Abstract
Cyclooxygenase-2 (COX-2) is overexpressed in most human cancers, but its precise regulatory mechanism in cancer cells remains unclear. The aims of this study are to discover and identify the new regulatory factors which bind to the COX-2 promoter and regulate COX-2 expression and cancer cell growth, and to elucidate the mechanisms of action of these factors in lung cancer. In this study, the COX-2 promoter-binding protein BPTF (bromodomain PHD finger transcription factor) was detected, identified and verified by biotin-streptavidin-agarose pulldown, mass spectrum analysis and chromatin immunoprecipitation (ChIP) in lung cancer cells, respectively. The expressions of COX-2 and BPTF in lung cancer cell lines, mouse tumor tissues and human clinical samples were detected by RT-PCR, Western blot and immunohistochemistry assays. The interaction of BPTF with NF-kB was analyzed by immunoprecipitation and confocal immunofluorescence assays. We discovered and identified BPTF as a new COX-2 promoter-binding protein in human lung cancer cells. Knockdown of BPTF inhibited COX-2 promoter activity and COX-2 expression in lung cancer cells in vitro and in vivo. We also found that BPTF functioned as a transcriptional regulator through its interaction with the p50 subunit of NF-kB. Knockdown of BPTF abrogated the binding of p50 to the COX-2 promoter, while the inhibition of p50 activity abolished the decreased trend of COX-2 expression and lung cancer cell proliferation caused by BPTF silencing. Moreover, we showed that the expressions of BPTF and COX-2 in tumor tissues of lung cancer patients were positively correlated, and high co-expression of BPTF and COX-2 predicted poor prognosis in lung cancer patients. Collectively, our results indicated that BPTF cooperated with p50 NF-κB to regulate COX-2 expression and lung cancer growth, suggesting that the BPTF/p50/COX-2 axis could be a potential therapeutic target for lung cancer.
Keywords: BPTF, p50 NF-κB, COX-2, lung cancer
Introduction
Inflammation is a hallmark of cancer [1]. As an important inflammatory factor, COX-2 (cyclooxygenase-2) has been demonstrated to play an important role in regulating the growth of cancer, including colon cancer, stomach cancer, esophageal cancer, lung cancer, breast cancer and skin cancer [2-10]. Inhibition of COX-2 expression by aspirin has been shown to suppress tumor growth [11-14]. COX-2 functions as an important cellular factor to regulate tumor growth via multiple molecular mechanisms [15-23]. It catalyzes the production of PGE2, which stimulates the EGFR-ERK pathway to promote tumor growth. It can also inhibit apoptosis of tumor cells by upregulating BCL-2 expression and downregulating the cleavage of caspases. In addition, COX-2 can restrain the immune system by controlling neutrophil infiltration and activation of macrophage. Although previous studies have indicated that COX-2 is highly expressed in many tumors and plays an important role in tumor growth, the precise regulatory mechanism of COX-2 in cancer cells remains unclear. The transcriptional factors such as SP1, AP-2, CBP and NF-κB have been shown to be involved in the regulation of COX-2 expression [24-28]. However, it is reasonable that besides these known transcriptional factors, there must be some other new tumor-specific transcriptional factors that can also bind to the promoter of COX-2 and regulate its expression in cancer cells to be further involved in cancer growth control. In this study, we identified BPTF (bromodomain PHD-finger transcription factor) as one of the new COX-2 promoter-binding proteins in human lung cancer cells using biotin-streptavidin agarose pulldown assay and proteomic technique.
BPTF is the largest unit of NURF (ATP-dependent nucleosome remodeling factor), which regulates chromatin remodeling. BPTF can recognize histone loci of methylation and acetylation [29,30]. Its PHD finger structural domain can specifically identify and bind H3K4me2/3, while its bromodomain can specifically bind the acetylation peptides of H4K12/16/20, thereby increasing the transcriptional activity. BPTF has been shown to promote the growth of lung cancer and melanoma [31-33]. Furthermore, BPTF is required for the transcriptional activity and in vivo tumorigenesis of c-myc [34-36]. In addition, the depletion of BPTF can enhance T-cell-mediated antitumor immunity [36-39]. In this study, we further investigated the roles of BPTF in the regulation of COX-2 expression and lung cancer cell growth, and also explored the molecular mechanism and the potential clinical significance of the BPTF/COX-2 signaling pathway in lung cancer.
Material and methods
Cell lines and cell culture
HLF (human normal lung cell line), A549 (pulmonary adenocarcinoma cell line), NCI-H460 (large cell lung carcinoma cell line), H322 (pulmonary adenocarcinoma cell line), and H1299 (non-small cell lung cancer cell line) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). HLF was cultured in DMEM medium. A549 was cultured in DMEM/F12 medium. H322, NCI-H460 and H1299 were cultured in RPMI-1640 medium. All the mediums were supplied with 10% fetal bovine serum. All cells were maintained in a humidified atmosphere and 5% CO2 at 37°C.
Streptavidin-agarose pulldown assay
The biotin-labeled double-stranded oligonucleotide probe was purchased from TAKARA Bio Inc. (Shiga, Japan), which corresponded to the -30~-508 fragment of COX-2 promoter sequence (sense: 5’-ACGTGACTTCCTCGACCCTC-3’; antisense: 5’-AAGACTGAAAACCAAGCCCA-3’). 400 μg nuclear protein, 4 μg double-stranded oligonucleotide probe, 500 μl PBS with proteinase inhibitor and 50 μl streptavidin-agarose beads solution were incubated on a rotating wheel at room temperature for 2 hours. Afterward, the DNA-protein complex was precipitated by centrifugation, and PBSI (PBS buffer with protease inhibitor) was used to wash the complex. The precipitated proteins were then denatured by boiling in 30 μl 2× SDS-PAGE loading buffer at 95°C for 5 min before analyzed by Western blot.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using the ChIP IT Express kit (Active Motif Company). Briefly, cells were treated with formaldehyde to fix protein/DNA interactions and then the fixed chromatin was sheared by sonication. The sheared chromatin was incubated with the antibody against BPTF, and antibody-bound protein/DNA complexes were pulled down by protein G-coupled magnetic beads. The immunoprecipitated chromatin was eluted after reverse crosslinking. The recovered DNA was analyzed by PCR to identify the DNA fragment of COX-2 promoter region associated with BPTF using pertinent primers (Primer 1: 5’-ACGTGACTTCCTCGACCCTC-3’, Primer 2: 5’-CAGGCGCACAGGTTTCCGCC-3’).
Luciferase assay
Cells were seeded in six-well plates and then BPTF siRNA and COX-2 promoter-driven luciferase plasmids were co-transfected into the cells by lipofectamine 3000 at the same time. 48 hours after transfection, the Luciferase Assay kit was applied to examine the luciferase expression in the cells according to the instructions.
Western blot analysis
Protein samples with proteinase/phosphatase inhibitors (Thermofisher, USA.) were diluted in SDS sample loading buffer and resolved by PAGE. 30-40 μg total protein was transferred to 0.22 μm or 0.45 μm PVDF (Millipore, Germany) overnight at 20-30 V. Membranes were blotted with antibodies of BPTF (Abcam, USA) at 1:2000, β-actin (Proteintech, USA) at 1:5000, GAPDH (Proteintech, USA) at 1:5000, overnight at 4°C, and then with HRP-conjugated secondary anti-rabbit or mouse antibodies (Proteintech, USA) at 1:5000-1:10000 dilutions for about 2 hours at room temperature in TBST. The protein bands were detected by ECL (Advansta, USA) according to manufacturer’s instructions.
RT-PCR
Total RNA was extracted and reversely transcribed into cDNA according to the instructions of RNAiso Plus kit and PrimeScript TM RT reagent Kit with gDNA Eraser (TaKaRa). The cDNA was amplified by PCR by the protocol of TaKaRa Taq TM Hot Start Version. The sequence of forward primer of BPTF was 5’-AATCGGAGAAGTCCAACGGG-3’; the sequence of reverse primer of BPTF was 5’-TTGCCCTATGTGATGCCCAG-3’. And the sequences of the primers of COX-2 were: 5’-TCACAGGCTTCATTGACCAG-3’ and 5’-CCGAGGCTTTCTACCAGA-3’. They were produced by life Technologies Company (Invitrogen, Carlsbad, USA).
Transient transfection of siRNA
siRNA was synthesized by Shanghai Genepharma Company (China). Their sequences were: nonspecific siRNA: 5’-UUCUCCGAACGUGUCACGUTT-3’ and 5’-ACGUGACACGUUCGGAGAATT-3’; BPTF-siRNA-1 (BPTF-homo-1550): 5’-GGUCCAACUUGCAGAAUUATT-3’ and 5’-UAAUUCUGCAAGUUGGACCTT-3’; BPTFsiRNA-2 (BPTF-homo-6959): 5’-GACCCAAACAACUGUUUCATT-3’ and 5’-UGAAACAGUUGUUUGGGUCTT-3’. The cells were cultured in six-well plate to 60% confluency when the mixture containing RNAiMax (Invitrogen, Carlsbad, USA) and BPTF siRNA were added. After 48 or 72 hours, cells were collected for analysis.
Immunohistochemistry (IHC) staining
The tissue microarray slides were deparaffinized in xylene and rehydrated through graded alcohol. Antigen retrieval was performed by incubating samples and with Target Retrieval Solution (pH 9; DakoCytomation) for 15 minutes using a pressure cooker. Then the slides were immersed in methanol containing 3% hydrogen peroxide for 20 minutes to block endogenous peroxidase activity. After preincubation in 2.5% blocking serum to reduce nonspecific binding, the sections were incubated overnight using primary antibody anti-BPTF (Bethyl, 1:50 dilution) or anti-COX-2 (Abcam diluted to 1:50) in a humidified container at 4°C. The tissue microarray slides were processed with horseradish peroxidase immunochemistry according to the manufacturer’s recommendations (DakoCytomation, CA). As a negative control, the staining procedure was performed with the primary antibody replaced by a normal rabbit IgG. Staining intensity was graded as: no staining as “-” for 0, weak as “+” for 1, moderate as “++” for 2, strong as “+++” for 3. Tissue staining percentage was calculated as follows: <25% positive cell staining scored for 1, 25%-50% positive cell staining for 2, 50%-75% positive cells staining for 3, and over 75% positive cells staining for 4. The total score of each sample is calculated by tissue staining intensity value multiplied by tissue staining percentage value. Therefore, the highest score is 12. Tissue protein expression was defined as high with a score >7, and low with a score ≤6. The result was analyzed independently by two senior pathologists.
Human lung adenocarcinoma tissue microarray
The human lung adenocarcinoma tissue microarray with 75 cases of lung adenocarcinoma and their adjacent non-malignant lung tissues was purchased from Shanghai Outdo Biotech (China). None of these cases got anticancer therapies before tumor resection. The detailed clinical and pathological information included age, sex, chief complaints, TNM stage, and overall survival (OS) duration.
Co-immunoprecipitation assay
The nuclear proteins were extracted. 500 μg nuclear proteins and 1-2 μg antibody were incubated respectively with a specific rabbit polyclonal antibody against BPTF or p50 or a nonspecific rabbit IgG in 500 μl ice-cold PBSi (PBS with protease inhibitor cocktail) at 4°C on a rotator for 4-6 hours, before the addition of 50 μl ProteinA/G agarose beads and further incubation at 4°C on a rotator overnight. The beads were then washed 3-5 times with PBSi buffer and suspended in 50 μl 2× SDS-PAGE loading buffer. The mixtures were boiled at 95°C for 5 min and centrifuged. The supernatant was collected and analyzed by Western blot.
Confocal immunofluorescence
The cells were seeded onto the cover slips in six-well plates and cultivated overnight. The cells were fixed by 4% paraformaldehyde for 15 minutes at room temperature, permeabilized by 0.5% Triton X-100 for 5 minutes, and blocked by 10% BSA for 30 minutes. The primary antibody against p50 (diluted to 1:50) or BPTF (diluted to 1:100) was then added and incubated overnight at 4°C. PBS was used to wash away the unbound primary antibody before the incubation with the secondary antibody conjugated with fluorescein isothiocyanate or rhodamine at room temperature for 1 hour in the dark room. After several washes with PBS, the slides were sealed by 20 μl anti-fade reagent, and the samples were observed using a Leica laser confocal microscope.
Xenograft mouse model and tumor processing
Animal experiments were conducted under the approval of APF laboratory Animal Center at Dalian Medical University according to the National Institute of Health Guide for the Care and Use of Laboratory Animal. A549 cells (5×106) were implanted subcutaneously in the left flank of nude mice. After 14 days, the tumors grew to approximately 5 mm×5 mm (length × width), and the nude mice were randomly divided into two groups with five in each group. The two groups of mice had no difference in size and tumor volumes. The first group of mice was injected with control shRNA, while the other group was injected with BPTF shRNA. The shRNA plasmids were transfected into tumors using lipofectamine 3000. 25 μg shRNA in 100 μl mixture liquid was injected once every 4 days for 28 days. The tumor volume was measured with digital calipers and calculated as volume = (width2 × length)/2. In the end, the mice were humanely sacrificed for further analysis of related proteins by Western blot.
Statistical analysis
To study the correlation between BPTF and COX-2 expression, χ2 test was used in the analysis of lung adenocarcinoma samples. The survival curves were generated by Kaplan-Meier method and analyzed by log rank test. SPSS 17.0 was used for statistical analysis. P<0.05 was considered to be statistically significant.
Results
BPTF was identified as a COX-2 promoter binding protein
In order to discover new specific regulatory factors of COX-2 transcription in lung cancer cells, streptavidin/biotin pull-down assay and mass spectrometry were used to identify the COX-2 promoter binding proteins in lung cancer cells compared with human lung fibroblast (HLF). As shown in Figure 1A, we detected a specific protein band with a molecular weight of about 300 kDa. Candidate targets were excised from gels and subjected to protein identification by undertaking the in-gel digestion approach and using mass spectrometric and proteomic tools. The identification result indicated that the protein band with the molecular weight of approximately 300 kDa is BPTF. We next verified BPTF as a COX-2 promoter-binding protein by immunoblot assay in the complex of the binding nuclear proteins pulled down by streptavidin-agarose beads coated by the biotin-labeled COX-2 promoter probe (Figure 1B). High level of BPTF protein was detected in the complexes pulled down from A549, H322, NCI-H460 and H1299 cell lines, whereas nearly no BPTF protein was found in the complexes from HLF cells. ChIP assay also validated that BPTF could bind to COX-2 promoter in lung cancer cells specifically (Figure 1C). These results show that BPTF can bind to COX-2 promoter in lung cancer cells and might further affect the expression of COX-2.
Figure 1.
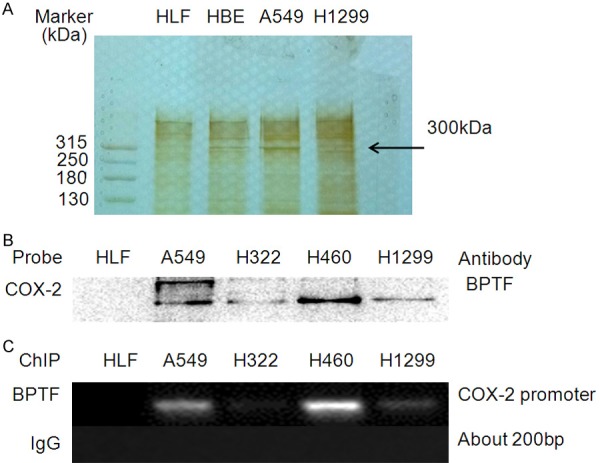
BPTF was identified as a COX-2 promoter binding protein. A. The COX-2 promoter-binding proteins were pulled down, separated by the SDS-PAGE, and stained with Coomassie Brilliant Blue. A tumor specific protein of ~300 kDa was indicated by arrow. B. The biotinylated COX-2 promoter probe was used to pull down its binding proteins. BPTF was identified to be one of the proteins by Western blot. C. Chromatin immunoprecipitation assay of COX-2 promoter in lung cancer cells.
BPTF regulated COX-2 expression transcriptionally
To consolidate the effect of BPTF on COX-2 promoter activity in lung cancer cells, we investigated the effect of BPTF on COX-2 promoter-driven luciferase and found that BPTF knockdown significantly inhibited COX-2 promoter driven luciferase expression, verifying the direct transcriptional activation of COX-2 by BPTF via anchoring at its promoter as transcriptional factor (Figure 2A, 2B). Next, we detected that BPTF knockdown considerably inhibited the expression of COX-2 at both mRNA (Figure 2C) and protein levels (Figure 2D). In addition, we validated the regulation of COX-2 expression by BPTF in a lung cancer mouse model in vivo. Ten nude mice were injected with A549 cells and the mice were randomly divided into 2 groups (BPTF shRNA group and control shRNA control) when the tumors grew to 5 mm×5 mm (length × width) after 2 weeks. The shRNAs were injected into the tumor tissue of the nude mice every 4 days. After 28 days, the nude mice were sacrificed and the levels of BPTF and COX-2 proteins in tumor tissues were detected by Western blot. As shown in Figure 2E, BPTF knockdown dramatically inhibited COX-2 expression in the mice treated with BPTF shRNA compared to the mice treated with the control shRNA. We also quantitatively analyzed the COX-2 levels in the tumor tissues of the mice, and the expression of COX-2 was significantly lower in the sh-BPTF group (Figure 2F). Together, BPTF could directly regulate COX-2 expression by binding to its promoter in lung cancer cells.
Figure 2.
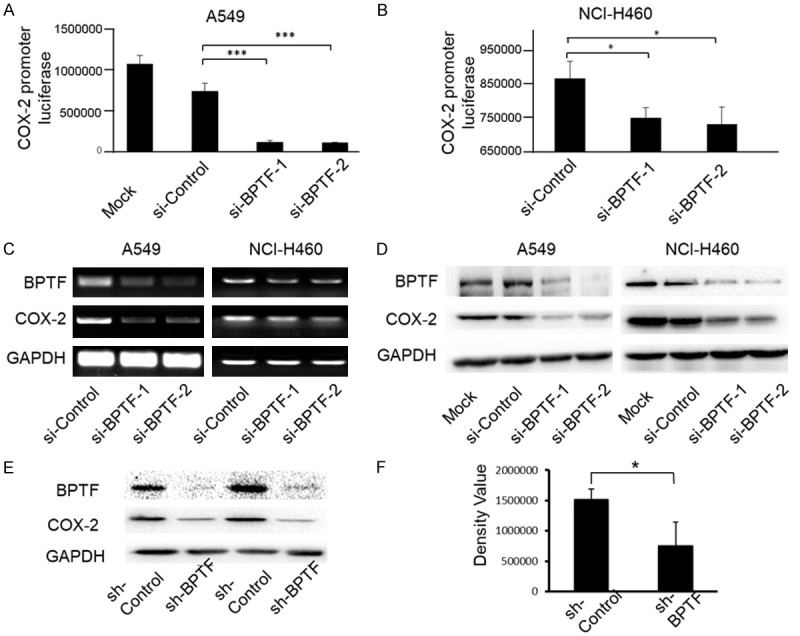
BPTF knockdown inhibited COX-2 promoter activity and COX-2 expression. A. A549 cells were co-transfected with BPTF siRNA and COX-2 promoter driven luciferase plasmid for 48 hours. Then the luciferase expression was analyzed by a microplate reader (***P<0.001). B. NCI-H460 cells were also co-transfected with BPTF siRNA and COX-2 promoter driven luciferase plasmid, and then analyzed by the microplate reader (*P<0.1). C. A549 and NCI-H460 cells were transfected with BPTF specific siRNA and the mRNA expression of BPTF and COX-2 was analyzed by RT-PCR. D. BPTF was knocked down by BPTF siRNA, and the protein expression of BPTF and COX-2 was detected by Western blot in A549 and NCI-H460 cells. E. Ten nude mice were injected with A549 cells and tumors grew to 5 mm×5 mm (length × width) after 2 weeks. Nude mice were randomly divided into 2 groups. BPTF shRNA and control shRNA were respectively injected into the tumors once every 4 days. After 28 days, the mice were sacrificed to examine the expression of BPTF and COX-2. Representative results from two pairs of nude mice were shown. F. Image Lab software was used to analyze the protein bands of BPTF and COX-2 in the two groups of nude mice. Bars represent the mean (*P<0.05).
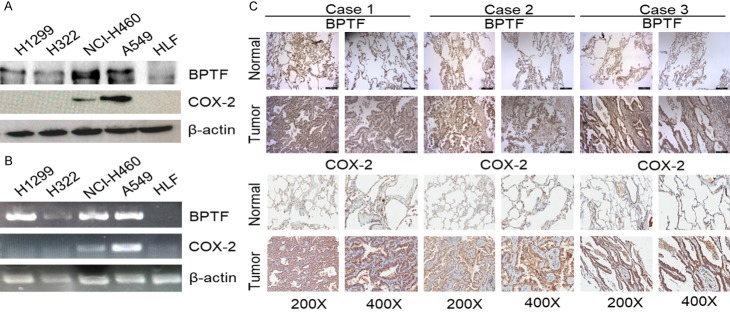
BPTF and COX-2 were highly expressed and positively correlated in lung tumor cells and human lung adenocarcinoma tissue samples
Firstly, we examined the expression of BPTF and COX-2 in NSCLC cell lines by Western blot and RT-PCR, and found that BPTF and COX-2 were both highly expressed in A549 and NCI-H460 lung cancer cell lines but not in normal HLF cells (Figure 3A, 3B). Moreover, to further confirm the regulation of COX-2 expression by BPTF, we analyzed their expression relationship in the tissue samples of patients with lung adenocarcinoma. A microarray with 75 paired lung adenocarcinoma tissues and their adjacent normal lung tissues was used, and the expression of BPTF and COX-2 was analyzed by immunohistochemistry. The result revealed that both BPTF and COX-2 had increased expression in tumor tissues (Figure 3C). Consistently, the summary of the clinicopathological features in Table 1 indicated that simultaneously high expression of BPTF and COX-2 was closely related with the metastasis to the lymph node, but not with age, sex or tumor volume. Moreover, among the 75 patients, 35 showed high expression of BPTF and COX-2, whereas 17 showed low expression of both BPTF and COX-2 (Figure 4A). The Pearson’s correlation coefficient between BPTF and COX-2 was 0.371 (P=0.001, χ2 test) (Figure 4B). In addition, we determined the relationship between the overall survival of the 75 patients and their BPTF/COX-2 expression by Kaplan-Meier analysis. The result demonstrated that patients with low expression of BPTF and COX-2 live longer than those with high expression (Figure 4C). Collectively, the above results suggested that BPTF and COX-2 were positively correlated in human lung adenocarcinoma tissues.
Figure 3.
BPTF and COX-2 were highly expressed in lung adenocarcinoma tissues and correlated with clinicopathological features. A. BPTF and COX-2 expression was detected in human normal lung cell line and NSCLC cell lines by Western blot. B. The expression of BPTF and COX-2 was analyzed by RT-PCR in normal lung cell line and NSCLC cell lines. C. IHC staining of BPTF and COX-2 was carried out in the tissue microarray of lung adenocarcinoma and adjacent non-malignant lung tissues.
Table 1.
Case material
| Total (n=53) | BPTF and COX-2 high expression (n=36) | BPTF and COX-2 low expression (n=17) | P | |
|---|---|---|---|---|
| Male | 29 | 18 (33.96%) | 11 (20.75%) | 0.24 |
| Female | 24 | 18 (33.96%) | 6 (11.32%) | |
| Age <60 y | 28 | 19 (35.85%) | 9 (16.98%) | 0.58 |
| Age ≥60 y | 25 | 17 (32.08%) | 8 (15.09%) | |
| T1+T2 | 46 | 30 (56.60%) | 16 (30.19%) | 0.269 |
| T3+T4 | 7 | 6 (11.32%) | 1 (1.89%) | |
| N0+N1 | 41 | 22 (71%) | 19 (30%) | 0.05 |
| N2+N3 | 6 | 6 (11.32%) | 0 (0%) | |
| Nx | 6 | 2 (3.77%) | 4 (7.55%) |
The association of BPTF and COX-2 expression with the clinicopathological features was analyzed in human lung adenocarcinoma samples.
Figure 4.
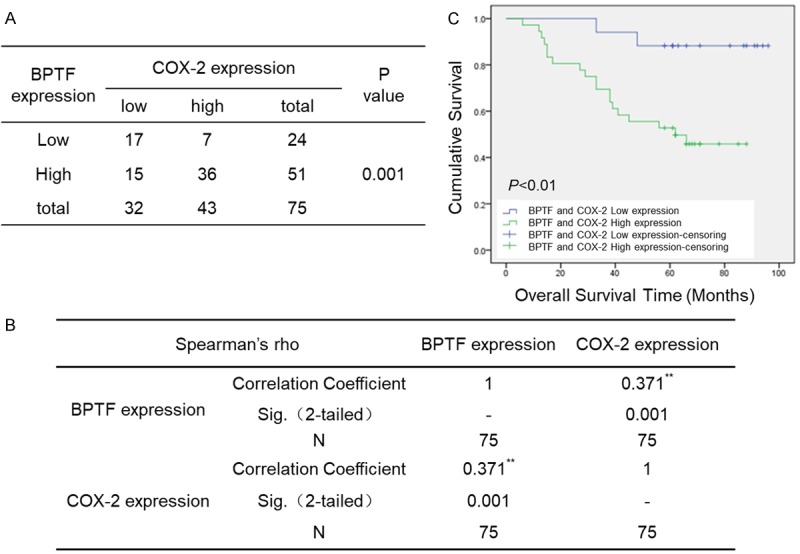
BPTF and COX-2 expression was positively correlated and highly co-expression of BPTF and COX-2 predicted poor prognosis in lung adenocarcinoma patients. A. The correlation between the protein expression of BPTF and COX-2 in lung adenocarcinoma tissues (P=0.001, χ2 test). B. The correlation coefficient of BPTF and COX-2 is 0.371. C. Kaplan-Meier analysis of overall survival with high and low expression of both BPTF and COX-2.
BPTF interacted with P50 to co-regulate COX-2 expression and lung cancer cell growth
It is well known that NF-κB functionalizes as the classic regulatory factors to control regulates COX-2 expression. Thus, we proposed that BPTF might interact with NF-κB p50/p65 to co-regulated COX-2 expression. To obtain insights into the role of p50/p65 in BPTF-mediated COX-2 expression regulation in lung cancer cells, we applied immunoprecipitation assay first to detect the possible interaction between BPTF and p50/p65. The result showed that p50 was one of the potential factors precipitated by BPTF antibody (Figure 5A), and reversibly, BPTF was also precipitated by p50 antibody (Figure 5B). We then examined the co-localization of BPTF and p50 in A549 and NCI-H460 cell lines by immunofluorescence analysis. As shown in Figure 5C, BPTF was mainly located in the nuclei of A549 and H460 cells, while p50 was mainly located in the nuclei of A549 cells, and in the cytoplasm of NCI-H460 cells. However, the distribution of these two proteins in the nuclei was almost identical. Additionally, we knocked down BPTF in A549 and NCI-H460 cells, and found that the binding of p50 to COX-2 promoter was also down-regulated accordingly (Figure 5D, 5E). Next, we used QNZ, an inhibitor of the transcription factor NF-κB, to treat lung cancer cells. The decreased expression of COX-2 was dramatically observed following QNZ treatment (Figure 5F). Moreover, BPTF knockdown could not further down-regulate COX-2 expression upon QNZ treatment. Similarly, BPTF silencing statistically led to significant suppression of proliferative capacity, whereas BPTF down-regulation could not further inhibit cell viability under the influence of QNZ (Figure 5G), indicating that NF-κB p50/p65 was pivotal for BPTF to regulate the expression of COX-2 and cancer cell proliferation. These results suggested that BPTF and p50 were co-localized in the nuclei of cells and interacted to co-regulate COX-2 expression in lung cancer cells.
Figure 5.
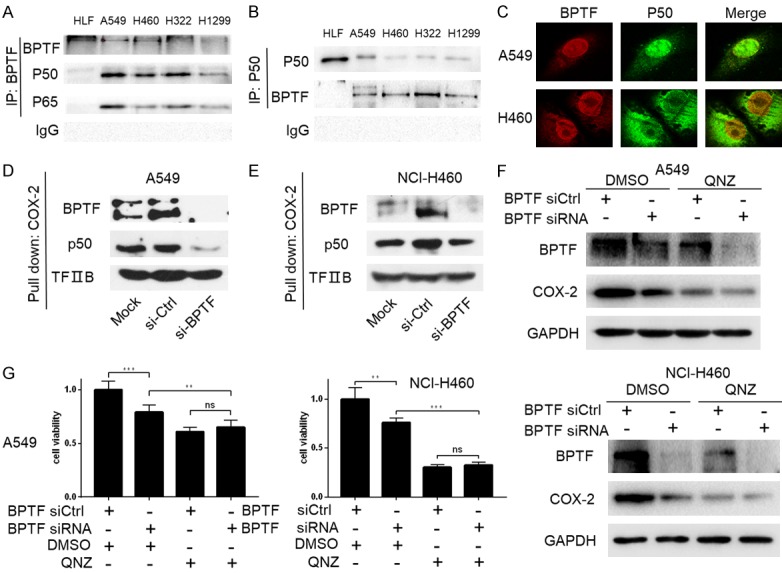
BPTF interacted with p50 to regulate COX-2 expression. A. Co-immunoprecipitation was performed to analyze the interaction between BPTF and p50/p65. BPTF antibody was used to precipitate p50/p65. B. P50 antibody was used to immunoprecipitate BPTF. C. The co-localization of BPTF and COX-2 was analyzed by Laser confocal microscope in A549 and NCI-H460 cells. D. BPFT was knocked down in A549. The nuclear proteins were pulled down by biotin-labeled COX-2 promoter probe and streptavidin agarose beads, and p50 was detected by Western blot. E. The NCI-H460 cells were transfected with BPTF siRNA. The biotin-streptavidin pulldown assay was used to evaluate the binding of p50 at the COX-2 promoter region. Normal IgG was used as a negative control. F. A549 and NCI-H460 cells transfected with BPTF specific siRNA or control siRNA for 48 hours were treated with or without QNZ (10 µM), and then BPTF and COX-2 expression were detected by western blot. G. A549 and NCI-H460 cells transfected with BPTF specific siRNA or control siRNA were treated with or without QNZ (10 µM), and then the cell viability was determined by MTT assay. (Error bars represent mean ± SEM. **P<0.01; ***P<0.001; ns, not significant).
Discussion
COX-2, an inducible cyclooxygenase, can promote the occurrence and development of colon cancer, stomach cancer, esophageal cancer, lung cancer, breast cancer, skin cancer and so on [6-14]. In vitro studies showed that tumor was dampened when COX-2 was suppressed by RNA interference or drug intervention. Recently, accumulating researches have focused on the discovery of transcriptional factors that regulate COX-2 expression, such as SP1, AP-2, CPSF4, CBP/P300 and NF-κB [24-28]. To discover novel tumor specific transcriptional factors of COX-2, we used streptavidin-agarose pulldown assay coupled with mass spectrometry, and found that BPTF was one of the transcriptional factors of COX-2 in lung cancers. It was found to specifically bind to COX-2 promoter region and regulated COX-2 promoter activity and its expression in vitro and in vivo. Notably, consistent with the expression of COX-2, BPTF was found to be lowly expressed in normal lung cells and tissues, but highly expressed in lung cancer cells and tissues. Furthermore, patients of lung adenocarcinomas with both high expression of BPTF and COX-2 embraced more metastasis to the lymph node and shorter overall survival, implying again the transcriptional regulation of COX-2 by BPTF in lung cancer and their significance in clinical diagnosis and therapy.
As the largest subunit of NURF that participates in chromatin remodeling, BPTF has two interesting structural components, the PHD finger and the bromodomain. The PHD finger domain can recognize and specifically bind dimethyl and trimethyl H3K4, while the adjacent bromodomain can mediate the acetylation of H4K12/16/20. In vivo, BPTF is enriched at H3K4me and H4K16ac [29,30]. CBP (CREB-binding protein) is a highly conservative transcriptional coactivator, which has a similar structure to p300. Both CBP and p300 can recruit various transcriptional factors to chromatin to regulate their binding to the promoter regions of different genes. Moreover, CBP has acetyl transferase activity for histone acetylation, and thus can increase chromatin accessibility, allowing transcriptional factors to bind to open promoters [38-40]. Interestingly, CBP also has PHD finger and bromodomain structures. Previous studies reported that BPTF was essential to the c-MYC-induced remodeling of target chromatin, and its silencing dampened the c-MYC transcriptional program [34,35] and/or modulate the activity of histone acetyltransferases (HATs) or histone deacetylases at gene promoters. A similar regulation was found in Drosophila melanogaster, where Nurf301 was required for the histone acetyl-transferase ATAC to access chromatin and maintain the decondensed architecture of the male X chromosome [41,42]. Accordingly, in this study, we have demonstrated a novel molecular mechanism by which BPTF regulates COX-2 and tumor growth via recruiting and interacting with NF-κB p50. BPTF silencing significantly suppressed the binding of p50 at COX-2 promoter region, illustrating the necessity of BPTF in the NF-κB-induced remodeling of target chromatin. Meanwhile, the activity inhibition of NF-κB not only remarkably suppressed COX-2 expression and cancer cell growth, but also abolished the significance of COX-2 expression decrease and cancer cell proliferation inhibition mediated by BPTF knockdown, from another perspective suggesting the important role of NF-κB activity in BPTF’s pro-oncogenic role in lung cancer progression.
Conclusion
BPTF cooperated with p50 NF-κB to regulate COX-2 expression and lung cancer growth. BPTF and COX-2 expression was positively correlated and highly co-expression of BPTF and COX-2 predicted poor prognosis in lung adenocarcinoma patients. These results indicated that the BPTF/p50/COX-2 axis might be a potential therapeutic target for lung cancer.
Acknowledgements
This work was supported by the funds National Natural Science Foundation of China (81572706).
Disclosure of conflict of interest
None.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Rahman M, Selvarajan K, Hasan MR, Chan AP, Jin C, Kim J, Chan SK, Le ND, Kim YB, Tai IT. Inhibition of COX-2 in colon cancer modulates tumor growth and MDR-1 expression to enhance tumor regression in therapy-refractory cancers in vivo. Neoplasia. 2012;14:624–633. doi: 10.1593/neo.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xin X, Majumder M, Girish GV, Mohindra V, Maruyama T, Lala PK. Targeting COX-2 and EP4 to control tumor growth, angiogenesis, lymphangiogenesis and metastasis to the lungs and lymph nodes in a breast cancer model. Lab Invest. 2012;92:1115–1128. doi: 10.1038/labinvest.2012.90. [DOI] [PubMed] [Google Scholar]
- 4.Axelsson H, Lönnroth C, Andersson M, Lundholm K. Mechanisms behind COX-1 and COX-2 inhibition of tumor growth in vivo. Int J Oncol. 2010;37:1143–1152. doi: 10.3892/ijo_00000766. [DOI] [PubMed] [Google Scholar]
- 5.Chan MW, Wong CY, Cheng AS, Chan VY, Chan KK, To KF, Chan FK, Sung JJ, Leung WK. Targeted inhibition of COX-2 expression by RNA interference suppresses tumor growth and potentiates chemosensitivity to cisplatin in human gastric cancer cells. Oncol Rep. 2007;18:1557–1562. [PubMed] [Google Scholar]
- 6.Yoshinaka R, Shibata MA, Morimoto J, Tanigawa N, Otsuki Y. COX-2 inhibitor celecoxib suppresses tumor growth and lung metastasis of a murine mammary cancer. Anticancer Res. 2006;26:4245–4254. [PubMed] [Google Scholar]
- 7.Qadri SS, Wang JH, Coffey JC, Alam M, O’Donnell A, Aherne T, Redmond HP. Surgically induced accelerated local and distant tumor growth is significantly attenuated by selective COX-2 inhibition. Ann Thorac Surg. 2005;79:990–995. doi: 10.1016/j.athoracsur.2004.07.042. discussion 990-995. [DOI] [PubMed] [Google Scholar]
- 8.Pun IH, Chan D, Chan SH, Chung PY, Zhou YY, Law S, Lam AK, Chui CH, Chan AS, Lam KH, Tang JC. Anti-cancer effects of a novel quinoline derivative 83b1 on human esophageal squamous cell carcinoma through down-regulation of COX-2 mRNA and PGE2. Cancer Res Treat. 2017;49:219–229. doi: 10.4143/crt.2016.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu HP, Shi LY, Lu WH, Su YH, Li YY, Xu SQ. Expression of cyclooxygenase-2 (COX-2) in human esophageal cancer and in vitro inhibition by a specific COX-2 inhibitor, NS-398. J Gastroenterol Hepatol. 2004;19:638–642. doi: 10.1111/j.1440-1746.2004.03345.x. [DOI] [PubMed] [Google Scholar]
- 10.Altorki N. COX-2: a target for prevention and treatment of esophageal cancer. J Surg Res. 2004;117:114–120. doi: 10.1016/j.jss.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Lee M, Gibbs P. RE: aspirin and COX-2 inhibitor use in patients with stage III colon cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nath N, Vassell R, Chattopadhyay M, Kogan M, Kashfi K. Nitro-aspirin inhibits MCF-7 breast cancer cell growth: effects on COX-2 expression and Wnt/beta-catenin/TCF-4 signaling. Biochem Pharmacol. 2009;78:1298–1304. doi: 10.1016/j.bcp.2009.06.104. [DOI] [PubMed] [Google Scholar]
- 13.Murashige N, Kami M, Ikeda M. Aspirin, COX-2, and the risk of colorectal cancer. N Engl J Med. 2007;357:824–825. author reply 824-825. [PubMed] [Google Scholar]
- 14.Kune GA. Colorectal cancer chemoprevention: aspirin, other NSAID and COX-2 inhibitors. Aust N Z J Surg. 2000;70:452–455. doi: 10.1046/j.1440-1622.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang SH, Wang SC, Chen PC, Wang ST, Liu YW. Induction of cyclooxygenase-2 gene by Candida albicans through EGFR, ERK, and p38 pathways in human urinary epithelium. Med Mycol. 2017;55:314–322. doi: 10.1093/mmy/myw082. [DOI] [PubMed] [Google Scholar]
- 16.McElroy SJ, Hobbs S, Kallen M, Tejera N, Rosen MJ, Grishin A, Matta P, Schneider C, Upperman J, Ford H, Polk DB, Weitkamp JH. Transactivation of EGFR by LPS induces COX-2 expression in enterocytes. PLoS One. 2012;7:e38373. doi: 10.1371/journal.pone.0038373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirota CL, Moreau F, Iablokov V, Dicay M, Renaux B, Hollenberg MD, MacNaughton WK. Epidermal growth factor receptor transactivation is required for proteinase-activated receptor-2-induced COX-2 expression in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2012;303:G111–119. doi: 10.1152/ajpgi.00358.2011. [DOI] [PubMed] [Google Scholar]
- 18.Melnick M, Abichaker G, Htet K, Sedghizadeh P, Jaskoll T. Small molecule inhibitors of the host cell COX/AREG/EGFR/ERK pathway attenuate cytomegalovirus-induced pathogenesis. Exp Mol Pathol. 2011;91:400–410. doi: 10.1016/j.yexmp.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YM, Park SY, Pyo H. Cyclooxygenase-2 (COX-2) negatively regulates expression of epidermal growth factor receptor and causes resistance to gefitinib in COX-2-overexpressing cancer cells. Mol Cancer Res. 2009;7:1367–1377. doi: 10.1158/1541-7786.MCR-09-0004. [DOI] [PubMed] [Google Scholar]
- 20.Menon DB, Gopalakrishnan VK. Terpenoids isolated from the shoot of plectranthus hadiensis induces apoptosis in human colon cancer cells via the mitochondria-dependent pathway. Nutr Cancer. 2015;67:697–705. doi: 10.1080/01635581.2015.1019631. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai T, Tamura K, Kogo H. Stimulatory effects of eicosanoids on ovarian angiogenesis in early luteal phase in cyclooxygenase-2 inhibitor-treated rats. Eur J Pharmacol. 2005;516:158–164. doi: 10.1016/j.ejphar.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Hamada T, Tsuchihashi S, Avanesyan A, Duarte S, Moore C, Busuttil RW, Coito AJ. Cyclooxygenase-2 deficiency enhances Th2 immune responses and impairs neutrophil recruitment in hepatic ischemia/reperfusion injury. J Immunol. 2008;180:1843–1853. doi: 10.4049/jimmunol.180.3.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranganathan PV, Jayakumar C, Mohamed R, Dong Z, Ramesh G. Netrin-1 regulates the inflammatory response of neutrophils and macrophages, and suppresses ischemic acute kidney injury by inhibiting COX-2-mediated PGE2 production. Kidney Int. 2013;83:1087–1098. doi: 10.1038/ki.2012.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi SY, Hwang JH, Park SY, Jin YJ, Ko HC, Moon SW, Kim SJ. Fermented guava leaf extract inhibits LPS-induced COX-2 and iNOS expression in Mouse macrophage cells by inhibition of transcription factor NF-kappaB. Phytother Res. 2008;22:1030–1034. doi: 10.1002/ptr.2419. [DOI] [PubMed] [Google Scholar]
- 25.Yang CM, Chen YW, Chi PL, Lin CC, Hsiao LD. Resveratrol inhibits BK-induced COX-2 transcription by suppressing acetylation of AP-1 and NF-kappaB in human rheumatoid arthritis synovial fibroblasts. Biochem Pharmacol. 2017;132:77–91. doi: 10.1016/j.bcp.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Wang J, Qin Y, Xuan Y, Jia Y, Hu W, Yu W, Dai M, Li Z, Yi C, Zhao S, Li M, Du S, Cheng W, Xiao X, Chen Y, Wu T, Meng S, Yuan Y, Liu Q, Huang W, Guo W, Wang S, Deng W. Ku80 cooperates with CBP to promote COX-2 expression and tumor growth. Oncotarget. 2015;6:8046–8061. doi: 10.18632/oncotarget.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konstantinopoulos PA, Vandoros GP, Sotiropoulou-Bonikou G, Kominea A, Papavassiliou AG. NF-kappaB/PPAR gamma and/or AP-1/PPAR gamma ‘on/off’ switches and induction of CBP in colon adenocarcinomas: correlation with COX-2 expression. Int J Colorectal Dis. 2007;22:57–68. doi: 10.1007/s00384-006-0112-y. [DOI] [PubMed] [Google Scholar]
- 28.Fu L, Chen W, Guo W, Wang J, Tian Y, Shi D, Zhang X, Qiu H, Xiao X, Kang T, Huang W, Wang S, Deng W. Berberine targets AP-2/hTERT, NF-kappaB/COX-2, HIF-1alpha/VEGF and cytochrome-c/caspase signaling to suppress human cancer cell growth. PLoS One. 2013;8:e69240. doi: 10.1371/journal.pone.0069240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai M, Lu JJ, Guo W, Yu W, Wang Q, Tang R, Tang Z, Xiao Y, Li Z, Sun W, Sun X, Qin Y, Huang W, Deng WG, Wu T. BPTF promotes tumor growth and predicts poor prognosis in lung adenocarcinomas. Oncotarget. 2015;6:33878–33892. doi: 10.18632/oncotarget.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dar AA, Nosrati M, Bezrookove V, de Semir D, Majid S, Thummala S, Sun V, Tong S, Leong SP, Minor D, Billings PR, Soroceanu L, Debs R, Miller JR 3rd, Sagebiel RW, Kashani-Sabet M. The role of BPTF in melanoma progression and in response to BRAF-targeted therapy. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dar AA, Majid S, Bezrookove V, Phan B, Ursu S, Nosrati M, De Semir D, Sagebiel RW, Miller JR 3rd, Debs R, Cleaver JE, Kashani-Sabet M. BPTF transduces MITF-driven prosurvival signals in melanoma cells. Proc Natl Acad Sci U S A. 2016;113:6254–6258. doi: 10.1073/pnas.1606027113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richart L, Carrillo-de Santa Pau E, Río-Machín A, de Andrés MP, Cigudosa JC, Lobo VJS, Real FX. BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat Commun. 2016;7:10153. doi: 10.1038/ncomms10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richart L, Real FX, Sanchez-Arevalo Lobo VJ. c-MYC partners with BPTF in human cancer. Mol Cell Oncol. 2016;3:e1152346. doi: 10.1080/23723556.2016.1152346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayes K, Alkhatib SG, Peterson K, Alhazmi A, Song C, Chan V, Blevins T, Roberts M, Dumur CI, Wang XY, Landry JW. BPTF depletion enhances T-cell-mediated antitumor immunity. Cancer Res. 2016;76:6183–6192. doi: 10.1158/0008-5472.CAN-15-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu B, Wang Y, Wang C, Wang GG, Wu J, Wan YY. BPTF is essential for T cell homeostasis and function. J Immunol. 2016;197:4325–4333. doi: 10.4049/jimmunol.1600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheikh BN. Crafting the brain - role of histone acetyltransferases in neural development and disease. Cell Tissue Res. 2014;356:553–573. doi: 10.1007/s00441-014-1835-7. [DOI] [PubMed] [Google Scholar]
- 39.Mannervik M. Control of Drosophila embryo patterning by transcriptional co-regulators. Exp Cell Res. 2014;321:47–57. doi: 10.1016/j.yexcr.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 41.Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The drosophila nucleosome remodeling factor NURF is required for ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon SY, Xiao H, Glover BP, Tjian R, Wu C, Badenhorst P. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]