Abstract
Objective
Peritoneal carcinomatosis (PC) occurs frequently in patients with gastric adenocarcinoma (GAC) and confers a poor prognosis. Multiplex profiling of primary GACs has been insightful but the underpinnings of PC’s development/progression remain largely unknown. We characterised exome/transcriptome/immune landscapes of PC cells from patients with GAC aiming to identify novel therapeutic targets.
Design
We performed whole-exome sequencing (WES) and whole transcriptome sequencing (RNA-seq) on 44 PC specimens (43 patients with PC) including an integrative analysis of WES, RNA-seq, immune profile, clinical and pathological phenotypes to dissect the molecular pathogenesis, identifying actionable targets and/or biomarkers and comparison with TCGA primary GACs.
Results
We identified distinct alterations in PC versus primary GACs, such as more frequent CDH1 and TAF1 mutations, 6q loss and chr19 gain. Alterations associated with aggressive PC phenotypes emerged with increased mutations in TP53, CDH1, TAF1 and KMT2C, higher level of ‘clock-like’ mutational signature, increase in whole-genome doublings, chromosomal instability (particularly, copy number losses), reprogrammed microenvironment, enriched cell cycle pathways, MYC activation and impaired immune response. Integrated analysis identified two main molecular subtypes: ‘mesenchymal-like’ and ‘epithelial-like’ with discriminating response to chemotherapy (31% vs 71%). Patients with the less responsive ‘mesenchymal-like’ subtype had high expression of immune checkpoint T-Cell Immunoglobulin And Mucin Domain-Containing Protein 3 (TIM-3), its ligand galectin-9, V-domain Ig suppressor of T cell activation (VISTA) and transforming growth factor-β as potential therapeutic immune targets.
Conclusions
We have uncovered the unique mutational landscape, copy number alteration and gene expression profile of PC cells and defined PC molecular subtypes, which correlated with PC therapy resistance/response. Novel targets and immune checkpoint proteins have been identified with a potential to be translated into clinics.
Keywords: cancer genetics, gastric cancer, gastrointestinal cancer, gene expression, mutations
Significance of this study.
What is already known on this subject?
Gastric adenocarcinoma (GAC) is rampant around the world. GAC is often diagnosed in advanced stages and peritoneal carcinomatosis (PC; malignant ascites or cells from implants) is common in patients with GAC affecting 45% of the population during the course of GAC.
Patients with PC have a very poor outcome with the current treatment options.
Although some genomic/transcriptomic profiles of primary GACs have been reported and characterised, the underlying molecular landscape of PC remains a mystery.
What are the new findings?
In this study, we sequenced whole exome and transcriptome of PC cells from 43 patients with GAC and performed integrative analyses to include clinical and histopathological data.
-
The significant new findings of our study included:
Identification of shared genomic alterations between primary GACs and PCs and demonstration of distinct genomic alterations in PC versus primary GACs, such as increased frequency of CDH1, TAF1 mutations in PC; increased proportion of ‘clock-like’ mutational signature and decreased levels of signatures associated with defective DNA mismatch repair and POLE mutations, and increased frequency of 6q loss and chromosome 19 gain.
Demonstration of the clonal and subclonal genomic architecture of PC, which revealed the complexity of intratumour heterogeneity: we identified four distinct clonal patterns.
Discovery of novel genomic and transcriptomic features that are associated with aggressive PC phenotypes, including higher frequency of TP53, CDH1, TAF1 and KMT2C mutations, increased proportion of ‘clock-like’ mutational signature, increase in whole-genome doubling events, and chromosomal instability, particularly copy number losses, reprogrammed PC microenvironment (tumour and immune cell contents and composition), enriched signalling pathways related to cell cycle, MYC activation and impaired immune response among others.
Immune profiling separated PC specimens into two main groups, the T-cell ’exclusive' and T- cell ’exhausted' subtypes.
The T-cell ’exhausted' subtype showed high levels of immune checkpoint TIM-3, its ligand galectin-9, VISTA and transforming growth factor-β (TGF-β1), while other classical checkpoints were low, suggesting potential therapeutic immune targets.
Defining novel molecular subtypes by integrative clustering of the genomic/transcriptomic/immune features, the ‘mesenchymal-like’ and ‘epithelial-like’ subtypes and demonstrated that the ‘mesenchymal-Iike’ subtype was associated with resistance to post-PC therapy (31% vs 71% compared with epithelial subtypes), while no association was observed between the traditional histopathology-based subtypes and therapy response.
Thus, molecular classification may be more meaningful for stratification of PCs for therapy decisions.
How might it impact on clinical practice in the foreseeable future?
Our results establish the molecular foundation for PC pathogenesis and progression and provide rationale for developing clinical trials with novel targeted agents and/or novel immune checkpoint inhibitors.
Our molecular classification may be more meaningful for stratification of PCs for therapy decisions and suggests that the less responsive ‘mesenchymal-like’ subtype exhibited T-cell exhaustion phenotype, with high expression of TIM-3, galectin-9, VISTA and TGF-β1, which could be targeted by immunotherapy.
Introduction
Gastric adenocarcinoma (GAC) imposes a significant global health burden1 2 and is frequently diagnosed in advanced stages with peritoneal carcinomatosis (PC) being a frequent site of metastasis.3 Patients with PC have a short overall survival, overwhelming symptoms, but limited and transiently palliative treatments.4 Patients are treated empirically and PC is often resistant to therapy,4 prompting an urgent clinical need to molecularly interrogate PC. However, our current knowledge of molecular or genomic makeup of PC is predominantly limited as TCGA and other similar analyses were carried out only on the primary GACs.5 6 It would be important to characterise GAC cells that migrate to populate the peritoneal cavity to form PC. A detailed genomics/transcriptomic/immune characterisation of PC could shed light on the driver alterations that promote PC development and progression.
GAC is highly heterogeneous, both phenotypically and genotypically. It can be classified morphologically into two major types: the diffuse and intestinal types.2 The diffuse-type GACs are histologically poorly differentiated and characterised by the presence of either isolated single tumour cells or small clusters of tumour cells in a rich fibrous stroma. Diffuse-type GAC is clinically aggressive and more frequently leads to PC than does the intestinal type.6 7 The presence of signet-ring cells (SRC) where the nucleus is pushed towards the cell wall by excessive cytoplasmic mucin is an established indicator of poor prognosis and resistance to therapy.8 9 Based on TCGA molecular classification of GACs,5 the diffuse-type maps to genomically stable genotype (GS), and the intestinal type can represent one of the other three genotypes: microsatellite instable (MSI), EBV+ and chromosomally instable (CIN). The MSI and EBV-related GACs often have an increased level of immune infiltration (have longer survival), considered more amenable to immune modulations. These two subtypes are rarely represented in patients with advanced GAC. Recurrent somatic alterations in TP53, CDH1 and RHOA have been reported in diffuse-type primary GACs.10–14 Most recently, Choi et al demonstrated that CMTM2 plays a role in diffuse-type GAC and lymph node metastases in a small patient cohort.11 Kurashige et al identified DDR2 as a potential driver gene and novel therapeutic target for those forming PC, by analysing four metastatic cell lines.15 Although the genomics of primary GACs have been well characterised, comprehensive molecular profiling of PC cells is yet to be conducted.
In this study, we performed integrative analyses of whole-exome sequencing (WES), whole transcriptome sequencing (RNA-seq), immune profile, clinical and pathological phenotypes of unique 43 patients with PC to characterise them molecularly and to identify novel targets and/or biomarkers. We identified shared genomic alterations between primary GACs and PC and discovered novel signatures that are unique to PC. Aggressive phenotype was associated with unique genomics/transcriptomic signatures and reprogrammed PC microenvironment. Novel molecular subtypes correlated with post-PC chemotherapy resistance/response. Patients with resistant ‘mesenchymal-like’ subtype had high expression of immune checkpoint TIM-3, its ligand galectin-9, VISTA and transforming growth factor-β (TGF-β) as potential therapeutic immune targets. Thus, we have established a molecular framework for better understanding of PC pathogenesis and progression that could potentially be exploited.
Materials and methods
Patient cohort, sample collection, DNA and RNA extraction
A total of 43 patients with GAC with documented PC were enrolled in this study. The detailed clinical and histopathological characteristics of this cohort are described in online supplementary table 1. Details of treatment (types of agents and duration) have been shown in online supplementary table 2. This project was in accordance with the policy advanced by the Declaration of Helsinki 1964 and later versions. The germline DNAs (gDNAs) were isolated using the QIAamp DNA Mini Kit (Qiagen) and the total RNAs were isolated using the miRNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Only samples passed sample intake quality check (gDNA: >200 ng; total RNA: RNA integrity number >7) were further processed for DNA and RNA-seq respectively.
gutjnl-2018-318070supp002.pdf (62.8KB, pdf)
gutjnl-2018-318070supp003.pdf (62.8KB, pdf)
More detailed materials and methods can be found in online supplementary materials.
gutjnl-2018-318070supp001.pdf (62.8KB, pdf)
Results
Patient characteristics
Specimens were collected from consented patients with GAC (including those with PC) who required a therapeutic procedure. All specimens were cytologically confirmed as having adenocarcinoma cells (PC) from unique 43 patients with GAC (44 samples). The clinical characteristics are shown in the online supplementary table 1. Among these 43 patients, blood for gDNA was collected from 15 patients. Among these, 8 had intestinal phenotype and 30 had diffuse phenotype by the Lauren classification.16 Eighteen had SRC, and others did not (non signet ring (NOS)). The tumour cell percentage in PC samples ranged from 5% to 95%; considerable variations in PC cell purity are known. H&E and epithelial cell adhesion molecule (EpCAM)/CD45 immunofluorescence staining were used to distinguish tumour and immune cell compartments, respectively (online supplementary figure 1).
Somatic mutations and signatures of PC cells varied by histology and location of the primary GAC
WES was performed on the genomic DNA of 34 PC specimens and 15 matched gDNA specimens. A mean of 134 non-synonymous mutations (range: 0–353) per case were identified, corresponding to 2.7 non-synonymous mutations per megabase (Mb) of the targeted DNA. The mutation frequencies varied slightly by phenotypes (online supplementary figure S2A). The overall median frequency was similar to TCGA primary GACs5 and oesophageal adenocarcinoma,17 but there was no MSI in any PC specimen.
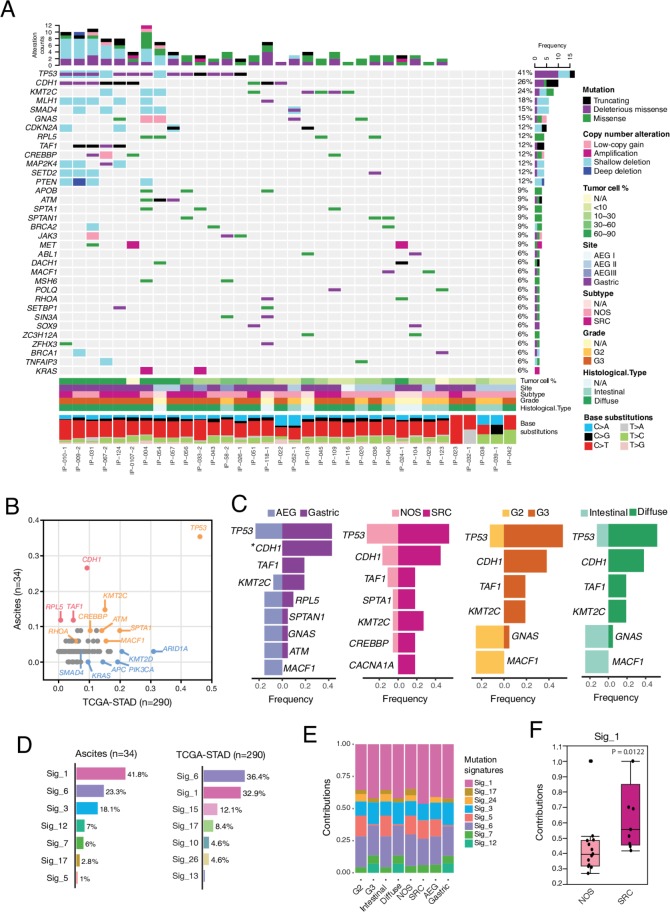
The most frequently altered genes in PC cells were TP53 then CDH1 altered at 41% and 26%, respectively (figure 1A). The frequency of TP53 mutation was similar to that in primary TCGA GACs while CDH1 was mutated at a much higher frequency in PC cells (26% vs 9%, figure 1B). Eight out of nine somatic mutations identified in CDH1 were either truncating or predicted to be deleterious missense by multiple algorithms (see ’Materials and methods' section), in the cadherin domain, suggesting potential loss-of-function. Notably, mutations of CDH1 were exclusively in tumours with the diffuse phenotype and were 2.5× more frequent in those with the SRC phenotype (figure 1C).
Figure 1.
The landscape of somatic mutations and deciphered mutational signatures of peritoneal carcinomatosis (PC) specimens. (A) Somatic genomic alterations identified in PC specimens. The middle panel shows somatic mutations and gene-level copy number alterations by patient (column) and by gene (row). The histogram on the top shows the number of alterations accumulated on 34 listed genes in each individual sample. The bottom tracks show histopathological characteristics, and the bottom histogram shows the mutational spectral. (B) Comparison of gene mutation frequencies between PC specimens in this study and primary gastric adenocarcinoma (GAC) from The Cancer Genome Atlas GAC cohort (TCGA-STAD).5 (C) Comparison of gene mutation frequencies by histopathological subtypes of PC specimens. The asterisk indicates a statistically significant difference was observed between two different subtypes. *Fisher’s exact test, p<0.05. (D) Decomposition of mutational signatures. All somatic substitutions identified in this study were included to decipher mutational signatures. Bar plots show the identified mutational signatures and their relative contributions to somatic mutations detected in ascites samples (left) and TCGA-STAD (right), respectively. Sig, signature. (E) The deciphered mutational signatures across histopathological subtypes. (F) Increased contribution of signature 1 to the signet-ring cell (SRC) subtype when compared with NOS. P value was calculated by non-parametric Mann-Whitney U test.
Other recurrently altered genes (>10%) included KMT2C, MLH1, SMAD4, GNAS, CDKN2A, RPL5, TAF1, CREBBP, MAP2K4, SETD2 and PTEN (figure 1A). Recurrent loss of heterozygosity was observed in MLH1, SMAD4, PTEN, CDKN2A, KMT2C and SETD2. Mutations of GNAS were more frequently seen in intestinal phenotype. Mutations in TAF1 and KMT2C were exclusive in diffuse phenotype and were twice more frequent in SRC versus NOS (figure 1C). The frequency of KMT2C mutation was similar to that of TCGA primary GACs, while TAF1 and RPL5 were mutated more frequently in PC specimens (figure 1B). Both RPL5 and TAF1 are PanCancer driver genes.18 RPL5 suppresses c-MYC expression through RNA-induced silencing complex for degradation.19 TAF1 encodes a transcription initiation factor that phosphorylates TP53 during G1 cell cycle progression20 and involves in apoptosis induction and cell cycle regulation. In PC, TAF1 mutations were either truncating or predicted to be functionally deleterious and they co-existed either with mutations in TP53 or CDH1 and all TAF1 mutations identified were clonal, suggesting that TAF1 mutation may confer additional selective advantages to TP53/CDH1-mutant cancer cells.
The C>T substitutions accounted for nearly 60% of somatic mutations, with transcription-induced mutational strand bias observed across all phenotypes (online supplementary figure S2B). Seven base-substitution signatures in PC specimens (figure 1D) were noted according to the catalogue of somatic mutations in cancer (COSMIC) nomenclature,21 including signature #1 (42%) which is associated with a clock-like mutational process, signature #3 (18%) which is associated with failure of DNA double-strand break-repair by homologous recombination and signature #6 (23%) which is associated with defective DNA mismatch repair (MMR). Compared with TCGA, the fraction of signature #6 was lower in PC specimens (23% vs 36%). In addition, signature #15 (12% in TCGA) and signature #26 (4.6% in TCGA) that are also associated with defective MMR, the signature #10 (4.6% in TCGA) that is associated with somatic hotspot mutations in POLE, were not observed in PC. This was consistent with the absence of MSI and POLE mutations in PC. A higher frequency of signature #1 (42% vs 33%) and presence of signature #3 were observed in PC. The mutational signatures across phenotypes are shown in figure 1E and signature #1 was significantly higher in SRC than in NOS (figure 1F). Signature #1, the ‘clock-like’ mutational signature is higher in cells with a higher turnover rate.22 This is consistent with PC specimens with SRCs.
Increased ploidy and chromosomal instability associated with aggressive phenotype
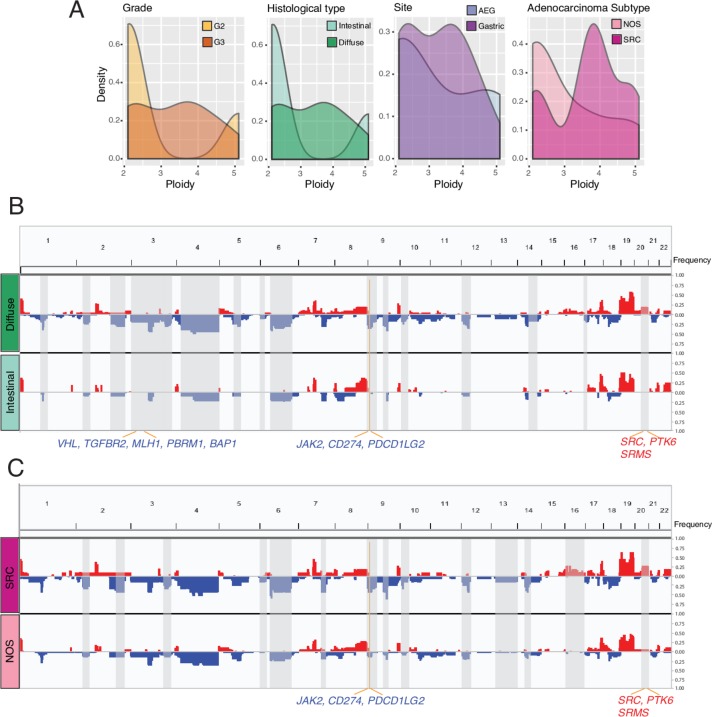
Most of the PC specimens had ploidy exceeding two (range 2–5), and more than half of them had ploidy exceeding three. The PC specimens with diffuse phenotype (and/or SRC) had a trend towards a higher ploidy than intestinal phenotype (NOS phenotype), notably the SRC phenotype (median 2.3 vs 3.8, p>0.05, Mann-Whitney U test) (figure 2A), same as TCGA with SRC features (median 2.1 vs 3.7, p>0.05) (online supplementary figure S3A). We noted whole-genome doubling (WGD) during PC progression. WGD promotes adaptability and diversity in proliferating cell lineages. The PanCancer aneuploidy analysis demonstrated that tumours with increased ploidy are more prone to additional aneuploidy events.23 Indeed, the phenotypes with high frequency of WGD had a higher degree of aneuploidy in the PC specimens, particularly the events of copy number losses (figure 2B-C).
Figure 2.
Increased ploidy and chromosomal instability in peritoneal carcinomatosis (PC) specimens with advanced stage and aggressive histological phenotypes. (A) Density plot showing the distribution of tumour ploidy by histopathological subtypes. (B, C) Composite of copy number profiles for ascites with diffuse or intestinal (B), signet-ring cell (SRC) or NOS subtypes (C), with gains in red and losses in blue. The regions that showed a difference in the frequency of copy number alterations between two subtypes were shaded in light gray rectangles, and within which the names of cancer-related or biologically important genes are labelled.
Frequent copy number alterations (CNAs) included loss of 3 p, chr4, 6q, 9 p, 18q and gain of 7 p, 8q, 17q and chr19, and 20q, with variability across the phenotypes (online supplementary figure S3B). All of them were also observed in TCGA5 but the frequencies of 6q loss and chromosomal 19 gain were much greater in the PC specimens. We compared CNAs across the phenotypes and noted genes located on the aneuploid regions (figure 2B-C). Compared with the intestinal phenotype, the diffuse phenotype had frequent heterozygous deletion of 3 p that encompasses multiple tumour suppressor genes (TSGs) including VHL, MLH1, PBRM1, TGFBR2 and BAP1, 9p24 involving JAK2, CD274 (PD-L1) and PDCD1LG2 (PD-L2), duplication of 20q that encompasses oncogenes SRC and PTK6 and tyrosine kinase SRMS. The SRC phenotype had more frequent deletion of 9 p and amplification of 20q, compared with NOS. We also noted more frequent deletion of 2 p, and 2q involving CASP8, the apoptosis inducing cysteine protease. Except TP53 and CASP8, the mRNA expression levels of other genes mentioned above decreased significantly in the PC specimens with hemizygous deletion of targeted genes compared with those without (online supplementary figure S4). Similarly, expression of SRC, SRMS and PTK6 increased in cases with copy number gain of the target genes versus those without. Co-deletion of JAK2/PD-L1/2 was recurrently reported (~2%) in various tumour types including non-small-cell lung carcinoma,24 in which JAK2 deficiency impairs interferon (IFN)-stimulated IRF1 expression and major histocompatibility complex class I antigen presentation on cancer cells. The reason for PD-L1/L2 deletion is unknown, but low PD-L1 expression associated with an unfavourable outcome in ovarian cancer.25
Together, our data suggested unique alterations in diffuse phenotype (and/or SRC) compared with the counterpart and a role of increased ploidy and CIN in driving PC progression.
Clonal and subclonal architecture showed the complexity of intratumoural heterogeneity
SciClone26 was applied to 15 PC specimens that had matched gDNA data to reconstruct the clonal and subclonal architecture. Figure 3A shows four clonality patterns: monoclonal—a single dominant clone, minor subclone—a minor subclone in addition to the dominant clone, biclonal—two clones were inferred and finally, complex—more than two clones were detected with a complex feature. The clonality patterns across the phenotypes and the clonal evolution during PC progression need to be investigated in a larger, longitudinal cohort, but our preliminary analysis revealed the complexity of both the inter- and intra-tumoral heterogeneity. We investigated clonality of recurrently mutated genes and found that mutations of TP53, CDH1 and TAF1 were located to the dominant founding clone (figure 3B), suggesting an earlier timing of mutation acquisition in these genes.
Figure 3.
The clonal and subclonal architecture inferred in peritoneal carcinomatosis (PC) specimens reflecting the complexity of intratumour heterogeneity. (A) Patterns of clonal and subclonal architecture inferred. Kernel density plots of variant allele frequency (VAF) across regions with copy number one, two, three or four, posterior predictive densities summed over all clusters for copy number neutral variants and posterior predictive densities for each cluster/component in 15 ascites samples with matched normal. (B) VAFs plotted vs read depth for each of the four copy number regions for three representative samples: IP-010–1, IP-022 and IP-031. Mutations in biologically important genes are labelled.
Signalling pathways and differentially expressed genes related to aggressiveness and biomarkers
RNA-seq was performed on 39 PC specimens. Among these, 21 had WES data. Unsupervised clustering analysis was performed on RNA-seq data to classify PC speciments based on gene expression profiles. Consensus clustering of the top 1000 most variable genes grouped them into two major clusters (online supplementary figure S5), and the grouping was strongly driven by tumour purity. However, unsupervised clustering of top variable genes, regardless of tumour purity, was still able to distinguish, in large part, the two phenotypes (online supplementary figure S6), suggesting an intrinsic PC characteristics and less due to purity. However, all subsequent analyses of the gene set enrichment analysis (GSEA) pathway enrichment and differentially expressed genes (DEGs) were stratified by tumour cell percentage—low, high or combined, respectively and across both phenotypes (figure 4).
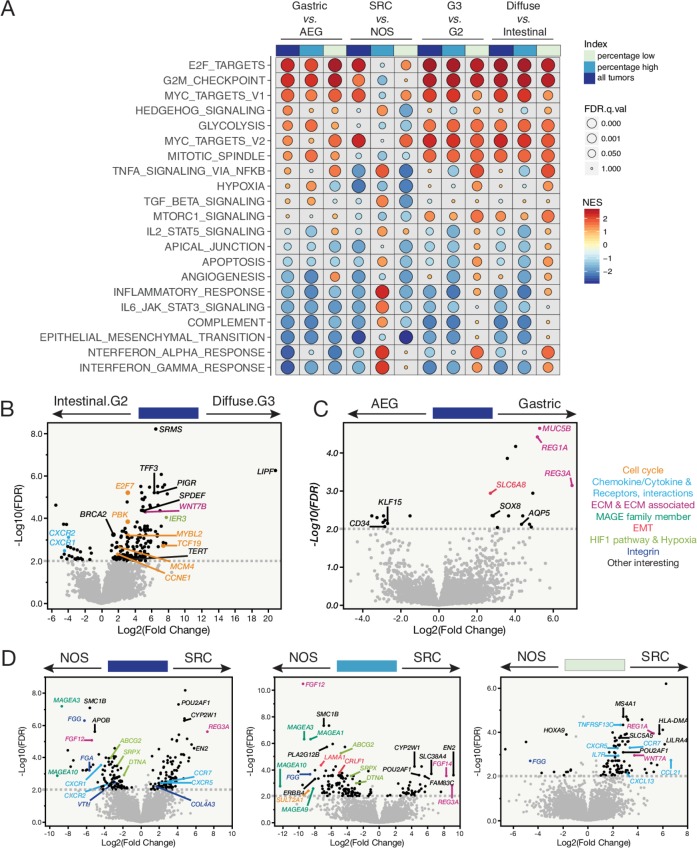
Figure 4.
Transcriptome profiling of peritoneal carcinomatosis (PC) specimens—enriched signalling pathways and differentially expressed genes (DEGs) associated with disease aggressiveness. (A) GSEA pathway analysis showing enriched signalling pathways by histological subtypes (eg, gastric vs adenocarcinoma of esophageal and gastric junction (AEG)) for all PC specimens together, PC specimens with higher tumour cell percentages and those with lower tumour cell percentages, respectively (top track). Each row represents a MSigDB Hallmark pathway. The size of the filled circle reversely reflects the FDR q values, the bigger the circle the smaller the q value. NES, normalised enrichment score. The colour tone of NES indicates the direction of enrichment, with warm colour denotes positive enrichments and cold colour denotes negative enrichment. (B) The DEGs identified in G3 diffuse against G2 intestinal subtypes. Dashed line indicates a cut-off of FDR q value≤0.01. Genes that are above the dashed line with a fold change≥2, cancer related or biologically important are labelled by their names in the volcano plot. (C) The DEGs identified in gastric against AEG subtypes of PC specimens. (D) The DEGs identified in signet-ring cell (SRC) vs NOS subtypes in all PC specimens together (left), PC specimens with higher (middle), or lower tumour cell percentage (right), respectively.
Overall, a number of cancer hallmark signalling pathways were nearly universally dysregulated at all levels of comparisons in diffuse phenotype (and/or SRC) compared with intestinal phenotype (NOS) (figure 4A), including marked enrichment of E2F targets, G2/M cell cycle checkpoint, mitotic spindle assembly, MYC targets and glycolysis, and depletion of IFN-α and IFN-γ responses, complement system, inflammatory response and apoptosis. Notably, enrichment of mammalian target of rapamycin complex (mTORC)1 signalling was only observed in the diffuse compared with intestinal phenotype consistent with reports on mTOR pathway in primary GACs, particularly, the phosphorylated mTOR (p-mTOR) was more frequent in diffuse than intestinal phenotype, and overexpression of p-mTOR was associated with aggressive biology.27 The profiles of high-purity and low-purity SRC specimens were different compared with NOS. For example, in the high-purity PC specimens, the activity of hedgehog, tumour necrosis factor via nuclear factor-κB and TGF-β signallings were higher in SRC than in NOS, while it was in the reverse direction in the low-purity specimens.
The DEG analysis suggested that important genes were associated with the phenotypes (figure 4B-D). Briefly, tyrosine kinase SRMS and the gastric lipase (LIPF) genes were dramatically upregulated in the diffuse compared with intestinal phenotype (figure 4B). SRMS has been suggested as a diagnostic biomarker for GAC.28 LIPF encodes an enzyme that digests triglycerides but its oncogenic role is unknown.
Several genes such as MUC5B, REG1A and AQP5 were highly expressed where the primary was located in the stomach than it was at the GE junction (figure 4C). Human mucin gene MUC5B is highly expressed in GACs and cell lines but absent in normal gastric mucosa.29 Aquaporin 5 (AQP5), a putative oncogene, promotes progression and invasion of several cancers including GAC.30 The regenerating protein 1 alpha (REG1A) was reported to regulate cell invasion, apoptosis and viability in GAC through activating PI3K/Akt-GSK3β signalling.31 REG Iα expression was related to nodal metastases and played a role in angiogenesis.32 REG Iα may be another target to treat PC.
When we compared SRC with NOS, nine genes were differentially expressed and shared between the combined cohort and the high-purity tumours (figure 4D). These genes included CYP2W1 and REG3A. Both were reported to be expressed highly in GACs than in normal tissues.33 34 Overexpression of REG3A accelerated pancreatic cancer growth via a REG3A-JAK2/STAT3 positive feedback loops,35 and promoted aggression in GACs. The upregulated DEGs that were unique to high-purity specimens included FAM83C, a recently identified oncogene in various human cancers36 REG1A overexpression was only observed in the low-purity SRC versus NOS. Altogether, we identified multiple biomarkers of aggressiveness and targets by the DEG analysis and functional studies are warranted.
Deconvolution of the cellular composition of PC specimens revealed reprogrammed TME
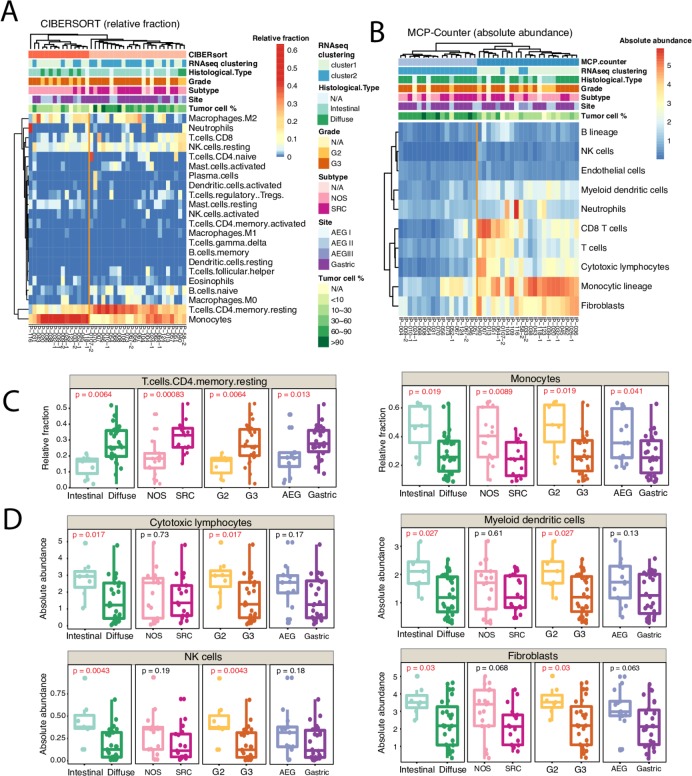
The tumour microenvironment (TME) of PC extends into the ascites, which acts as a reservoir for cytokine, growth factors and cellular components that provides a tumour-promoting context. Two deconvolution approaches were applied—CIBERSORT37 to estimate the relative cellular fraction of 22 immune cell types (figure 5A), and MCP-counter38 to produce the absolute abundance scores for eight major immune cell types, endothelial cells and fibroblasts (figure 5B). Unsupervised clustering of both the cellular fractions and the abundance scores grouped the specimens into two major clusters with distinct signatures, and the expression profiles from both approaches agreed in large part with each other.
Figure 5.
The immune and stromal cell composition of peritoneal carcinomatosis (PC) specimens. (A) Relative fraction of 22 immune cell types inferred by CIBERSORT. (B) Absolute abundance of 8 immune cell types, endothelial cells and fibroblasts inferred by MCP-counter. (C) Box plots showing difference in the relative fractions of T cells CD4 memory resting cells (left) and monocytes (right) between histological subtypes. (D) Box plots showing difference in the absolute abundance of cytotoxic lymphocytes (top left), myeloid dendritic cells (top right), natural killer (NK) cells (bottom left) and fibroblasts (bottom right), between different histology types. P values were calculated by non-parametric Mann-Whitney U test.
Among 22 inferred immune cell types by CIBERSORT, the cellular fractions of monocytes and resting memory CD4+ T cells varied across the phenotypes (figure 5C), especially the resting memory CD4+ T cells. A higher fraction of resting memory CD4+ T cells associated with more aggressive phenotype. In agreement with this, it is reported that mice with resting memory CD4+ T cells had faster tumour growth.39 Instead, recent evidence suggested that the resting CD4+ T cells can be converted to regulatory T cells that promote tumour immune escape in target tissues.40–42 The abundance scores produced by MCP-counter showed abundance of cytotoxic lymphocytes, natural killer (NK) cells, myeloid dendritic cells (a major stimulator of T cells) and normal peritoneal fibroblasts were significantly lower in the diffuse phenotype (figure 5D). Together, our data suggested that tumour cells might have reprogrammed the TME to facilitate PC progression.
Immune profiling of PC specimens identified novel targets for immunotherapy
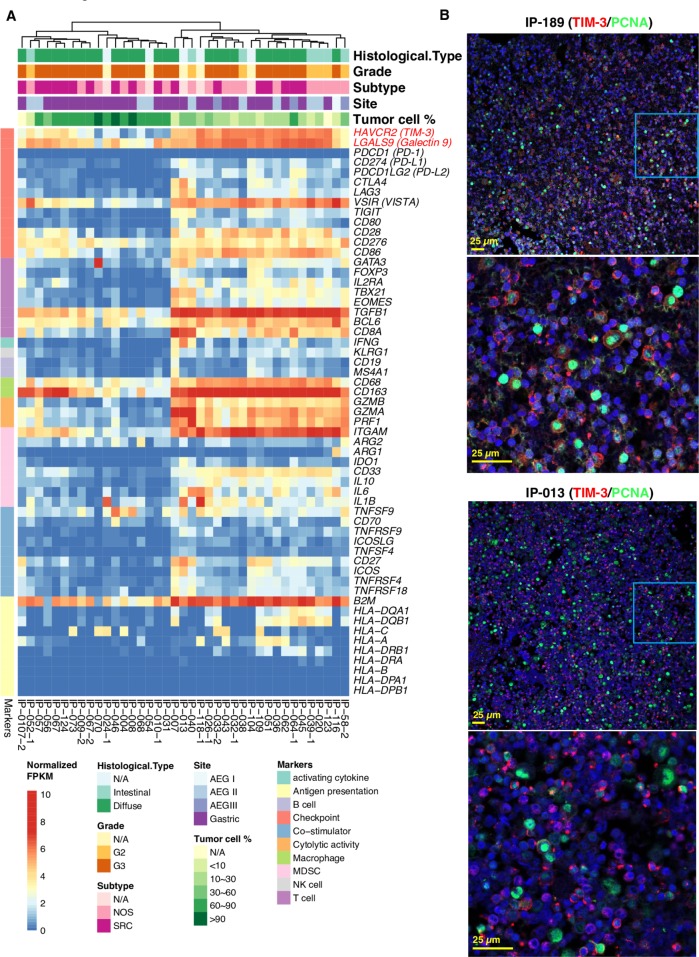
We performed a more comprehensive immune profiling of specimens using our curated immune gene panel43 including markers for antigen presentation, B/T/macrophage/NK/MDSC cell lineages, co-stimulatory and co-inhibitory immune checkpoint and receptors, cytolytic activity and activating cytokines (figure 6A). Unsupervised hierarchical clustering of immune gene expression data separated PC specimens into two main groups: T-cell exclusive and T-cell exhausted. The T-cell exclusive subtype exhibited low expression level of CD8 T-cell marker CD8A, cytolytic markers GZMA, GZMB, PRF1, IFNG, checkpoint inhibitors and other immune-related genes. In contrast, the T-cell exhausted subtype had high expression of most of the immune-related genes, particularly CD8A, cytolytic markers GZMA, GZMB, PRF1, pan-macrophage marker CD68 and M2 marker CD163. While the expression of PD-1, PD-L1/L2, CTLA-4, LAG-3, IDO1, TIGIT was low in all specimens. Instead, the immune checkpoint TIM-3, its ligand galectin-9 and VISTA were highly expressed in the T-cell exhausted subtype, as well as TGF-β1 suggesting an immune suppressive microenvironment. To examine TIM-3 expression at protein level, we performed dual immunofluorescent staining of TIM-3 and proliferating cell nuclear antigen (PCNA) and confirmed a high expression of TIM-3 (figure 6B). Interestingly, TIM-3 was expressed on both immune cells and some PC cells. Together, we note novel immunotherapy targets, particularly for the T-cell exhausted phenotype of PC specimens.
Figure 6.
Immune profiling of peritoneal carcinomatosis (PC) specimens identified novel immune checkpoint targets. (A) Profiling of PC specimens using a curated panel of immune-related genes. Unsupervised hierarchical clustering was performed on the normalised RNA expression data of these immune-related genes. (B) Dual immunofluorescent staining of TIM-3 and PCNA, a known tumour cell nuclear proliferating antigen in PC specimen. (anti-TIM-3 antibody was from Cell Signaling Technology, Danvers, Massachusetts, USA, 1:200; anti-PCNA antibody was from Cell Signaling Technology, 1:200). Representative images on expression of TIM-3 in red and PCNA in green of two patients are shown. Scale bar: 25 µm.
Integrative classification of PCs identified ‘mesenchymal-like’ and ‘epithelial-like’ subtypes associated with response
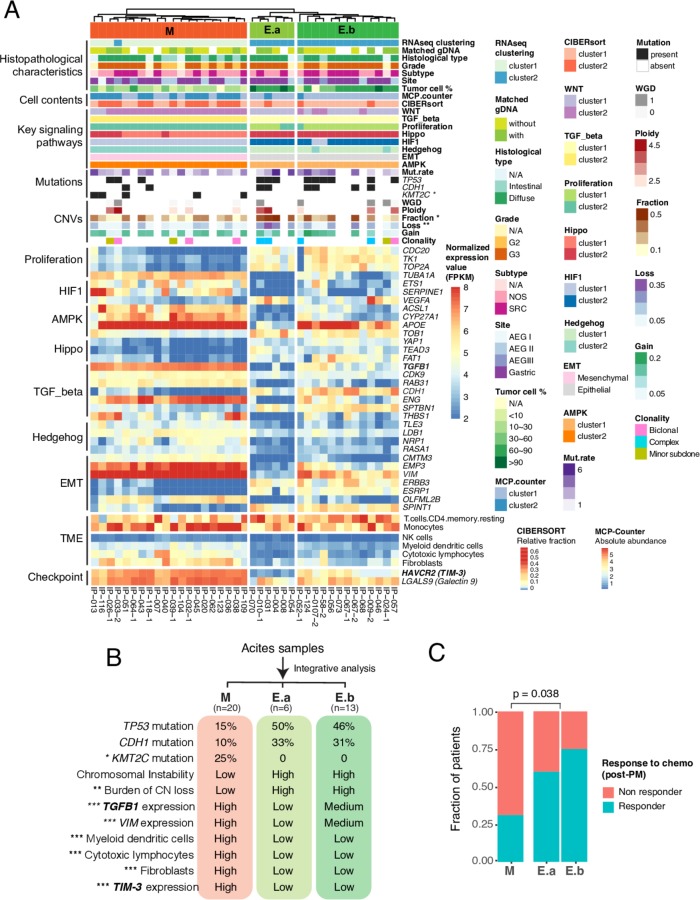
We profiled the expression signatures of key signalling pathways involved in GAC as documented in our review paper,2 including the epithelial mechachemal transition (EMT), TGF-β, Wnt, Hippo, hedgehog, stemness, hypoxia-inducible factor (HIF1), amp activated protein kinase (AMPK), chromatin remodelling pathways and the proliferation signature (online supplementary figure S7). The most significant DEGs of each pathway were then selected and their normalised expression data were used for unsupervised clustering of the PC specimens, together with the profiles of somatic mutation, CNA, and immune composition (figure 7A).
Figure 7.
Integrative clustering of clinical, genomic and expression features of peritoneal carcinomatosis (PC) specimens defines three molecular phenotypes that correlated with post-PC chemotherapy response. (A) Heatmap of histopathological and molecular features of three defined clusters. Top track: molecular phenotypes: M, ‘mesenchymal-like’ phenotype; E.a, ‘epithelial-like’ phenotype a; E.b, ‘epithelial-like’ phenotype b. Heatmap showing normalised expression values in FPKM for 32 most variable differentially expressed genes (DEGs) selected from 7 key signalling pathways. The asterisk indicates statistically significant difference between M and E phenotypes (*p<0.05; **p<0.01). Fisher’s exact test and non-parametric Mann-Whitney U test were used to calculate p values. (B) Genomic and molecular characteristics of three molecular subgroups. CIN, chromosomal instability; CN, copy number; exp, expression. (C) Correlation of defined molecular phenotypes with post-PC chemotherapy response. Compared with the epithelial phenotypes (E.a and E.b), the mesenchymal phenotype is less likely respond to chemotherapy. P value was calculated by the non-parametric Mann-Whitney U test. Chemo, chemotherapy.
The integrative clustering of specimens identified three groups—’mesenchymal-like (M)', ‘epithelial-like, a (E.a)' and ‘epithelial-like, b (E.b)’ with distinct features (figure 7B). Compared with the E subtypes, the M subtype had higher expression of mesenchymal signature genes (VIM and EMP3) and TGF-β pathway genes (ENG and TGFB1), lower expression of epithelial markers, more frequent KMT2C mutation, less frequent mutation of TP53 and CDH1, a lower level of CIN, increased proportion of fibroblasts and cytolytic cells (online supplementary figure S8), a T-cell exhausted phenotype, with significantly elevated expression of TGFB1,44 and immune checkpoint TIM-3, its ligand galectin-9 and VISTA. The two E subgroups exhibited similar profiles in somatic mutation, CNA, and TME with differences in expression of VIM, EMP3, TGFB1, APOE and TIM-3 (figure 7A-B). We further correlated the molecular-based classification of PC specimens with the patient prognosis and as expected, no significance was observed as all patients in this cohort had a short survival. We observed significant difference in response to post-PC therapy between the M and Esubgroups (figure 7C). Patients with an E genotype were more responsive than the M genotype (E:60%, E:75% vs M:31%), while no correlation was noted between the phenotypes and response. This observation needs validation in a larger cohort.
Discussion
To the best of our knowledge, this is the first study to comprehensively characterise the genomic/immune landscape of PC cells from patients with GAC. Our study provides novel insights into PC’s molecular landscape and identified novel targets for potential exploitation. The significance of these findings can only be explained in the clinical context of our patients who have suffered enormously from PC and the glaring paucity of effective therapeutic options we have for them. In the clinic, the patients with advanced GAC (including those with PC) have the tumour tissue assessed for three biomarkers (Her2 that is overexpressed in ~10% and when positive, the use of trastuzumab has a marginal benefit,2 microsatellite status, however, MSI high status is very rare in patients with advanced GAC,2 and PD-L-1 expression in tumour/immune cells but the anti-PD-1 or anti-PD-L-1 therapy has marginal effect45). Then another serious issue is that the higher the line of therapy (eg, third-line or fourth-line therapy), the less effective it is but more resistant becomes the cancer and more symptomatic as well as physically deteriorated is the patient. Thus, the current armamentarium to treat patients with advanced GAC is highly deficient. Our results, in this context provide a detailed view of advanced GAC in a molecular dimension that has been lacking.
Analysis of PC specimens revealed shared mutations with the primary GACs in common cancer drivers5 6 13: TP53, ATM, RHOA, CDH1 and KMT2C, but with distinct mutational profiles. For example, inactivating CDH1 and TAF1 mutations are more frequent in PC cells versus primary and associated with aggressive diffused phenotype. Intriguingly, all CDH1 mutations were clonal and they had a higher VAF than that of TP53 in more than half of the specimens with both gene mutations, indicating that they are initiating mutations acquired in early stage of the disease. All CDH1 mutations in this study were co-occurring with mutation of TP53, and TAF1 mutation in a subset. TAF1 protein phosphorylates p53 during G1 cell cycle progression and is considered as a member of p53 signalling pathway.20 Earlier evidence suggested that the loss of CDH1 is only beneficial to cancer cells with abnormal p53 pathway.46 Therefore, the co-occurrence of TP53 and CDH1 mutations is less likely an accidental phenomenon; instead, they might have been under positive selection during GAC progression. The co-inactivation of CDH1, TAF1 and TP53 might be involved in PC development and progression. E-cadherin loss might create vulnerabilities with the potential to be targeted in the clinic. A most recent study demonstrated that loss of E-cadherin hyperactivates the insulin-like growth factor 1 receptor pathway, which can be targeted.47 Although, our entire cohort is small and only four patients had not received prior systemic therapy, our analyses showed no significant influence of prior therapy in the molecular patterns of PC samples. Clearly, more specimens will need to be analysed for a definite answer.
Additionally, we identified unique genomic alterations in aggressive phenotype: more frequent mutation of TP53, CDH1, TAF1, and KMT2C. We identified a number of DEGs that are associated with disease aggressiveness, such as LIPF, REG3A, CYP2W1 and FAM83C.48 49 Further functional studies and validation of these unique genes in aggressive phenotypes are under investigation. Analysis of CNAs in PC samples, revealed increased ploidy and CIN, and uncovered unique gains or losses in aggressive phenotype, such as amplification of 20q that encompasses oncogenes Src, PTK6 and SRMS were exclusively observed in the diffused/SRC phenotype. By integration with RNA-seq data, we noted that the expression these genes (eg, SRMS) was also upregulated in more aggressive phenotype. Interestingly, these oncogenes are non-receptor tyrosine kinases and have been reported to involve in cancer progression and metastases.50 These tyrosine kinases are worth exploring in PC as their inhibitors are already in clinical trials (eg, NCT02389309 and NCT01015222). A limitation of our study is that we could not assess the corresponding primary GACs because all PC specimens were collected during a therapeutic procedure, collection of primary tumour was not clinically justifiable and patients with PC who are quite ill cannot be subjected to a non-standard invasive procedure. Another deficiency is that the timing of collection cannot be planned and are clinical context dependent.
The two E genotypes are genomically and chromosomally more unstable than the M genotytype. Notably, no correlation was observed between phenotypes and genotypes, E subtypes were more responsive to therapy consistent with prior observation.51 The M subtype had high expression of TGF-β1, and immune checkpoint TIM-3, its ligand galectin-9, and another immune checkpoint VISTA, high proportion of monocytes, increased proportion of M2 macrophage, and enriched abundance of fibroblast in the TME, thus uncovering novel targets for immunotherapy. Although several studies have demonstrated that TIM-3, its ligand galectin-9 and VISTA are dysregulated in GAC tumour tissues and pointed their potential clinical implication,52–56 there are some limitations to these studies due to the study design and was limited to primary tumour tissues and PC cells were not included. However, our discovery on TIM-3/galectin-9 and other immune suppression molecules-VISTA and TGF-β1 were derived from a comprehensive profiling of patients’ PC samples. Therefore, targeting these immune checkpoint-TIM-3/galectin-9, VISTA and TGF-β1 could be particularly promising for M subtype of patients with PC. We acknowledge that more cases need to be studied to understand if the location of primary has a definite influence on molecular profiling of PC. In addition, comparison of PC specimen with primary specimen from the same patient would be of considerable interest. Finally, emerging technologies such as circulating tumour DNA will need to be evaluated simultaneously.
In summary, our analyses have uncovered unique mutational landscape, CNAs, and gene expression profiles in PC cells and molecularly defined different phenotypes. Novel targets have been emerged. Our results should widely stimulate analysis of many more patients with GAC with PC and doing so will lead to uncovering of enormous wealth of new knowledge to much better understand the biology of PC to develop more effective therapies through rational clinical trials.
Acknowledgments
The authors would like to thank patients with gastric cancer who have donated their specimen and time to this study.
Footnotes
RW, SS and KH contributed equally.
Contributors: SS, LW and JAA conceived and jointly supervised the study. RW conducted the major bioinformatics and biostatistics analysis of the multiplatform data, generated figures and tables for the manuscript. JZ, XS and XM contributed to raw WES data processing. GH and SZ assisted with WES and RNA-seq analysis. BB, MM, FGA, SC and JE assisted with clinical samples consent, collection and pathological evaluation. KH took charge of ascites cells isolations, DNA/RNA extraction and coordinated with DNA core facility for WES and RNA-seq and gathered all patients’ clinical information. YX, MP, JJ, YW, AS, LM, LH, NS, MK, JV, JG, PG, J-SL and ZW assisted and coordinated for tissues and ascites cells processing and other related analyses and experiments. LW, RW, SS, KH and JA wrote the manuscript. LW, JAA, RW, SS, KH, GZ, SH, ZW and AF revised the manuscript.
Funding: This study was supported in part by the National Cancer Institute awards CA129906, CA127672, CA138671 and CA172741 and the DOD grants: CA150334 and CA162445 to JAA and DOD grants CA160433 and CA170906 to SS, and the start-up research funds provided to LW by the Department of Genomic Medicine, Division of Cancer Medicine of MD Anderson Cancer Center. This study was also supported by SMF Core grant CA016672 (SMF).
Competing interests: None declared.
Ethics approval: This project was approved by the institutional review committee.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: All WES and RNA-Seq data have been deposited at the European Genome-phenome Archive (EGA). The datasets can be fully accessed under the accession number EGAS00001003180. Further information about EGA can be found on https://ega-archive.org (’the EGA of human data consented for biomedical research': http://www.nature.com/ng/journal/v47/n7/full/ng.3312.html).
Author note: Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to JA (jajani@mdanderson.org), SS (ssong@mdanderson.org) or LW (lwang22@mdanderson.org).
Patient consent for publication: Not required.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2. Ajani JA, Lee J, Sano T, et al. Gastric adenocarcinoma. Nat Rev Dis Primers 2017;3:17036 10.1038/nrdp.2017.36 [DOI] [PubMed] [Google Scholar]
- 3. Shiozaki H, Elimova E, Slack RS, et al. Prognosis of gastric adenocarcinoma patients with various burdens of peritoneal metastases. J Surg Oncol 2016;113:29–35. 10.1002/jso.24087 [DOI] [PubMed] [Google Scholar]
- 4. Gill RS, Al-Adra DP, Nagendran J, et al. Treatment of gastric cancer with peritoneal carcinomatosis by cytoreductive surgery and HIPEC: a systematic review of survival, mortality, and morbidity. J Surg Oncol 2011;104:692–8. 10.1002/jso.22017 [DOI] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. 10.1038/nm.3850 [DOI] [PubMed] [Google Scholar]
- 7. Sohn BH, Hwang JE, Jang HJ, et al. Clinical significance of four molecular subtypes of gastric cancer identified by the cancer genome atlas project. Clin Cancer Res 2017:4441–9. 10.1158/1078-0432.CCR-16-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charalampakis N, Nogueras González GM, Elimova E, et al. The proportion of signet ring cell component in patients with localized gastric adenocarcinoma correlates with the degree of response to pre-operative chemoradiation. Oncology 2016;90:239–47. 10.1159/000443506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chirieac LR, Swisher SG, Correa AM, et al. Signet-ring cell or mucinous histology after preoperative chemoradiation and survival in patients with esophageal or esophagogastric junction adenocarcinoma. Clin Cancer Res 2005;11:2229–36. 10.1158/1078-0432.CCR-04-1840 [DOI] [PubMed] [Google Scholar]
- 10. Cho SY, Park JW, Liu Y, et al. Sporadic early-onset diffuse gastric cancers have high frequency of somatic CDH1 alterations, but low frequency of somatic RHOA mutations compared with late-onset cancers. Gastroenterology 2017;153:536–49. 10.1053/j.gastro.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi JH, Kim YB, Ahn JM, et al. Identification of genomic aberrations associated with lymph node metastasis in diffuse-type gastric cancer. Exp Mol Med 2018;50:6 10.1038/s12276-017-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583–7. 10.1038/ng.2984 [DOI] [PubMed] [Google Scholar]
- 13. Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573–82. 10.1038/ng.2983 [DOI] [PubMed] [Google Scholar]
- 14. Nishizawa T, Nakano K, Harada A, et al. DGC-specific RHOA mutations maintained cancer cell survival and promoted cell migration via ROCK inactivation. Oncotarget 2018;9:23198–207. 10.18632/oncotarget.25269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurashige J, Hasegawa T, Niida A, et al. Integrated molecular profiling of human gastric cancer identifies DDR2 as a potential regulator of peritoneal dissemination. Sci Rep 2016;6:22371 10.1038/srep22371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Research Network Analysis Working Group: Asan University BC Cancer Agency Brigham and Women’s Hospital Broad Institute Brown University Case Western Reserve University Dana-Farber Cancer Institute Duke University Greater Poland Cancer Centre Harvard Medical School Institute for Systems Biology KU Leuven Mayo Clinic Memorial Sloan Kettering Cancer Center National Cancer Institute Nationwide Children’s Hospital Stanford University University of Alabama University of Michigan University of North Carolina University of Pittsburgh University of Rochester University of Southern California University of Texas MD Anderson Cancer Center University of Washington Van Andel Research Institute Vanderbilt University Washington University Genome Sequencing Center: Broad Institute Washington University in St. Louis Genome Characterization Centers: BC Cancer Agency Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University University of Southern California Epigenome Center Genome Data Analysis Centers: Broad Institute Brown University: University of California Santa Cruz Biospecimen Core Resource: International Genomics Consortium Research Institute at Nationwide Children’s Hospital Tissue Source Sites: Analytic Biologic Services Asan Medical Center Asterand Bioscience Barretos Cancer Hospital BioreclamationIVT Botkin Municipal Clinic Chonnam National University Medical School Christiana Care Health System Cureline Emory University Erasmus University Indiana University School of Medicine Institute of Oncology of Moldova International Genomics Consortium Invidumed Israelitisches Krankenhaus Hamburg Keimyung University School of Medicine National Cancer Center Goyang Ontario Tumour Bank Peter MacCallum Cancer Centre Pusan National University Medical School Ribeirão Preto Medical School St. Joseph’s Hospital &Medical Center St. Petersburg Academic University Tayside Tissue Bank University of Dundee University of Kansas Medical Center University of North Carolina at Chapel Hill University of Pittsburgh School of Medicine Disease Working Group: Duke University Yonsei University College of Medicine Data Coordination Center: CSRA Inc. Project Team: National Institutes of Health. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–75. 10.1038/nature20805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371–85. 10.1016/j.cell.2018.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao JM, Zhou X, Gatignol A, et al. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 2014;33:4916–23. 10.1038/onc.2013.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang J, Wu LY, Zhang XS, et al. Discovery of co-occurring driver pathways in cancer. BMC Bioinformatics 2014;15:271 10.1186/1471-2105-15-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402–7. 10.1038/ng.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor AM, Shih J, Ha G, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 2018;33:676–89. 10.1016/j.ccell.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen T, Chen Z, Zhao ZJ, et al. Genetic defects of the IRF1-mediated major histocompatibility complex class I antigen presentation pathway occur prevalently in the JAK2 gene in non-small cell lung cancer. Oncotarget 2017;8:60975–86. 10.18632/oncotarget.17689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aust S, Felix S, Auer K, et al. Absence of PD-L1 on tumor cells is associated with reduced MHC I expression and PD-L1 expression increases in recurrent serous ovarian cancer. Sci Rep 2017;7:42929 10.1038/srep42929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller CA, White BS, Dees ND, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014;10:e1003665 10.1371/journal.pcbi.1003665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu G, Wang J, Chen Y, et al. Overexpression of phosphorylated mammalian target of rapamycin predicts lymph node metastasis and prognosis of chinese patients with gastric cancer. Clin Cancer Res 2009;15:1821–9. 10.1158/1078-0432.CCR-08-2138 [DOI] [PubMed] [Google Scholar]
- 28. Yoo MW, Park J, Han HS, et al. Discovery of gastric cancer specific biomarkers by the application of serum proteomics. Proteomics 2017;17:1600332 10.1002/pmic.201600332 [DOI] [PubMed] [Google Scholar]
- 29. Perrais M, Pigny P, Buisine MP, et al. Aberrant expression of human mucin gene MUC5B in gastric carcinoma and cancer cells. Identification and regulation of a distal promoter. J Biol Chem 2001;276:15386–96. 10.1074/jbc.M010534200 [DOI] [PubMed] [Google Scholar]
- 30. Woo J, Lee J, Chae YK, et al. Overexpression of AQP5, a putative oncogene, promotes cell growth and transformation. Cancer Lett 2008;264:54–62. 10.1016/j.canlet.2008.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qiu YS, Liao GJ, Jiang NN. DNA methylation-mediated silencing of regenerating protein 1 Alpha (REG1A) Affects Gastric Cancer Prognosis. Med Sci Monit 2017;23:5834–43. 10.12659/MSM.904706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hara K, Fukui H, Sun C, et al. Effect of REG Iα protein on angiogenesis in gastric cancer tissues. Oncol Rep 2015;33:2183–9. 10.3892/or.2015.3878 [DOI] [PubMed] [Google Scholar]
- 33. Aung PP, Oue N, Mitani Y, et al. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene 2006;25:2546–57. 10.1038/sj.onc.1209279 [DOI] [PubMed] [Google Scholar]
- 34. Chen ZF, Huang ZM, Xue HB, et al. REG3A promotes the proliferation, migration, and invasion of gastric cancer cells. Onco Targets Ther 2017;10:2017–23. 10.2147/OTT.S131443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu X, Wang J, Wang H, et al. REG3A accelerates pancreatic cancer cell growth under IL-6-associated inflammatory condition: Involvement of a REG3A-JAK2/STAT3 positive feedback loop. Cancer Lett 2015;362:45–60. 10.1016/j.canlet.2015.03.014 [DOI] [PubMed] [Google Scholar]
- 36. Cipriano R, Miskimen KL, Bryson BL, et al. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res 2014;12:1156–65. 10.1158/1541-7786.MCR-13-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Flynn S, Stockinger B. Tumor and CD4 T-cell interactions: tumor escape as result of reciprocal inactivation. Blood 2003;101:4472–8. 10.1182/blood-2002-10-3030 [DOI] [PubMed] [Google Scholar]
- 40. Valzasina B, Piconese S, Guiducci C, et al. Tumor-induced expansion of regulatory T cells by conversion of CD4+CD25- lymphocytes is thymus and proliferation independent. Cancer Res 2006;66:4488–95. 10.1158/0008-5472.CAN-05-4217 [DOI] [PubMed] [Google Scholar]
- 41. Olkhanud PB, Damdinsuren B, Bodogai M, et al. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4⁺ T cells to T-regulatory cells. Cancer Res 2011;71:3505–15. 10.1158/0008-5472.CAN-10-4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Su S, Liao J, Liu J, et al. Blocking the recruitment of naive CD4+ T cells reverses immunosuppression in breast cancer. Cell Res 2017;27:461–82. 10.1038/cr.2017.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cancer Genome Atlas Research Network. Electronic address wbe, cancer genome atlas research n. comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taieb J, Moehler M, Boku N, et al. Evolution of checkpoint inhibitors for the treatment of metastatic gastric cancers: Current status and future perspectives. Cancer Treat Rev 2018;66:104–13. 10.1016/j.ctrv.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 46. Gao D, Inuzuka H, Korenjak M, et al. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol Biol Cell 2009;20:3305–16. 10.1091/mbc.e09-01-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagle AM, Levine KM, Tasdemir N, et al. Loss of E-cadherin Enhances IGF1-IGF1R Pathway Activation and Sensitizes Breast Cancers to Anti-IGF1R/InsR Inhibitors. Clin Cancer Res 2018;24 10.1158/1078-0432.CCR-18-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X, Ni Z, Duan Z, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One 2015;10:e0116979 10.1371/journal.pone.0116979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L, Hou Y, Ashktorab H, et al. The impact of C-MYC gene expression on gastric cancer cell. Mol Cell Biochem 2010;344:125–35. 10.1007/s11010-010-0536-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ito K, Park SH, Nayak A, et al. PTK6 Inhibition suppresses metastases of triple-negative breast cancer via SNAIL-Dependent E-Cadherin Regulation. Cancer Res 2016;76:4406–17. 10.1158/0008-5472.CAN-15-3445 [DOI] [PubMed] [Google Scholar]
- 51. Oh SC, Sohn BH, Cheong JH, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 2018;9:9 10.1038/s41467-018-04179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiang J, Jin MS, Kong F, et al. Decreased galectin-9 and increased Tim-3 expression are related to poor prognosis in gastric cancer. PLoS One 2013;8:e81799 10.1371/journal.pone.0081799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang J, Zhu L, Cai Y, et al. Role of downregulation of galectin-9 in the tumorigenesis of gastric cancer. Int J Oncol 2014;45:1313–20. 10.3892/ijo.2014.2494 [DOI] [PubMed] [Google Scholar]
- 54. Wang Y, Zhao E, Zhang Z, et al. Association between Tim‑3 and Gal‑9 expression and gastric cancer prognosis. Oncol Rep 2018;40:2115–26. 10.3892/or.2018.6627 [DOI] [PubMed] [Google Scholar]
- 55. Wang Z, Yin N, Zhang Z, et al. Upregulation of T-cell Immunoglobulin and Mucin-Domain Containing-3 (Tim-3) in Monocytes/Macrophages Associates with Gastric Cancer Progression. Immunol Invest 2017;46:134–48. 10.1080/08820139.2016.1229790 [DOI] [PubMed] [Google Scholar]
- 56. Böger C, Behrens HM, Krüger S, et al. The novel negative checkpoint regulator VISTA is expressed in gastric carcinoma and associated with PD-L1/PD-1: A future perspective for a combined gastric cancer therapy? Oncoimmunology 2017;6:e1293215 10.1080/2162402X.2017.1293215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2018-318070supp002.pdf (62.8KB, pdf)
gutjnl-2018-318070supp003.pdf (62.8KB, pdf)
gutjnl-2018-318070supp001.pdf (62.8KB, pdf)