros B., Monzon A.M. et al.. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018; 46:D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A.. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019; 47:D1038–D1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fukuchi S., Amemiya T., Sakamoto S., Nobe Y., Hosoda K., Kado Y., Murakami S.D., Koike R., Hiroaki H., Ota M.. IDEAL in 2014 illustrates interaction networks composed of intrinsically disordered proteins and their binding partners. Nucleic Acids Res. 2014; 42:D320–D325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varadi M., De Baets G., Vranken W.F., Tompa P., Pancsa R.. AmyPro: a database of proteins with validated amyloidogenic regions. Nucleic Acids Res. 2018; 46:D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miskei M., Antal C., Fuxreiter M.. FuzDB: database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res. 2017; 45:D228–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K.. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003; 53:566–572. [DOI] [PubMed] [Google Scholar]
- 34. Wootton J.C. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem. 1994; 18:269–285. [DOI] [PubMed] [Google Scholar]
- 35. Putnam A., Cassani M., Smith J., Seydoux G.. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019; 26:220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsang B., Arsenault J., Vernon R.M., Lin H., Sonenberg N., Wang L.-Y., Bah A., Forman-Kay J.D.. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. U. S. A. 2019; 116:4218–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hyman A.A., Weber C.A., Julicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell. Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 38. Brady J.P., Farber P.J., Sekhar A., Lin Y.H., Huang R., Bah A., Nott T.J., Chan H.S., Baldwin A.J., Forman-Kay J.D. et al.. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E8194–E8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D. et al.. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018; 174:688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin Y.H., Chan H.S.. Phase separation and single-chain compactness of charged disordered proteins are strongly correlated. Biophys. J. 2017; 112:2043–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das R.K., Pappu R.V.. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sawle L., Ghosh K.. A theoretical method to compute sequence dependent configurational properties in charged polymers and proteins. J. Chem. Phys. 2015; 143:085101. [DOI] [PubMed] [Google Scholar]
- 43. Lin Y.H., Forman-Kay J.D., Chan H.S.. Sequence-specific polyampholyte phase separation in membraneless organelles. Phys. Rev. Lett. 2016; 117:178101. [DOI] [PubMed] [Google Scholar]
- 44. Lin Y.H., Song J.H., Forman-Kay J.D., Chan H.S.. Random-phase-approximation theory for sequence-dependent, biologically functional liquid–liquid phase separation of intrinsically disordered proteins. J. Mol. Liq. 2017; 228:176–193. [Google Scholar]
- 45. McCarty J., Delaney K.T., Danielsen S.P.O., Fredrickson G.H., Shea J.E.. Complete phase diagram for liquid–liquid phase separation of intrinsically disordered proteins. J. Phys. Chem. Lett. 2019; 10:1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orlando G., Raimondi D., Tabaro F., Codice F., Moreau Y., Vranken W.. Computational identification of prion-like RNA-binding proteins that form liquid phase-separated condensates. Bioinformatics. 2019; doi:10.1093/bioinformatics/btz274. [DOI] [PubMed] [Google Scholar]
- 47. Vernon R.M., Chong P.A., Tsang B., Kim T.H., Bah A., Farber P., Lin H., Forman-Kay J.D.. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018; 7:e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vernon R.M., Forman-Kay J.D.. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019; 58:88–96. [DOI] [PubMed] [Google Scholar]