Abstract
We developed a new method of glucose sensing using an inactive form of glucose oxidase from Aspergillus niger. Glucose oxidase was rendered inactive by removal of the FAD cofactor. The resulting apo-glucose oxidase still binds glucose as observed from a decrease in its intrinsic tryptophan fluorescence. 8-Anilino-1-naphthalenesulfonic acid (ANS) was found to bind spontaneously to apo-glucose oxidase as seen from an enhancement of the ANS fluorescence. The steady state intensity of the bound ANS decreased 25% upon binding of glucose, and the mean lifetime of the bound ANS decreased about 40%. These spectral changes occurred with a midpoint from 10 to 20 mM glucose, which is comparable to the KD of holo-glucose oxidase. These results suggest that apo-glucose oxidase can be used as a reversible nonconsuming sensor for glucose.
Keywords: glucose oxidase, fluorescence spectroscopy, glucose estimation, diabetes, time-resolved fluorescence
Close regulation of blood glucose is essential for diabetics to minimize the long term adverse health consequences of diabetes [1–2]. Measurement of blood glucose requires a finger stick, which is done infrequently by most diabetics. As a result there is a substantial worldwide effort to develop non-invasive and minimally invasive methods for frequent or continuous monitoring of blood glucose. A wide variety of methods have been tested, including optical rotation [3–4], near infrared absorbance and Raman scattering [5–7] and the design and synthesis of glucosespecific fluorescence probes [8–9]. Because of its simplicity and sensitivity, fluorescence has been extensively evaluated for use in glucose sensing. Most of these studies used the binding of a glucosespecific protein such as concanavalin A to glucose or some polysaccharide. This work has its origins with the studies of Schultz and co-workers [10–12] who developed a competitive glucose assay which does not require substrates and does not consume glucose. This assay used fluorescence resonance energy transfer (RET) between a fluorescence donor and an acceptor, each covalently linked to concanavalin A (ConA) or dextran. In the absence of glucose the binding between ConA and dextran resulted in a high RET efficiency. The addition of glucose resulted in its competitive binding to ConA, ConA displacement of ConA from the labeled dextran, and a decrease in the RET efficiency.
In this laboratory we also studied the ConA-dextran system for glucose sensing. In our case we chose to use fluorescence decay times in place of the fluorescence intensities. We chose lifetime-based sensing because the lifetimes are mostly independent of the total intensity [13–15]. We use ConA-dextran to develop lifetime sensors using both visible and NIR fluorophores [16–17] and a long-lifetime metal-ligand complexes as the donor [18]. While the ConA-dextran system worked in the laboratory, the sensors were only partially reversible, and they become increasingly irreversible with time following mixing of the ConA and dextran. We often observed precipitation. We reasoned these effects were due to binding of the multi-valent ConA with the multi-valent dextran and subsequent cross-linking. To circumvent this problem we developed a sensor based on ConA and a small protein labeled with a single covalently attached glucose residue [18], but this sensor also proved to be only partially reversible.
To circumvent the difficulties of lack of reversibility we decided to explore the use of proteins which bind glucose, such as those present in bacteria. We reasoned that a mono-valent glucose binding protein interacting with a monomeric sugar would be completely reversible. In our initial studies we used the glucosegalactose binding protein (GGBP) from E. coli, which was labeled with a fluorophore at a single genetically cysteine residue at position 26 [19]. This labeled protein displayed spectral changes in response to micro-molar concentrations of glucose.
In the present report we extend the use of glucose sensing with proteins to a another protein which binds glucose. Glucose oxidase (EC 1.1.3.4) from Aspergillus niger (GO) is an oxidoreductase enzyme that catalyzes the conversion of β-D-glucose and oxygen to D-glucono-1,5-lactone and hydrogen peroxide. This GO is a flavoprotein, highly specific for β-D-glucose [20], and is widely used to estimate glucose concentration in blood or urine samples through the formation of colored dyes by the hydrogen peroxide produced in the reaction [21–22]. Because glucose is consumed this enzyme cannot be used as a reversible sensor.
To prevent glucose oxidation we removed the FAD cofactor which is required for the reaction. We found that the resulting apo-glucose oxidase still bound glucose with an affinity comparably to the holo enzyme. Importantly, the intrinsic tryptophan fluorescence of apo-glucose oxidase was sensitive to glucose binding. Additionally, apo-glucose oxidase non-covalently bound 8-anilino-1-naphthalene sulfonic acid (ANS). The ANS bound to glucose oxidase displayed decreases in both intensity and lifetime upon addition of glucose. These results suggested a new approach to obtaining protein sensors for glucose, these being glucose-converting enzymes rendered inactive by removal of necessary cofactors.
MATERIALS AND METHODS
Glucose and all other chemicals were of reagent grade and purchased from Sigma. 8-Anilino-1-naphthalenesulfonic acid (ANS) and glucose oxidase from Aspergillus niger were also obtained from Sigma. Steady state fluorescence measurements were carried out on a SLM AMINCO spectrofluorometer at room temperature, by using quartz cuvettes.
Frequency-domain intensity decay measurements were performed using instrumentation described previously [23]. The intensity decay data were analyzed in terms of the multi-exponential model
| [1] |
In this expression the αi,j values are the initial amplitudes of the component with a decay time τi. The subscript j refers to different concentrations of glucose. The data were analyzed globally with the decay times (τi) global for all concentrations of glucose (αi,j). The mean lifetime at each glucose concentration was calculated using
| [2] |
Preparation of apo-glucose oxidase.
A saturated ammonium sulfate solution at 25% was acidified to pH 1.4 with H2SO4. 5 mg of glucose oxidase in 0.5 ml solution was added drop-wise with stirring to 5.0 ml of the acidified salt solution at 4°C. The FAD was split off from the enzyme, and the yellow supernatant was removed after centrifugation at 13,000 rpm for 15 min [24]. The precipitate was re-dissolved and neutralized by adding of sodium acetate. The neutralized solution was subjected two more cycles of acidified salt treatment, centrifugation and neutralization. Finally, the protein was washed three times on Centricon tube (Amicon) in 10 mM sodium phosphate, pH 6.8. The apo-GO retained its capacity for reactivation by FAD for several weeks when stored at 0°C, and pH 6.8.
In order to exchange the original buffer with 10 mM phosphate buffer, pH 6.8 the enzyme solution was passed through to a PD-10 gel filtration column (Pharmacia). The obtained enzyme was used in our experiments.
RESULTS AND DISCUSSION
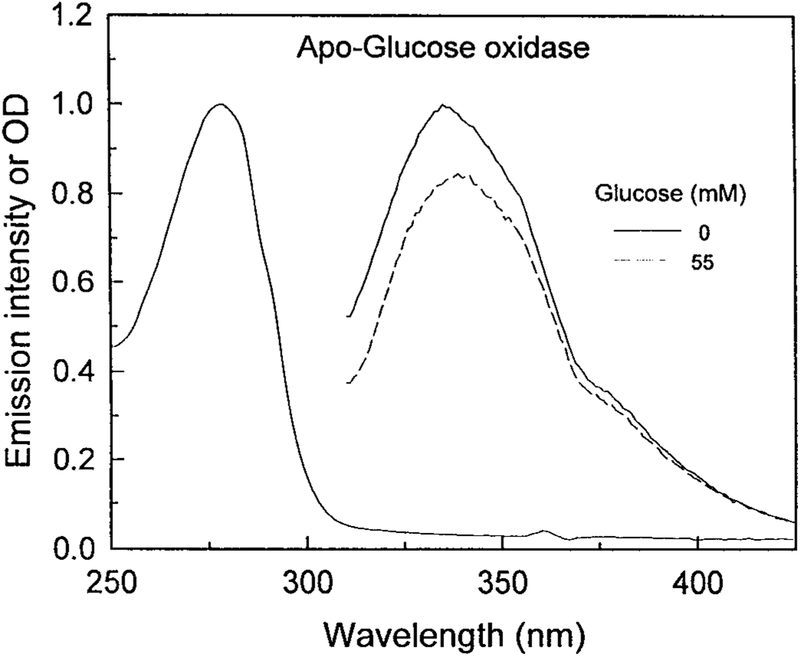
Our approach to estimate the glucose concentration is based on fluorescence spectral changes which do not require the glucose oxidation. We prepared the FAD-free GO. The apo-enzyme is known to bind the glucose, but it cannot oxide the sugar [25]. Figure 1 shows the absorption and emission spectra of apo-GO. The absorbance spectrum shows the characteristic shape of the coenzyme-free proteins, with a maximum of absorbance at 278 nm due to the aromatic amino acid residues. The absence of absorption at wavelengths above 300 nm indicates the FAD has been completely removed. The fluorescence emission spectrum of apo-GO at room temperature upon excitation at 298 nm displays an emission maximum at 340 nm, which is characteristic of partially shielded tryptophan residues. The addition of 20 mM glucose to the enzyme solution resulted in a quenching of the tryptophanyl fluorescence emission about 18%. This result indicates that the apo-GO is still able to bind glucose. The observed fluorescence quenching may be mainly ascribed to the tryptophanyl residue 426. In fact, as shown by X-ray analysis and molecular dynamics simulations analyses the glucose-binding site of GO is formed by Asp 584, Tyr 515, His 559 and His 516. Moreover, Phe 414, Trp 426 and Asn 514 are in locations where they might form additional contacts to the glucose [26–28].
FIG. 1.
Absorption and emission spectra of apo-glucose oxidase. Excitation at 298 nm. The protein concentration was 0.05 mg/ml.
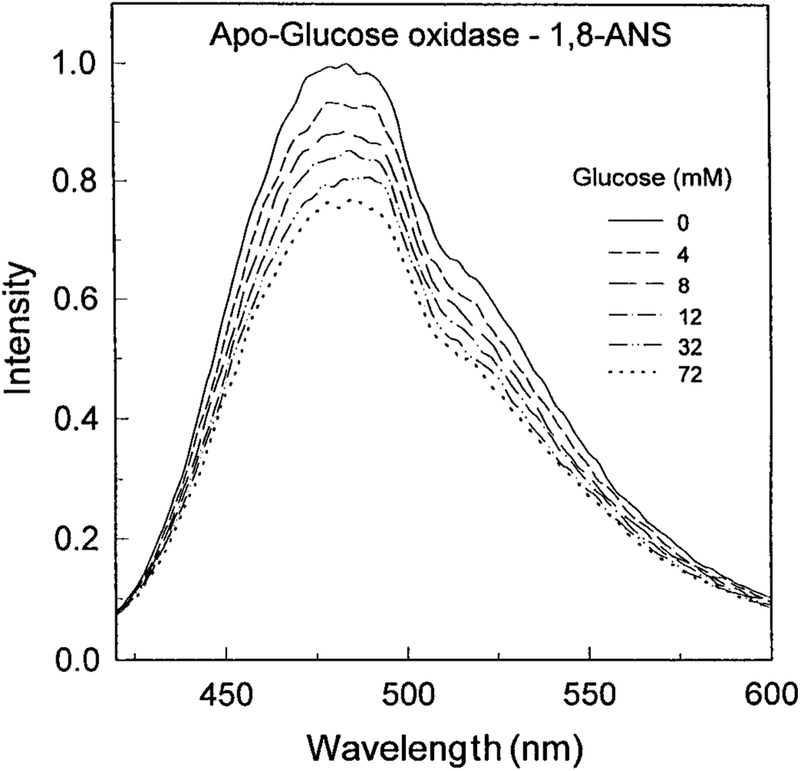
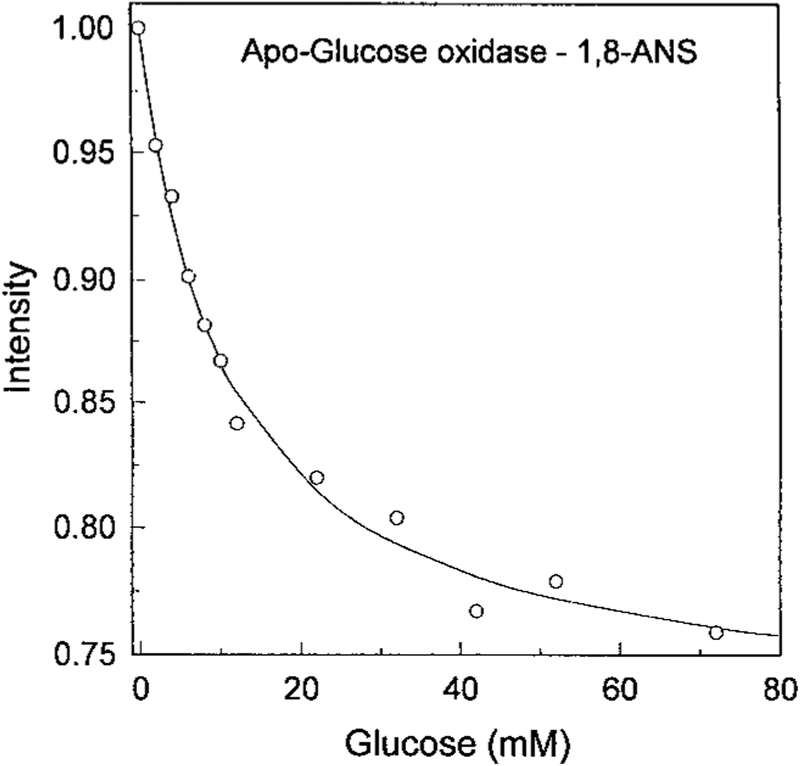
The intrinsic fluorescence from proteins is usually not useful for clinical sensing because of the need for complex or bulky light sources and the presence of numerous proteins in most biological samples. In an attempt to obtain a glucose sensor with longer excitation and emission wavelengths we studied whether ANS would bind to apo-GO. Addition of apo-GO to an ANS solution resulted in an approximate 30-fold increase in the ANS intensity. Importantly, the intensity of the ANS emission was sensitive to glucose, decreasing approximately 25% upon glucose addition (Fig. 2). The ANS was not covalently bound to the protein. Addition of glucose resulted in a progressive decrease in the ANS fluorescence intensity. This suggests that the ANS is being displaced into a more polar environment upon binding glucose. The decreased ANS intensity occurred with a glucose-binding constant near 10 mM (Fig. 3), which is comparable to the KD of the holoenzyme. Since the binding affinity has not changed significantly, one can suggest that the binding is still specific for glucose.
FIG. 2.
Emission spectra 5 μM 1,8-ANS in the presence of 3 μM apo-glucose oxidase. Excitation at 325 nm.
FIG. 3.
Glucose-dependent emission intensity of 1,8-ANS bound to apo-glucose oxidase. Excitation at 325 nm, emission at 480 nm.
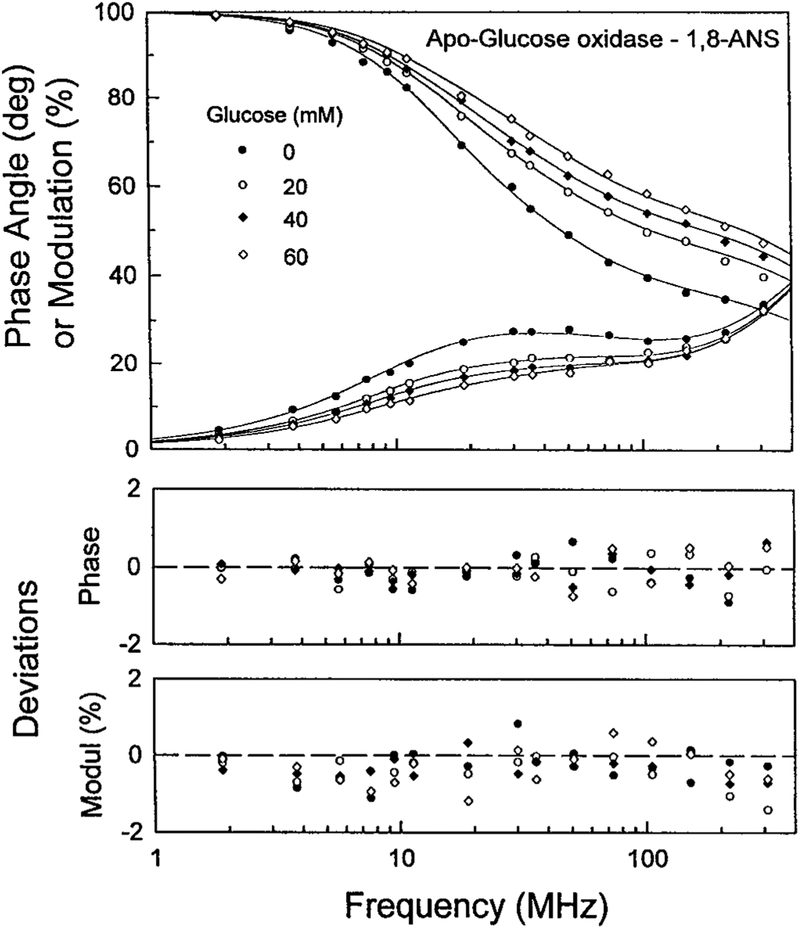
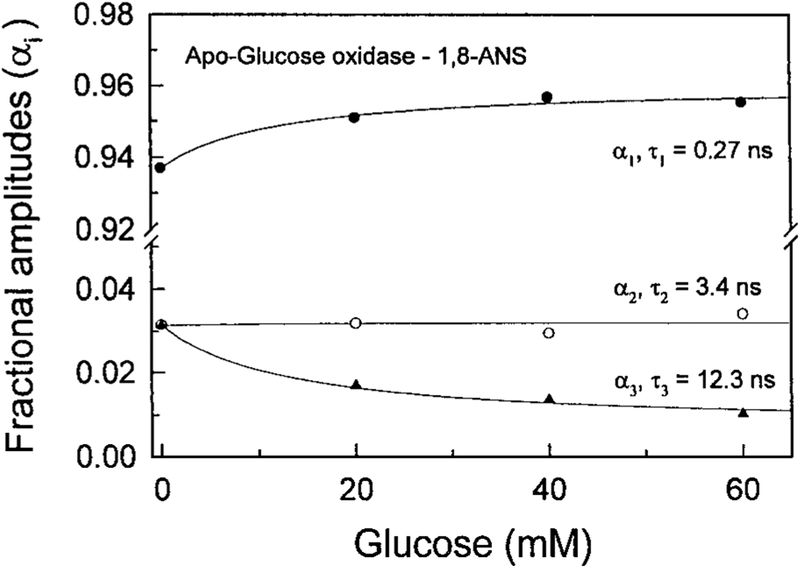
Recognizing the favorable attributes of lifetime-based sensing [13–15] we measured the intensity decay of ANS-labeled apo-GO in the presence of increasing concentrations of glucose (Fig. 4). Addition of glucose shifts the frequency responses to higher frequencies, which is due to a decreased ANS lifetime. The shorter lifetimes of ANS apo-GO in the presence of glucose is consistent with our suggestion that glucose displaces the ANS to a more polar environment. The FD data were analyzed globally in terms of the multi-exponential model (Fig. 5). The lifetimes were held global at all glucose concentrations. These results show that addition of glucose resulted in a decreased amplitude of the long component with a decay time of 12.3 ns. The decreased amplitude of this component is accompanied by an increased amplitude of the short component with a lifetime of 0.27 ns. These results are consistent with our suggestion that glucose displaces ANS from a hydrophobic to a more polar environment.
FIG. 4.
Frequency-domain intensity decays of 1,8-ANS-apoglucooxidase in the presence of increasing concentrations of glucose. Excitation was 335 nm (DCM dye laser), emission was observed through interference filter 535/50 nm and two Corning 3–71 cut-off filters.
FIG. 5.
Glucose-dependent lifetimes and pre-exponential factors from the lifetime-global analysis.
Addition of glucose does not affect the amplitude of the middle component with a decay time of 3.4 ns. This suggests that the 3.4 ns component is due to ANS which is bound to the protein but at a site distant from the glucose binding site. Additional studies are required to confirm or refute this speculation.
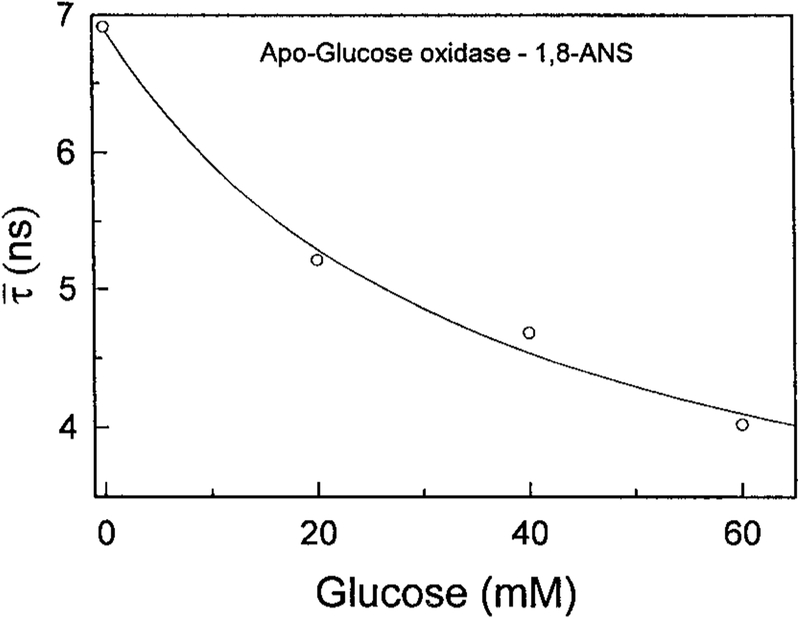
And finally, Fig. 6 shows the mean lifetime of ANS apo-GO in the presence of increasing glucose concentrations. The mean lifetime decreases by over 40% upon addition of glucose. These results demonstrate that apo-glucose oxidase, when labeled with suitable fluorophores, can serve as a protein sensor for glucose.
FIG. 6.
Effect of glucose on the mean decay time of 1,8-ANS labeled apo-glucose oxidase.
ACKNOWLEDGMENT
This work was supported by a Grant from the NIH, National Center for Research Resources, RR-08119.
Abbreviations used:
- ANS
8-anilino-1-naphthalenesulfonic acid
- ConA
concanavalin A
- GO
glucose oxidase
- NIR
near infrared
- RET
resonance energy transfer
REFERENCES
- 1.The Diabetes Control and Complications Trial Research Group (1997) Diabetes 4, 271–286. [PubMed] [Google Scholar]
- 2.The Diabetes Control and Complications Trial Research Group (1993) N. Engl. J. Med 329, 977–986. [DOI] [PubMed] [Google Scholar]
- 3.Rabinovitch B, March WF, and Adams RL (1982) Diabetes Care 5(3), 254–258. [DOI] [PubMed] [Google Scholar]
- 4.March WF, Rabinovitch B, Adams R, Wise JR, and Melton M (1982) Trans. Am. Soc. Artif. Intern. Organs 28, 232–235. [PubMed] [Google Scholar]
- 5.Amato I (1992) Science 258, 892–892.1439802 [Google Scholar]
- 6.Robinson MR, Eaton RP, Haaland DM, Koepp GW, Thomas EV, Stallard BR, and Robinson PL (1992) Clin. Chem 38(9), 1618–1622. [PubMed] [Google Scholar]
- 7.Yu N-T, Krantz BS, Eppstein JA, Ignotz KD, Samuels MA, Long JR, and Price JF (1996) J. Biomed. Optics 1(3), 280–288. [DOI] [PubMed] [Google Scholar]
- 8.James TD, Shinmori H, Takeuchi M, and Shinkai S (1996) Chem. Commun pp. 705–706. [Google Scholar]
- 9.James TD, Shinmori H, and Shinkai S (1997) Chem. Commun pp. 71–72. [Google Scholar]
- 10.Schultz JS, and Sims G (1979) Biotech. Bioeng. Symp 9, 65–71. [PubMed] [Google Scholar]
- 11.Schultz J, Mansouri S, and Goldstein IJ (1982) Diabetes Care 5(3), 245–253. [DOI] [PubMed] [Google Scholar]
- 12.Meadows D, and Schultz JS (1988) Talanta 35(2), 145–150. [DOI] [PubMed] [Google Scholar]
- 13.Szmacinski H, and Lakowicz JR (1994) in Topics in Fluorescence Spectroscopy (Lakowicz JR, Ed.), Vol. 4, pp. 295–334, Plenum Press, New York. [Google Scholar]
- 14.Szmacinski H, and Lakowicz JR (1995) Sensors and Actuators B 29, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lippitsch ME, Draxler S, and Kieslinger D (1997) Sensors and Actuators B 38–39, 96–102. [Google Scholar]
- 16.Lakowicz JR, and Maliwal BP (1993) Anal. Chim. Acta 271, 155–164. [Google Scholar]
- 17.Tolosa L, Malak H, Rao G, and Lakowicz JR (1997) Sensors and Actuators 45, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolosa L, Szmacinski H, Rao G, and Lakowicz JR (1997) Anal. Biochem 250, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolosa L, Gryczynski I, Eichhorn LR, Dattelbaum JD, Castellano FN, Rao G, and Lakowicz JR (1999) Anal. Biochem 267, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manstein DJ, Pai EF, Schopfer LM, and Massey V (1986) Biochemistry 25, 22, 6807–6816. [DOI] [PubMed] [Google Scholar]
- 21.Teng FY, Punla O, and Koos BJ (1995) J. Soc. Gynecol. Invest 2, 618–622. [DOI] [PubMed] [Google Scholar]
- 22.Miyasaka T, Taniyama Y, Sakai K, and Yoshim Y (1997) ASAIO J. 43, M505–M509. [PubMed] [Google Scholar]
- 23.Lakowicz JR, and Gryczynski I (1991) in Topics in Fluorescence Spectroscopy, Vol. 1: Techniques (Lakowicz JR, Ed.), pp. 293–355, Plenum Publishing, New York. [Google Scholar]
- 24.Swoboda BEP (1969) Biochim. Biophys. Acta 175, 364–379. [DOI] [PubMed] [Google Scholar]
- 25.Morgan HW, and Corke CT (1977) Canadian Journal of Microbiol. 23(9), 1109–1117. [DOI] [PubMed] [Google Scholar]
- 26.Hecht HJ, Kalisz HM, Hendle J, Schmid RD, and Schomburg D (1993) J. Mol. Biol 229(1), 153–172. [DOI] [PubMed] [Google Scholar]
- 27.Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisz HM, and Hecht HU (1999) Acta. Crystallogr. D. Biol. Crystallogr 55(5), 967–977. [DOI] [PubMed] [Google Scholar]
- 28.Meyer M, Wohlfahrt G, Knablein J, and Schomburg D (1998) J. Comp. Aided Molec. Des 12(5), 425–440. [DOI] [PubMed] [Google Scholar]