Abstract
Previous efforts to evaluate the detection of human papilloma viral (HPV) DNA in whole saliva as a diagnostic measure for HPV-associated oropharyngeal cancer (HPV-OPC) have not shown sufficient clinical performance. We hypothesize that salivary exosomes are packaged with HPV-associated biomarkers, and efficient enrichment of salivary exosomes through isolation can enhance diagnostic and prognostic performance for HPV-OPC. In this study, an acoustofluidic (the fusion of acoustics and microfluidics) platform was developed to perform size-based isolation of salivary exosomes. These data showed that this platform is capable of consistently isolating exosomes from saliva samples, regardless of viscosity variation and collection method. Compared with the current gold standard, differential centrifugation, droplet digital RT-PCR analysis showed that the average yield of salivary exosomal small RNA from the acoustofluidic platform is 15 times higher. With this high-yield exosome isolation platform, we show that HPV16 DNA could be detected in isolated exosomes from the saliva of HPV-associated OPC patients at 80% concordance with tissues/biopsies positive for HPV16. Overall, these data demonstrated that the acoustofluidic platform can achieve high-purity and high-yield salivary exosome isolation for downstream salivary exosome–based liquid biopsy applications. Additionally, HPV16 DNA sequences in HPV-OPC patients are packaged in salivary exosomes and their isolation will enhance the detection of HPV16 DNA.
Although therapy for oropharyngeal cancer (OPC) has progressed, the 5-year survival rate for OPC remains below 50% and has not shown significant improvement over the past decade.1,2 Poor prognoses can be attributed to the lack of early detection methods because clinical symptoms tend to appear in later stages of the disease.3,4 Screening based on risk factors, such as excess smoking, offers one approach to identify high-risk OPC patients for early diagnostic testing. Recently, human papillomavirus (HPV) has become the prominent associative factor of OPCs, surpassing conventional risk factors. This rise indicates a shift toward younger and healthier populations, who will only seek medical attention after apparent regional cervical metastasis indicative of a late-stage prognosis.5 Unfortunately, even if medical visits occurred more often, clinical examination alone cannot identify early-stage OPC premalignant lesions.6,7 Thus, it is critical to develop methods for the early detection of OPC.
Saliva harbors tumor biomarkers that can enable early detection, noninvasive screening, and risk assessment of OPCs.8 Wang et al9 reported that when analyzing the whole saliva from HPV-OPC–positive patients with droplet digital PCR (ddPCR), they were able to identify traces of pathogenic HPV16 DNA. Unfortunately, HPV16 DNA was only detectable in 40% of cases. Exosomes are enriched vesicular sources of tumor-specific genomic and proteomic molecular targets.10 Therefore, isolating tumor-specific exosomes from saliva and identifying their molecular signatures can be a viable strategy for early detection of HPV-OPC. Although there is no reported evidence of exosomal existence of HPV DNA, it was hypothesized that HPV16 DNA sequences in HPV-OPC patients are packaged in salivary exosomes, and exosome isolation will enhance the detection of HPV16 DNA.11
Conventional isolation approaches such as differential centrifugation and immune capture require a large sample volume, is time-consuming, and has a low yield and purity.12 Because high centrifugal forces and/or multiple washing steps are required, conventional methods may also compromise the integrity of isolated exosomes. Variation in the physical properties of saliva due to factors such as health status and stimulation during collection complicates exosome isolation protocols.13,14 For example, the viscosity of saliva ranges from 1.10 to 2.30 mPa · s in contrast to plasma (viscosity 1.10 to 1.30 mPa · s) which typically has much more stable physical properties. To achieve salivary exosome isolation for liquid biopsies, the isolation technology must not only have high yield, high purity, and high biocompatibility, but it must also achieve stable performance on samples with vastly different physical properties.
Previous efforts yielded an acoustofluidic platform (the fusion of acoustics and microfluidics) that uses standing surface acoustic waves (SAW) to isolate exosomes from undiluted blood samples specifically.15 However, the differences in the viscosities of saliva samples can yield unpredictable exosome isolation results, because viscosity-induced drag force is important during size-based isolation.13,14 In this study, the acoustofluidic platform was optimized for exosome isolation in saliva samples with different physical qualities, including a wide range of viscosities. It was also determined that the isolation of exosomes can improve the efficiency of HPV16 DNA detection in patients with HPV-associated oropharyngeal cancer (HPV-OPC).
Materials and Methods
Simulation
Simulation of forces and motion of particles in the channel was derived by COMSOL Multiphysics software version 5.4 (COMSOL, Burlington, MA). Particles primarily experienced an acoustic radiation force arising from the SAW field and a drag force from the liquid. The acoustic radiation force Fr is described by Equations 1 and 2:
| (1) |
| (2) |
In these equations, p0, Vp, λ, k, x, ρp, ρf, βp, and βf represent acoustic pressure, volume of the particle, wavelength, wave number, distance from a pressure node, density of the particle, density of the fluid, compressibility of the particle, and compressibility of the fluid, respectively.
The drag force, named Stokes drag force, is represented by Equation 3:
| (3) |
In the equation, η, Rp, up, and uf are viscosity of the fluid, radius of the particle, velocity of the particle, and velocity of the fluid, respectively.
Device Fabrication
Fabrication of acoustofluidic devices follows our previously described procedures.15 Two pairs of interdigital transducers (IDTs) that generated 20 MHz and 40 MHz SAW were deposited on a Y+128° X-propagation lithium niobate (LiNbO3) substrate per a standard photolithography process. The IDTs' shapes were acquired by an MA/BA6 mask aligner (SUSS MicroTec, Garching, Germany) and SPR3012 photoresist (MicroChem, Westborough, MA). After CD26 developing solution (MicroChem) removed unwanted photoresist, an e-beam evaporator (Semicore Equipment, Livermore, CA) was used to deposit a metal double layer (Cr/Au, 50 Å/500 Å) on the substrate. PRS3000 resist stripper (VWR) was applied to form IDTs with electrode widths of 50 μm and 25 μm on the substrate. Standard soft lithography using SU-8 photoresist (MicroChem) created a silicone mold. Sylgard 184 Silicone Elastomer Curing Agent and Base (Dow Corning, Midland, MI) were solidified on the mold to form a 100-μm–height and 800-μm–width polydimethylsiloxane (PDMS) microchannel. The microchannel and LiNbO3 substrate were bonded after oxygen plasma coating and incubation at 65°C overnight.
In brief, two pairs of IDTs that generate 20-MHz and 40-MHz SAWs were bonded with a PDMS microchannel of 100-μm height and 800-μm width.
Saliva Collection
Healthy volunteers were instructed to refrain from eating, drinking, smoking, or oral hygiene procedures for at least 1 hour before saliva collection using three different methods. Method one: saliva samples were collected and centrifuged at 2600 × g for 15 minutes at 4°C to become cell-free saliva. Method two: samples were collected by SuperSAL saliva collectors (Oasis Diagnostics, Vancouver, WA). Method three: whole saliva samples were collected without further treatment.16 All collected samples were stored at −80°C and transported on dry ice.
HPV-OPC Patient Samples
All 10 HPV-OPC patients were recruited in the multicenter De-Escalate trial (collected REC 11/WM/0381; covered by REC 16/NW/0265). The samples were collected and stored using institutional review board–approved protocols from the Institute for Head and Neck studies and Education at the University of Birmingham, United Kingdom. Each patient was asked to rinse their mouth and gargle with normal saline (0.9% NaCl) for 2 minutes before spitting into a collection pot. The oral fluid samples were then spun at 1500 × g for 10 minutes. The supernatant was removed and stored at −80°C, and the pellets were resuspended in 1 mL phosphate-buffered saline (PBS) and stored at −80°C for further exosome isolation and ddPCR analysis.
Acoustofluidic Separation Experimental Setup
Polystyrene particles of diameters 50 nm, 500 nm, and 1 μm (Bangs Laboratory, Fishers, IN) were spiked into deionized water and 30% sucrose solution in deionized water, which represent low- and high-viscosity solutions. The acoustofluidic chip was placed on a TEC1-12730bPeltier cooling system (Hebei IT, Shanghai, China) and powered by a TP1505D variable DC power supply (Tekpower, Montclair, CA). The sample fluid and sheath fluid were individually controlled by neMESYS syringe pumps (CETONI, Korbussen, Germany). The optimized flow rates of the sample, upper sheath flow, lower sheath flow, and second separation module sheath flow were 6 μL/minute, 9 μL/minute, 2 μL/minute, and 11 μL/minute, respectively. The upright BX51WI microscope (Olympus, Tokyo, Japan) combined with a CoolSNAP HQ2CCD camera (Photometrics, Huntington Beach, CA) were used for recording the separation process. IDTs on acoustofluidic chip were driven by an E4422B function generator (Agilent, Santa Clara, CA) and a 100A250A amplifier (Amplifier Research, Souderton, PA). Solutions of 1 μm and 500 nm beads were used for validating the performance of 20-MHz IDTs and 40-MHz IDTs, respectively. Isolated exosome samples, submicrometer-sized waste, and micrometer-sized waste were collected. Particle size distributions were analyzed by Zetasizer Nano (Malvern Panalytica, Malvern, UK) and nanoparticle tracking analysis (Nanosight LM10; Malvern Panalytica) systems.
Transmission Electron Microscopy
The isolated exosome sample was fixed by 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO). A 300-mesh copper grid support film (Electron Microscopy Sciences, Hatfield, PA) covered a 100-μL drop of isolated exosome sample for absorption. The grid was then washed with distilled water, followed by uranyl–acetate negative staining (Electron Microscopy Sciences). Finally, the grid was washed again using distilled water and left to air dry at room temperature. The sample was then observed under the electron microscope (FEI, Hillsboro, OR).
Differential Centrifugation
Saliva samples were processed by centrifugation at 3000 × g for 15 minutes at 8°C. Supernatants were diluted with PBS to 2.3 mL and spun at 120,000 × g in TLS-55 tubes for 45 minutes at 8°C. The supernatants were then discarded, and the pellets were resuspended with 2.3 mL of PBS for another 120,000 × g differential centrifugation in TLS-55 tubes (Beckman Coulter, Brea, CA) for 45 minutes at 8°C. The pellets were then resuspended in 100 μL of PBS and stored at −80°C.
Western Blot Analysis
Isolated products (including exosomes, submicrometer-sized waste, and micrometer-sized waste) from original samples of similar volumes were processed by differential centrifugation. Twenty microliters of each sample were lysed in Pierce Cell Lysis Buffer (Thermo Fisher Scientific, Waltham, MA) with Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). Lysates were processed by SDS/PAGE and transferred to a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA). Primary antibodies, including mouse anti-CD63 (Santa Cruz Biotechnology, Dallas, TX), mouse anti-HSP90, and rabbit anti-TSG101 (Abcam, Cambridge, UK), were separately used for incubating the membrane for 12 hours at 4°C. Appropriate horseradish peroxidase secondary antibody incubation, including goat anti-mouse IgG and goat anti-rabbit IgG (Abcam), was used for 1 hour incubation at room temperature. ChemiDoc XRS+ (Bio-Rad) was used to characterize protein expression levels.
Nucleic Acid Extraction and ddPCR
The exosomal RNA extraction from optimized acoustofluidic separation and differential centrifugation methods was performed with miRNeasy micro kit (QIAGEN, Hilden Germany) according to the manufacturer's instructions. The reverse transcription was conducted with a TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). DNA was extracted from salivary microvesicles and exosomes from HPV-OPC patients with Quick-gDNA MiniPrep kit (Zymo Research, Irvine, CA). The ddPCR analysis was performed by using the QX100 Droplet Digital PCR system (Bio-Rad). The following components were well-mixed: 20 μL of a PCR reaction containing 10 μL of 2× ddPCR supermix (Bio-Rad), 1 μL of 20× TaqMan probes/primers, 4 μL of sample DNA or cDNA, and 5 μL of distilled water. The small RNA TaqMan assay for miR148a and piR014923 were ordered from Thermo Fisher Scientific.17 For the HPV16 assay, primer sequences were adapted from the literature and the probe sequence: 5′-FAM/TGCGTACAAAGCACACACGTAGACATTC/IBFQ-3′.9 The PCR reactions were transferred into sample wells of an eight-channel, disposable droplet generator cartridge (Bio-Rad). An additional 70 μL of droplet generation oil was loaded into the oil well for each channel. After the droplet generation, droplets were gently pipetted to a 96-well PCR plate. The plate was heat-sealed and then placed in a thermal cycler for PCR with the following cycling conditions: 95°C × 10 minutes (1 cycle), 40 cycles of 94°C × 30 seconds and 58°C × 1 minute, and 12°C hold. Finally, the PCR plate was read on the droplet reader, followed by data analysis with QuantaSoft software version 1.7 (Bio-Rad).
Theory and Mechanism
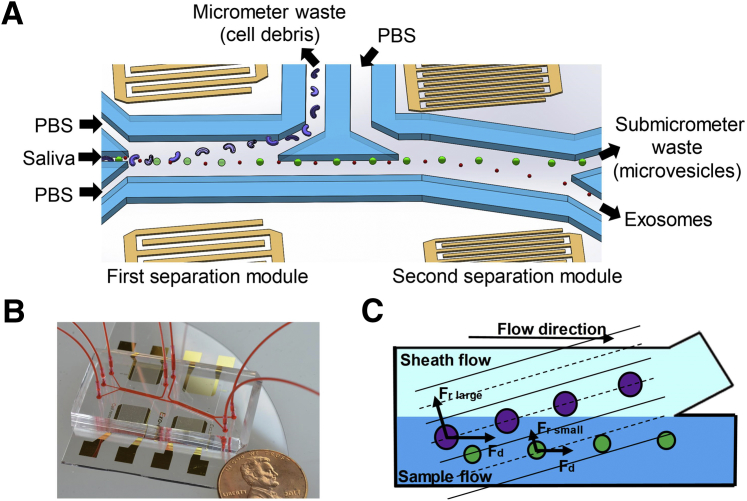
Continuous exosome isolation is achieved by the acoustofluidic device with two separation modules (Figure 1). In each module, the acoustic radiation force Fr pushes particles from areas with high acoustic pressure to pressure nodes that are parallel to the IDT electrodes. Because the acoustic radiation force is proportional to the volume of the particle, larger particles experience a greater acoustic radiation force, which more effectively moves them in the direction of the pressure nodes. Due to the tilted angle between the microchannel and the pressure nodes, larger particles are directed to the waste outlet, whereas smaller particles remain in the original sample flow. Particles larger than 1 μm are removed by the first separation module into the micrometer waste to avoid entering the second separation module. The second module generates a very large acoustic radiation force on micrometer-sized particles and can capture them, thereby blocking the channel. Therefore, it is critical that the first module remove the majority of micrometer-sized particles to allow the device to operate continuously. Particles smaller than 1 μm, but larger than exosomes (30 to 150 nm), will then be removed by the second separation module into the submicrometer waste, leaving only exosomes and other small particles in the final collected samples.
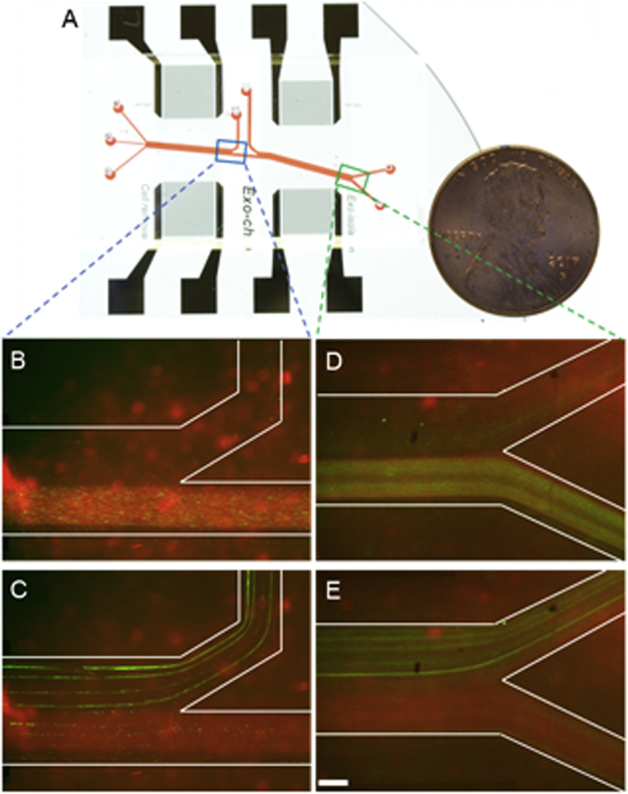
Figure 1.
Schematics and mechanism of the device. A: Schematic of the acoustofluidic device for salivary exosome separation. The device has two modules using 20-MHz and 40-MHz surface acoustic waves (SAWs) for micrometer and submicrometer particle separation. B: An optical image of the integrated acoustofluidic device (penny shown for size comparison). C: Size-based separation occurs in each module. Due to the acoustic radiation force (Fr) induced by a SAW field and a drag force induced by fluid (Fd) large particles are separated into a sheath flow, whereas smaller particles remain in the primary sample flow. PBS, phosphate-buffered saline.
Results
Acoustofluidic Separation in Fluids with Variable Viscosities
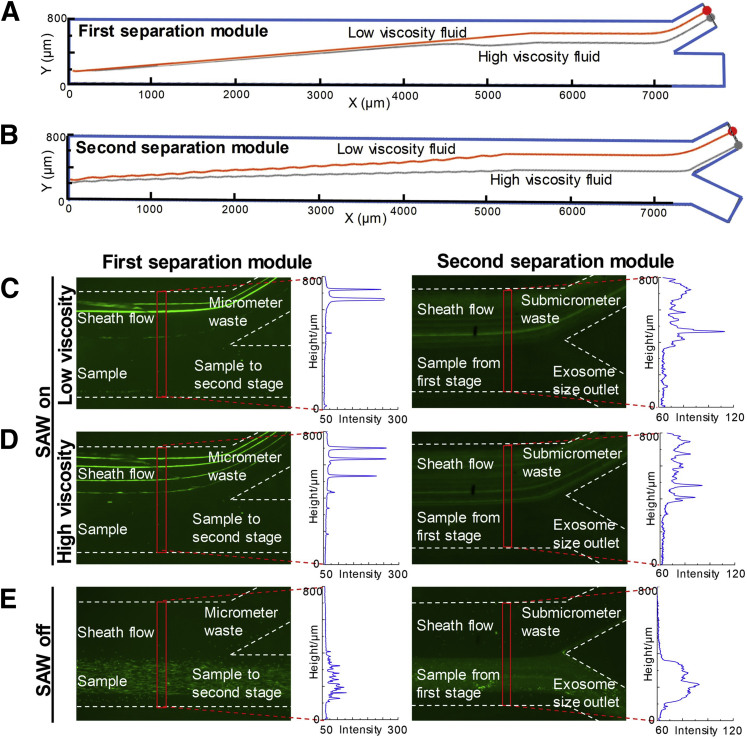
During acoustofluidic separation, the Stokes drag force produces a negative effect on particle isolation, because it can impede the movement of particles along the pressure nodes. Because patient saliva samples can have highly variable viscosities, the influence of fluid viscosity on acoustofluidic system performance was evaluated. Because saliva viscosity was reported to range from 1.10 to 2.30 mPa · s, deionized water (with viscosity of 0.89 mPa · s) and 30% sucrose-water solution (2.65 mPa · s at room temperature) were used to represent different viscosity conditions.18 Simulation and experimental results demonstrate that the acoustofluidic chip's performance was not significantly influenced by changes in viscosity. Although simulations on the trajectories of particles show that higher viscosity fluids make it more difficult to push large particles to the waste outlet, both separation modules still achieved isolation in simulation results (Figure 2, A and B). Experimental results demonstrate the same conclusion. When the SAW was turned on, both separation modules successfully pushed large particles (2-μm or 500-nm polystyrene particles) to the waste outlets in both low- and high-viscosity conditions (Figure 2, C and D), whereas those beads remained in the sample flow when the SAW was turned off (Figure 2E). Particle distributions in the vertical direction of the channel are demonstrated by fluorescent intensities. These results show that particle movement in high-viscosity fluids is lower than in low-viscosity fluids, which indicates that variable viscosity interferes with isolation performance but cannot induce isolation failure. Particles smaller than 150 nm (representing exosomes) cannot be manipulated by either separation modules, meaning these particles will follow the primary streamline and flow to the exosome outlet (Supplemental Figure S1).
Figure 2.
Influence of fluid viscosity on particle separation. A: Simulation of 2-μm particle trajectories in the first separation module with 0.89- and 2.65-mPas viscosity fluids. B: Simulation of 500-nm particle trajectories in the second separation module with 40-MHz surface acoustic waves (SAW) in 0.89- and 2.65-mPas viscosity fluids. C and D: Experimental results for acoustofluidic separation of 2-μm and 500-nm particles in low-viscosity (C) and high-viscosity (D) fluids. Compared with low viscosity, high viscosity causes particles to move closer to the sample outlets. Therefore, under the right experimental conditions, high viscosity interferes with separation but will not cause failure separation of 2-μm or 500-nm particles to the micrometer waste outlet or submicrometer waste outlet when SAW is on. E: When both the separation modules were deactivated, 2-μm or 500-nm particles could not be unseparated out from the sample flow. C–E: Optical images and the results from nanoparticle tracking analysis, which is used to measure particle size distributions.
Salivary Exosome Isolation and Validation
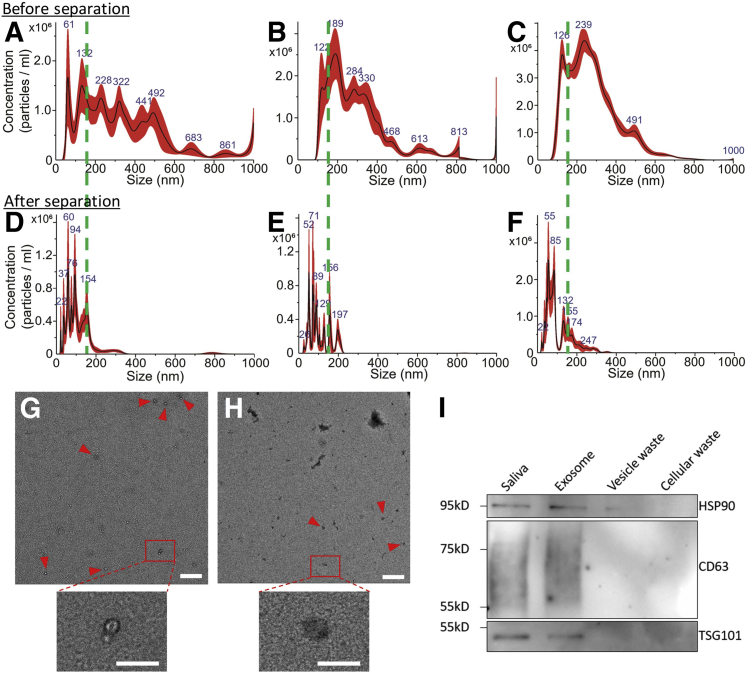
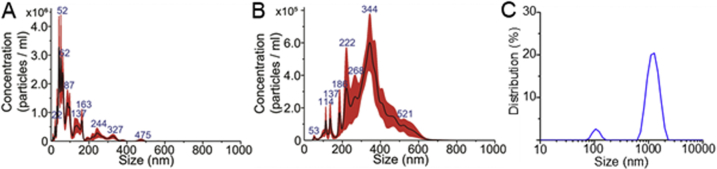
Following simulation and experimental validation using polystyrene particles, acoustofluidic chip performance was validated using saliva samples collected by three different approaches. Because collection methods can potentially influence the composition of saliva samples, centrifugation processed cell-free saliva, saliva collected by SuperSAL saliva collectors, and untreated whole saliva were examined. Nanoparticle tracking analysis was used to measure particle size distributions of different saliva samples after being processed by the acoustofluidic chip. Compared with samples before isolation (Figure 3, A–C) that contained a high ratio of particles larger than 150 nm, the samples processed by the acoustofluidic chip (Figure 3, D–F) demonstrated successful elimination of waste particles larger than 150 nm. Size distributions of products from exosome, submicrometer waste (cell debris), and micrometer waste (microvesicles) outlets showed that each product contained particles of different size ranges (Supplemental Figure S2). Further evaluation of isolated exosome morphology was processed by transmission electron microscopy images. Particles with diameters around 100 nm with cup-like concavity, which is consistent with the established morphology of exosomes, are also shown (Figure 3, G and H); these were isolated from whole saliva samples. Transmission electron microscopy images of isolated exosome samples lacked aggregated exosomes and irregular-shaped components, indicating a high-performance, biocompatible exosome isolation. Exosomal biomarkers CD63, TSG101, and HSP90 were found in the isolated samples but are absent in waste, and the abundance of targeted proteins was similar to proteins from an equal volume of the original sample, indicating that the isolated samples preserved exosomal proteins with a high yield (Figure 3I).
Figure 3.
Characterization of acoustofluidic chip exosome isolation. A–F: Particle size distributions of original samples of cell-free (A), Oasis-collected (B), and whole (C) and acoustofluidic-isolated cell-free (D), Oasis-collected (E), and whole (F) saliva. The acoustofluidic-isolated samples show low concentrations of particles bigger than 200 nm compared with the original samples. Dashed green lines show the 150 nm position on the x axis of the size distribution results. G and H: A transmission electron microscopy image demonstrates an isolated, whole saliva sample that contains fewer contaminants (G) than the original sample (H). Particles with exosomal morphology are tagged by red arrowheads. Boxed areas in top panels are shown at higher magnification in the lower panels. I: Western blot analysis for an exosomal protein biomarker. Only isolated exosome products demonstrate exosomal biomarkers with counts similar to the original sample. Scale bars: 500 nm (G and H, upper panels); 200 nm (G and H, lower panels).
Analysis of Isolation Yield
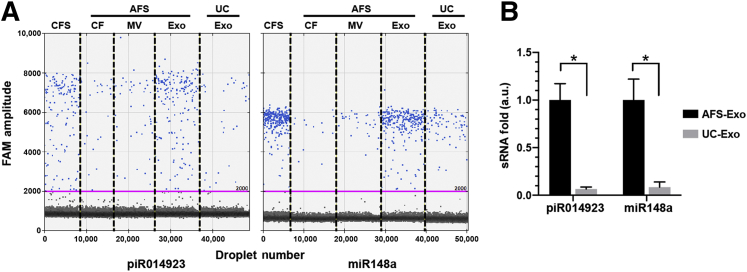
It was further investigated how the yield of exosomes from this acoustofluidic isolation platform compared with the standard method of differential centrifugation. It has been previously demonstrated that the majority of small RNAs including miR148a and piR014923 are packaged in salivary exosomes.17 These two small RNAs were used as markers to compare the yields of exosomes from different methods with identical saliva samples. The RNA was extracted from exosomes isolated from differential centrifugation and different vesicle fractions from the acoustofluidic platform. ddPCR was performed to compare the identical number of miR148a and piR014923. Both miR148 and piR014923 are predominantly located in the exosome fraction compared with other fractions (microvesicles and cell debris fractions) from an acoustofluidic platform (Figure 4A). These results are consistent with previously published results,17 and they demonstrate that the acoustofluidic platform can efficiently separate different salivary extracellular vesicles. Based on the measurements of these two small RNAs, the average yield of exosome from acoustofluidic platform is 15.18 times (9.37- to 18.94-fold, SD ± 4.48) higher than that from a differential centrifugation method (Figure 4B).
Figure 4.
Comparison of yields of exosomes from acoustofluidic separation (AFS) technology and differential centrifugation. A: DNA samples extracted from cell-free saliva (CFS), different acoustofluidic isolated fractions, and exosomes from differential ultracentrifugation (UC) were tested with digital droplet RT-PCR assays for piR014923 and miR148a, which were reported to be predominately located in salivary exosomes. The pink lines show the threshold of positive droplets detection at 2000 amplitude. B: The yield (expressed as a relative fold difference) of individual small RNA in isolated salivary exosomes with acoustofluidic separation (AFS-Exo) technology and differential ultracentrifugation (UC-Exo) method. *P < 0.05 (t-test). a.u., arbitrary units; CF, cell debris fraction; Exo, exosome; MV, microvesicle.
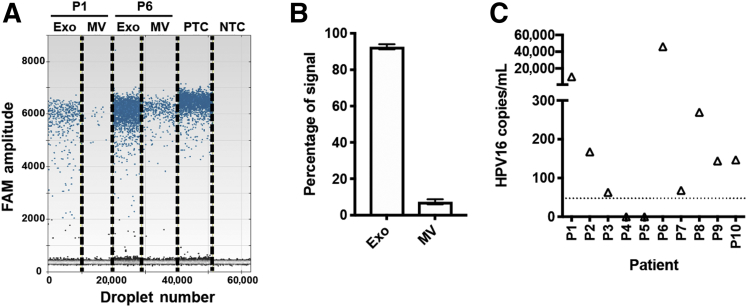
Detection of HPV16 Viral DNA in Exosomes of Saliva from HPV-OPC Patients
To validate whether the acoustofluidic-isolated salivary exosomes are effective for HPV-associated OPC screening and subtype analysis, the detection of HPV16 DNA was explored for early detection/risk assessment of HPV-OPC patients. To determine the distribution of HPV16 DNA in different extracellular vesicles (exosomes, microvesicles), acoustofluidic separation of different vesicles was conducted with HPV-OPC patients' saliva samples, and DNA was extracted from each extracellular vesicle fraction. A ddPCR assay for the detection of HPV16 DNA was designed and tested on DNA extracted from acoustofluidic-isolated vesicles. The HPV16 ddPCR assay showed high reproducibility with relative SD for the copy number of 5.41%. The limit of detection of the assay is 47.8 copies of HPV16 DNA/mL sample based on the SD of the response and the slope in the linear region. The turnaround time of the ddPCR assay is 4 hours.
Ninety-two percent of the total HPV16 signals were concentrated in the exosome fraction, which is roughly 12 times higher than in microvesicles (Figure 5, A and B). HPV16 DNA can be reliably detected in the exosome fractions from HPV-OPC patients. HPV16 DNA was further tested in acoustofluidic-isolated saliva exosomes with a 10 HPV-OPC patient cohort. Eight of 10 patients (80%) tested positive for HPV16 DNA with the HPV16 ddPCR assay (Figure 5C). These data demonstrate that partnering acoustofluidic isolation technology with ddPCR assay can detect HPV16 DNA in 80% of HPV+ OPC patients. Furthermore, the majority of circulating HPV16 DNA is contained in the exosomal fraction.
Figure 5.
Detection of HPV16 DNA in acoustofluidic-isolated salivary exosomes of HPV16-positive oropharyngeal cancer (OPC) patients. A: Distribution of HPV16 DNA in exosomes (Exo) and microvesicles (MV) isolated from saliva of HPV16+ OPC by acoustofluidic technology. Representative results from two HPV16 exosome–positive patients (P1 and P6). Droplet digital PCR (ddPCR) HPV16 DNA detection of different vesicle fractions isolated by acoustofluidic device. B: HPV16 signals in different vesicle fractions. C: Results of HPV16 ddPCR assay for saliva exosomes of HPV-OPC patients. Eight of 10 patients (80%) were positive for HPV16 DNA. Dashed line shows a threshold of detection at 47.8 copies/mL saliva. Exo, exosomes; MV, microvesicles; NTC, negative control; PTC, positive control with synthetic HPV E7 gene.
Discussion
Multiple studies have demonstrated that salivary exosomes are suitable candidates for liquid biopsy.19, 20, 21 Because exosomes are released through the fusion between endosomal membrane compartments and plasma membranes, both extracellular and intracellular biomarkers can reveal the type and status of the originating cell. Tumor cell–derived exosomes contain specific messages that can activate tumor progression and metastasis, and have been investigated in diagnostic and therapeutic fields.22,23 Previous studies have shown that exosome isolation enriches salivary miRNA biomarkers of tumor cells.24 However, the challenge of exosome isolation has been a limiting factor. Saliva is composed of a complex mixture of secretory products from multiple tissues, and salivary components other than exosomes can interfere with the detection of exosomal cancer biomarkers.
The acoustofluidic platform has high tolerance for sample viscosity fluctuations and multiple saliva collection protocols. Differing from blood, which has a relatively stable composition, components in saliva show dramatic differences related to health status.25,26 Composition change leads to dramatic viscosity differences: compared with the viscosity of plasma, saliva viscosity has a much broader range, which can cause unpredictable fluidic drag force interference on acoustofluidic exosome isolation.13,14 For OPC patients, physical and mental stress during salivary collection or radiation-induced xerostomia can alter salivary properties and affect viscosity.27 Successful performance in fluids with different viscosity and reproducible isolation of saliva collected by multiple approaches shows the acoustofluidic platform's high adaptability, which is important for clinical applications.
The acoustofluidic platform performs biocompatible exosome isolation with high purity and yield. Particles in isolated saliva samples contain established exosomal protein biomarkers and match the expected exosomal size and morphology, indicating the isolated samples contain high-purity exosomes. Transmission electron microscopy images showed that isolated particles did not aggregate or burst, indicating that the integrity of isolated exosomes was protected (Figure 3, G and H). Through quantitative ddPCR for miRNAs, the amount of exosomal miRNA in acoustofluidic-isolated samples was determined to be higher than in the sample isolated by differential centrifugation isolation, indicating that acoustofluidic exosome isolation can achieve a higher isolation yield. We suspect that the higher yield is due to the precise and continuous nature of acoustofluidic exosome isolation. By contrast, differential centrifugation involves multiple steps of discarding supernatant, which may lead to inadvertent loss of exosomes. Furthermore, when processing small-volume samples by differential centrifugation, pellets containing exosomes are difficult to visualize and manually recover. Because exosomes in OPC patients' saliva have complex origins, and the sample volume is often low, the reproducible, biocompatible, high-purity and high-yield nature of this acoustofluidic approach will significantly benefit the development of a salivary exosome–based liquid biopsy.
With this platform, HPV16 DNA was successfully detected in salivary exosomes of HPV-OPC patients with 80% concordance with tissues positive for HPV16. Salivary HPV DNA biomarkers could evaluate the risk of HPV-OPC. Studies have also shown that tumor progression is promoted by HPV16 DNA integration, which inactivates p53 and the retinoblastoma tumor suppressor gene.28 One caveat is that in the United States, high-risk HPV is only present in 4% of adults aged 18 to 69 years. Accordingly, the panel of biomarkers screened by a liquid biopsy assay must be expanded.29 Driver mutations such as TP53, PIK3CA, CDKN2A, FBXW7, HRAS, and NRAS should be interrogated alongside key oncogene regions in HPV16 and -18.9 As demonstrated by the increased detectability of HPV16 DNA, it is plausible that these driver mutations are also enriched in the salivary exosome fraction.
Developing a salivary exosome isolation method for liquid biopsy meets the urgent clinical requirement for efficient detection and monitoring approaches for HPV-related OPCs. Head and neck cancer is the sixth most common cancer worldwide, with an annual incidence of around 600,000 cases with poor treatment outcome.30 OPC is a subtype of head and neck cancer that starts inside the part of the throat directly behind the nose. The most common type of OPC is squamous cell cancer of the oropharynx, which is difficult to detect clinically.7,31 Traditional biopsy can provide the histologic definition and show a genetic profile of the cancer, which determines the appropriate therapy and precise prognosis by a single snapshot of the lesion.32 However, current clinical examination approaches, mostly visual examination and palpation, cannot identify premalignant lesions in oral cavities.31 Although conventional clinical examinations are already inefficient, an increasing number of cases of HPV-OPCs, which lack antecedent risk factors for current OPC risk evaluation, bring further challenges for early detection of OPC. Furthermore, continuous monitoring cannot be achieved by traditional biopsy but is necessary for solving progressing tumor heterogeneity, which causes a diverse response of cancer to therapy. Continuous measurements can be made with liquid biopsy, which can screen tumors through genomic and proteomic molecular targets in saliva.33 Avoiding rebiopsy significantly extends liquid biopsy's applicable situations, which makes early detection and monitoring possible.
Acoustofluidics is an efficient, biocompatible platform for sample isolation.34, 35, 36 With a single device, exosome isolation is automated and avoids multistep protocols that require several instruments and trained technicians. Shortened isolation time (10 to 20 minutes of turnaround time) and low sample consumption enhance biosafety and allow high-throughput screening in a large population of patients. A high tolerance to physical property fluctuation of samples further demonstrates the platform's potential in clinical settings. Because isolation derived by low power intensity SAW is potentially gentler than long-term exposure to a high centrifugal force, the platform has the potential to isolate structurally intact and biologically active exosomes. Furthermore, the high purity and yield properties allow the investigation of rare exosomal miRNAs and protein signatures in OPC patients' saliva, which can significantly improve genomic and proteomic profiling efficiency by liquid biopsy.
OPC has an approximate incidence of 115,000 cases per year worldwide and is one of the fastest-rising cancers in Western countries, due to increasing HPV-related incidence.37 With the increasing incidence of HPV-OPC, it is paramount that surveillance methods are developed to improve early detection and outcomes. Considering these facts, the successful detection of HPV16 from salivary exosomes isolated by our acoustofluidic platform offers distinct advantages that will benefit salivary exosome–based liquid biopsy detection of HPV16 including early detection, risk assessment, and screening for HPV-OPC.
Footnotes
Supported by NIH grants UG3 TR002978 (D.T.W.W.), UH3 TR000923 (D.T.W.W.), U01 CA233370 (D.T.W.W.), UH2 CA206126 (D.T.W.W.), R01GM132603 (T.J.H.), R01 HD086325 (T.J.H.), and R21 CA239052 (D.T.W.W. and F.L.); United States Army Medical Research Acquisition Activity W81XWH-18-1-0242 (T.J.H); the Canadian Institute of Health (CIHR) Doctoral Foreign Student Award (J.C.); the Tobacco-Related Disease Research Program (TRDRP) Predoctoral Fellowship (J.C.); and the Queen Elizabeth Hospital Birmingham (QEHB) Charity UK and the Get-A-Head charity UK support the InHANSE (H.M.).
Z.W. and F.L. contributed equally to this work.
Disclosures: T.J.H. has four US patents (patent numbers: 8,573,060; 9,608,547; 9,606,086; and 9,757,699) related to acoustofluidics. T.J.H. also cofounded a start-up company, Ascent Bio-Nano Technologies Inc., to commercialize technologies involving acoustofluidics.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.08.004.
Contributor Information
David T.W. Wong, Email: dtww@ucla.edu.
Tony Jun Huang, Email: tony.huang@duke.edu.
Supplemental Data
Supplemental Figure S1.
Acoustofluidic-based particle isolation using polystyrene particles of different sizes. A: Photo of the acoustofluidic platform. Squares demonstrate areas of microscopy capture. Blue boxed area is shown at higher magnification in B and C; green boxed area is shown at higher magnification in D and E. B: Mixture of 2-μm (green) and 50-nm (red) particles after the first separation module without surface acoustic waves (SAW). Both large and small particles flowed to the second separation module. C: Mixture of 2-μm (green) and 50-nm (red) particles after the first separation module with a SAW field in the channel. Two-micron particles were pushed to the waste outlet, whereas 50-nm particles continuously went to the second separation module. D: Mixture of 500-nm (green) and 50-nm (red) particles after the second separation module without SAW. Both large and small particles flowed to the exosome outlet. E: Mixture of 500-nm (green) and 50-nm (red) particles after the first separation module with SAW field in the channel. The 500-nm particles are pushed to the waste outlet, whereas 50-nm particles continuously go to the exosome outlet. Scale bar = 200 μm.
Supplemental Figure S2.
Size distribution of saliva products from different outlets after acoustofluidic separation. A: Size distribution [obtained via nanoparticle tracking analysis (NTA)] of isolated saliva components from the exosome outlet. Most particles were smaller than 200 nm. B: Size distribution (obtained via NTA) of isolated saliva components from the submicrometer waste outlet. Most particles were between 200 and 600 nm. C: Size distribution (obtained via dynamic light scattering) of isolated saliva components from the micrometer waste outlet. Most particles were about 1 μm.
References
- 1.Wang M.B., Liu I.Y., Gornbein J.A., Nguyen C.T. HPV-positive oropharyngeal carcinoma: a systematic review of treatment and prognosis. Otolaryngol Head Neck Surg. 2015;153:758–769. doi: 10.1177/0194599815592157. [DOI] [PubMed] [Google Scholar]
- 2.Sedghizadeh P.P., Billington W.D., Paxton D., Ebeed R., Mahabady S., Clark G.T., Enciso R. Is p16-positive oropharyngeal squamous cell carcinoma associated with favorable prognosis? A systematic review and meta-analysis. Oral Oncol. 2016;54:15–27. doi: 10.1016/j.oraloncology.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Goy J., Hall S.F., Feldman-Stewart D., Groome P.A. Diagnostic delay and disease stage in head and neck cancer: a systematic review. Laryngoscope. 2009;119:889–898. doi: 10.1002/lary.20185. [DOI] [PubMed] [Google Scholar]
- 4.McGurk M., Scott S. The reality of identifying early oral cancer in the general dental practice. Br Dent J. 2010;208:347–351. doi: 10.1038/sj.bdj.2010.345. [DOI] [PubMed] [Google Scholar]
- 5.Zhan K.Y., Eskander A., Kang S.Y., Old M.O., Ozer E., Agrawal A.A., Carrau R.L., Rocco J.W., Teknos T.N. Appraisal of the AJCC 8th edition pathologic staging modifications for HPV− positive oropharyngeal cancer, a study of the National Cancer Data Base. Oral Oncol. 2017;73:152–159. doi: 10.1016/j.oraloncology.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe A., Taniguchi M., Tsujie H., Hosokawa M., Fujita M., Sasaki S. The value of narrow band imaging endoscope for early head and neck cancers. Otolaryngol Head Neck Surg. 2008;138:446–451. doi: 10.1016/j.otohns.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 7.Brocklehurst P., Kujan O., O'Malley L.A., Ogden G., Shepherd S., Glenny A.M. Screening programmes for the early detection and prevention of oral cancer. Cochrane Database Syst Rev. 2013;2013(11):CD004150. doi: 10.1002/14651858.CD004150.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu S., Arellano M., Boontheung P., Wang J., Zhou H., Jiang J., Elashoff D., Wei R., Loo J.A., Wong D.T. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Springer S., Mulvey C.L., Silliman N., Schaefer J., Sausen M., James N., Rettig E.M., Guo T., Pickering C.R., Bishop J.A., Chung C.H., Califano J.A., Eisele D.W., Fakhry C., Gourin C.G., Ha P.K., Kang H., Kiess A., Koch W.M., Myers J.N., Quon H., Richmon J.D., Sidransky D., Tufano R.P., Westra W.H., Bettegowda C., Diaz L.A., Jr., Papadopoulos N., Kinzler K.W., Vogelstein B., Agrawal N. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 11.Borsetto D., Cheng J., Payne K., Nankivell P., Batis N., Rao K., Bhide S., Li F., Kim Y., Mehanna H., Wong D. Surveillance of HPV-positive head and neck squamous cell carcinoma with circulating and salivary DNA biomarkers. Crit Rev Oncog. 2018;23:235–245. doi: 10.1615/CritRevOncog.2018027689. [DOI] [PubMed] [Google Scholar]
- 12.Taylor D.D., Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015;87:3–10. doi: 10.1016/j.ymeth.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Késmárky G., Kenyeres P., Rábai M., Tóth K. Plasma viscosity: a forgotten variable. Clin Hemorheol Microcirc. 2008;39:243–246. [PubMed] [Google Scholar]
- 14.Christersson C.E., Lindh L., Arnebrant T. Film-forming properties and viscosities of saliva substitutes and human whole saliva. Eur J Oral Sci. 2000;108:418–425. doi: 10.1034/j.1600-0722.2000.108005418.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu M., Ouyang Y., Wang Z., Zhang R., Huang P.-H., Chen C., Li H., Li P., Quinn D., Dao M. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci U S A. 2017;114:10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y.-H., Zhou H., Reiss J.K., Yan X., Zhang L., Chia D., Wong D.T. Direct saliva transcriptome analysis. Clin Chem. 2011;57:1295–1302. doi: 10.1373/clinchem.2010.159210. [DOI] [PubMed] [Google Scholar]
- 17.Bahn J.H., Zhang Q., Li F., Chan T.-M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telis V., Telis-Romero J., Mazzotti H., Gabas A. Viscosity of aqueous carbohydrate solutions at different temperatures and concentrations. Int J Food Prop. 2007;10:185–195. [Google Scholar]
- 19.Azmi A.S., Bao B., Sarkar F.H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camussi G., Deregibus M.C., Bruno S., Cantaluppi V., Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 21.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Mark M.T., Molina H., Kohsaka S., Di Giannatale A., Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahlert C., Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michael A., Bajracharya S.D., Yuen P.S., Zhou H., Star R.A., Illei G.G., Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epstein J.B., Thariat J., Bensadoun R.J., Barasch A., Murphy B.A., Kolnick L., Popplewell L., Maghami E. Oral complications of cancer and cancer therapy: from cancer treatment to survivorship. CA Cancer J Clin. 2012;62:400–422. doi: 10.3322/caac.21157. [DOI] [PubMed] [Google Scholar]
- 26.Dawes C. Salivary flow patterns and the health of hard and soft oral tissues. J Am Dent Assoc. 2008;139 Suppl:18S–24S. doi: 10.14219/jada.archive.2008.0351. [DOI] [PubMed] [Google Scholar]
- 27.Dirix P., Nuyts S., Van den Bogaert W. Radiation-induced xerostomia in patients with head and neck cancer: a literature review. Cancer. 2006;107:2525–2534. doi: 10.1002/cncr.22302. [DOI] [PubMed] [Google Scholar]
- 28.Gasco M., Crook T. The p53 network in head and neck cancer. Oral Oncol. 2003;39:222–231. doi: 10.1016/s1368-8375(02)00163-x. [DOI] [PubMed] [Google Scholar]
- 29.McQuillan G., Kruszon-Moran D., Markowitz L.E., Unger E.R., Paulose-Ram R. Prevalence of HPV in adults aged 18-69: United States, 2011-2014. NCHS Data Brief. 2017;280:1–8. [PubMed] [Google Scholar]
- 30.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 31.Neville B.W., Day T.A. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52:195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 32.Lingen M.W., Kalmar J.R., Karrison T., Speight P.M. Critical evaluation of diagnostic aids for the detection of oral cancer. Oral Oncol. 2008;44:10–22. doi: 10.1016/j.oraloncology.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardelli A., Pantel K. Liquid biopsies, what we do not know (yet) Cancer Cell. 2017;31:172–179. doi: 10.1016/j.ccell.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Wu M., Huang P.H., Zhang R., Mao Z., Chen C., Kemeny G., Li P., Lee A.V., Gyanchandani R., Armstrong A.J. Circulating tumor cell phenotyping via high-throughput acoustic separation. Small. 2018;14:e1801131. doi: 10.1002/smll.201801131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren L., Yang S., Zhang P., Qu Z., Mao Z., Huang P.H., Chen Y., Wu M., Wang L., Li P. Standing surface acoustic wave (SSAW)-based fluorescence-activated cell sorter. Small. 2018;14:1801996. doi: 10.1002/smll.201801996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding X., Peng Z., Lin S.-C.S., Geri M., Li S., Li P., Chen Y., Dao M., Suresh S., Huang T.J. Cell separation using tilted-angle standing surface acoustic waves. Proc Natl Acad Sci U S A. 2014;111:12992–12997. doi: 10.1073/pnas.1413325111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaturvedi A.K., Engels E.A., Pfeiffer R.M., Hernandez B.Y., Xiao W., Kim E., Jiang B., Goodman M.T., Sibug-Saber M., Cozen W. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]