Abstract
The Escherichia coli SecA ATPase motor protein is essential for secretion of proteins through the SecYEG translocon into the periplasmic space. Its function relies upon interactions with the surrounding lipid bilayer as well as SecYEG translocon. That negatively charged lipids are required for bilayer binding has been known for more than 25 years, but little systematic quantitative data is available. We have carried out an extensive investigation of SecA partitioning into large unilamellar vesicles (LUV) using a wide range of lipid and electrolyte compositions, including the principal cytoplasmic salt of E. coli, potassium glutamate, which we have shown stabilizes SecA. The water-to-bilayer transfer free energy is about −7.5 kcal mol–1 for typical E. coli lipid compositions. Although it has been established that SecA is dimeric in the cytoplasm, we find that the most widely cited dimer form (PDB 1M6N) binds only weakly to LUVs formed from E. coli lipids.
Keywords: membrane partitioning, potassium glutamate, lipid-protein interactions, protein secretion
1. Introduction
Escherichia coli and other Gram-negative bacteria require the cytoplasmic SecA ATPase motor protein in order to secrete periplasm-destined proteins across the inner membrane. Since the discovery of SecA in 1982 [1, 2], much has been learned about SecA-driven secretion of signal sequence-bearing preproteins through the membrane-embedded SecYEG (reviewed by Crane and Randall [3]). Because SecA (901 amino acids, MW = 102 kDa) is a soluble protein that associates with cytoplasmic membranes [4, 5], the nature of the association has long been of interest (see review [6]), and it has been recently proposed that SecA gains access to the SecYEG complex via a lipid-bound intermediate state [7]. In vivo, de Vrije et al. [8] showed in 1988 that negatively charged membrane lipids are involved in protein translocation across E. coli inner membranes, which led to several studies of SecA interactions with lipid monolayers [9] and bilayers [10–12]. These studies revealed the necessity of anionic lipids for SecA binding to both monolayers and bilayers and that some portion of SecA penetrates deeply into lipid bilayers. Both the N- and C-terminal regions of SecA have been shown to play a role in lipid interactions [13, 14], but it appears that the N-terminal domain is more important for initial interactions of SecA with membranes [15]. Recently, Findik et al. [16] showed that the ten N-terminal residues partition into LUV formed from E. coli lipids as an amphipathic helix that penetrates about 8 Å into the membrane interface. This is similar to the behavior of the 26-residue amphipathic helix of the bee venom melittin [17] and a designed 18-residue amphipathic helix [18].
A notable feature of SecA in solution is the formation of homodimers, as first shown by Akita et al. [19] using size-exclusion chromatography. Subsequently, Driessen [20] found the homodimers observed in his experiments to be functional. The structure of SecA homodimers has been examined extensively by electron cryo-microscopy [21] and especially analytical ultracentrifugation [22, 23]. Five different X-ray crystal structures of dimeric SecA from different bacterial species have suggested several possible dimer interfaces [24–28]. Exactly which, if any, of the observed dimer interfaces represents the functional dimer in vivo has been controversial, but a consensus seems to be that the antiparallel dimer structure determined for Bacillus subtilis SecA [24] (PDB 1M6N) represents the dominant dimer in vitro (Figure 1B) as determined by analytical ultracentrifugation [22, 23] and Förster resonance energy transfer (FRET) [29]. A site-specific cross-linking study suggested that the 1M6N dimer promotes active protein translocation in vivo [30]. Although 1M6N might be the dominant dimer form both in vivo and in vitro, Zimmer et al. [27] found that SecA likely has several different dimeric states that are in equilibrium with one another and with the monomeric state.
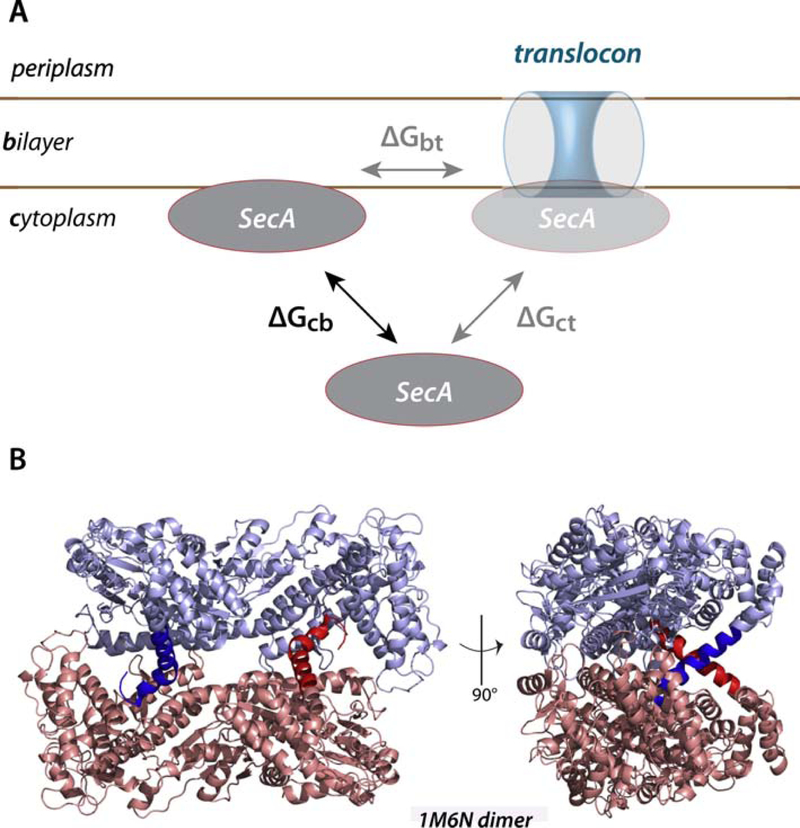
Figure 1. A thermodynamic scheme for the interaction of SecA with the SecYEG translocon and membrane lipid bilayer.
(panel A) and the molecular structure of the SecA dimer (PDB 1M6N) (panel B). SecA is known to bind to both negatively charged lipid bilayers and SecYEG translocon, but the relationship between the binding events is uncertain. We suggest that the relationship can be clarified by determining the relative free energies of transfer of SecA from the cytoplasm to the bilayer (ΔGcb) and to the translocon (ΔGct). The difference between these free energies(ΔGbt) should provide insights into the role of the lipid bilayer in the binding of SecA to the translocon. The general belief is that cytoplasmic SecA exists as a dimer represented by 1M6N. In the images in panel B, the monomeric protomers are colored blue and pink. The 21-residue N-terminal helices, highlighted by dark blue and dark red, are thought to be responsible for binding SecA to the bilayer. Note that the conformations of these helices suggest that they are also important in dimer formation.
Given that SecA interacts strongly with lipid bilayers and with the SecYEG translocon, a fundamental question is the distribution of SecA between the aqueous cytoplasm, the bilayer membrane, and the membrane embedded translocon [4, 31]. A related question is the whether the bilayer-associated form is dimeric or monomeric. When bound to the translocon, some data suggests that SecA is monomeric [32, 33] while other data suggest that dimers can interact as well [34, 35]. An x-ray crystal structure shows SecA bound to SecYEG as a monomer [36], although the presence of detergents and high salt concentrations during crystal preparation could have affected the results [37]. Because long-chain phospholipid analogs promote dissociation of SecA dimers [38], it may be that partitioning of SecA dimers into bilayers leads to dissociation into monomers that then bind to SecYEG with little further contact with lipids [39, 40]. However, a single-molecule study using dual-color fluorescence-burst analysis (DCFBA) indicated that SecA is active in translocation as a dimer [37].
A simplified view of the problem of describing SecA interactions with bilayers and the SecYEG translocon is shown in Figure 1A. What matters are the equilibrium free energies of transfer of SecA from the cytoplasm to the bilayer phase (ΔGcb) and from the cytoplasm to the translocon (ΔGct). Knowing these two numbers, the transfer free energy from bilayer to translocon (ΔGbt) can be calculated. Relevant to ΔGct, Kusters et al. [37] reported a dissociation constant Kct of 3.6 ± 1.2 nM based using their DCFBA approach. This dissociation constant can be converted to free energy if the proper standard state is established [41]. As discussed by White et al. [42], the standard biochemical dissociation-constant approach can be problematic, because the hydrophobic and electrostatic interactions that drive most protein-bilayer interactions arise from the collective properties and behavior of the lipids in the bilayer. It is better to think of partitioning between two phases: the aqueous phase and the bilayer phase [42]. We use that approach here to determine the free energy of transfer of SecA from water to bilayer, ΔGwb, under a wide range of lipid and salt conditions including potassium glutamate (KGlu), which is the principal cytoplasmic salt of E. coli that is known to stabilize monomeric and dimeric SecA [43–45]. We also examine the partitioning of two disulfide-stabilized dimers based upon the 1M6N dimer. The stabilized dimers partition weakly, suggesting that our partitioning free energies are due either to partitioning of monomers or a dimer other than 1M6N.
2. Materials and Methods
2.1. Materials
All phospholipids were purchased form Avanti Polar Lipids (Alabaster, AL): E. coli total extract (catalog number 100500), 1-palmitoyl-2-oleoyl-glycero-3-phosphatiylcholine (POPC, 850457), 1-palmytoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE, 850757), 1-palmioyl-2-oleoyl-sn-glycero-3-phospho-(1’rac-glycerol) (POPG, 840457), and cardiolipin (841199).
2.2. Construction of the single-cysteine C403-SecA mutant
Starting from a cysteine-less construction (pT7-secAC4, gift from Prof. Donald Oliver), single amino acid substitution mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene) using the following primer to change Serine 403 to a cysteine: 5’-CCGAAGCTTTCGAATTTTGCTCAATCTACAAGCTGGATACCG-3’. Mutation was confirmed via DNA sequence analysis.
2.3. SecA Protein production
WT-SecA, C11+C661-SecA, or C403-SecA were obtained from E. coli BL21 coco cells carrying the corresponding secA gene with a C-terminal His6-tag under the control of the T5 promotor. Cells were grown in LB medium at 37°C with constant shaking. Log-phase cultures (OD 0.8) were stimulated with IPTG (1 mM) for 2 hours at 30°C. Cells were then harvested by centrifugation at 4,000 rpm for 15 minutes and the resulting pellet stored at −20°C until needed.
2.4. SecA Protein purification
All protein purification steps and centrifugations were performed at 4°C. Bacterial pellets (from 400 mL culture) were dispersed in 48 mL of Buffer A (50 mM Hepes-NaOH pH 7.4, 10 mM imidazole and 50 mM KCl) for 10 minutes at room temperature. Protease inhibitor cocktail (Roche) was added before the cell suspension was passed through a French Pressure Cell (SLM-Amico) at 10,000 lb/in2. The resulting suspension was then centrifuged at 13,000 g for 15 minutes to pellet the membrane fraction. The supernatant was loaded onto a Talon-Resin column (1.5 ˣ 5 cm) previously equilibrated with 25 mL of Buffer A. His-tagged SecA protein was then eluted with Buffer B (50 mM Hepes-NaOH pH 7.4, 500 mM imidazole, 50 mM KCl, 1 mM DTT). Two-mL fractions were collected, and the protein profile analyzed using SDS-PAGE. Fractions containing 100 kDa SecA were then pooled, concentrated, and loaded onto a Superdex 200 increase 10/300 GL equilibrated in 50 mM Hepes-NaOH pH 7.4, 1 mM DTT and 50 mM KCl). SecA was then eluted at a flow rate of 0.5 mL/minute and monitored by optical absorbance at 280 nm. 500-µL fractions were collected and the protein profile analyzed using SDS-PAGE. Fractions containing the SecA protein were pooled and the protein concentration was estimated using the BioRad Assay with BSA as a reference and a molecular weight of 102 kDa.
For convenience, we used the His-tagged protein in our measurements, because we found no difference in the partitioning of His-tagged protein compared to untagged protein (Figure S1), as expected over this pH range where we should have about 25 times more unprotonated histidine residues than protonated as described by the Henderson-Hasselbalch equation. Additionally, Bauer et al. [46] found that a modified version of SecA in which the N-terminal 20 residues were replaced by 6 His residues did not support translocation in the absence of Ni-NTA lipids in the bilayer.
2.5. N-terminus peptide preparation
SecA2–23 with sequence LIKLLTKVFGSRNDRTLRRMRKV was provided by BioMatik USA (Wilmington, Delaware) and the purity (>93 %) was verified by SDS-PAGE. The peptide (10 mg) was solubilized in 10 mM Hepes-NaOH pH 7 as recommended by the manufacturer to a final concentration of 400 µM. Sonication was briefly used (20 seconds, duty cycle 50%, output 40%) to ensure complete solubilization. Peptide concentration for CD measurement was 40 µM, and the lipid concentration varied between 0 and 4 mM.
2.6. Liposome preparation
Phospholipids dissolved in chloroform were dried under a stream of nitrogen and further dried under vacuum overnight. Lipids were then suspended in 25 mM Hepes-NaOH pH 7.4 and vortexed 15 minutes. Large unilamellar vesicles (LUVs) were prepared by extrusion through a membrane (100 nm pore diameter). Lipid concentrations were determined according to the procedure of Bartlett [47].
2.7. Tryptophan fluorescence
Fluorescence spectra were recorded using an OLIS-modified SLM-Aminco 8100 steady-state fluorescence spectrometer (Jobin Yvon, Edison, NJ, formerly SLM/Aminco, Urbana, IL) equipped with double-grating excitation and single-grating emission monochromators. All measurements were made in 2 mm × 10 mm cuvettes using an excitation wavelength of 295 nm. Excitation slits were not wider than 8 nm; emission slits were 4 nm. Unless otherwise indicated, the emission polarizer was oriented at 0° relative to the vertical and the excitation polarizer at 90°. Spectra were collected in the region of 310–400 nm in increments of 1 nm. Generally, 10 or more spectra were averaged to achieve an adequate signal-to-noise ratio. In all cases, a SecA monomeric concentration of 4 μM was prepared in 50 mM Hepes-NaOH pH 7.4, 1 mM EDTA, 2 mM MgCl2, 1 mM DTT, and 50 mM KCl. Fluorescence intensities using excitation at 295 nm (1 nm slit) were recorded between 310 and 400 nm (4 nm slit). The same conditions were used for recording scans in buffer alone, which were then subtracted from the appropriate protein spectra.
2.8. Data analysis: The partition coefficient
The association of proteins with bilayers is best treated as partitioning between two immiscible phases, water and bilayer [42]. The chemical potential (partial molar free energy) of a solute in a phase (bilayer or water) is given by:
| (1) |
where the composition of the phase is described by the solute activity a0, which equals 1 in the standard state. Several different units, such as mole fraction or volume fraction can be used to describe solute activities. We prefer, as did Tanford [48] and Lewis & Randall [49], to use mole fraction x as the measure of composition. Again, following those authors, we take infinite dilution of the solute as the standard state, i.e. a/x → 1 as x → 0.
At equilibrium, the chemical potentials of the peptide in the bilayer (bil) and in water (w) must be equal, i.e. μbil −μw = 0. The standard free energy of transfer of x from water to bilayer is thus
| (2) |
where the subscripts x mean that mole-fraction units are being used and Kx is the mole-fraction partition coefficient given by
| (3) |
In this equation, [P]bil and [P]w are the bulk molar concentrations of protein attributable to protein in the bilayer and water phases, respectively, and [L] and [W] are the concentrations of lipid and water. It will always be true that [W] = 55.3 M is much, much greater than [P]w. Further, because the concentration of the protein in the bilayer is kept as low as possible to avoid concentration-dependent partition coefficients, it is also generally true that [L] >>[P]bil. One may thus write with high accuracy that
| (4) |
In the experiments described here, we measure partitioning by titrating a peptide or protein solution of fixed concentration with lipid vesicles and calculating the fraction fp of the total amount of peptide or protein partitioned into the lipid vesicles as a function of lipid concentration [L]. Given that [P]total = [P]bil + [P]w, one can show that
| (5) |
Note in this equation, which assumes a fixed amount of protein in the aqueous phase during titration, that Kx is constant and the only variable is the lipid concentration. Thus, as [L] increases through titration, fP increases.
2.9. Data analysis: Measuring fp
Given eq. (5) and its underlying assumptions, the problem of measuring Kx, and thus , reduces to measuring the fraction of peptide bound as the lipid concentration is increased through titration of the protein solution with lipid vesicles. The principal experimental problem is to measure fp using some kind of detection system in which the detector’s response to partitioning is linear in fp. The two principal methods used in our lab are intrinsic tryptophan fluorescence and circular dichroism spectroscopy. The requirements for using these two methods as a linear fp response function has been discussed extensively elsewhere [42, 50]. At high lipid concentrations, light scattering induced by the increasing concentration vesicles increases more rapidly than the increases in fluorescence [50]. The fluorescence signals must therefore be corrected for light scattering at each lipid concentration [L] by using intensity data from the non-partitioning Trp zwitterion (20 µM) under the same experimental conditions as follows:
| (6) |
In this equation, cor means ‘corrected’ and buf ‘buffer’. The fraction of membrane-bound protein was determined by titration using fluorescence measurements, as described by White et al. (1998) [42]:
| (7) |
Notice the appearance in eq. (7) of the term (Kx[L])/([W] + Kx[L]), which is fp. That is, Icor is a linear function of fp. The fitting of eq. (7) to the experimental data by non-linear least-squares (NLLS) yields Kx from which the free energy of transfer of SecA from water-to-bilayer was calculated from the mole-fraction partition coefficient Kx using:
| (8) |
where R is the gas constant (1.987 × 10−3 kcal K−1mol−1) and T is the temperature in Kelvin. All NLLS fits of experimental data to obtain Kx and I∞ were performed with KaleidaGraph 4.5.0.
2.10. Circular dichroism spectroscopy
CD measurements were performed with a JASCO J-810 spectropolarimeter, using a protein concentration of 4 μM and a 0.1 cm cell path length. The buffer was 10 mM Hepes-NaOH pH 7.4, 2 mM MgCl2, 1 mM DTT, 50 mM KF, and increasing concentrations of lipids. Spectra were acquired at 37 °C at a scan speed of 20 nm min−1, with a 0.2 nm data pitch, using a 1 nm bandwidth and a 4s digital integration time. The spectra were averaged after four accumulations and corrected by subtraction of the buffer spectrum obtained under the same conditions.
2.11. Preparation of crosslinked species for titration
Proteins (C11-C661 or C403) were purified in the presence of 0.4 mM reduced glutathione (GSH). The formation of disulfide bridge was induced by the addition of 4 mM of oxidized glutathione (GSSG) for at least one hour at room temperature. Samples were then concentrated and loaded onto a Superdex 200 increase 10/300 GL previously equilibrated with adequate buffer containing 1 mM GSSH (no GSH). SecA was then eluted at a flow rate of 0.5 mL/min and the optical absorbance at 280 nm was used to monitor protein elution. 500 µL fractions were collected and the protein profile analyzed using SDS-PAGE without DTT. Fractions containing the cross-linked species (MW ~ 200 kDa) were then pooled and the protein concentration was estimated using the BioRad Assay using BSA as a reference.
2.12. Crosslinking of cysteine-mutants on liposomes
Proteins (C11-C661 or C403) were purified in the presence of 0.4 mM reduced glutathione (GSH). After binding to lipid vesicles (30 minutes), GSH concentration was reduced to 0.1 mM and 4 mM oxidized glutathione was introduced by dialysis. The formation of disulfide bridge was induced for 4 hours before running the sample on a SDS-PAGE gel in the absence of DTT.
3. Results
We had three goals when we began these experiments. First, we wanted to obtain accurate quantitative data on the interactions of SecA with LUV membranes of different lipid compositions. Second, in light of the fact that potassium glutamate (KGlu)—the principal cytoplasmic salt [43, 44] of E. coli—significantly enhances the thermal stability of SecA monomers and dimers in solution [45], we wished to establish how KGlu affects the partitioning of SecA into membranes. Third, we wished to explore whether SecA partitions into membranes as a monomer or as a dimer, specifically the 1M6N dimer.
3.1. SecA binds strongly to liposomes made from E. coli lipids
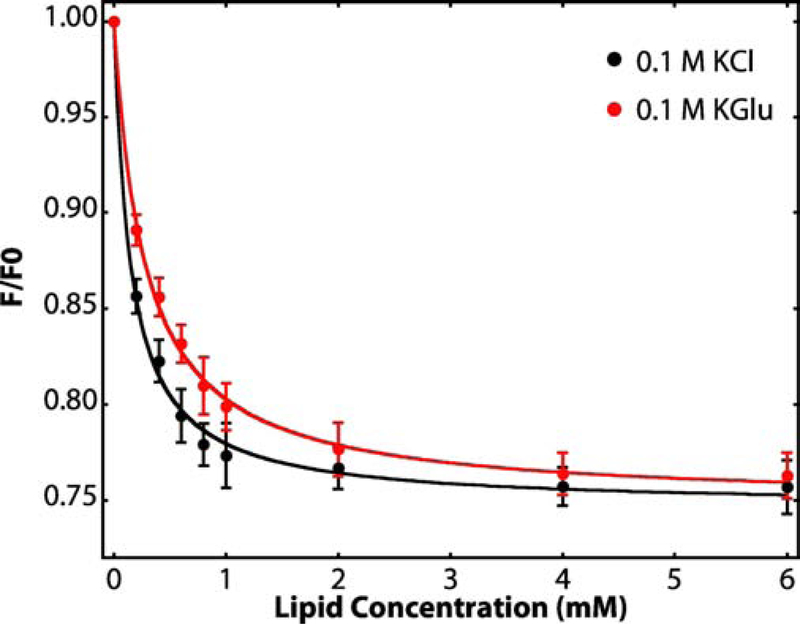
SecA contains 7 tryptophan residues whose fluorescence provides a convenient indicator of the partitioning of SecA into membranes [12]. Presumably due to small structural rearrangements upon binding, the net fluorescence at 340 nm is reduced [12]. Following the protocols of Ladokhin et al. [50], we titrated SecA solutions of fixed concentration with large unilamellar vesicles (LUVs) formed from E. coli lipids at 37°C, the optimal growth temperature of E. coli, and analyzed partitioning according to White et al. [42] (see Materials and Methods section). The titration curves resulting from titration of SecA with LUVs formed from E. coli lipids in the presence of either KCl or KGlu are shown in Figure 2. The free energy of transfer of SecA from water to bilayer (ΔGwb) at 37° C in 0.1 M KCl (black line) is −7.8 ± 0.1 kcal mol−1 whereas in the presence of 0.1 M KGlu, the free energy of transfer of −7.4 ± 0.1 kcal mol−1.
Figure 2. Titration of SecA solutions with large unilamellar vesicles (LUV) reveals strong interactions of SecA with LUV formed from E. coli lipids.
Relative tryptophan fluorescence intensity changes (F/F0) accompanying the titration of aqueous solutions of SecA (4 μM) with large unilamellar vesicles (LUV) formed from E. coli lipids at 37 °C. The black curve shows partitioning data for SecA in 0.1 M KCl. The red curve shows the partitioning of SecA in 0.1 M KGlu (potassium glutamate). We showed earlier [45] that KGlu stabilizes SecA against thermal unfolding. F0 is the florescence intensity in the absence of vesicles and F the intensity in the presence of vesicles. The intensities are corrected for light scattering effects [50] (Equation 1, see Methods). The curves are non-linear least-squares fits of Equation 2 to the data from which the mole-fraction partition coefficients Kx are derived. Free energies of transfer ΔGwb of SecA from the aqueous phase to lipid vesicles can be calculated from Equation 3. Fluorescence intensities were recorded between 310 and 400 nm (4 nm slit) using excitation at 295 nm (1 nm slit). For determination of F/F0, we used the fluorescence intensity at 340 nm [50]. Three independent sets of experiments were performed for each electrolyte condition. The error bars represent the standard errors of the mean (SEM) resulting from these independent measurements. Values of Kx and ΔGwb determined from these measurements are summarized in Table 1.
3.2. Parameters affecting SecA partitioning
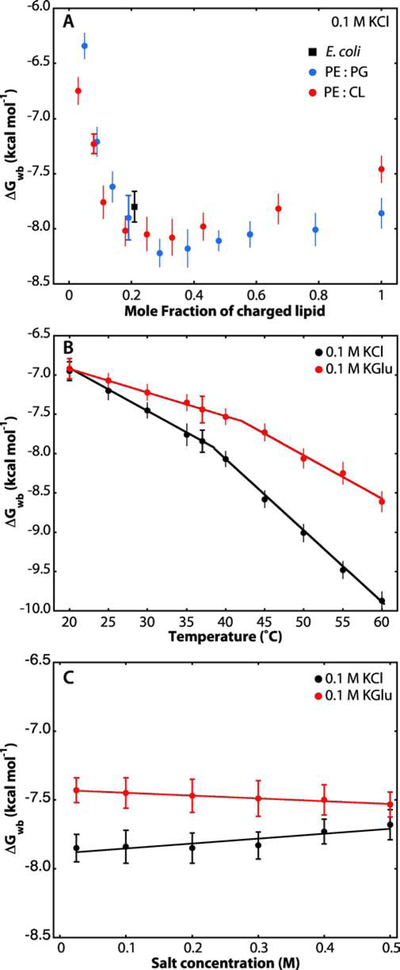
As well established [10–12], SecA function requires the presence of anionic lipids. The dependence of ΔGwb on LUV lipid composition supports this conclusion (Figure S2). SecA does not partition significantly into LUVs formed only from phosphatidylcholine (PC) (Table 1). Surprisingly, however, SecA partitions weakly into LUVs formed from E. coli phosphatidylethanolamine (PE) and PC (1:1). It was previously reported that SecA can weakly interact with PE [9], probably by interacting with the ethanolamine headgroup that contains a primary amine that can form hydrogen bonds [51]. Otherwise, SecA partitions about the same (≈ −7.8 kcal mol−1) into all mixtures containing the anionic lipids phosphatidylglycerol (PG, 20%) and/or cardiolipin (CL, 20%). The partitioning of SecA into membranes is sensitive to the fraction of negatively charged lipids in the membrane, as observed by the increase of ΔGwb of two-orders of magnitude when going from 0 to 30% of either POPG or CL (Figure 3A). Very high concentrations of negatively charged lipids seemed to reduce slightly the binding of SecA to the bilayer, perhaps because of electrostatic repulsion between the negatively charged lipids and the 16 amino-acid excess of acidic amino acids in SecA [52].
Table 1.
Summary of SecA partitioning free energies at 37°C. Titration curves from which these data are obtained are provided in Figure 2 and Figure S2.
| aLipid | bComposition | cKx ×10−3 | dΔGwb 0.1 M KCl | cKx ×10−3 | dΔGwb 0.1 M KGlu |
|---|---|---|---|---|---|
| E. coli extract | — | 325 ± 22 | −7.8 ± 0.1 | 198 ± 21 | −7.4 ± 0.1 |
| PC | — | en.o. | en.o. | en.o. | en.o. |
| PE:PC | 50:50 | 8.5 ± 2.3 | −5.2 ± 0.2 | 9.4 ± 4.1 | −5.6 ± 0.3 |
| PE:PG | 80:20 | 305 ± 20 | −7.8 ± 0.2 | 235 ± 22 | −7.6 ± 0.1 |
| PE:CL | 80:20 | 347 ± 18 | −7.9 ± 0.1 | 245 ± 20 | −7.6 ± 0.1 |
PC, palmitoyloleoylphosphatidylcholine (POPC); PE, palmitolyloleoylphosphatidylethanolamine (POPE); PG, palmitolyloleoylphosphatidylglycerol (POPG); CL, cardiolipin.
mole ratio.
mole-fraction partition coefficient (see Methods)
free energy of transfer, water to bilayer, kcal mol−1
not observable
Figure 3. LUV lipid composition, temperature, and electrolyte concentration affect the partitioning of SecA into LUVs.
Data points with horizontal caps represent the SEMs for experiments performed in triplicate. Data points lacking horizontal caps indicate single titration experiments. In that case, the vertical error bars represent estimates of uncertainties determined from non-linear least-squares fits. (A) Negatively charged lipids (POPG or CL) strongly improve SecA partitioning into lipid vesicles, as established earlier by other laboratories [9, 12]. Free energies of transfer from water to bilayer were determined from the partitioning of SecA at 37°C into LUVs containing an increasing fraction of POPG (blue) or cardiolipin (red) monitored by Trp fluorescence. As a reference, partitioning into LUV formed from E. coli lipids is also plotted (black square). (B) Influence of temperature on the partitioning of SecA to liposomes made from E. coli lipids in the presence of 0.1 M KCl (black) or 0.1 M KGlu (red). The increases in the magnitudes of ΔGwb with temperature are likely due to partial thermal unfolding of SecA, which leads to exposure of otherwise buried hydrophobic residues. The temperature effect is smaller in KGlu compared to KCl, as expected from the stabilization of SecA by glutamate [45]. (C) Effect of electrolyte concentration on free energies of transfer of SecA at 37 °C. The free energies in KGlu are slightly lower than in KCl, likely due to glutamate stabilization that reduces exposure of normally-buried hydrophobic residues [45]. Linear fits to the data of panel C are shown. It remains to be determined why the straight-line fits to the points have opposite slopes and why they converge to ΔGwb = −7.6 kcal mol−1 at 0.8 M salt concentration. Linear-fit parameters: KCl, intercept at 0 mM salt is −7.9 with a slope of +0.36. KGlu, intercept at 0 mM salt is −7.4 with a slope of −0.20
We showed earlier that KGlu stabilizes SecA against thermal unfolding based upon both tryptophan fluorescence and CD measurements [45]. Specifically, for KCl solutions the denaturation midpoints determined by Trp fluorescence and CD spectroscopy were 38.2 ± 0.4 °C and 43.6 ± 0.5 °C, respectively, independent of KCl concentration. Trp fluorescence is very sensitive to tertiary structure; as the protein starts to ‘open up’, Trp residues become more water exposed without significant changes in secondary structure. That is, the tertiary structure of a protein can be denatured by heat while some or all of the secondary structure elements—alpha-helices and beta-sheet—persist until higher temperatures are reached. CD, on the other hand is sensitive to secondary structure, which is more stable than tertiary structure. Consequently, ‘unfolding’ as measured by CD occurs at higher temperatures than tertiary structural changes. Glutamate stabilizes both tertiary and secondary structure. In KGlu solutions, stabilization was also concentration dependent, but reaching 42 °C by Trp fluorescence and 48 °C by CD spectroscopy (KGlu = 300 mM). Measurements of partitioning free energies to liposomes made from E. coli lipids reveal a similar stabilizing effect (Figure 3B). As expected, in KCl, as the protein begins to unfold at 38°C, a break is observed in the ΔGwb due to greater exposure of SecA’s hydrophobic core to the lipid bilayer (black line). This exposure is apparently reduced for KGlu due to the glutamate stabilization effect (red line). At 43 °C, ΔGwb is little changed in KGlu whereas in KCl it increases to about −8.2 kcal mole−1.
Salt-dependent titration curves show that the free energies of transfer decrease slightly with increasing KCl concentration (Figure 3C, black), probably because of the shielding of charges on the surface of the bilayer or the protein. However, at very high salt concentration (500 mM), where SecA has been described as a monomer in solution [22, 23], the ΔGwb was similar to the value at low salt concentrations. Surprisingly, ΔGwb increases slightly with increasing KGlu concentration (Figure 3C, red), but the effect is not large given the experimental uncertainties.
3.3. Potassium glutamate stabilizes SecA on the membrane
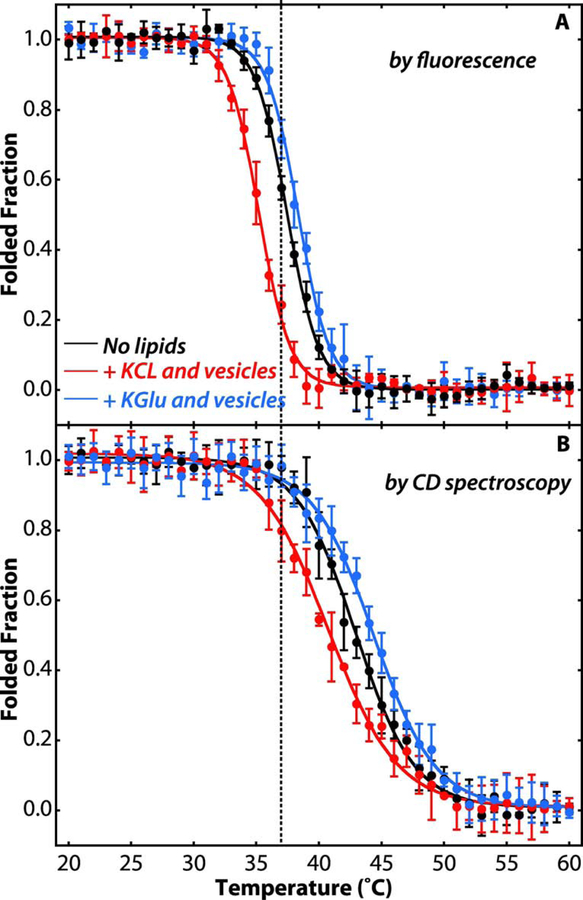
Denaturation curves determined using Trp fluorescence showed that, in the presence of KCl, the partitioning of SecA into the membrane destabilizes the tertiary structure of the protein (Figure 4A, red line) with the denaturation midpoint moving from 37.8±0.4 °C in solution to 35.0 ± 0.5°C with 4 mM LUVs. Similar results have been previously reported using SUVs [10] and micelles [38]. In the presence of LUVs, the addition of KGlu caused a significant increase in stability with a denaturation midpoint of 38.3 ± 0.3°C (blue line). A similar stabilizing effect was observed when monitoring the stability of the secondary structure by circular dichroism (Figure 4B). These results indicate that, in addition to stabilizing SecA in solution [45], glutamate also stabilizes membrane-bound SecA. How this stablization on the membrane might affect the interaction with translocons, the pre-protein, and consequently the translocation process remains to be determined.
Figure 4. Potassium glutamate stabilizes SecA on the membrane.
(A) Temperature dependence of the unfolding of SecA tertiary structure as indicated by Trp fluorescence monitored at 340 nm (excitation at 295 nm). (B) Temperature dependence of the unfolding of SecA secondary structure as indicated molar ellipticity at 222 nm. Folded fractions were determined by assuming SecA (4 μM) was fully folded in its native state at 20°C and fully unfolded at 60°C. The folded fraction was determined from either the Trp fluorescence at 340 nm or the molar ellipticity at 222 nm, normalized by setting the intensities of the native and fully unfolded SecA to be 1 and 0 after correction for the temperature dependence of the fluorescence intensity. Black curves were determined for SecA in 100 mM KCl in the absence of lipid vesicles. The red curves show the thermal unfolding in 100 mM KCl and LUVs formed from E. coli lipids (4 mM). Blue curves are for 100 mM KGlu and LUVs formed from E. coli lipids (4 mM). When vesicles are present, approximately 95–98% of SecA is partitioned into them (see Figure 2). In KCl solutions, SecA bound to LUVs is less stable, as indicated by the shift of the curves to lower temperatures (red). In KGlu solutions, on the other hand, the stability of LUV-bound SecA is increased, as indicated by shifts of the unfolding curves to higher temperatures (blue). Experiments were carried out in triplicate. The error bars represent the standard errors of the mean (SEM) resulting from the three independent measurements.
3.4. The N-terminus plays a key role in membrane partitioning
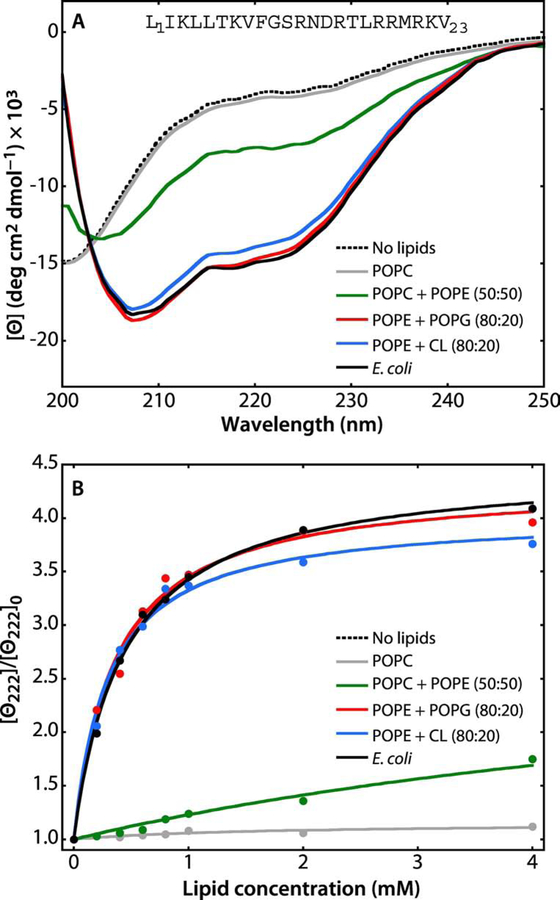
Given the results of Findik et al. [16] who showed that the ten N-terminal residues of intact SecA partition into LUVs formed from E. coli lipids as an amphipathic helix, we determined the partitioning free energy a synthetic SecA N-terminal peptide, L1IKLLTKVFGSRNDRTLRRMRKV23. Because the peptide has no tryptophan, we instead measured partitioning using CD spectroscopy. In the absence of lipid, the spectrum was typical of an unordered peptide (Figure 5A, black dashed line), while in the presence of 2 mM LUVs made from E. coli lipids, the signal corresponds to that of an alpha-helix signature (black solid line). Measuring the ellipticity at 222 nm as a function of lipid concentration yielded the lipid titration curve shown in Figure 5B. From these data, we found ΔGwb = −7.7 ± 0.1 kcal mol−1, virtually identical to the value for SecA (Table 1, Table 2). The partitioning of the N-terminus peptide into lipid vesicles is also dependent on the LUV composition and requires the presence of negatively charged lipids (Table 2). In the presence of CL (blue line) or PG (red line), the peptide partitions into the bilayer resulting in an alpha-helix signature. No partitioning was observed using LUVs formed from PC alone (gray line, Figure 5A). These results suggest that the N-terminus of SecA is highly exposed to the aqueous phase in a manner that mimics the behavior of the synthetic N-terminal peptide free in solution.
Figure 5. A synthetic peptide corresponding to the N-terminal 23 residues of SecA partition strongly into LUV.
The N-terminal domain of SecA has long been known to be critical for the interaction of SecA with lipids [54, 55] and it has been shown that the first ten residues form an amphipathic helix in the membrane interface [16]. Because the peptide contains no Trp residue, partitioning was determined by measuring molar ellipticity at 222 nm. The general behavior of the peptide in the absence and presence of lipid vesicles is very similar the behavior of the melittin 26-residue amphiphilic peptide [56–58]. For all measurements, the peptide concentration was 40 μM. (A) In the absence of LUV (dotted black curve) or in the presence POPC LUVs (gray curve), the spectra are those expected for unfolded/unbound peptides. There is weak binding of the peptide to POPC:POPE (50:50) LUV (green curve), indicated by the appearance of a weak α-helical signal at 222 nm. The α-helicity of the peptide increases dramatically in the presence of POPE:POPG (80:20, red curve), POPE:CL (80:20, blue curve), or E. coli lipids (black curve). (B) Titration curves obtained by titrating the 40 μM peptide solution with LUVs of varying composition at 37 °C monitored by the change in molar ellipticity ([Θ]) at 222 nm. The color codes are the same as in panel A. The titration curves show that the peptide binds most strongly to E. coli lipids. The partition coefficients (Kx) and free energies of transfer (ΔGwb) determined from these curves are shown in Table 2.
Table 2.
Summary partitioning free energies at 37°C of the 21-residue N-terminus peptide. Titration curves from which these data are obtained are provided in Figure 5B.
| aLipid | bComposition | cKx ×10−3 | dΔGwb |
|---|---|---|---|
| E. coli extract | — | 224 ± 19 | −7.6 ± 0.1 |
| PC | — | en.o. | en.o. |
| PE:PC | 50:50 | 13 ± 3.2 | −5.8 ± 0.2 |
| PE:PG | 80:20 | 252 ± 29 | −7.7 ± 0.1 |
| PE:CL | 80:20 | 281 ± 32 | −7.7 ± 0.1 |
PC, palmitoyloleoylphosphatidylcholine (POPC); PE, palmitolyloleoylphosphatidylethanolamine (POPE); PG, palmitolyloleoylphosphatidylglycerol (POPG); CL, cardiolipin.
mole ratio.
mole-fraction partition coefficient (see Methods)
free energy of transfer, water to bilayer, kcal mol−1
not observable
3.5. SecA does not bind to membranes as the 1M6N dimer
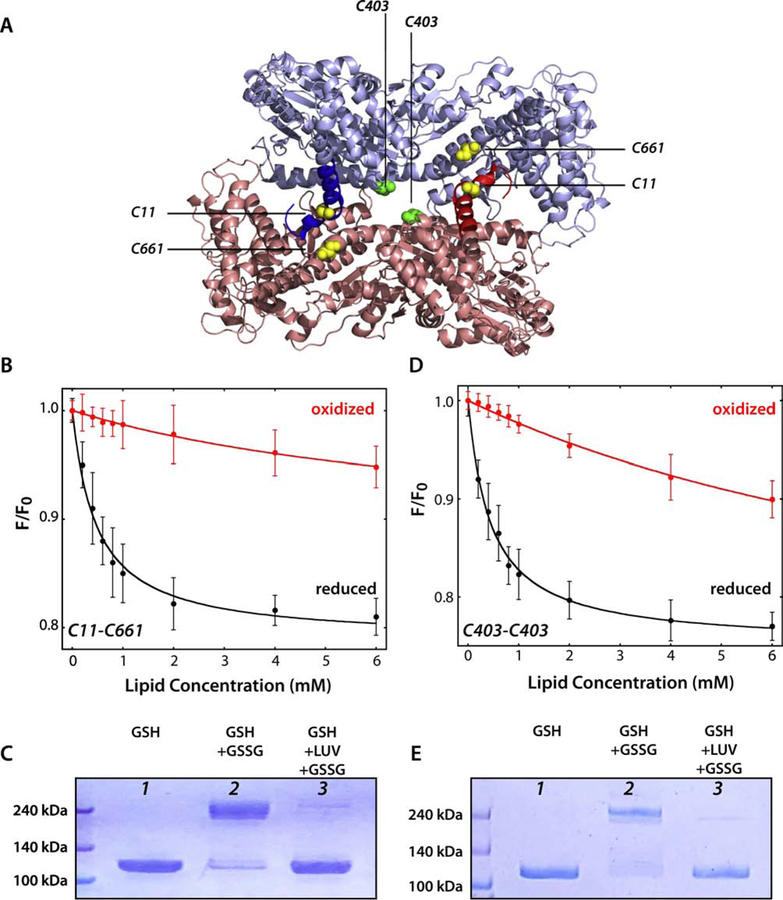
At this point, our data could not distinguish between SecA partitioning into LUVs as a monomer or as a dimer. Indeed, the titration of SecA with LUVs at very high salt concentration (> 400 mM), where the monomer-dimer equilibrium should be toward the monomer [22], resulted in transfer free energies similar to those observed at low salt concentration where SecA should exist as a dimer (Figure 3C). This suggested that the dimeric state is not required for membrane partitioning. We therefore examined the partitioning of two disulfide-stabilized dimers based upon the frequently assumed physiological dimer (PDB 1M6N). One of the dimers was formed by cross linking Cys11-Cys661, which was previously studied by Jilaveanu and Oliver [35]. In this construction, C11 is located on the N-terminus of one protomer and can be cross-linked with C661 on the rigid helical scaffold domain of the second protomer. This results in a stabilized head-to-tail dimer with two disulfide bridges (Figure 6A, yellow pair). The second dimer was designed using site-directed mutagenesis to mutate Serine 403 to cysteine, a position on one of the helices of the Nucleotide Binding Domain 1, resulting in the formation of a unique disulfide bridge (Figure 6A, green pair). Both SecA cysteine-mutants were expressed and purified as described in the Material and Methods section with the exceptions that oxygen was introduced during the purification steps, dithiothreitol was left out, and oxidized glutathione (GSSG) was introduced to favor disulfide bond formation. Only fractions containing the dimer (≈ 200 kDa) after the gel filtration were pooled together.
Figure 6. SecA binds weakly to LUV as the 1M6N dimer.
Cysteines were engineered into a Cys-free SecA construct to stabilize the 1M6N dimer under oxidizing conditions. (A) 3D structure of the 1M6N dimer in a head-to-tail disposition. The stabilizing pairs of cysteines for mutant C11-C661 and C403 are highlighted in yellow and green, respectively. (B) Titration with E. coli LUVs of the C11-C661 dimer (4 µM) under oxidizing (red curve) or reducing conditions (black curve). The Cys-stabilized dimer binds very weakly compared to the monomer (Table 3). (C) Coomassie blue-stained gels of C11–661-SecA protein under different conditions. Lanes 1 and 2 show, respectively, that SecA is monomeric under reducing conditions and dimeric under oxidizing conditions. For lane 2, an excess of oxidized glutathione was introduced by dialysis. In lane 3, SecA was incubated with LUV (E. coli lipids, 4 mM) for 30 minutes under reducing conditions to allow equilibrium binding of ~95% of SecA. Then, oxidized glutathione was introduced by dialysis to cross link any SecA dimers that existed on the membrane. The great preponderance of SecA migrated on the gel as a monomer, indicating that SecA either partitioned as a monomer or as a dimer different from 1M6N formed upon partitioning. (D) Titration of C403 SecA with E. coli LUVs under oxidizing (red curve) or reducing conditions (black curve). The Cys-stabilized dimer binds very weakly compared to the monomer (Table 3). (E) Coomassie blue-stained gels of C403-SecA protein using the same protocol as in panel C. The results are consistent with membrane-bound SecA being monomeric, as in panel C.
We examined the partitioning at 37°C of the reduced and oxidized forms of both proteins (Figure 6 B and D). Neither of the oxidized dimeric forms partitioned strongly into LUVs formed from E. coli lipids whereas both reduced forms partitioned strongly. The difference between the partition coefficients of oxidized or reduced SecA from water to bilayer is roughly two orders of magnitude (Table 3), meaning that the partitioning of reduced SecA is about a hundred times greater than the partitioning of the oxidized form. This suggests that the ability of the homodimer to dissociate or to assume a different dimer structure to assure full exposure of the N-terminus is important for the binding of SecA to lipid membranes.
Table 3.
Summary of SecA partitioning free energies into lipid vesicles formed from E. coli lipids at 37 °C for reduced and oxidized dimer constructs.
To determine if SecA maintains its physiological quaternary structure (1M6N) upon membrane binding under reducing conditions, we added an oxidizer to allow the formation of the disulfide bridge (C11-C661 or C403-C403) (Figure 6 C and E). In the absence of lipid vesicles, the introduction of the oxidizer (oxidized glutathione, GSSG) resulted in the formation of the dimeric form of SecA (Lane 2). However, when the oxidizer is added after the partitioning of SecA onto lipid vesicles (Lane 3), the protein is monomeric on SDS gels despite the presence of the oxidizer. This supports the idea that that the protein either monomerizes upon membrane binding, as previously proposed [32, 33] or that a dimer other than 1M6N is formed upon partitioning.
4. Discussion
We have examined the partitioning of SecA into large unilamellar vesicles formed from various lipids with a focus on E. coli lipids. Our systematic measurements (Figure 3A, Table 1) expand upon earlier studies showing the necessity for negatively charged lipids for the membrane binding of SecA [9] [10–12]. Because we now know that the principal cytoplasmic salt of E. coli is KGlu [43–45] and that KGlu stabilizes SecA against thermal unfolding in solution [45], we compared membrane partitioning of SecA in KCl and KGlu aqueous solutions (Figure 2, Figure 3C) and examined the thermal stability of SecA bound LUVs under the two salt conditions (Figure 4). At 37°C, the free energy of transfer (ΔGwb) of SecA to LUV formed from E. coli lipids is somewhat more favorable in KCl solutions (−7.8 ± 0.1 kcal mol−1) than in KGlu solutions (−7.4 ± 0.1 kcal mol−1). Using a very simple in vitro model membrane system and pure SecA, we have thus shown that SecA binds avidly to membranes containing negatively charged lipids. However, the implications for SecA binding in vivo in E. coli, where both the cytoplasm and membrane are crowded, remain to be determined.
The electrolyte concentration-dependence of SecA partitioning into E. coli LUVs is also affected by whether KCl or KGlu solutions are used (Figure 3C). As for SecA in solution [45], KGlu stabilizes SecA against thermal unfolding when membrane bound (Figure 4). Interestingly, the magnitude of ΔGwb increases linearly with increasing KGlu concentration whereas the magnitude decreases linearly with increasing KCl concentrations. Linear extrapolations of the data suggest that the values of ΔGwb intersect at salt concentrations of about 0.8 M. The reason for this behavior is presently not clear, but it is likely due to subtle differences in entropy and enthalpy of SecA upon partitioning. Plotting ΔGwb against temperature shows that the free energy of partitioning is persistently lower in KGlu than in KCl between 20° and 35°C (Figure 3B). We suggest that this is due to the greater thermal stability of SecA in KGlu, which reduces exposure of buried hydrophobic amino acid sidechains. Consistent with this conclusion, the magnitude of ΔGwb increases dramatically above the thermal melting temperatures determined in Figure 4.
There is general agreement in the literature that the N-terminus of SecA is required for membrane partitioning [6, 15, 16]. In confirmation, we examined the partitioning of a synthetic 21 residue N-terminal sequence into LUV formed from various lipid mixtures (Figure 5). The free energies of transfer for the synthetic peptide are virtually identical to the values for SecA (Table 3). This suggests that the N-terminus of SecA might be unfolded and highly exposed when free in solution.
Finally, we examined the question of whether SecA partitions as a dimer [21–23], believed by many to be represented by the 1M6N crystal structure of B. subtilis SecA [22, 23, 29, 30]. We did this by creating 1M6N-based dimers by engineering Cys residues into the dimer interface and examining SecA partitioning under reducing and oxidizing conditions. The results showed that the cross-linked dimer binds only weakly to E. coli LUV (Figure 6B, 6D) and that SecA partitioned into the LUV cannot be significantly cross-linked into the 1M6N dimer under oxidizing conditions (Figure 6C, 6E). These observations suggest three possibilities. (i) The weak binding of the 1M6N-dimer SecA is sufficient for its key role in translocation. (ii) SecA binds the membrane as a dimer other than 1M6N. No fewer than five different SecA crystal structures with different subunit interfaces have been reported [24–28]. The binding to the membrane might induce the rearrangement of the quaternary structure of SecA, resulting in a dimer different than 1M6N, as suggested by Gouridis et al. [53] who reported the co-existence of interchangeable dimer conformers in vitro, where the two protomers of dimeric SecA can move relative to one another without dissociating. (iii) Finally, SecA might dissociate into monomers upon interaction with E. coli membranes, consistent with an earlier study by Or et al. [32]. In any case, the crucial question is the role of membrane partitioning in the binding of SecA to the SecYEG translocon.
Supplementary Material
Highlights.
Partitioning of SecA into lipid vesicles of various compositions has been measured
Negatively charged lipids are required for strong partitioning, as expected
The natural E. coli cytoplasmic salt potassium glutamate stabilizes vesicle-bound SecA
Dimeric SecA correponding to the 1M6N crystallographic structure partitions weakly
Acknowledgements
This work was supported by NIH grant GM074637. We are grateful to Donald Oliver for providing plasmids for Cys-free SecA constructs, Dr. Eric Lindner for his advice on protein expression and purification, and Dr. Gargi Dasgupa for outstanding technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Oliver DB, Beckwith J, The identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli, J. Bacteriol, 150 (1982) 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Oliver DB, Beckwith J, Regulation of a membrane component required for protein secretion in Escherichia coli, Cell, 30 (1982) 311–319. [DOI] [PubMed] [Google Scholar]
- [3].Crane JM, Randall LL, The Sec system: Protein export in Escherichia coli, EcoSalplus, (2017) 10.1128/ecosalplus.ESP-0002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W, The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane, Cell, 63 (1990) 269–279. [DOI] [PubMed] [Google Scholar]
- [5].Cabelli RJ, Dolan KM, Qian L, Oliver DB, Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli, J. Biol. Chem, 266 (1991) 24420–24427. [PubMed] [Google Scholar]
- [6].van Voorst F, de Kruijff B, Role of lipids in the translocation of proteins across membranes, Biochem. J, 347 (2000) 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Koch S, de Wit JG, Vos L, Birkner JP, Gordiichuk P, Herrmann A, van Oijen AM, Driessen AJM, Lipids activate SecA for high affinity binding to the SecYEG complex, J. Biol. Chem, 291 (2016) 22534–22543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B, Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes, Nature, 334 (1988) 173–175. [DOI] [PubMed] [Google Scholar]
- [9].Breukink E, Demel RA, de Korte-Kool G, de Kruijff B, SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study, Biochemistry, 31 (1992) 1119–1124. [DOI] [PubMed] [Google Scholar]
- [10].Ulbrandt ND, London E, Oliver DB, Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding, J. Biol. Chem, 267 (1992) 15184–15192. [PubMed] [Google Scholar]
- [11].Keller RCA, Snel MME, de Kruijff B, Marsh D, SecA restricts, in a nucleotide-dependent manner, acyl chain mobility up to the center of a phospholipid bilayer, FEBS Lett, 358 (1995) 251–254. [DOI] [PubMed] [Google Scholar]
- [12].Ahn T, Kim J-S, Lee B-C, Yun C-H, Effects of lipids on the interaction of SecA with model membranes, Arch. Biochem. Biophys, 395 (2001) 14–20. [DOI] [PubMed] [Google Scholar]
- [13].Rajapandi T, Oliver D, Integration of SecA protein into the Escherichia coli inner membrane is regulated by its amino-terminal ATP-binding domain, Mol. Microbiol, 20 (1996) 43–51. [DOI] [PubMed] [Google Scholar]
- [14].Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B, The C terminus of SecA is involved in both lipid binding and SecB binding, J. Biol. Chem, 270 (1995) 7902–7907. [DOI] [PubMed] [Google Scholar]
- [15].Dapic V, Oliver D, Distinct membrane binding properties of N- and C-terminal domains of Escherichia coli SecA ATPase, J. Biol. Chem, 275 (2000) 25000–25007. [DOI] [PubMed] [Google Scholar]
- [16].Findik BT, Smith VF, Randall LL, Penetration into membrane of amino-terminal region of SecA when associated with SecYEG in active complexes, Protein Sci, 27 (2018) 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hristova K, Dempsey CE, White SH, Structure, location, and lipid perturbations of melittin at the membrane interface, Biophys. J, 80 (2001) 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hristova K, Wimley WC, Mishra VK, Anantharamaiah GM, Segrest JP, White SH, An amphipathic a-helix at a membrane interface: A structural study using a novel x-ray diffraction method, J. Mol. Biol, 290 (1999) 99–117. [DOI] [PubMed] [Google Scholar]
- [19].Akita M, Shinkai A, Matsuyama S.-i., Mizushima S, SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer, Biochem. Biophys. Res. Commun, 174 (1991) 211–216. [DOI] [PubMed] [Google Scholar]
- [20].Driessen AJM, SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer, Biochemistry, 32 (1993) 13190–13197. [DOI] [PubMed] [Google Scholar]
- [21].Chen Y, Pan X, Tang Y, Quan S, Tai PC, Sui S-F, Full-length Escherichia coli SecA dimerizes in a closed conformation in solution as determined by cryo-electron microscopy, J. Biol. Chem, 283 (2008) 28783–28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wowor AJ, Yu D, Kendall DA, Cole JL, Energetics of SecA dimerization, J. Mol. Biol, 408 (2011) 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wowor AJ, Yan Y, Auclair SM, Yu D, Zhang J, May ER, Gross ML, Kendell DA, Cole JL, Analysis of SecA dimerization in solution, Biochemistry, 53 (2014) 3248–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J, Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA, Science, 297 (2002) 2018–2026. [DOI] [PubMed] [Google Scholar]
- [25].Vassylyev DG, Mori H, Vassylyev MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K, Crystal structure of the translocation TPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer, J. Mol. Biol, 364 (2006) 248–258. [DOI] [PubMed] [Google Scholar]
- [26].Papanikolau Y, Papadovasilaki M, Ravelli RBG, McCarthy AA, Cusack S, Economou A, Petratos K, Structure of dimeric SecA, the Escherichia coli preprotein translocase motor, J. Mol. Biol, 366 (2007) 1545–1557. [DOI] [PubMed] [Google Scholar]
- [27].Zimmer J, Li W, Rapoport TA, A novel dimer interface and conformational changes revealed by an x-ray structure of B. subtilis SecA, J. Mol. Biol, 364 (2006) 259–265. [DOI] [PubMed] [Google Scholar]
- [28].Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR Jr, Sacchettini JC, Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Auclair SM, Oliver DB, Mukerji I, Defining the solution state dimer structure of Escherichia coli SecA using Förster resonance energy transfer, Biochemistry, 52 (2013) 2388–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Banerjee T, Lindenthal C, Oliver D, SecA functions in vivo as a discrete anti-parallel dimer to promote protein transport, Mol. Microbiol, 103 (2017) 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hendrick JP, Wickner W, SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane, J. Biol. Chem, 266 (1991) 24596–24600. [PubMed] [Google Scholar]
- [32].Or E, Navon A, Rapoport T, Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane, EMBO J, 21 (2002) 4470–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Or E, Rapoport T, Cross-linked SecA dimers are not functional in protein translocation, FEBS Lett, 581 (2007) 2616–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].de Keyzer J, van der Sluis EO, Spelbrink REJ, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJM, Covalently dimerized SecA is functional in protein translocation, J. Biol. Chem, 280 (2005) 35255–35260. [DOI] [PubMed] [Google Scholar]
- [35].Jilaveanu LB, Oliver D, SecA dimer cross-linked at its subunit interface is functional for protein translocation, J. Bacteriol, 188 (2006) 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zimmer J, Nam Y, Rapoport TA, Structure of a complex of the ATPase SecA and the protein-translocation channel, Nature, 455 (2008) 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level, Structure, 19 (2011) 430–439. [DOI] [PubMed] [Google Scholar]
- [38].Benach J, Chou Y-T, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF, Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA, J. Biol. Chem, 278 (2003) 3628–3638. [DOI] [PubMed] [Google Scholar]
- [39].Joly JC, Wickner W, The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids, EMBO J, 12 (1993) 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].van Voorst F, van der Does C, Brunner J, Driessen AJM, de Kruijff B, Translocase-bound SecA is largely shielded from the phospholipid acyl chains, Biochemistry, 37 (1998) 12261–12268. [DOI] [PubMed] [Google Scholar]
- [41].Janin J, For Guldberg and Waage, with love and cratic entropy, Proteins, 24 (1996) i–ii. [DOI] [PubMed] [Google Scholar]
- [42].White SH, Wimley WC, Ladokhin AS, Hristova K, Protein folding in membranes: Determining energetics of peptide-bilayer interactions, Methods Enzymol, 295 (1998) 62–87. [DOI] [PubMed] [Google Scholar]
- [43].Culham DE, Shkel IA, Record MT Jr., Wood JM, Contributions of coulombic and Hofmeister effects to the osmotic activation of Escherichia coli transporter ProP, Biochemistry, 55 (2016) 1301–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cheng X, Guinn EJ, Buechel E, Wong R, Sengupta R, Shkel IA, Record MT Jr., The basis of protein stabilization by K glutamate: Unfavorable interactions with carbon, oxygen groups, Biophys. J, 111 (2016) 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Roussel G, Lindner E, White SH, Stabilization of SecA ATPase by the Primary cytoplasmic salt of Escherichia coli, Protein Sci, 28 (2019) 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bauer BW, Shemesh T, Chen Y, Rapoport TA, A “push and slide” mechanism allows sequence-insentive translocation of secretory proteins by the SecA ATPase, Cell, 157 (2014) 1416–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bartlett GR, Phosphorus assay in column chromatography, J. Biol. Chem, 234 (1959) 466–468. [PubMed] [Google Scholar]
- [48].Tanford C, The Hydrophobic Effect: Formation of Micelles and Biological Membranes, 1 ed., John Wiley & Sons, New York, 1973. [Google Scholar]
- [49].Lewis GN, Randall M, Thermodynamics, 2 ed., McGraw-Hill, New York, 1961. [Google Scholar]
- [50].Ladokhin AS, Jayasinghe S, White SH, How to measure and analyze tryptophan fluorescence in membranes properly, and why bother?, Anal. Biochem, 285 (2000) 235–245. [DOI] [PubMed] [Google Scholar]
- [51].Boggs JM, Lipid intermolecular hydrogen bonding: Influence on structural organization and membrane function, Biochim. Biophys. Acta, 906 (1987) 353–404. [DOI] [PubMed] [Google Scholar]
- [52].Schmidt MG, Rollo EE, Grodberg J, Oliver DB, Nucleotide sequence of the secA gene and secA(Ts) mutations preventing export in Escherichia coli, J. Bacteriol, 170 (1988) 3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gouridis G, Karamanou S, Sardis MF, Schärer MA, Capitani G, Economou A, Quaternary dynamics of the SecA motor drive translocase catalysis, Mol. Cell, 52 (2013) 655–666. [DOI] [PubMed] [Google Scholar]
- [54].Das S, Stivision E, Folta-Stogniew E, Oliver D, Reexamination of the role of the amino terminus of SecA in promoting its dimerization and functional state, J. Bacteriol, 190 (2008) 7302–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Breukink E, Keller RC, de Kruijff B, Nucleotide and negatively charged lipid-dependent vesicle aggregation caused by SecA. Evidence that SecA contains two lipid-binding sites., FEBS Lett, 331 (1993) 19–24. [DOI] [PubMed] [Google Scholar]
- [56].Vogel H, Incorporation of melittin into phosphatidylcholine bilayers: Study of binding and conformational changes, FEBS Lett, 134 (1981) 37–42. [DOI] [PubMed] [Google Scholar]
- [57].Ladokhin AS, White SH, Folding of amphipathic a-helices on membranes: Energetics of helix formation by melittin, J. Mol. Biol, 285 (1999) 1363–1369. [DOI] [PubMed] [Google Scholar]
- [58].Ladokhin AS, Fernández-Vidal M, White SH, CD spectroscopy of peptides and proteins bound to large unilamellar vesicles, J. Membr. Biol, 236 (2010) 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.