Abstract
The objective of the present study was to investigate the mechanism whereby long-chain non-coding RNA (LncRNA) antisense non-coding RNA (ANRIL) in the INK4 locus promotes angiogenesis and thrombosis by the miR-99a and miR-449a interventional autophagy pathway. The expression of LncRNA ANRIL, autophagy-related gene beclin1, and miR-99a and miR-449a in human umbilical vein endothelial cells (HUVECs) was determined by qRT-PCR. Thrombomodulin expression was examined by Western blotting assays. The levels of autophagy-related factors were determined by ELISA. CCK-8 assays were used to assess cell viabilities. Apoptosis was detected by flow cytometry via annexin V-FITC/propidium iodide double labeling and TUNEL assays. The interaction between ANRIL, miR-99a and miR-449a was studied using luciferase reporter assays. The role of ANRIL in autophagy was assessed in rats. Our data revealed that ANRIL and beclin-1 were highly expressed, while miR-99a and miR-449a were down-regulated in HUVECs serum of the autophagy model. Luciferase reporter assays, in vitro rescue assays, and Matrigel assays demonstrated that ANRIL increased beclin-1 expression via miR-99a and miR-449a sponges to upregulate thrombomodulin and promote angiogenesis. In addition, in vivo experiments confirmed that knockdown of ANRIL reduced thrombosis in rats. In conclusion, ANRIL promotes angiogenesis and thrombosis by upregulating the expression of miR-99a and miR-449a during autophagy.
Keywords: LncRNA ANRIL, autophagy, angiogenesis, venous thrombosis
Introduction
Prevention and treatment of blood clots can reduce the risk of stroke, heart attack and pulmonary embolism. There is strong evidence that miRNAs play an important role in cellular physiology and pathology, and are involved in the regulation of gene expression in major diseases such as cardiovascular disease, metabolic syndrome and pulmonary embolism [1]. MiR-99a and miR-449a belong to the miR-99 and miR-449 families, respectively. In the human genome, miR-99 is located on human chromosome 9 q22.32 and belongs to the intron sequence. The miR-449 family is located on human chromosome 19 p13.13 and belongs to the intergenic sequence. MiR-99 and miR-449 are widely present in a variety of mammalian tissues, and are especially highly expressed in cardiovascular tissues. Studies have found that when vascular endothelial cells undergo oxidative stress, the expression level of miR-24 is significantly upregulated [2]. It is suggested that miR-99 and miR-449 are closely related to the functions of vascular endothelial cells.
Atherosclerosis is a vascular arteriosclerotic disease. Chronic ischemic heart disease is often caused by atherosclerosis, especially in the coronary arteries, which causes myocardial undernutrition over time. This disease is characterized by formation of plaques in the inner walls of blood vessels. Those plaques are composed of cholesterol, lipids, cells such as monocytes/macrophages, and calcium. Once these plaques fall off, they will block the flow of blood. Atherosclerosis leads to reduced blood flow and ischemia, and is often the cause of heart attacks, strokes and gangrene. Menghini et al. showed that miR-99a can regulate ox-LDL-induced autophagy in human endothelial cells by directly inhibiting beclin-1. They also found that ATG5 levels were also reduced, but luciferase assays did not confirm that ATG5 was directly targeted by miR-99a. Overexpression of miR-99a inhibits autophagy, leading to accumulation of oxLDL and increased monocyte adhesion which suggests a potential role for plaque formation in early atherosclerosis. In addition, downregulation of miR-99a induces autophagy in human endothelial cells and provides protection against ox-LDL.
Atherosclerosis is closely related to cerebrovascular accidents such as stroke. Wang et al. have found that inhibition of miR-30a attenuates death/injury associated with cerebral ischemia and protects mice from neurological deficits. The other two miRNAs, miR-449a and miR-352, play important roles in cerebral ischemia by regulating autophagy. Both miRNAs are reportedly downregulated after ischemia, and are capable of inhibiting lysosomal-associated membrane proteins (LAMP). Tao et al. showed that upregulation of miR-449a reduced the number of lysosomes and autophagosomes and increased the number of autophagic vacuoles, indicating that miR-449a can prevent autophagosome maturation and degradation. MiR-449a mimics reduce infarct size in ischemic injury, suggesting that miR-449a prevents autophagic cell death.
Autophagy is a process in which lysosomes degrade and transform their own damaged proteins and aging organelles into energy and raw materials to maintain the metabolic balance of the cells. Autophagy of endothelial cells is highly correlated with the physiological and pathological processes of vascular diseases. Studies have found that a large number of autophagosomes are found and beclin-1 expression is significantly enhanced in the atherosclerotic plaque [3].
The antisense non-coding RNA (ANRIL) in the INK4 locus is a 3.8 kb long non-coding RNA (lncRNA) that is widely expressed in mammalian tissues or organs such as the lungs and the liver [4,5]. Previous preliminary studies have found that lncRNA ANRIL is highly expressed in a human umbilical vein endothelial cell (HUVEC) autophagy model and participates in the expression of thrombomodulin (TM). It is unclear whether lncRNA ANRIL regulates the expression of TM by regulating deubiquitination. Based on bioinformatics analysis, we found that lncRNA ANRIL shares miRNA response elements with TM in miR-99a and miR-449a. An abnormal decrease in renal miR-99a and miR-449a was observed in rapamycin-induced autophagy mice [6]. However, little is known about the expression and role of miR-99a and miR-449a in autophagy models.
Therefore, we hypothesized that lncRNA ANRIL acts as a miRNA sponge in the autophagy pathway, regulates angiogenesis via cavernous miR-99a and miR-449a, and upregulates TM expression to promote thrombosis.
Materials and methods
Samples
A total of 25 patients with thrombosis and 25 healthy volunteers were included in the study. Fasting blood samples were collected from all participants and then centrifuged at 3,000 g for 10 minutes at 4°C. Serum samples were then collected and examined for expression levels of ANRIL, miR-99, miR-449, TM, beclin-1, IL-18, TNF-α and IL-6 using qRT-PCR analysis and ELISA.
The study was approved by the ethics committee of the hospital and informed consent was obtained from all participants.
Methods
Cells and rapamycin-induced autophagy model
HUVECs, purchased from the European Collection of Authenticated Cell Cultures (ECACC, Porton Down, UK), were cultured in Dulbecco’s modified eagle medium/Ham’s F12 (DMEM/F12; Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and 100 mg/mL penicillin/streptomycin (Life Technologies) in a humidified atmosphere containing 5% CO2 at 37°C. HUVECs were treated with 1 mM rapamycin and after 24 hours, the expression levels of ANRIL, miR-99, miR-449, TM, beclin-1, IL-18, TNF-α and IL-6.pcDNA3 were determined. 1-ANRIL (ANRIL), pcDNA3.1 empty vector (Vector), small interfering RNA (siRNA) targeting ANRIL (si-ANRIL), disrupting negative control siRNA (si-Ctrl), miR-99 and miR-449 simulation, simulated competition for negative control (simulated NC), miR-99 and miR-449 inhibitors, inhibitor competing for negative control (inhibitor NC), siRNA beclin-1 (si-beclin-1), and competing for negative control siRNA (si-Ctrl) were purchased from GenePharma Co., Ltd. HUVECs were transfected prior to rapamycin treatment using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions and further studies were performed 48 hours after transfection.
Animal experiments
All animal experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee of Wuhan University People’s Hospital. Male Sprague-Dawley (SD) rats, weighing 160-180 g each, were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and individually housed in an air-conditioned room at 22°C ± 2°C with a 12 hour light/dark cycle and a regular rodent diet or a high adenine diet. All rats were randomly divided into 3 groups (6 in each group), namely, the control group, the autophagy model group, and the si-ANRIL injection group. The model rats returned to the normal diet 20 days after the high adenine diet; the rats in the si-ANRIL group were intraperitoneally injected with si-ANRIL once weekly for 4 weeks. All rats were sacrificed 4 weeks later and lumen formation of HUVECs in each group was examined by Matrigel assays.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from serum and HUVECs using Trizol reagent (Life Technologies) and reverse transcribed into cDNA using the PrimeScript RT kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. Relative expression levels of ANRIL, miR-99, miR-449 and TM, and beclin-1 were determined using an SYBR Premix Ex Taq (TaKaRa) on an ABI 7500 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA).
Western blotting assays
To analyze TM and beclin-1 expression in HUVECs and kidney tissues, we extracted total cellular protein using radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China) and the lysate was clarified by centrifugation at 14,000 g for 15 minutes at 4°C. The proteins in the lysate were resolved on 10% SDS-PAGE, transferred to a PVDF membrane and blocked with Tris buffered saline (TBST) containing 5% skim milk for 2 hours at room temperature. The blot was probed overnight at 4°C with primary antibodies against TM (1:1000; Abcam, Cambridge, UK) and beclin1 (1:1000; Cell Signaling Technology, Boston, MA, USA), with horseradish peroxide Enzyme incubation (HRP)-conjugated goat anti-rabbit IgG (1:1000; Cell Signaling Technology) for 1 hour at room temperature. Protein bands were visualized using ECL (Thermo Scientific, Shanghai, China) in a Molecular Imager Chem-iDoc XRS system (Bio-Rad Laboratories, Hercules, CA, USA). β-actin was used as an internal reference.
Annexin V-FITC/propidium iodide double labeling flow cytometry
Apoptosis was analyzed by flow cytometry using the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences; San Jose, CA, USA) according to the manufacturer’s protocol. HUVECs after transfection and rapamycin treatment were harvested, washed with cold PBS, and stained with binding buffer containing annexin V-FITC and propidium iodide (PI) for 15 minutes at 4°C in the dark. Finally, cells were sorted by flow cytometry (Beckman Coulter, Fullerton, CA, USA).
ELISA
The levels of IL-1b, IL-8, TNF-α and IL-6 in the serum or cell culture supernatant were measured with appropriate ELISA kits (R & D Systems, Minneapolis, MN, USA). Optical density (OD) was read at 540 mm according to the manufacturer’s instruction.
Histopathological examination
Formalin-fixed and paraffin-embedded renal tissue sections (5 mm) were stained with hematoxylin and eosin (H&E) and Mason trichrome staining (Sigma-Aldrich, St. Louis, MO, USA) by routine procedure. Histopathological changes of blood vessels were assessed by staining.
TUNEL assays
Paraffin embedded renal tissue sections were incubated with recombinant terminal deoxynucleotidyltransferase (rTdT) solution for 1 hour at 37°C in the dark and stained with 3,30-diaminobenzidine (DAB; Roche, Shanghai, China). TUNEL-stained cells were observed under a fluorescence microscope (Olympus). The apoptotic index was calculated as the percentage of TUNEL positive cells divided by the total number of cells.
Statistical analysis
Data were expressed as mean ± standard deviation (SD) and analyzed by SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). Significant differences were analyzed using t-test or analysis of variance (ANOVA), and P values less than 0.05 were considered to be statistically significant.
Results
Serum ANRIL and TM overexpression in patients with thrombosis
In order to determine if ANRIL and TM proteins were altered in the thrombus, we first tested the serum levels of ANRIL and TM in thrombotic patients and age-matched healthy subjects. QRT-PCR analysis showed that serum ANRIL and TM contents were elevated in patients with thrombosis compared with the normal control group (Figure 1A). In addition, in order to confirm the release of pro-inflammatory cytokines in the thrombus, serum IL-1b, IL-18, TNF-α and IL-6 levels were measured by ELISA. The results showed that serum IL-1b, IL-18, TNF-α and IL-6 levels were significantly higher in thrombosis patients than the normal group (Figure 1B), suggesting ANRIL-mediated thrombotic response.
Figure 1.
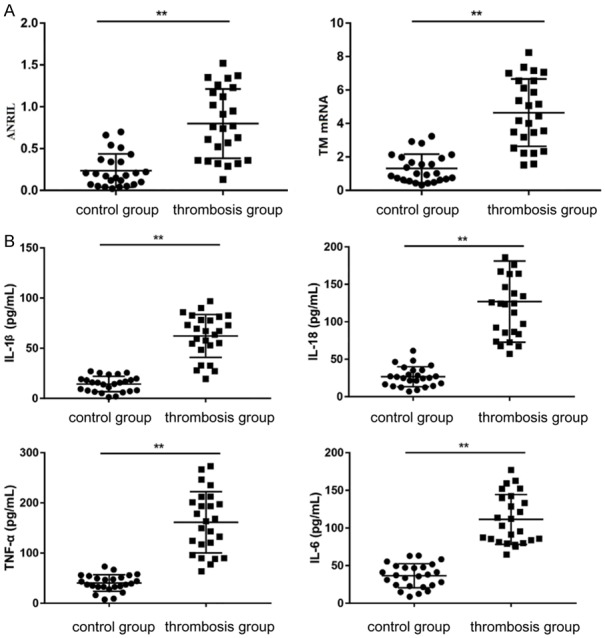
Expression of ANRIL, TM and inflammatory cytokines in UAN patients. Note: *P < 0.05 compared with the normal group, **P < 0.01 compared with the normal group.
ANRIL promotes TM activation in HUVECs
Rapamycin increased the expression levels of ANRIL and TM (Figure 2A, 2B). TM levels (Figure 2C) in HUVECs transfected with the ANRIL expression vector were significantly upregulated after 24 hours of rapamycin treatment. On the contrary, siRNAs silenced ANRIL and inhibited TM expression. These data indicated that ANRIL triggered downregulation of TM in HUVECs.
Figure 2.
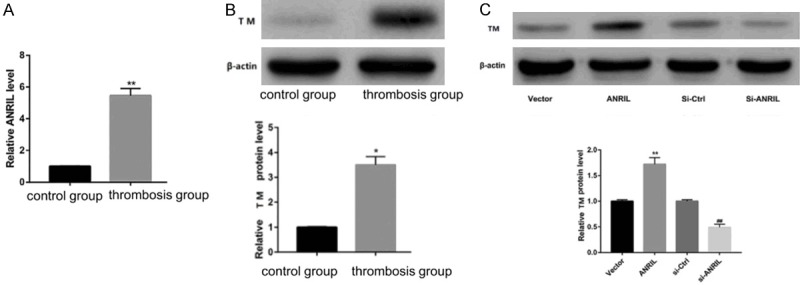
Effect of ANRIL on the expression levels of TM in vitro. Note: *P < 0.05 compared with the normal group, **P < 0.01 compared with the normal group.
ANRIL acts as a miRNA sponge, up-regulating the expression of beclin-1 via miR-99a and miR-449a sponge
QRT-PCR and Western blot analysis further showed that expression of miR-99a and miR-449a was decreased, and beclin-1 expression was elevated in the serum of thrombosis patients (Figure 3A). The results of rapamycin treated HUVECs are consistent with serum results (Figure 3B). A large body of evidence suggested that lncRNAs may act as competitive endogenous RNAs (ceRNAs), possibly downregulating miRNA expression, thereby regulating miRNA target inhibition at the posttranscriptional level [7]. Therefore, this study explored whether ANRIL had a similar mechanism in HUVECs. Through bioinformatics analysis, it was found that ANRIL and beclin-1 shared the regulatory sites of miR-99a and miR-449a. ANRIL expression and mRNA and protein expression levels of beclin-1 in HUVECs transfected with miR-99a and miR-449a mimics, miR-99a and miR-449a inhibitors or respective controls were assessed by qRT-PCR and Western blotting assays. As shown in Figure 3C and 3D, miR-99a and miR-449a overexpression inhibited ANRIL and beclin-1 expression levels, while miR-99a and miR-449a silencing significantly promoted ANRIL and beclin-1 expression. In conclusion, ANRIL positively regulated beclin-1 by ejecting miR-99a and miR-449a in HUVECs.
Figure 3.
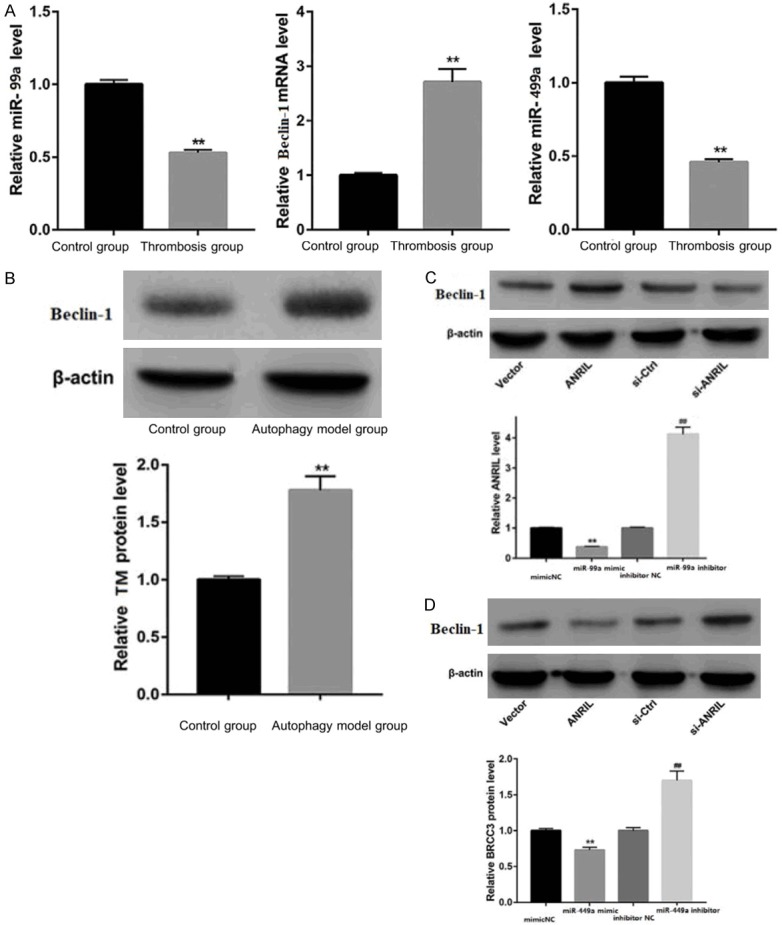
Interaction between ANRIL, miR-99a, miR-449a and beclin-1. Note: **P < 0.01 compared to the normal group.
ANRIL-induced upregulation of TM is mediated through miR-99a and miR-449a
In order to further understand the effect of ANRIL on the expression and function of miR-99a and miR-449a, HUVECs were co-transfected with miR-99a and miR-449a or mock NC and pcDNA-ANRIL vectors prior to rapamycin treatment. The results showed that overexpression of miR-99a and miR-449a significantly reduced TM levels. In contrast, ANRIL overexpression significantly increased TM levels (Figure 4).
Figure 4.
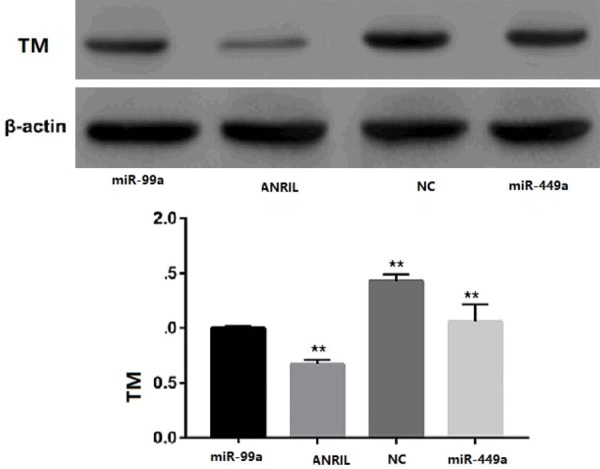
Effect of ANRIL on the expression and function of miR-99a and miR-449a.
Silenced ANRIL improves blood vessel formation in a rat model of thrombosis
In order to confirm the effect of ANRIL on angiogenesis in autophagy rats, SD rats were randomly divided into the control group, the autophagy model group and the si-ANRIL injection group. Matrigel assays were used to detect lumen formation of HUVECs in each group. The results showed that compared with the blank control group, the number of lumen formation in the silent ANRIL group was relatively small, and most of the lumens were smaller than the control group. However, HUVECs in the autophagy model group had a tendencyto aggregate, but did not form a distinct lumen-like network (Figure 5).
Figure 5.
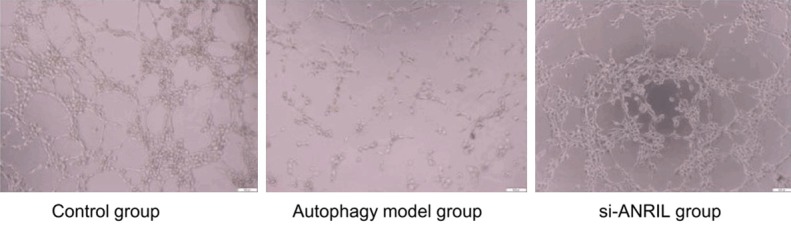
Formation of blood vessels in each group.
Discussion
LncRNA ANRIL can regulate mir-99a and mir-449a. The regulatory pathways of IncRNA for miRNA can be divided into 6 aspects: 1) lncRNA can degrade lncRNA by interacting with specific miRNA; 2) lncRNA can be used as a lure for miRNA to inhibit binding of miRNA to target gene mRNA; 3) lncRNA can play the role of endogenous miRNA sponge and inhibit the expression of miRNA; 4) lncRNA indirectly inhibits the negative regulation of target gene mRNA by competing with miRNA to bind to the 3’UTR of target gene mRNA; 5) lncRNA can produce miRNA and regulate the target gene mr-na; 6) lncRNA can also serve as the competing binding site of the same miRNA by ceRNA and other RNA transcripts, so as to degrade target gene mRNA. Among them, the most important is the ceRNA pathway, that is, lncRNA can act as ceRNA to competitively bind to miRNA regulating gene with the same MRE through miRNA response element (MRE), which affects cellular function.
Our current work revealed that lncRNA ANRIL promoted the activation of TM by increasing miR-99a and miR-449a, thus providing new insights into the pivotal role of lncRNA ANRIL in the pathogenesis of thrombosis. LncRNA is defined as a novel RNA transcript of more than 200 nucleotides with narrow protein-encoding functions and is involved in regulating cellular processes such as cell growth, metabolism, differentiation and apoptosis [8]. Recent studies have revealed an important role of lncRNA in vascular disease and may open up opportunities for the development of new therapeutic targets. For example, a recent report by Hu et al. provided evidence of the pathogenic role of metastasis-associated transcript 1 (MALAT1) in patients with pulmonary embolism [9-12]. Dun and colleagues found that taurine up-regulated gene 1 (TUG1) acts as a mediator of extracellular matrix accumulation in DN progression via the miR-377/PPARg axis [13-16]. In our study, we demonstrated that lncRNA ANRIL was highly expressed in thrombosis patient sera and human renal proximal tubular epithelial cells treated with rapamycin. In addition, lncRNA ANRIL promoted the activation of beclin-1. A broader understanding of the lncRNA mechanism may open a new way to study the mechanism whereby autophagy pathways induce thrombosis.
LncRNAs may act as ceRNAs, and regulate the distribution of miRNAs on their targets. LncRNA ANRIL has been reported to regulate tumor proliferation, migration and invasion by miR-99a and miR-449a as ceRNAs in oral cancer [17-20]. Therefore, in this study, we investigated whether lncRNA ANRIL acted as a ceRNA in thrombus progression. By using bioinformatics software, we found that lncRNA ANRIL contained several target binding sites for miR-99a and miR-449a. Luciferase activity assays confirmed the binding relationship between lncRNA ANRIL, miR-99a and miR-449a. Based on the data, it was concluded that ANRIL acted as a miR-99a and miR-449a ceRNA in the thrombus. Overexpression of miR-99a and miR-449a inhibited ANRIL and beclin-1 expression levels, while miR-99a and miR-449a silencing significantly promoted ANRIL and TM expression. In conclusion, ANRIL positively regulated beclin-1 by ejecting miR-99a and miR-449a in HUVECs.
In the present study, TM was shown to be a direct target of miR-99a and miR-449a in thrombus. Further exploration indicated that lncRNA ANRIL acted as an endogenous miR-99a and miR-449a sponge that upregulated TM expression. The number of lumen formation in the silent ANRIL group was relatively small, and most of the lumens were smaller than those in the control group. However, HUVECs in the autophagy model group had a tendency to become aggregated, but did not form a distinct lumen-like network. The effect of lncRNA ANRIL on inhibition of angiogenesis and promotion of thrombosis was confirmed.
Disclosure of conflict of interest
None.
References
- 1.Cai B, Pan Z, Lu Y. The roles of microRNAs in heart diseases: a novel important regulator. Curr Med Chem. 2010;17:407–411. doi: 10.2174/092986710790226129. [DOI] [PubMed] [Google Scholar]
- 2.Lorenzen JM, Kaucsar T, Schauerte C, Schmitt R, Rong S, Hubner A, Scherf K, Fiedler J, Martino F, Kumarswamy R, Kolling M, Sorensen I, Hinz H, Heineke J, van Rooij E, Haller H, Thum T. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol. 2014;25:2717–2729. doi: 10.1681/ASN.2013121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinet W, De Bie M, Schrijvers DM, De Meyer GR, Herman AG, Kockx MM. 7-ketocholesterol induces protein ubiquitination, myelin figure formation, and light chain 3 processing in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2004;24:2296–2301. doi: 10.1161/01.ATV.0000146266.65820.a1. [DOI] [PubMed] [Google Scholar]
- 4.Kang YH, Kim D, Jin EJ. Down-regulation of phospholipase D stimulates death of lung cancer cells involving up-regulation of the long ncRNA ANRIL. Anticancer Res. 2015;35:2795–2803. [PubMed] [Google Scholar]
- 5.Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, Yin L, Zhang EB, De W, Shu YQ. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell proliferation by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8:57. doi: 10.1186/s13045-015-0153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CG, Kim JG, Kim HJ, Kwon HK, Cho IJ, Choi DW, Lee WH, Kim WD, Hwang SJ, Choi S, Kim SG. Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int. 2014;86:943–953. doi: 10.1038/ki.2014.117. [DOI] [PubMed] [Google Scholar]
- 7.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Zhao JC. Functional analysis of long noncoding RNAs in development and disease. Adv Exp Med Biol. 2014;825:129–158. doi: 10.1007/978-1-4939-1221-6_4. [DOI] [PubMed] [Google Scholar]
- 9.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, Chen L, Lv Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J Cell Mol Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ, Yu DM. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARgamma in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:598–604. doi: 10.1016/j.bbrc.2017.01.145. [DOI] [PubMed] [Google Scholar]
- 11.Chai L, Yuan Y, Chen C, Zhou J, Wu Y. The role of long non-coding RNA ANRIL in the carcinogenesis of oral cancer by targeting miR-125a. Biomed Pharmacother. 2018;103:38–45. doi: 10.1016/j.biopha.2018.01.105. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 13.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 15.Abderrazak A, Syrovets T, Couchie D, El Hadri K, Friguet B, Simmet T, Rouis M. NLRP3 inflammasome: from a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biol. 2015;4:296–307. doi: 10.1016/j.redox.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero CA, Remor A, Latini A, De Paul AL, Torres AI, Mukdsi JH. Uric acid activates NRLP3 inflammasome in an in-vivo model of epithelial to mesenchymal transition in the kidney. J Mol Histol. 2017;48:209–218. doi: 10.1007/s10735-017-9720-9. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, Zhou R. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160:62–73. doi: 10.1016/j.cell.2014.11.047. [DOI] [PubMed] [Google Scholar]
- 18.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, Han L, Jiang G, Zhang L, Gao C, Zhao W. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:13727. doi: 10.1038/ncomms13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphries F, Bergin R, Jackson R, Delagic N, Wang B, Yang S, Dubois AV, Ingram RJ, Moynagh PN. The E3 ubiquitin ligase Pellino2 mediates priming of the NLRP3 inflammasome. Nat Commun. 2018;9:1560. doi: 10.1038/s41467-018-03669-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog. 2010;6:e1000967. doi: 10.1371/journal.ppat.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]


