Abstract
Cyclin-dependent kinase 1 (CDK1) has a unique role in cell cycle regulation, as it is crucial for cell cycle progression and cell division. The aim of the present study was to use a combination of various detection methods to examine the expression and clinical significance of CDK1 in thyroid cancer (THCA). We used in-house tissue microarrays, immunohistochemistry, public RNA-sequencing, gene microarrays, and meta-analyses to conduct a comprehensive analysis of the role of CDK1 in the occurrence and development of THCA. CDK1 protein expression was notably higher in THCA tissues than in non-cancer tissues as evidenced by the in-house tissue microarrays. The expression of CDK1 protein was also significantly higher in pathologic T3-T4 than in T1-T2 samples. The pooled standardized mean difference (SMD) for CDK1 was 0.71 (95% CI, 0.46-0.95) including a total of 931 THCA and 585 non-cancerous thyroid tissue samples. An aggregation of the immunohistochemistry results and the RNA-sequencing/microarray findings gave a pooled SMD for CDK1 expression of 2.13 (95% CI, 1.30-2.96). The final area under curve (AUC) for the summarized receiver operating characteristic (sROC) was 0.7941 using all 1102 cases of THCA and 672 cases of controls. KEGG analysis with the co-expressed genes of CDK1 in THCA demonstrated the top enriched pathways to be the cell cycle, thyroid hormone synthesis, autoimmune thyroid disease, etc. In summary, we reveal the overexpression of CDK1 in THCA based on multiple detection methods that combine independent cohorts. However, further studies are required to elucidate the molecular mechanisms of CDK1 that promotes the biological aggressiveness of THCA cells.
Keywords: Cyclin-dependent kinase 1, thyroid cancer, tissue microarray, immunohistochemistry, RNA-sequencing, gene microarray
Introduction
Regulation of the cell division cycle is obligatory in proliferating cells; therefore, cancer cell malignancy is inextricably linked to changes in cell cycle regulation. Cell cycle progression is determined by cyclin-dependent kinases (CDKs), and the activity of CDKs is precisely controlled by their cooperation with cyclins. CDKs belong to the family of serine/threonine kinases that comprise catalytic kinase subunits. Numerous cyclins in human cells are expressed and degraded in a manner that is stringently coordinated with cell cycle progression [1]. Human cells have more than 13 diverse CDKs and more than 25 cyclins; these can form multiple CDK-cyclin complexes at distinct stages of the cell cycle to exert specific effects on cell cycle progression [2].
Among all the CDK family members, only CDK1, 2, 4, and 6 are directly involved in cell cycle modification. CDK2, 4, and 6 are not critical for the cell cycle, whereas CDK1 has a unique role in cell cycle regulation, as it is crucial for cell cycle progression and cell division [3]. CDK1 regulates the G2 phase as part of a complex with cyclin A and participates in G2/M conversion by forming a complex with cyclin B. Silencing of CDK1 incr eases G2/M arrest in the cell cycle and can lead to the production of polyploid cells [4]. CDK1 is an important modulator of various mitotic processes, including cytoskeletal reorganization, chromosome segregation, and daughter cell formation and isolation. Deregulation of CDK1 activity leads to serious defects in these procedures [5-7].
CDK1 expression is upregulated in several malignancies. The upregulation of CDK1 expression or its activation has been documented in glioma [8], oral squamous cell carcinoma [9], ovarian cancer [10], colorectal cancer [11] and prostate cancer [12]. Therefore, CDK1 expression is remarkably correlated with the occurrence and progression of cancers, making this kinase a prospective target for targeted cancer therapy aimed at preventing the progression of the cell cycle and inducing apoptosis in various cancers. Several CDK inhibitors have been developed, and their effects have also been tested [13,14]. However, the expression level of CDK1 in thyroid cancer (THCA) and the function and potential of CDK1 for targeted therapy in THCA have not been resolved.
By far, the clinical value of CDK1 expression in THCA remains largely unclarified. Only two groups have reported findings, but these are inconsistent and based on small numbers of clinical samples. Ito et al. were the first and the only group to determine the protein level of CDK1 in THCA using immunohistochemistry [15]. That group reported CDK1 upregulation in six (17.1%) of 35 follicular thyroid cancer (FTC) samples, in five (17.2%) of 29 papillary thyroid cancer (PTC) samples, and in 16 (76.2%) of 21 undifferentiated anaplastic thyroid cancer (ATC) samples. The CDK1 upregulation was also closely correlated to cancer differentiation. Fluge et al. [16] used a cDNA microarray to determine the gene expression profile in 10 differentiated (classic) PTCs and 6 aggressive PTCs. They found a 1.3-fold change in CDK1 mRNA in classic PTC compared to controls and one of the 10 cases had a greater than 4-fold change. A 5.9-fold change in CDK1 was detected in the aggressive PTCs, with 4 of 6 cases showing a greater than 4-fold change. Later, three groups re-analyzed the limited number of microarrays and concluded that CDK1 was differentially expressed in ATCs [17-19]; however, the actual expression level of CDK1 was not calculated.
More suitable high-throughput data on other subtypes of THCA are now available for data mining. In the past 30 years, thyroid cancer has become one of the most common malignant tumors in endocrine-related systemic tumors, and its incidence is still increasing. The histopathological types of THCA include PTC, FTC, medullary thyroid cancer (MTC), and ATC. Of these various subtypes, PTC is the most widespread. PTC has a five-year survival that exceeds 95%, but some PTCs have a high risk of recurrence. The other types of THCA have a poorer prognosis.
The molecular mechanisms that regulate the development and progression of THCA have not yet been elucidated. In view of the status and potential therapeutic effects of CDK1 in other malignant tumors, the aim of the present study was to use a combination of various detection methods to examine the expression and clinical significance of CDK1 in THCA. We used tissue microarray, immunohistochemistry, RNA-sequencing (RNA-seq), gene microarray, and meta-analyses to conduct a comprehensive analysis of the role of CDK1 in the occurrence and development of THCA. In addition, signaling pathways related to CDK1 were studied to reveal the underlying molecular mechanism by which CDK1 may function in THCA, with the goal of providing a new perspective for clinical THCA screening and exploitation of new targeted drugs.
Materials and methods
Integrated assessment of CDK1 expression in THCA
In-house immunohistochemical detection of CDK1 protein
Two tissue microarrays (THC961 and THC1021) were obtained from Fanpu Biotech, Inc. (Guilin, China), these contained 23 samples of non-cancerous thyroid tissue and 125 samples of THCA (including PTC, FTC, MTC, and ATC) tissues. Another 64 non-cancerous thyroid tissues and 46 THCA cases were collected from the Department of Pathology, First Affiliated Hospital of Guangxi Medical University, from March 1 to December 1, 2018. The study was approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University. In total, this immunohistochemistry (IHC) study contained 87 non-cancer controls and 171 THCA cases. All patients provided written informed consent for use of their samples in the study. Two pathologists (Wei-Jia Mo and Gang Chen) independently assessed all slides without prior knowledge of the clinical outcome using a semi-quantitative scoring system to classify the staining intensity and the percentage of positive tumor cells. Staining intensity was scored as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). Staining proportion was scored as 0 (<10%), 1 (11-25%), 2 (26-50%), 3 (51-75%), or 4 (76-100%). The final immunoreactivity scores were determined by combining the intensity and proportion scores [20,21]. Enumeration data acquired from IHC detection were used to evaluate the relation between CDK1 protein expression and THCA by comparison to adjacent non-cancerous tissues. In addition, differential expressions of CDK1 were calculated between two independent clinicopathological parameter groups. Student’s independent t-test was performed in these evaluations using SPSS 23.0 software, and P<0.05 was considered statistically significant. All data were calculated twice.
Expression of CDK1 protein in THCA tissues based on data from The Human Protein Atlas (THPA)
THPA is a project for examining normal and carcinoma tissues based on antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology antibody proteomics [22,23]. Immunohistochemical results for CDK1 from THPA were downloaded to verify the CDK1 protein expression level in THCA and were compared with our immunohistochemical experimental results.
Evaluation of CDK1 mRNA expression in THCA based on RNA-seq and microarray data
The Cancer Cell Line Encyclopedia (CCLE) contains over 1400 cancer cell lines and provides the expression data for genes in these cell lines. The CDK1 expression data for different cancer types and different cancer cell lines in the CCLE were retrieved and compared.
The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) provided RNA-Seq data for genes in cancer tissues and normal tissues. Expression data for CDK1 and clinical parameter information were downloaded and the value of Transcripts Per Million (TPM) was transferred. Subsequently, RNA-seq or microarray datasets were screened from the Gene Expression Omnibus (GEO), ArrayExpress, Sequence Read Archive (SRA), and Oncomine databases. The following keywords were used to retrieve relative data: (thyroid) AND (cancer OR carcinoma OR tumo* OR neoplas* OR malignan*) AND (CDK1 OR “Protein Kinase, CDC2” OR “Cdk1 Protein Kinase” OR “Protein Kinase, Cdk1” OR “cdk1 Kinase” OR “cdc2 + Protein” OR “Cyclin-Dependent Kinase 1” OR “Cyclin Dependent Kinase 1” OR “p34cdc2 Protein” OR “Protein p34cdc2” OR “p34cdc2, Protein” OR “Histone Kinase p34 (cdc2)”). The entry type was limited to “series”, and the organism was filtered by “homo sapiens”. The PubMed, Web of Sciences, EMBASE, Google Scholar, Wanfang, Qvip, and CNKI databases were also searched for articles published before August 1, 2019 containing RNA-seq or microarray data.
The obtained RNA-seq and microarray data were then converted to box plots using GraphPad 8.0 software to compare the CDK1 expression level in THCA and adjacent non-cancerous tissues, and receiver operating characteristic (ROC) curves were also generated. The gathered data were statistically analyzed using Student’s t-test in SPSS 23.0 to compare the differences in CDK1 expression among the various groups. All expression values were presented as mean ± standard deviation (Mean ± SD). Forest plots with pooled standardized mean difference (SMD) and 95% confidential interval (CI), funnel plots, and sensitivity analyses were generated using STATA 12.0 in a random effects model for different subtypes. The True positive (TP), False positive (FP), False negative (FN), and True negative (TN) values of all studies were then applied in MetaDiSc 1.4 to analyze the sensitivity, specificity, positive likelihood ratios, negative likelihood ratios, and diagnostic odds ratio. A summarize ROC (sROC) curve was generated for a second evaluation of the difference in CDK1 expression between different subtypes of THCA and control samples, as well as between DTC and ATC. The prognostic value of CDK1 expression was assessed by both univariate and multivariate analyses with hazard ratio (HR) being computed.
Integrated assessment of CDK1 expression combining tissue microarray, RNA-seq and gene microarray datasets
The CDK1 protein data in the tissue arrays were added to the SMD and sROC computations of the RNA-seq and gene microarray datasets to strengthen the reliability of the results. The analysis was performed using STATA 12.0 and Meta-DiSc v.1.4, as mentioned above.
Potential mechanism of CDK1 upregulation in THCA
Identification of CDK1 co-expressed genes in THCA based on Pearson correlation coefficient calculation
The potential molecular mechanism is closely related to the co-expressed genes; therefore, the correlation coefficient indexes between CDK1 and all other genes in each dataset were calculated using the correlation coefficient calculation method based on R. The CDK1 co-expressed genes were selected if |Pearson’s r| ≥0.5 and P<0.05. Since this study contained 14 datasets (13 gene microarrays and one RNA-seq data), the co-expressed genes appearing more than twice were selected as subjects for subsequent work.
Identification of differentially expressed genes (DEGs) in THCA
If the genes co-expressed with CDK1 also have roles in the carcinogenesis of THCA, they have more possibilities to work together with CDK1 to influence the biological function of THCA. For this reason, we also drew a comprehensive roadmap of the DEGs in THCA, based on all available sources. Apart from the 14 datasets containing CDK1 expression, another three gene microarray datasets (GSE6339, GSE9115, and GSE50901) were available for inclusion to identify the DEG signature in THCA. The DEGs between THCA and non-cancer thyroid tissues were analyzed with the R-based limma method for all gene microarray data and edgR was used for RNA-seq data. The cut-off value for this step was |log2FC| >1, FDR <0.05. We collected the upregulated or downregulated genes that appeared in at least two independent datasets as the first part of the DEGs. Robust rank aggregation (RRA) was used for the collection of the second part of the DEGs; RRA uses a probabilistic model for aggregation that is robust to noise and calculates the significance probabilities for all the elements in the final ranking [24-29]. The cut-off P-value was set at 0.05 for RRA. The second part of the DEGs from all gene microarray datasets and TCGA/GTEx RNA-seq data were obtained using the RRA method. Eventually, the merged first and second parts were regarded as the final candidates for DEGs in THCA for the next step.
Related pathways of the interaction of CDK1 co-expressed genes and DEGs in THCA
The genes co-expressed with CDK1 were interacted with the DEGs in THCA, followed by gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. GO enrichment analysis was performed using the online Metascape [30] to identify the gene annotations of biological processes (BP), cellular components (CC), and molecular function (MF). KEGG pathway analysis was carried out using ClueGo in Cytoscape. Protein-protein interaction (PPI) enrichment analysis was performed, and a PPI network was constructed in Metascape.
Results
Integrated assessment of CDK1 expression in THCA
Expression of CDK1 protein based on tissue microarray in-house and from THPA
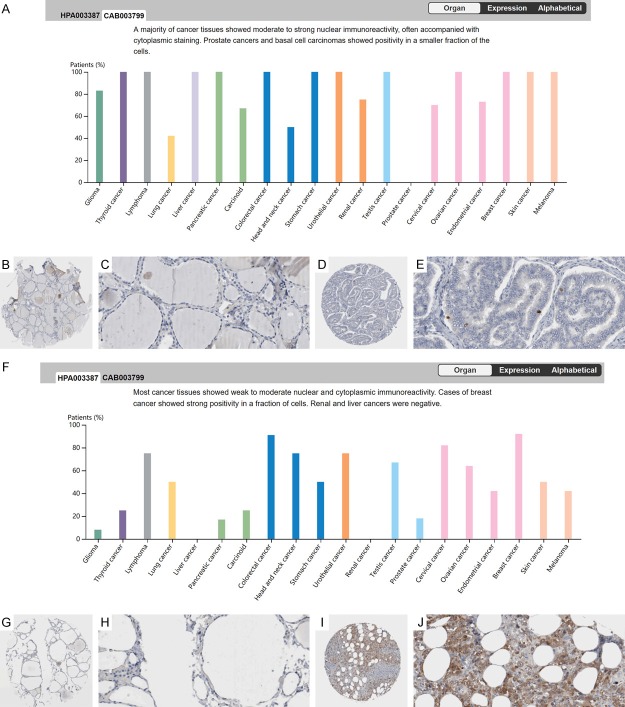
CDK1 protein expression was notably higher in THCA tissues than in non-cancer tissues (Figure 1A-H). The relationships between CDK1 expression and clinical parameters are shown in Table 1. The expression of CDK1 protein was also significantly higher in pathologic T3-T4 than in T1-T2 samples (Figure 1I, 1J). The expression of CDK1 protein using the antibodies of CAB003799 and HPA003387 from THPA also showed a concordant upregulated pattern (Figure 2). However, due to the limited sample size, a statistical analysis could not be performed with the data from THPA.
Figure 1.

CDK1 protein overexpression in thyroid cancer (THCA) tissues assessed by in-house tissue microarrays. (A) Normal thyroid tissue (200×); (B, C) THCA (200×); (D) Normal thyroid tissue (400×); (E, F) THCA (400×). Immunohistochemistry, 3,3’-Diaminobenzidine (DAB) staining. Expression level of CDK1 protein between THCA and non-cancer controls (G, H) and between patients in T1, T2 and T3, T4 stages (I, J).
Table 1.
Relationship between CDK1 protein expression and clinical parameters in THCA detected by in-house immunohistochemistry with tissue microarrays
| Clinicopathological features | n | CDK1 expression | |||
|---|---|---|---|---|---|
|
| |||||
| Mean ± SD | t/F value | p value | |||
| Tissue | THCA | 171 | 7.08 ± 0.109 | 29.331a | <0.001 |
| Non-cancer | 87 | 1.10 ± 0.189 | |||
| Age | <45 | 83 | 7.18 ± 1.27 | 0.934a | 0.351 |
| ≥45 | 88 | 6.98 ± 1.55 | |||
| Gender | Male | 54 | 7.04 ± 1.43 | -0.243a | 0.808 |
| Female | 117 | 7.09 ± 1.43 | |||
| Pathological stage I | Stage 1 | 84 | 7.07 ± 0.144 | F=1.242b | 0.296 |
| Stage 2 | 52 | 6.85 ± 0.203 | |||
| Stage 3 | 29 | 7.38 ± 0.245 | |||
| Stage 4 | 6 | 7.67 ± 1.085 | |||
| Pathological stage II | Stage 1-2 | 136 | 6.99 ± 0.118 | 1.652a | 0.100 |
| Stage 3-4 | 35 | 7.43 ± 0.267 | |||
| Pathological T stage I | T1 | 14 | 6.86 ± 0.345 | F=1.660b | 0.178 |
| T2 | 95 | 6.91 ± 0.143 | |||
| T3 | 56 | 7.36 ± 0.175 | |||
| T4 | 6 | 7.67 ± 1.085 | |||
| Pathological T stage II | T1-T2 | 109 | 6.90 ± 0.132 | -2.180a | 0.031 |
| T3-T4 | 62 | 7.39 ± 0.186 | |||
| Pathological N stage | N0 | 129 | 7.01 ± 1.44 | -1.101a | 0.273 |
| N1 | 42 | 7.29 ± 1.37 | |||
Student’s 2-sample independent t-test was applied.
One-way analysis of variance (ANOVA) test was conducted.
THCA: thyroid cancer.
Figure 2.
Overexpression of CDK1 protein stained by CAB003799 and HPA003387 antibodies in thyroid cancer (THCA) from the Human Protein Atlas (THPA). (A) The expression pattern of CDK1 stained by the antibody CAB003799 in multiple cancers. Negative staining of CDK1 in non-cancerous thyroid epithelium (B: ×40, C: ×400). Weak positive staining of CDK1 in THCA tissues (D: ×40, E: ×400). (F) The expression pattern of CDK1 stained by the antibody HPA003387 in multiple cancers. Negative staining of CDK1 in non-cancerous thyroid epithelium (G: ×40, H: ×400). Strong positive staining of CDK1 in THCA tissues (I: ×40, J: ×400). Immunohistochemistry, 3,3’-Diaminobenzidine (DAB) staining.
Upregulation of CDK1 in THCA and its clinicopathological characteristics with RNA-seq data
According to the data from CCLE, CDK1 was expressed in different cancer types, including THCA cells (Supplementary Figure 1). Unfortunately, no normal thyroid epithelial cells were available for the comparison. A better understanding of the clinical role of CDK1 in THCA was obtained by extracting the data on CDK1 mRNA level from 512 THCA and 316 non-cancerous thyroid tissues from the TCGA and GTEx projects. The expression of CDK1 was apparently higher in THCA tissues than in non-cancerous thyroid tissues (1.8525 ± 0.74073 vs. 1.1769 ± 0.71166; P<0.0001; Figure 3A; Table 2). AUC of CDK1 mRNA expression was 0.7665 (Figure 3B, P<0.0001). Regarding the relationships between CDK1 and clinicopathological features, a total of 504 patients with complete clinical data were obtained. CDK1 mRNA levels were obviously higher in tissues from the cases who aged under 45 (1.9702 ± 0.68887), compared to those over 45 (1.7535 ± 0.75714; P=0.001, Figure 3C, 3D; Table 2). Next, we inspected the prognostic significance of CDK1 expression. The results of Kaplan-Meier (K-M) survival curve analysis indicated that the THCA cases with higher CDK1 levels tended to have a slightly worse disease-free survival (DFS) (HR=1.3; Figure 3E, 3F) than those with lower expression levels. Nevertheless, the log-rank P was 0.35 and the ability of CDK1 to predict the survival needs to be further validated.
Figure 3.
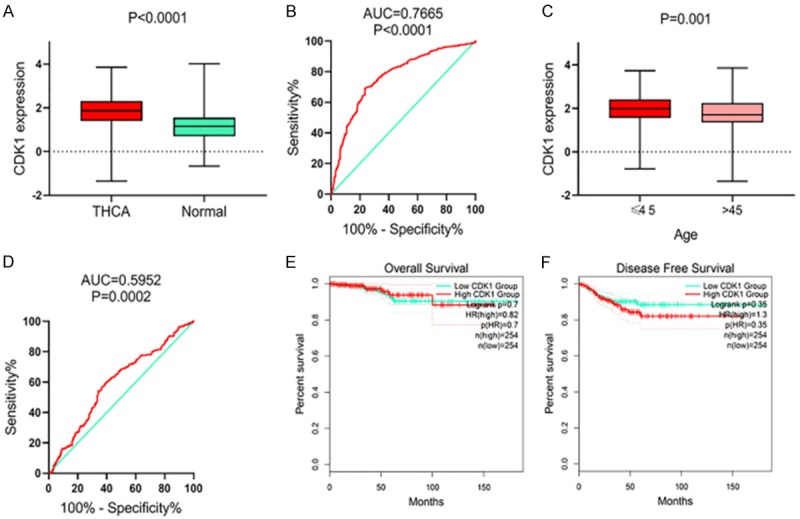
Clinical role of upregulation of CDK1 mRNA level in thyroid cancer (THCA) based on RNA-sequencing data. Expression level of CDK1 mRNA between THCA and non-cancer thyroid controls (A, B) and between patients with different ages (C, D). Prognostic value of CDK1 mRNA in THCA: overall survival (E), disease free survival (F).
Table 2.
Expression of CDK1 in THCA based on the TCGA and GTEx RNA-seq data
| Clinicopathological features | n | CDK1 expression (log2 (TPM +0.001)) | |||
|---|---|---|---|---|---|
|
| |||||
| Mean ± SD | t/F value | p value | |||
| Tissue | THCA | 512 | 1.8525 ± 0.74073 | 12.941a | <0.001 |
| Normal | 316 | 1.1769 ± 0.71166 | |||
| Gender | Male | 136 | 1.9056 ± 0.76938 | 0.926a | 0.355 |
| Female | 368 | 1.8375 ± 0.71935 | |||
| Age | ≤45 | 238 | 1.9702 ± 0.68887 | 3.347a | 0.001 |
| >45 | 266 | 1.7535 ± 0.75714 | |||
| Racec | Asia | 51 | 1.9268 ± 0.71100 | 0.519b | 0.596 |
| Black/African American | 27 | 1.7537 ± 0.82126 | |||
| White | 333 | 1.8856 ± 0.72397 | |||
| Pathologic T staged | T1-T2 | 308 | 1.8853 ± 0.70984 | 1.190a | 0.235 |
| T3-T4 | 194 | 1.8054 ± 0.76813 | |||
| Pathologic N stagee | N0 | 229 | 1.8098 ± 0.76498 | -1.569a | 0.117 |
| N1 | 225 | 1.9185 ± 070954 | |||
| Pathologic M stagef | M0 | 281 | 1.8742 ± 0.72134 | -0.879a | 0.380 |
| M1 | 9 | 2.0900 ± 0.84321 | |||
| Pathologic TNM stagesg | I-II | 335 | 1.8906 ± 0.69509 | 1.519a | 0.129 |
| III-IV | 167 | 1.7851 ± 0.80503 | |||
Student’s 2-sample independent t-test was applied.
One-way analysis of variance (ANOVA) test was conducted.
93 samples were not identified.
2 TX samples were excluded.
50 NX samples were excluded.
213 MX samples were excluded and 1 sample was not identified.
2 samples were not identified.
Upregulated CDK1 in THCA via gene microarray data
We sought to strengthen the observation of overexpression level of CDK1 detected with RNA-sequencing data by looking for other independent cohorts that included CDK1 expression data for THCA. Evaluation of the possible high throughput data sources revealed 16 gene microarrays, and CDK1 expression information could be extracted from 13 of them (GSE3467, GSE3678, GSE6004, GSE27155, GSE29265, GSE29315, GSE33630, GSE53072, GSE53157, GSE35570, GSE58545, GSE60542, and GSE65144). These 13 microarrays contained 419 THCA samples and 269 non-cancerous thyroid tissues (Supplementary Figure 2; Table 3). The expression levels of CDK1 were largely higher in the cancerous cases than in the non-cancerous cases in GSE27155 (2.7388 ± 0.2178 vs. 2.5732 ± 0.1096, P=0.0011), GSE29265 (5.4644 ± 1.3456 vs. 4.4615 ± 0.4694, P=0.0025), GSE33630 (7.0142 ± 1.295 vs. 6.1941 ± 0.9184, P=0.0005), GSE35570 (4.777 1 ± 0.9207 vs. 4.1095 ± 1.1762, P=0.0013), GSE53072 (9.518 ± 0.98401 vs. 6.1622 ± 0.20926, P=0.0003), GSE60542 (6.1474 ± 0.8125 vs. 5.7416 ± 0.9439, P=0.0321), and GSE65144 (10.3965 ± 1.0618 vs. 8.0477 ± 0.7913, P<0.0001) (Figure 4). The capability of CDK1 to distinguish cancerous from non-cancerous thyroid samples was further identified by generating ROC curves. Favorable AUCs for CDK1 were obtained from some microarray data, including GSE27155 (AUC=0.7643), GSE29265 (AUC=0.8172), GSE33630 (AUC=0.7330), GSE53157 (AUC=0.8750), GSE35570 (AUC=0.7884), GSE53072 (AUC=1.0000), GSE60542 (AUC=0.6749), and GSE65144 (AUC=0.9551) (all P<0.05) (Figure 5).
Table 3.
Expression of CDK1 in THCA and non-cancerous thyroid tissues based on microarray data
| Accession | Year | Exp n | Exp mean | Exp sd | Ctrl n | Ctrl mean | Ctrl sd |
|---|---|---|---|---|---|---|---|
| GSE3467 | 2005 | 9 | 6.652 | 0.4539 | 9 | 6.5136 | 0.2191 |
| GSE3678 | 2006 | 7 | 6.167 | 0.1814 | 7 | 6.0522 | 0.3852 |
| GSE6004 | 2006 | 14 | 5.6423 | 0.1763 | 4 | 5.591 | 0.5144 |
| GSE27155 | 2011 | 78 | 2.7388 | 0.2178 | 21 | 2.5732 | 0.1096 |
| GSE29265 | 2012 | 29 | 5.4644 | 1.3456 | 20 | 4.4615 | 0.4694 |
| GSE29315 | 2012 | 31 | 2.9157 | 0.2545 | 40 | 2.8367 | 0.2348 |
| GSE33630 | 2012 | 60 | 7.0142 | 1.295 | 45 | 6.1941 | 0.9184 |
| GSE53072 | 2013 | 5 | 9.518 | 0.9840 | 4 | 6.1622 | 0.2093 |
| GSE53157 | 2013 | 24 | 7.0131 | 0.6983 | 3 | 6.4275 | 0.1235 |
| GSE35570 | 2015 | 65 | 4.7771 | 0.9207 | 51 | 4.1095 | 1.1762 |
| GSE58545 | 2015 | 27 | 2.87 | 0.7067 | 18 | 2.8148 | 0.771 |
| GSE60542 | 2015 | 58 | 6.1474 | 0.8125 | 34 | 5.7416 | 0.9439 |
| GSE65144 | 2015 | 12 | 10.3965 | 1.0618 | 13 | 8.0477 | 0.7913 |
Exp: Experimental group, Ctrl: Control group, sd: standard deviation.
Figure 4.
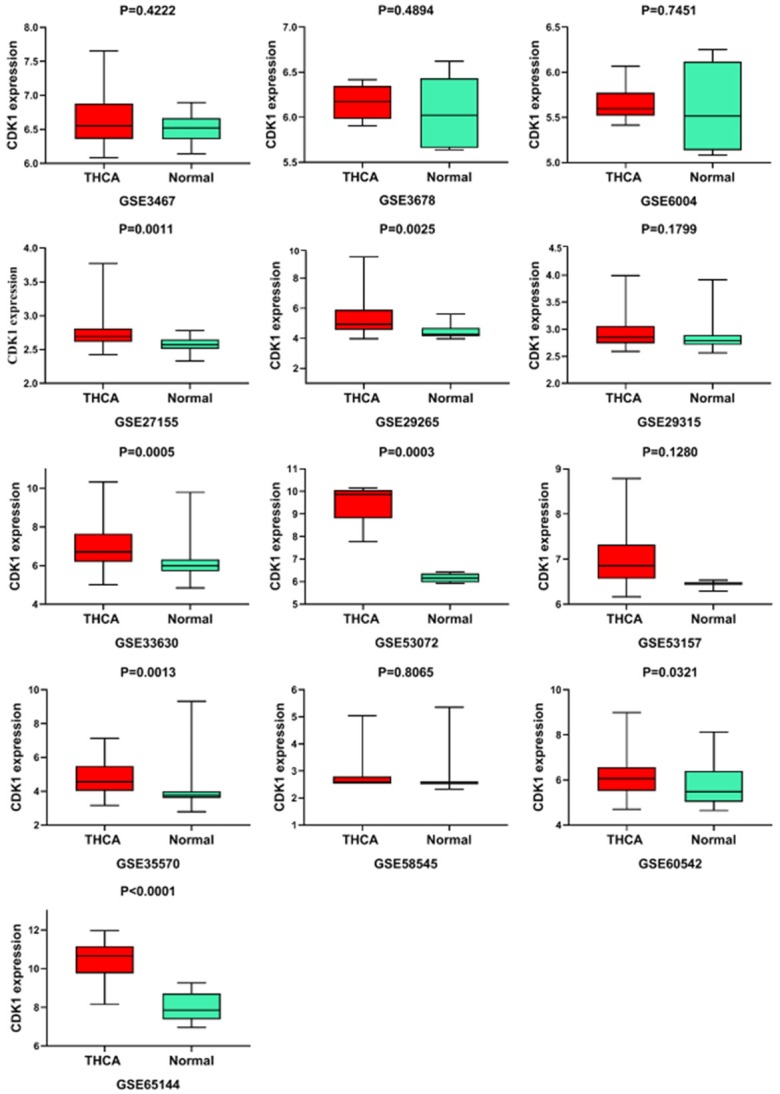
Box plots of CDK1 mRNA expression in thyroid cancer (THCA) based on 13 GEO gene microarrays.
Figure 5.

ROC curves of CDK1 mRNA expression in thyroid cancer (THCA) based on 13 gene microarrays.
Further verification of CDK1 upregulation in THCA via integrated assessment
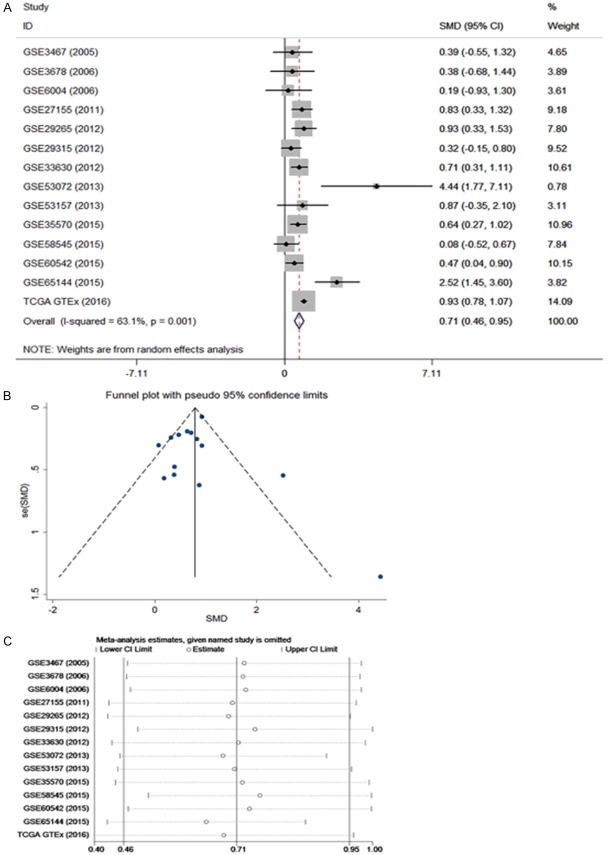
Some of the results from the RNA-sequencing and gene microarray dataset analyses were inconsistent; therefore, we performed an integrated assessment to determine the overall CDK1 mRNA expression from the TCGA/GTEx and microarray databases; these included a total of 931 THCA and 585 non-cancerous thyroid tissue samples. The pooled SMD for CDK1 was 0.71 (95% CI, 0.46-0.95) according to the random effects model (Figure 6). The pooled sensitivity of CDK1 was 0.82, specificity 0.62, positive likelihood ratio 2.09, negative likelihood ratio 0.30, and diagnostic odds ratio 7.53 (Supplementary Figure 3). Most importantly, the AUC of CDK1 reached 0.7959 (Supplementary Figure 3), confirming an oncogenic role for CDK1 in THCA and its ability to discriminate THCA from non-cancerous tissues.
Figure 6.
Integrated assessment of the CDK1 mRNA level in thyroid cancer (THCA) based on gene microarray and RNA-seq data. A: Forest plot; B: Funnel plot; C: Sensitivity analysis.
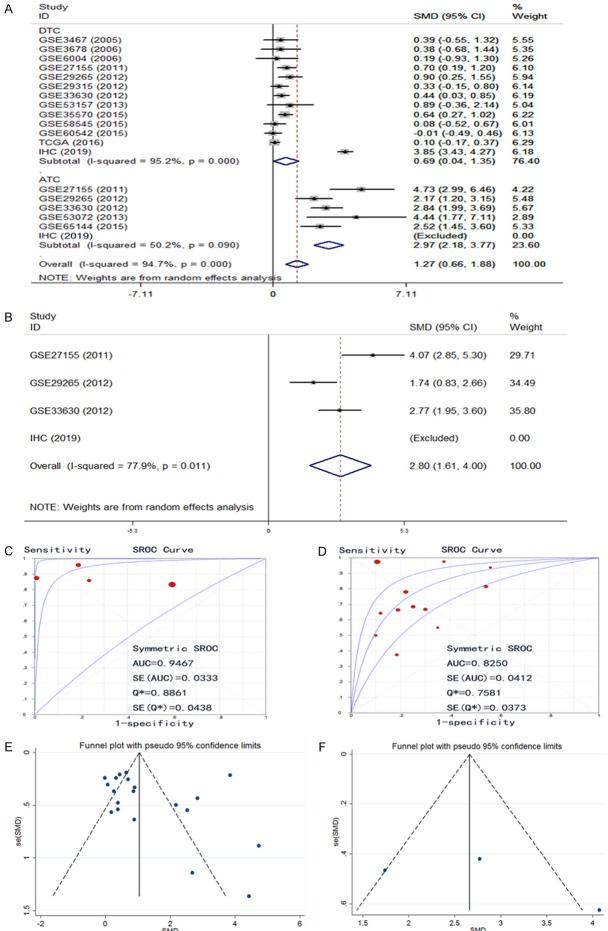
After we obtained the protein and mRNA levels for CDK1 in THCA, we also wished to have a comprehensive overview of the CDK1 level combining all data. An aggregation of the immunohistochemistry results and the RNA-sequencing/microarray findings gave a pooled SMD for CDK1 expression of 2.13 (95% CI, 1.30-2.96) by the random effects model (Supplementary Figure 4). The final AUC for the sROC increased to 0.8372, the pooled sensitivity of CDK1 was 0.74, specificity 0.78, positive likelihood ratio 2.72, negative likelihood ratio 0.32, and diagnostic odds ratio 10.41 (Figure 7) using all 1102 cases of THCA and 672 cases of controls; this value was slightly higher than the AUC generated using only mRNA expression data. We also compared the CDK1 expression between DTC and ATC. The results showed that the SMD of CDK1 in DTC was 0.69 (0.04, 1.35) and in ATC it was greatly higher (2.97 (2.18, 3.77)) (Figure 8A). Using the three datasets containing both DTC and ATC information, we also made a comparison. The direct SMD between ATC and DTC was 2.80 (1.61, 4.00), which supported the above findings and previous reports (Figure 8B). The AUCs of sROC were 0.9467 and 0.8250 for ATC and DTC, respectively (Figure 8C, 8D). We further compared the influence of histological subtypes on CDK1 expression. The SMDs were also different according to the histology subtypes (Figure 9), being 0.72, 1.64 and 2.97, and the AUCs were 0.8440, 0.9440 and 0.9673 for PTC, FTC and ATC, respectively. Considering the prognostic value of CDK1 expression in THCA, no sufficient data were available for a comprehensive meta-analysis to calculate a pooled HR. Based on 507 cases from RNA-seq, the HR was 1.493 (P=0.26 by univariate analysis), which suggests that higher CDK1 had a trend to cause poorer survival (Data not shown). However, larger sample sizes in other cohorts are needed to validate this finding.
Figure 7.
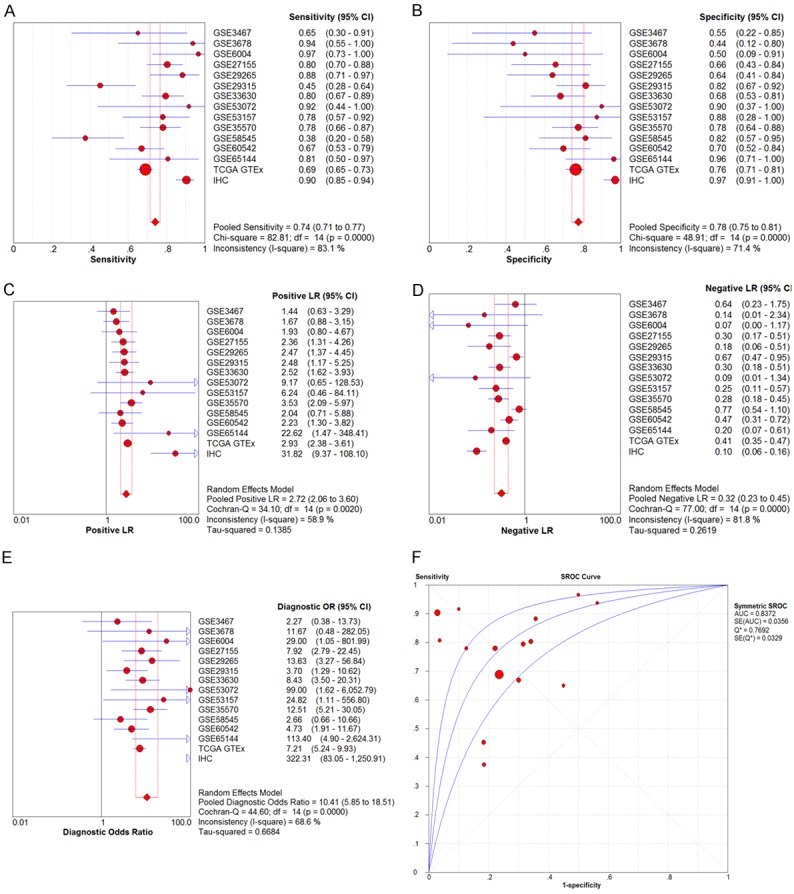
Expression level of CDK1 confirmed using sROC curves including all available data. A: Sensitivity; B: Specificity; C: Positive likelihood ratios; D: Negative likelihood ratios; E: Diagnostic odds ratio; F: AUC of sROC.
Figure 8.
Expression level of CDK1 between anaplastic thyroid cancer (ATC) and differentiated thyroid cancer (DTC). A: Subgroup analysis forest plot of ATC and DTC; B: Forest plot by comparing ATC and DTC; C: SROC of ATC; D: SROC of DTC; E: Funnel plot of subgroup analysis about ATC and DTC; F: Funnel plot by comparing ATC and DTC.
Figure 9.
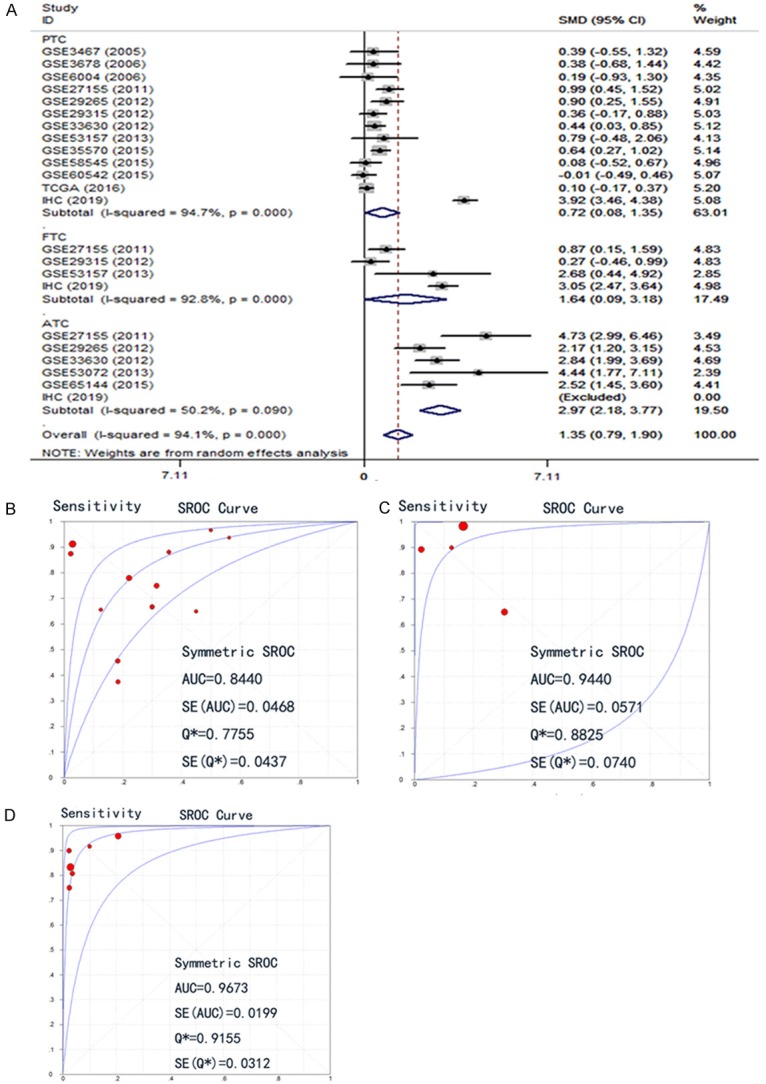
Expression level of CDK1 in different histology subtypes among papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), anaplastic thyroid cancer (ATC). A: Subgroup analysis forest plot. B: SROC of PTC; C: SROC of FTC; D: SROC of ATC.
The potential functions and pathways of CDK1 in THCA
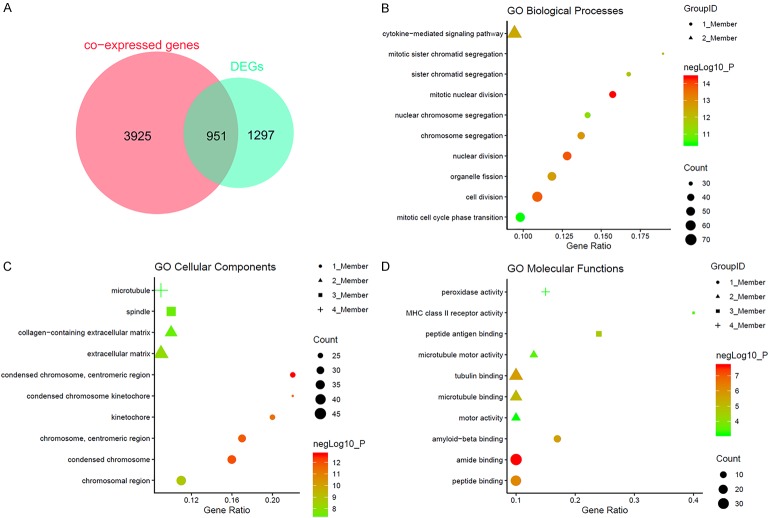
Ranking the CDK1 co-expressed genes from each dataset gave 4876 genes that were present in more than two datasets. The process of screening data with gene expression in THCA retrieved 16 microarrays plus TCGA/GTEx. Supplementary Figure 5 shows the top ten DEGs from each microarray dataset and Supplementary Figure 6 displays the top 10 from the TCGA/GTEx projects. The DEGs for analysis in the next step were selected from those that appeared at least twice in the 17 datasets, for a total of 2248 genes. The reliability of the DEG identification was increased by the conduction of RRA, which identified 212 DEGs. Interestingly, these 212 DEGs were all included in the 2248 candidates assessed in the previous step. Interaction of the 4876 co-expressed genes with the 2248 DEGs in THCA led to a final total of 951 genes (Figure 10A).
Figure 10.
The potential mechanism of CDK1 in thyroid cancer (THCA) assessed by GO annotation. A: Interaction of CDK1 co-expressed genes and differentially expressed genes (DEGs) in THCA; B-D: GO analyses.
Subsequently, GO enrichment showed enrichment of mitotic nuclear division, nuclear division, and cell division in BP (Figure 10B). Condensed chromosome, centromeric region, condensed chromosome and chromosome, and centromeric region were observed in CC (Figure 10C), while amide binding, peptide binding, and tubulin binding were found in MF (Figure 10D; Table 4). KEGG analysis demonstrated the top enriched pathways to be the cell cycle, thyroid hormone synthesis, autoimmune thyroid disease, rheumatoid arthritis, viral myocarditis, phagosome, leishmaniasis, Staphylococcus aureus infection, and tuberculosis (Table 4). The PPI networks for the first four KEGG pathways were also generated (Figure 11).
Table 4.
Top 10 significant GO terms and KEGG pathways of the 951 overlapped co-expressed genes of CDK1 in THCA
| Ontology classification/KEGG | Description | negLog10_P | Gene count | Gene ratio | Top 10 gene symbols |
|---|---|---|---|---|---|
| BP | mitotic nuclear division | 14.36 | 42 | 0.16 | BIRC5, BMP7, BUB1, BUB1B, CDC6, CDC20, CENPF, CHEK1, EDN3, KIF11 |
| BP | nuclear division | 13.83 | 52 | 0.13 | BIRC5, BMP7, BRCA2, BUB1, BUB1B, CDC6, CDC20, CDC25B, CENPF, CHEK1 |
| BP | cell division | 13.72 | 65 | 0.11 | BIRC5, BRCA2, BUB1, BUB1B, CDC6, CDC20, CDC25B, CENPA, CENPF, CKS2 |
| BP | chromosome segregation | 12.82 | 44 | 0.14 | BIRC5, BUB1, BUB1B, CDC6, CDC20, CENPF, FEN1, KIFC1, MAD2L1, MKI67 |
| BP | organelle fission | 12.67 | 53 | 0.12 | BIRC5, BMP7, BRCA2, BUB1, BUB1B, CDC6, CDC20, CDC25B, CENPF, CHEK1 |
| BP | cytokine-mediated signaling pathway | 12.47 | 75 | 0.09 | ALOX5, ANXA2, BIRC5, BST2, CASP1, CD86, CD74, CCR1, COL1A2, EGR1 |
| BP | mitotic sister chromatid segregation | 12.19 | 29 | 0.19 | BUB1, BUB1B, CDC6, CDC20, CENPF, KIFC1, MAD2L1, NEK2, TTK, PRC1 |
| BP | sister chromatid segregation | 11.85 | 32 | 0.17 | BUB1, BUB1B, CDC6, CDC20, CENPF, FEN1, KIFC1, MAD2L1, NEK2, TOP2A |
| BP | nuclear chromosome segregation | 11.27 | 37 | 0.14 | BUB1, BUB1B, CDC6, CDC20, CENPF, FEN1, KIFC1, MAD2L1, NEK2, TOP2A |
| BP | mitotic cell cycle phase transition | 10.42 | 58 | 0.10 | AIF1, BCAT1, BID, BUB1, BUB1B, CDC6, CDC20, CDC25B, CDKN2A, CDKN2C |
| CC | condensed chromosome, centromeric region | 12.69 | 26 | 0.22 | BIRC5, BUB1, BUB1B, CENPA, CENPF, MAD2L1, NEK2, AURKA, AURKB, NDC80 |
| CC | condensed chromosome | 11.99 | 35 | 0.16 | ADD3, BIRC5, BRCA2, BUB1, BUB1B, CENPA, CENPF, CHEK1, MAD2L1, MKI67 |
| CC | chromosome, centromeric region | 11.79 | 32 | 0.17 | BIRC5, BUB1, BUB1B, CENPA, CENPF, HELLS, MAD2L1, NEK2, AURKA, TTK |
| CC | kinetochore | 11.32 | 26 | 0.20 | BIRC5, BUB1, BUB1B, CENPA, CENPF, MAD2L1, NEK2, TTK, AURKB, NDC80 |
| CC | condensed chromosome kinetochore | 11.25 | 23 | 0.22 | BIRC5, BUB1, BUB1B, CENPA, CENPF, MAD2L1, NEK2, NDC80, SPAG5, KIF2C |
| CC | chromosomal region | 8.80 | 39 | 0.11 | BIRC5, BRCA2, BUB1, BUB1B, CENPA, CENPF, CHEK1, EZH2, FEN1, HELLS |
| CC | extracellular matrix | 8.09 | 49 | 0.09 | ANXA2, BMP7, C1QA, C1QB, C1QC, CDH13, CTSC, CLU, COL1A2, COL5A2 |
| CC | spindle | 7.39 | 36 | 0.10 | BIRC5, BUB1B, CDC6, CDC20, CDC25B, CENPF, KIF11, KIFC1, MAD2L1, NEK2 |
| CC | collagen-containing extracellular matrix | 7.36 | 40 | 0.10 | ANXA2, BMP7, C1QA, C1QB, C1QC, CDH13, CTSC, CLU, COL1A2, COL5A2 |
| CC | microtubule | 6.73 | 39 | 0.09 | BIRC5, KIF11, KIFC1, STMN1, NEK2, AURKA, TBCE, TUBA1A, PRC1, AURKB |
| MF | amide binding | 7.65 | 38 | 0.10 | ACACB, ACADL, C1QA, CD14, CD74, CLU, CRYAB, EPHA4, FOLR1, GPR37 |
| MF | peptide binding | 6.21 | 31 | 0.10 | C1QA, CD14, CD74, CLU, CRYAB, EPHA4, GPR37, GSTM1, GSTM2, GSTM3 |
| MF | tubulin binding | 5.74 | 32 | 0.10 | BIRC5, BRCA2, CRYAB, GAPDH, KIF11, KIFC1, STMN1, LYN, S100A9, TBCE |
| MF | amyloid-beta binding | 5.65 | 14 | 0.17 | C1QA, CD74, CLU, CRYAB, EPHA4, ITGAM, ITGB2, MSR1, TLR2, FZD5 |
| MF | microtubule binding | 5.11 | 25 | 0.10 | BIRC5, CRYAB, GAPDH, KIF11, KIFC1, S100A9, PRC1, VAPB, MAP4K4, KIF20A |
| MF | peptide antigen binding | 4.61 | 8 | 0.24 | HLA-B, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, TAP1, SLC7A5 |
| MF | microtubule motor activity | 3.50 | 11 | 0.13 | KIF11, KIFC1, KIF20A, KIF2C, KIF4A, DNAH7, KIF15, WDR78, KIF18A, DYNLRB2, KIF18B |
| MF | MHC class II receptor activity | 3.45 | 4 | 0.40 | HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA |
| MF | peroxidase activity | 3.16 | 8 | 0.15 | ALOX5AP, GPX1, GPX3, GSTM2, TPO, PXDN, IPCEF1, IYD |
| MF | motor activity | 3.15 | 14 | 0.10 | KIF11, KIFC1, MYO1B, DNALI1, KIF20A, KIF2C, KIF4A, DNAH7, KIF15, WDR78 |
| KEGG | Cell cycle | 0.00 | 21 | 0.17 | BUB1, BUB1B, CCNB2, CCNE2, CDC20, CDC25B, CDC45, CDC6, CDC7, CDK1 |
| KEGG | Thyroid hormone synthesis | 0.00 | 14 | 0.19 | CREB3L4, DUOXA2, GPX1, GPX3, HSP90B1, HSPA5, ITPR2, IYD, LRP2, PAX8, SLC26A4, TG, TPO, TSHR |
| KEGG | Autoimmune thyroid disease | 0.00 | 11 | 0.21 | CD86, FAS, HLA-B, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, TG, TPO, TSHR |
| KEGG | Rheumatoid arthritis | 0.00 | 18 | 0.20 | ACP5, ATP6V0E2, CCL20, CD86, CXCL5, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1 |
| KEGG | Viral myocarditis | 0.00 | 12 | 0.20 | BID, CD86, HLA-B, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, ICAM1, ITGB2, RAC2, SGCD |
| KEGG | Phagosome | 0.00 | 24 | 0.16 | ATP6V0E2, CD14, CLEC7A, CTSS, CYBB, FCGR1A, FCGR2A, FCGR3A, HLA-B, HLA-DPA1 |
| KEGG | Leishmaniasis | 0.00 | 16 | 0.22 | CYBB, FCGR1A, FCGR2A, FCGR3A, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, ITGAM |
| KEGG | Staphylococcus aureus infection | 0.00 | 19 | 0.34 | C1QA, C1QB, C1QC, C3AR1, C5AR1, CFB, FCGR1A, FCGR2A, FCGR3A, FPR1 |
| KEGG | Tuberculosis | 0.00 | 23 | 0.13 | BID, CD14, CD74, CLEC7A, CTSS, FCER1G, FCGR1A, FCGR2A, FCGR3A, HLA-DPA1 |
GO: Gene Ontology, KEGG: Kyoto Encyclopedia of Genes and Genomes, BP: Biological Process, CC: Cellular Component, MF: Molecular Function.
Figure 11.

The potential mechanism of CDK1 in thyroid cancer (THCA) assessed by KEGG analysis. A: The KEGG pathway annotations for the 951 overlapped co-expressed genes of CDK1. PPI networks for the genes in the top KEGG pathways; B: Cell cycle; C: Thyroid hormone synthesis; D: Rheumatoid arthritis; E: Staphylococcus aureus infection.
Discussion
This study is divided into essentially two parts. The aim of the first part was to synthesize all available sources, including in-house tissue microarrays, public gene microarrays, and RNA-sequencing data, to verify the overexpression CDK1 at both the protein and mRNA levels. The goal of the second part was to conduct a preliminary examination of the possible mechanism of CDK1 in THCA through CDK1-related genes. The findings of the current study can provide a new orientation for further research in THCA.
The expression levels of CDK1 protein and mRNA have previously been detected by different methods. However, the sample sizes were small and the findings were not consistent. Furthermore, no integrated analysis that combines all available data has been attempted. Among the previous reports, Ito et al. [15] performed immunohistochemistry on 35 FTC, 29 PTC, and 21 ATC samples, and they found positive ratios for CDK1 of 17.1% for FTC, 17.2% for PTC, and 76.2% for the undifferentiated subtype, ATC. By contrast, in non-cancerous controls, the positive ratio for CDK1 was only 8.3% in 13 cases of follicular adenomas, as only one case showed positivity. Furthermore, Ito et al. [15] divided the THCA cases into undifferentiated, poorly differentiated, and well-differentiated groups. The positive CDK1 protein expression ratios in these three groups were 76.2%, 33.3%, and 9.3%, respectively, indicating that CDK1 overexpression could play a pivotal role in the differentiation of THCA. This previous study, at present, is the only one to use immunohistochemistry to study the clinical role of CDK1 in THCA tissues. In the current study, using a larger size (n=171) for the protein level detection, we confirmed the upregulation of CDK1 in THCA tissues. We also found that larger tumors had higher CDK1 expression, suggesting that CDK1 may also accelerate tumor growth.
Fluge et al. [16] performed their own microarray with 10 differentiated (classic) PTCs and 6 aggressive PTCs (poorly differentiated PTC and those with extensive local invasion or synchronous distant metastases). A modified Eberwine protocol for the amplification of antisense RNA (aRNA) followed by aminoallyl-dUTP incorporation in cDNA and subsequent Cy3- or Cy5-labeling was carried out. They consistently reported that CDK1 mRNA had an upregulated trend in PTCs, as the fold change of CDK1 mRNA in classic PTC was 1.3 times higher than non-cancer controls and a 5.9-fold change was achieved in aggressive PTCs. However, this microarray data has not been uploaded to with GEO or ArrayExpress. Recently, several papers re-calculated the available microarrays and documented that CDK1 was found to be dys-expressed in ATCs [17-19]; however, a detailed evaluation of CDK1 expression levels was not presented. In fact, more appropriate high-throughput data of THCA are available for data mining, including the RNA-sequencing data from TCGA and other microarrays included in the current study. Herein, one RNA-sequencing study and 16 microarrays containing CDK1 expression data were recruited and set for multiple integrated analyses. We obtained an inclusive indication of the CDK1 level by combining all data with in-house immunohistochemistry and public RNA-sequencing/microarray sources and then computing the pooled SMD of CDK1 expression with all 1102 cases of THCA and 672 cases of controls; the SMD reached 2.13 (95% CI, 1.30-2.96). More interestingly, the final AUC of the sROC reached 0.8372, also supported an upregulation of CDK1. We also confirmed that CDK1 expression level was greatly higher in ATC than in DTC, which verified the correlation between CDK1 and the differentiation of THCA. Together with previous findings based on small sample sizes, we confirmed the upregulation of CDK1 at both the protein and mRNA levels in THCA tissues. The overexpression of CDK1 may be the cause of the tumorigenesis, indicating the potential use of CDK1 as an indicator in THCA screening and as a target therapy candidate, especially in ATC.
A previous study has also pointed out a relationship between CDK1 variation in THCA and exposure to Chernobyl radiation [31]. The tissues from 33 PTC cases exposed to Chernobyl radiation (ECR) and 32 PTC non-ECR controls were used in a 3’-oligonucleotide microarray study. A total of 239 genes were differentially expressed between these two groups, including CDK1, which was further confirmed to show lower expression in ECR than non-ECR groups by quantitative RT-PCR in an independent cohort. This phenomenon could be explained by an impaired repair of the radiation-induced DNA damage in ECR patients related to the simultaneous lower expression of CDK1. However, due to the special circumstances caused by Chernobyl radiation, this hypothesis has not been verified by other groups.
CDK1 has been well documented as a G2-M boundary modulator of the cell cycle, suggesting that it can influence tumor origin and progression. However, depending on the tumor type, CDK1 can function through different molecular mechanisms. CDK1 acts as a representative cyclin-dependent kinase in THCA, and an interaction between P-ERK and cyclin A, cyclin B1, or CDK1 has been reported in THCA [15,32]. CDK1 is also regulated by miR-7 in THCA [33]. However, no specific molecular machinery for CDK1 in THCA has been comprehensively reported. We explored this issue by collecting data from 17 high-throughput datasets and integrating the differentially expressed genes (DEGs) and co-expressed genes of CDK1 in THCA. These genes, which interact with CDK1, were enriched in some pathways specific to tumorigenesis or to the thyroid organ itself. In the KEGG results, the appearance of the cell cycle pathway is not surprising and confirms the correctness of our research method, as CDK1 is a typical representative of the cell cycle. At the same time, the pathways of thyroid hormone synthesis and autoimmune thyroid disease were also evident. These pathways and their related genes may provide new research directions for revealing the mechanism underlying the effects of CDK1 expression in THCA.
The important role played by CDK1 in the cell cycle makes this gene an excellent target for cancer treatment. Some reports on THCA treatment regard the CDK1/CyclinB signaling pathway as a potential target of intervention. Different drugs and other treatment methods can inhibit the growth of THCA cells by reducing the expression of CDK1 [34-36]. More interestingly, CDK inhibitors are now being developed and tested [13,14]. One of these CDK1 inhibitors, dinaciclib, has been verified as highly effective in MTC cells. More CDK1 inhibitors are expected to be tested in pre-clinical and clinical phases.
There are several limitations of the current study, which could be improved in the future work. The prognostic role of CDK1 expression needs to be verified by multi-centered data. The biological function and importance of CDK1 in different subtypes of THCA remain to be explored. Furthermore, the prospective molecular mechanisms of CDK1 in THCA also need more evidences and clinical validation, in vitro and in vivo work have been expected.
Conclusions
In summary, we demonstrate overexpression of CDK1 in THCA, based on multiple detection methods that combine independent cohorts. However, further studies are required to elucidate the molecular machinery of CDK1 that promotes the biological aggressiveness of THCA cells.
Acknowledgements
The study was supported by the Fund of Natural Science Foundation of Guangxi, China (2017GXNSFAA198253), Guangxi Degree and Postgraduate Education Reform and Development Research Projects, China (JGY2019050), Guangxi Medical University Training Program for Distinguished Young Scholars, Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, Guangxi Zhuang Autonomous Region Health and Family Planning Commission Self-financed Scientific Research Project (Z20180979) and the Fund of Future Academic Star of Guangxi Medical University (WLXSZX19050). The authors thank all the public data sources involved in the current study.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Petrone A, Adamo ME, Cheng C, Kettenbach AN. Identification of candidate cyclin-dependent kinase 1 (Cdk1) substrates in mitosis by quantitative phosphoproteomics. Mol Cell Proteomics. 2016;15:2448–2461. doi: 10.1074/mcp.M116.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao Y, Feng Y, Shen J, Hornicek FJ, Duan Z. The roles and therapeutic potential of cyclin-dependent kinases (CDKs) in sarcoma. Cancer Metastasis Rev. 2016;35:151–163. doi: 10.1007/s10555-015-9601-1. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Xue K, Li Z, Zheng W, Dong W, Song J, Sun S, Ma T, Li W. c-Myc regulates the CDK1/cyclin B1 dependentG2/M cell cycle progression by histone H4 acetylation in Raji cells. Int J Mol Med. 2018;41:3366–3378. doi: 10.3892/ijmm.2018.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Han S, Qian W, Gu Y, Li X, Yang K. Metformin induces miR-378 to downregulate the CDK1, leading to suppression of cell proliferation in hepatocellular carcinoma. Onco Targets Ther. 2018;11:4451–4459. doi: 10.2147/OTT.S167614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Pan Y, Fu H, Zhang J. Nucleolar and spindle associated protein 1 (NUSAP1) inhibits cell proliferation and enhances susceptibility to epirubicin in invasive breast cancer cells by regulating cyclin D kinase (CDK1) and DLGAP5 expression. Med Sci Monit. 2018;24:8553–8564. doi: 10.12659/MSM.910364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prevo R, Pirovano G, Puliyadi R, Herbert KJ, Rodriguez-Berriguete G, O’Docherty A, Greaves W, McKenna WG, Higgins GS. CDK1 inhibition sensitizes normal cells to DNA damage in a cell cycle dependent manner. Cell Cycle. 2018;17:1513–1523. doi: 10.1080/15384101.2018.1491236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Z, Pan Y, Ling G, Wang S, Huang M, Jiang X, Ke Y. Escape of U251 glioma cells from temozolomide-induced senescence was modulated by CDK1/survivin signaling. Am J Transl Res. 2017;9:2163–2180. [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Huang Q, Zhai DZ, Dong J, Wang AD, Lan Q. CDK1 expression and effects of CDK1 silencing on the malignant phenotype of glioma cells. Zhonghua Zhong Liu Za Zhi. 2007;29:484–488. [PubMed] [Google Scholar]
- 9.Chen X, Zhang FH, Chen QE, Wang YY, Wang YL, He JC, Zhou J. The clinical significance of CDK1 expression in oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20:e7–12. doi: 10.4317/medoral.19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LL, Sun KX, Wu DD, Xiu YL, Chen X, Chen S, Zong ZH, Sang XB, Liu Y, Zhao Y. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21:3055–3065. doi: 10.1111/jcmm.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung WW, Lin YM, Wu PR, Yen HH, Lai HW, Su TC, Huang RH, Wen CK, Chen CY, Chen CJ, Yeh KT. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer. 2014;14:951. doi: 10.1186/1471-2407-14-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsaur I, Makarević J, Hudak L, Juengel E, Kurosch M, Wiesner C, Bartsch G, Harder S, Haferkamp A, Blaheta RA. The cdk1-cyclin B complex is involved in everolimus triggered resistance in the PC3 prostate cancer cell line. Cancer Lett. 2011;313:84–90. doi: 10.1016/j.canlet.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 13.Liao H, Ji F, Ying S. CDK1: beyond cell cycle regulation. Aging (Albany NY) 2017;9:2465–2466. doi: 10.18632/aging.101348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang M, Zhao T, Ma L, Guo Y. CHK1 inhibition sensitizes pancreatic cancer cells to gemcitabine via promoting CDK-dependent DNA damage and ribonucleotide reductase downregulation. Oncol Rep. 2018;39:1322–1330. doi: 10.3892/or.2017.6168. [DOI] [PubMed] [Google Scholar]
- 15.Ito Y, Yoshida H, Nakano K, Takamura Y, Kobayashi K, Yokozawa T, Matsuzuka F, Matsuura N, Kuma K, Miyauchi A. Expression of G2-M modulators in thyroid neoplasms: correlation of cyclin A, B1 and cdc2 with differentiation. Pathol Res Pract. 2002;198:397–402. doi: 10.1078/0344-0338-00272. [DOI] [PubMed] [Google Scholar]
- 16.Fluge Ø, Bruland O, Akslen LA, Lillehaug JR, Varhaug JE. Gene expression in poorly differentiated papillary thyroid carcinomas. Thyroid. 2006;16:161–175. doi: 10.1089/thy.2006.16.161. [DOI] [PubMed] [Google Scholar]
- 17.Yu JW, Mai W, Cui YL, Kong LY. Genes and pathways identified in thyroid carcinoma based on bioinformatics analysis. Neoplasma. 2016;63:559–568. doi: 10.4149/neo_2016_409. [DOI] [PubMed] [Google Scholar]
- 18.Pan Z, Li L, Fang Q, Qian Y, Zhang Y, Zhu J, Ge M, Huang P. Integrated bioinformatics analysis of master regulators in anaplastic thyroid carcinoma. Biomed Res Int. 2019;2019:9734576. doi: 10.1155/2019/9734576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Liao Y, Chen L. Identification of key pathways and genes in anaplastic thyroid carcinoma via integrated bioinformatics analysis. Med Sci Monit. 2018;24:6438–6448. doi: 10.12659/MSM.910088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu M, Sun Q, Mo CH, Pang JS, Hou JY, Pang LL, Lu HP, Dang YW, Fang SJ, Tang D, Chen G, Feng ZB. Prospective molecular mechanism of COL5A1 in breast cancer based on a microarray, RNA sequencing and immunohistochemistry. Oncol Rep. 2019;42:151–175. doi: 10.3892/or.2019.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Mo W, Fang Y, Wei G, Wei M, Dang Y, Chen G, Hu K, Wei D. Up-regulation of Polo-like Kinase 1 in nasopharyngeal carcinoma tissues: a comprehensive investigation based on RNA-sequencing, gene chips, and in-house tissue arrays. Am J Transl Res. 2018;10:3924–3940. [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 23.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstrom H, Glimelius B, Sjöblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 24.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrakopoulos C, Hindupur SK, Häfliger L, Behr J, Montazeri H, Hall MN, Beerenwinkel N. Network-based integration of multi-omics data for prioritizing cancer genes. Bioinformatics. 2018;34:2441–2448. doi: 10.1093/bioinformatics/bty148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan YJ, Ma JY, Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19:183. doi: 10.1186/s12935-019-0905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao H, Xu D, Chen P, Zeng G, Wang X, Zhang X. Identification of five genes as a potential biomarker for predicting progress and prognosis in adrenocortical carcinoma. J Cancer. 2018;9:4484–4495. doi: 10.7150/jca.26698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song ZY, Chao F, Zhuo Z, Ma Z, Li W, Chen G. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging (Albany NY) 2019;11:4736–4756. doi: 10.18632/aging.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Zhu J, Chen F, Ma L. Weighted gene coexpression network analysis identifies key genes and pathways associated with idiopathic pulmonary fibrosis. Med Sci Monit. 2019;25:4285–4304. doi: 10.12659/MSM.916828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handkiewicz-Junak D, Swierniak M, Rusinek D, Oczko-Wojciechowska M, Dom G, Maenhaut C, Unger K, Detours V, Bogdanova T, Thomas G, Likhtarov I, Jaksik R, Kowalska M, Chmielik E, Jarzab M, Swierniak A, Jarzab B. Gene signature of the post-Chernobyl papillary thyroid cancer. Eur J Nucl Med Mol Imaging. 2016;43:1267–1277. doi: 10.1007/s00259-015-3303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YY, Liu ZB, Ye XG, Ren WM. Iodine regulates G2/M progression induced by CCL21/CCR7 interaction in primary cultures of papillary thyroid cancer cells with RET/PTC expression. Mol Med Rep. 2016;14:3941–3946. doi: 10.3892/mmr.2016.5686. [DOI] [PubMed] [Google Scholar]
- 33.Hua K, Jin J, Zhang H, Zhao B, Wu C, Xu H, Fang L. MicroRNA-7 inhibits proliferation, migration and invasion of thyroid papillary cancer cells via targeting CKS2. Int J Oncol. 2016;49:1531–1540. doi: 10.3892/ijo.2016.3660. [DOI] [PubMed] [Google Scholar]
- 34.Erinjeri NJ, Nicolson NG, Deyholos C, Korah R, Carling T. Whole-exome sequencing identifies two discrete druggable signaling pathways in follicular thyroid cancer. J Am Coll Surg. 2018;226:950–959. e5. doi: 10.1016/j.jamcollsurg.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 35.Lin SF, Lin JD, Hsueh C, Chou TC, Wong RJ. A cyclin-dependent kinase inhibitor, dinaciclib in preclinical treatment models of thyroid cancer. PLoS One. 2017;12:e0172315. doi: 10.1371/journal.pone.0172315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Cheng X, Gao Y, Bao J, Guan H, Lu R, Yu H, Xu Q, Sun Y. Induction of ROS-independent DNA damage by curcumin leads to G2/M cell cycle arrest and apoptosis in human papillary thyroid carcinoma BCPAP cells. Food Funct. 2016;7:315–325. doi: 10.1039/c5fo00681c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.