Abstract
The Orange Carotenoid Protein (OCP) is a structurally and functionally modular photoactive protein involved in cyanobacterial photoprotection. Recently, based on bioinformatic analysis and phylogenetic relationships, new families of OCP have been described, OCP2 and OCPx. The first characterization of the OCP2 showed both faster photoconversion and back-conversion, and lower fluorescence quenching of phycobilisomes relative to the well-characterized OCP1. Moreover, OCP2 is not regulated by the fluorescence recovery protein (FRP). In this work, we present a comprehensive study combining ultrafast spectroscopy and structural analysis to compare the photoactivation mechanisms of OCP1 and OCP2 from Tolypothrix PCC 7601. We show that despite significant differences in their functional characteristics, the spectroscopic properties of OCP1 and OCP2 are comparable. This indicates that the OCP functionality is not directly related to the spectroscopic properties of the bound carotenoid. In addition, the structural analysis by X-ray footprinting reveals that, overall, OCP1 and OCP2 have grossly the same photoactivation mechanism. However, the OCP2 is less reactive to radiolytic labeling, suggesting that the protein is less flexible than OCP1. This observation could explain fast photoconversion of OCP2.
Keywords: OCP1, OCP2, photoactivation, X-ray footprinting, ultrafast spectroscopy
Introduction
Photosynthetic organisms have developed photoprotective mechanisms allowing them to cope with high light conditions when their antenna systems absorb excess energy that cannot be utilized for charge separation in reaction centers. Mechanisms of this non-photochemical quenching (NPQ) vary, but the essential NPQ agents are almost exclusively carotenoids [1–6]. In plants and algae, the NPQ site(s) are located in antenna proteins embedded in thylakoid membrane [7], while in cyanobacteria, which utilize phycobilisomes as an antenna system, the Orange Carotenoid Protein (OCP) is essential for photoprotection [1, 8, 9].
The OCP is a 35 kDa water soluble protein binding a single carotenoid. The first crystal structure of the OCP [10] revealed two discrete structural domains: an all-helical N-terminal domain (NTD), composed of two discontinuous four-helix bundles, and a C-terminal domain (CTD) with a mixed α-β fold. The NTD (pfam09150) is found only in cyanobacteria, whereas the CTD is a member of the ubiquitous Nuclear Transport Factor-2 (NTF2) superfamily (pfam02136). The carotenoid spans the two protein domains, through the largest interdomain interface (the major interface) which is stabilized by a salt bridge (R155-E244). The binding of a carotenoid containing a conjugated keto group (a keto–carotenoid) involves two hydrogen bonds to absolutely conserved aromatic residues, Y201 (Y203 for Tolypothrix OCP1) and W288 (W290 for Tolypothrix OCP1), in the CTD. OCP-associated keto-carotenoids include 3’hydroxyechinenone (hECN) [10] (ECN) or canthaxanthin (CAN) [11–13], OCP isolated from an axenic cyanobacterial culture can contain a mixture of carotenoids [11]; Kerfeld et al., have proposed that the identity of the carotenoid contributes specificity to functionality [9, 14]. The OCP is photoactive [15]; upon blue-green light illumination the OCP converts to an active form that binds to phycobilisome and induces antenna quenching [2]. The active form has red-shifted absorption spectrum therefore, it is denoted as OCP-red (OCPR) while the inactive, orange form is referred to as OCPO. The OCPO-OCPR conversion involves a global structural change: the CTD-and NTD of OCP become separated and the carotenoid moves 12 Å, to fully span the NTD [13, 16]. This large structural rearrangement is reversible as the OCPR-OCPo conversion occurs even in dark and the reversion can be facilitated by the fluorescence recovery protein (FRP) [17, 18].
The presence of the conjugated keto-group at the carotenoid terminal ring buried in the CTD is a necessary structural feature to achieve OCPO-OCPR conversion. OCP mutants containing carotenoids lacking the keto-group, such as e.g. zeaxanthin, are not able to undergo conversion to OCPR and remain locked in the OCPO state [11]. On the other hand, mutations of W288, one of the residues involved in hydrogen bonding to the carotenoid keto-group, results in OCP having absorption spectrum resembling that of OCPR [19–21]. These mutants with red-shifted absorption spectra, W288F or W288A, also persistently quench phycobilisomes [20, 21], indicating that these mutations likely lock OCP in the active OCPR state.
The mechanism of the light-induced OCPO-OCPR conversion remains elusive. Especially because very low OCPO-OCPR conversion efficiency is required, which is less than 1% [15, 22]. Yet, a few recent studies shed some light on this process. It is clear that light absorption by the carotenoid bound to OCP triggers a sequence of structural arrangements resulting in separation of the NTD and CTD and migration of the carotenoid into the NTD [22, 23]. Using nanosecond flash photolysis and monitoring Trp fluorescence, Maksimov et al. [20] identified an intermediate which they assigned to an OCP state in which the carotenoid still spans the C-and N-terminal domains, but the hydrogen bonds to the carotenoid keto group are broken [20]. Recently, Konold et al. [22] used femtosecond-to-microsecond visible and infrared transient absorption spectroscopy to identify two additional spectroscopic OCP states corresponding to structural intermediates in the OCPO to OCPR pathway [22].
The primary trigger inducing the chain of reactions eventually leading to OCPR is likely disruption of hydrogen bonds between Trp and Tyr and keto oxygen of the carotenoid [11, 13, 24]. However, what exactly causes this bond breaking remains unclear. It has been proposed that light absorption induces rotation of β-ring nested in the C-terminal domain resulting in breaking the hydrogen bonds [20, 25]. On the other hand, Bandara et al. proposed, based on results of dynamic crystallography, that a transient shift in keto-enol equilibrium disrupts the hydrogen bonds [19]. Recently, femtosecond transient absorption data suggested that a specific carotenoid state, typically denoted as the S* state, is the key state leading to the disruption of the hydrogen bonds [22]. The S* state of long (having more than 11 conjugated C=C bonds) carotenoids, such as ECN or CAN typically bound to OCP, is likely related to a hot carotenoid ground state that dissipates its energy to the immediate surrounding of the carotenoid [26]. It is thus possible that energy absorbed by the carotenoid in OCP is transferred to vibrational modes of the amino acids nearby leading to the disruption of Trp and Tyr hydrogen bonds. Similar mechanism of H-bond breaking is known for example in water, in which the intermolecular vibrational modes coupled to H-bond causes its breaking [27], and transfer of vibrational energy from retinal to protein has also been reported for bacteriorhodopsin [28].
Ultrafast transient spectroscopy was also used to characterize spectroscopic properties of the carotenoid bound to both OCPO, OCPR or RCP (i.e. the isolated NTD binding carotenoid). The first ultrafast experiment showed that the lifetime of the lowest excited state of hECN in OCPO, 3.3 ps, is significantly shorter than 6 ps observed in solution, following the change of the β-ring configuration. In OCPO it takes the s-trans configuration while s-cis is the lowest energy configuration in solution, resulting in a change in effective conjugation length of the carotenoid and consequently also of the S1 lifetime [29]. About the same lifetimes were reported for RCP or OCPR [30–32] suggesting that even when the carotenoid is shifted to the NTD, one of the β-rings is in s-trans configuration. These results were obtained for OCPs containing either hECN [31, 32] or CAN [33].
Ultrafast spectroscopy data further provided information about intramolecular charge transfer (ICT) state of the carotenoid bound to OCP. The ICT state is a characteristic feature of certain keto-carotenoids and is readily observed in transient absorption spectra as a separate band appearing only in polar solvents [34, 35]. Yet, keto-carotenoids with long (N>11) conjugation and with a conjugated keto-group located at the terminal ring, such as ECN or CAN, typically do not exhibit such a band [30]. It has been demonstrated, however, that the ICT band may appear upon binding these carotenoids to OCP, indicating that the OCP binding pocket may activate the ICT state [32, 33]. The spectroscopic features related to the ICT were reported to be even more pronounced in OCPR [32] or RCP [33], thus the ICT state was suggested to play a key role in the quenching [32].
Besides the role of the ICT state, spectroscopic data recorded during the past few years also revealed spectral heterogeneity of OCPO. Slight differences in spectral and dynamical response obtained after excitation of OCPO at 495 and 530 nm were reported already in 2005 [29]. These differences were later explained as due to a small fraction of OCPR present in OCPO sample [31], but resonance Raman data suggested that spectral heterogeneity occurs also within the OCPO itself. This was further confirmed by transient absorption data measured on OCP binding zeaxanthin that is incapable of OCPO-OCPR conversion [24]. Even for this OCP, which is locked in OCPO state, two spectroscopically distinct zeaxanthins were identified [33]. The origin of this heterogeneity is unclear, but studies carried out by Maksimov et al. [25] suggested that there are two additional metastable configurations of the terminal ring which retain the hydrogen bonding to Trp and Tyr. As the change of torsional angle of the terminal ring may vary spectroscopic properties substantially [36, 37], a fraction of these configurations may explain the observed spectral heterogeneity of OCPO.
The vast majority of these studies have been carried out with the OCP derived from the unicellular model cyanobacterium Synechocystis PCC 6803. However, in the past five years there has been a significant increase in the number of available cyanobacterial genome sequences and a recent detailed bioinformatic analyses of these data revealed new types of OCPs together with new families of homologs to the NTD, Helical Carotenoid Proteins (HCPs), and to the CTD homologs (CTDHs) [38]. Both HCPs and CTDHs bind carotenoid [21, 39]. In addition, besides the canonical, well characterized OCP found in Arthrospira or Synechocystis (hereafter OCP1), two other OCPs, denoted as OCP2 and OCPx were identified in some cyanobacteria.[40] Considering the principles of protein evolution combined with the taxonomic species distribution, it has been hypothesized that an ancestral OCP was derived from an HCP/CTDH fusion event [9, 10, 38, 39]. OCP2 appears to be more primitive than OCP1 [8], reflecting an intermediate stage in the evolution of photoprotection in cyanobacteria. To-date, all of the studies detailing the OCP photocycle have been confined to the canonical OCP1.
The filamentous cyanobacterium Tolypothrix sp. PCC 7601 (hereafter Tolypothrix) encodes OCP1 and OCP2. The paralogs when expressed in a CAN producing E. coli strain, readily bound CAN [40]. Although OCP1 and OCP2 have similar absorption spectra, their functional properties are markedly different. OCP2 is expressed only under high irradiance and exhibits faster OCP2O↔OCP2R turnover than OCP1 [40]. Moreover, back OCP2R-OCP2O conversion is not controlled by FRP and OCP2 is less efficient in phycobilisome quenching than OCP1 [40].
Here we combine two techniques, the ultrafast transient absorption spectroscopy and X-ray Footprinting with mass spectrometry (XFMS), an ideal technique for revealing protein conformational dynamics at the single residue level, to characterize structural events in photoactivation of OCP1 and OCP2. Additionally, in contrast to previous experiments, we have expanded the spectral window used in transient absorption spectroscopy to near-IR spectral region to better monitor spectral features related to the carotenoid ICT state. Further, an experimental setup allowing for continuous side illumination of the sample enabled to collect data directly on OCPR. We show that despite significantly different in photoactivation characteristics of OCP1 and OCP2, the spectroscopic properties of these two OCP types are comparable, showing that the difference is not directly related to the spectroscopic properties of the bound carotenoid. Structurally, we find that OCP2 is less flexible than OCP1, suggesting that the differences in the photoactivation mechanism are due to the protein rigidity.
Materials and Methods
1. Expression and purification of OCP-CAN holoproteins in E. coli
To express OCP binding CAN in E. coli, C-terminal 6× His-tagged Tolypothrix ocp1 or ocp2 genes were cloned in a pET28 expression vector (Novagene). Then, BL21(DE3) cells were transformed simultaneously with the pET28-OCP containing either ocp1 or ocp2 gene from Tolypothrix, and a pAC-CANTHipi plasmid (Addgene plasmid 53301) producing of CAN. OCP-CAN holoprotein (OCP1 and OCP2) expressed in E. coli was isolated according to the method described previously.[8] These samples, binding exclusively CAN, were used for all spectroscopic and XFMS measurements.
2. Overexpression and purification of OCP1 and OCP2 in Tolypothrix
To construct an overexpression vector of OCP1 and OCP2–7x His-tagged, the gene was PCR-amplified from genomic DNA of Tolypothrix, cloned between the constitutively expressed apcA promoter [41] and a sequence coding for HisTag, and OCP1 was expressed on a pDU1-based replicating plasmid [42] and OCP2 was expressed on a RSF1010 replicating plasmid (pSL80). The resulting plasmid (pHB4 and pHB5) was introduced into WT Tolypothrix strain SF33, a shortened filament strain [43], by conjugation, using the conjugative plasmid pRL443 [44] and the methylating plasmid pSL1 [45]. The OCP1 and OCP2 overexpression strains were grown in buffered BG-11 pH 8.0 medium supplemented with 25 μg/ml of kanamycin. The purification of both proteins expressed in Tolypothrix was done according to the method described previously [45].
3. Extraction and analysis of carotenoids by LC-MS/MS
In order to determine carotenoid composition in a native Tolypothrix OCPs, we determined the carotenoid content according to the following protocol. The Tolypothrix OCP1 and OCP2 overexpression strains were grown under red-enriched light with 3% of CO2. Samples of purified OCP1 and OCP2 were extracted with acetone at −20 °C for 30 min. After centrifugation (21,000 g for 10 min at 4 °C), the supernatant was incubated at −20 °C for 30 min and then centrifuged. This procedure was repeated several times until a white pellet was obtained, which indicated the absence of pigment in the protein sample. The detection of the carotenoids was done according to the method described previously.[45] Analysis by LC-MS/MS showed that OCP2 naturally bound mainly (77%) the keto-carotenoid CAN, with minor contribution of ECN (22%) and β-carotene (1%). OCP1 bound nearly exclusively CAN (96%) with a minor fraction of ECN (4%).
4. Crystallization of OCP1
A 4 mg/ml solution of Tolypothrix OCP1 spontaneously crystallized in 0.75 M potassium phosphate pH 7.0 at 4 °C. Crystals were stabilized by adding 30% of glycerol, mounted in a nylon loop (CrystalCap ALS HT, Hampton Research), and then frozen in liquid nitrogen. X-ray diffraction was measured at beamline 5.0.2 of the Advanced Light Source (ALS) at Lawrence Berkeley National Lab.
Diffraction data were integrated with XDS [46] and scaled with SCALA (CCP4).[47] The OCP1 structure was solved by molecular replacement using 3MG1 with phenix.phaser [48]. The resulting solution was refined and rebuilt using phenix.refine [48] and COOT [49]. Statistics for diffraction data collection, structure determination and refinement are summarized in Table 1.
Further analysis of the structure was performed using the following software: PDBePISA at the EBI (http://www.ebi.ac.uk/pdbe/pisa/) and Profunc (https://www.ebi.ac.uk/thorntonsrv/databases/profunc/) [47].
Structural coordinates have been deposited in the Protein Data Bank (http://www.wwpdb.org/) under the accession code 6PQ1.
5. XFMS measurement, mass spectrometry and data analysis
OCP1 and OCP2 (expressed in E. coli) was exchanged into 10 mM potassium phosphate (pH 7.0), 100 mM NaCl by Size Exclusion Chromatography on a Superdex-75 10/300 GL column before XFMS experiments. Before X-ray irradiation of OCPR, the sample syringe was cooled with an ice pack and illuminated with a blue LED array (470 nm Luxeon Rebel; Philips Lumileds). Protein samples were irradiated in the microsecond time range at beamline 3.2.1 at the Advanced Light Source. All samples, including the control (no X-ray irradiation), were subjected to cysalkylation and desalting before overnight trypsin and endoproteinase Glu-C digestions at pH 8.0 and 37 °C. Proteolyzed samples were analyzed in an Agilent 6550iFunnel Q-TOF mass spectrometer (Agilent Technologies) coupled to an Agilent 1290 LC system (Agilent) using Sigma-Aldrich As-centis Peptides ES-C18 reverse phase column (2.1 mm×100 mm,2.7-μm particle size; Sigma-Aldrich) as it has been previously described in [Ref. 16].
6. Ultrafast spectroscopy
For all experiments, a 2 mm path length quartz cuvette with a magnetic stirrer, placed into a temperature-controlled sample holder (Quantum Northwest) was used. The sample temperature was kept at 10°C to prevent degradation and to minimize OCPO↔OCPR conversion during the experiment. For measurements on OCPR samples, a side entrance of the cuvette holder was used to continuously illuminate the sample by a LED diode. Absorption spectra of all samples were measured before and after experiments to ensure the integrity of samples during the transient absorption experiments.
Transient absorption experiments were performed on a home built set-up installed at the ELI Beamlines laser facility. The system is based on 1 kHz fs Titanium Sapphire amplifier (Astrella, Coherent co.) generating 34 fs femtosecond pulses centered at 800 nm. The main beam was split into excitation and probe beams by a dielectric beam splitter. The excitation beam was used to pump optical parametric amplifier (Topas Prime, Light Conversion) generating pulses of < 50 fs duration at desired wavelengths. The probe beam pumped another optical parametric amplifier (Topas Twins, Light Conversion) generating < 50 fs pulses centered at 1400 nm and used to generate white-light supercontinuum in a 2 mm thick CaF2 plate that was continuously translated in the beam to ease the heat deposition in the generation spot. A dichroic mirror with 350 – 1064 nm reflectivity (Semrock) was used to partially separate supercontinuum from the strong driving energy at 1400 nm. The resulting supercontinuum probe had detectable intensity in the entire recorded range from 450 to 1200 nm. Spectrum was resolved by a home-build quartz prism spectrograph that allowed image the entire probe spectra on the detector array without overlapping higher order diffractions that would happen with a grating spectrograph and otherwise prevented reliable recording spectra beyond the full octave range (e.g. second order diffraction at 500 nm overlaps with first order diffraction at 1000 nm). Excitation and probe pulses were focused and overlapped on the sample with spot sizes measured to be about 40 μm for probe and about 120 μm for pump. Temporal delay between pump and probe pulses was adjusted by remote controlled optical delay line inserted in the excitation path.
Transient absorption signal was measured by a CCD camera (Entwicklungsbuero Stresing) operated in full vertical binning achieving shot-to-shot detection at 1 kHz. Two optical choppers were used: 500 Hz in the probe path and 250 Hz in pump path for on-fly background correction. Chopper phases were directly fed into detection system as a TTL signal to align the data flow with the four-pulse sequence. The dwell time at each time delay was 2 s but several scans were collected to record statistics for the data quality assessment.
The obtained data were fitted using CarpetView global fitting analysis software (Light Conversion) under a sequential exponential decay scheme resulting in evolution-associated difference spectra (EADS) that correspond to the spectra of each spectral component in the sequential scheme.
Results
1. Steady-state absorption spectra
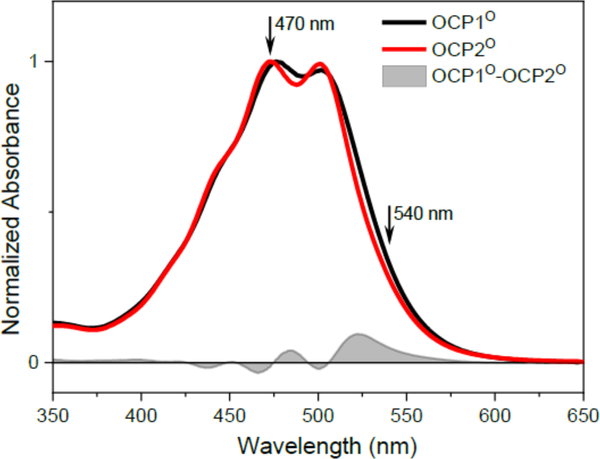
Absorption spectra of the dark-adapted OCP1 and OCP2 are shown in Fig. 1. Although the spectra of both paralogs are similar, there are subtle differences indicating distinct spectroscopic properties of both OCP1 and OCP2. The vibronic bands are more pronounced in OCP2, which may result from tighter locking the particular carotenoid conformation in the binding site. The absorption spectrum of OCP2 is also slightly blue-shifted from that of OCP1: the OCP2 absorption bands peaks at 473 nm (maximum) and 501 nm (the lowest energy band), while for OCP1 these peaks are at 477 and 502 nm. However, the major difference between the two paralogs is the increased absorption in the red part of the OCP1 absorption spectrum.
Figure 1:
Absorption spectra of OCP1O (black) and OCP2O (red) from Tolypothrix. The spectra are normalized to maximum. The grey are shows the OCP1O-OCP2O difference spectrum. Excitation wavelength used in transient absorption experiments are denoted by arrows.
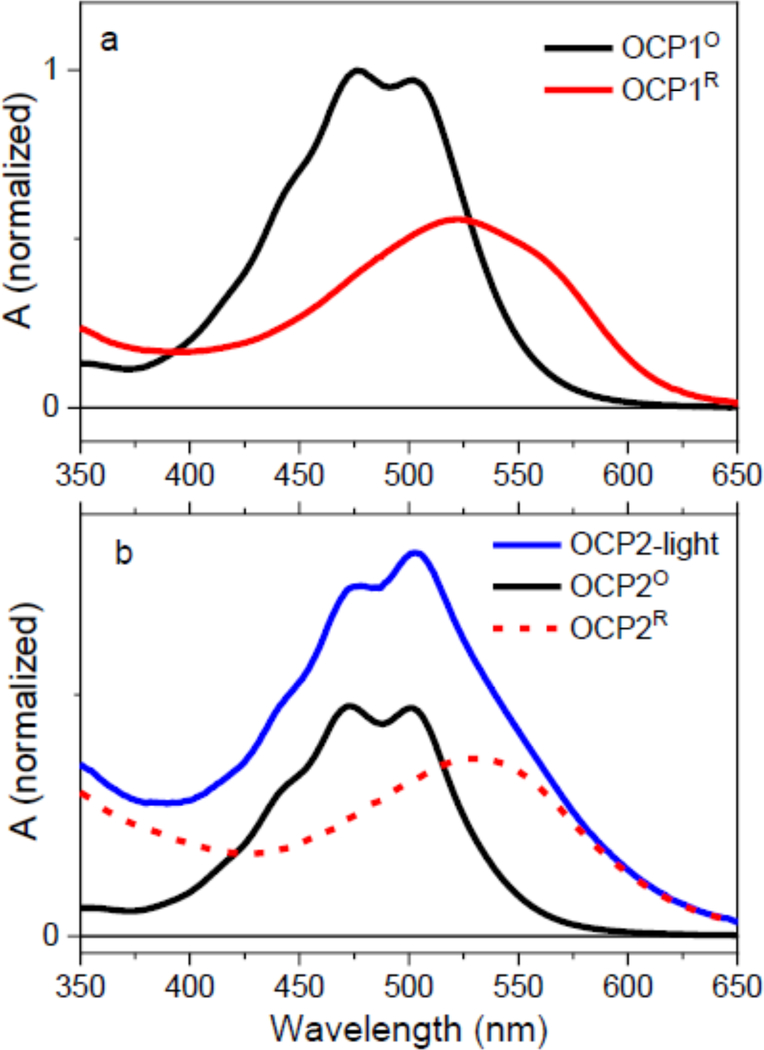
Upon illumination, both paralogs undergo transition to the activated form (OCPR) as shown in Fig. 2. The samples were illuminated continually and absorption spectra of the illuminated samples in Fig. 2 were measured after 10–15 minutes of illumination. We have tested that after this time an equilibrium was established and no further changes in absorption spectra were observed. This is important for transient absorption measurements, because the data collection in the transient absorption experiment takes approximately 60–90 minutes. The OCP1 converted completely to OCP1R as evidenced by the significant change in absorption spectrum that loses the resolution of vibronic bands and shifts to 525 nm (Fig. 2a). In OCP2 however, the illumination produces an equilibrium between OCP2O and OCP2R forms (spectrum denoted as OCP2-light in Fig. 2b). This is in agreement with earlier report [8], showing that both OCP2O to OCP2R conversion and back recovery are much faster than the corresponding reactions in OCP1. To estimate the spectrum of pure OCP2R, we subtracted a scaled OCP2O spectrum from the spectrum obtained after continuous illumination of OCP2. The resulting OCP2R spectrum has a maximum at 530 nm, matching the OCP2R spectrum reported earlier [8].
Figure 2:
OCPO to OCPR transition under illumination of OCP1 (a) and OCP2 (b) at 10 °C. While OCP1O is completely converted to OCPR, illumination of OCP2 leads to an equilibrium between OCP2O and OCP2R forms (OCP2-light, blue). The absorption spectrum of OCP2R (red) was estimated by substraction of scaled OCP2O spectrum (black) from the experimental data (blue).
2. Transient absorption spectroscopy
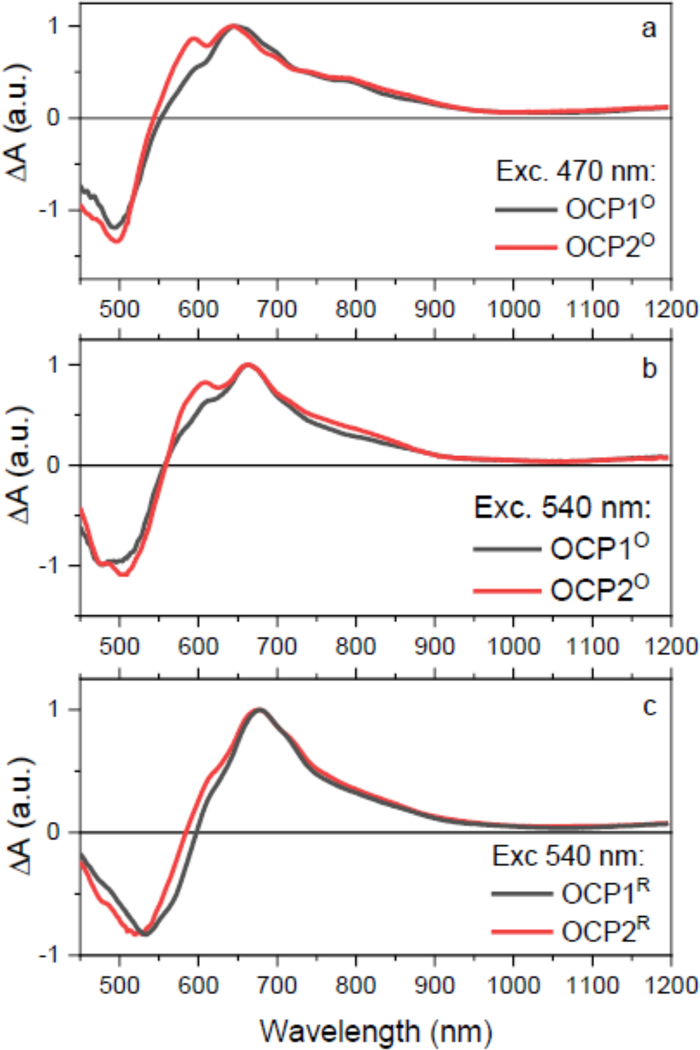
For the transient absorption measurements we have chosen two excitation wavelengths, 470 and 540 nm to excite blue and red parts of the absorption spectrum (see the arrows in Fig. 1), because it has been demonstrated that even the OCPO contains two spectral forms [33]. The transient absorption data of activated OCPR paralogs were collected under continuous side illumination of the sample to keep the OCP paralogs in the activated OCPR form under the whole course of the experiment. The OCPR paralogs were excited at 540 nm. Note that for OCP2, the illuminated complex inevitably contains a mixture of OCP2O and OCP2R, but at 540 nm the dominating species (70%) is the OCP2R (Fig. 2b).
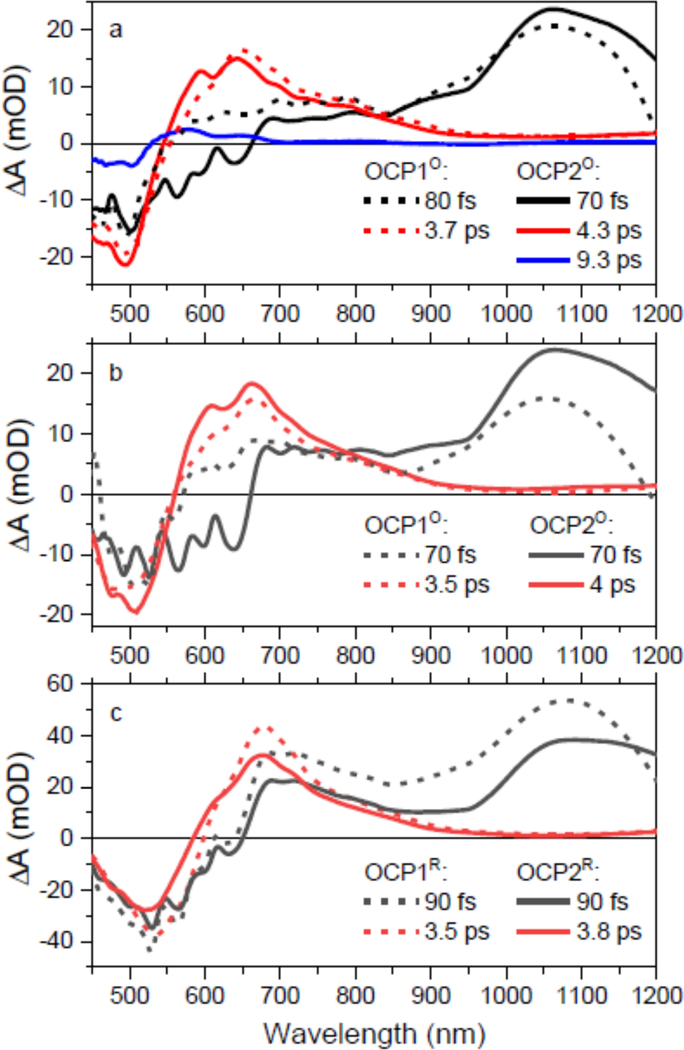
Fig. 3 depicts transient absorption spectra of OCP1 and OCP2 paralogs at 1 ps after excitation. At this delay time, most of the relaxation processes associated with S2-S1/ICT internal conversion and hot S1/ICT relaxation are over, so the spectra represent the response of the system when it is in a relaxed S1/ICT state. All transient absorption spectra shown in Fig. 3 consist of a negative band peaking around 500 nm due to the ground state bleaching and a positive signal due to excited state absorption (ESA) from the S1/ICT state. Some keto-carotenoids in polar environment allow to identify contributions from the S1 and ICT part of the coupled S1/ICT state as the S1-Sn and ICT-SN transitions are well separated [35]. Yet, this is not the case of CAN in solution, for which there is only one ESA band [50]. When bound to OCP, however, the binding site increases the degree of charge transfer character of the S1/ICT state, making the ICT-SN transition visible in transient absorption spectrum. This has been observed for hECN in OCP [32] and data in Fig. 3 shows it occurs also for an OCP bound CAN.
Figure 3:
Transient absorption spectra of OCP1 (balck) and OCP2 (red) paralogs. The spectra were measured at 1 ps after excitation of OCPO at 470 nm (a) and 540 nm (b). Panel (c) shows comparison of OCP1R and OCP2R after excitation at 540 nm. The data in panel (c) were recorded under continuous side illumination of the sample. All spectra are normalized to maximum.
While various transient absorption spectra of OCP1 reported earlier were typically limited to the 450–750 nm spectral window, our extension of the spectral window to 450–1200 nm allows for better identification of the features in transient absorption spectra. The main ESA band peaking at about 650 nm is due to the S1-Sn transition. At 600 nm, the S1-Sn band has a shoulder that is observed in a number of carotenoids and is typically denoted as the S* signal [51]. The ICT-like band is associated with the signal around 800 nm, which is exclusively observed when CAN is bound to OCP; in solution, the CAN ESA signal drops to zero around 750 nm [50]. The magnitude of the ICT-like band does not exceed 50% of the main S1-Sn ESA band, suggesting that even though the charge-transfer character of the coupled S1/ICT state has been enhanced by binding to OCP, it remains moderate. This is further demonstrated by absence of a negative band associated with an ICT stimulated emission that is readily observed in the 900–1100 nm spectral region in some keto-carotenoids [35]. Here, the shape of the transient absorption signal around 1000 nm is suggestive of a weak negative band, but it is only visible as a broad dip on the positive ESA background.
Comparison of data taken for OCP1 and OCP2 (Fig. 3) does not show any significant difference between the two, except the more pronounced 600 nm shoulder in OCP2O. The more pronounced structure of the S1-Sn band in OCP2O reflects the difference observed in absorption spectra (Fig. 1). The ICT-like bands in the 800–1100 nm region do not exhibit any difference between the two paralogs. The same is observed when the activated forms, OCP1R and OCP2R, are compared in Fig. 3c. The OCP2R has slightly blue-shifted bleaching signal, which is most likely due to a fraction of OCP2O that contributes to the signal after 540 nm excitation (Fig. 2b), but the S1-like and ICT-like bands are essentially identical in OCP1R and OCP2R.
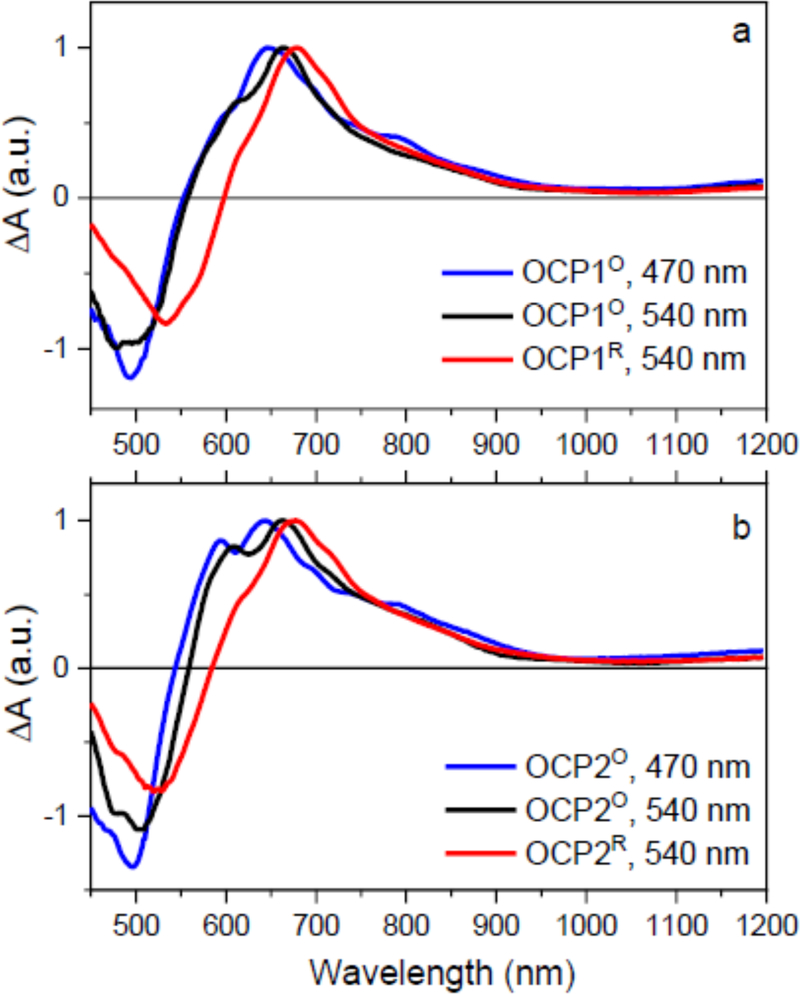
Spectroscopic differences between OCPO and OCPR are shown in Fig.4. For both OCP1 and OCP2, the OCPR form has significantly red shifted S1-Sn band, which peaks at 680 nm. The position of the S1-Sn maximum is also sensitive to excitation wavelength when dark-adapted form OCPO is excited. Excitation at 470 nm generates a blue-shifted S1-Sn band (645 nm in both paralogs) than excitation at 540 nm (663 nm in both paralogs). This observation confirms a spectral heterogeneity of OCPO that must contain at least two spectral forms [33, 52] Interestingly, the spectral heterogeneity of OCPO and difference between OCPO and OCPR is pronounced only in the S1-Sn band, while the ICT part of the transient absorption spectrum (800–1100 nm) remains nearly independent of excitation wavelength. Moreover, the ICT bands are not enhanced even upon OCPO to OCPR conversion, indicating the ICT state is not enhanced in the activated form of OCP. This observation is valid for both OCP1 and OCP2.
Figure 4:
Comparison of transient absorption spectra of OCP1 (a) and OCP2 (b) paralogs. OCPO form was excited either at 470 nm (blue) or at 540 nm (black). The activated OCPR form (red) was excited at 540 nm. The spectra recorded at 1 ps after excitation and are normalized to maximum.
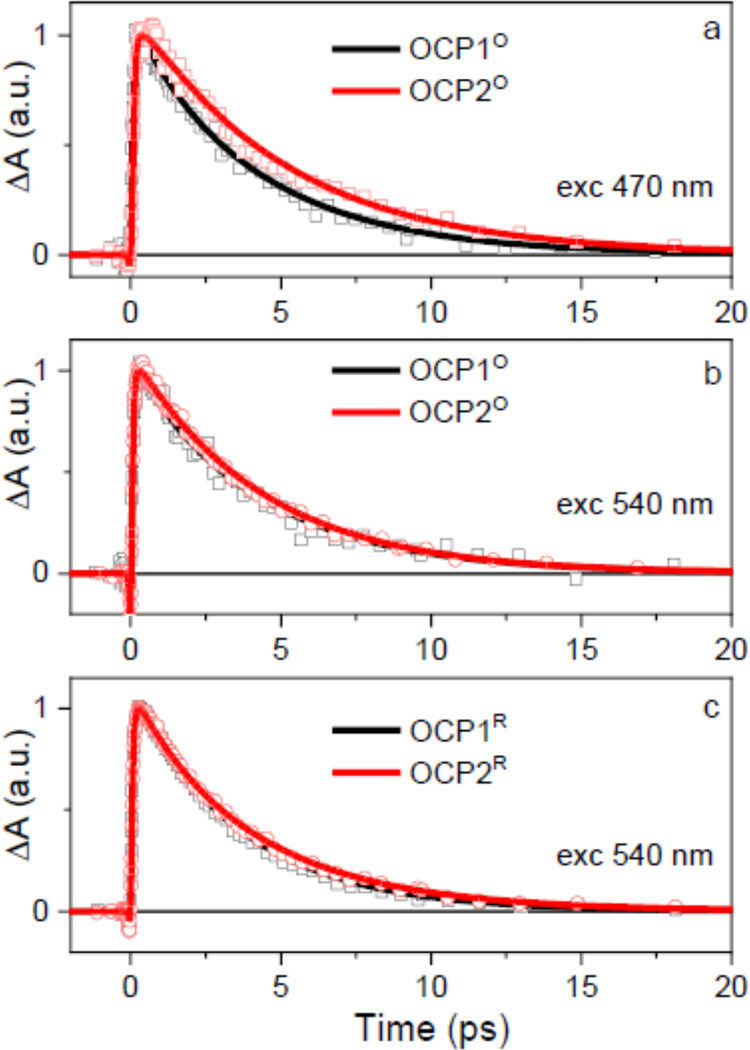
The spectroscopic similarity between the OCP1 and OCP2 is further demonstrated by their similar dynamics. Exemplary kinetics measured at the maximum of the S1-Sn band of both paralogs are shown in Fig. 5. The decay of the S1/ICT state is nearly identical in OCP1 and OCP2 except for excitation of OCPO at 470 nm, for which the OCP2O exhibits slower decay. To extract the lifetimes we have applied a global fitting and results are summarized in Fig. 6. We have employed the simplest possible model that contains two excited states, the initially excited S2 state and the coupled S1/ICT state. This model provided satisfactory fits for all measured datasets, except for OCP2O excited at 470 nm, which required one extra component to fit the data. In all datasets, the first EADS (black) corresponds to the initially excited S2 state. The spectral profile of the first EADS is dominated by ESA signal in the 900–1200 nm due to the S2-Sm transition [53]. The decay of this signal (see Supporting Info for kinetics) provides the S2 lifetime that is in the 70–90 fs range for all paralogs, in agreement with earlier reports [29, 32, 53]. The first EADS decays to form the spectrum corresponding to the S1/ICT state whose lifetime is 3.5–3.7 ps for OCP1O and 4.0–4.3 ps for OCP2O, the actual value depending on excitation wavelength. The activated OCP1R and OCP2R forms have essentially the same S1/ICT lifetimes as their non-activated counterparts, yielding 3.5 ps and 3.8 ps for OCP1R and OCP2R, respectively.
Figure 5:
Kinetics measured at the S1-Sn maximum for OCP1 (black) and OCP2 (red). Solid lines are fits.
Figure 6:
EADS obtained from global fitting the data recorded for OCP1 (dashed lines) and OCP2 (solid lines). (a) OCPO excited at 470 nm. (b) OCPO excited at 540 nm. (c) OCPR excited at 540 nm.
Thus, comparing OCP1 and OCP2, the S1/ICT dynamics is systematically slower for OCP2, but the difference is rather negligible. The same is valid for differences induced by change of the excitation wavelength: for both OCP1 and OCP2, OCPO excited at 470 nm gives longer S1/ICT lifetime than that excited at 540 nm, yet the difference is marginal. However, the important alteration of dynamics is identified in OCP2O excited at 470 nm, for which one extra fitting component having a lifetime of 9.3 ps is required. The spectrum of the 9.3 ps EADS (Fig. 6a) is reminiscent of the S* spectrum reported for a number of carotenoids both in solution and bound to proteins. We note that if we use an additional component to fit the data from other experiments, we can force the global fitting to generate an EADS with S*-like shape, but amplitude of such component is always less than 5% with a rather undefined lifetime as it may vary from 5–100 ps. Only for the OCP2O excited at 470 nm this EADS has appreciable amplitude with a well-defined lifetime (Fig. 6a).
3. Comparison of the structural details of Tolypothrix OCP1 and OCP2 Photoactivation
The 1.6 Å resolution structure of the Tolypothrix OCP1 was solved by molecular replacement using pdb 3MG1 as a search model. The Tolypothrix OCP1 (pdb 6PQ1) shows the canonical OCP1 structure first described in 2003 [10] and crystallizes as an anti-parallel dimer. It superimposes on the structure of Synechocystis 6803 OCP1 (pdb 4XB5) with a root mean square deviation of 0.54 Å over 266 aligned α-carbon atoms). The carotenoid in the OCP1 structure is identified as CAN (Supplementary Figure 1) and its conformation is identical and to CAN in Synechocystis 6803 (pdb 4XB5) and ECN in Synechocystis 6803 (pdb 3MG1) (Supplementary Figure 1).
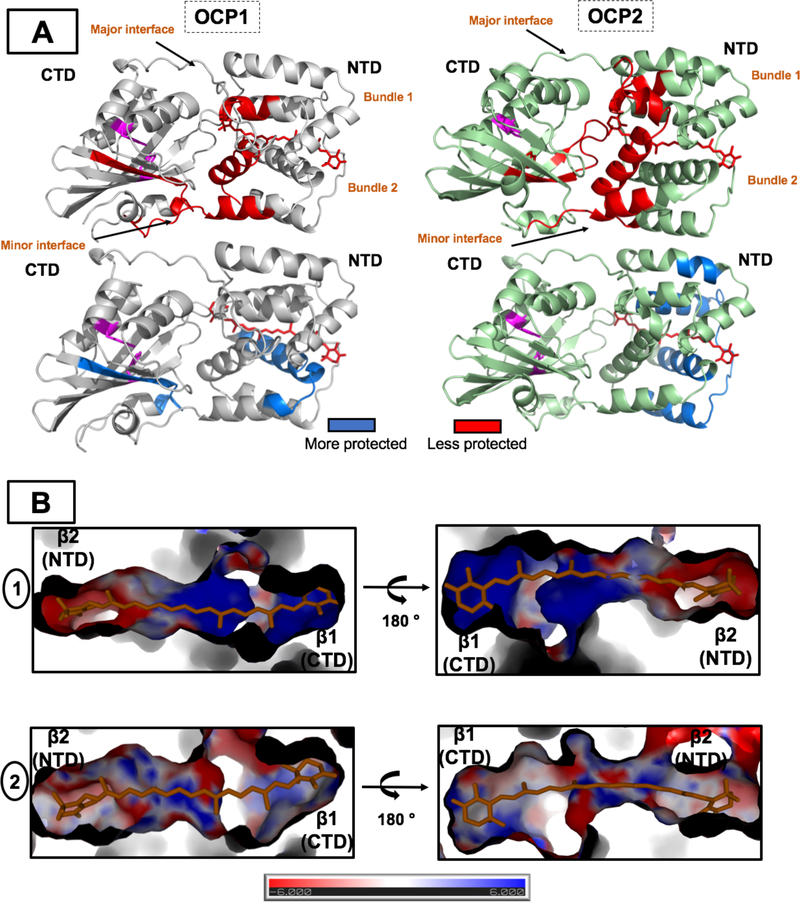
The structure of Tolypothrix OCP2 was modeled using Phyre2 [54]. The 36 residues associated with the carotenoid, within 4 Å distance, in either of the carotenoid-protein configurations are generally conserved between OCP1 and OCP2 (see Figure 2 in Ref. [8]) with seven residues differing in between OCP2 (OCP1:OCP2: I40L, I53P, A114G, I151L, L205M, M284I and the critical major interface residue, R155Q). The electrostatic surface of the interior of the carotenoid-binding pocket between OCP1 and OCP2 shows pronounced differences (Figure 7B). These differences are largest in the vicinity of the terminal rings of the carotenoid. Specifically, the region encompassing the β1 ring is positively charged in OCP1 and negatively charged in OCP2. This region contains the conserved Y201 and W288 that form hydrogen bonds to the carotenoid. It has been described in Synechocystis 6803 OCP1 that the backbone of the Y201 is hydrogen-bonded to the backbone of T197 and L205, and also the backbone of W288 forms hydrogen bonds with the backbone of V269 [11]. The structure of Tolypothrix OCP1 shows the same hydrogen bonds network: Y203-T199 (2.83 Å), Y203-I200 (3.37 Å) and W290-V271 (2.83 Å). However, the homology model of OCP2 presents an additional hydrogen bond between the backbone of Y201 and the backbone of V198 (3.3 Å). This could potentially contribute to the rigidity in the resting form of OCP2.
Figure 7: A. Local structural changes in the Tolypothrix OCP1 and OCP2 upon light activation as measured by XFMS.
The changes in solvent accessibility (> 1.5-fold changed) upon light activation are plotted on the OCP1 crystal structure (PDB 6PQ1) and on the OCP2 homology model showing regions becoming less protected (red) and more protected (blue). The carotenoid in OCPR form is shown in red sticks. The domain separation is not shown. The conserved Y201 and W288 are highlighted in magenta. B. Charge distribution of the carotenoid tunnel in OCP1 (1) and OCP2 (2). β1 and β2 regions are highlighted. It is colored by electrostatic potential from −6kT/e (red) to 6kT/e (blue).
To identify residue-level structural differences between OCP1 and OCP2 photoactivation, we carried out XFMS analysis at 10 °C, same temperature as for the ultrafast spectroscopy analysis. In this method, a high flux density X-ray beam ionizes water to produce hydroxyl radicals in situ and covalently modifies amino acid side chains on the microsecond time scale [55, 56]. The samples after irradiation were subjected to a bottom-up liquid chromatography LC-MS analysis using trypsin and Glu-C digestion obtaining a sequence coverage greater than 90%. The peptides detected by XFMS and the ratio of hydroxyl radical reactivity for red versus orange form are listed in Supplementary Tables S1 (OCP1) and S2 (OCP2). The rate of side-chain labeling is governed by both the intrinsic reactivity of the amino acid and the solvent accessibility to the side-chain.
In general, the changes in solvent accessibility showed that Tolypothrix OCP1 and OCP2 have a similar photoactivation mechanism as has been described previously for Synechocystis PCC 6803 OCP1 [13, 16]. However, overall, the reactivity of OCP2 is lower than OCP1. The degree of the rate of the labeling reflects the degree of accessibility of the residue. For instance, the rate of the peptide 97–106 (major interface) under the dark-adapted state in OCP2 is 0.07 s−1 versus 2.0 s−1 in OCP1. Upon illumination, this region increased its solvent accessibility in both photoactivated OCP paralogs. Nevertheless, OCP2 is still less reactive than OCP1 (0.17 s−1 versus 3.3 s−1). Another example is the N-terminal helix (α-helix A, peptide 15–26 for OCP2 and 20–27 for OCP1), which presents a rate of 0.87 s−1 for OCP2 and 3.1 s−1 for OCP1 under the dark-adapted state. Upon photoactivation to OCPR, this region became less protected in both OCPs. However, OCP2 N-terminal helix presented less reactivity (2.86 s−1) than OCP1 (7.1 s−1). These results suggest that OCP2 is structurally more rigid than OCP1.
We observed large increases in solvent accessibility by XFMS in similar regions of the OCP1 and OCP2 proteins (Figure 7A). As it has been previously shown for Synechocystis 6803 OCP1,[13, 16] these regions are the major interface between NTD-CTD and the minor interface of the N-terminal helix (α-helix A) and CTD. At the major interface, the largest increase (3.92–fold in OCP1 and 5.71-fold in OCP2) was observed in the peptides that contain the conserved residue R155 in OCP1, which is substituted by Q155 in OCP2 (residues 144–160 and residues144–171, respectively) (Supplementary Table S1 and S2). R155 is a key residue in the OCP1 structure, forming a salt bridge with E244 on the CTD in the OCPO [10, 11]. Like R155, Q155 also increases solvent accessibility upon photoactivation, confirming separation of the CTD and NTD domains in both OCP1R and OCP2R.
The largest solvent accessibility decrease upon photoactivation are observed in the NTD region (Figure 7A). In OCP1 the more protected regions are α-B and α-C (peptide 28–49 in Supplementary Table 2), these helices are found in only one of the two four-helix bundles. In contrast, in OCP2 four α-helices are protected: α-B (peptide 35–46 in Supplementary Table 2), α-D (peptide 67–72 in Supplementary Table 2), α-F (peptide 107–118 in Supplementary Table 2) and α-G (peptide 119–143 in Supplementary Table 2); in the OCP2 both four-helix bundles become less solvent accessible upon photoactivation. This can be interpreted as a reflection the rigidity of OCP2. When the carotenoid translocates in OCP2, residues in both bundles show more protection, indicating a more compact structure than OCP1, in which only residues in one of the bundle were more protected after photoconversion.
Discussion
The first characterization of the new OCP family, OCP2, pointed out functionally relevant differences with respect to the canonical OCP1.[8] One is its faster photoconversion and back-reversion. To probe these the differences in photoactivation/conversion this study combines two different approaches that helped us to understand the distinct contribution of the carotenoid (ultrafast transient spectroscopy) and the contribution of the protein structure (XFMS).
The data presented in Section 1 and 2 were based on few different comparisons that generated an experimental landscape allowing identification of spectroscopic properties of various forms of OCPs and relating these properties to possible functions. First, we compared two paralogs OCPs, OCP1 and OCP2, which exhibited significantly different dynamics of OCPO↔OCPR conversion [8]. Second, for OCPO we used two excitation wavelengths to identify possible spectral heterogeneity that has been reported for OCPO from Synechocystis PCC 6803 or Anabaena PCC 7120 [33, 52]. Third, using an experimental arrangement enabling to continuously illuminate OCP during transient absorption experiment, we obtained ultrafast spectroscopic data on OCPR under ambient temperature.
Spectral heterogeneity of OCPO
Both OCP1O and OCP2O exhibit certain heterogeneity as excitations at 470 and 540 nm and generate slightly different transient absorption spectra (Fig. 4). The structural origin of the spectral heterogeneity remains unclear, but it is likely related to hydrogen bonding to the keto-oxygen of CAN. It has been hypothesized that any OCP sample inevitably contains both OCPO and OCPR in equilibrium, resulting in observed spectral heterogeneity.[31] Yet, spectral heterogeneity was reported even for OCPs that are not able of OCPO→OCPR conversion, such as OCP binding the carotenoid zeaxanthin [12]. In addition, resonance Raman spectroscopy showed that the frequency of the characteristic Raman band, corresponding to C=C stretch, is different for Raman pump tuned to the red part of OCPO and for OCPR [52]. These results clearly point to an inherent spectral heterogeneity of the OCPO itself.
It is known that mutants replacing the tryptophan and tyrosine forming the H-bonds to the carotenoid keto-oxygen by amino acid preventing H-bonding, such as phenylalanine or alanine, OCP exhibits red-shifted absorption spectrum resembling that of OCPR [19–21, 24]. It is therefore obvious that red-shift of absorption spectrum can occur even without significant structural changes accompanying the OCPO→OCPR conversion [13]. Thus, the spectral heterogeneity of OCPO could be related to slight differences in the H-bonding pattern to the carotenoid keto-oxygen. Loosening tight H-bonding in some OCPO will lead to a red shift of absorption spectrum and to a less fixed position of the terminal ring containing the keto-oxygen, resulting in broader absorption spectrum. We note, however, that H-bonding cannot be the only reason for the observed spectral heterogeneity as OCP binding zeaxanthin, which lacks the C=O group and cannot form the H-bonds, also exhibit two spectral forms [33].
Comparing OCP1O and OCP2O, the red part of absorption spectrum is more pronounced in OCP1O, in line with less pronounced vibrational bands in absorption spectrum (Fig. 1). This suggests that CAN in OCP1O has a less constrained position in the binding pocket than in OCP2O. We considered that this might be due to differences in the hydrogen bonding network to the backbone of the Tyr and Trp that H-bond the keto-oxygen of the CAN. The observed differences are slight, the OCP2 has one additional backbone hydrogen bond. On the other hand, the spectral heterogeneity in transient absorption spectra (Fig. 4) is larger for OCP2O, implying that even though there is less red absorbing OCP2O, the spectroscopic difference between blue and red absorbing OCP2O is larger than for OCP1O. The possible reason for this difference could be the extra hydrogen bond between Y201 and V198 predicted by homology modeling of OCP2. It must be noted though, that the spectroscopic differences are rather small and essentially limited to the shape of transient absorption spectrum. The lifetime of the S1/ICT state is nearly the same for 470 and 540 nm excitations, meaning that the spectral heterogeneity does not affect the effective conjugation length of CAN. Such change, induced for example by significant torsion of the terminal ring must be accompanied by changes in S1/ICT lifetime, which is obviously not the case here. Thus, the spectral heterogeneity of OCPO must be rather related to changes in carotenoid-protein interaction (H-bonds, distances, electrostatics) than to significant changes in carotenoid conformation.
Role of the ICT state
Covering the spectral window extending up to 1200 nm allowed monitoring full ICT band that for CAN in OCP appears in the 700–900 nm (ICT excited state absorption) and 900–1100 nm (ICT stimulated emission) spectral region. The increase of the charge transfer character of the S1/ICT state, seen as an increase of magnitude of the ICT band, upon binding to OCP is ascribed to twist of the terminal ring having the keto-oxygen buried in the C-terminal domain. In contrast to solution, where this ring is in s-cis configuration, the binding to C-terminal domain forces the ring to adopt s-trans configuration. Apart from prolonging the effective conjugation length [36], this change also increases the influence of the keto group on spectroscopic properties, leading to the observed increase of the CT character of the S1/ICT state.
An increase of the CT character of the S1/ICT state was reported also upon OCPO→OCPR conversion. Indeed, significant increase of the ICT band magnitude was reported for OCPR from Synechocystis binding ECN [32], and similar result was obtained for ECN bound to RCP [33]. These observations led to a hypothesis that the ICT state is the quencher state in OCP-induced quenching of phycobilisomes [32]. Here, however, the transient absorption spectra of OCPR and OCPO are nearly identical in the 750–1100 nm spectral region, implying that the ICT state of CAN is not sensitive to OCPO↔OCPR conversion in both OCP1 and OCP2. Yet, OCP2 is significantly less efficient in phycobilisome quenching [8], challenging the importance of the ICT state for the quenching mechanism.
The insensitivity of the ICT state to type (OCP1 vs. OCP2), state (OCPO vs. OCPR) and even to the excitation wavelength (470 vs. 540 nm) shows that the degree of charge transfer character of the S1/ICT state plays no role in physiological function of OCPs from Tolypothrix. Even though OCP1 and OCP2 exhibit very different dynamics of OCPO↔OCPR conversion and very different capability of phycobilisome quenching [8], these differences are not reflected in spectroscopic properties of the S1/ICT state. Comparing our data with results reported earlier, it is very likely that the magnitude of the ICT band is controlled only by the carotenoid structure and is not related to the physiological functions. The increase of the ICT band magnitude upon OCPO→OCPR conversion has been observed only for OCPs binding exclusively ECN or hECN [32, 33], thus carotenoids having only one keto group positioned asymmetrically at the end of the conjugated system. Since it is known that an asymmetric conjugated keto group is the key structural factor determining the magnitude of the ICT band [57], the OCPO→OCPR conversion likely enhances the asymmetry by changes in the local environment of the conjugated keto group. However, for CAN, which has two keto groups positioned symmetrically at both terminal rings, the asymmetry is not introduced resulting in essentially identical magnitudes of the ICT bands in OCPO and OCPR.
The minor role of the ICT state is further underlined by comparable S1/ICT lifetimes of CAN in OCPO and OCPR. While OCPR or RCP binding only ECN need an extra time component to fit the data in the spectral region of the ICT band [32, 33], no such component is required here (Fig. 6). The similarity of the S1/ICT lifetimes in OCPO and OCPR in both OCP1 and OCP2 also implies comparable effective conjugation lengths. This is in agreement with structural data showing that both OCPO [10] and RCP [13] have one terminal ring in s-trans configuration. The observed lifetimes of 3.5 – 4.3 ps supports this observation as the lifetimes are shorter than those in solution, but longer than the ~1.5 ps lifetime expected for fully extended (both rings in s-trans) configuration.[50] The S1/ICT lifetimes of CAN in OCP2 are systematically longer than the lifetimes measured in OCP1, suggesting slightly different terminal ring torsions in these two OCPs.
Protein structure features influencing the different dynamics of OCPO↔OCPR conversion in OCP1 and OCP2
Besides the different contributions of blue and red absorbing forms of OCPO described above there is another notable difference between OCP1 and OCP2. Excited-state dynamics of CAN in OCP2O requires additional decay component whose spectral and dynamical properties resembles those known for the S* signal. We note the S* component is required only after excitation at 470 nm (Fig. 6), implying that the blue form of OCP2O is more prone to generate the S* signal. This is interesting especially in the light of recent report showing that the S* may be the initial state of the OCP photocycle [22], because then the presence of S* signal in OCP2 correlates with faster OCPO↔OCPR conversion.
Since recent theoretical treatment of the fate of absorbed energy in long carotenoids assigned the S* signal to a hot ground state [26], it is possible that even the first step of the photocycle – breaking the H-bonds to Trp and Tyr in the C-terminal domain – is a ground state reaction. The large amount of energy by CAN (~21,000 cm-1 for 470 nm excitation) is quickly (~4 ps) dumped to the ground state forming a hot ground state population that will promptly transfer the excess energy to the first solvation shell [26], which in the case of OCP are the amino acids nearby. Thus, the energy initially stored in the CAN ground state vibrational modes is exploited to break Trp and Tyr H-bonds, initializing the photocycle.
The question remains why the initialization and the whole photocycle is faster in OCP2 than in OCP1. The origin of this difference may be traced in different carotenoid binding pocket in OCP1 and OCP2. Even though most of the key amino acids determining the carotenoid protein interaction are conserved, some substitutions in OCP2 modify charge distribution along the carotenoid tunnel [8]. While in OCP1O the binding pocket in the C-terminal domain (containing the Trp and Tyr residues involved in H-bonding) is positively charged, the same binding pocket in OCP2O exhibits negative charge (Figure 7B). This difference may affect the initial phases of the OCPO↔OCPR conversion leading to a release of the carotenoid from the C-terminal binding pocket.
We note here that we used OCPs binding exclusively CAN for our spectroscopic and XFMS studies, but there is a precedent for Synechocystis PCC 6803 OCP1 to bind a range of carotenoids [10, 13]. Thus, we have also analyzed native OCP1 and OCP2 from Tolypothrix and, based on the crystallization of OCP1 and the purification of OCP2 and its domain homologs from Tolypothrix [45], we show that CAN is the predominant carotenoid bound to the OCP. Since the dominating native carotenoid of both Tolypothrix OCP1 and OCP2 is CAN, the functional and spectroscopic differences must arise from the contribution of the protein.
Time-resolved spectroscopy showed that the first intermediates in the photocycle have an absorption spectrum reminiscent of photoactivated OCP, even without separation of NTD and CTD domains [20, 22]. Similarly, some OCP mutants preventing formation of the hydrogen bond between keto-oxygen of carotenoid and W290 are locked in a state having absorption spectrum characteristic of OCPR [19, 21]. Thus, the faster OCP2O↔OCP2R turnover could be also explained by an unfinished photocycle, in which OCP2 reaches only the intermediate state with red-shifted absorption spectrum, but with not fully separated NTD and CTD. To test this hypothesis we used the XFMS analysis of the OCP1 and OCP2 photoactivation. It shows grossly the same solvent accessibility changes, indicating a similar mechanism, which includes the separation of the NTD and CTD. Thus, poor domain separation in OCP2 cannot be the reason for fast OCP2O↔OCP2R turnover. Instead, replacement of the key R155 residue in OCP1 by Q155 in OCP2, preventing to form a salt bridge between NTD and CTD domain, is likely the driving force behind faster OCP2O↔OCP2R turnover. Similar behavior, including insensitivity of back OCPR-OCPO conversion to FRP, has been reported for OCPs mutants having positively charged R155 replaced by a neutral or negatively charged residue [58]. However, since the R155-Q155 replacement in OCP2 also decreases the quenching capability, its physiological role remains elusive.
Despite the overall similarity of the photoactivation process in OCP1 and OCP2, there are some differences in the regions more protected after photoactivation (Figure 7). Overall, the reactivity of the OCP2 is slightly lower than that of OCP1, with the exception of the C-terminus and a region near the β2 ring. The two bundles of OCP2 decreased the solvent accessibility (more protected), suggesting a contraction of these two bundles which might act like a ‘clamp’ for the carotenoid. However, in contrast to what has been hypothesized previously [8], the XFMS reactivity data shows that, in general, OCP2 is less reactive than OCP1, indicating that the protein is more rigid. The rigidity of OCP2 is in good agreement with the results presented in the discussion about the spectral heterogeneity of OCPO, where it has been suggested that the carotenoid in OCP1 is less constrained in the binding pocket than in OCP2.
Conclusions
We compared properties of OCP1 and OCP2 from Tolypothrix by combination of structural and ultrafast spectroscopic methods. We show that although OCP1 and OCP2 exhibit very different dynamics of the OCPO-OCPR photocycle and efficiency of phycobilisome quenching, their initial photophysics is nearly identical. Our results show that the key determinants of quenching capability of OCPs thus lie in particular amino acid sequences, while the spectroscopic properties of the bound carotenoids plays only a minor role. The small difference in initial carotenoid excited-state dynamics is attributed to changes in carotenoid-protein interactions involving the keto-oxygen of CAN in CTD. Homology modeling of the OCP2 structure revealed one extra hydrogen bond to the CAN keto-oxygen, which could responsible for more constrained structure of CAN in OCP2 and consequently more structured spectral bands in both absorption and transient absorption spectra. We identified two spectral forms of OCP1O and OCP2O, while the blue form of OCP2O is the only form having appreciable amplitude of the S* signal that has been suggested as the trigger of OCP photocycle [22]. Time-resolved spectroscopy on photoactivated OCP1 and OCP2 showed that the charge transfer character of the S1/ICT state does not change upon photoactivation, thus the ICT state play essentially no role in quenching mechanism.
Supplementary Material
Acknowledgments
The authors thank Prof. Beronda Montgomery for helpful discussions. We also thank Dr. Sigal Lechno-Yossef for her help to design the cloning strategy in Tolypothrix. We thank the Metabolomic facility core at Michigan State University. This work was supported by the National Science Foundation (IOS 1557324) and the Office of Science of the U.S. Department of Energy DE-FG02-91ER20021. M.A.D.M was supported by a Marie Curie Individual Global Fellowship within the Horizon 2020 European Framework Program (project Photo-cy-apps, no. 795070). T.P. thanks the Czech Science Foundation (18-21631S) for financial support. We thank Arnel Mariano for assisting sample preparation at Advanced Light Source Beamline 3.2.1. The XFMS was supported in part by NIH 1R01GM126218, and conducted at the Advanced Light Source and Joint BioEnergy Institute, supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. DOE under contract DE-AC02-05CH11231.
Footnotes
Conflicts of interest statement
No conflict of interest.
References
- [1].Kirilovsky D, Kerfeld C, Cyanobacterial photoprotection by the orange carotenoid protein, Nature plants, 2 (2016) 16180. [DOI] [PubMed] [Google Scholar]
- [2].Wilson A, Ajlani G, Verbavatz JM, Vass I, Kerfeld CA, Kirilovsky D, A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria, The Plant cell, 18 (2006) 992–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Derks A, Schaven K, Bruce D, Diverse mechanisms for photoprotection in photosynthesis. Dynamic regulation of photosystem II excitation in response to rapid environmental change, Bba-Bioenergetics, 1847 (2015) 468–485. [DOI] [PubMed] [Google Scholar]
- [4].Erickson E, Wakao S, Niyogi KK, Light stress and photoprotection in Chlamydomonas reinhardtii, Plant J, 82 (2015) 449–465. [DOI] [PubMed] [Google Scholar]
- [5].Niyogi KK, Truong TB, Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis, Curr Opin Plant Biol, 16 (2013) 307–314. [DOI] [PubMed] [Google Scholar]
- [6].Grossman AR, Schaefer MR, Chiang GG, Collier JL, The phycobilisome, a light-harvesting complex responsive to environmental conditions, Microbiol Rev, 57 (1993) 725–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Allorent G, Petroutsos D, Photoreceptor-dependent regulation of photoprotection, Curr Opin Plant Biol, 37 (2017) 102–108. [DOI] [PubMed] [Google Scholar]
- [8].Bao H, Melnicki M, Pawlowski EG, Sutter M, Lechno-Yossef S, Montgomery BL, Kerfeld CA, Functional characterization of a new family of Orange Carotenoid Proteins, Nature plants, (2017). [DOI] [PubMed] [Google Scholar]
- [9].Kerfeld CA, Melnicki MR, Sutter M, Dominguez-Martin MA, Structure, function and evolution of the cyanobacterial orange carotenoid protein and its homologs, New Phytol, 215 (2017) 937–951. [DOI] [PubMed] [Google Scholar]
- [10].Kerfeld C, Sawaya M, Brahmandam V, Cascio D, Ho K, Trevithick-Sutton C, Krogmann D, Yeates T, The crystal structure of a cyanobacterial water-soluble carotenoid binding protein, Structure, 11 (2003) 55–65. [DOI] [PubMed] [Google Scholar]
- [11].Wilson A, Kinney JN, Zwart PH, Punginelli C, D’Haene S, Perreau F, Klein MG, Kirilovsky D, Kerfeld CA, Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria, The Journal of biological chemistry, 285 (2010) 18364–18375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Punginelli C, Wilson A, Routaboul J, Kirilovsky D, Influence of zeaxanthin and echinenone binding on the activity of the Orange Carotenoid Protein, Biochimica et Biophysica Acta-Bioenergetics, 1787 (2009) 280–288. [DOI] [PubMed] [Google Scholar]
- [13].Leverenz RL, Sutter M, Wilson A, Gupta S, Thurotte A, Bourcier de Carbon C, Petzold CJ, Ralston C, Perreau F, Kirilovsky D, Kerfeld C.a., De Carbon C. Bourcier, Petzold CJ, Ralston C, Perreau F, Kirilovsky D, Kerfeld CA, A 12 Å carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection, Science, 348 (2015) 1463–1466. [DOI] [PubMed] [Google Scholar]
- [14].Dominguez-Martin MA, Kerfeld CA, Engineering the orange carotenoid protein for applications in synthetic biology, Curr Opin Struct Biol, 57 (2019) 110–117. [DOI] [PubMed] [Google Scholar]
- [15].Wilson A, Punginelli C, Gall A, Bonetti C, Alexandre M, Routaboul J-M, Kerfeld C.a., Grondelle RV, Robert B, Kennis JTM, Kirilovsky D, van Grondelle R, Robert B, Kennis JTM, Kirilovsky D, A photoactive carotenoid protein acting as light intensity sensor, Proc Natl Acad Sci U S A, 105 (2008) 12075–12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gupta S, Guttman M, Leverenz R, Zhumadilova K, Pawlowski E, Petzold C, Lee K, Ralston C, Kerfeld C, Local and global structural drivers for the photoactivation of the orange carotenoid protein, Proc Natl Acad Sci USA, 112 (2015) E5567–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Boulay C, Wilson A, D’Haene S, Kirilovsky D, Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria, Proc Natl Acad Sci USA, 107 (2010) 11620–11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sutter M, Wilson A, Leverenz RL, López-Igual R, Thurotte A, Salmeen AE, Kirilovsky D, Kerfeld CA, Crystal structure of the FRP and identification of the active site for modulation of OCP-mediated photoprotection in cyanobacteria, Proc Natl Acad Sci USA, 110 (2013) 10022–10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bandara S, Ren Z, Lu L, Zeng X, Shin H, Zhao KH, Yang X, Photoactivation mechanism of a carotenoid-based photoreceptor, Proc Natl Acad Sci U S A, 114 (2017) 6286–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Maksimov EG, Sluchanko NN, Slonimskiy YB, Slutskaya EA, Stepanov AV, Argentova-Stevens AM, Shirshin EA, Tsoraev GV, Klementiev KE, Slatinskaya OV, Lukashev EP, Friedrich T, Paschenko VZ, Rubin AB, The photocycle of orange carotenoid protein conceals distinct intermediates and asynchronous changes in the carotenoid and protein components, Sci Rep, 7 (2017) 15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sluchanko N, Klementiev K, Shirshin E, Tsoraev G, Friedrich T, Maksimov E, The purple Trp288Ala mutant of Synechocystis OCP persistently quenches phycobilisome fluorescence and tightly interacts with FRP, Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1858 (2017) 1–11. [DOI] [PubMed] [Google Scholar]
- [22].Konold PE, van Stokkum IHM, Muzzopappa F, Wilson A, Groot ML, Kirilovsky D, Kennis JTM, Photoactivation Mechanism, Timing of Protein Secondary Structure Dynamics and Carotenoid Translocation in the Orange Carotenoid Protein, J Am Chem Soc, 141 (2019) 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gupta S, Sutter M, Remesh SG, Dominguez-Martin MA, Bao H, Feng XA, Chan LG, Petzold CJ, Kerfeld CA, Ralston CY, X-ray radiolytic labeling reveals the molecular basis of orange carotenoid protein photoprotection and its interactions with fluorescence recovery protein, J Biol Chem, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wilson A, Punginelli C, Couturier M, Perreau F, Kirilovsky D, Essential role of two tyrosines and two tryptophans on the photoprotection activity of the Orange Carotenoid Protein, Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1807 (2011) 293–301. [DOI] [PubMed] [Google Scholar]
- [25].Maksimov EG, Shirshin EA, Sluchanko NN, Zlenko DV, Parshina EY, Tsoraev GV, Klementiev KE, Budylin GS, Schmitt FJ, Friedrich T, Fadeev VV, Paschenko VZ, Rubin AB, The Signaling State of Orange Carotenoid Protein, Biophys J, 109 (2015) 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Balevicius V, Wei TJ, Di Tommaso D, Abramavicius D, Hauer J, Polivka T, Duffy CDP, The full dynamics of energy relaxation in large organic molecules: from photo-excitation to solvent heating, Chemical Science, 10 (2019) 4792–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Steinel T, Asbury JB, Zheng JR, Fayer MD, Watching hydrogen bonds break: A transient absorption study of water, Journal of Physical Chemistry A, 108 (2004) 10957–10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nogly P, Weinert T, James D, Carbajo S, Ozerov D, Furrer A, Gashi D, Borin V, Skopintsev P, Jaeger K, Nass K, Bath P, Bosman R, Koglin J, Seaberg M, Lane T, Kekilli D, Brunle S, Tanaka T, Wu WT, Milne C, White T, Barty A, Weierstall U, Panneels V, Nango E, Iwata S, Hunter M, Schapiro I, Schertler G, Neutze R, Standfuss J, Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser, Science, 361 (2018). [DOI] [PubMed] [Google Scholar]
- [29].Polivka T, Kerfeld CA, Pascher T, Sundstrom V, Spectroscopic properties of the carotenoid 3 ‘-hydroxyechinenone in the orange carotenoid protein from the cyanobacterium Arthrospira maxima, Biochemistry, 44 (2005) 3994–4003. [DOI] [PubMed] [Google Scholar]
- [30].Chábera P, Durchan M, Shih P, Kerfeld C, Polívka T, Excited-state properties of the 16 kDa red carotenoid protein from Arthrospira maxima, Biochimica Biophysica Acta 1807(1) (2011) 30–35. [DOI] [PubMed] [Google Scholar]
- [31].Polivka T, Chabera P, Kerfeld CA, Carotenoid-protein interaction alters the S(1) energy of hydroxyechinenone in the Orange Carotenoid Protein, Biochim Biophys Acta, 1827 (2013) 248–254. [DOI] [PubMed] [Google Scholar]
- [32].Berera R, van Stokkum IHM, Gwizdala M, Wilson A, Kirilovsky D, van Grondelle R, The Photophysics of the Orange Carotenoid Protein, a Light-Powered Molecular Switch, Journal of Physical Chemistry B, 116 (2012) 2568–2574. [DOI] [PubMed] [Google Scholar]
- [33].Slouf V, Kuznetsova V, Fuciman M, de Carbon CB, Wilson A, Kirilovsky D, Polivka T, Ultrafast spectroscopy tracks carotenoid configurations in the orange and red carotenoid proteins from cyanobacteria, Photosynthesis Research, 131 (2017) 105–117. [DOI] [PubMed] [Google Scholar]
- [34].Frank HA, Bautista JA, Josue J, Pendon Z, Hiller RG, Sharples FP, Gosztola D, Wasielewski MR, Effect of the solvent environment on the spectroscopic properties and dynamics of the lowest excited states of carotenoids, Journal of Physical Chemistry B, 104 (2000) 4569–4577. [Google Scholar]
- [35].Zigmantas D, Hiller RG, Sharples FP, Frank HA, Sundstrom V, Polivka T, Effect of a conjugated carbonyl group on the photophysical properties of carotenoids, Physical Chemistry Chemical Physics, 6 (2004) 3009–3016. [Google Scholar]
- [36].Llansola-Portoles MJ, Sobotka R, Kish E, Shukla MK, Pascal AA, Polivka T, Robert B, Twisting a beta-Carotene, an Adaptive Trick from Nature for Dissipating Energy during Photoprotection, Journal of Biological Chemistry, 292 (2017) 1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fuciman M, Kesan G, LaFountain AM, Frank HA, Polivka T, Tuning the Spectroscopic Properties of Aryl Carotenoids by Slight Changes in Structure, Journal of Physical Chemistry B, 119 (2015) 1457–1467. [DOI] [PubMed] [Google Scholar]
- [38].Melnicki M, Leverenz R, Sutter M, Lopez-Igual R, Wilson A, Pawlowski E, Perreau F, Kirilovsky D, Kerfeld C, Structure, Diversity, and Evolution of a New Family of Soluble Carotenoid-Binding Proteins in Cyanobacteria, Mol Plant, 9 (2016) 1379–1394. [DOI] [PubMed] [Google Scholar]
- [39].Lechno-Yossef S, Melnicki MR, Bao H, Montgomery BL, Kerfeld CA, Synthetic OCP heterodimers are photoactive and recapitulate the fusion of two primitive carotenoproteins in the evolution of cyanobacterial photoprotection, Plant J, 91 (2017) 646–656. [DOI] [PubMed] [Google Scholar]
- [40].Bao H, Melnicki MR, Kerfeld CA, Structure and functions of Orange Carotenoid Protein homologs in cyanobacteria, Curr Opin Plant Biol, 37 (2017) 1–9. [DOI] [PubMed] [Google Scholar]
- [41].Singh SP, Montgomery BL, Regulation of BolA abundance mediates morphogenesis in Fremyella diplosiphon, Front Microbiol, 6 (2015) 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wolk CP, Fan Q, Zhou R, Huang G, Lechno-Yossef S, Kuritz T, Wojciuch E, Paired cloning vectors for complementation of mutations in the cyanobacterium Anabaena sp. strain PCC 7120, Arch Microbiol, 188 (2007) 551–563. [DOI] [PubMed] [Google Scholar]
- [43].Cobley JG, Zerweck E, Reyes R, Mody A, Seludo-Unson JR, Jaeger H, Weerasuriya S, Navankasattusas S, Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium, Fremyella diplosiphon, Plasmid, 30 (1993) 90–105. [DOI] [PubMed] [Google Scholar]
- [44].Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP, Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120, J Bacteriol, 179 (1997) 1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dominguez-Martin MA, Polivka T, Sutter M, Ferlez B, Lechno-Yossef S, Montgomery BL, Kerfeld CA, Structural and spectroscopic characterization of HCP2, Biochim Biophys Acta Bioenerg, 1860 (2019) 414–424. [DOI] [PubMed] [Google Scholar]
- [46].Kabsch W, Integration, scaling, space-group assignment and post-refinement, Acta Crystallogr D Biol Crystallogr, 66 (2010) 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments, Acta Crystallogr D Biol Crystallogr, 67 (2011) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr D Biol Crystallogr, 68 (2012) 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr D Biol Crystallogr, 60 (2004) 2126–2132. [DOI] [PubMed] [Google Scholar]
- [50].Chabera P, Fuciman M, Hribek P, Polivka T, Effect of carotenoid structure on excited-state dynamics of carbonyl carotenoids, Physical Chemistry Chemical Physics, 11 (2009) 8795–8803. [DOI] [PubMed] [Google Scholar]
- [51].Polivka T, Sundstrom V, Dark excited states of carotenoids: Consensus and controversy, Chemical Physics Letters, 477 (2009) 1–11. [Google Scholar]
- [52].Kish E, Pinto MMM, Kirilovsky D, Spezia R, Robert B, Echinenone vibrational properties: From solvents to the orange carotenoid protein, Biochimica et Biophysica Acta-Bioenergetics, 1847 (2015) 1044–1054. [DOI] [PubMed] [Google Scholar]
- [53].Niedzwiedzki DM, Liu HJ, Blankenship RE, Excited State Properties of 3 ‘-Hydroxyechinenone in Solvents and in the Orange Carotenoid Protein from Synechocystis sp PCC 6803, Journal of Physical Chemistry B, 118 (2014) 6141–6149. [DOI] [PubMed] [Google Scholar]
- [54].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis, Nat Protoc, 10 (2015) 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gupta S, Chai J, Cheng J, D’Mello R, Chance MR, Fu D, Visualizing the kinetic power stroke that drives proton-coupled zinc(II) transport, Nature, 512 (2014) 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Xu G, Chance MR, Hydroxyl radical-mediated modification of proteins as probes for structural proteomics, Chem Rev, 107 (2007) 3514–3543. [DOI] [PubMed] [Google Scholar]
- [57].Enriquez MM, Fuciman M, LaFountain AM, Wagner NL, Birge RR, Frank HA, The Intramolecular Charge Transfer State in Carbonyl-Containing Polyenes and Carotenoids, Journal of Physical Chemistry B, 114 (2010) 12416–12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wilson A, Gwizdala M, Mezzetti A, Alexandre M, Kerfeld CA, Kirilovsky D, The Essential Role of the N-Terminal Domain of the Orange Carotenoid Protein in Cyanobacterial Photoprotection: Importance of a Positive Charge for Phycobilisome Binding, Plant Cell, 24 (2012) 1972–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.