Abstract
MicroRNA (miRNA) biogenesis is a tightly controlled multi-step process operated in the nucleus by the activity of the Microprocessor and its associated proteins. Through high resolution mass spectrometry (MS)- proteomics we discovered that this complex is extensively methylated, with 84 methylated sites associated to 19 out of its 24 subunits. The majority of the modifications occurs on arginine (R) residues (61), leading to 81 methylation events, while 30 lysine (K)-methylation events occurs on 23 sites of the complex. Interestingly, both depletion and pharmacological inhibition of the Type-I Protein Arginine Methyltransferases (PRMTs) lead to a widespread change in the methylation state of the complex and induce global decrease of miRNA expression, as a consequence of the impairment of the pri-to-pre-miRNA processing step. In particular, we show that the reduced methylation of the Microprocessor subunit ILF3 is linked to its diminished binding to the pri-miRNAs miR-15a/16, miR-17–92, miR-301a and miR-331. Our study uncovers a previously uncharacterized role of R-methylation in the regulation of miRNA biogenesis in mammalian cells.
INTRODUCTION
MicroRNAs (miRNAs) are short non-coding RNA molecules that regulate gene expression at the post-transcriptional level (1–4). They interact with target mRNAs by pairing with the corresponding miRNA-binding sites, typically located in the 3′ untranslated regions (3′UTRs), and promote their translational repression and/or degradation (5). MicroRNAs are preferentially transcribed by RNA Polymerase II into long primary transcripts, called pri-miRNAs, that possess the 7-methyl-guanosine cap at the 5′-end, the poly-A tail at the 3′-end and the stem-loop structures, where the mature miRNA sequences are embedded (6–8). Genes encoding miRNAs are located in different genomic regions: intergenic miRNAs are transcribed as separated transcriptional units, while intragenic miRNAs are transcribed together with their ‘host’ gene, the majority encoded within introns and a few deriving from exons. Interestingly, miRNA loci located in close proximity are often co-transcribed as unique transcripts, giving rise to polycistronic units, composed of 2–19 individual miRNA hairpins (6,8). In the nucleus, the Microprocessor complex, which comprises the type-III RNase Drosha and two molecules of DGCR8, processes pri-miRNAs into shorter stem-loop molecules of 60–70 nucleotides, called precursor miRNAs (pre-miRNAs) (2,9,10). The DGCR8 dimer binds to the pri-miRNA through its double strand RNA-binding domain and favors the correct positioning of Drosha on the stem-loop (4,11–13), which is a crucial step for the subsequent pri-miRNA cleavage and determination of the guide and passenger miRNA strands (14–17). Pre-miRNAs are then exported in the cytoplasm by the exportin-5 (XPO5)- RAN- GTP complex and processed by the Dicer/Trbp complex into small RNA duplexes, about 22nt-long (18–21). These duplexes are finally loaded into the RNA-Induced Silencing Complex (RISC), where the dsRNA is unwound, the passenger strand is removed and degraded, while the guide strand is retained and used for the recognition of the miRNA-binding site within the mRNA targets (22,23).
The tight control of microRNA biogenesis at multiple steps ensures the production of the correct levels of miRNA molecules that, in turn, fine-tune gene expression. Aberrant miRNA levels have been, in fact, observed in several pathologies, including cancer (24,25). An important mechanism to regulate miRNA biogenesis is represented by the modulation of the Microprocessor activity, which is rate-limiting for the whole process (26). The expression and activity of the Microprocessor is controlled in multiple ways. First, Drosha and DGCR8 protein levels are tightly regulated by a double-negative feedback loop, whereby DGCR8 stabilizes Drosha protein level, which, in turn, promotes the degradation of DGCR8 transcript by cleaving two hairpins located in its 5′UTR (27,28). Second, although the Microprocessor alone can complete the pri-miRNAs cleavage reaction, there is evidence that various accessory proteins associate to it and regulate its catalytic activity. In fact, 22 co-factors have been described to interact with the Microprocessor (Corum database Complex ID number 1332 and 3082 (29)). We refer to this set of Drosha/DGCR8 associated proteins as the Large Drosha Complex (LDC), in line with previous reports (30). Accessory proteins comprise mainly RNA binding proteins (RBPs), such as the DEAD-box helicases DDX5 and DDX17, a number of heterogeneous ribonucleoproteins (hnRNPs), the FET proteins (FUS, EWSR1, TAF15) and other factors (2,31,32). They modulate the catalytic activity and define the substrate specificity of the Microprocessor, in various ways (2,31,33–35). DDX5 and DDX17, for instance, are required for the recognition and processing of specific secondary structures within a subset of pri-miRNAs (33,34). TAR DNA Binding Protein (TARDBP) has a dual effect on the Microprocessor activity by both facilitating the binding and cleavage of specific pri-miRNAs and protecting Drosha protein from proteasome-dependent degradation (36,37). Interleukin Enhancer Binding Factor 2 (ILF2, also known as NF45) and the splicing isoform known as NF90 of Interleukin Enhancer Binding Factor 3 (ILF3) were initially considered negative regulators of miRNA biogenesis, being shown to sequester some pri-miRNAs (e.g. pri-let-7a and pri-miR-21) from the Microprocessor when overexpressed (38,39). More recent experimental evidences based on gene knockdown experiment have, instead, demonstrated that basal ILF3 stabilizes specific pre- and mature miRNAs, thus exerting a positive regulation on the biogenesis of some miRNAs (40).
The LDC can also be regulated by post-translational modifications (PTMs), as recently reviewed (4). For instance, whereas phosphorylation stabilizes the levels of DGCR8 (41), Drosha phosphorylation by the MAPK p38 promotes its cytoplasmic export and degradation (42). Moreover, Histone deacetylase 1 (HDAC1) enhances microRNA processing via deacetylation of DGCR8 (43), while Drosha acetylation blocks its ubiquitination, thus preventing its degradation by the ubiquitin-proteasome pathway (44).
In 2013, our group discovered for the first time that several LDC subunits are methylated on arginine (R) residues (30), an observation that was then confirmed by a following global methyl-proteomic study (45). However, besides the high frequency of this modification on the complex, its functional impact on the LDC activity, and consequently on miRNA biogenesis, remains elusive.
A family of 9 protein R-methyltransferases (PRMTs) can catalyze R-methylation in mammals. They are grouped in 3 classes based on the specific reaction that they can catalyze: type-I enzymes generate both mono-methylation and asymmetric di-methylation, whereby asymmetric di-methyl arginine (ADMA) presents 2 methyl-groups added to the same terminal ω-guanidino nitrogen atom; type-II enzymes catalyze symmetric di-methylation at distinct terminal ω-guanidino nitrogen atoms of R (SDMA); type-III group comprises only PRMT7 that is capable of generating exclusively mono-methyl arginine (MMA) (46,47).
Protein Arginine N-methyltransferases 1 (PRMT1) is the most active type-I PRMT, responsible of more than 85% of the annotated R-methylations in the mammalian proteome (48,49), and it primarily modifies arginines located within the glycine- and arginine-rich sequences (RGG/RG) (50,51). Besides the well-known target arginine 3 on histone H4 (H4R3me2a) (52,53), several non-histone proteins are substrates of PRMT1, such as the transcription factor RUNX1 (54); the transcription elongation factor SPT5 (55); some enzymes involved in the DNA damage response like MRE11, 53BP1 and BRCA1 (56–59); and several RNA binding proteins, including also the Microprocessor-associated proteins ILF3, EWSR1, FUS and TAF15 (48,60–63). It has been shown in various studies that PRMT1-dependent methylation can regulate proteins by affecting their subcellular localization, or interaction with both other proteins and nucleic acids, in particular RNAs (61,62,64–67).
Prompted by our initial evidence that LDC is hyper-methylated (30), we set to investigate the possible role of this modification in regulating both the composition and function of the complex and, consequently, miRNA biosynthesis.
Overall, this study describes for the first time that the extensive R-methylation of the LDC is largely dependent on PRMT1 and it modulates the activity of the complex, thus regulating miRNA biogenesis and levels.
MATERIALS AND METHODS
Cell culture, heavy methyl SILAC and SILAC labeling of cells
HeLa and HEK293 cells were grown in DMEM (Lonza, LA-0009E) supplemented with 10% FBS (Euroclone, ECSO182L), 1% glutamine (Lonza, BE17605E) and 100 U/ml Penicillin and Streptomycin (Euroclone, ECB3001D).
HeLa Flp-In T-REx cells were in-house generated according to the guidelines of Thermo Fisher Scientific and grown in DMEM supplemented with 10% FBS Tetracycline free (Euroclone, ECS01822), 1% glutamine and 100 U/ml Penicillin and Streptomycin.
For Heavy methyl SILAC labeling, HeLa S3 cells were cultured in ‘Light’ and ‘Heavy’ SILAC media (PAA, custom) depleted of lysine, arginine and methionine and supplemented with l-arginine (Sigma Aldrich, A6969) l-lysine (Sigma Aldrich, L8662), and either l-[13CD3]-methionine (Met-4, heavy, Sigma Aldrich, 299154) or l-[12CH3]-methionine (Met-0, light, Sigma Aldrich M5308), as previously described (76).
For standard SILAC labeling, HeLa cells were grown in ‘‘Light’’ and ‘‘Heavy’’ SILAC DMEM (Thermo Fisher Scientific, 88420) supplemented with either L-arginine and L-lysine, or their heavy isotope-counterparts l-arginine-13C6, 15N4 hydrochloride (Arg10, Sigma, 608033) and L-lysine-13C6, 15N2 hydrochloride (Lys 8, Sigma, 608041) (76).
All media were supplemented with 10% dialyzed FBS (26400-044 Gibco, Life Technology), 1% glutamine, 100 U/ml Penicillin and 100 mg/ml Streptomycin.
Antibodies and chemical compounds
The following antibodies were used for immuno-precipitation and Western Blot experiments, according to the manufacturer's instruction: DGCR8 (Abcam ab90579), EWSR1 (Abcam ab54708), FUS (Bethyl A300–293A), DDX5 (Abcam ab126730), Drosha (Santa Cruz sc-33778), Vinculin (VCL) (Millipore 06–866), PRMT1 (Abcam ab73246), ASYM24 (Millipore), SYM10 (Millipore), Mono-Methyl Arginine (R*GG) (D5A12) (Cell Signaling Technology 8711), LAMIN A/C (sc-6215), Lamin B1 (Abcam ab16048), GAPDH (Abcam ab9484), H3 (Abcam ab1791), H4 (Abcam ab7311), H4R3me2a (Active Motif 39705), TAF15 (Bethyl Laboratories A300–308A), ILF3 (Bethyl A303–615A), ILF2 (sc-271718), DDX17 (sc-130650), HDAC1 (Abcam ab7028) and HA.11 (Biolegend 901513).
MS023 and MS094 compounds were kindly provided by the SGC Toronto—Structural Genomic Consortium (http://www.thesgc.org/scientists/groups/toronto). Compounds were dissolved in DMSO and used at a final concentration of 10 μM for the indicated time intervals.
Cloning strategies and generation of stable cell lines
PRMT1 knockdown (KD) cells were produced using a second-generation pLKO lentiviral vectors, in which specific sh-RNA targeting PRMT1 were cloned. The pLKO-Empty vector was used as control.
To overexpress PRMT1, the cDNA of the v2 isoform was amplified starting from HeLa cDNA, using the following primers:
FW 5′-GGGGATCCCCCGGGCTGCAGATGGCGGCAGCCGAGGCCGCGAACTGCA-3′
REV 5′-GAGGTTGATTGTCGACTCAGCGCATCCGGTAGTCGGTGG-3′
The cDNA of PRMT1 was cloned into the pCCLsin.CPPT.PGK.GFP.WPRE lentiviral vector after plasmid linearization with PstI and SalI using the in-fusion HD EcoDry cloning plus, according to the manufacturer's instructions (Clontech Laboratories, Inc., CA, USA). The GFP cassette was removed from the vector upon digestion with XhoI.
To obtain both PRMT1 KD and PRMT1 overexpressing (OE) cells, HeLa cells were transduced using lentiviruses whose stocks were produced by transient transfection of HEK293 cells with the packaging plasmid pCMV-DR8.74, the envelope plasmid pMD2G-VSVG and the respective transfer gene- carrying vector.
The human coding sequence of ILF3 (HsCD00439701) purchased from the DNASU plasmid repository was cloned into a pCDNA 5.0 FRT TO plasmid using the Gateway cloning strategy (Thermo Fisher Scientific). ILF3 point mutations R609A and R609K were introduced in the plasmid through the Q5 Site-directed Mutagenesis kit (NEB, E0554S) and verified by Sanger sequencing. For the generation of HeLa Flp-In T-REx (Thermo Fisher Scientific) inducible cells, the pCDNA 5.0 FRT TO plasmids containing the coding sequences for ILF3, ILF3 R609A and ILF3 R609K were co-transfected with the plasmid expressing the pOG44 Flp-Recombinase (Thermo Fisher Scientific) using the ViaFect transfection reagent (Promega, E4981). After transfection, cells were growth in medium supplemented with 1μg/mL Blasticidine S hydrochloriode (Sigma-Aldrich, 15205) and 200ng/mL Hygromycin B (Roche, 10843555001) to select only cells which have the pCDNA 5.0 FRT TO plasmid integrated in the genome. The expression of ILF3 proteins was obtained by the administration of 1μg/mL doxycycline for 24h in the cell culture medium.
Cell lysis and sub-cellular fractionation
For preparation of whole cell extracts, cell pellets were lysed in 3 volumes of SDS Lysis Buffer (0.1 M Tris–HCl pH 7.5, 4% SDS), previously warmed to 95°C. Lysates were then sonicated, centrifuged 15 min at 13 000 rpm to precipitate cell debris and then loaded on SDS-PAGE for subsequent protein separation.
For the preparation of cytoplasmic and nuclear extracts, cells were harvested, washed once with PBS and resuspended in 2 volumes of Lysis Buffer A (10 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2% NP-40, 1X Roche Protease Inhibitors, 1 U/μl NEB RNAse Inhibitors). After 20 strokes with a dounce homogenizer, cells were centrifuged 15 min at 3750 rpm. The supernatant (representing the cytoplasmic extract) was collected and the pellet (corresponding to crude nuclei) was washed twice with PBS and re-suspended in 2 volumes of Buffer C (420 mM NaCl, 0.1% NP40, 0.5 mM DTT, 20 mM HEPES–KOH pH 7.9, 2 mM MgCl2, 0.2 mM EDTA and 20% glycerol, 1X Roche Protease Inhibitors, 1 U/μl NEB RNAse Inhibitors). The suspension was rocked 1 h at 4°C, then ultracentrifuged at 33 000 rpm for 1 h and the supernatant representing the nuclear extract was collected and quantified.
For the sub-cellular fractionation into cytosol, nucleosol and chromatin, cell lysates were obtained according to the protocol described in (68). Briefly, cells were resuspended in Buffer A (10 mM HEPES, [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT and 1X Roche Protease Inhibitors) complemented with Triton X-100 (0.1%) and incubated for 5 min on ice, followed by low-speed centrifugation (4 min, 1300 × g, 4°C). The cytosolic fraction (supernatant) was further clarified by high-speed centrifugation (15 min, 20 000 × g, 4°C) to remove cell debris and insoluble aggregates. Nuclei (pellets) were washed once in Buffer A and then lysed in Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1X Roche Protease Inhibitors as described above) for 30 min at 4°C. Insoluble chromatin was collected by centrifugation (4 min, 1700 × g, 4°C), washed once in Buffer B, and centrifuged again under the same conditions. The final chromatin pellet was resuspended in SDS-containing Buffer and sonicated.
For subsequent small RNA analysis from the same fractions, the chromatin pellet was washed with Buffer C, resuspended in Ripa Buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% SDS, 1% Triton, 1 mM EDTA, 0.1% Na deoxycholate) and centrifuged for 5 min at 13 000 rpm, at 4°C.
Protein co-immunoprecipitation and western blot analysis
Experiments of protein co-IP were performed starting from 1 to 2 mg of whole cell extract in Ripa Buffer supplemented with fresh PMSF and 1× Roche protease inhibitors. When using nuclear extracts as input, 1–2 mg nuclear extracts were diluted with Ripa Buffer lacking NaCl, in order to decrease NaCl concentration to 150 mM. The extracts were pre-cleared 3 times with 50 μl of Dynabeads protein G (Thermo Fisher Scientific) and then the specific antibody was added and incubated overnight on a rotating wheel at 4°C. The following day, 50 μl of Dynabeads protein G pre-equilibrated in PBS supplemented with 0.5% BSA, were added to the extract and incubated for 2 h on a rotating wheel at 4°C. Beads were then washed 3 times with Ripa Buffer and then bound proteins were eluted by incubation with LSD Sample Buffer (NuPAGE) supplemented with 100 mM DTT, at 95°C for 5 min. Samples were then loaded on SDS-PAGE for subsequent WB analysis.
In-gel digestion of immunoprecipitated proteins
In gel digestion of gel-separated proteins with Trypsin, prior to MS analysis, was carried out as previously described (69). After digestion and extraction from the gel pieces, the digested peptides were desalted and concentrated by reversed-phase chromatography onto micro-column C18 Stage Tips (70). Peptides were then eluted from the tips with high organic solvent (80% ACN, 0.5% acetic acid), lyophilized, re-suspended in 1% TFA and subjected to LC–MS/MS analysis.
Liquid chromatography and tandem mass spectrometry (LC–MS/MS)
Peptide mixtures were analyzed by online nano-flow liquid chromatography tandem mass spectrometry using an EASY-nLC™ 1000 (Thermo Fisher Scientific) connected to a quadrupole-Orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific) through a nanoelectrospray ion source. The nano-LC system was operated in one column set up with a 25-cm analytical column (75 μm inner diameter, 350 μm outer diameter), packed with C18 reversed-phase resin (ReproSil, Pur C18AQ 1.9 μm, Dr Maisch, Germany) configuration. Solvent A was 0.1% formic acid (FA) and 5% ACN in ddH2O and solvent B was 80% ACN with 0.1% FA. Peptides were injected at a flow rate of 500 nl/min and separated with a gradient of 5–40% solvent B over 90 min, followed by a gradient of 40–60% for 10 min and 60–80% over 5 min at a flow rate of 250 nl/min. The Q-Exactive was operated in the data-dependent mode (DDA). HCD-fragmentation method when acquiring MS/MS spectra consisted of an Orbitrap full MS scan followed by up to 10 MS/MS experiments (Top10) on the most abundant ions detected in the full MS scan. Mass spectrometer conditions for all experiments were as follows: full MS (AGC 3e6; resolution 70 000; m/z range 300–1650; maximum ion time 20 ms); MS/MS (AGC 17 500; maximum ion time 50 ms; isolation width 1.8 Da with a dynamic exclusion time of 20 s). Singly charged ions and ions for which no charge state could be determined were excluded from the selection. Normalized collision energy was set to 28%; spray voltage was 2.2 kV; no sheath and auxiliary gas flow; heated capillary temperature was 275°C; S-lens RF level of 60%.
Assignement of hmSILAC/SILAC peptide sequences using MaxQuant and hmSEEKER
Acquired Raw data were analyzed with the integrated MaxQuant software v.1.5.5.1 and v.1.6.0.16, using the Andromeda search engine (71,72). The January 2016 version (UniProt Release 2016_01) of the Uniprot sequence was used for peptide identification. Enzyme specificity was set to Trypsin/P, meaning that trypsin cleavage occurs also in the presence of proline, after lysine or arginine residues. In MaxQuant, the estimated false discovery rate (FDR) of all peptide identifications was set to a maximum of 1%. The main search was performed with a mass tolerance of 6 ppm. A maximum of three missed cleavages were permitted, and the minimum peptide length was fixed at seven amino acids. Carbamidomethylation of cysteine was set as a fixed modification.
To assign hmSILAC peptide sequences, we defined new modifications in MaxQuant (v1.5.5.1 and v.1.6.0.16) with the mass increment and residue specificities corresponding to the heavy versions of mono-methylated K/R, di-methylated K/R, and tri-methylated K. Additionally, we defined new variable modifications for heavy methionine (Met4) and oxidized heavy methionine (Met4ox). To reduce the complexity of the search, and given the computational resources available, each experimental set of raw data was analysed three times using three distinct sets of variable modifications, namely: (1) Met4, Met4ox, oxidation, mono-methyl-K/R, mono-methyl4-K/R; (2) Met4, Met4ox, oxidation, di-methyl-K/R, di-methyl4-K/R; (3) Met4, Met4ox, oxidation, tri-methyl-K, tri-methyl4-K. Identification of high-confidence methylated sites was carried out with hmSEEKER, an in-house developed Perl pipeline that processes MaxQuant output files to find doublets of heavy and light hmSILAC peptides, by integrating the information contained in msms and allPeptides output files (73). HmSEEKER enables the retrieval of heavy/light methyl-peptide pairs whereby one of the two counterparts are not MS/MS identified. HmSEEKER performs the following steps: methyl-peptides identified in the evidence file are first filtered to remove: (i) all contaminants and decoy peptides; (ii) all peptides with single charge and (iii) all peptides bearing simultaneous heavy and light modifications. Then each peptide is associated to its corresponding MS1 peak in the allPeptides file. Finally, the H or L counterpart of each peak is searched among other peaks detected in the same raw data file. Because the pair is searched in msmsScans, hmSEEKER can find doublets even when one of the two counterparts has not been MS/MS sequenced, thus not appearing in the msms file. Assuming that the H and L counterpart must co-elute, undergo the same ionization process and differ for a specific mass, we considered true positives the heavy/light peptide pairs that satisfied the following criteria: same charge, retention time difference between the two peptides <2 min and difference between observed and expected mass shift <10 ppm. In addition, hmSEEKER was used to automatically filter out all methyl-peptides carrying a modification with a localization probability <0.75. No Andromeda Score filtering was applied.
Analysis of identified SILAC peptides
SILAC peptide and protein quantification was performed automatically with MaxQuant (v.1.5.5.1 and v.1.6.0.16) using the default settings parameters. N-terminal acetylation of protein, methionine oxidation mono-methyl-K/R, di-methyl-K/R and tri-methyl-K were set as variable modifications in MaxQuant. Outputs from MaxQuant were manually filtered to accept proteins identified with at least two peptides of which one unique; quantified proteins were considered for further analysis only if they had ratio count (RC) ≥1.
A data analysis pipeline, written in Perl, was developed in-house to process MaxQuant output. In this pipeline, the evidence.txt file was first filtered: potential contaminants and reverse sequences were removed. No Andromeda score or PTM localization probability cut-off was imposed. For quantitative analysis, methyl-peptides ratios were normalised on protein-level H/L information within each IP experiment and the median ratio of redundant methyl-peptides was calculated. To define significantly regulated methyl-peptides upon PRMT1 KD and overexpression (OE), for each IP experiment, we calculated the mean and standard deviation of the unmodified peptidome distributions. We then calculated the average ratios and standard deviations among biological replicates for each IP experiment and used these parameters to define statistically significant regulated peptides.
Localization analyses of methylated peptides were performed using the Pfam database (http://pfam.xfam.org/), the Eukaryotic Linear Motif (ELM) resource (http://elm.eu.org/) or SMART database (http://smart.embl-heidelberg.de/smart/).
Finally, we employed an in-house developed Perl tool, named hmLINKER, to intersect the SILAC dataset and the previously acquired hmSILAC dataset. For each methyl-peptide identified in the SILAC experiment, hmLINKER checks if a peptide with the same sequence is present in the hmSILAC dataset. In case a match is not found, it then tries to validate the individual modification sites.
Total and small-RNA extraction
Total and small-RNA were prepared using mirVANA™ miRNA Isolation kit (Ambion), according to the manufacturer's specification. For the analysis of small-RNAs in distinct cellular compartments, RNA was isolated from the chromatin fraction, the nucleosol and the cytosol by using Trizol (Trizol LS Reagent, Ambion). DNAse I (Zymo Research) treatment of RNA was performed before reverse transcription. For qPCR analyses of LDC transcriptome, RNA was extracted using the RNA extraction kit (Zymo research) following manufacturer's instruction.
cDNA synthesis and qPCR
The cDNA for mRNA, pri-miRNA, pre-miRNA and miRNA profiling was produced using the reverse-transcriptase miScript II RT Kit (Qiagen) according to the manufacturer's specifications. One tenth of the reaction was used for qPCR reactions in a 7500HT Fast Real-Time PCR System. miScript SYBR Green PCR Kit (Qiagen) was used for miRNA and pre-miRNA analysis, following manufacturer's instruction, while mRNA and pri-miRNA were analyzed with FAST Sybr Green Master Mix (Life Technologies).
Primers used for qPCR
MicroRNAs were amplified using forward primers from miScript primer assays (Qiagen) and reverse universal primers from miScript SYBR Green PCR Kit (Qiagen). For precursor miRNAs analysis, we adapted the miScript Qiagen strategy also to precursor amplification using custom forward primers specific for the stem-loop sequences of each pre-miRNAs of interest and the reverse universal primers from miScript SYBR Green PCR Kit (Qiagen). For primary miRNAs, primers were designed to amplify the region upstream the first stem loop of the miRNA cluster.
DGCR8: Fwd 5′-AAAACTTGCGAAGAATAAAGCTG-3′,
DGCR8: Rev 5′-TCTGTTTAACAAAGTCAGGGATGA-3′
pri-miR-15a/16-1: Fwd 5′-GCCCTGTTAAGTTGGCATAGC-3′,
pri-miR-15a/16-1: Rev 5′-ACTGAAGTCCATTCTGTGCCC-3′
pri-miR-17-92: Fwd 5′-TGCCACGTGGATGTGAAGAT-3′,
pri-miR-17-92: Rev 5′-GGCCTCTCCCAAATGGATTGA-3′
pri-miR-301a: Fwd 5′-GTCATCAATAAGCAACATCAC-3′
pri-miR-301a: Rev 5′-CACAAAAGCATCTTGCATCGGTTG-3′
pri-miR-331: Fwd 5′-GTTACCTTCTGTTCATACCATGAC-3′
pri-miR-331: Rev 5′-GGGAGTGCTTTCAGCTCAGGTAG-3′
pre-miR-15: Fwd 5′-CCTTGGAGTAAAGTAGCAGCACA– 3′
pre-miR-16: Fwd 5′-CAGTGCCTTAGCAGCACGTA-3′
pre-miR-17: Fwd 5′-AAAGTGCTTACAGTGCAGGTAGT-3′
pre-miR-18: Fwd 5′-GGTGCATCTAGTGCAGATAGTGA-3′
pre-miR-19a: Fwd 5′-GTCCTCTGTTAGTTTTGCATAGTTG-3′
pre-miR-19b: Fwd 5′-GTTAGTTTTGCAGGTTTGCATCC-3′
pre-miR-20: Fwd 5′-GTAGCACTAAAGTGCTTATAGTGCAGG-3′
pre-miR-92: Fwd 5′-CTACACAGGTTGGGATCGGT-3′
pre-miR-301a: Fwd 5′-ACTGCTAACGAATGCTCTGAC-3′
pre-miR-331: Fwd 5′-GAGTTTGGTTTTGTTTGGGTTTG-3′
Taqman array human microRNAs
TaqMan Array Human microRNA A+B Cards (Applied Biosystems) were used for global miRNAs analysis, following manufacturer’ specifications. Data were normalized on the geometric mean of two housekeeping genes (MammU6, U6snRNA). MicroRNAs were considered significantly up/down-regulated when miRNA expression in PRMT1 KD cells relative to the control was greater/lower than 1.5-fold changes, respectively.
Small-RNA sequencing
Total RNA extracted from PRMT1 KD (sh-1) and control cells was used as input for libraries preparation with the TruSeq Small RNA Library Prep Kit (Illumina), following manufacturer's instructions. Briefly, RNA 3′ and RNA 5′ adapters were sequentially ligated to the RNA. Reverse transcription followed by PCR was used to create cDNA constructs based on the small RNA ligated with the adapters. The resulting cDNA constructs were gel-purified, eluted and concentrated by ethanol precipitation. The DNA fragment library was quantified with Bioanalyzer (Agilent Technologies) and sequencing was performed on an Illumina HiSeq2000 at 50bp single-read mode and 20 million read depth. Data analysis was performed with the IsomiRage workflow, as previously described (Mueller et al. 2014). Data normalization was performed after reads mapping, assignment and filtering. Normalization of the data was performed with a reads-per-million (RPM) normalization, using small nucleolar RNAs (snoRNA) reads in each sample as normalizer.
UV-crosslinked RNA immunoprecipitation (UV-RIP)
UV-RIP protocol was performed as previously described (74,75). Briefly, HeLa cells were harvested and UV-crosslinked with 2 cycles of irradiations at 100 000 μJ/cm2. Cells were lysed with Lysis Buffer (0.5% NP-40, 0.5% NaDeoxycholate, 1× Roche protease inhibitors mixture (04693116001 MERCK), 25 U/ml Superase-RNAse inhibitor (Thermo Fisher Scientific) in PBS for 30 min at 4°C and then treated with 30 U of Turbo DNAseI (Thermo Fisher Scientific) for 30 min at 37°C. An aliquot (10%) of DNA-digested lysates was used as input while the remaining protein extract (90%) was split in two fractions and incubated overnight at 4°C with either IgG (Millipore), anti-DGCR8 (Abcam), or anti-ILF3 (Bethyl) rabbit antibodies. For the HA-RIP the anti-HA.11 (BioLegend) mouse antibody was used. The day after, protein G dynabeads (Thermo Fisher Scientific) were added and samples rocked for additional 3 h at 4°C. Afterwards, the dynabeads were washed 4 times with Washing Buffer I (PBS supplemented with 1% NP-40, 0.5% NaDeoxycholate, 300 mM NaCl, 1× Roche protease inhibitor mix and 25 U/ml Superase-RNAse inhibitor), resuspended in RNAse-free water and treated again with Turbo DNAse I (Thermo Fisher Scientific) for 30 min at 37°C. The input material was treated in parallel in the same manner. The dynabeads were then washed 4 times with Washing Buffer II (PBS supplemented with 1% NP-40, 0.5% NaDeoxycholate, 300 mM NaCl, 10 mM EDTA, 1× Roche protease inhibitor and 25 U/ml Superase-RNAse inhibitor). Finally, the RNA was eluted from the beads with the Elution Buffer (100 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM EDTA, 500 μg Proteinase K, 0.5% SDS), for 1 h at 55°C. Beads were then pelleted, the supernatants containing the first RNA eluted fraction collected in a clean Eppendorf tube and the RNAs were extracted with the RNA-extraction kit (Zymo Research), retro-transcribed to cDNA using the miSCRIPT II RT kit (Qiagen) and analysed by qPCR, as above described. The values obtained for each immunopurification were normalized over their respective input material and plotted in a histogram, as relative fold enrichment over the RNA not immuno-purified.
Immunofluorescence
Cells were plated on glass coverslips, fixed in 4% paraformaldehyde for 20 min at room temperature (RT) and permeabilized with PBS-0.1% Triton X-100 for 2 min on ice. Subsequently, cells were initially incubated with PBS-2% BSA for 30 min at RT and then with the following antibodies (diluted in PBS-2% BSA) for 2 h: anti-EWS (5μg/ml); anti-TAF15 (1:500). After being washed, cells were stained with the respective Alexa Fluor 488 secondary antibody (Molecular Probes, Eugene, OR, USA) diluted 1:400 in PBS–2% BSA for 1 h at RT. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were obtained with a Leica TCS SP2 (Leica Microsystems, Heerbrugg, Switzerland).
RESULTS
In-depth characterization of the LDC methyl-proteome
To obtain in-depth annotation of the methylations occurring on the LDC, we combined the affinity-enrichment of the complex with heavy methyl (hm)SILAC-labeling and MS-proteomics. Methionine is an essential amino acid that, in the cell, is the precursor of S-adenosyl methionine (SAM), the sole donor of methyl-groups in enzymatic methylation reactions. In a hmSILAC context, cells are grown in media containing the heavy isotopic variant of methionine, 13CD3-methionine (Met4). Upon uptake, Met4 is intracellularly converted to 13CD3-SAM, which serves as donor of heavy methyl-groups for lysine- (K) and arginine- (R) methyltransferases so that substrate proteins become heavy methyl-labelled. When coupled to high resolution MS, this strategy allows distinguishing with high confidence in vivo methylation events on R and K from false positive identifications, such as chemical methylation, or amino acid substitutions that are isobaric to this modification (76). In fact, in the MS spectra heavy methylated peptides result as isotopic peptide-pairs, where the heavy and the light peaks differ of unique mass-differences (deltamass, Δmass), on the basis of the number of methyl-groups added.
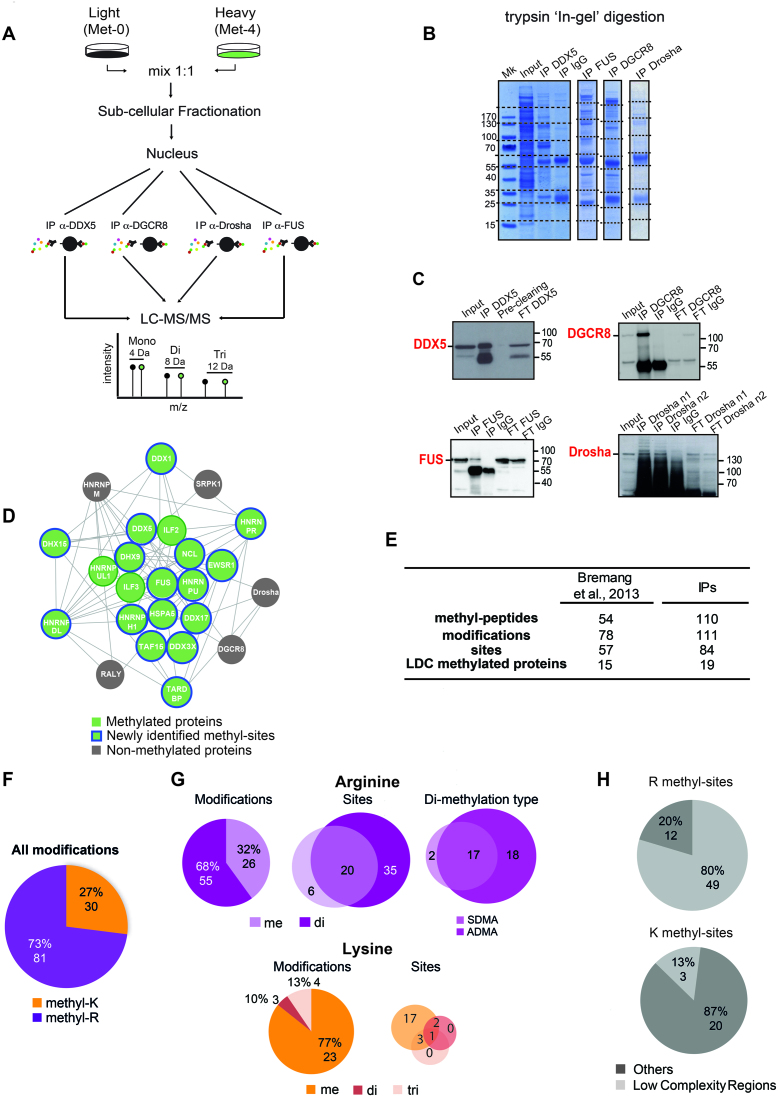
The experimental workflow designed for the characterization of the LDC methyl-proteome is summarized in Figure 1A. Based on the preferential localization of the LDC in the nucleus, the nuclear extract of hmSILAC-labeled cells was used as input for the immuno-precipitation (IP) of the complex (Supplementary Figure S1A). In order to maximize the LDC protein coverage, we performed four independent co-immunoprecipitations (co-IPs) using as bait four different LDC subunits: DDX5, DGCR8, Drosha and FUS (Figure 1B). Upon protein separation, digestion and MS analysis of the input and each IP fraction, raw data were analysed using MaxQuant algorithm (71). In the nuclear input, we identified 2808 proteins with at least two peptides of which 1 was unique and -among them- all the subunits of the LDC (Supplementary Table S1). In each IP, we retrieved 1172, 750, 564, and 1013 proteins co-purified with DDX5, DGCR8, Drosha and FUS, respectively; and each bait was efficiently precipitated and together with it, all the LDC proteins were enriched over the nuclear proteome used as input, confirming the IP efficiency (Figure 1C, Supplementary Figure S1B). All subunits were reproducibly identified in the immuno-precipitated material, with an average sequence coverage of more than 30% (Supplementary Figure S1C). The MaxQuant output data was further analysed using the hmSEEKER pipeline, in-house developed to improve the identification of genuine methyl-sites from hmSILAC MS data. HmSEEKER searches for the isotopic peptide-pairs (doublets) among all detected MS1 peaks, allowing the identification of methyl-doublets even if only one of the two peak-counterparts is sequenced at the MS2 level (73). Through this analytical pipeline, we identified with high confidence a total of 432 methylated peptides, of which 110 belong to the LDC subunits (Supplementary Table S2), thus significantly extending our previous annotation of the LDC methyl-proteome (30). The modified peptides correspond to 84 distinct R- and K- sites in total, associated to 19 out of 24 subunits of the LDC (Figure 1D and E). Notably, the majority (73%) of these modifications occurs on R-residues, of which 32% are mono- and 68% di-methylated (Figure 1F and G). In total, 61 distinct R-sites are modified, of which 35 are exclusively di-methylated, 6 only mono-methylated and 20 both mono- and di-methylated (Figure 1G). Since arginine can be either asymmetrically (ADMA) or symmetrically (SDMA) di-methylated, we discriminated between these two di-methylation types by MS, by visual inspection of the neutral loss of heavy-dimethylamine [NH(13CD3)2] and heavy-monomethylamine (NH213CD3), generated in MS2 spectra when ADMA and SDMA are present in the peptide sequence, respectively (77). We found that 35 R-methyl-peptides produced the neutral loss of heavy di-methylamine, while 19 generated the neutral loss of heavy–mono-methylamine; this indicates that the LDC complex is predominantly asymmetrically di-methylated (Figure 1G). Further individual IPs of four selected proteins of the LDC, followed by WB profiling using anti-pan-methyl-R antibodies confirmed that they exist mainly in the R-mono- and asymmetric R-di-methylated forms, while symmetrical di-methylated arginines are less represented (Supplementary Figure S1D). Interestingly, only 23 lysines of the LDC resulted methylated, of which 77% are mono-methylated, 10% di-methylated and 13% tri-methylated. All together 17 K-sites are exclusively mono-methylated; 2 K-sites can exist as both mono- and di-methylated; 3 K-sites are mono- and tri-methylated and only 1 K-site exists in all the 3 methylated forms (Figure 1G). Overall, the data indicate a prevalence of arginine methylation within the regulatory Microprocessor-associated proteins, while neither of the two subunits of the core catalytic complex, Drosha and DGCR8, result modified.
Figure 1.
Characterization of the LDC methylation pattern by MS. (A) Workflow of the hmSILAC/co-IP approach applied to characterize the LDC methyl-proteome. The methylation degree can be identified based on the mass difference between the light and heavy peptide (4 Da = mono-methylation, 8 Da = di-methylation, 12 Da = tri-methylation). (B) Coomassie-stained gels of the immuno-precipitated material in-gel digested with trypsin and the analyzed by LC–MS/MS; dashed lines correspond to the individual gel slices processed and MS- analyzed. Mk = protein molecular marker. (C) Validation of the IP efficiency of the four LDC proteins used as baits, by WB analysis. IP with Immunoglobulin G (IgG) was used as negative control. FT = IP flow-through. Pre-clearing = unspecific proteins that bind magnetic beads when incubated with cell extract, prior to the IP. (D) Graphical representation of the LDC complex using Cytoscape with methylated LDC proteins displayed in green, non-methylated LDC proteins in grey; blue circles indicate proteins bearing the newly annotated methyl-sites. (E) Summary of the annotated LDC methyl-proteome, compared to the one previously published (30). Methyl-peptides: number of identified peptides harboring one or multiple methylation events; modifications: total number of mono-, di- and tri-methylation events occurring on both K- and R- residues identified by hmSILAC/co-IP; sites: number of methylated residues; LDC methylated proteins: number of LDC proteins found methylated by hmSILAC/co-IP. (F) Summary of all arginine (R) and lysine (K) methylations on the LDC. The number of identified modifications and the corresponding percentage are reported. (G) Upper panel left: summary of mono- and di-methyl-arginines identified by hmSILAC/protein IP. Upper panel middle: Venn diagram of the R-sites identified as either mono- or di-methylated. Upper panel right: Venn diagram of di-methylated peptides carrying either asymmetrically di-methylated arginine (ADMA) or symmetrically di-methylated arginine (SDMA). Lower panel left: summary of the mono-, di- and tri-methyl-lysines identified by hmSILAC/co-IP. Lower panel right: Venn diagram of the K-sites identified as mono-, di- and tri-methylated. The number of identified modifications and the corresponding percentage are reported. (H) Localization analysis of the R-methyl-sites (upper panel) and K-methyl-sites (lower panel) identified according to the SMART domain database (113). The number of identified modifications and the corresponding percentage are reported.
Interestingly, 80% of the annotated R-methylated sites occur within Low Complexity (LC) regions (Figure 1H), which have been often described to be involved in protein-protein and protein-RNA/DNA interactions (78–80). Conversely, 87% of K-methylated sites are located outside of LC regions. This suggests that R-methylation may affect either the protein composition of the complex or the binding of the Microprocessor-associated proteins with substrate miRNAs.
PRMT1 down-regulation impairs global miRNA expression
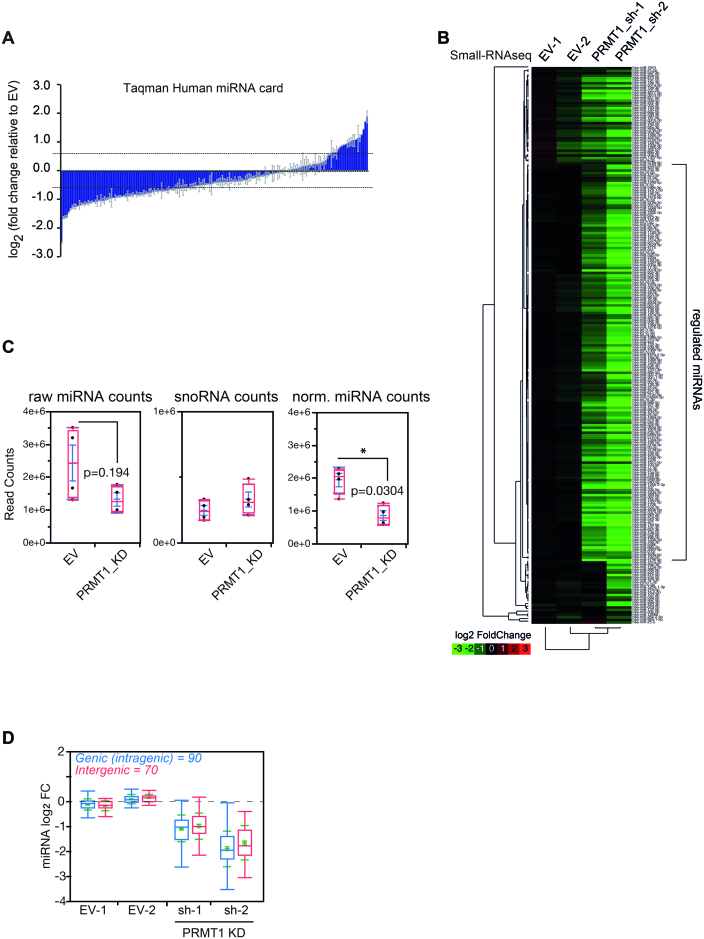
To assess whether asymmetric R-di-methylation regulates the LDC activity, we investigated if changes of this PTM could affect miRNA biogenesis. Focusing on PRMT1 in light of its predominant activity within type-I PRMT family, we generated knockdown (KD) HeLa cells for this enzyme by RNA interference using two distinct short hairpin RNAs (shRNAs) inserted in lentiviral constructs. Reduction of the protein level was observed upon cell transduction with the two shRNAs (Supplementary Figure S2A). Up to 96 h post-infection, the growth of PRMT1-depleted cells was similar to control cells, transduced with the empty vector (EV) (Supplementary Figure S2B), whereas at later time points KD cells showed a progressive growth reduction until full arrest, in accordance with the reported embryonic lethality due to PRMT1 loss, in mice (49). To investigate the effect of PRMT1 depletion on miRNA expression levels, we first monitored the levels of the two clusters miR-15a/16 and miR-17–92 (81,82), whose deregulation was already mechanistically linked to the lack of some Microprocessor-associated proteins (83–85). We observed the down-regulation of the mature miRNAs from both clusters, with this effect peaking at 72 h post-infection (Supplementary Figure S2C). We then globally profiled miRNA levels in control and PRMT1 KD cells, 72 h post-infection, by both Small-RNAseq and quantitative PCR using the Taqman human miRNA cards (Supplementary Tables S3 and S4). We observed a pervasive miRNA down-regulation, with only a minor proportion of unchanged or up-regulated miRNAs (Figure 2A, B and Supplementary Figure S2D). This effect on miRNAs was not mirrored by a change in small nucleolar RNAs (snoRNAs) (Figure 2C), suggesting that PRMT1 depletion does not impact on other cellular ncRNAs.
Figure 2.
PRMT1 down-regulation impairs mature miRNA expression, globally. (A) Global expression analysis of miRNAs upon PRMT1 depletion by PRMT1 sh-1, performed with TaqMan Array Human miRNA Card. Data are displayed as log2 fold changes and are normalized on the geometric mean of a panel of housekeeping genes (mammU6, RNU44, RNU46, U6snRNA). Histograms represent mean ± SEM of miRNAs quantified in 2 technical replicates. Regulated miRNAs were considered significant when their fold change was greater/lower than 1.5. (B) Heat map displaying log2 fold changes of miRNAs identified in control (EV) and PRMT1 KD achieved with the sh-1 and sh-2 shRNA constructs. Data are normalized over the small nucleolar RNAs. The average of 2 technical replicates is shown for each sample. (C) Box-plot of the distribution of miRNA and small nucleolar RNA (snoRNA) raw counts, and of miRNA normalized over snoRNA counts. Statistical analyses were performed using non-parametric Wilcoxon test. (D) Box-plot shows the distributions of intragenic and intergenic miRNAs upon PRMT1 KD, obtained from miRiad (114). Only guide miRNAs were considered for the analysis.
To understand whether PRMT1 affects the biosynthesis of specific classes of miRNAs, we first focused on the analysis of intronic and intergenic miRNAs, whereby the former derive from the processing of introns and require spliceosomal components for their biogenesis, while the latter are transcribed as independent transcription units (86,87). Observing no differential expression between the two miRNA types in PRMT1 deficient cells (Figure 2D), we concluded that the reduction in miRNA processing does not depend on their genomic origin. Second, we analyzed the effect of PRMT1 on the positioning and orientation of Drosha cleavage, elaborating on the fact that miRNAs can originate from the 5″ (5p) or 3″ (3p) arm of the pri-miRNA hairpin depending on Drosha activity (16). The observation that 5p and 3p miRNAs are equally regulated suggests that PRMT1 depletion does not affect directly the catalytic activity of Drosha towards one of the two arms (Supplementary Figure S2E).
PRMT1 depletion impairs the processing of primary-to-precursor miRNAs
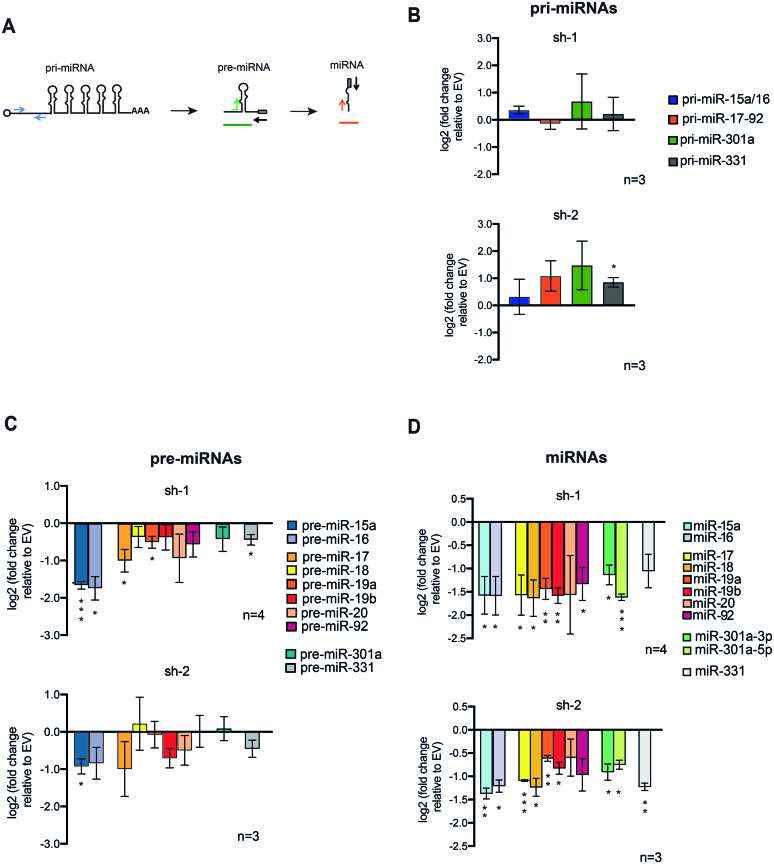
The primary function of the LDC is to cleave nuclear pri-miRNAs into shorter pre-miRNAs. Thus, we sought to investigate whether the global miRNA down-regulation observed upon PRMT1 depletion is caused by the specific impairment of this catalytic step. We designed quantitative real time PCR (qPCR) primers that allow distinguishing pri- from pre- and mature miRNAs (Figure 3A). We analysed not only miR-15a/16 and miR-17–92 clusters, whose mature levels where previously assessed, but also two of the most down-regulated miRNAs, miR-301a and miR-331, based on the Small-RNAseq and Taqman analyses (Supplementary Figure S3A). The levels of the four pri-miRNAs were unchanged, or even increased, upon PRMT1 depletion (Figure 3B), whereas pre- and mature miRNAs were significantly reduced (Figure 3C and D). To corroborate this evidence, we also profiled the pri- and pre-miRNAs from the miR-15a/16 and 17–92 clusters upon further fractionation of nuclei into chromatin and nucleosol, since pri-miRNAs are more associated to chromatin, while pre-miRNA are released in the nucleosol prior to their cytoplasmic export. By profiling pri-miRNAs in the chromatin fraction and pre-miRNAs in the nucleosol, we confirmed that pre-miRNAs are down-regulated upon PRMT1 KD, while the corresponding pri-miRNAs are unchanged (Supplementary Figure S3B).
Figure 3.
The processing of primary-to-precursor miRNAs is impaired upon PRMT1 depletion. (A) Schematic representation of the qPCR primers designed for the selective detection of pri-, pre- and mature miRNAs. The arrows indicate the binding region of the primers used to amplify by qPCR each miRNA isoform. The grey rectangle indicates the binding region of the Qiagen universal reverse primer (represented by a black arrow) which was used to amplify the same 3′end region of pre- and mature miRNAs, according to the vendor's protocol. (B) qPCR profiling of pri-miRNAs of miR-15a/16, miR-17–92, miR-301a and miR-331, carried out upon PRMT1 KD using sh-1 and sh-2 constructs in HeLa cells. Analyses were performed 72 h post infection. Histograms represent mean ± SEM of the log2 fold change of PRMT1-depleted cells over EV control cells from 3 biological experiments (n = 3). Statistical analysis was performed using the one sample T-test. * = Values with P-value < 0.05. (C) qPCR profiling of the pre-miRNAs originating miR-15a/16, miR-17–92, miR-301a and miR-331 upon PRMT1 KD with sh-1 and sh-2 in HeLa cells. Analyses were performed 72 h post infection. Histograms represent the mean ± SEM of the log2 fold change over EV control cells from the averages of four biological replicates for sh-1 (n = 4, upper panel) and three biological replicates for sh-2 (n = 3, lower panel). Statistical analysis was performed using the one sample t-test. * = Values with P-value < 0.05. ** = values with P-value ≤0.01 and *** = values with P-value ≤0.001. (D) qPCR profiling of mature miRNAs derived from miR-15a/16 and miR-17–92 clusters as well as miR-301a-5p, miR-301a-3p and miR-331 upon PRMT1 KD with sh-1 and sh-2 in HeLa cells. Analyses were performed 72 h post infection. Histograms represent the mean ± SEM of the log2 fold change of KD over EV control cells. The results are the average of four biological replicates for sh-1 (n = 4, upper panel) and 3 biological replicates for sh-2 (n = 3, lower panel). Statistical analysis was performed using the one sample t-test. * = Values with P-value < 0.05. ** = values with P-value ≤0.01 and *** = values with P-value ≤0.001.
Overall, these findings demonstrate a mechanistic link between PRMT1 expression levels and the reaction specifically catalyzed by the LDC.
Modulation of PRMT1 strongly affects the methylation state of the Microprocessor-associated proteins
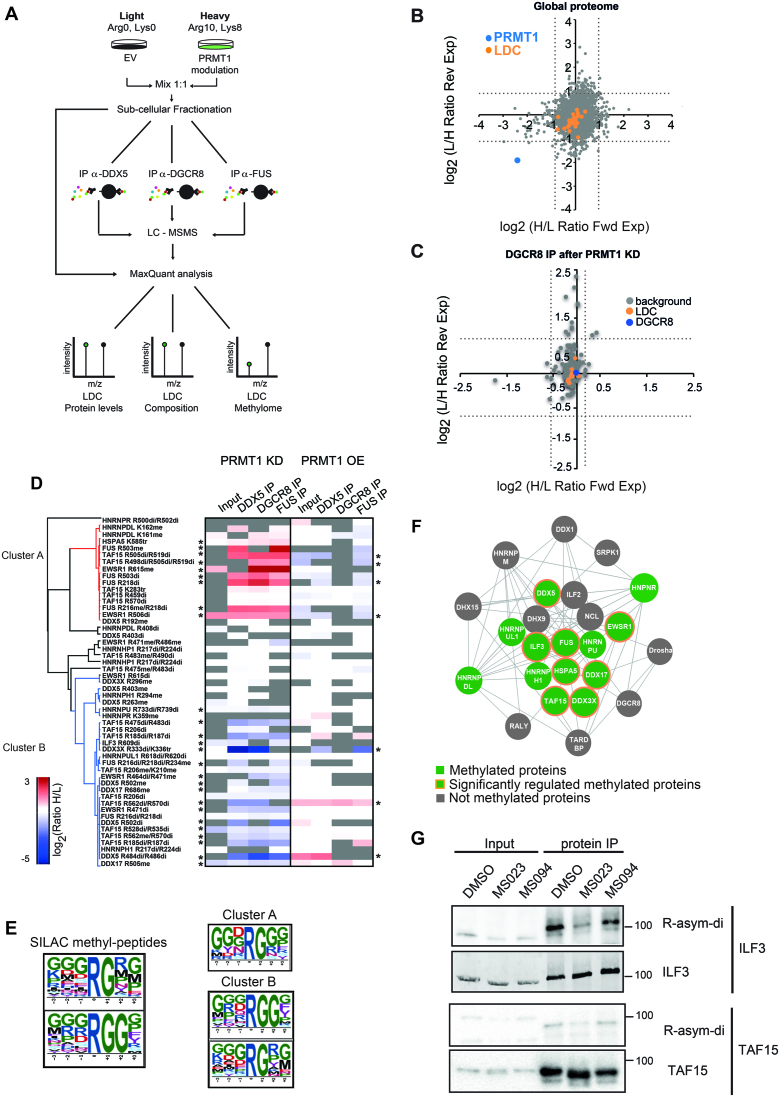
To gain insights in the mechanism of PRMT1 regulation of the LDC activity, we characterized the expression, composition and methylation state of the complex by combining SILAC-based proteomics with the modulation of PRMT1 expression levels (Figure 4A). We first chose the optimal time-point to carry out these analyses, profiling by WB the global level of R mono- and asymmetric di-methylated proteins in PRMT1 KD and overexpressing (OE) cells, compared to the respective controls (Supplementary Figure S4A, C). When PRMT1 was depleted, we observed a reduction in the level of asymmetric di-methyl arginine (ADMA), mirrored by an increase of mono-methyl arginine (MMA) over time, with a peak between 72 and 96 h after cell infection (Supplementary Figure S4A), in line with previous studies (88). On the contrary, when PRMT1 was overexpressed, we observed a positive correlation between the enzyme levels and that of ADMA and MMA, starting from 48 h, compared to EV control cells (Supplementary Figure S4C). Overall, both PRMT1 KD and OE cells showed the strongest changes in global protein methylation 72 h upon modulation of PRMT1 expression. Interestingly, the timing of these changes corresponds to the one when the peak of miRNA down-regulation was observed (Supplementary Figure S2C). Hence, 72 h post infection was chosen as the time point for the MS-proteomics experiment (Figure 4A). HeLa cells were metabolically labeled with either the light or the heavy isotopic variants of lysine and arginine (Arg0 and Lys0, and Arg10 and Lys8 for the light (L) and heavy (H) channels, respectively). In the ‘Forward’ setting, EV control cells were cultured in the L medium and PRMT1 KD/OE cells in the H medium, while in the ‘Reverse’ experiment, the L and H channels were swapped. Once harvested and mixed in 1:1 ratio, cells were fractionated into nuclear and cytoplasmic extracts (Supplementary Figure S4B and D) and a minor fraction of the nuclear extract was directly subjected to MS analysis for protein profiling of LDC (Figure 4A). The rest was used as input for the LDC immuno-enrichment, carrying out three co-IPs in parallel, using DDX5, DGCR8 and FUS as baits (Supplementary Figure S4B and D). Each IP was performed in three biological replicates.
Figure 4.
PRMT1 does not change LDC expression and composition but modulates LDC methylation. (A) Workflow of the experimental setup designed for proteomics analysis of LDC expression and composition, upon PRMT1 modulation. Co-IP of the LDC using nuclear extracts from a 1:1 mix of SILAC-labelled HeLa cells, depleted of- or overexpressing PRMT1 (KD, sh-1 and PRMT1 OE), or infected with the EV (control). Each immuno-enrichment of the complex with individual baits was carried out in three independent biological replicates (n = 3), with SILAC channels swapping (Forward and Reverse experiments). (B) LDC expression analysis: scatterplot of all proteins identified and quantified in the SILAC nuclear inputs of HeLa cells, depleted or not of PRMT1. The scatterplot displays the log2 SILAC distribution of the quantified proteins in the Forward (Fwd) and Reverse (Rev) experiments (H/L ratio for the Fwd and L/H ratio for the Rev experiment). The dot corresponding to PRMT1 is highlighted in blue; LDC subunits are displayed in orange and all remaining proteins are in grey. Dashed lines indicate the μ ± 2б cut-off, calculated from the whole peptide SILAC ratio distribution in both Fwd and Rev experiments, that separate significant outliers form the unchanging population. (C) LDC protein composition analysis: scatterplots of all proteins identified and quantified in the DGCR8 co-IPs, analyzed as described in B; the dot corresponding to DGCR8 is displayed in blue; all LDC proteins are indicated in orange, the remaining proteins are in grey. Dashed lines represent the μ±2б cut-off calculated on the protein ratio distribution, and define significantly changing from unchanging proteins. (D) Heat map display of the unsupervised clustering analysis of methylated peptides identified and quantified in the SILAC/co-IP experiments upon PRMT1 KD and PRMT1 OE. Only peptides identified in at least two co-IP experiments of either the PRMT1 KD or PRMT1 OE were included in the analysis. Data are displayed as the average from three independent IPs for each bait and expressed as log2 ratio of PRMT1 KD/OE cells, compared to control cells. The heat map shows an unsupervised clustering of the data, generated using Pearson correlation and data average analysis. The type of methylation identified for each methyl-peptides is displayed (me = mono-methylation; di = di-methylation; tr = tri-methylation), as well as the respective methyl-sites. The 2 main clusters identified are indicated with differential color code (cluster A in red and cluster B in blue). * = Methylated peptides significantly regulated in the SILAC experiments upon a μ ± 1б cut-off calculated from the unmodified peptide distribution of each IP experiment. (E) Motif enrichment analysis of all the SILAC-quantified methyl-peptides using ScanX software (115). The following parameters were imposed: R central character; width = 7; occurrences = 10; significance = 0.000001 (right panel). The same analysis was performed also on the methyl-peptides identified in the cluster A and cluster B, separately (right panel). (F) LDC complex representation with Cytoscape of the proteins identified in the SILAC/co-IP experiments: proteins found methylated are displayed in green. Methylated proteins whose methylation state is significantly changing upon PRMT1 modulation are highlighted with orange circles. Non-methylated proteins are displayed in gray. (G) Immuno-enrichment of ILF3 and TAF15 from HeLa cells treated with DMSO, the MS023 inhibitor and the MS094 control compound, used at 10 μM concentration for 16 h. WB profiling using anti-ADMA, anti-ILF3 and anti-TAF15 to assess the level of di-methylation and unmodified ILF3 and TAF15 protein levels, respectively.
In the input, 945 nuclear proteins were identified in all the three biological replicates, with at least two peptides of which one was unique (Supplementary Table S5). Interestingly, the SILAC ratios of the nuclear proteome upon PRMT1 KD did not indicate significant differences in the expression levels of the LDC subunits, compared to EV control (Figure 4B), a result that was further confirmed by WB analysis of various LDC proteins carried out on whole cell lysate of PRMT1 depleted cells (Supplementary Figure S5A). In line with these results, no major changes were detected in the mRNA expression of most LDC subunits, except for HNRNPR, HNRNPDL, SRPK1 and TAF15, whose transcript levels appeared slightly reduced upon PRMT1 knockdown, a change which was however not statistically significant and not mirrored by a corresponding decrease of the respective protein level (Supplementary Figure S5B and Table S5).
The stability of the LDC protein levels upon PRMT1 depletion also correlated with the evidence that the overall composition of the complex, independently evaluated upon DGCR8, DDX5 and FUS co-IPs, remained substantially unaltered (Figure 4C and Supplementary Figure S5C). We profiled 231, 162 and 210 proteins in the DDX5, DGCR8 and FUS co-IPs respectively (Supplementary Table S5) and found that the SILAC ratios of the LDC proteins within each co-IP were similar to 1, indicating that they are co-immuno-precipitated with the same efficiency upon PRMT1 depletion, even using different baits. In light of the lack of effect of PRMT1 inhibition on LDC expression and composition, we hypothesized that the PRMT1-dependent impairment of miRNA biogenesis could be due to the alteration of the complex methylation state.
We therefore analysed the methyl-proteome of the LDC upon PRMT1 KD and OE by quantitatively profiling the methyl-peptides by SILAC, in both the nuclear extract and the immuno-enriched complex. For the analysis of the LDC methylation state, we considered only methyl-peptides which were reproducibly identified in at least two out of three co-IPs (Figure 4D) and identified 52 methyl-peptides displaying differences between PRMT1-modulated and control cells, of which 26 were statistically significant (Figure 4D and Supplementary Table S6). The number of regulated methyl-peptides is higher in the KD than in the OE cells, probably due to the fact that the basal activity of the endogenous PRMT1 is already saturating the methylation level for most of the R-sites of the Microprocessor-associated proteins, which are therefore not free for further modification by the mild overexpression of the exogenous enzyme. In line with this, we did not observe significant changes in miR-15a and miR-17 levels upon PRMT1 overexpression, confirming the hypothesis that the activity of the endogenous PRMT1 may be already saturating the LDC methylation sites. Hence, the upregulation of the enzyme (two/three-fold compared to endogenous PRMT1) does not further boost the LDC activity, in this model system (Supplementary Figure S6A).
The regulated methyl-sites are high-quality identifications, since 78% was orthogonally-validated by hmSILAC (Supplementary Figure S6B, upper part). Interestingly, the number of hmSILAC-validated methyl-peptides increases when only the significantly regulated peptides are considered (92%) (Supplementary Figure S6B, lower part), which confirms on the one hand that the dynamic change of methyl-peptides is a good indicator of in vivo methylation and, on the other hand, that our high-confidence methyl-proteome is not yet saturated.
Unsupervised clustering analysis of the PRMT1-dependent LDC methyl-proteome showed the existence of two methyl-peptide clusters. Cluster A collects peptides that are hyper-methylated upon PRMT1 depletion and hypo-methylated in PRMT1 overexpressing cells. Among these peptides, we found seven di-methylated peptides, three mono-methylated peptides and one peptide carrying one di-methyl and one mono-methyl site, and which may be, in principle, targets of other PRMTs responsible of the scavenging effect in absence of the major PRMT, as previously described (88). However, upon in-depth inspection, we observed that two out of four up-regulated mono-methylated sites (corresponding to R216 of FUS and R615 of EWSR1) were down-regulated in their di-methylated version. This could be explained by the conversion of di- to mono-methylation in the absence of PRMT1 activity, as result of higher turnover of the modification by a yet uncharacterized arginine-de-methylase. This is an interesting observation, which could represent one of the very first evidence of R-de-methylation in vivo.
Cluster B includes peptides hypo-methylated in PRMT1 KD cells and whose methylation increased when PRMT1 was upregulated (Figure 4D and Supplementary Table S6). The fact that the majority of these peptides are di-methylated and that their levels positively correlate with PRMT1 expression suggests they are direct targets of the enzyme. Motif enrichment analysis of all significantly regulated methyl-peptides displayed a strong enrichment of both the RGG and RG motifs, where the former is more specific for PRMT1 and the latter is a more general target sequence for the whole PRMT family (47,89) (Figure 4E, left panel). However, when we carried out the motif analysis separately on the peptides from the two clusters, cluster B displayed the enrichment of both motifs, whereas cluster A showed the enrichment of the sole RG motif (Figure 4E, right panel and Supplementary Table S6). The observation that cluster B displays both the expected trend of variation in dependence of PRMT1 and the RGG consensus motif, more specific for PRMT1, corroborate the hypothesis that the corresponding methyl-sites are genuine PRMT1 targets. On the contrary, the peptides in cluster A, hyper-methylated upon PRMT1 depletion and enriched for the more generic consensus sequence, may be substrates of other PRMTs when the major enzyme of the family is lacking. In cluster B, we found DDX17, DDX3X, DDX5 and HNRNPH1 proteins, hereby considered novel PRMT1 targets (Supplementary Figure S6C). Overall by quantitative proteomics we identified and characterized the exact methyl-sites of 13 subunits of LDC, 8 of which are also significantly modulated in dependence of PRMT1 (Figure 4F).
To confirm the MS-data, we used the type-I PRMTs inhibitor MS023 and its homologous inactive counterpart MS094 (90). Treatment with MS023 led to a reduction of ADMA and a parallel increase of MMA, while MS094 did not induce methyl-proteome changes (Supplementary Figure S6D). When we carried out protein-IP of ILF3 and TAF15, followed by WB profiling with pan-R-methyl antibodies we observed that the basal di-methylation level of both proteins was reduced upon MS023, while remained unaltered by MS094, in agreement with the proteomic data (Figure 4G). With the same approach, we could confirm that DDX17 is solely R-mono-methylated and this modification is slightly reduced by MS023 (Supplementary Figure S6E).
These results collectively show that LDC methylation is significantly affected by the modulation of PRMT1 expression or catalytic activity and hint towards a key role of this modification in regulating the function of the complex and, consequently, miRNA biosynthesis.
Inhibition of PRMT1 catalytic activity impairs miRNA biogenesis by reducing the interaction of the Microprocessor-associated proteins with target pri-miRNAs
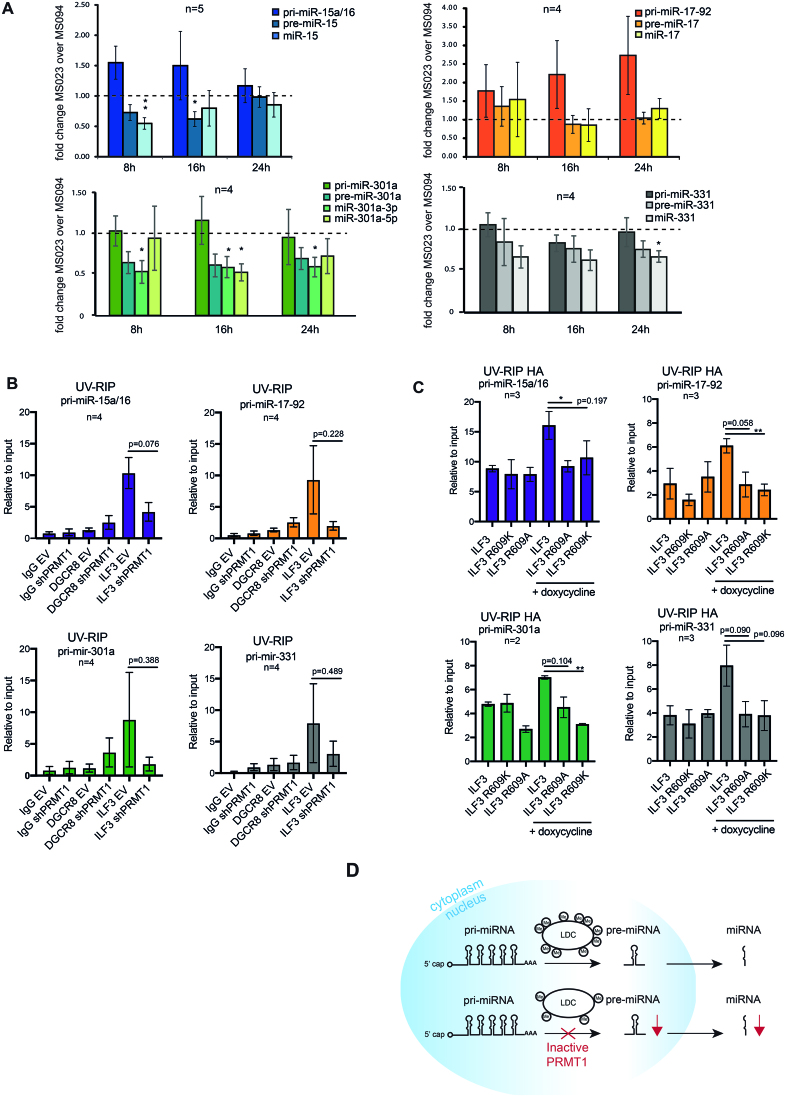
To confirm that PRMT1 enzymatic activity, rather than its expression, is crucial for the correct processing of miRNAs, we assessed the levels of the intermediate products of miRNA biogenesis upon HeLa cells treatment with MS023 and MS094. A time-course inhibition experiment showed that MS023 exerts its effect on global ADMA/MMA levels already 8 hours upon drug administration (Supplementary Figure S6F). We thus profiled the expression of primary, precursor and mature forms of the previously profiled miRNAs in cells treated with the drugs at different time points and observed that the levels of pre- and mature miRNAs were reduced while the pri-miRNAs were unchanged at 8 and 16 h post inhibition (Figure 5A). This confirms that the specific impairment of the pri-to-pre-miRNA processing step is due to the inhibition of the LDC catalytic activity.
Figure 5.
Pharmacological inhibition of PRMT1 inhibits the pri-to-pre miRNA processing step and affects ILF3 interaction with pri-miRNA targets. (A) qPCR analysis of pri-, pre- and mature miRNAs from the miR-15a/16 and miR-17–92 clusters and miR-301a and miR-331 transcripts, upon treatment with MS023 and MS094 (10 μM) in HeLa cells. Histograms represent the mean ± SEM of the fold change of MS023 over MS094 treated cells, calculated from five biological replicates for pri-, pre- and mature miR-15a/16 (n = 5) and four biological replicates for the other displayed pri-, pre- and mature miRNAs (n = 4). Statistical analysis was performed using the one sample t-test. * = Values with P-value < 0.05. ** = values with P-value ≤0.01. The dashed line is set to 1 which is the theoretical level in which the expression of the transcripts is equal in both MS023 and MS094 treated cells. (B) UV-RIP of ILF3 and DGCR8 in HeLa cells expressing either the empty vector (EV) or the shRNA specific for PRMT1 (sh-1). IgG were used as mock control for the IP. The data shown are the average of 4 biological replicate experiments (n = 4) and are represented as ratio over the input, thus indicating the fold enrichment. UV-RIP of ILF3 with pri-miR-15a/16 and pri-miR-17–92 are shown in the upper bar-graphs; while the one with pri-miR-301a and pri-miR-331 are in the bottom bar graphs. Statistical analysis was performed using non-parametric, 2-tailed t-Test. * = Values with a significant P value < 0.05. (C) UV-RIP of HA-ILF3 wild type, HA-ILF3 R609A and HA-ILF3 R609K mutants, which were expressed in HeLa cells upon doxycycline administration. UV-RIP of HA-tagged proteins with pri-miR-15a/16 and pri-miR-17–92 are displayed in the upper bar-graphs, while the UV-RIP with pri-miR-301a and pri-miR-331 are shown in the bottom bar-graphs. The data shown are the average of three biological replicate experiments (n = 3), except for pri-miR-301a (n = 2), and are represented as ratio over the input, thus indicating the fold enrichment. Statistical analysis was performed using non-parametric, two-tailed t-Test. * = Values with a significant P value < 0.05. ** = values with P-value ≤0.01. (D) Model of the regulatory role of PRMT1 in miRNA biogenesis, through the methylation of the Microprocessor associated proteins.
Most of the LDC proteins identified as PRMT1 substrates by proteomics are RBPs. Moreover, their methylations occur within the RGG-rich sequences comprised in low complexity regions known to be involved in protein-RNA interactions (91). Thus, we hypothesized that PRMT1-dependent R-methylation could modulate the interaction of some of the Microprocessor-associated proteins with their pri-miRNA targets. We focused on ILF3 because this protein emerged as a genuine PRMT1 target by MS, with a single di-methylated site on R609 which was significantly down-regulated upon PRMT1 KD, and orthogonally validated by hmSILAC (Supplementary Figure S7A). ILF3 is predominantly expressed in the nucleus of HeLa cells and its cellular distribution was not affected by PRMT1 depletion (Supplementary Figure S7B). We set up an experiment of UV-crosslinking followed by RNA-immunoprecipitation (UV-RIP) using ILF3 and DGCR8 as bait, in control and PRMT1 KD cells, and assessed their binding to the pri-miRNA-15a/16, pri-miRNA-17–92, pri-miRNA-301a and pri-miRNA-331. Both ILF3 and DGCR8 specifically bind to all pri-miRNAs assessed. However, ILF3 binding to the pri-miRNAs is strongly reduced upon PRMT1 depletion, while DGCR8 binding is increased (Figure 5B). The observation of the reduced binding of ILF3 to all the tested pri-miRNAs in dependence of R-methylation was corroborated by the generation of isogenic inducible HeLa Flp-In T-REx cells expressing either the HA-tagged wild-type version of ILF3 or the respective R609A and R609K mutants that both mimic the absence of asymmetric R-di-methylation. First, we confirmed that both the exogenous wt and mutants ILF3 show the same cellular distribution of the endogenous one, with a predominant expression in the nucleus (Supplementary Figure S7C). Then, we observed that wild-type HA-ILF3 binds specifically to the pri-miRNA-15a/16, pri-miRNA-17–92, pri-miRNA-301a and pri-miRNA-331, with an affinity similar to the endogenous protein, while both mutants display a reduced binding (Figure 5C).
Overall, these data demonstrate that R-methylation of ILF3, which is targeted by PRMT1 in HeLa cells, directly modulated its interaction to the pri-miRNA substrates, likely disturbing their processing by the Microprocessor complex and, hence, their biogenesis. While it remains to be assessed whether this regulatory mechanism applies also to other Microprocessor-associated proteins, the data collected so far allow formulating a model whereby PRMT1 regulates miRNA biogenesis through the extensive R-methylation of the Microprocessor-associated proteins, which probably regulates the loading of pri-miRNAs into the complex. Consequently, a reduction of the activity of PRMT1 leads to the aberrant modification of the LDC, which ultimately affects miRNAs biogenesis (Figure 5D).
DISCUSSION
In this study, we took advantage of quantitative MS-based proteomics to investigate the extent, dynamicity and functional impact of R-methylation within the Large Drosha Complex. We initially combined MS-analysis with the affinity-enrichment of the complex to carry out a thorough characterization of its in vivo methylations. MS-analysis of protein methylation is particularly challenging both because this modification is isobaric to various amino acid substitutions and since chemical methylations introduced during sample preparation can be mis-assigned to the in vivo enzymatic modification. Thus, it has been demonstrated that label-free approaches for global analysis of methylation by MS can lead to high false discovery rate (FDR) in the absence of orthogonal validation strategies (92,93). Yet, the generation of robust and reliable methyl-sites dataset is crucial for biological and functional follow up studies. We took advantage of the hmSILAC labeling- currently considered the golden standard strategy to reduce the FDRs in global MS-based identification of methyl-sites (93)- to generate the largest high-quality LDC methyl-proteome annotated so far.
Non-histone protein methylation, likewise other PTMs, is sub-stoichiometric and thus its analysis requires enrichment steps prior to MS. This is achieved by affinity purification strategies using pan-methyl antibodies, recently developed for global protein methylation analysis (30,92,94,95). The methyl-proteomes annotated so far using this strategy indicate MMA as the most abundant methylation type present globally. While this may truly reflect the larger extent of this methylation degree over the others, it is still difficult to rule out whether, instead, it is consequence of the better performance of the anti-pan-MMA antibody (95). As a matter of fact, the quantification of the different types of methylation carried out in MEFs in the absence of affinity-enrichment steps indicated ADMA as the predominant modification (88). In line with this evidence, we found that 68% of the methyl-arginines within the LDC are di-methylated. This suggests that pan-methyl-antibodies may introduce biases in methyl-proteomics analyses and that the protein co-IP in combination with hmSILAC-MS may lead to more realistic pictures of the extent and composition of sub-methyl-proteomes.
Upon PRMT1 modulation, we observed significant changes in the methylation state of various Microprocessor-associated proteins that were thus identified for the first time as PRMT1 substrates, when their methylation state positively correlated with the enzyme level and occurred within the PRMT1-specific consensus motif RGG. In addition, our quantitative proteomic analysis also showed some peptides whose methylation was unaffected by changes of PRMT1 expression: they may be either targets of other PRMTs, or constitutively methylated peptides possibly required for the correct assembly or functionality of the complex. Another subset of peptides was up-regulated in the absence of PRMT1 and enriched in the RG-motif and this includes both mono-methylated and di-methylated peptides: these may be substrates of other PRMTs, in line with the scavenging effect previously described upon PRMT1 ablation (88). Interestingly, 2 methyl-peptides were found down-regulated in their di-methylated form and conversely up-regulated in their mono-methylated version, suggesting a putative modulation of R-methylation by an R-de-methylase which converts di-methylated peptides into mono-methylated ones in the absence of PRMT1. So far, the members of the JmjC Nϵ-methyl-lysine de-methylases (KDMs) were reported to be able to catalyze methyl-R de-methylation on both histones and non-histone proteins, such as hnRNPK and 53BP1 (96–98), even if the existence of a R-de-methylation process in cells remains controversial (99). For instance, Jmjd6, the leading member of KDMs that was proposed as R-demethylases, was reported to act as a lysine hydroxylases in cells, but its de-methylation activity on arginines could not be fully verified (100). In this context, starting from our result suggestive of the existence of a R-de-methylase activity in this model system, future experiments will be devoted to corroborate this evidence, investigating the identity of these putative R-de-methylases and assessing their role in defining the physiological methylation state of the LDC.
When analyzing the LDC methyl-proteome, we noticed that none of the identified methylation events occur in the nuclear localization signals (NLSs) of EWSR1, FUS and TAF15 that were previously reported to modulate their cytoplasm-to-nucleus translocation (61,101,102). In line with this, HeLa treatment with MS023 and MS094 did not affect the subcellular localization of these proteins, confirming that the methyl-sites involved in the LDC function are different from the ones defining the protein localization (Supplementary Figure S8A).
Conversely, we discovered that 80% of R-methylations occur on LC regions enriched in RG- sequences and typically involved in RNA-interactions. According to the fact that the majority of the Microprocessor-associated proteins are RNA-binding proteins, we linked the wide-spread down-regulation of miRNAs upon PRMT1 depletion to a possible role of R-methylation in regulating the RNA-binding properties of the LDC subunits. Our hypothesis was confirmed by the observed reduced binding of ILF3 to pri-miRNAs upon PRMT1 depletion that was paralleled by an increase binding of DGCR8. The stronger association of DGCR8 upon PRMT1 depletion may be, indeed, related to a defective recognition of specific RNA secondary structures by the hypo-methylated Microprocessor-associated proteins and the consequent reduction of the catalytic activity of the Microprocessor which -in turn- remains associated longer to the pri-miRNAs.
Besides ILF3, protein-R-methylation may regulate the RNA-binding ability of other Microprocessor-associated proteins that possess RG-rich sequences, as well as other RBPs in general. Even if our findings on ILF3 suggest that R-methylation promotes protein-RNA interactions, we cannot exclude that this modification may exert an opposite effect on other components of the complex, as already described, for instance, for hnRNPA1 (103). In fact, although methylation does not change the net charge of the arginine guanidine group, it enhances its hydrophobicity, thus enabling two possible effects: on the one hand, it could interfere with the hydrogen bonds established between arginine residues and the negatively charged phosphate backbone of the RNA, thus introducing steric hindrance for protein-RNA binding (104); on the other hand, methylation could facilitate the stacking of methylated arginines within the bases of RNA, hereby promoting protein-RNA interactions (50).
The emerging role of PRMT1-dependent R-methylation on miRNA biogenesis is similar to that reported for other PTMs decorating the LDC, i.e. phosphorylation and acetylation, which regulate the intracellular levels of miRNAs in response to intra- and external- stimuli (41,42,44). Interestingly, PRMTs are de-regulated in several types of cancer (46) and in particular PRMT1 overexpression was shown to positively correlate with tumor progression (105,106). Also, a recent study has showed that the overexpression of type-I PRMTs, including PRMT1, promotes mammary gland tumorigenesis in mice (107). Similarly, microRNA expression is often altered in cancer, where these molecules can display either oncogenic or tumour-suppressive functions (108–110). In addition, proteins involved in miRNA biogenesis, such as Drosha and DGCR8, have also been found mutated and de-regulated in tumors (111), confirming the connection between aberrant LDC function, altered miRNA levels and cancer progression (23).
In this scenario, the fact that the miRNA biogenesis depends on R-methylation and can be modulated by PRMT inhibitors allow speculating on the possibility of using the pharmacological inhibition of PRMTs to tackle tumor vulnerabilities linked to miRNA processing and regulation, offering novel therapeutic options for cancer treatment.
DATA AVAILABILITY
MS-proteomics data have been deposited in the ProteomeXchange Consortium via PRIDE (112) partner repository with the dataset identifier PXD011617.
The hmSEEKER code is available on Bitbucket (https://bit.ly/2scCT9u). Perl codes used to analyze the SILAC data and to intersect it with the hmSEEKER output are available upon request.
Small-RNAseq data have been deposited in GEO with the dataset identifier GSE136418 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136418).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Structural Genomic Consortium (SGC, Toronto, Canada) for providing the MS023 and MS094 compounds, E. Guccione for providing the lentiviral vectors for PRMT1 KD, Anna De Antoni for the generation of the HeLa Flp-In T-REx cells and all the members of the T. Bonaldi's group for helpful discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (AIRC) [IG21834 1 to T.B., IG18774 to F.N.]; Consiglio Nazionale delle Ricerche (CNR) [EPIGEN flagship project to T.B.]; Italian Ministry of Health [GR-2011-02347880 to T.B.]; Italian Foundation for Cancer Research (F.I.R.C.) [‘Leonino Fontana e Maria Lionello’ fellowship to V.S.]; Fondazione CRUI – PhD ITalents programme [144770571 to R.G.]; Cariplo [2015-0590 to F.N.]. Funding for open access charge: Italian Association for Cancer Research (AIRC) [IG21834].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 2. Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R.. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004; 432:235–240. [DOI] [PubMed] [Google Scholar]
- 3. Treiber T., Treiber N., Meister G.. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019; 20:5–20. [DOI] [PubMed] [Google Scholar]
- 4. Michlewski G., Caceres J.F.. Post-transcriptional control of miRNA biogenesis. RNA. 2019; 25:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bethune J., Artus-Revel C.G., Filipowicz W.. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012; 13:716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 7. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S. et al.. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003; 425:415–419. [DOI] [PubMed] [Google Scholar]
- 8. Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N.. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004; 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J.. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004; 432:231–235. [DOI] [PubMed] [Google Scholar]
- 10. Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S.. Functional Anatomy of the Human Microprocessor. Cell. 2015; 161:1374–1387. [DOI] [PubMed] [Google Scholar]
- 11. Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N.. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004; 18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blaszczyk J., Tropea J.E., Bubunenko M., Routzahn K.M., Waugh D.S., Court D.L., Ji X.. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001; 9:1225–1236. [DOI] [PubMed] [Google Scholar]
- 13. Kwon S.C., Baek S.C., Choi Y.G., Yang J., Lee Y.S., Woo J.S., Kim V.N.. Molecular basis for the single-nucleotide precision of primary microRNA processing. Mol. Cell. 2019; 73:505–518. [DOI] [PubMed] [Google Scholar]
- 14. Zeng Y., Yi R., Cullen B.R.. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005; 24:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis B.N., Hata A.. Regulation of MicroRNA biogenesis: a miRiad of mechanisms. Cell Commun. Signal. 2009; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartel D.P. Metazoan MicroRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yi R., Qin Y., Macara I.G., Cullen B.R.. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003; 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U.. Nuclear export of microRNA precursors. Science. 2004; 303:95–98. [DOI] [PubMed] [Google Scholar]
- 20. Bohnsack M.T., Czaplinski K., Gorlich D.. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004; 10:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park J.E., Heo I., Tian Y., Simanshu D.K., Chang H., Jee D., Patel D.J., Kim V.N.. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011; 475:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gregory R.I., Shiekhattar R.. MicroRNA biogenesis and cancer. Cancer Res. 2005; 65:3509–3512. [DOI] [PubMed] [Google Scholar]
- 24. Mendell J.T., Olson E.N.. MicroRNAs in stress signaling and human disease. Cell. 2012; 148:1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peng Y., Croce C.M.. The role of MicroRNAs in human cancer. Signal. Transduct. Target Ther. 2016; 1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reichholf B., Herzog V.A., Fasching N., Manzenreither R.A., Sowemimo I., Ameres S.L.. Time-resolved small RNA sequencing unravels the molecular principles of MicroRNA homeostasis. Mol. Cell. 2019; 75:756–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siomi H., Siomi M.C.. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010; 38:323–332. [DOI] [PubMed] [Google Scholar]
- 28. Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N.. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009; 136:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Ruepp A.. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019; 47:D559–D563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bremang M., Cuomo A., Agresta A.M., Stugiewicz M., Spadotto V., Bonaldi T.. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. Biosyst. 2013; 9:2231–2247. [DOI] [PubMed] [Google Scholar]
- 31. Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N.. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp. Cell Res. 2007; 313:4196–4207. [DOI] [PubMed] [Google Scholar]
- 32. Newman M.A., Hammond S.M.. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010; 24:1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mori M., Triboulet R., Mohseni M., Schlegelmilch K., Shrestha K., Camargo F.D., Gregory R.I.. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014; 156:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong S., Noh H., Chen H., Padia R., Pan Z.K., Su S.B., Jing Q., Ding H.F., Huang S.. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary microRNAs. Sci. Signal. 2013; 6:ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Connerty P., Ahadi A., Hutvagner G.. RNA binding proteins in the miRNA pathway. Int. J. Mol. Sci. 2015; 17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawahara Y., Mieda-Sato A.. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Carlo V., Grossi E., Laneve P., Morlando M., Dini Modigliani S., Ballarino M., Bozzoni I., Caffarelli E.. TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol. Neurobiol. 2013; 48:952–963. [DOI] [PubMed] [Google Scholar]
- 38. Sakamoto S., Aoki K., Higuchi T., Todaka H., Morisawa K., Tamaki N., Hatano E., Fukushima A., Taniguchi T., Agata Y.. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell Biol. 2009; 29:3754–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Higuchi T., Todaka H., Sugiyama Y., Ono M., Tamaki N., Hatano E., Takezaki Y., Hanazaki K., Miwa T., Lai S. et al.. Suppression of MicroRNA-7 (miR-7) Biogenesis by Nuclear Factor 90-Nuclear Factor 45 Complex (NF90-NF45) Controls Cell Proliferation in Hepatocellular Carcinoma. J. Biol. Chem. 2016; 291:21074–21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nussbacher J.K., Yeo G.W.. Systematic discovery of RNA binding proteins that regulate MicroRNA levels. Mol. Cell. 2018; 69:1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Herbert K.M., Pimienta G., DeGregorio S.J., Alexandrov A., Steitz J.A.. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep. 2013; 5:1070–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Q., Li W., She H., Dou J., Duong D.M., Du Y., Yang S.H., Seyfried N.T., Fu H., Gao G. et al.. Stress induces p38 MAPK-mediated phosphorylation and inhibition of Drosha-dependent cell survival. Mol. Cell. 2015; 57:721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wada T., Kikuchi J., Furukawa Y.. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 2012; 13:142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang X., Wen S., Zheng D., Tucker L., Cao L., Pantazatos D., Moss S.F., Ramratnam B.. Acetylation of drosha on the N-terminus inhibits its degradation by ubiquitination. PLoS One. 2013; 8:e72503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Larsen S.C., Sylvestersen K.B., Mund A., Lyon D., Mullari M., Madsen M.V., Daniel J.A., Jensen L.J., Nielsen M.L.. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016; 9:rs9. [DOI] [PubMed] [Google Scholar]
- 46. Yang Y., Bedford M.T.. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013; 13:37–50. [DOI] [PubMed] [Google Scholar]
- 47. Bedford M.T., Clarke S.G.. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009; 33:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang J., Frankel A., Cook R.J., Kim S., Paik W.K., Williams K.R., Clarke S., Herschman H.R.. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 2000; 275:7723–7730. [DOI] [PubMed] [Google Scholar]
- 49. Yu Z., Chen T., Hebert J., Li E., Richard S.. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell Biol. 2009; 29:2982–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bedford M.T., Richard S.. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005; 18:263–272. [DOI] [PubMed] [Google Scholar]
- 51. Wada K., Inoue K., Hagiwara M.. Identification of methylated proteins by protein arginine N-methyltransferase 1, PRMT1, with a new expression cloning strategy. Biochim. Biophys. Acta. 2002; 1591:1–10. [DOI] [PubMed] [Google Scholar]
- 52. Strahl B.D., Briggs S.D., Brame C.J., Caldwell J.A., Koh S.S., Ma H., Cook R.G., Shabanowitz J., Hunt D.F., Stallcup M.R. et al.. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001; 11:996–1000. [DOI] [PubMed] [Google Scholar]
- 53. Wang H., Huang Z.Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B.D., Briggs S.D., Allis C.D., Wong J., Tempst P. et al.. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001; 293:853–857. [DOI] [PubMed] [Google Scholar]
- 54. Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H. et al.. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008; 22:640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kwak Y.T., Guo J., Prajapati S., Park K.J., Surabhi R.M., Miller B., Gehrig P., Gaynor R.B.. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003; 11:1055–1066. [DOI] [PubMed] [Google Scholar]
- 56. Boisvert F.M., Rhie A., Richard S., Doherty A.J.. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005; 4:1834–1841. [DOI] [PubMed] [Google Scholar]
- 57. Boisvert F.M., Dery U., Masson J.Y., Richard S.. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005; 19:671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dery U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., Masson J.Y.. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell Biol. 2008; 28:3058–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guendel I., Carpio L., Pedati C., Schwartz A., Teal C., Kashanchi F., Kehn-Hall K.. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLoS One. 2010; 5:e11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belyanskaya L.L., Gehrig P.M., Gehring H.. Exposure on cell surface and extensive arginine methylation of ewing sarcoma (EWS) protein. J. Biol. Chem. 2001; 276:18681–18687. [DOI] [PubMed] [Google Scholar]
- 61. Araya N., Hiraga H., Kako K., Arao Y., Kato S., Fukamizu A.. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem. Biophys. Res. Commun. 2005; 329:653–660. [DOI] [PubMed] [Google Scholar]
- 62. Tradewell M.L., Yu Z., Tibshirani M., Boulanger M.C., Durham H.D., Richard S.. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum. Mol. Genet. 2012; 21:136–149. [DOI] [PubMed] [Google Scholar]
- 63. Scaramuzzino C., Monaghan J., Milioto C., Lanson N.A. Jr., Maltare A., Aggarwal T., Casci I., Fackelmayer F.O., Pennuto M., Pandey U.B.. Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS One. 2013; 8:e61576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dammer E.B., Fallini C., Gozal Y.M., Duong D.M., Rossoll W., Xu P., Lah J.J., Levey A.I., Peng J., Bassell G.J. et al.. Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One. 2012; 7:e38658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Destouches D., El Khoury D., Hamma-Kourbali Y., Krust B., Albanese P., Katsoris P., Guichard G., Briand J.P., Courty J., Hovanessian A.G.. Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS One. 2008; 3:e2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Blackwell E., Ceman S.. Arginine methylation of RNA-binding proteins regulates cell function and differentiation. Mol. Reprod. Dev. 2012; 79:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stetler A., Winograd C., Sayegh J., Cheever A., Patton E., Zhang X., Clarke S., Ceman S.. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006; 15:87–96. [DOI] [PubMed] [Google Scholar]
- 68. Mendez J., Stillman B.. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell Biol. 2000; 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 70. Rappsilber J., Mann M., Ishihama Y.. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007; 2:1896–1906. [DOI] [PubMed] [Google Scholar]
- 71. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 72. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011; 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- 73. Massignani E., Cuomo A., Musiani D., Jammula S., Pavesi G., Bonaldi T.. hmSEEKER: Identification of hmSILAC Doublets in MaxQuant Output Data. Proteomics. 2019; 19:e1800300. [DOI] [PubMed] [Google Scholar]
- 74. Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D.. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012; 149:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jeon Y., Lee J.T.. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011; 146:119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ong S.E., Mittler G., Mann M.. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004; 1:119–126. [DOI] [PubMed] [Google Scholar]
- 77. Musiani D., Bok J., Massignani E., Wu L., Tabaglio T., Ippolito M.R., Cuomo A., Ozbek U., Zorgati H., Ghoshdastider U. et al.. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 2019; 12:eaat8388. [DOI] [PubMed] [Google Scholar]
- 78. Cumberworth A., Lamour G., Babu M.M., Gsponer J.. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem. J. 2013; 454:361–369. [DOI] [PubMed] [Google Scholar]
- 79. Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Calabretta S., Richard S.. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem. Sci. 2015; 40:662–672. [DOI] [PubMed] [Google Scholar]
- 81. Pekarsky Y., Croce C.M.. Role of miR-15/16 in CLL. Cell Death Differ. 2015; 22:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Olive V., Sabio E., Bennett M.J., De Jong C.S., Biton A., McGann J.C., Greaney S.K., Sodir N.M., Zhou A.Y., Balakrishnan A. et al.. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. Elife. 2013; 2:e00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang D., Huang J., Hu Z.. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol. Cell Proteomics. 2012; 11:M111 011932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Allegra D., Bilan V., Garding A., Dohner H., Stilgenbauer S., Kuchenbauer F., Mertens D., Zucknick M.. Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia. Leukemia. 2014; 28:98–107. [DOI] [PubMed] [Google Scholar]
- 85. Pickering B.F., Yu D., Van Dyke M.W.. Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J. Biol. Chem. 2011; 286:44095–44103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lin S.L., Miller J.D., Ying S.Y.. Intronic microRNA (miRNA). J. Biomed. Biotechnol. 2006; 2006:26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lin S.L., Chang D., Wu D.Y., Ying S.Y.. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochem. Biophys. Res. Commun. 2003; 310:754–760. [DOI] [PubMed] [Google Scholar]
- 88. Dhar S., Vemulapalli V., Patananan A.N., Huang G.L., Di Lorenzo A., Richard S., Comb M.J., Guo A., Clarke S.G., Bedford M.T.. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 2013; 3:1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 90. Eram M.S., Shen Y., Szewczyk M., Wu H., Senisterra G., Li F., Butler K.V., Kaniskan H.U., Speed B.A., Dela Sena C. et al.. A potent, selective, and Cell-Active inhibitor of human type I protein arginine methyltransferases. ACS Chem. Biol. 2016; 11:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ozdilek B.A., Thompson V.F., Ahmed N.S., White C.I., Batey R.T., Schwartz J.C.. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017; 45:7984–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Geoghegan V., Guo A., Trudgian D., Thomas B., Acuto O.. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat Commun. 2015; 6:6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hart-Smith G., Yagoub D., Tay A.P., Pickford R., Wilkins M.R.. Large Scale Mass Spectrometry-based Identifications of Enzyme-mediated Protein Methylation Are Subject to High False Discovery Rates. Mol. Cell Proteomics. 2016; 15:989–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sylvestersen K.B., Horn H., Jungmichel S., Jensen L.J., Nielsen M.L.. Proteomic analysis of arginine methylation sites in human cells reveals dynamic regulation during transcriptional arrest. Mol. Cell Proteomics. 2014; 13:2072–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K.A., Yang V., Aguiar M., Kornhauser J., Jia X. et al.. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteomics. 2014; 13:372–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chang B., Chen Y., Zhao Y., Bruick R.K.. JMJD6 is a histone arginine demethylase. Science. 2007; 318:444–447. [DOI] [PubMed] [Google Scholar]
- 97. Poulard C., Rambaud J., Hussein N., Corbo L., Le Romancer M.. JMJD6 regulates ERalpha methylation on arginine. PLoS One. 2014; 9:e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Walport L.J., Hopkinson R.J., Chowdhury R., Schiller R., Ge W., Kawamura A., Schofield C.J.. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 2016; 7:11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bottger A., Islam M.S., Chowdhury R., Schofield C.J., Wolf A.. The oxygenase Jmjd6–a case study in conflicting assignments. Biochem. J. 2015; 468:191–202. [DOI] [PubMed] [Google Scholar]
- 100. Webby C.J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M.L., Schmitz C., Butler D.S., Yates J.R. 3rd et al.. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009; 325:90–93. [DOI] [PubMed] [Google Scholar]
- 101. Hung C.J., Lee Y.J., Chen D.H., Li C.. Proteomic analysis of methylarginine-containing proteins in HeLa cells by two-dimensional gel electrophoresis and immunoblotting with a methylarginine-specific antibody. Protein J. 2009; 28:139–147. [DOI] [PubMed] [Google Scholar]
- 102. Jobert L., Argentini M., Tora L.. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp. Cell Res. 2009; 315:1273–1286. [DOI] [PubMed] [Google Scholar]
- 103. Rajpurohit R., Paik W.K., Kim S.. Effect of enzymic methylation of heterogeneous ribonucleoprotein particle A1 on its nucleic-acid binding and controlled proteolysis. Biochem. J. 1994; 304:903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chong P.A., Vernon R.M., Forman-Kay J.D.. RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 2018; 430:4650–4665. [DOI] [PubMed] [Google Scholar]
- 105. Baldwin R.M., Morettin A., Paris G., Goulet I., Cote J.. Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells. Cell Cycle. 2012; 11:4597–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yoshimatsu M., Toyokawa G., Hayami S., Unoki M., Tsunoda T., Field H.I., Kelly J.D., Neal D.E., Maehara Y., Ponder B.A. et al.. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer. 2011; 128:562–573. [DOI] [PubMed] [Google Scholar]
- 107. Bao J., Di Lorenzo A., Lin K., Lu Y., Zhong Y., Sebastian M.M., Muller W.J., Yang Y., Bedford M.T.. Mouse models of overexpression reveal distinct oncogenic roles for different type I protein arginine methyltransferases. Cancer Res. 2019; 79:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. et al.. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838. [DOI] [PubMed] [Google Scholar]
- 109. Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M. et al.. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005; 353:1793–1801. [DOI] [PubMed] [Google Scholar]
- 110. Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M.. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006; 20:2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lin S., Gregory R.I.. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015; 15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al.. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Letunic I., Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018; 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hinske L.C., Franca G.S., Torres H.A., Ohara D.T., Lopes-Ramos C.M., Heyn J., Reis L.F., Ohno-Machado L., Kreth S., Galante P.A.. miRIAD-integrating microRNA inter- and intragenic data. Database. 2014; 2014:bau099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chou M.F., Schwartz D.. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics. 2011; doi:10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS-proteomics data have been deposited in the ProteomeXchange Consortium via PRIDE (112) partner repository with the dataset identifier PXD011617.
The hmSEEKER code is available on Bitbucket (https://bit.ly/2scCT9u). Perl codes used to analyze the SILAC data and to intersect it with the hmSEEKER output are available upon request.
Small-RNAseq data have been deposited in GEO with the dataset identifier GSE136418 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136418).