Synopsis
The TenA protein family occurs in prokaryotes, plants, and fungi; it has two subfamilies, one (TenA_C) having an active-site cysteine, the other (TenA_E) not. TenA_C proteins participate in thiamin salvage by hydrolysing the thiamin breakdown product 4-amino-5-aminomethyl-2-methylpyrimidine (amino-HMP) to 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP); the function of TenA_E proteins is unknown. Comparative analysis of prokaryote and plant genomes predicted (i) that TenA_E has a salvage role similar but not identical to that of TenA_C, and (ii) that TenA_E and TenA_C also have non-salvage roles since they occur in organisms that cannot make thiamin. Recombinant Arabidopsis and maize TenA_E proteins (At3g16990, GRMZM2G080501) hydrolysed amino-HMP to HMP and, far more actively, hydrolysed the N-formyl derivative of amino-HMP to amino-HMP. Ablating the At3g16990 gene in a line with a null mutation in the HMP biosynthesis gene ThiC prevented its rescue by amino-HMP. Ablating At3g16990 in the wild type increased sensitivity to paraquat-induced oxidative stress; HMP overcame this increased sensitivity. Furthermore, the expression of TenA_E and ThiC genes in Arabidopsis and maize was inversely correlated. These results indicate that TenA_E proteins mediate amidohydrolase and aminohydrolase steps in the salvage of thiamin breakdown products. As such products can be toxic, TenA_E proteins may also pre-empt toxicity.
Keywords: 4-amino-5-aminomethyl-2-methylpyrimidine, N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine, Arabidopsis, comparative genomics, thiaminase II, Zea mays
INTRODUCTION
Thiamin, in its diphosphate form, is an essential cofactor for central metabolic enzymes such as transketolase and the pyruvate and α-ketoglutarate dehydrogenase complexes [1]. Thiamin consists of 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) and 4-methyl-5-(2-hydroxyethyl)thiazole (THZ) moieties (Figure 1A). Plants, fungi, and most prokaryotes synthesise both these moieties de novo and couple them together to give thiamin [2]. These organisms can also salvage HMP and THZ derived from thiamin breakdown for re-use in thiamin synthesis [2,3], while certain prokaryotes rely totally on salvage of exogenous HMP and/or THZ because they do not make these compounds [4]. Even in organisms that do make HMP and THZ, salvage can be crucial to the thiamin economy because thiamin is chemically and metabolically labile [5,6] and HMP and THZ are costly to make [2,7].
Figure 1. Thiamin synthesis, degradation, and salvage routes.
(A) Bacterial thiamin synthesis and salvage pathways. The plant pathways are the same except that the THZ moiety is synthesised by a single enzyme (THI4) from NAD and glycine, and ThiL and ThiK are absent. AIR, 5-aminoimidazole ribotide; DHGIy, dehydroglycine; DXP, deoxy-D-xylulose 5-phosphate; HMP, 4-amino-5-hydroxymethyl-2-methylpyrimidine; -P, phosphate; -PP, diphosphate; ThiS-COSH, ThiS thiocarboxylate; THZ, 4-methyl-5-(2-hydroxyethyl)thiazole.
(B) Selected chemical and enzymatic thiamin degradation reactions. The sequential actions of YlmB (formylamino-HMP amidohydrolase) and TenA_C (amino-HMP aminohydrolase) convert the breakdown product formylamino-HMP to HMP, which can re-enter the synthesis pathway as shown in part A (HMP is highlighted in parts A and B to show this connection). Other reactions (not shown) include oxidation of the alcohol groups of oxothiamin, THZ, and HMP to the respective acids, cleavage of oxothiamin to HMP and the oxo derivative of THZ, and cleavage of thiamin thiol form to amino-HMP and a thioketone. Various other uncharacterised breakdown products have been reported, including some that retain both rings or large parts thereof [11,52].
Thiamin biosynthesis is now largely understood in bacteria, plants, and yeast [1–3] (Figure 1A). Thiamin salvage is less understood, in part because thiamin breakdown products are numerous, incompletely characterised, and vary with environmental conditions [5,8–12] (Figure 1B). What is clear so far is that: (i) thiamin can be split into its HMP and THZ moieties chemically, photochemically or enzymatically; (ii) these moieties – separately or as parts of the thiamin molecule – undergo various degradation reactions; (iii) the HMP moiety is generally more stable than the THZ moiety; and (iv) intact HMP and THZ can be phosphorylated and then re-enter the synthesis pathway [2,4,6,13,14] (Figure 1A). In bacteria, yeast, and plants, HMP is converted to its mono- and diphosphates by HMP (phosphate) kinase (ThiD), and THZ is phosphorylated by THZ kinase (ThiM) [2,15,16].
Among the things that are unclear about breakdown and salvage is how HMP is formed. One route involves hydrolysis of intact thiamin by the thiaminase II activity of microbial TenA family proteins [17–19]. However, recent work in bacteria and yeast indicates that a more important source of HMP in vivo is hydrolysis of 4-amino-5-aminomethyl-2-methylpyrimidine (amino-HMP) by the amino-HMP hydrolase activity of TenA proteins [8,19]. Amino-HMP itself can come from deconstructing thiamin, after damage to its THZ ring, via a route whose last intermediate is the N-formyl derivative of amino-HMP (formylamino-HMP) and whose last enzyme is formylamino-HMP amidohydrolase (YlmB) [8] (Figure 1B). Breakdown products that retain more of the THZ ring than formylamino-HMP (some of which are shown in Figure 1B) could also be TenA substrates, but this has yet to be proven [8,20].
Also unclear is which TenA-family proteins have amino-HMP aminohydrolase and thiaminase II activities. TenA proteins fall into two widely distributed subfamilies: one has an active-site cysteine residue, the other does not but often has two conserved glutamates [21]. We term these subfamilies TenA_C and TenA_E, respectively, based on the one-letter codes for cysteine and glutamate. Crystal structures are available for representatives from both families [17,21–24]. TenA_C proteins from Bacillus subtilis [8,17], yeast [19], and Helicobacter pylori [20] have been assayed for enzyme activities; all three have amino-HMP aminohydrolase activity, while the first two also have low thiaminase II activity. In contrast, no TenA_E proteins seem to have been tested for activity in published literature. An activity related to thiamin salvage is, however, implied by the HMP [23,25] or HMP-phosphate [24] ligands that co-purified with the Pyrococcus furiosus and Arabidopsis TenA_E proteins and were visualised in their crystal structures.
If the TenA_E protein encoded in the Arabidopsis genome (At3g16990) [21,23] has such an activity, it could explain why Arabidopsis mutants blocked in HMP synthesis can be rescued by amino-HMP [26,27], because such rescue requires amino-HMP aminohydrolase activity. Arabidopsis encodes a TenA_C domain in another protein (At5g32470) [3] that alternatively could account for salvage of amino-HMP. However, this protein has a haloacid dehalogenase family domain fused to its TenA_C domain, unlike other members of the TenA family, and this unusual structure creates uncertainty as to the enzymatic function of this fusion protein.
In this study, we present a comparative genomic analysis of the TenA family in prokaryotes and plants that predicts a thiamin salvage function for the TenA_E subfamily. We then validate this prediction by demonstrating that: (i) Arabidopsis TenA_E and its maize ortholog have amino-HMP aminohydrolase activity; (ii) both proteins also have high formylamino-HMP amidohydrolase activity; and (iii) the Arabidopsis and maize TenA_E genes are strongly expressed when the HMP biosynthesis gene ThiC is not. The comparative genomic analysis also suggests that, besides functioning in thiamin salvage, TenA_E and TenA_C proteins can pre-empt metabolic damage from thiamin breakdown products.
EXPERIMENTAL
Bioinformatics and gene expression analysis
Protein sequences were taken from GenBank, MaizeSeqence.org, and the SEED database [28]. Comparative analyses of prokaryotic and plant genomes were made using SEED tools [28]; full results of this analysis are encoded in the SEED subsystem named ‘TenA’, available at http://pubseed.theseed.org//SubsysEditor.cgi?page=ShowSpreadsheet&subsystem=TenA. Sequences were aligned using Muscle [29]. Phylogenetic trees were constructed by the neighbor-joining method using MEGA5 [30]. Arabidopsis microarray gene expression data were taken from CSB.DB [31] and maize RNA-seq data were taken from qTeller http://qteller.com/ [32]. Gene expression data for B73 endosperm tissue harvested at various times were obtained by qRT-PCR as described [33], and were expressed relative to the value at 14 days after pollination (= 1.0).
Chemicals
Thiamin hydrochloride, thiamin monophosphate chloride dihydrate, thiamin pyrophosphate, oxythiamin chloride hydrochloride, and thiochrome were obtained from Sigma-Aldrich (St. Louis, MO). HMP, amino-HMP, and oxothiamin (3-[(4-amino-2-methyl-5-pyrimidinyl)methyl]-5-(2-hydroxyethyl)-4-methyl-2(3H)-thiazolone) were from Toronto Research Chemicals Inc. (Toronto, ON, Canada). Thiamin disulfide hydrate and 1H-benzotriazole-1-carboxaldehyde were from TCI (Tokyo, Japan). Oxy-HMP (5-(hydroxymethyl)-2-methylpyrimidin-4(1H)-one) was from CGeneTech (Indianopolis, IN). Formylamino-HMP was prepared from amino-HMP as described [8]. The preparation was 92.7% pure and contained 0.4% amino-HMP, as judged from HPLC analysis. Desthiothiamin was prepared as described [34]. Briefly, 10 ml of a 1.5 M solution of thiamin (15 mmol) in 3 M sodium hydroxide was mixed with 7.5 ml of a 1.55 M solution of glycine (12 mmol) in 1.55 M sodium hydroxide, and stirred at room temperature for 3 days. White crystals were obtained, and washed with cold ethanol. The preparation was 95.8% pure, and contained ≤0.02% thiamin, as judged from HPLC analysis.
Constructs for expression in Escherichia coli
The At3g16990 (AtTenA_E) and GRMZM2G080501 (ZmTenA_E) coding sequences were amplified by PCR from leaf cDNA libraries using Phusion DNA polymerase (New England BioLabs). For At3g16990, the amplicons were treated with Taq DNA polymerase to add A-overhangs before cloning into pYES2.1 TOPO (Invitrogen) and this construct served as PCR template for subsequent cloning into pET28b (Novagen). BSU11650 (BsTenA_C) was amplified by colony PCR using Phusion DNA polymerase. All three amplified coding sequences were cloned into pET28b between the Ncol and Notl sites, which adds a C-terminal hexahistidine tag. All constructs were sequence-verified. The primers used are given in Supplementary Table S2.
Production and purification of recombinant proteins
The AtTenA_E and BsTenA_C pET28b constructs were introduced into E. coli strain BL21-Codon-Plus (DE3)-RIPL and the ZmTenA_E pET28b construct was introduced into Rosetta-gami 2. Cultures (250 ml for AtTenA_E and BsTenA_C, 2 liters for ZmTenA_E) were grown at 37°C in LB medium containing 50 μg ml−1 kanamycin. When A600 reached 0.6, isopropyl β-d-1-thiogalactopyranoside (final concentration, 1 mM) was added; incubation was continued for 3 h at 37°C for BsTenA_C and AtTenA_E. For ZmTenA_E, induction was preceded by a 30-min ethanol shock (final concentration, 5% vol/vol), and incubation was continued overnight at 22°C. Subsequent steps were at 4°C. Cells were harvested by centrifugation, resuspended in 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0, and sonicated. To the cleared supernatant, 0.5 ml of Ni2+-nitrilotriacetic acid agarose 50% slurry (Qiagen) was added, followed by rotary shaking for 1 h at 4°C. The mixture was then poured into a column and allowed to drain by gravity. After washing with 16 ml of 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0, proteins were eluted with 2 ml of this buffer containing 250 mM imidazole, desalted on PD-10 columns (GE Healthcare) equilibrated in 50 mM Tris-HCl (pH 7.5), 1 mM DTT and 500 mM glycinebetaine, and concentrated with an Amicon Ultra- 0.5 mL 10K unit (Millipore). Purified proteins were frozen in liquid N2 and stored at −80°C. Protein was estimated by dye binding [35] with bovine serum albumin as standard. As pilot tests showed that very little ZmTenA_E could be obtained in soluble form, AtTenA_E was used for characterisation work; BsTenA_C served as a benchmark.
Enzyme assays
Assays were routinely made in triplicate in 100-μl reaction mixtures containing 45 mM Tris-HCl (pH 7.5), 0.9 mM DTT, 450 mM glycinebetaine, and the specified concentrations of substrates. Assays were run at 30°C for 30-300 min and stopped on ice, and then deproteinised at 4°C using Amicon Ultra-0.5 mL 10K units. Samples (typically 40 μl) of the flow-through were analyzed by HPLC with UV detection (Waters 2695 Separation module, Waters 2998 PDA detector). HPLC analysis used a C18 column (ACE Excel SuperC18, 5 μm, 250 × 4.6 mm) with a column guard, equilibrated with 100 mM K-phosphate, pH 6.6. The elution gradient (1 ml min−1) was as follows, steps being linear transitions except where noted: 0-4 min – 20% K-phosphate (100 mM, pH 6.6), 80% water; 4-10 min – 20% K-phosphate, 7% methanol, 73% water; 10-15 min – held at 20% K-phosphate, 7% methanol, 73% water; 15-20 min – 10% K-phosphate, 60% water, 30% methanol; 20-30 min – 20% K-phosphate, 7% methanol, 73% water; 30-35 min – 20% K-phosphate, 80% water. Detection was by absorbance at 235 nm. Values of kcat and Km were estimated by nonlinear fitting using GraphPad Prism version 6.00 (GraphPad Software, San Diego CA).
Liquid chromatography-mass spectrometry
The amino-HMP and HMP reaction products of AtTenA_E action on formylamino-HMP were analyzed using a Thermo LTQ Velos mass spectrometer with an Accela 600 UPLC apparatus and Accela open autosampler (Thermo Fisher). The spectrometer was operated in positive heated-electrospray mode under the following conditions: 3,500 V for the spray needle, 325°C for the source temperature, flow rates 40 arbitrary units sheath gas, 10 arbitrary units auxiliary gas, and 325°C capillary temperature. Gradient elution was employed on the above C18 column equilibrated with 100 mM ammonium acetate, pH 6.6, using a flow rate of 1 ml min−1. The flow was split to 0.3 ml min−1 after separation to avoid salt build-up on the ion source. The elution gradient was as follows: 0-4 min – 20% ammonium acetate buffer (100 mM, pH 6.6), and 80% water; 4-14 min – 10% ammonium acetate, 30% water, and 60% methanol; 14-18 min – 10% ammonium acetate, 30% water and 60% methanol; 18-25 min – 20% ammonium acetate and 80% water; 25-28 min – 20% ammonium acetate and 80% water. The injections were of 5 μl and 1 μl for enzyme assays and standards diluted in enzyme assay buffer, respectively. Full scan spectra were collected from m/z 100 to 600.
Experiments with Arabidopsis mutants
Seeds of the wild type Columbia, the SALK-062985 homozygous At3g16990 knockout line, and the py-1 (ThiC) mutant (stock number CS3491) were obtained from the Arabidopsis Biological Resource Center. The TenA_E knockout was verified by PCR, using gene-specific primers located 5’ or 3’ of the T-DNA and a T-DNA-specific primer. Amplicons were sequenced to confirm the insertion site. Semi-quantitative RT-PCR was performed as follows: Total RNA was extracted from triplicate samples of knockout and wild type leaves using RNeasy kits (Qiagen) and treated with DNase (DNA-free™ kit, Ambion). Total RNA was used for first-strand cDNA synthesis using M-MuLV (New England BioLabs), followed by PCR with primers designed to amplify a fragment of the TenA_E transcript or a fragment of the actin transcript. Amplicons were analyzed by agarose-gel electro-phoresis. The primers used are given in Supplementary Table S2. Seeds of the py-1 mutant line were grown on 1× MS medium [36] plates supplemented with 2% sucrose, 100 mg l−1 inositol, 0.5 mg l−1 nicotinic acid and 0.5 mg l−1 pyridoxine but not thiamin, and mutant plants were selected based on their phenotype [27] and crossed with the TenA_E knockout line. The resulting F1 plants were selfed to generate the double homozygous mutant. Seed of the wild type, single, and double mutants were cultured on MS medium as above with or without 100 μM thiamin, HMP, or amino-HMP. The plated seeds were vernalised at 4°C for 5 days, germinated at 21 °C in continuous light (100 μmol.m−2.s−1), and imaged at two weeks. For paraquat (methyl viologen) stress experiments, wild type and TenA_E knockout seeds were grown on MS medium as above containing 0.1 μM paraquat or 0.1 μM paraquat supplemented with the specified concentrations of HMP or thiamin. Seeds were vernalised and grown as above except that plates were arranged vertically. Total root length was measured at 9 days. Stress experiments were performed with five to seven technical replications, each containing six seedlings per line, and were repeated at least three times.
Molecular modeling
The crystal structure of Arabidopsis TenA_E complexed with HMP at 2.1 Å resolution (PDB ID 2F2G) [23] was used as a template for modeling, and the structure of HMP already present in the active site of Arabidopsis TenA_E was used as a starting point for modeling the conformations of formylamino-HMP and amino-HMP. The program COOT [37] was used to manually dock the formylamino-HMP or amino-HMP molecules in the active site of Arabidopsis TenA_E based on alignment of the aromatic moiety of HMP as a reference. The most favorable conformations visualised for formylamino-HMP and amino-HMP in Arabidopsis TenA_E are shown. Version 1.6 of the PyMOL Molecular Graphics System (Schrödinger LLC) was used to generate all molecular graphics images presented in this paper. The same approach was used to manually dock a molecule of formylamino-HMP in the active site of B. subtilis TenA_C; in this case the crystal structure of the complex of B. subtilis TenA_C with HMP (PDB ID 1YAK) [17] was used as a template for modeling.
qRT-PCR analysis of maize gene expression
For analysis of maize TenA_E (GRMZM2G080501) and ThiC (GRMZM2G027663) expression, kernels from field-grown W22 inbred plants (Citra, Florida, spring 2013) at the stage indicated were harvested and their endosperms were immediately frozen in liquid N2 and held at −80°C until use. After grinding in liquid N2, total RNA was extracted with RLT buffer (2 ml per 20 mg) and purified using plant RNeasy kits. RNAs were quantified using a NanoDrop 1000 (Thermo Fisher Scientific), and 5.5 μg of RNA from each sample was treated with RQ1 RNase-free DNase (Promega). Negative RT controls were confirmed that there was no carryover of genomic DNA. For quantitative PCR, a Power SYBR green RNA-to-CT 1-step kit (Applied Biosystems) was used with an iCycler iQ real-time PCR detection system (Bio-Rad). The 18S rRNA was used as an internal reference for relative quantitative expression analysis [38]. The primers used are given in Supplementary Table S2.
RESULTS
Sequence and phylogenetic analysis of TenA proteins
We first verified that simple presence/absence of a conserved active-site cysteine residue (C135 in B. subtilis TenA) splits a full range of TenA proteins into two biologically meaningful subfamilies because the correlation with the presence of cysteine had previously been proposed based on analysis of just a small set of TenA sequences [21]. A set of 39 diverse TenA proteins from bacteria, archaea, plants, and yeast was identified using Blastp and Conserved Domain tools at NCBI. These sequences were separated into plus-cysteine (TenA_C) and minus-cysteine (TenA_E) classes, aligned, and subjected to phylogenetic analysis (Supplementary Figure S1). The plus- and minus-cysteine sequences fell neatly into two different clades, even when they came from the same organism. This result supports the active-site cysteine as a convenient marker that distinguishes two distinct subfamilies. It also suggests that these subfamilies are anciently diverged and are likely be functionally distinct. A further point worth noting is that the second Arabidopsis TenA protein (At5g32470) and its maize orthologues [3] are of the TenA_C class, so that plants are among the organisms that encode both TenA_E and TenA_C proteins. Plant TenA_E and TenA_C proteins are phylogenetically closest to the respective cyanobacterial proteins.
Comparative genomics connects both TenA_E and TenA_C with thiamin metabolism
We next surveyed the distribution of TenA family genes among >12,000 prokaryote genomes, using the SEED database and its tools [28]. This survey showed that TenA proteins are common in Cren- and Euryarchaeota, Bacteroidetes, Cyanobacteria, Firmicutes, and Proteobacteria, and occur at least occasionally in all other major taxa except Aquificae, Acidobacteria, and Thermotogae. A subset of 345 high-quality genomes from taxonomically and ecologically diverse organisms was then chosen for further analysis. Results are summarised below, and are available in full in the SEED database (http://pubseed.theseed.org//SubsysEditor.cgi?page=ShowSpreadsheet&subsystem=TenA).
Among the 345 selected genomes, 40% had at least one TenA gene: 27% had only TenA_C, 3% had only TenA_E, and 10% had both (Supplementary Table S1). TenA_E is thus less common than TenA_C, and occurs much more often with TenA_C than alone. This pattern reinforces the inference from sequence and phylogeny that the two subfamilies do not have identical functions. Consistent with functions in HMP salvage, TenA_C and TenA_E are found more than twice as often in organisms whose thiamin synthesis pathway depends on salvaging HMP from exogenous sources than in those that can make HMP, i.e. in organisms that, respectively, lack or have ThiC, and have otherwise complete suites of thiamin synthesis genes.
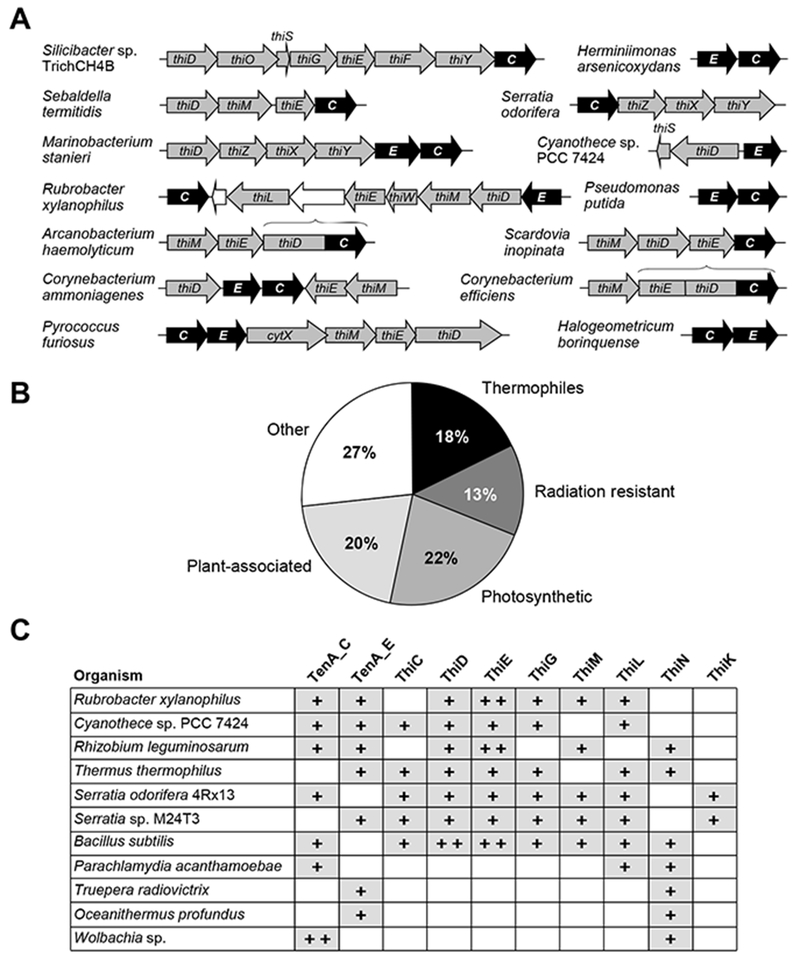
Further evidence that TenA_E and TenA_C have non-identical functions in thiamin salvage comes from gene-clustering and gene-fusion analyses. Genes of each subfamily commonly cluster on the chromosome with genes for thiamin synthesis and salvage enzymes or transporters, often in predicted operons. Furthermore, TenA_E and TenA_C quite commonly cluster with each other, again in putative operons (Figure 2A). The co-regulation of TenA_E and TenA_C implied by these operonic structures connotes non-redundant functions even more strongly than co-occurrence in the same genomes. Genes encoding TenA_C are fused to thiD, or thiD and thiE, in certain Actinobacteria (Figure 1A), fungi [19], protozoa (e.g. Perkinsus marinus) and green algae (e.g. Chlorella variabilis) (not shown). Genes coding for TenA_E seem not to be fused to other genes in any sequenced genome.
Figure 2. Comparative genomic analysis of the TenA family in prokaryotes.
(A) Chromosomal clustering of genes encoding TenA_C (C) and TenA_E (E) with thiamin synthesis and salvage genes, and genes encoding components of formylamino-HMP transporter ThiXYZ [8] or predicted thiamin salvage-related transporters ThiW and CytX [4]. Genes are shown as arrows pointing in the direction of transcription; overlaps denote translational coupling. Colour code: black, TenA protein genes; grey, thiamin-related genes; white, other genes. Horizontal braces mark gene fusions. The 14 species used as examples represent seven major taxa: Actinobacteria, Cyanobacteria, Euryarcheota, Fusobacteria, and α-, β-, and γ-Proteobacteria.
(B) Categorisation of prokaryotes whose genomes encode TenA_E (without or with TenA_C) according to their physiology and ecology. Forty-five organisms (Supplementary Table S1) were sorted into five classes: (i) thermophiles; (ii) resistant to ultraviolet or ionizing radiation, or colonizing light-exposed surfaces; (iii) photosynthetic; (iv) plant symbionts or plant biomass degraders; (v) other lifestyles. The chart shows the percentage of each class.
(C) The distribution of genes coding for TenA_C, TenA_E, and thiamin synthesis and salvage enzymes. The top eight rows show typical patterns for organisms that can make thiamin, either entirely de novo or from HMP and THZ precursors. The bottom four rows show patterns for organisms that are completely unable to make thiamin but have genes for TenA_C or TenA_E. Plus signs in grey boxes indicate presence of a gene or genes; open boxes indicate absence of a gene.
To get clues to the functional differences between TenA_E and TenA_C, we compared the lifestyles of the prokaryotes in which they occur. For TenA_C, no lifestyle bias was evident, but TenA_E was concentrated in thermophiles, phototrophs, plant-associated bacteria, and organisms resistant to UV or ionizing radiation (Figure 2B). Given the established lability of thiamin to heat, light, and radiation, and that these stresses lead to products that can differ from each other and from those formed metabolically or at high pH, [5,10–14], the lifestyle evidence raises the possibility that TenA_E proteins prefer as substrates certain thermo-, photo-, or radiodecomposition products formed from thiamin either intra- or extracellularly.
Comparative genomics points to non-salvage roles for TenA proteins
TenA genes usually occur in organisms that can make thiamin from HMP and THZ moieties, based on their having genes encoding ThiD, ThiE, ThiG or ThiM, and ThiL or ThiN (Figure 2C). However, TenA_E or TenA_C genes also occur in a small, disparate group of bacteria that have no biosynthetic genes except those for ThiN, or ThiN plus ThiL, and therefore have no thiamin biosynthesis capacity and require preformed thiamin (or its monophosphate) (Figure 2C). This group includes the radiation-resistant extremophile Truepera radiovictrix and the hydrothermal vent thermophile Oceanithermus profundus, which have TenA_E, as well as strains of the obligate intracellular bacteria Wolbachia and Parachlamydia acanthamoebae, which have TenA_C. Wolbachia genomes are highly reduced in size [39] and P. acanthamoebae genomes are somewhat reduced [40]. The presence of TenA_E or TenA_C in diverse organisms that cannot re-use thiamin breakdown products for thiamin biosynthesis implies that these enzymes have some function other than thiamin salvage. This implication is particularly strong for the obligate intracellular bacteria, in which genome reduction has jettisoned many other enzyme genes. One possible non-salvage function might be simply to catabolise thiamin breakdown products (as discussed below).
Enzymatic activities of recombinant plant TenA_E proteins
Among the organisms with TenA_E, only plants such as Arabidopsis and maize are genetically tractable and have experimentally characterised thiamin synthesis pathways for which mutants are available [3]. We therefore targeted Arabidopsis TenA_E (At3g16990) and its maize ortholog (GRMZM2G080501) for experimental characterisation. These proteins are predicted to be cytosolic and have not been detected in organelles [41]. Both proteins were expressed in E. coli in His-tagged form and isolated by Ni2+-affinity chromatography (Supplementary Figure S2), along with the well-studied B. subtilis TenA_C protein as a benchmark. All three proteins were then tested for activity against most of the thiamin degradation products shown in Figure 1B, as well as against thiamin and its phosphates (Table 1).
Table 1. Substrate ranges of Arabidopsis TenA_E proteins and B. subtilis TenA_C.
Substrate concentrations were 1 mM. Activities were determined at 30°C in 45 mM Tris-HCl buffer, pH 7.5, containing 0.9 mM DTT and 0.45 M glycinebetaine. Results are means ± S.E.M. of six replicates.
| Substrate | Specific activity (nmol min−1 mg−1) |
|
|---|---|---|
| Arabidopsis TenA_E | B. subtilis TenA_C | |
| Amino-HMP | 0.34 ± 0.05 | 292 ± 34 |
| Formylamino-HMP | 6.52 ± 0.31 | <0.05 |
| Thiamin | <0.05 | 162 ± 18 |
| Thiamin monophosphate | <0.05 | <0.05 |
| Thiamin diphosphate | <0.05 | <0.05 |
| Oxothiamin | <0.05 | 91 ± 20 |
| Oxythiamin | <0.05 | 205 ± 30 |
| Thiamin disulfide | <0.05 | <0.05 |
| Desthiothiamin | <0.05 | <0.05 |
| Thiochrome | <0.05 | <0.05 |
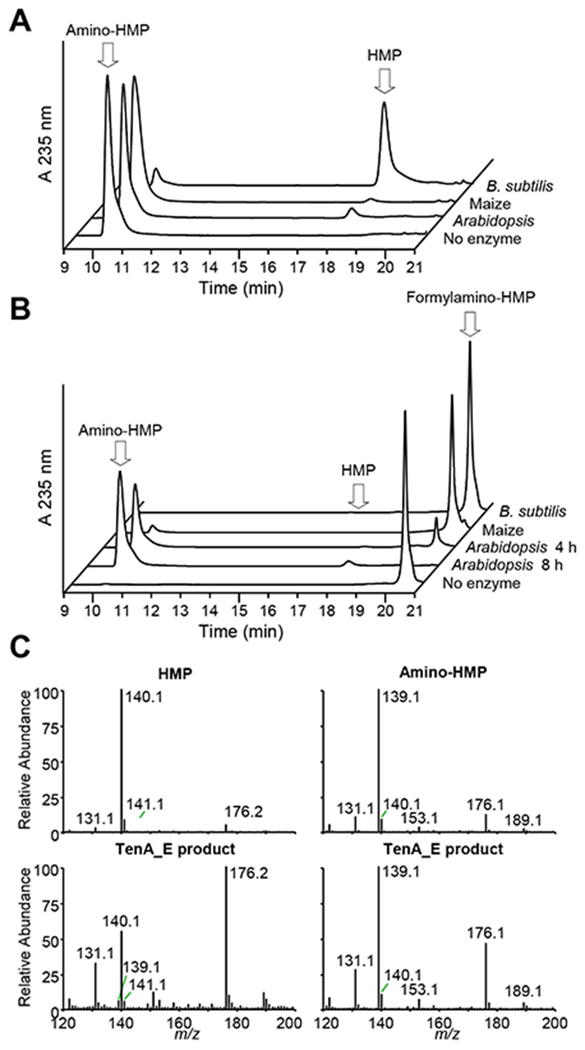
As expected for TenA_C proteins [2,19]. B. subtilis TenA_C hydrolyzed amino-HMP, thiamin, oxythiamin, and oxothiamin, but not formylamino-HMP or thiamin phosphates, thus validating our pro-cedures. In contrast, while both plant TenA_E proteins had some activity against amino-HMP, they had 20-fold more activity against formylamino-HMP, and none against the other compounds tested (Table 1; Figures 3A and 3B). The initial product of plant TenA_E action on formylamino-HMP was amino-HMP, which was then slowly hydrolyzed to HMP (Figure 3B). The identities of the successive reaction products were confirmed by mass spectrometry (Figure 3C). These results demonstrate that plant TenA_E proteins are bifunctional enzymes with formylamino-HMP amidohydrolase and amino-HMP aminohydrolase activities, and that they lack detectable thiaminase II activity. Kinetic characterisation of the formylamino-HMP amidohydrolase and amino-HMP aminohydrolase activities of the Arabidopsis enzyme showed that formylamino-HMP was very strongly preferred over amino-HMP, as demonstrated by a 19-fold higher kcat value and a 26-fold lower Km value (Table 2). However, even for the preferred substrate formylamino-HMP, the kcat value was quite low for a metabolic enzyme (2.81 × 10−3 s−1).
Figure 3. Evidence that plant TenA_E proteins have amino-HMP aminohydrolase and formylamino-HMP amidohydrolase activities.
(A) HPLC analyses of reaction mixtures (40 μl) in which amino-HMP (100 nmol) was incubated for 4 h at 30°C with Arabidopsis or maize TenA_E (50 μg), or B. subtilis TenA_C (0.5 μg) as a benchmark. A control incubated without enzyme is included.
(B) HPLC analyses of reaction mixtures (40 μl) in which formylamino-HMP (100 nmol) was incubated for 4 h at 30 °C with Arabidopsis or maize TenA_E (50 μg), or B. subtilis TenA_C (0.5 μg). The Arabidopsis reaction mixture was also incubated for 8 h. A control incubated without enzyme is included. Note that the formylamino-HMP preparation contained a trace (0.4 %) of amino-HMP.
(C) Electrospray mass spectra of authentic HMP (molecular ion [M+H+] = m/z 140.1) and amino-HMP (molecular ion [M+H+] = m/z 139.1) (upper panels), and the two products formed by Arabidopsis TenA_E from formylamino-HMP (lower panels). The signals at m/z 176 are solvent-related.
Table 2. Kinetic constants of the Arabidopsis TenA_E protein.
Results are means ± S.E.M. of three replicates.
| Substrate | 10−3 × kcat (s−1) | Km (μM) | kcat/Km (s−1 M−1) |
|---|---|---|---|
| Formylamino-HMP | 2.81 ± 0.23 | 70 ± 0.06 | 40.9 |
| Amino-HMP | 0.15 ± 0.007 | 1816 ± 200 | 0.08 |
Arabidopsis TenA_E acts as an amino-HMP aminohydrolase in planta
To test the in vivo function of TenA_E in thiamin salvage, we first identified a homozygous TenA_E knockout line (062985.53.80.x) in the Salk Arabidopsis T-DNA mutant collection. Plants of this mutant line were confirmed to have a T-DNA insertion in exon two and to lack detectable TenA_E mRNA, i.e. to be knockouts (Supplemental Figure S3). The TenA_E mutant was then crossed with a ThiC (py) mutant (which cannot synthesise the thiamin HMP moiety) [27] to give the double mutant.
The growth of the single and double mutants and of the wild type (Col-0) was then compared on MS medium alone, or supplemented with thiamin, HMP, or amino-HMP (Figure 4A). The growth of the TenA_E mutant was indistinguishable from that of the wild type on all four media. The HMP-supplemented medium gave slightly poorer growth than the other three, possibly because HMP is phosphorylated by ThiD and the resulting phospho-HMP, an analog of the cofactor pyridoxal 5’-phosphate, can be toxic at high levels [42]. As expected [27], the ThiC single and TenA_E ThiC double mutants showed little or no growth on MS medium alone, and fairly good growth on MS medium containing HMP. Most importantly, the ThiC mutant grew well on medium containing amino-HMP, whereas the double mutant hardly grew at all on this medium. This result demonstrates that TenA_E is required for hydrolysis of amino-HMP to HMP in vivo. This requirement prevented use of the TenA_E mutant to test whether TenA_E is also needed for formylamino-HMP hydrolysis because this step is upstream of amino-HMP hydrolysis.
Figure 4. Genetic evidence that Arabidopsis TenA_E can convert amino-HMP to HMP in vivo.
(A) Wild type (WT), TenA_E single mutant, ThiC single mutant, and TenA_E ThiC double mutant plants were cultured for two weeks on MS medium alone or containing 100 μM thiamin, HMP, or amino-HMP. Nine plants of each genotype occupy a quadrant on each plate. That the double mutant showed somewhat more growth in the absence of thiamin than the ThiC single mutant may be attributable to differences in thiamin content of the seeds, i.e. the amount of thiamin received from the maternal parent plants, which were irrigated with a 0.01% thiamin solution.
(B) Wild type and TenA_E single mutant seeds were germinated on MS medium containing 0.1 μM paraquat. Seedlings were grown vertically, and root length was measured after nine days. Results are means of 12-21 replicate plants ± S.E.M. Root lengths of wild type and TenA_E mutant plants grown on MS medium without paraquat were 42.5 ± 8.7 and 44.0 ± 6.7 mm, respectively. **, Difference between wild type and TenA_E mutant significant at P <0.002.
Because paraquat-induced oxidative stress and thiamin status are intertwined [43] and thiamin can be oxidatively degraded (Figure 1B), we compared the effect of paraquat on wild type and TenA_E mutant plants by measuring root growth, which paraquat strongly inhibits [43]. The TenA_E mutant was found to be significantly more sensitive to paraquat than the wild type (Figure 4B). Supplementation of the medium with 25 μM HMP eliminated this difference, as did supplementation with 25 μM thiamin (Figure 4B). However, while increasing the HMP concentration to 50 or 100 μM had no further effect on the mutant or wild type, increasing the thiamin concentration caused a proportional increase in growth, as noted previously for wild type roots [43]. Collectively, these data are consistent with salvage of the thiamin HMP moiety by TenA_E. Thus, oxidative stress is likely to promote thiamin degradation, so that salvage of the degradation products becomes a larger factor in maintaining thiamin pool levels, and TenA_E-mediated reclamation of HMP becomes more crucial. The observation that thiamin increases growth of both wild-type and mutant, while HMP does not, suggests that THZ becomes more limiting than HMP during oxidative stress, which fits with the greater stability of the latter compound [5].
Modeling of formylamino-HMP and amino-HMP in the active site of Arabidopsis TenA_E and Bacillus subtilis TenA_C
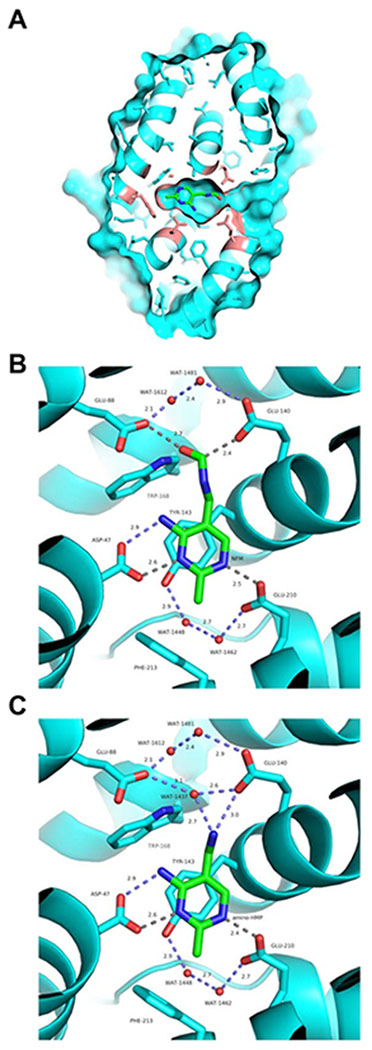
Because a crystal structure is available for Arabidopsis TenA_E with bound HMP [23,25], it was straightforward to manually model formylamino-HMP (Figure 5A) and amino-HMP (Figure 5B) in the active site. As for other TenA proteins [17,20,24], the active site of Arabidopsis TenA_E is largely sequestered from the bulk solvent and only a very narrow passage into the active site is observed when calculating the solvent accessible surface of the protein (Figure 5A). This ground-state structure implies that the enzyme is likely to have to have to undergo a conformational fluctuation of substantial magnitude in order to bind the substrate and release the product. Such a requirement could explain the relatively low turnover rates (kcat values) exhibited by the enzyme (Table 2).
Figure 5. Formylamino-HMP or amino-HMP modeled in the active site of Arabidopsis TenA_E.
The indicated HMP derivatives were manually docked into the experimentally determined crystal structure of Arabidopsis TenA_E containing bound HMP that co-purified with the enzyme (PDB ID 2F2G).
(A) The solvent-accessible surface of AtTenA_E with formylamino-HMP docked in the active site. The protein (cyan) is represented in both cartoon and stick formats. Note the narrow channel into the active site on the right.
(B) The environment of formylamino-HMP docked in the active site of Arabidopsis TenA_E. Residues within 4 Å of the formylamino-HMP molecule are shown in stick representation. Blue dotted lines show possible hydrogen bond interactions, while gray dotted lines show other interatomic distances (in Ångströms). Carbon atoms are green, nitrogen atoms are blue, and oxygen atoms are red. Crystallographically ordered water (WAT) molecules are depicted as red spheres.
(C) The environment amino-HMP docked in the active site of Arabidopsis TenA_E, displayed in an equivalent manner to panel A.
The structural models preserve the previously observed pi-pi interactions between Y143, F50 and the pyrimidine ring, and the hydrogen bonds (H-bonds) between D47 and the N4 and amino groups of the pyrimidine [23,25]. In the case of formylamino-HMP (Figure 5B), our model shows that the carboxylate groups of residues E88 and E140 are located within hydrogen-bonding distances of the carbonyl moiety of the formyl group of this ligand. This stereochemistry would enable E88 and E140 to participate in the amidohydrolase reaction mechanism and potentially act as general base catalysts. Although neither of these glutamate residues is conserved in all TenA_E orthologs, one or the other is always present (Supplemental Figure S1A). Furthermore, a pair of crystallographically ordered water molecules (wat-1612 and wat-1481) proximal to residues E88 and E140 in the active site could also participate in the amidohydrolase reaction mechanism.
These water molecules are preserved in their locations in our manual model with bound amino-HMP (Figure 5C), which also contains an additional crystallographically ordered water molecule (wat-1437) that had to be removed to model formylamino-HMP molecule in the active site. This third water molecule could play a functional role in the amidohydrolase reaction, because it makes strong interactions with E88/E140 and also the amino group of the aminomethyl moiety of amino-HMP.
Because B. subtilis TenA_C cannot hydrolyse formylamino-HMP but can hydrolyse the larger molecule thiamin (Table 1), it was of interest to model formylamino-HMP in the TenA_C active site. Manual modeling of formylamino-HMP into the HMP-bound structure of B. subtilis TenA_C (17) showed that it is possible to form a good H-bond between the formyl group of formylamino-HMP and the hydroxyl group on the side chain of residue Y163. Formation of this H-bond results in the nitrogen atom of formylamino-HMP being positioned 4.5 Å away from sulfhydryl group of the catalytic cysteine residue C135 (Supplementary Figure S4), which is likely to be too far away to catalyze cleavage. If this modeled structure is stable, formylamino-HMP could act as a competitive inhibitor of amino-HMP or thiamin hydrolysis by TenA_C enzymes. (No equivalent in silico docking exercise could be performed for a plant TenA_C because a structure has not yet been determined for any of these enzymes.)
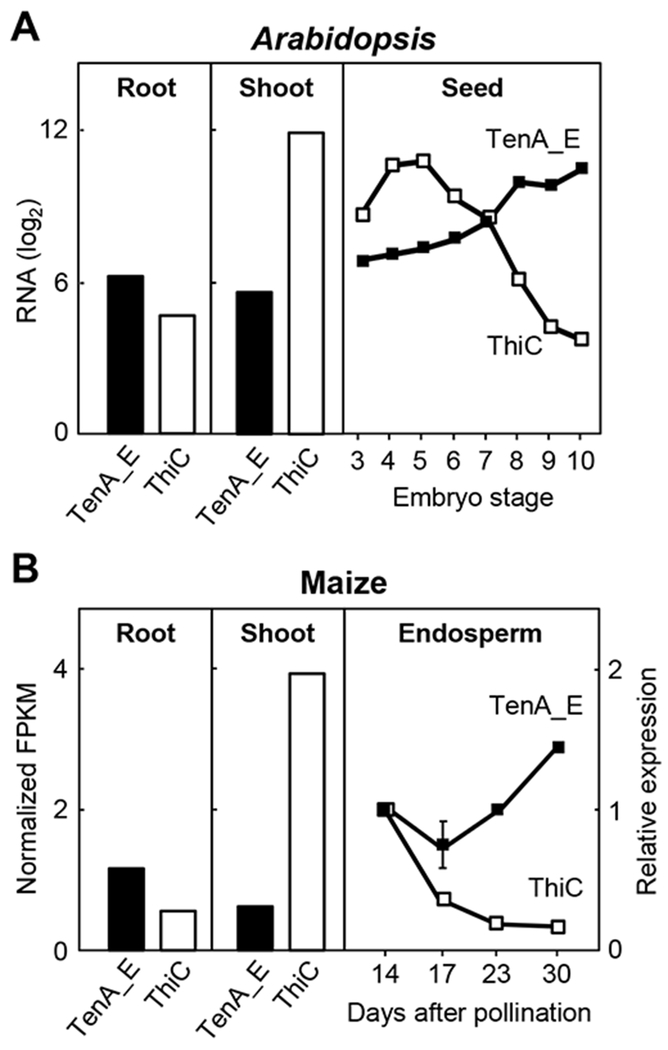
Complementary expression patterns of TenA_E and ThiC genes in Arabidopsis and maize
Because salvage of formylamino-HMP and amino-HMP via TenA_E is a potential alternative to de novo HMP synthesis via ThiC, and because ThiC expression varies greatly between tissues [44,45], we investigated whether the expression of TenA_E is inversely correlated with that of ThiC. We used CSB.DB microarray data for Arabidopsis [31], and a combination of qTeller RNA-seq [32] data and our own qRT-PCR data for maize. The Arabidopsis data showed that ThiC is expressed far more strongly in shoots than in roots, as previously reported [44], and that TenA_E is expressed modestly more in roots than in shoots (Figure 5A). The same pattern was evident in maize shoots and roots (Figure 6B). During Arabidopsis seed development, ThiC expression declined markedly whereas TenA_E expression rose (Figure 6A), and the same pattern was seen in maize endosperm (Figure 6B). These opposite trends in TenA_E and ThiC expression in various organs are consistent with the idea that TenA_E has an in-vivo salvage function that complements – and may sometimes partially replace – the capacity for de novo HMP synthesis.
Figure 6. Complementary expression patterns of plant TenA_E and ThiC genes.
(A) Expression of TenA_E and ThiC (At2g29630) genes in roots and shoots (above ground tissues) of seven-day-old plants, and developing seeds of Arabidopsis. Data were extracted from the CSB.DB microarray database and are expressed on a log2 scale.
(B) Expression of TenA_E and ThiC (GRMZM2G027663) genes in roots and shoots of seedlings, and developing endosperm of maize. FPKM (fragments per kilobase of exon per million fragments mapped) values were from seedling root and shoot libraries in the qTeller RNA-seq database and were normalised to the mean value for each gene in 25 qTeller datasets derived from diverse maize tissues. Endosperm data were obtained by qRT-PCR and are expressed relative to the value at 14 days after pollination (= 1.0). Data points are means ± S.E.M. for three replicates. Where no error bars appear they were smaller than the symbols.
DISCUSSION
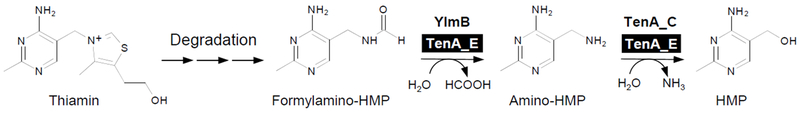
To our knowledge, the study reported in this paper is the first to demonstrate a biochemical function for the TenA_E subfamily of TenA proteins. The plant TenA_E proteins we tested exhibit dual formylamino-HMP amidohydrolase and amino-HMP aminohydrolase activities. Therefore, they are able to carry out two successive steps in the salvage of the thiamin breakdown product formylamino-HMP, whereas two separate enzymes, the amidohydrolase YlmB and the aminohydrolase TenA_C, mediate these steps in Bacillus species [8] (Figure 7). The amidohydrolase and aminohydrolase reactions catalyzed by plant TenA_E proteins are chemically quite distinct, as further discussed below.
Figure 7. Salvage of the HMP moiety of thiamin in plants and Bacillus species.
Formylamino-HMP derived from thiamin degradation is hydrolysed first to amino-HMP, and then to HMP, by different enzymes in plants and Bacillus species. In plants, the TenA_E protein catalyzes both steps. In Bacillus, the YlmB protein mediates the first (amidohydrolase) step, while the TenA_C protein mediates the second (aminohydrolase) step (8).
Although both plant TenA_E proteins prefer formylamino-HMP to amino-HMP in vitro, the Arabidopsis mutant data show that the activity against amino-HMP is physiologically significant. Thus, that the TenA_E ThiC double mutant can be rescued by HMP but not amino-HMP proves that the amino-HMP aminohydrolase activity of TenA_E is functionally important in vivo, at least when HMP synthesis is blocked by a ThiC mutation. That TenA_E functions similarly during normal development – waxing important in HMP salvage when HMP synthesis wanes – is suggested by the opposing expression patterns of TenA_E and ThiC. Because Arabidopsis has TenA_C as well as TenA_E, the failure of amino-HMP to rescue the double mutant also indicates that TenA_C cannot replace TenA_E amino-HMP aminohydrolase activity. Whether this is because Arabidopsis TenA_C lacks aminohydrolase activity, lacks expression, or lacks access to exogenous amino-HMP remains to be tested.
Our comparative analysis of prokaryotic genomes predicted the function of TenA_E so neatly that it could serve as a textbook example. Thus, the analysis predicted a role for TenA_E in thiamin salvage that is similar but not identical to that of TenA_C, and this exactly matches the experimental findings that plant TenA_E proteins have both aminohydrolase activity (like TenA_C) and amidohydrolase activity (unlike TenA_C). This correspondence between prediction and observation makes it probable that prokaryotic TenA_E proteins are also formylamino-HMP amidohydrolases. TenA_E proteins may therefore be the ‘missing’ amidohydrolase [8] that substitutes for YlmB in the many prokaryotes whose genomes encode TenA but not YlmB. If prokaryotic TenA_E proteins are amidohydrolases, and TenA_C proteins aminohydrolases (as in B. subtilis and H. pylori [8,20]), then the co-occurrence and chromosomal clustering of TenA_E and TenA_C genes would be explicable based on their specifying adjacent steps in thiamin salvage. Our data may also explain why the yeast TenA_E protein Pet18 can functionally replace E. coli ThiC when cells are cultured on minimal agar medium [46]. Agar has been shown to contain traces of HMP moieties [47], and these could be salvaged by amidohydrolase and/or aminohydrolase activities of Pet18.
Besides predicting a role for TenA_E in thiamin salvage, comparative genomic analysis pointed to connections between TenA_E and prokaryote lifestyles involving light, heat, ionizing radiation, or plants. If prokaryotic TenA_E proteins have activities like those of their plant counterparts, these connections are readily explained because: (i) UV photolysis [5] and probably thermolysis [13] of thiamin produce amino-HMP; (ii) Radiolysis produces a formylamino-HMP analog [48,49]; and (iii) Amino-HMP and formylamino-HMP from plants could be available to phytobacteria. Comparative genomics also pointed to a non-salvage role for TenA_E (and TenA_C) proteins because they occur in thiamin auxotrophs that rely on thiamin uptake from the environment or host. A plausible rationale for such occurrences is that some thiamin breakdown products are toxic and TenA renders them harmless. Amino-HMP is known to inhibit the thiamin transporter in yeast [50]; were this true of thiamin auxotrophs, hydrolysing amino-HMP could clearly benefit them. Whatever the case, as amino-HMP and formylamino-HMP are analogues of thiamin and of pyrimidine nucleobases, they are potential metabolic inhibitors. It is therefore reasonable to infer that TenA proteins can serve a damage pre-emption function [51] by hydrolyzing products that would otherwise do harm.
Although formylamino-HMP and amino-HMP were the only TenA_E substrates found in the panel of thiamin degradation products tested (Figure 1B), there is a wide range of other potential products, of which many may have a corrupt THZ moiety but an intact – and therefore salvageable – HMP moiety [5,8,11]. Such products could also be TenA_E substrates. Unfortunately they are complex, poorly known, and in some cases reactive, making them problematic to test in enzymatic assays in vitro.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. T.P. Begley for insightful discussion and Dr. Y. Kamiyoshihara for help with translation from Japanese.
FUNDING
Supported by U.S. National Science Foundation [grant numbers MCB-1153413 and IOS-1025398 to A.D.H., and MCB-0236210 to D.K.S.], by an endowment from the C.V. Griffin Sr. Foundation, by a General Research Fund grant from the Oregon State University Research Office [to A.G.], by N.I.H. to the Southeast Center for Integrated Metabolomics (U24 DK097209-01A1; T.J.G.), and by the Northeast Structural Genomics Consortium [2U54GM75026; J.F.H. and J.B.].
Abbreviations used:
- amino-HMP
4-amino-5-aminomethyl-2-methylpyrimidine
- formylamino-HMP
N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine
- HMP
4-amino-5-hydroxymethyl-2-methylpyrimidine
- THZ
4-methyl-5-(2-hydroxyethyl)thiazole
REFERENCES
- 1.Goyer A (2010) Thiamine in plants: aspects of its metabolism and functions. Phytochemistry 71, 1615–1624. [DOI] [PubMed] [Google Scholar]
- 2.Jurgenson CT, Begley TP and Ealick SE (2009) The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem 78, 569–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes S, Lerma-Ortiz C, Frelin O, Seaver SM, Henry CS, de Crécy-Lagard V and Hanson AD (2012) Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J. Exp. Bot 63, 5379–5395. [DOI] [PubMed] [Google Scholar]
- 4.Rodionov DA, Vitreschak AG, Mironov AA and Gelfand MS (2002) Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem 277, 48949–48959. [DOI] [PubMed] [Google Scholar]
- 5.Dwivedi BK and Arnold RG (1973) Chemistry of thiamine degradation in food products and model systems: a review. J. Agric. Food Chem 21, 54–60. [DOI] [PubMed] [Google Scholar]
- 6.McCourt JA, Nixon PF and Duggleby RG (2006) Thiamin nutrition and catalysis-induced instability of thiamin diphosphate. Br. J. Nutr 96, 636–638. [PubMed] [Google Scholar]
- 7.Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE and Begley TP (2011) Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins AH, Schyns G, Potot S, Sun G and Begley TP (2007) A new thiamin salvage pathway. Nat. Chem. Biol 3, 492–497. [DOI] [PubMed] [Google Scholar]
- 9.Pribat A, Blaby IK, Lara-Núñez A, Jeanguenin L, Fouquet R, Frelin O, Gregory JF 3rd, Philmus B, Begley TP, de Crécy-Lagard V and Hanson AD (2011) A 5-formyltetrahydrofolate cycloligase paralog from all domains of life: comparative genomic and experimental evidence for a cryptic role in thiamin metabolism. Funct. Integr. Genomics 11, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amos WH Jr. and Neal RA (1970) Isolation and identification of 3-(2’-methyl-4′-amino-5′-pyrimidylmethyl)-4-methylthiazole-5-acetic acid (thiamine acetic acid) and 2-methyl-4-amino-5-formylaminomethylpyrimidine as metabolites of thiamine in the rat. J. Biol. Chem 245, 5643–5648. [PubMed] [Google Scholar]
- 11.Chijate C, Albarran G and Negron-Mendoza A (1998) Radiolysis of aqueous solutions of thiamine. Radiat. Phys. Chem 52, 401–404. [Google Scholar]
- 12.Parkhomenko IM, Stepuro II, Donchenko GV and Stepuro VI (2012) Oxidized derivatives of thiamine: formation, properties, biological role. Ukr. Biokhim. Zh 84, 5–24. [PubMed] [Google Scholar]
- 13.Hartman GJ, Carlin JT, Scheide JD and Ho CT (1984) Volatile products formed from the thermal degradation of thiamin at high and low moisture levels. J. Agric. Food Chem 32, 1015–1018. [Google Scholar]
- 14.Vaid FHM, Shaikh RH, Ansari IA and Ahmad I (2006) Chromatographic study of the photolysis of aqueous thiamine hydrochloride and ascorbic acid solutions in the presence and absence of riboflavin. J. Chem. Soc. Pakistan 28, 461–464. [Google Scholar]
- 15.Yazdani M, Zallot R, Tunc-Ozdemir M, de Crécy-Lagard V, Shintani DK and Hanson AD (2013) Identification of the thiamin salvage enzyme thiazole kinase in Arabidopsis and maize. Phytochemistry 94, 68–73. [DOI] [PubMed] [Google Scholar]
- 16.Rapala-Kozik M, Olczak M, Ostrowska K, Starosta A and Kozik A (2007) Molecular characterization of the thi3 gene involved in thiamine biosynthesis in Zea mays: cDNA sequence and enzymatic and structural properties of the recombinant bifunctional protein with 4-amino-5-hydroxymethyl-2-methylpyrimidine (phosphate) kinase and thiamine monophosphate synthase activities. Biochem. J 408, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toms AV, Haas AL, Park JH, Begley TP and Ealick SE (2005) Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II. Biochemistry 44, 2319–2329. [DOI] [PubMed] [Google Scholar]
- 18.Haas AL, Laun NP and Begley TP (2005) Thi20, a remarkable enzyme from Saccharomyces cerevisiae with dual thiamin biosynthetic and degradation activities. Bioorg. Chem 33, 338–344. [DOI] [PubMed] [Google Scholar]
- 19.Onozuka M, Konno H, Kawasaki Y, Akaji K and Nosaka K (2008) Involvement of thiaminase II encoded by the THI20 gene in thiamin salvage of Saccharomyces cerevisiae. FEMS Yeast Res. 8, 266–275. [DOI] [PubMed] [Google Scholar]
- 20.Barison N, Cendron L, Trento A, Angelini A and Zanotti G (2009) Structural and mutational analysis of TenA protein (HP1287) from the Helicobacter pylori thiamin salvage pathway - evidence of a different substrate specificity. FEBS J. 276, 6227–6235. [DOI] [PubMed] [Google Scholar]
- 21.French JB, Begley TP and Ealick SE (2011) Structure of trifunctional THI20 from yeast. Acta Crystallogr. D. Biol. Crystallogr 67, 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itou H, Yao M, Watanabe N and Tanaka I (2004) Structure analysis of PH1161 protein, a transcriptional activator TenA homologue from the hyperthermophilic archaeon Pyrococcus horikoshii. Acta Crystallogr. D. Biol. Crystallogr 60, 1094–1100. [DOI] [PubMed] [Google Scholar]
- 23.Blommel PG, Smith DW, Bingman CA, Dyer DH, Rayment I, Holden HM, Fox BG and Phillips GN Jr. (2004) Crystal structure of gene locus At3g16990 from Arabidopsis thaliana. Proteins 57, 221–222. [DOI] [PubMed] [Google Scholar]
- 24.Benach J, Edstrom WC, Lee I, Das K, Cooper B, Xiao R, Liu J, Rost B, Acton TB, Montelione GT and Hunt JF (2005) The 2.35 A structure of the TenA homolog from Pyrococcus furiosus supports an enzymatic function in thiamine metabolism. Acta Crystallogr. D. Biol. Crystallogr 61, 589–598. [DOI] [PubMed] [Google Scholar]
- 25.Levin EJ, Kondrashov DA, Wesenberg GE and Phillips GN (2007) Ensemble refinement of protein crystal structures: validation and application. Structure 15, 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rédei GP (1965) Genetic blocks in the thiamine synthesis of the angiosperm Arabidopsis. Am. J. Bot 52, 834–841. [Google Scholar]
- 27.Li SL and Rédei GP (1969) Thiamine mutants of the crucifer, Arabidopsis. Biochem. Genet 3, 163–170. [DOI] [PubMed] [Google Scholar]
- 28.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M and Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinhauser D, Usadel B, Luedemann A, Thimm O and Kopka J (2004) CSB.DB: a comprehensive systems-biology database. Bioinformatics 20, 3647–3651. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS and Deng XW (2009) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21, 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ and McCarty DR (2012) Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 160, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurata G-I, Sakai T and Miyara T (1968) Studies on antagonists of thiamine : (XVIII) Reaction condition in formation of desthiothiamine from alkaline thiamine solution with amino acids. Vitamins 37, 398–402. [Google Scholar]
- 35.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 36.Murashige T and Skoog F (1962) A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 15, 473–497. [Google Scholar]
- 37.Emsley P and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bordenstein SR, Paraskevopoulos C, Dunning Hotopp JC, Sapountzis P, Lo N, Bandi C, Tettelin H, Werren JH and Bourtzis K (2009) Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol. Biol. Evol 26, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, et al. (2011) Unity in variety--the pan-genome of the Chlamydiae. Mol. Biol. Evol 28, 3253–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD and van Wijk KJ (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37, D969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haughton BG and King HK (1958) Toxo-pyrimidine phosphate as an inhibitor of bacterial enzyme systems that require pyridoxal phosphate. Biochem. J 70, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tunc-Ozdemir M, Miller G, Song L, Kim J, Sodek A, Koussevitzky S, Misra AN, Mittler R and Shintani D (2009) Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol. 151, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raschke M, Bürkle L, Müller N, Nunes-Nesi A, Fernie AR, Arigoni D, Amrhein N and Fitzpatrick TB (2007) Vitamin B1 biosynthesis in plants requires the essential iron sulfur cluster protein, THIC. Proc. Natl. Acad. Sci. U. S. A 104, 19637–19642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bocobza SE, Malitsky S, Araújo WL, Nunes-Nesi A, Meir S, Shapira M, Fernie AR and Aharoni A (2013) Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis. Plant Cell 25, 288–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morett E, Korbel JO, Rajan E, Saab-Rincon G, Olvera L, Olvera M, Schmidt S, Snel B and Bork P (2003) Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat. Biotechnol 21, 790–795. [DOI] [PubMed] [Google Scholar]
- 47.Karunakaran R, Ebert K, Harvey S, Leonard ME, Ramachandran V and Poole PS (2006) Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol 188, 6661–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xuetong F (2012) Radiation chemistry of food components In Food Irradiation Research and Technology (Fan X and Sommers CH, ed.), pp. 75–98, Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 49.Kawasaki C (1964) Modified thiamine compounds In Vitamins and Hormones, Vol. 21 (Harris RS, Wool IG. and Loraine JA, ed.), pp. 69–111, Academic Press, New York: [PubMed] [Google Scholar]
- 50.Iwashima A Kawasaki Y and Kimura Y (1990) Transport of 2-methyl-4-amino-5-hydroxymethylpyrimidine in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1022, 211–214. [DOI] [PubMed] [Google Scholar]
- 51.Linster CL, Van Schaftingen E and Hanson AD (2013) Metabolite damage and its repair or pre-emption. Nat. Chem. Biol 9, 72–80. [DOI] [PubMed] [Google Scholar]
- 52.Balaghi M and Pearson WN (1967) Comparative studies of the metabolism of 14C-pyrimidine-labeled thiamine, 14C-thiazole-labeled thiamine and 35S-labeled thiamine in the rat. J. Nutr 91, 9–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.