Abstract
Breast cancer is a prevalent malignancy in women and its incidence is increasing at an alarming rate. Breast cancer treatment is mainly obstructed by late diagnosis due to lack of biomarkers, dearth of therapeutic targets, constant relapses, drug resistance, and adverse effects of the chemotherapy. MicroRNAs (miRs) have shown exceptional potential to act as therapeutic agents for treatment of cancer. This study was designed to investigate the role and therapeutic implications of miR-151-5p in breast cancer. The results of the present study revealed that miR-151-5p is significantly downregulated in breast cancer tissues and cell lines. Overexpression of miR-151-5p in SK-BR-3 and CAMA-1 cells inhibits their proliferation and colony formation. Wound heal and transwell assays showed that miR-151-5p inhibits the migration and invasion of the SK-BR-3 and CAMA-1 breast cancer cells. TargetScan analysis followed by the dual luciferase assay confirmed SOCS5 to be the target of miR-151-5p in breast cancer. The expression of SOCS5 was upregulated in all the breast cancer tissues and cell lines. Silencing of SOC5 caused significant inhibition in the proliferation, migration and invasion of the SK-BR-3 cells. Nonetheless, overexpression of SCOS5 could avoid the growth inhibitory effects of miR-151-5p on the breast cancer cells. To conclude, miR-151-5p acts as a tumor suppressor in breast cancer and may exhibit therapeutic implications.
Keywords: Proliferation, microRNA-151, migration, invasion, SOCS5
Introduction
Being the most prevalent type of cancer in women, breast cancer is one of the principal health problems in women accounting for significant mortality. Annually, 1.3 million new cases of breast cancer and 0.4 million mortalities due to breast cancer are reported [1]. Breast cancer has been reported to be remarkably heterogeneous as far as its pathological characteristics are considered. While it has been reported to be very aggressive in some cases, in other it may be slow growing [2]. Global breast cancer statistics has shown that the incidence of breast cancer is increasing significantly. Around 18% increase was observed for breast cancer incidence and mortality from 2008 to 2012. In US, 1 in every 8 women will develop breast cancer during her life [3]. It is believed that by 2050, there will be 3.2 million new cases of breast cancer annually [4]. All these statistics, suggest that there is urgent need to curb the growing incidence of breast cancer and to develop novel and efficient treatment regimes for the treatment of breast cancer. It has been reported that breast cancer treatment is mainly obstructed by late diagnosis due to lack of biomarkers, dearth of therapeutic targets, constant relapses, drug resistance, and adverse effects of the chemotherapy [5]. Recent studies have shown that microRNAs (miRs) may serve as vital biomarkers and therapeutic targets for the treatment of deadly diseases such as cancer. The miRs are non-coding RNAs consisting of 19-25 nucleotides produced from the endogenous hairpin transcripts. In human miRs are highly conserved and exhibit multiple isoforms [6]. They negatively control the expression of human genes and it has been reported that around 60% of the human protein-coding genes harbour miR-binding sites [7]. Since, miRs control vital cellular processes in human, it is not surprising that they control the development of diseases such as cancer [8]. The dysregulation of several miRs may contribute to the development of cancer was for the first time discovered in 2002 when deletion of miR-15 and miR-16 were reported to be involved in the development of lymphocytic leukemia [9]. MicroRNA-151-5p (now onwards referred as miR-151) is one of the less studied miRs and there is only one study wherein miR-151 has been shown to play a role in the proliferation of gastric cancer [10]. However, the role and therapeutic implications of miR-151 have not been explored in breast cancer. This study for the first time reports that miR-151 acts a tumor suppressor in breast cancer. Moreover, miR-151 regulates the proliferation, migration and invasion of breast cancer. Taken together, this study indicates that miR-151 may exhibit therapeutic implications in breast cancer.
Materials and methods
Breast cancer tissues, cell lines and culture conditions
Breast cancer tissue specimen and normal adjacent tissues of 15 breast cancer patients who underwent surgical resection in the Department of breast surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin, China, from June 2018 to December 2018 were included. Patients were aged 35-74 years with an average age of 46.31 years. The breast cancer cell lines (MDA-MB-231, MDA-MB-436, SK-BR-3, and CAMA-1) and three normal breast cell line (MB 157) were procured from Type Culture Collection of Chinese Academy of Sciences, Shanghai, China. The cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum, 100 μg/ml streptomycin and 100 U/ml penicillin and a humidified atmosphere containing 5% CO2.
cDNA synthesis and qRT-PCR
The TRIzol reagent (Invitrogen/Life Technologies, Carlsbad, CA, USA) was used for the extraction of RNA form the tissues and cell lines. Subsequently, the total RNA was reverse-transcribed by RevertAid cDNA synthesis kit (Fermentas). Thereafter the quantitative RT-PCR was performed by using SYBR Green master mix (Applied Biosystems; Foster City, Calif, USA) on an ABI 7900HT system. All reactions were carried out in triplicates and the gene expression was determined by the 2-ΔΔCt method.
Cell transfection
The miR-151 mimics and miR-NC were ordered from RiboBio (Guangzhou, China). Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was then used to perform the transfections in accordance with the manufacturer’s guidelines. As the SK-BR-3 and CAMA-1 cells reached 80%, the appropriate concentrations of miR-151 mimics or miR-NC was transfected into these cells.
The WST-1 assay
The proliferation rate of SK-BR-3 and CAMA-1 cells was monitored by WST-1 assay. In brief, SK-BR-3 and CAMA-1 cells were cultured in 96 well plates at the density of 2 × 105 cells/well. The cells were then transfected with miR-NC or miR-151 mimics and again incubated for 24 h at 37°C. This was followed by the incubation of the cells with WST-1 at 37°C for 4 h. The absorbance was then taken at 450 nm using a victor 3 microplate reader to determine the proliferation rate at 0, 12, 24, 48 and 96 h time intervals.
Wound heal assay
The transfected SK-BR-3 and CAMA-1 cells were placed in twelve well plates with approximately 1 × 105 cells per well. A scratch was made by a pipette tip at 24 h after transfection. The cells were PBS washed and fresh media was added. Following 24 h incubation at 37°C, the cells were subjected to fixation with methanol. The initial wound width and final wound width was determined from photomicrographs.
Cell invasion assay
Transwell chambers with Matrigel employed to monitor invasion of the SK-BR-3 and CAMA-1. In brief, the cells were transfected with appropriate constructs and 48 h post-transfection, the cells were harvested and suspended in fresh media. While as 200 µL of the cell suspension containing approximately 5 × 104 cells was placed onto the upper compartment, a fresh 500 µmedia was placed in the lower compartment. After 24 hours cells present at the upper compartment were removed by swabbing while cells that invaded to the lower surface were fixed and then subsequently stained with 0.05% crystal violet. Finally, ten random fields were selected to determine the invasion under the light microscope.
Target identification and dual-luciferase reporter assay
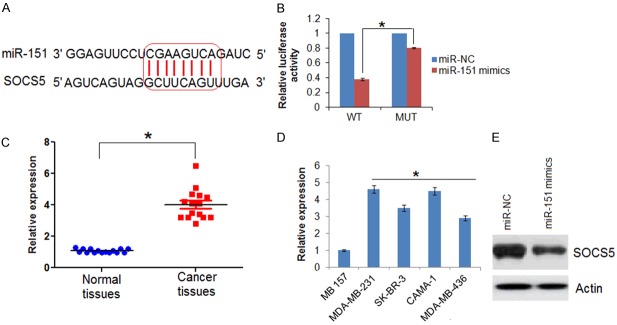
The target of miR-151 was identified by TargetScan online software (http://www.targetscan.org). The miR-151 mimics or miR-NC were co-transfected with Plasmid pGL3-SOCS5-3’-UTR-WT or pGL3-SOCS5-3’-UTR-MUT into SK-BR-3 cells. Dual-luciferase reporter assay (Promega) was performed at 48 h post-transfection and Renilla luciferase utilised for normalization.
Western blotting
The breast cancer tissues and cell lines were lysed and the protein concentration in each sample was measured by Bradford assy. Equal concentrations of the proteins from each sample were loaded on 10% SDS polyacrylamide gels (SDS-PAGE) and then followed by shifting to polyvinylidene fluoride membranes. Blocking of the membrane was then performed by fat-free milk (5%) in TBST. This was followed by incubation with a primary antibody for 24 h at 4°C. Subsequently, a secondary antibody was added at 25°C for about 2 h. The bands of interest were finally observed by chemiluminescence.
Statistical analysis
The experiments were carried out in three biological replicates. The data are presented as the mean the replicates ± SD. The statistical analysis was carried out by student’s t test or one way ANOVA followed by Tukeys test using GraphPad prism 7 software. P < 0.05 was considered as the statistically significant difference.
Results
miR-151 is downregulated in breast cancer
The expression prolife of miR-151 was examined in fifteen breast cancer and fifteen normal adjacent tissues. The results showed that the expression of miR-151 was significantly downregulated in the breast cancer tissues relative to the expression of miR-151 in normal adjacent tissues. Upto 8 fold downregulation of miR-15 was observed in breast cancer tissue (Figure 1A). The expression of miR-151 was also examined in four different breast cancer cell lines as well as the normal cell line by qRT-PCR. Results showed that miR-151 was significantly suppressed in the breast cancer cells relative to its expression in normal cells (Figure 1B). The expression of miR-151 was observed to be 4.5 folds lower in the breast cancer cells.
Figure 1.
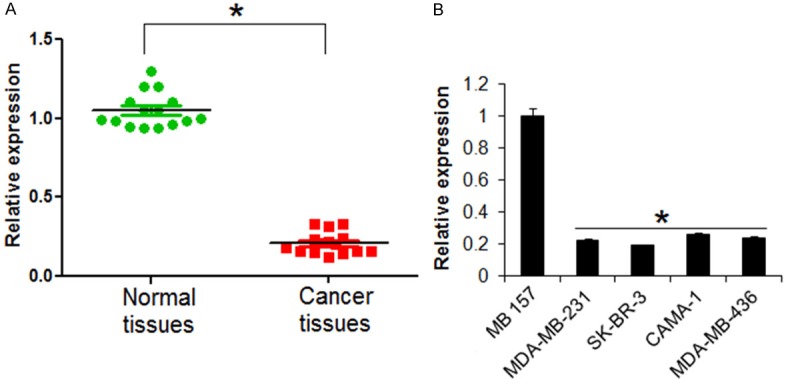
The miR-151 is downregulated in breast cancer. A. Expression of miR-151 in normal and breast cancer tissues. B. Expression of miR-151 in normal and breast cancer cell lines. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
miR-151 acts a tumor suppressor in breast cancer
To ascertain the role of miR-151 in the proliferation of the breast cancer SK-BR-3 and CAMA-1 cells, the cells were transfected with miR-151 mimics. The overexpression of miR-151 in SK-BR-3 and CAMA-1 cells was validated by qRT-PCR which showed 8.3 and 10 fold increase in the miR-151 expression respectively (Figure 2A). Next the proliferation rate of miR-151 overexpressing SK-BR-3 and CAMA-1 cells was monitored at different time periods. The results showed that miR-151 overexpression resulted in significant decrease in the proliferation rate of the SK-BR-3 and CAMA-1 breast cancer cells (Figure 2B). The effects of miR-151 overexpression were also assessed on the colony formation potential of the breast cancer SK-BR-3 and CAMA-1 cells. The results revealed that miR-151 overexpression caused significant decrease in the colony formation of the breast cancer SK-BR-3 and CAMA-1 (Figure 2C).
Figure 2.
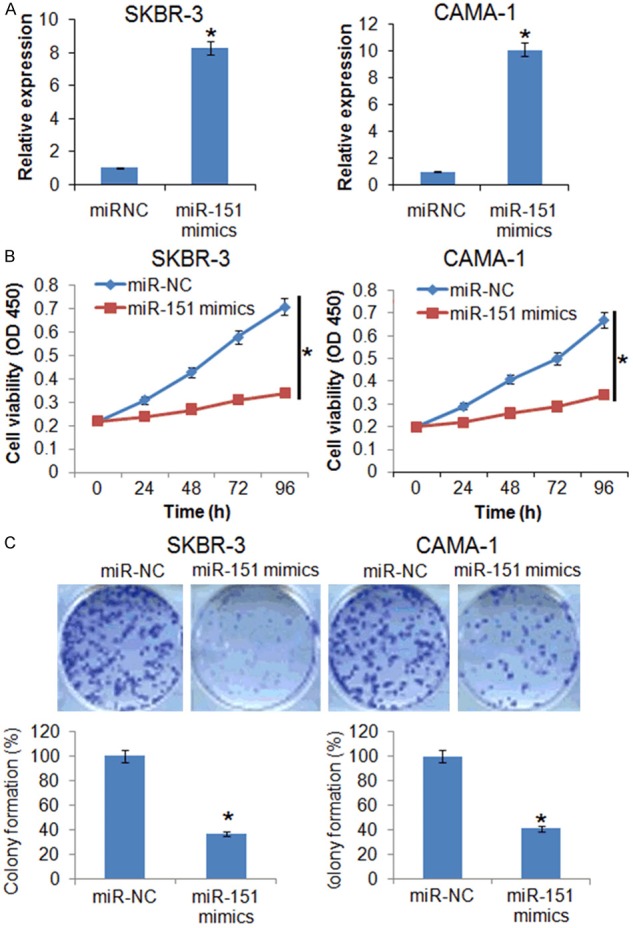
The miR-151 inhibits the proliferation of the breast cancer cells. A. Expression of miR-151 in miR-NC and miR-151 mimics transfected miR-NC or miR-151 mimics transfected SKBR-3 and CAMA-1 cells. B. WST-1 assay showing the viability of the miR-NC or miR-151 mimics transfected SKBR-3 and CAMA-1 cells. C. Colony formation assay of the miR-NC or miR-151 mimics transfected SKBR-3 and CAMA-1 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
miR-151 suppresses the invasion of breast cancer cells
The effects of miR-151 on the migration of the SK-BR-3 cells were determined by wound heal assay. The results showed that miR-15 caused significant decrease in the migration of the SK-BR-3 and CAMA-1 cells as evident from the wound width (Figure 3A). Transwell assay was used to investigate the cell invasion of the SK-BR-3 and CAMA-1 cells and the results showed that the migration of the SK-BR-3 and CAMA-1 was significantly inhibited. The migration in case in case of miR-151 overexpressing SK-BR-3 and CAMA-1 cells was found to be 65 and 57% respectively (Relative to control) (Figure 3B).
Figure 3.
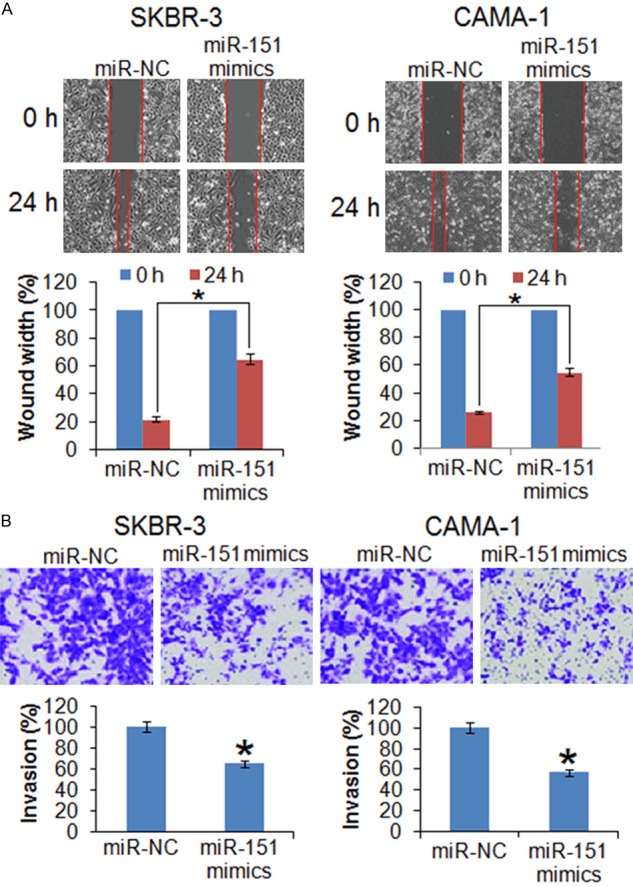
The miR-151 inhibits the migration and invasion of the breast cancer cells. (A) Wound heal assay showing migration in the miR-NC and miR-151 mimics transfected SK-BR-3 and CAMA-1 cells and (B) Transwell assay showing invasion in the miR-NC and miR-151 mimics transfected SK-BR-3 and CAMA-1 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
miR-151 targets SOCS5 in breast cancer cells
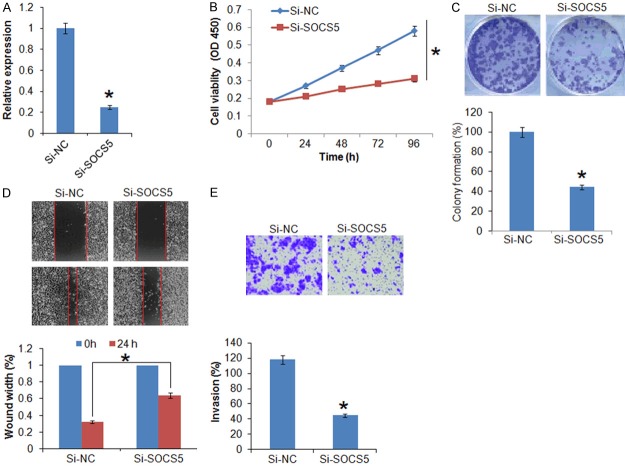
The TargetScan revealed that suppressor of cytokine signaling 5 (SOCS5) acts as the target of miR-151 in breast cancer cells (Figure 4A, Supplementary Table 1). SOCS5 was further confirmed as the target of miR-151 by dual luciferase assay (Figure 4B). The qRT-PCR revealed that the SOCS5 was significantly overexpressed in the all the breast cancer tissues and cell lines (Figure 4C and 4D). Nonetheless, overexpression of miR-151 resulted in suppression of SOCS5 expression in SK-BR-3 cells (Figure 4E), confirming SOCS5 as the target of miR-151. Next to assess the effects of SOCS5 on the proliferation of the SK-BR-3 breast cancer cells, its expression was silenced in SK-BR-3 breast cancer cells (Figure 5A). The results showed that silencing of SOCS5 resulted in decrease of the SK-BR-3 cell proliferation and colony formation (Figure 5B and 5C). Additionally, wound heal and transwell assay showed that miR-151 overexpression also suppressed the migration and invasion of the breast cancer cells (Figure 5D and 5E).
Figure 4.
miR-151 targets SCOCS5 in breast cancer. A. TargetScan analysis showing SOCS5 as the target in miR-151. B. Dual luciferase showing interaction between miR-155 and SOCS5. C. SOCS5 expression in normal and adjacent cancer tissues. D. Expression of SOCS5 in normal and different breast cancer cell lines. E. Western blot analysis showing the expression of SOCS5 in miR-NC and miR-151 mimics transfected SK-BR-3 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
Figure 5.
Silencing of SOCS5 inhibits the proliferation, migration and invasion of breast cancer cells. (A) Expression of SOCS5 in si-NC and si-SOCS5 transfected SK-BR-3 cells. (B) WST-1 showing the viability of the si-NC and si-SOCS5 transfected SK-BR-3 cells. (C) Colony formation of the si-NC and si-SOCS5 transfected SK-BR-3 cells. (D) Wound heal assay showing migration in the si-NC and si-SOCS5 transfected SK-BR-3 cells and (E) Transwell assay showing the invasion of the si-NC and si-SOCS5 transfected SK-BR-3 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
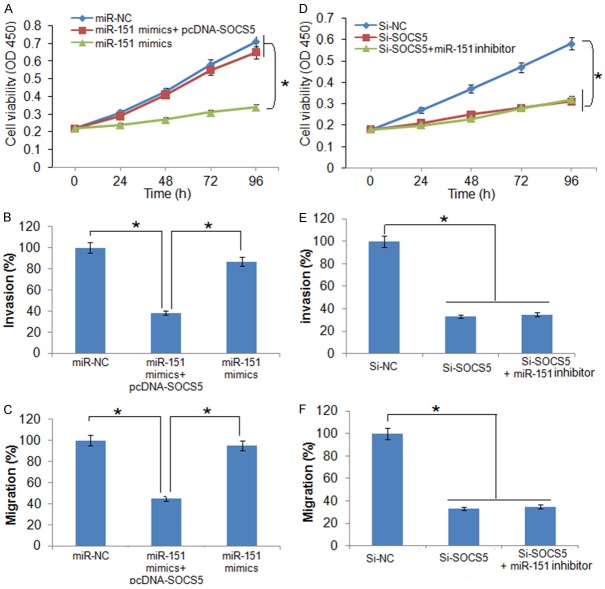
Additionally, it was found that overexpression of SOCS5 in SK-BR-3 cells could promote the growth, migration and invasion of the SK-BR-3 cells and nullified the growth inhibitory effects of miR-151 overexpression on SK-BR-3 cell proliferation (Figure 6A-C). However, miR-151 inhibition had no effects on the proliferation, migration and invasion of the Si-SOCS5 transfected SK-BR-3 cells (Figure 6A-D).
Figure 6.
Effect of SOCS5 overexpression on (A) viability, (B) invasion, and (C) migration of the miR-151 mimics transfected SK-BR-3 cells. Effect of miR-151 inhbition on the (D) viability, (E) migration, and (F) invasion of the si-SOCS5 transfected SK-BR-3 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
Discussion
The incidence of breast cancer is increasing at an alarming rate and there is pressing need to identify the therapeutic targets to enable efficient treatment of breast cancer cells [11]. Recent studies have provided ample evidence that miRs may severe as important biomarkers and therapeutic targets for the treatment of human diseases including cancer [12]. Due to the involvement of miRs in diverse cellular processes, they are involved in the development and progression of deadly disease like cancer [13]. In this study we for the first time demonstrated that miR-151-5p is significantly repressed in breast cancer tissues and cell lines. The dysregulation of miRs has also been reported previously. While as some miRs have been shown to be downregulated, many others have been reported to be upregulated [14,15]. For example, miR-145 has been shown to be downregulated in breast cancer tissues [14] and miR-155 is upregulated in breast cancer [15]. Our result that miR-151 is downregulated in breast cancer tissues and cell lines suggest that miR-151 may exhibit the potential to act as a biomarker for breast cancer. However, this is an initial study and more studies will be required to establish miR-151 as biomarker for breast cancer. To decipher the role of miR-151 in breast cancer, it was overexpressed in two breast cancer cell lines SK-BR-3 and CAMA-1 and the proliferation of these two cell lines was examined by WST-1 assay. The results showed that the proliferation of both these breast cancer cell lines was significantly inhibited, suggesting the tumor-suppressive effects of miR-151. Several miRs have been shown to act as tumor suppressors in breast cancer for example; miR-520/373 family acts as tumor suppressor in breast cancer [16]. Similarly, miR-34a has been shown to inhibit the growth of breast cancer and therefore acts as tumor suppressor [17]. The effects of miR-151 overexpression were investigated on the migration and invasion of the SK-BR-3 and CAMA-1 breast cancer cells. The results showed that upon overexpression of miR-151 in SK-BR-3 and CAMA-1, their migration and invasion was significantly inhibited. These results provide a preliminary clue that miR-151 may also have a role in the metastasis of the breast cancer. The results of the present investigation are in confirmation with previous studies wherein several of the microRNAs have been shown to regulate the migration and invasion of the breast cancer cells. For example, miR-30a has been shown to regulate the migration and invasion of the breast cancer cells [18]. All miRs exert their effects via post-transcriptional suppression of the target genes [19]. It is therefore important to identify the targets of the tumor-suppressive miRs. Therefore, we carried out TargetScan analysis and suppressor of cytokine signaling 5 (SOCS5) was found to be the potential target of miR-151. Several miRs such as miR-432 have been shown target SOCS5 in cancer cells [20]. The expression analysis showed that the expression of SOCS5 to be significantly upregulated in breast cancer tissues and cell lines. The expression of SOCS5 has also been reported to be dysregulated prostate cancer [21]. Silencing of SOCS5 in breast cancer also caused suppression of the proliferation migration and invasion of the breast cancer cells. Nonetheless, the overexpression of SOCS5 in SK-BR-3 cells could nullify the effects of miR-151 overexpression on the proliferation, migration and invasion of the breast cancer cells. This in confirmation with previous studies wherein SOC5 has been shown to pay a role in the tumorigenisis of liver cancer [22].
In conclusion, miR-151 is significantly upregulated in breast cancer tissues and cell lines. Moreover, miR-151 acts as a tumor suppressor and regulates the migration and invasion of the breast cancer cells by inhibiting the expression of SOCS5. Taken together miR-151 may exhibit therapeutic implications in the treatment of breast cancer.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Tao Z, Shi A, Lu C, Song T, Zhang Z, Zhao J. Breast cancer: epidemiology and etiology. Cell Biochem Biophy. 2015;72:333–338. doi: 10.1007/s12013-014-0459-6. [DOI] [PubMed] [Google Scholar]
- 2.McPherson K, Steel C, Dixon JM. Breast cancer-epidemiology, risk factors, and genetics. BMJ. 2000;321:624–628. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelsey JL. Breast cancer epidemiology: summary and future directions. Epidemiol Rev. 1993;15:256–263. doi: 10.1093/oxfordjournals.epirev.a036112. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Robbins AS, Lin CC, Flanders WD, DeSantis CE, Ward EM, Freedman RA. Factors that contributed to black-white disparities in survival among nonelderly women with breast cancer between 2004 and 2013. J. Clin. Oncol. 2017;36:14–24. doi: 10.1200/JCO.2017.73.7932. [DOI] [PubMed] [Google Scholar]
- 5.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and microRNAs in cancer. Int J Mol Sci. 2018;19:459. doi: 10.3390/ijms19020459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev. 2016;17:43–46. doi: 10.7314/apjcp.2016.17.s3.43. [DOI] [PubMed] [Google Scholar]
- 8.Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and microRNAs in cancer. Int J Mol Sci. 2018;19:459. doi: 10.3390/ijms19020459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji W, Sun B, Su C. Targeting microRNAs in cancer gene therapy. Genes. 2017;8:21. doi: 10.3390/genes8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Ann Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 11.Hsu KW, Fang WL, Huang KH, Huang TT, Lee HC, Hsieh RH, Chi CW, Yeh TS. Notch1 pathway-mediated microRNA-151-5p promotes gastric cancer progression. Oncotarget. 2016;7:38036. doi: 10.18632/oncotarget.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turashvili G, Lightbody ED, Tyryshkin K, SenGupta SK, Elliott BE, Madarnas Y, Ghaffari A, Day A, Nicol CJ. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. FASEB J. 2018;32:5937–5954. doi: 10.1096/fj.201800120R. [DOI] [PubMed] [Google Scholar]
- 13.Hosseinahli N, Aghapour M, Duijf PH, Baradaran B. Treating cancer with microRNA replacement therapy: a literature review. J Cellu Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 14.Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, Nam SJ, Chun KH. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011;155:427–434. doi: 10.1016/j.jconrel.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 16.Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T, Schneeweiss A, Sahin O, Wiemann S, Tschulena U. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. 2012;31:4150. doi: 10.1038/onc.2011.571. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–17. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 18.Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012;134:1081–93. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 19.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Sharma N, Kumawat KL, Rastogi M, Basu A, Singh SK. Japanese encephalitis virus exploits the microRNA-432 to regulate the expression of suppressor of cytokine signaling (SOCS) 5. Sci Rep. 2016;6:27685. doi: 10.1038/srep27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu JG, Dai QS, Han ZD, He HC, Mo RJ, Chen G, Chen YF, Wu YD, Yang SB, Jiang FN, Chen WH. Expression of SOCSs in human prostate cancer and their association in prognosis. Mol Cellular Biochem. 2013;381:51–9. doi: 10.1007/s11010-013-1687-6. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Mejias A, Kwon J, Chew XH, Siemens A, Sohn HS, Jing G, Zhang B, Yang H, Tay Y. A novel SOCS5/miR-18/miR-25 axis promotes tumorigenesis in liver cancer. Int J Cancer. 2019;144:311–321. doi: 10.1002/ijc.31857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.