Abstract
Introduction
Nephrotic syndrome (NS) is a kidney disease that affects both children and adults. Glucocorticoids have been the primary therapy for >60 years but are ineffective in approximately 20% of children and approximately 50% of adult patients. Unfortunately, patients with steroid-resistant NS (SRNS; vs. steroid-sensitive NS [SSNS]) are at high risk for both glucocorticoid-induced side effects and disease progression.
Methods
We performed proton nuclear magnetic resonance (1H NMR) metabolomic analyses on plasma samples (n = 86) from 45 patients with NS (30 SSNS and 15 SRNS) obtained at initial disease presentation before glucocorticoid initiation and after approximately 7 weeks of glucocorticoid therapy to identify candidate biomarkers able to either predict SRNS before treatment or define critical molecular pathways/targets regulating steroid resistance.
Results
Stepwise logistic regression models identified creatinine concentration and glutamine concentration (odds ratio [OR]: 1.01; 95% confidence interval [CI]: 0.99–1.02) as 2 candidate biomarkers predictive of SRNS, and malonate concentration (OR: 0.94; 95% CI: 0.89–1.00) as a third candidate predictive biomarker using a similar model (only in children >3 years). In addition, paired-sample analyses identified several candidate biomarkers with the potential to identify mechanistic molecular pathways/targets that regulate clinical steroid resistance, including lipoproteins, adipate, pyruvate, creatine, glucose, tyrosine, valine, glutamine, and sn-glycero-3-phosphcholine.
Conclusion
Metabolomic analyses of serial plasma samples from children with SSNS and SRNS identified elevated creatinine and glutamine concentrations, and reduced malonate concentrations, as auspicious candidate biomarkers to predict SRNS at disease onset in pediatric NS, as well as additional candidate biomarkers with the potential to identify mechanistic molecular pathways that may regulate clinical steroid resistance.
Keywords: biomarkers, metabolomics, nephrotic syndrome, steroid resistance
See Commentary on Page 1
Glomerular disease is the third leading cause of end-stage kidney disease in the United States and its related health care costs are approximately $4.1 billion annually. Immunosuppressive drugs are the primary therapies for NS, as well as most glomerular diseases, although approximately 20% to 50% of patients with NS fail to achieve clinical remission in response to glucocorticoid (GC) therapy. Unfortunately, in the absence of biomarkers to predict treatment responsiveness, many patients receive prolonged yet ineffective GC therapy, leaving them at high risk for both toxic side effects and disease progression. However, we still do not have an adequate understanding of the prognostic factors and specific molecular pathways that are the most critical regulators of NS.
Potential strategies to develop biomarkers able to predict clinical steroid responsiveness in glomerular disease include the use of either directed approaches using gene arrays (gene expression), enzyme-linked immunosorbent assay testing (protein expression), or unbiased methodologies, such as proteomics, metabolomics, or transcriptomics.1, 2, 3, 4, 5, 6 Because the kidney is the major organ regulating the composition of urine and blood, kidney diseases are uniquely suited to be studied by metabolomics. Indeed, some of the earliest metabolic profiling studies were undertaken to identify uremic toxins,7 and metabolomics has since been widely applied to kidney diseases.8, 9, 10, 11, 12 However, only a limited number of studies have focused on glomerular disease and/or NS,1,2,13, 14, 15, 16, 17, 18 and these studies have focused primarily on urinary metabolomic profiling of cases and controls, or disease severity in adults.
NS is a kidney disease that affects both children and adults. The annual incidence in children is 2 to 7 new cases, and the prevalence is 16 cases per 100,000 children.19, 20, 21 Although GC treatment has been the mainstay of therapy for >60 years, approximately 20% of children and approximately 50% of adults with NS fail to enter complete remission after 6 to 8 weeks of oral GC therapy, at which point children are then referred to as having SRNS. This distinction is critical, because patients who present with or develop SRNS have a dramatically higher risk (approximately 50%) for developing progressive kidney disease or end-stage kidney disease within the following 5 years,22 unlike patients who enter complete remission following GC treatment (SSNS) and are at low risk.
Unfortunately, no validated biomarkers have yet been identified that are reliably able to predict SRNS, leaving many patients at high risk for both toxic side effects of GC treatment, as well as disease progression. Thus, the identification and validation of biomarkers able to predict the clinical response to GC at disease onset could avoid significant GC-induced drug toxicity by enabling physicians to more rapidly initiate alternative treatments more likely to induce disease remission and delay or prevent disease progression. In addition, the identification of specific molecular pathways and targets responsible for NS and/or GC resistance could enable the development of more effective and less toxic targeted future therapies for NS.
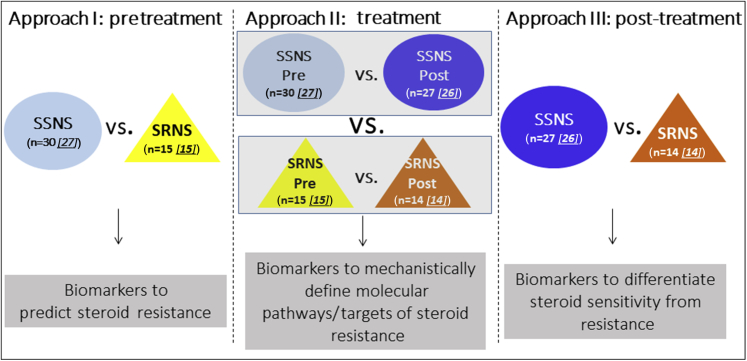
The present studies were designed to test the hypotheses that metabolomic analyses of paired plasma samples from children with SSNS and SRNS can be used to identify biomarkers able to (i) predict clinical steroid resistance, (ii) define specific molecular pathways or targets mechanistically associated with clinical steroid resistance, and (iii) differentiate steroid sensitivity from steroid resistance in NS (see Figure 1). To test these hypotheses, we analyzed paired plasma biosamples collected from 2008 to 2014 through the Midwest Pediatric Nephrology Consortium from children with NS that were obtained both at the time of disease presentation (before initiation of GC therapy) and after an average of approximately 7 weeks of GC treatment, when the clinical determination of SRNS versus SSNS was made by the treating nephrologist.
Figure 1.
Study hypothesis and design. The present studies were designed to test the hypothesis that metabolomic analyses of paired plasma samples from children with steroid-sensitive nephrotic syndrome (SSNS) and steroid-resistant nephrotic syndrome (SRNS) can be used to identify biomarkers able to (approach I) predict clinical steroid resistance, (approach II) define specific mechanistic molecular pathways or targets associated with clinical steroid resistance, and (approach III) differentiate clinical steroid sensitivity from steroid resistance. SSNS Pre, light blue circle; SRNS Pre, yellow triangle; SSNS Post, dark blue circle; SRNS Post, orange triangle. Starting patient samples: n = 86 (SSNS Pre = 30, SSNS Post = 27, SRNS Pre = 15, SRNS Post = 14). Approach I: Thirty SSNS patient samples were subjected to 1H NMR and 27 were analyzed after removing 2 outliers and 1 steroid-dependent [SD] patient; 15 SRNS patient samples were subjected to HNMR and were analyzed. Approach II: Twenty-seven SSNS pre-patient samples were analyzed after removing 2 outliers and 1 SD patient; 26 SSNS post-patient samples were analyzed after removing 1 SD patient; 24 sample pairs were analyzed for relative concentration data; 15 SRNS pre-patient samples were analyzed; 14 SRNS post-patient samples were analyzed; and 14 pairs were analyzed for relative concentration data. Approach III: Twenty-six SSNS post-patient samples were analyzed after removing 1 SD patient; 14 SRNS post-patient samples were analyzed.
Methods
Study Approval and Ethics Statement
All research protocols and consent documents were approved by the institutional review board of Nationwide Children's Hospital as the coordinating center (approval numbers IRB07–00400, IRB12–00039 and IRB05–00544), as well as by each of the other participating centers of the Midwest Pediatric Nephrology Consortium. Informed written consent (and assent, where appropriate) was obtained from the parents of all participants before samples were collected, in accordance with the Declaration of Helsinki.
Pediatric Patients With NS and Plasma Collection
Pediatric patients with NS aged between 18 months and 18 years were included in this study if they exhibited 3+ proteinuria and edema. The clinical response of each patient to GC (i.e., SRNS or SSNS) was assessed approximately 6 to 10 weeks after initial presentation and is also shown in Table 1. Fifty percent of the patients enrolled were excluded from this study because of our inability to confirm with certainty that they had not received even a single dose of GC therapy before beginning the study, or before the first (pre-steroid) sample collection. Paired plasma samples were collected for each patient, first sample “pre” at the time of disease presentation before even a single dose of GC, and second sample “post” after 6 to 10 weeks of GC therapy. There were 4 patients (3 SSNS and 1 SRNS) who did not have the second sample collected. Broad-spectrum NMR data were acquired for 86 patient samples and the distribution of sample sizes is described in detail in Figure 1 and provided in figure legends and tables. One patient was identified as steroid-dependent and was excluded from all further analyses. Please see Supplementary Material for additional details.
Table 1.
Demographic of patients with nephrotic syndrome
| Patient attributes | Steroid-resistant (n = 15) |
Steroid-sensitive (n = 30) |
P |
|---|---|---|---|
| Mean (SD) or count (%) | Mean (SD) or count (%) | ||
| Male | 5 (33.3%) | 13 (43.3%) | 0.57 |
| Female | 9 (60%) | 16 (53.3%) | |
| Unknown | 1 (6.7%) | 1 (3.3%) | |
| White | 6 (40%) | 13 (43.3%) | 0.28 |
| Black/African American | 7 (46.7%) | 7 (23.3%) | |
| Othera | 0 (0%) | 7 (23.3%) | |
| Unknownb | 2 (13.3%) | 3 (10%) | |
| Body mass index percentile | 88.7 (12.0) | 85.3 (20.4) | 0.50 |
| Subjects with nonmissing data | 13 | 27 | |
| Age (yr) | 9.5 (3.8) | 5.8 (3.7) | 0.0006 |
| Subjects with nonmissing data | 14 | 28 | |
| Weeks to post-treatment sample | 7.0 (2.2) | 6.9 (2.8) | 0.84 |
| Subjects with nonmissing data | 14 | 27 |
Demographics of the phenotypic groups indicating count (%) and P value from a χ2 test of males, females only, or white, black/African American only for categorical variables. The mean (SD) and P value from a t test with Satterthwaite approximation for unequal variances are reported for continuous variables. Bold value is statistically significant.
Includes Asian and Native American persons.
Race not disclosed or missing.
1H NMR Metabolomics
1H NMR spectra of diluted plasma samples were acquired with a 1D Carr-Purcell-Meiboom-Gill pulse sequence (cpmgpr1d) on a Bruker (Billerica, MA) Avance III 700 MHz NMR spectrometer (located at the David H. Murdock Research Institute at Kannapolis, NC) using a 3-mm cryogenically cooled CRYO QNP probe and ambient temperature of 25 °C. Representative 1H NMR spectra can be found in Supplementary Figure S1 and further procedural details are outlined in the Supplementary Material.
Statistical Methods
For univariate statistics, the count (%) and P value from a χ2 test is reported for categorical variables (demographics). The mean (SD) and P value from a t test with Satterthwaite approximation for unequal variances are reported for continuous variables (demographics, bins, and concentrations). A P value less than 0.05 was considered significant, and P values were not adjusted for multiple testing because this was an exploratory analysis. Statistical analyses were conducted using SAS 9.4 (SAS Institute Inc., Cary, NC). Unsupervised (principal component analysis) and supervised (orthogonal partial least-squares discriminate analysis [OPLS-DA]) multivariate analyses were completed in SIMCA 14.1 (Umetrics, Malmo, Sweden) using mean centering and pareto scaling to reduce the dimensionality of the binned data and visualize the variation in the data.23,24 OPLS-DA allows visualization of the separation of the study groups, and the variable influence on projection (VIP) statistic provides information on the importance of a bin in differentiating these groups with higher VIP values, indicating greater importance. Bins important to differentiating the phenotype had either VIP ≥1 where the jack-knifed CI does not include 0 OR a P < 0.1 from a t test with a Satterthwaite approximation when paired samples were treated as independent, to be consistent with the comparison of 2 classes in the binned analyses.
Logistic regression models for the odds of steroid resistance among subjects were created using a stepwise selection procedure with candidate predictors entering the model at P < 0.05 and exiting the model at P < 0.05. Three models were created: (i) demographics alone, (ii) metabolites (relative concentration) alone, and (iii) metabolites plus demographics. The candidate predictors for the demographics-only model were age (years), body mass index percentile, and dichotomized race/ethnicity (African American or white/other). The concentration fit metabolites were candidate predictors for the metabolites-only model. The final model including demographics and metabolites always included the final result from the demographics model, and the model selection algorithm chose the metabolites. Continuous variables were centered, and the fit of each model was assessed using the Hosmer-Lemeshow goodness of fit test. Receiver operating characteristics curve and area under the curve (AUC) analyses were used to assess each model’s ability to predict clinical steroid resistance. Subjects identified as outliers (n = 2) or who were missing age (n = 3) were excluded from the modeling.
Modeling was completed for 2 sets of samples: (i) samples from subjects of all ages (n = 39) and (ii) samples from subjects older than 3 years (n = 28).
Results
Patients
Eighty-eight pediatric patients were enrolled from 2008 to 2014, although only approximately 50% of these patients (n = 45) were able to be enrolled before receiving even a single dose of steroids. All 45 patients who had not received any steroids before the first sample collection (pretreatment) were included in the metabolomic analyses, and the other patients were excluded. Detailed clinical data were obtained from all patients and included in the analysis (Table 1). Of these, paired samples were used from 41 patients (27 SSNS and 14 SRNS), and pretreatment only samples were used from 4 additional patients (3 SSNS and 1 SRNS). Patients clinically phenotyped as SSNS achieved complete remission of proteinuria within an average of approximately 7 weeks of steroid therapy, whereas patients who did not achieve remission during this time frame were phenotyped as SRNS. Children with SSNS differed from those with SRNS in age, as SRNS patients presented (as expected) at a later age than SSNS (9.5 vs. 5.8 years).25 To account for differences in the pharmacodynamics of steroids in children with SSNS versus SRNS due to differences in weight, height, and body mass index, we calculated the average prescribed steroid dosage in a representative subset cohort of these 2 groups. The differences in steroid dosage (SSNS, 1.81 ± 0.23 mg/kg per day vs. SRNS, 1.24 ± 0.14 mg/kg per day) were found to be not significantly different between the 2 groups by nonparametric Mann-Whitney test (P > 0.05). Moreover, SRNS was associated with a trend toward a higher percentage of African American patients (46.7% of SRNS patients) than in SSNS (23.3% of total SSNS patients).
Metabolite Biomarkers to Predict Steroid Resistance
NMR metabolomics data quality and metabolic profiling are described in the Supplementary Material and presented in Supplementary Figure S2. The various approaches to analyze the data are outlined in Figure 1 and the results described later in this article. Although NMR data were collected from all the patient samples, 2 SSNS pretreatment samples were identified as outliers and excluded from all analyses for predictive markers and for paired analyses. In addition, 1 patient identified as having steroid-dependent NS was also excluded from all future analyses.
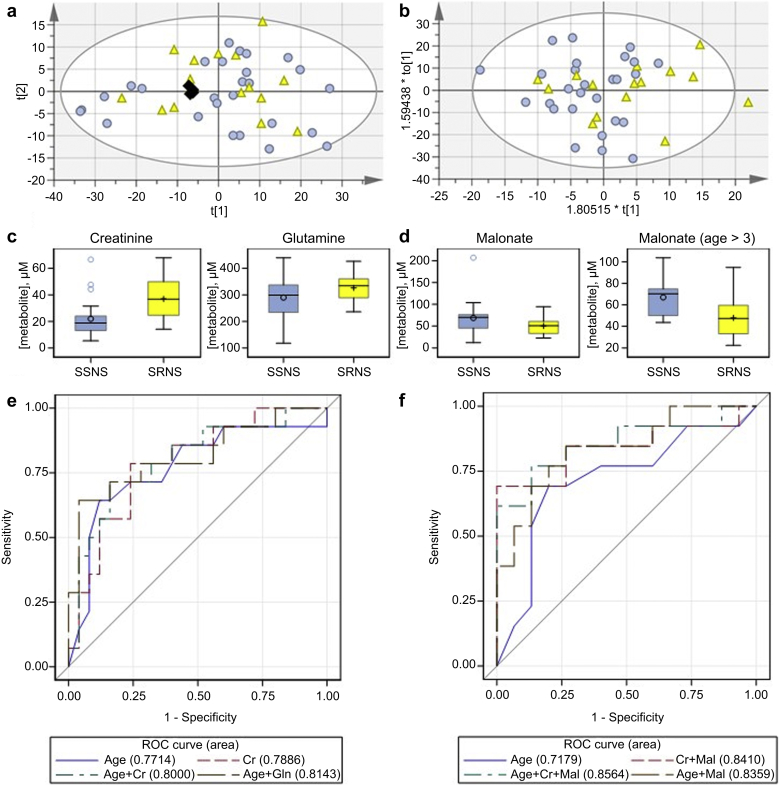
Multivariate analysis using both principal component analysis and OPLS-DA of binned NMR data (Figure 2a and b) from only pretreatment samples (approach I, Figure 1) did not distinguish the SSNS and SRNS patients’ baseline samples. Concentration fitting, followed by comparison of concentrations in the pretreatment samples, found only creatinine to be significantly different between the SRNS versus SSNS groups (Supplementary Table S1). Logistic regression modeling (25 SSNS, 14 SRNS) (after deleting 2 outliers, 1 steroid-dependent patient and 2 with missing ages from initial 30 SSNS and deleting 1 with missing age from 15 SRNS patients) using the relative concentration data for 22 metabolites also selected creatinine (Figure 2c and e; AUC = 0.79, OR: 1.07; OR 95% CI: 1.02–1.12; goodness of fit χ2 = 4.23, df = 8, P = 0.84) as a metabolite able to predict the subsequent response to GC treatment. Creatinine is a well-known marker of kidney function and has previously been correlated with the known steroid resistance risk factor, age at presentation. Age alone predicted SRNS (OR: 1.33; OR 95% CI: 1.09–1.64; goodness of fit χ2 = 5.42, df = 6, P = 0.49). When age was included in the stepwise selection algorithm for metabolites, age (OR: 1.38; OR 95% CI: 1.10–1.73) and glutamine (OR: 1.01, OR 95% CI: 1.00–1.02), but not creatinine, were selected as predictors of response to GC treatment (goodness of fit χ2 = 5.69, df = 8, P = 0.68). The AUC for the age and glutamine model was 0.81, compared with 0.77 in a model using age alone (Figure 2e). With one exception, SRNS cases in this cohort were observed beginning at age 4. Therefore, a similar modeling approach was completed for samples with age >3 years only (n = 28, 15 SSNS and 13 SRNS) for which age was more likely to misclassify samples. Using age as a predictor in the logistic regression model (OR: 1.3; OR 95% CI: 1.00–1.71; goodness of fit χ2 = 4.80, df = 5, P = 0.44) resulted in an AUC = 0.72. Creatinine (OR: 1.08; OR 95% CI: 1.00–1.16) and malonate (OR: 0.93; OR 95% CI: 0.88–0.99) were selected for the metabolites-only model (goodness of fit χ2 = 12.69, df = 7, P = 0.08, AUC = 0.84). When age was included in the model and the selection algorithm was used to select metabolites, the OR for age was 1.36 (OR 95% CI: 0.99–1.86) and malonate was selected (OR: 0.94; OR 95% CI: 0.89–1.00). The model with age and malonate (goodness of fit χ2 = 4.26, df = 7, P = 0.75) had an AUC of 0.84, compared with 0.72 for age alone (Figure 2d and f).
Figure 2.
Metabolite biomarkers to predict steroid resistance. Score plots for binned data comparing steroid-sensitive nephrotic syndrome (SSNS) (n = 27) and steroid-resistant nephrotic syndrome (SRNS) (n = 15) pretreatment samples: (a) Principal component analysis, R2X(cum) 0.904; Q2(cum) 0.803. (b) Orthogonal partial least-squares discriminate analysis scores plot R2X(cum) 0.719; R2Y(cum) 0.16; Q2(cum) −0.0467; SSNS Pre, light blue circle; SRNS Pre, yellow triangle; pools, black diamond. (c) Box plots showing the pretreatment concentration by phenotype of metabolites selected by stepwise selection algorithms for inclusion in a logistic regression model of the odds of SRNS among subjects of all ages. (d) Box plots showing the pretreatment concentration by phenotype of malonate, which was significantly different between patients >3 years of age with SSNS versus SRNS. (e) A logistic regression model comparing the known risk factor age with metabolites for prediction of SRNS. (f) A logistic regression model comparing the known risk factor, age, with metabolites for prediction of SRNS in patients >3 years of age. ROC, receiver operating characteristic.
Metabolite Biomarkers to Define Mechanistic Molecular Pathways/Targets of Steroid Resistance
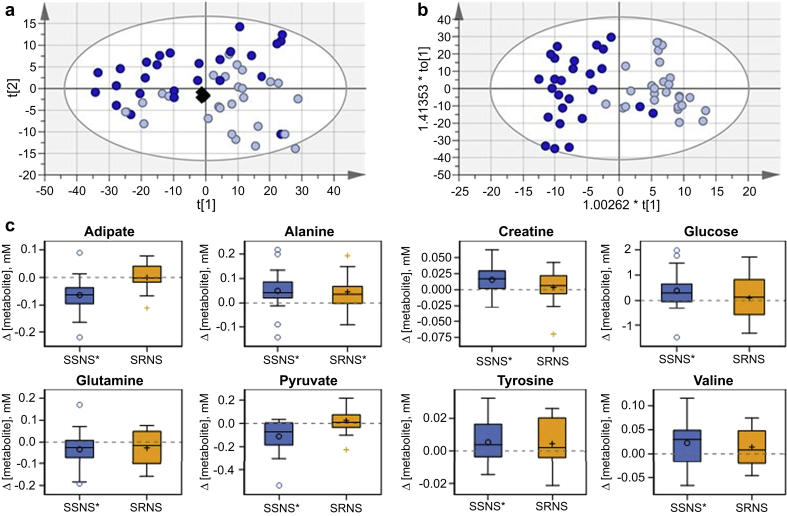
Differentiation of the SSNS samples (approach II, Figure 1) in the first 2 components of the unsupervised multivariate analysis (principal component analysis) showed that GC treatment perturbed the metabolomic profiles of post-treatment samples compared with pretreatment samples. Supervised analysis (OPLS-DA) found a model meeting guidelines for acceptability23 (Figure 3b, R2X(cum) = 0.911; R2Y(cum) = 0.734; Q2(cum) = 0.595). Supplementary Table S2 compares pre- and post-treatment SSNS samples and provides the VIP statistic, P value, and fold change for bins with their associated metabolites that had either a VIP ≥ 1 with a jack-knifed CI that did not include 0 or a P < 0.1. Important bins for differentiating the pre- and post-treatment samples indicated changes in circulating lipoproteins, plasma glucose concentrations, and creatine. Lipoproteins and collection tube additives were large contributors to the total spectra, and their influence on the quantitation of lower concentration metabolites was of concern. Individual metabolites with characteristic NMR peaks were therefore fit, and the concentration determined relative to the 1-mM format reference standard. Concentration fitting, followed by 1-sample t tests, of the concentration difference between pre- and post-treatment samples identified adipate, alanine, creatine, glucose, glutamine, pyruvate, tyrosine, and valine as significantly affected by treatment in SSNS samples (Figure 3c, Supplementary Table S3). Results from these same metabolites in SRNS pretreatment versus post-treatment samples are also shown for comparison purposes and are described as follows.
Figure 3.
Metabolite changes with steroid therapy in steroid-sensitive nephrotic syndrome (SSNS). Multivariate analysis scores plots for comparing SSNS pre- (n = 27) and post-treatment (n = 26) samples: (a) SSNS-only principal component analysis; R2X(cum) 0.937; Q2(cum) 0.85. (b) SSNS-only orthogonal partial least-squares discriminate analysis; R2X(cum) 0.911; R2Y(cum) 0.734; Q2(cum) 0.595; SSNS Pre, light blue circle; SSNS Post, dark blue circle; pools, black diamond. (c) Box plots showing the concentration difference by phenotype, where Δ = post-treatment – pretreatment concentrations (i.e., values above the dotted 0 line imply GC-induced increases in concentration). Metabolites displayed were significantly changed between paired pretreatment and post-treatment SSNS samples. Metabolite changes in Δ SRNS are also shown for comparison and described below. Phenotype is marked with * if P < 0.05 from a paired t test of paired pretreatment and post-treatment samples.
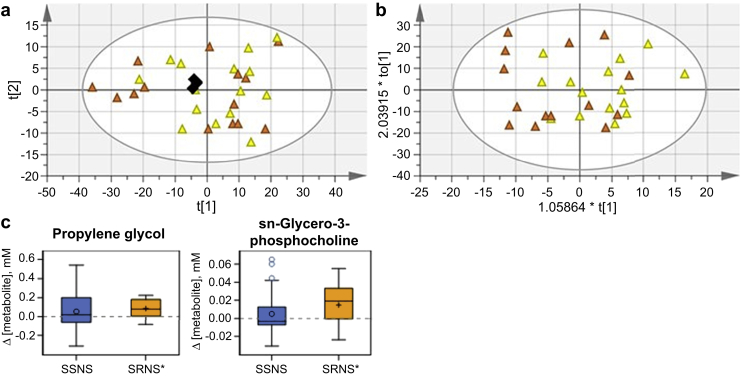
Using the same multivariate modeling approach, the SRNS samples (approach II, Figure 1) did not show differentiation in the unsupervised or supervised score plots. No model could be fit that met the criteria for acceptance (Figure 4a and b), indicating that the overall metabolomics profiles between these 2 groups were not significantly perturbed. However, concentration fitting followed by 1-sample t tests of the concentration difference between pre- and post-treatment SRNS samples did identify alanine, propylene glycol, and sn-glycero-3-phosphocholine as affected by GC treatment in SRNS samples (Figure 4c, Supplementary Table S3). Alanine concentration was also perturbed in the SSNS group, and thus may be interpreted as resulting from GC treatment. This is an important proof-of-concept that the metabolites of children with SRNS are in general not being perturbed in response to GC treatment to the same extent as the metabolites of children with SSNS. When post-treatment concentration changes in children with SSNS were compared with those with SRNS (approach II, Figure 1), Δ[adipate] (P = 0.002) and Δ[pyruvate] (P = 0.004) were significantly lower (Figure 3c, Supplementary Table S3).
Figure 4.
Metabolite changes with steroid therapy in steroid-resistant nephrotic syndrome (SRNS). Scores plots for comparing SRNS pre- (n = 15) and post-treatment (n = 14) samples: (a) SRNS-only principal component analysis; R2X(cum) 0.964; Q2(cum) 0.845. (b) SRNS-only orthogonal partial least-squares discriminate analysis; R2X(cum) 0.75; R2Y(cum) 0.229; Q2(cum) −0.586; SRNS Pre, yellow triangle; SRNS Post, orange triangle; pools, black diamond. (c) Box plots showing the concentration difference by phenotype, where Δ = post-treatment – pretreatment concentration. Metabolites are displayed if significantly changed between paired pre- and post-SRNS samples and not previously displayed in Figure 2. Phenotype is marked with * if P < 0.05 from a paired t test of paired pre- and post-samples.
Metabolite Biomarkers to Differentiate Steroid Sensitivity From Resistance
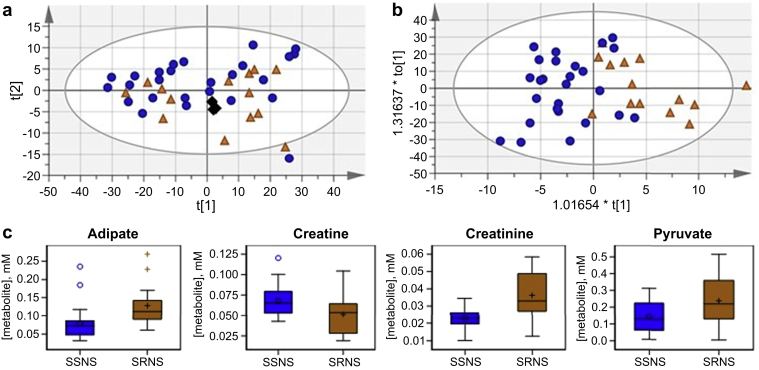
SSNS could be distinguished from SRNS post-treatment (approach III, Figure 1, Figure 5a and b; principal component analysis R2X[cum] 0.897; OPLS-DA R2Y[cum] 0.53; Q2[cum] 0.214). Supplementary Tables S4 and S5 compare post-treatment SSNS and post-treatment SRNS samples and provides the VIP statistic, P value, and fold change for bins with their associated metabolites that had either a VIP ≥ 1 with a jack-knifed CI that did not include 0 or a P < 0.1. Important bins for differentiation of the SSNS and SRNS phenotypic groups post-treatment were nearly identical to those important for differentiation of the SSNS pre- and post-treatment samples. Comparison of concentrations in the post-treatment samples found adipate, creatine, creatinine, and pyruvate to be significantly different between the SRNS and SSNS groups (Figure 5c, Supplementary Table S4).
Figure 5.
Metabolite biomarkers to differentiate steroid sensitivity from resistance. Scores plots for comparison of steroid-sensitive nephrotic syndrome (SSNS) (n = 26) and steroid-resistant nephrotic syndrome (SRNS) (n = 14) post-treatment: (a) principal component analysis, R2X(cum) 0.966; Q2(cum) 0.858; (b) orthogonal partial least-squares discriminate analysis, R2X(cum) 0.897; R2Y(cum) 0.53; Q2(cum) 0.214; SSNS Post, dark blue circle; SRNS Post, orange triangle; pools, black diamond. (c) Box plots showing the concentration by phenotype for metabolites with P < 0.05 from a t test with Satterthwaite approximation for unequal variances between SSNS and SRNS samples post-treatment.
Detailed analyses for metabolomics methodology validation are presented in the Supplementary Material (Supplementary Figures S3 and S4, and Supplementary Tables S6–S8).
Conclusion
NS is a kidney disease that affects both children and adults, although approximately 20% of children and approximately 50% of adults present with or subsequently develop steroid resistance, with its greatly increased risks for both treatment side effects and disease progression. The current study tested the hypothesis that paired plasma metabolomic sample analyses could identify candidate biomarkers able to either predict steroid resistance before GC treatment or define critical mechanistic molecular pathways/targets that regulate steroid resistance (see Figure 1 and Table 2). We used a metabolomic discovery approach starting with 86 samples, and identified a small group of candidate biomarkers predictive of steroid resistance, such as glutamine and malonate, after controlling for age. Paired-sample analyses also enabled us to identify a larger group of candidate metabolite biomarkers potentially able to define specific mechanistic molecular pathways/targets of steroid resistance, including lipoproteins, adipate, pyruvate, creatine, glucose, tyrosine, valine, and glutamine. A single endogenous metabolite, sn-glycero-3-phosphocholine, was also identified as perturbed by GC treatment in children with SRNS, but not in those with SSNS. Together, these findings have identified several specific candidate metabolite biomarkers able to predict SRNS at disease onset, as well as potential therapeutic target pathways that could enable the future development of more targeted and effective treatments for NS.
Table 2.
Candidate metabolite biomarkers to predict, define, or differentiate steroid resistance in pediatric nephrotic syndrome
| Metabolite or bin | Predictive biomarkerI |
Mechanistic biomarkerII |
Differentiating biomarkerIII |
||
|---|---|---|---|---|---|
| SSNS Pre vs. SRNS Pre | SSNS Pre vs. SSNS Post | SRNS Pre vs. SRNS Post | Δ SSNS vs. Δ SRNS |
SSNS Post vs. SRNS Post | |
| Creatinine | X | X | |||
| Glutamine | X | X | |||
| Malonate (age >3) | X | ||||
| Adipate | X | X | X | ||
| Pyruvate | X | X | X | ||
| Creatine | X | X | |||
| Lipoproteins (bins) | X | X | |||
| Glucose | X | ||||
| Tyrosine | X | ||||
| Valine | X | ||||
| sn-Glycero-3-phosphocholine | X | ||||
SSNS, steroid-sensitive nephrotic syndrome; SRNS, steroid-resistant nephrotic syndrome.
Approaches defined in Figure 1.
GCs are known to have significant metabolic effects, including (i) increased mobilization of amino acids and increased urinary nitrogen, with an overall increase in conversion of protein to glycogen storage; (ii) stimulation of gluconeogenesis and reduced insulin response, leading to increased plasma glucose levels; and (iii) increased lipoprotein concentrations, with redistribution of fat from the extremities to the trunk and face.26 The increase in glucose bins and [glucose] in post- versus pretreatment SSNS in our study likely resulted from this gluconeogenesis and reduction in insulin sensitivity. Consistently, [alanine], [tyrosine], and [valine] all were similarly increased with GC treatment. [Alanine] also increased in the SRNS group, implying that this pathway may be a generic GC-responsive pathway, and not necessarily associated with GC efficacy. In contrast, [Glycerophosphocholine] increased after GC treatment in SRNS, but not in SSNS. Glycerophosphocholine is a major renal osmolyte,27 so this finding suggests that GC may significantly influence osmotic stress, even in the absence of clinical efficacy. GCs are also known to augment the biosynthesis of phosphatidylcholine for lung surfactant,28 so the metabolic fate of choline also may be affected by the response to GC. In addition, GCs modulate the activity of the Na+/H+ exchanger in renal proximal tubular cells,29 suggesting that GC could alternatively affect osmotic stress through this mechanism.
Some of the metabolic GC responses observed in this study have been described in studies of adult patients, or with animal models.28,29 We observed a decrease in [adipate], adipate bins, and lipoprotein bins with GC treatment in SSNS samples, and in post-treatment SSNS versus SRNS samples. Hyperlipidemia is a well characterized finding in NS, in addition to albuminuria and the associated decrease in oncotic pressure.30 The observed changes in [adipate], adipate bins, and lipoprotein bins in SSNS patients may be related to the improvement in hyperlipidemia associated with GC-induced remission. Serum creatinine is a standard marker of renal function, and by both 1HNMR and clinical assay, [Creatinine] was higher in SRNS vs. SSNS samples, both pre- and post-treatment with GC. In contrast, [Creatine] was higher after GC treatment in SSNS versus SRNS samples. Creatine is nonenzymatically converted to creatinine, thus changes in creatine concentration may be anticipated with a change in creatinine homeostasis.31
It is worth noting that 2 studies have recently attempted to differentiate the major histological subtypes of glomerular disease, focal segmental glomerulosclerosis, minimal change disease, IgA nephropathy, and membranous nephropathy, from each other and from healthy controls in adults, based on urinary metabolite profiles.1,15 In contrast to these, the current study used unique paired plasma samples from children that were obtained both pretreatment and post-treatment with GC, when the clinical distinction was made between SSNS versus SRNS. This approach enabled us to identify specific metabolites with the potential ability to differentiate SSNS from SRNS, both at the time of disease onset (before GC treatment) as well as after GC therapy. Although our study uniquely characterized plasma in a pediatric cohort, lower pyruvate concentrations in SSNS versus SRNS post-treatment samples reinforced recent findings that pyruvate can distinguish focal segmental glomerulosclerosis from minimal change disease and membranous nephropathy in the urine of adults.1,15 One of these studies also speculated regarding potential effects on renal proximal tubules in addition to the glomerulus in pathologies such as focal segmental glomerulosclerosis,15 although it is well-established that NS is a glomerular disease with very few cases of accompanying renal tubular dysfunction.32,33
Pyruvate-metabolizing enzymes, including pyruvate carboxylase, are also regulated by GC.34,35 Pyruvate also may be an important mediator or marker of kidney injury, as has been reported in acute kidney injury36 and diabetic nephropathy models.37,38 There are therefore reasonable mechanisms by which GC treatment may regulate [pyruvate] and kidney function, and conversely where a lack of response to GC may prevent recovery of [pyruvate] and kidney function.
Glutamine is metabolized by the kidney for the maintenance of pH homeostasis. During acidosis, glutamine is catabolized to 2 molecules of ammonium and α-ketoglutarate, which then enter the tricarboxylic acid cycle and gluconeogenesis. Malonate is involved in fatty acid biosynthesis39 and is protective in cases of ischemia reperfusion,40 although its regulation by the kidney is less well understood.
The role of podocyte energy metabolism and its role in different nephropathies has been an active area of research in recent years.41, 42, 43 It is possible that the glomerular dysfunction seen in NS induces systemic metabolic perturbations, which may in turn drive metabolic effects in podocytes that could affect response to GC therapy and/or disease progression.
This study had several limitations and strengths. Although most similar biomarker and metabolomics studies have been carried out using urine, which is noninvasive, our studies used paired plasma samples in addition to pre- and post-treatment samples. However, the identification of these markers in plasma is highly clinically and biologically relevant, because NS is a glomerular disease with systemic involvement, and plasma metabolites directly reflect their concentrations in the blood, following renal tubular reabsorption and/or secretion. It will be of significant interest to further study and validate our identified biomarkers in urine samples from a larger and separate childhood NS cohort in the future, and such studies are already under way. Furthermore, SRNS typically presents in children at older ages and thus higher weights than in children with SSNS. Because this is a typical generalized difference between the 2 clinically defined groups, accounting for these as confounders to the current studies was not found to be essential or clinically relevant. Predictive models were however adjusted for age (except where noted as excluded), whereas other analyses were not. In addition, we acknowledge the possibility that some of the SRNS patients may have had an underlying genetic cause of disease. However, because the sample collections began more than a decade ago, there were no provisions to screen them for monogenetic causes at that time. Despite this limitation, this study emphasizes differences between children with SRNS and SSNS that are clearly detectable at the time of clinical presentation, regardless of any underlying genetic causes. However, future metabolomics studies that could differentiate SRNS due to known genetic versus nongenetic causes are currently being planned in a broader patient cohort.
One of the major strengths of this study was the evaluation of paired samples from most patients, both before any GC treatment and following an average of approximately 7 weeks of daily oral GC. This enabled us to use multiple approaches (see Figure 1) to analyze and interpret the dataset, in terms of predicting, mechanistically defining, and differentiating biomarkers in children with SSNS versus SRNS. Notably, this exact same group of patient samples also has undergone plasma proteomics analyses,44 and we are now initiating studies to identify additional relevant molecular pathways and biomarkers of steroid resistance in NS using approaches to integrate the proteomics and metabolomics datasets.
Technically, NMR may not capture the richest possible dataset for this analysis because of its comparatively lower sensitivity. However, it offers a technical strength due to its nondestructive nature, superior stability, reproducibility, and chemical structure content of the spectra compared with other metabolomics techniques, and these factors were considered to be more important for the current studies. Another strength of the metabolomics approach is its ability to capture the emergent properties of a system (phenotype), because metabolism integrates inputs from genetics, regulatory or compensatory responses to stimuli, environmental exposures, and more.45 However, this pathway integration could result in metabolomic analyses being somewhat less specific than some other -omics methods regarding the mechanism of changes or identifying specific therapeutic targets.
In summary, the current studies identified several specific metabolite differences between children with SSNS versus SRNS at the time of disease onset, as well as following GC therapy, by using paired plasma samples obtained both before and after an average of approximately 7 weeks of daily oral GC. Because steroid resistance is a clinical feature also seen in many other diseases, further validation of our study results could greatly improve our ability to predict the risk of clinical steroid resistance at disease onset for patients presenting with any of the many diseases for which steroids are a primary treatment, including asthma, rheumatoid arthritis, autoimmune hepatitis, and other inflammatory conditions. In addition, these findings could improve our understanding of the molecular pathways and mechanisms that regulate the clinical response to GC, and thus help identify potential future molecular targets to improve the treatment of NS as well as other conditions treated with steroids.
Appendix
Members of the Midwest Pediatric Nephrology Consortium
Drs. John Mahan, Hiren Patel, and Richard F. Ransom (NCH, Columbus, OH, USA); Cynthia Pan (Medical College of Wisconsin, Milwaukee, WI, USA); Denis F. Geary (The Hospital for Sick Children, Toronto, ON, Canada); Myra L. Chang (West Virginia University, Charleston, WV, USA); Keisha L. Gibson (University of North Carolina, Chapel Hill, NC, USA); Franca M. Iorember (Louisiana State University, New Orleans, LA, USA); Patrick D. Brophy (Children’s Hospital, University of Iowa, Iowa City, IA, USA); Tarak Srivastava (Children’s Mercy Hospital, Kansas City, MO, USA); and Larry A. Greenbaum (Emory University School of Medicine, Atlanta, GA, USA).
Disclosure
All the authors declared no competing interests.
Acknowledgments
We thank the Midwest Nephrology Consortium, its participating centers, physicians, and study and nurse coordinators for their contributions toward the collection of the plasma samples used in this study. These include Drs. John Mahan, Hiren Patel, and Richard F. Ransom (NCH, Columbus, OH, USA); Cynthia Pan (Medical College of Wisconsin, Milwaukee, WI, USA); Denis F. Geary (The Hospital for Sick Children, Toronto, ON, Canada); Myra L. Chang (West Virginia University, Charleston, WV, USA); Keisha L. Gibson (University of North Carolina, Chapel Hill, NC, USA); Franca M. Iorember (Louisiana State University, New Orleans, LA, USA); Patrick D. Brophy (Children’s Hospital, University of Iowa, Iowa City, IA); Tarak Srivastava (Children’s Mercy Hospital, Kansas City, MO, USA); and Larry A. Greenbaum (Emory University School of Medicine, Atlanta, GA, USA). We also thank the Biopathology Core at Nationwide Children’s Hospital for storing and maintaining the sample biorepository.
We particularly thank and acknowledge the late Dr. Jason Burgess for his invaluable contributions to this work. Dr. Burgess was the Program Coordinator for the National Institutes of Health Common Fund Eastern Regional Metabolomics Resource Core before his passing on January 14, 2018. He made significant contributions to this project through acquiring NMR data and overseeing components of the data analysis.
This study was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) U24DK097193 to SJS, the National Institute of General Medical Sciences (NIGMS) K01GM109320 to JRG NIDDK-NIH (DK110077 to WES, SJS, and JBK) and by internal funds from Nationwide Children’s Hospital (NCH to WES and SA).
Author Contributions
JRG and SA conceptualized and designed the studies, performed experiments, analyzed and interpreted the data, prepared the figures and tables, and drafted and edited the manuscript. SM and ZA analyzed the data, SM edited the manuscript, and ZA prepared the figures and tables. MLM and JBK interpreted the data and edited the manuscript. WES and SJS conceptualized and designed the study, analyzed and interpreted the data, and edited the manuscript. All the authors approved of the final version of the manuscript.
Data Sharing
The data obtained in this study was deposited into the NIH Common Fund’s Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) Web site, http://www/metabolomicsworkbench.org, where it has been assigned a Metabolomics Workbench Project ID: PR000648. The data will be directly accessible at http://www.metabolomicsworkbench.org/data/DRCCMetadata.php?Mode=Project&ProjectID=PR000648.
Footnotes
Figure S1. Representative NMR spectra and method illustration.
Figure S2. 1H NMR metabolomics data quality.
Figure S3. Validation of NMR method against gold-standard serum creatinine clinical assay.
Figure S4. Patient subset analysis and comparison.
Table S1. Differences in pretreatment concentrations (mM) of fitted metabolites between children with SRNS versus SSNS.
Table S2. NMR bins important for differentiating pretreatment (n = 27) and post-treatment (n = 26) SSNS samples.
Table S3. Change in concentrations (mM) of fitted metabolites between disease onset (pretreatment) and follow-up (post-treatment).
Table S4. Differences in post-treatment concentrations (mM) of fitted metabolites between children with SRNS versus SSNS.
Table S5. NMR bins important for differentiating SRNS (n = 14) and SSNS (n = 26) in post-treatment samples.
Table S6. Metabolite differences between SRNS and SSNS at presentation (pre) in the smaller subset.
Table S7. metabolite changes between presentation (pre) and after therapy (post) in the smaller subset.
Table S8. Metabolite differences between SRNS and SSNS after therapy (post) in the smaller subset.
Contributor Information
Shipra Agrawal, Email: Shipra.agrawal@nationwidechildrens.org.
William E. Smoyer, Email: William.smoyer@nationwidechildrens.org.
Susan J. Sumner, Email: susan_sumner@unc.edu.
The Midwest Pediatric Nephrology Consortium:
John Mahan, Hiren Patel, Richard F. Ransom, Cynthia Pan, Denis F. Geary, Myra L. Chang, Keisha L. Gibson, Franca M. Iorember, Patrick D. Brophy, Tarak Srivastava, and Larry A. Greenbaum
Supplementary Material
References
- 1.Hao X., Liu X., Wang W. Distinct metabolic profile of primary focal segmental glomerulosclerosis revealed by NMR-based metabolomics. PloS One. 2013;8 doi: 10.1371/journal.pone.0078531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalantari S., Nafar M., Samavat S. 1 H NMR-based metabolomics study for identifying urinary biomarkers and perturbed metabolic pathways associated with severity of IgA nephropathy: a pilot study. Magn Reson Chem. 2017;55:693–699. doi: 10.1002/mrc.4573. [DOI] [PubMed] [Google Scholar]
- 3.Hanna M.H., Dalla Gassa A., Mayer G. The nephrologist of tomorrow: towards a kidney-omic future. Pediatr Nephrol. 2017;32:393–404. doi: 10.1007/s00467-016-3357-x. [DOI] [PubMed] [Google Scholar]
- 4.Thongboonkerd V. Biomarker discovery in glomerular diseases using urinary proteomics. Proteomics Clin Appl. 2008;2:1413–1421. doi: 10.1002/prca.200800036. [DOI] [PubMed] [Google Scholar]
- 5.Tryggvason S.H., Guo J., Nukui M. A meta-analysis of expression signatures in glomerular disease. Kidney Int. 2013;84:591–599. doi: 10.1038/ki.2013.169. [DOI] [PubMed] [Google Scholar]
- 6.Bennett M.R., Pleasant L., Haffner C. A novel biomarker panel to identify steroid resistance in childhood idiopathic nephrotic syndrome. Biomark Insights. 2017;12 doi: 10.1177/1177271917695832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niwa T. Mass spectrometry in the search for uremic toxins. Mass Spectrom Rev. 1997;16:307–332. doi: 10.1002/(SICI)1098-2787(1997)16:6<307::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 8.Sekula P., Goek O.N., Quaye L. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol. 2016;27:1175–1188. doi: 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee E.P., Clish C.B., Ghorbani A. A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol. 2013;24:1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blydt-Hansen T.D., Sharma A., Gibson I.W. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am J Transplant. 2014;14:2339–2349. doi: 10.1111/ajt.12837. [DOI] [PubMed] [Google Scholar]
- 11.Mussap M., Noto A., Fanos V. Emerging biomarkers and metabolomics for assessing toxic nephropathy and acute kidney injury (AKI) in neonatology. BioMed Res Int. 2014;2014:602526. doi: 10.1155/2014/602526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes E., Nicholls A.W., Lindon J.C. Development of a model for classification of toxin-induced lesions using 1H NMR spectroscopy of urine combined with pattern recognition. NMR Biomed. 1998;11:235–244. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<235::aid-nbm507>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 13.Gao X., Chen W., Li R. Systematic variations associated with renal disease uncovered by parallel metabolomics of urine and serum. BMC Syst Biol. 2012;6(Suppl 1):S14. doi: 10.1186/1752-0509-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia L., Wang C., Zhao S. Metabolomic identification of potential phospholipid biomarkers for chronic glomerulonephritis by using high performance liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860:134–140. doi: 10.1016/j.jchromb.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 15.Lee J.E., Lee Y.H., Kim S.Y. Systematic biomarker discovery and coordinative validation for different primary nephrotic syndromes using gas chromatography-mass spectrometry. J Chromatogr A. 2016;1453:105–115. doi: 10.1016/j.chroma.2016.05.058. [DOI] [PubMed] [Google Scholar]
- 16.Psihogios N.G., Kalaitzidis R.G., Dimou S. Evaluation of tubulointerstitial lesions' severity in patients with glomerulonephritides:an NMR-based metabonomic study. J Proteome Res. 2007;6:3760–3770. doi: 10.1021/pr070172w. [DOI] [PubMed] [Google Scholar]
- 17.Romick-Rosendale L.E., Brunner H.I., Bennett M.R. Identification of urinary metabolites that distinguish membranous lupus nephritis from proliferative lupus nephritis and focal segmental glomerulosclerosis. Arthritis Res Ther. 2011;13:R199. doi: 10.1186/ar3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sui W., Li L., Che W. A proton nuclear magnetic resonance-based metabonomics study of metabolic profiling in immunoglobulin a nephropathy. Clinics (Sao Paulo) 2012;67:363–373. doi: 10.6061/clinics/2012(04)10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark A.G., Barratt T.M. Steroid-responsive nephrotic syndrome. In: Barratt T.M., Avner E.D., Harmon W.E., editors. Pediatric Nephrology. 4th ed. Lippincott Williams & Wilkins; Baltimore: 1998. pp. 731–747. [Google Scholar]
- 20.McEnery P.T., Strife C.F. Nephrotic syndrome in childhood. Management and treatment in patients with minimal change disease, mesangial proliferation, or focal glomerulosclerosis. Pediatr Clin North Am. 1982;89:875–894. [PubMed] [Google Scholar]
- 21.Nash M.A., Edelmann C.M.J., Bernstein J. The nephrotic syndrome. In: Edelmann C.M.J., editor. Pediatric Kidney Disease. 2nd ed. Little, Brown, and Company; Boston: 1992. pp. 1247–1266. [Google Scholar]
- 22.Gipson D.S., Chin H., Presler T.P. Differential risk of remission and ESRD in childhood FSGS. Pediatr Nephrol. 2006;21:344–349. doi: 10.1007/s00467-005-2097-0. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson L., Byrne T., Johansson E. MKS Umetrics AB; Malmö, Sweden: 2013. Multi- and Megavariate Data Analysis Basic Principles and Applications. 33–54, 215. [Google Scholar]
- 24.Trygg J., Holmes E., Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.S., Bellew C.A., Silverstein D.M. High incidence of initial and late steroid resistance in childhood nephrotic syndrome. Kidney Int. 2005;68:1275–1281. doi: 10.1111/j.1523-1755.2005.00524.x. [DOI] [PubMed] [Google Scholar]
- 26.McKay L.I., Cidlowski J.A. Physiologic and pharmacologic effects of corticosteroids. In: Kufe D.W., Pollock R.E., Weichselbaum R.R., editors. Holland-Frei Cancer Medicine. 6th edition. BC Decker; Hamilton, ON: 2003. https://www.ncbi.nlm.nih.gov/books/NBK13780/ Accessed October 30, 2019. [Google Scholar]
- 27.Zablocki K., Miller S.P., Garcia-Perez A. Accumulation of glycerophosphocholine (GPC) by renal cells:osmotic regulation of GPC: choline phosphodiesterase. Proc Natl Acad Sci U S A. 1991;88:7820–7824. doi: 10.1073/pnas.88.17.7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torday J.S., Smith B.T., Giroud C.J. The rabbit fetal lung as a glucocorticoid target tissue. Endocrinology. 1975;96:1462–1467. doi: 10.1210/endo-96-6-1462. [DOI] [PubMed] [Google Scholar]
- 29.Bidet M., Merot J., Tauc M. Na+-H+ exchanger in proximal cells isolated from kidney. II. Short-term regulation by glucocorticoids. Am J Physiol. 1987;253:F945–F951. doi: 10.1152/ajprenal.1987.253.5.F945. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler D.C., Bernard D.B. Lipid abnormalities in the nephrotic syndrome: causes, consequences, and treatment. Am J Kidney Dis. 1994;23:331–346. doi: 10.1016/s0272-6386(12)80994-2. [DOI] [PubMed] [Google Scholar]
- 31.Eggert W., Syllm-Rapoport I., Daniel A. [Behavior of creatine in red blood cells and in plasma of children with chronic kidney insufficiency] Helv Paediatr Acta. 1983;38:281–290. [PubMed] [Google Scholar]
- 32.McVicar M., Exeni R., Susin M. Nephrotic syndrome and multiple tubular defects in children:an early sign of focal segmental glomerulosclerosis. J Pediatr. 1980;97:918–922. doi: 10.1016/s0022-3476(80)80420-3. [DOI] [PubMed] [Google Scholar]
- 33.Praga M., Andres A., Hernandez E. Tubular dysfunction in nephrotic syndrome: incidence and prognostic implications. Nephrol Dial Transplant. 1991;6:683–688. doi: 10.1093/ndt/6.10.683. [DOI] [PubMed] [Google Scholar]
- 34.Frawley T.F. The role of the adrenal cortex in glucose and pyruvic acid metabolism in man including the use of intravenous hydrocortisone in acute hypoglycemia. Ann N Y Acad Sci. 1955;61:464–493. doi: 10.1111/j.1749-6632.1955.tb42498.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z., Iwasaki Y., Zhao L.F. Hormonal regulation of glycolytic enzyme gene and pyruvate dehydrogenase kinase/phosphatase gene transcription. Endocr J. 2009;56:1019–1030. doi: 10.1507/endocrj.k09e-178. [DOI] [PubMed] [Google Scholar]
- 36.Zager R.A., Johnson A.C., Becker K. Renal cortical pyruvate depletion during AKI. J Am Soc Nephrol. 2014;25:998–1012. doi: 10.1681/ASN.2013070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laustsen C., Ostergaard J.A., Lauritzen M.H. Assessment of early diabetic renal changes with hyperpolarized [1-(13) C]pyruvate. Diabetes Metab Res Rev. 2013;29:125–129. doi: 10.1002/dmrr.2370. [DOI] [PubMed] [Google Scholar]
- 38.Ju K.D., Shin E.K., Cho E.J. Ethyl pyruvate ameliorates albuminuria and glomerular injury in the animal model of diabetic nephropathy. Am J Physiol Renal Physiol. 2012;302:F606–F613. doi: 10.1152/ajprenal.00415.2011. [DOI] [PubMed] [Google Scholar]
- 39.Witkowski A., Thweatt J., Smith S. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J Biol Chem. 2011;286:33729–33736. doi: 10.1074/jbc.M111.291591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojtovich A.P., Brookes P.S. The endogenous mitochondrial complex II inhibitor malonate regulates mitochondrial ATP-sensitive potassium channels:implications for ischemic preconditioning. Biochim Biophys Acta. 2008;1777:882–889. doi: 10.1016/j.bbabio.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imasawa T., Obre E., Bellance N. High glucose repatterns human podocyte energy metabolism during differentiation and diabetic nephropathy. FASEB J. 2017;31:294–307. doi: 10.1096/fj.201600293R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozawa S., Ueda S., Imamura H. Glycolysis, but not Mitochondria, responsible for intracellular ATP distribution in cortical area of podocytes. Sci Rep. 2015;5:18575. doi: 10.1038/srep18575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi W., Keenan H.A., Li Q. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med. 2017;23:753–762. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal S., Merchant M.L., Kino J. Predicting and defining steroid resistance in pediatric nephrotic syndrome using plasma proteomics. Kidney Int Rep. 2020;5:66–80. doi: 10.1016/j.ekir.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beger R.D., Dunn W., Schmidt M.A. Metabolomics enables precision medicine: "A White Paper, Community Perspective. Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.