Abstract
The peptidoglycan fragments γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) and muramyl-dipeptide (MDP) are microbial-specific metabolites that activate intracellular pattern recognition receptors and stimulate immune signaling pathways. While extensive structure-activity studies have demonstrated that these bacterial cell wall metabolites trigger NOD1- and NOD2-dependent signaling, their direct binding to these innate immune receptors or other proteins in mammalian cells has not been established. To characterize these fundamental microbial metabolite-host interactions, we synthesized a series of peptidoglycan metabolite photoaffinity reporters and evaluated their crosslinking to NOD1 and NOD2 in mammalian cells. We show that active iE-DAP and MDP photoaffinity reporters selectively crosslinked NOD1 and NOD2, respectively, and not their inactive mutants. We also discovered MDP reporter crosslinking to Arf GTPases, which interacted most prominently with GTP-bound Arf6 and co-immunoprecipitated with NOD2 upon MDP stimulation. Notably, MDP binding to NOD2 and Arf6 was abrogated with loss-of-function NOD2 mutants associated with Crohn’s disease. Our studies demonstrate peptidoglycan metabolite photoaffinity reporters can capture their cognate immune receptors in cells and reveal unpredicted ligand-induced interactions with other cellular cofactors. These photoaffinity reporters should afford useful tools to discover and characterize other peptidoglycan metabolite-interacting proteins.
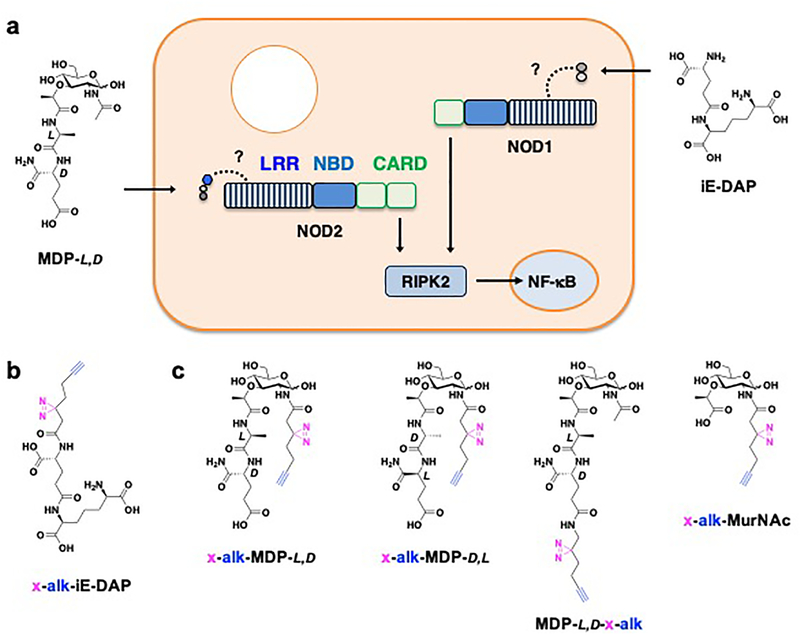
Peptidoglycan is a complex glycopeptide polymeric network that governs the structural integrity and shape of bacteria.1 The structure of peptidoglycan is composed of repeating disaccharide units of N-acetylglucosamine-β−1,4-muramic acid crosslinked via variable peptide stems, which differ between bacterial species and are remodeled during bacterial growth and division.1 As a highly conserved and unique feature of bacteria, peptidoglycan is a target of antibiotics and specifically recognized by plant, insect and animal immune systems for host defense and symbiosis.2, 3 Mammals have evolved a number of peptidoglycan recognition proteins and processing enzymes to detect and target bacteria in different tissues and cells.2, 3 Notably, the discovery of nucleotide-binding oligomerization domain-containing protein 1 and 2 (NOD1 and NOD2) revealed the key intracellular pattern recognition receptors in mammals that sense peptidoglycan fragments.4, 5 Structure-activity studies have shown that NOD1 and NOD2 are activated by γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) and muramyl-dipeptide (MDP),6 respectively (Figure 1a), which revealed an important link to earlier fractionation studies and active components of Freund’s adjuvant.7 The detection of iE-DAP and MDP is proposed to induce the oligomerization of NOD1 and NOD2, respectively, recruit RIPK2 binding and activate phosphorylation of downstream signaling factors to trigger the expression of NF-κB and MAP kinase-dependent genes involved in host immunity and pathogen clearance.4, 5
Figure 1. Photoaffinity reporters for investigating peptidoglycan metabolite interactions with intracellular pattern recognition receptors in mammalian cells.
(a) Proposed interactions of iE-DAP and MDP with NOD1 and NOD2, respectively. NOD1 and NOD2 contain leucine-rich repeat (LRR) domains (stripped rectangle), nucleotide-binding domain (NBD) domains (blue rectangle) and caspase recruitment domain (CARD) (green rectangles) required for activation of NF-κB-dependent genes involved in immune signaling and pathogen clearance. Dotted lines highlight proposed binding of peptidoglycan metabolites with LRRs. (b) Structure of iE-DAP photoaffinity reporter. (c) Structures of MDP photoaffinity reporters. Stereochemistry of key amino acids in MDP are indicated. Diazirine crosslinker and alkyne detection tag are highlighted in pink and blue, respectively.
NOD1 is ubiquitously expressed in many cell types and plays an important role in sensing iE-DAP that is most prevalent in Gram-negative bacteria.4, 5 Alternatively, NOD2 is selectively expressed in activated epithelial cells, Paneth cells, Lrg5+ intestinal stem cells, macrophages and dendritic cells and senses MDP, a conserved feature of Gram-negative and Gram-positive bacteria as well as mycobacteria.4, 5 Indeed, genetic ablation of Nod2 in mice revealed defects in both innate and adaptive immune responses.8 Moreover, intestinal inflammation was also observed in the absence of pathogenic microbes, which suggests NOD2 may also sense commensal bacteria from the gut microbiota.9 For example, our recent studies show that specific species of commensal bacteria such as Enterococcus faecium encode a secreted peptidoglycan hydrolase that can remodel peptidoglycan fragments and confer protection against intestinal pathogens in vivo,10 which requires the expression of NOD2 in mice11. Importantly, genetic analysis of inflammatory bowel disease patients has revealed several loss-of-function polymorphisms of NOD2 that are associated with Crohn’s disease,12, 13 suggesting NOD2 is crucial for maintaining intestinal barrier function and host immunity to invasive and inflammatory microbes.14 Alternatively, a constitutively active R334W mutant in the nucleotide-binding domain is associated with the inflammatory disorder Blau syndrome.15
Extensive structure-activity studies of iE-DAP and MDP have demonstrated their selective activation of NOD1- and NOD2-expressing cells, respectively.16–21 Nonetheless, the direct binding of iE-DAP and MDP to NOD1 and NOD2, respectively, as well as other proteins in mammalian cells has not been investigated (Figure 1a). Biotinylated analogs of MDP have suggested direct binding to NOD2 from cell lysates by streptavidin-bead pulldown22 or recombinant protein by surface plasma resonance (SPR) studies23–26. Similar SPR studies have also been performed for iE-DAP analogs.27 However, the use of biotinylated iE-DAP and MDP derivatives may affect cellular uptake and/or activity, which precluded their analysis with NOD1 and NOD2 in mammalian cells. To characterize the direct interaction of iE-DAP and MDP with NOD1 and NOD2 in living cells, we synthesized a series of peptidoglycan metabolite photoaffinity chemical reporters containing a diazirine for photocrosslinking in cells and an alkyne tag for bioorthogonal detection of covalently-labeled proteins (Figure 1b,c). The analysis of these peptidoglycan metabolite photoaffinity reporters demonstrate that iE-DAP and MDP can directly bind to NOD1 and NOD2 in mammalian cells. The crosslinking of these peptidoglycan metabolite photoaffinity reporters only occur with active variants of NOD1 and NOD2. Beyond direct binding to NOD2, the active MDP photoaffinity reporter selectively crosslinked Arf GTPases in NOD2-expressing cells and did so most prominently with GTP-bound Arf6. Additional labeling and co-precipitation experiments suggest that MDP induces a complex between NOD2 and GTP-Arf6. This study highlights the utility of peptidoglycan metabolite photoaffinity reporters for characterizing their direct binding to intracellular pattern recognition receptors and identifying unpredicted cellular cofactors. The applications of these microbial-specific metabolite reporters should enable the discovery of other receptors, transporters and regulatory enzymes in host cells and microbes.
RESULTS AND DISCUSSION
Analysis of peptidoglycan metabolite photoaffinity reporters with NOD1 and NOD2 in mammalian cells.
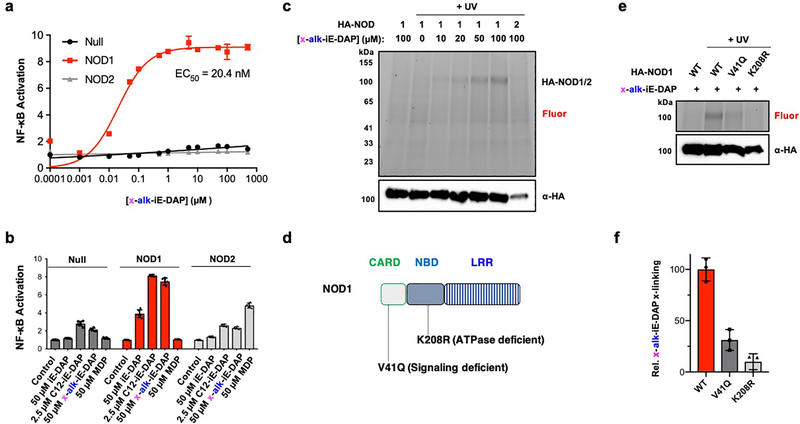
To evaluate potential covalent and non-covalent interactions of peptidoglycan metabolites, we synthesized bifunctional iE-DAP and MDP photoaffinity chemical reporters bearing a diazirine for photocrosslinking and a terminal alkyne for bioorthogonal detection (Figures 1b,c, S1 and S2), inspired by previous studies from our laboratory28 and others29–32. For iE-DAP photoaffinity reporter, we appended the diazirine and alkyne on the N-terminus of D-glutamate, which has been shown to tolerate functionalization with long-chain fatty acids that improve its activity towards NOD1 in cells.33 Indeed, x-alk-iE-DAP activated NOD1, but not NOD2, in HEK293T cells in a dose-dependent manner similar to iE-DAP derivatives (Figures 2a,b). NOD1-expressing HEK293T cells treated with x-alk-iE-DAP were then subjected to in-cell UV-crosslinking, and total cell lysates were reacted with azide-rhodamine and analyzed by in-gel fluorescence profiling (Figure S3). We observed UV- and dose-dependent crosslinking of x-alk-iE-DAP with HA-tagged NOD1 in total cell lysates (Figure 2c) and not with HA-NOD2, which expressed at lower levels. The UV-crosslinking of x-alk-iE-DAP only occurred with active NOD1 (Figure 2e,f) and was abrogated with inactivating mutations in the ATPase (K208R) and CARD (V41Q) domains that are proposed to mediate oligomerization and downstream signaling (Figure 2d)34, respectively.
Figure 2. iE-DAP photoaffinity chemical reporter reveals activity-dependent crosslinking with NOD1 in cells.
(a) NOD1-specific and x-alk-iE-DAP concentration-dependent activation of NF-κB reporter in HEK293T cells. (b) Activity of x-alk-iE-DAP in comparison to known NOD1 (iE-DAP and N-fatty-acylated C12-iE-DAP) and NOD2 (MDP) agonists. (c) UV- and concentration-dependent crosslinking of x-alk-iE-DAP to HA-NOD1, but not HA-NOD2, in HEK293T cells. Total cell lysates were reacted with az-rhodamine via click chemistry and visualized by in-gel fluorescence profiling. Anti-HA western blot was also performed to evaluate HA-NOD1/2 expression levels. (d) Human NOD1 is 953 amino acids in length composed of CARD, NBD and LRR domains. Key inactive mutants are highlighted. (e) In-gel fluorescence profile of total cell lysates from x-alk-iE-DAP (50 μM) photocrosslinking with HA-NOD1 mutants in HEK293T cells. (f) Quantitation of x-alk-iE-DAP crosslinking with HA-NOD1 variants by ImageJ analysis. Data represent average and standard deviation of three independent replicates in (e).
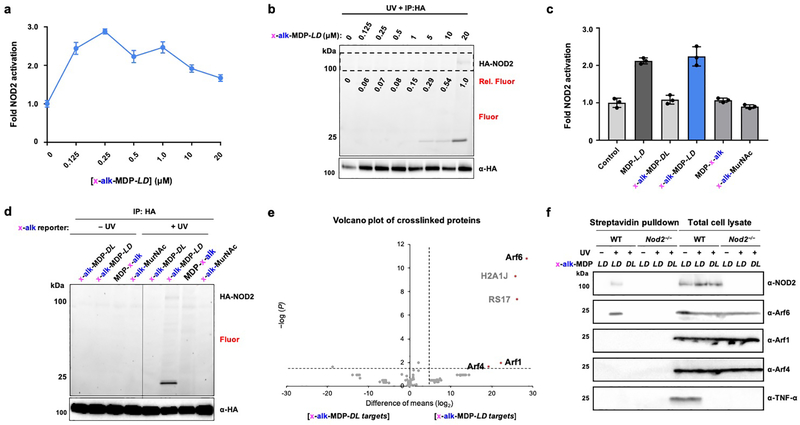
To investigate MDP interactions with NOD2 in cells, we installed the bifunctional alkyne and diazirine crosslinker on the N-acyl group of MDP (x-alk-MDP), which is known to tolerate modifications,6 and evaluated its activity as well as protein crosslinking in NOD2-expressing HEK293T cells. This bifunctional MDP reporter activated NOD2 in a dose-dependent manner (Figure 3a) and crosslinked immunoprecipitated HA-NOD2 (Figure 3b). While x-alk-MDP activation of NOD2 saturated at approximately 200 nM, robust photocrosslinking by in-gel fluorescence was only detected with 20 μM of the photoaffinity reporter (Figure 3b), which likely reflects the differential sensitivity of direct metabolite-protein crosslinking versus enzyme-mediated transcriptional reporter assays that we also observed with x-alk-iE-DAP and NOD1 (Figure 2a,c). Unlike x-alk-iE-DAP crosslinking of NOD1, x-alk-MDP crosslinking to HA-NOD2 was not readily observed in total cell lysates (Figure S4a), perhaps due to lower levels of HA-NOD2 expression (Figure 2c) and/or MDP reporter uptake, affinity and crosslinking efficiency. Interestingly, x-alk-MDP also crosslinked proteins around 20 kilodaltons (kDa) in the HA-NOD2 immunoprecipitated samples (Figure 3b) and total cell lysates (Figure S4a), which could be competed away with MDP as well as GlcNAc-MDP (Figure S4b,c). The ~20 kDa protein crosslinking was specific to active x-alk-MDP-L,D and did not occur with the inactive x-alk-MDP-D,L diastereomer, the peptide stem-modified MDP-x-alk isomer or muramic acid derivative (x-alk-MurNAc) at the 20 μM in HA-NOD2 immunoprecipitated samples w(Figure 3c,d) or total cell lysates (Figure S4d). Notably, x-alk-MDP-L,D protein crosslinking was not observed with NOD1 and TLR2 expression and activation (Figure S5a,b), demonstrating x-alk-MDP crosslinking to the 20 kDa proteins is specific to NOD2 expression and not due to indirect activation of NF-κB. These results provide direct evidence for the binding of active iE-DAP and MDP analogs to NOD1 and NOD2 in mammalian cells.
Figure 3. MDP photoaffinity chemical reporters reveals activity-dependent crosslinking with NOD2 and Arf GTPases in mammalian cells.
(a) Analysis of NOD2 activation by x-alk-MDP in HA-NOD2-transfected HEK293T cells. (b) In-gel fluorescence analysis and click chemistry labeling with az-rhodamine of immunoprecipitated HA-NOD2 after x-alk-MDP photocrosslinking in HEK293T cells. (c) NOD2 activation by MDP and different x-alk-MDP photoaffinity reporters at 20 μM. (d) In-gel fluorescence analysis and click chemistry labeling with az-rhodamine of immunoprecipitated HA-NOD2 after photocrosslinking with different x-alk-MDP photoaffinity reporter (20 μM) in HEK293T cells. (e) Volcano-plot analysis of ~20 kDa proteins recovered from NOD2-transfected HEK293T cells crosslinked with active x-alk-MDP-L,D versus inactive D,L-diastereomer, reacted with az-biotin, enriched with streptavidin beads and separated by SDS-PAGE. (f) Western blot analysis of endogenous Arf6, 4 and 1 in primary murine bone marrow-derived macrophages from wild-type and Nod2−/− mice, treated with x-alk-MDP (20 μM), + or - UV crosslinking. The cell lysates were then reacted with az-biotin, enriched with streptavidin beads and separated by SDS-PAGE. TNF-α expression in total cell lysates were also analyzed in parallel to evaluate activation of endogenous Nod2 in primary murine bone marrow-derived macrophages.
Discovery of muramyl-dipeptide photoaffinity reporter crosslinking to Arf GTPases.
To identify the ~20 kDa x-alk-MDP-crosslinked proteins in NOD2-expressing HEK293T cells (Figure 3d), the corresponding total cell lysates were reacted with azide-biotin and affinity purified with streptavidin beads. Our initial analysis of trypsin-digested streptavidin beads did not reveal any selective x-alk-MDP-crosslinked proteins (data not shown). The streptavidin-captured proteins were therefore eluted, separated by SDS-PAGE, and polypeptides around 100 and 20 kDa were processed for identification by mass spectrometry (Figures S6a). LC-MS/MS analysis of the gel slices around 100 kDa did not reveal any tryptic peptides for NOD2 or other proteins specifically crosslinked by x-alk-MDP-L,D reporter (Figures S6b,c and Tables S1 and S2). However, the comparative analysis of two different control samples (no UV-crosslinking and inactive D,L-diastereomer) with four independent replicates by label-free quantitation revealed that Arf GTPases (6, 4 and 1) were candidate x-alk-MDP-crosslinked proteins around 20 kDa (Figure 3e, S6d and Tables S3 and S4). Western blot analysis of the biotinylated and affinity-purified proteins showed that endogenous Arf6 was indeed captured by x-alk-MDP-crosslinking in HEK293T cells (Figure S6e). Our x-alk-MDP reporter also crosslinked endogenously expressed NOD2 and Arf6 and stimulated TNF-α expression in primary bone marrow-derived macrophages from wild-type (Nod2+/+) but not Nod2−/− mice (Figure 3f and Figure S7), suggesting MDP interacts with Arf6 and NOD2 expressed at physiological levels.
Specificity of muramyl-dipeptide photoaffinity reporter crosslinking to Arf GTPases.
The Arf GTPases are important regulators of membrane trafficking in eukaryotic cells.35 These small GTPases are targeted to cellular membranes by N-terminal myristoylation and are regulated by guanine nucleotide loading, which alters their conformation and recruits specific protein effectors responsible for membrane fusion and vesicle trafficking.36 The guanine nucleotide state is regulated by guanine nucleotide exchange factors (GEFs) that afford GTP-bound Arfs that are hydrolyzed to their guanosine-5’-diphosphate (GDP)-bound forms by GTPase-activating proteins (GAPs).35 As key regulators of membrane trafficking, the Arf GTPases have not only been implicated in cellular homeostasis,35 but also play important roles in controlling the turnover of cell surface receptors and uptake of antigens involved in host immunity37 and specifically targeted by microbial pathogens38. Arf6 in particular has been suggested to regulate the endocytosis of Toll-like receptors (4 and 9) and affect their downstream signaling.39–42 We therefore further characterized NOD2-dependent MDP interaction with Arf GTPases.
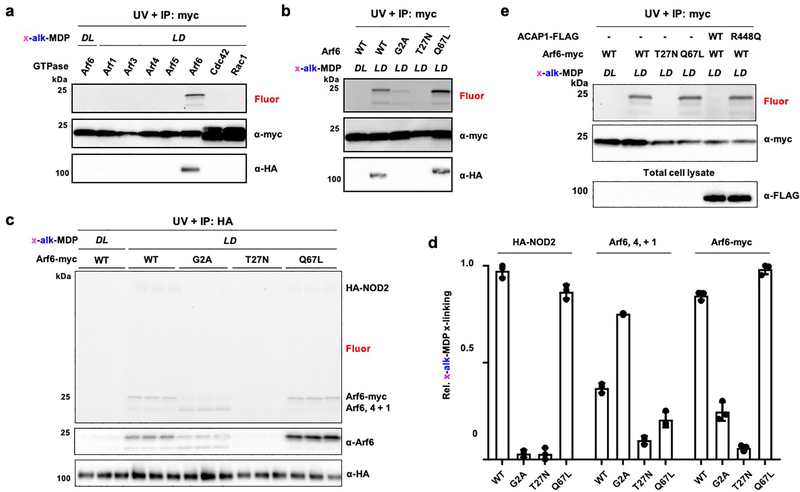
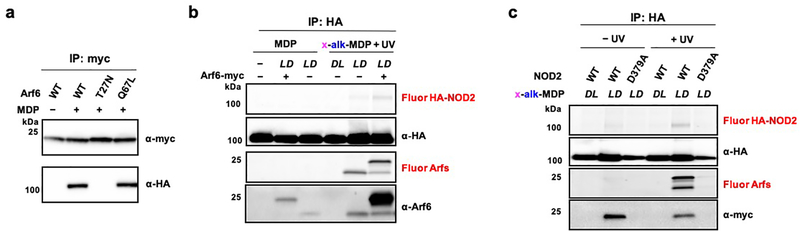
To validate and determine the specificity of x-alk-MDP crosslinking, we evaluated the expression and immunoprecipitation of C-terminal myc-tagged GTPases. Indeed, x-alk-MDP selectively crosslinked myc-tagged Arf6 (Figure 4a), but not Arfs (1, 3, 4 and 5) or other prominent small GTPases Cdc42 or Rac1, which have been suggested to modulate NOD1 and NOD2 activity.43 Our x-alk-iE-DAP reporter also did not photocrosslink Cdc42, Rac1 or RhoA in HA-NOD1 expressing cells (Figure S8), nor did we observe co-immunoprecipitation of x-alk-iE-DAP-crosslinked NOD1 with any GTPase. In contrast, immunoprecipitation of x-alk-MDP-crosslinked Arf6-myc with anti-myc beads readily recovered HA-NOD2, which did not occur with the inactive x-alk-MDP diastereomer (Figure 4a). The x-alk-MDP crosslinking appears to be selective for the membrane-associated and GTP-bound form of Arf6, as the N-myristoylation mutant (G2A) and GDP-bound Arf6 (T27N) were both not crosslinked compared to the wild-type and GTP-bound Arf6 (Q67L) (Figure 4b). Interestingly, we observed two fluorescent crosslinked bands upon transfection with Arf6-myc, suggesting that labeling of endogenous Arfs can still be observed in the presence of over-expressed Arf6-myc (Figure 4a,b).
Figure 4. x-alk-MDP selectively crosslinks to GTP-Arf6.
(a) In-gel fluorescence analysis and click chemistry labeling with az-rhodamine of anti-Myc immunoprecipitated C-terminal myc-tagged small GTPases after x-alk-MDP (20 μM) crosslinking in HA-NOD2 co-transfected HEK293T cells. (b) x-alk-MDP (20 μM) crosslinking and anti-myc immunoprecipitation of wild-type and Arf6 mutants with HA-NOD2 from HEK293T cells, as performed in (a). (c) In-gel fluorescence profile and click chemistry labeling with az-rhodamine of anti-HA immunoprecipitated HA-NOD2 and Arf6-myc variants as well as endogenous Arf6, 4 and 1 after x-alk-MDP (20 μM) crosslinking in co-transfected HEK293T cells. (d) Quantification of x-alk-MDP crosslinking Arf6-myc variant in (c) by ImageJ analysis. Data represent average and standard deviation of independent replicates shown in (c). (e) In-gel fluorescence profile of x-alk-MDP (20 μM) crosslinking with endogenous Arfs in HEK293T cells co-transfected with HA-NOD2 and Arf6-myc variants, wild-type or inactive ACAP1 (Arf6-GAP).
We then explored the effects of Arf6 variants on x-alk-MDP crosslinking and observed that expression of the wild-type Arf6 and GTP-Arf6 (Q67L) mutant afforded similar levels of x-alk-MDP-NOD2 crosslinking, which also competed for the crosslinking of endogenously expressed Arf6, 4 and 1 (Figure 4c,d). The N-myristoylation mutant Arf6 mutant (G2A) was only modestly crosslinked by x-alk-MDP and did not significantly compete for crosslinking of endogenous Arfs (Figure 4c,d). In contrast, expression of the GDP-Arf6 (T27N) mutant was not crosslinked by x-alk-MDP and even abrogated crosslinking with endogenously expressed Arf6, 4 and 1 (Figure 4c,d). To independently confirm that GDP-Arf6 interferes with x-alk-MDP crosslinking of NOD2 and Arf GTPases, we expressed the Arf GTPase-activating protein ACAP144 and also observed that x-alk-MDP crosslinking to NOD2 and endogenous Arf6, 4 and 1 were abrogated (Figure 4e). This effect was ACAP1 activity-dependent as the catalytically inactive ACAP1-R448Q mutant44 did not diminish x-alk-MDP crosslinking to HA-NOD2 or endogenous Arf-GTPases (Figure 4e).
MDP induces the co-immunoprecipitation of NOD2 and Arf6 GTPase.
In addition to x-alk-MDP crosslinking, only wild-type and GTP-bound Arf6 (Q67L) immunoprecipitated HA-NOD2 in the presence of unmodified MDP without UV-crosslinking (Figure 5a), suggesting MDP forms a non-covalent, ternary complex with GTP-Arf6 and NOD2 in cells. Inverse immunoprecipitation of HA-NOD2 also recovered endogenous and transfected Arf6-myc only upon MDP treatment (Figure 5b), further supporting the formation of a NOD2:MDP:Arf6 ternary complex. Co-immunoprecipitation of Arf6 was also observed with x-alk-MDP photocrosslinking, but was not with its inactive diastereomer or an inactive ATPase-deficient NOD2 mutant (D379A) (Figure 5c), underscoring the chemical and functional selectivity of the complex.
Figure 5. MDP induces co-immunoprecipitation NOD2 and Arf6.
(a) Western blot analysis of anti-myc immunoprecipitation of wild-type and Arf6 mutants in HA-NOD2-transfected HEK293T cells ± 1 μM MDP. (b) In-gel fluorescence analysis and click chemistry labeling with az-rhodamine of anti-HA immunoprecipitated HA-NOD2 from HEK293T cells co-transfected with Arf6-myc ± MDP (1 μM) or x-alk-MDP (20 μM) with UV-crosslinking. Western blot analysis of HA-NOD2 and Arf6 expression levels. (c) In-gel fluorescence analysis and click chemistry labeling with az-rhodamine of immunoprecipitated HA-NOD2 (wt and D379A mutant) from HEK293T cells co-transfected with wt Arf6-myc ± x-alk-MDP (20 μM) crosslinking.
MDP interacts with Arf4 and 1 in the absence of Arf6.
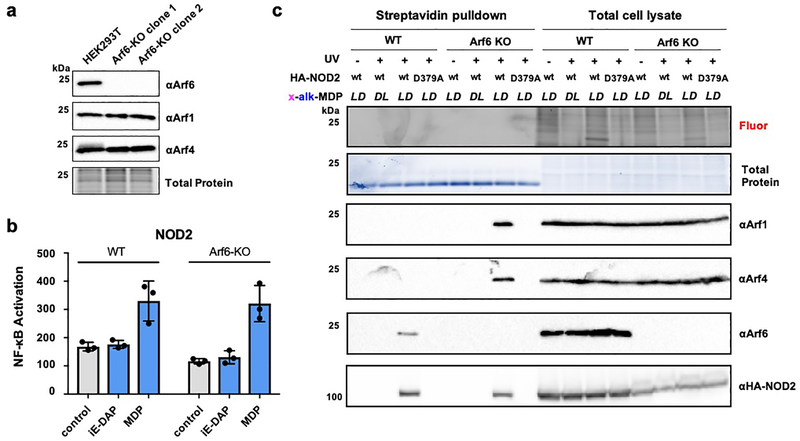
To explore the effects of GTP-Arf6 on NOD2 activity, we generated Arf6-KO HEK293T cells by CRISPR-Cas9 gene editing (Figure 6a). Surprisingly, the Arf6-KO cells afforded similar levels of NOD2 activation compared to wild-type HEK293T cells (Figure 6b), even at higher doses of MDP (Figure S9). We then performed further crosslinking experiments in the Arf6-KO cells and discovered that x-alk-MDP crosslinked to NOD2 and Arf4 and 1 in the absence of Arf6 (Figure 6c), which suggests Arf4 and 1 binding may compensate for Arf6 depletion and regulate NOD2 activity. Indeed, our proteomic data indicated that x-alk-MDP did crosslink to endogenous Arf4 and 1, but at lower levels compared to Arf6 (Figure 3e, S6d and Table S3). These results and the analysis of transfected Arf4 and 1 (Figure 4a) suggest MDP selectively binds to NOD2 and GTP-Arf6 but may also interact with Arf4 and 1 in the absence of Arf6, which can be disrupted with increasing levels of GDP-Arf6.
Figure 6. MDP binds to NOD2, Arf4 and 1 in the absence of Arf6.
(a) Western blot analysis of endogenous Arf6, Arf4 and Arf1 in wt and Arf6-KO HEK293T cell lines. (b) NF-κB luciferase activity in wt and Arf-KO HEK293T cell lines transfected with HA-NOD2 and treated with iE-DAP (20 μM) or MDP (20 μM). (c) In-gel fluorescence analysis (reacted with az-rhodamine) and western blot analysis of wt and Arf6-KO HEK293T cells treated with x-alk-MDP (20 μM) reporters, + or - UV-crosslinking. For streptavidin pulldown, total cell lysates were reacted with az-biotin, incubated with streptavidin beads and eluted for western blot analysis with antibodies to endogenous Arf6, Arf4, Arf1 and HA-NOD2.
MDP binding to the NOD2 and Arf6 is associated with NOD2 disease mutations.
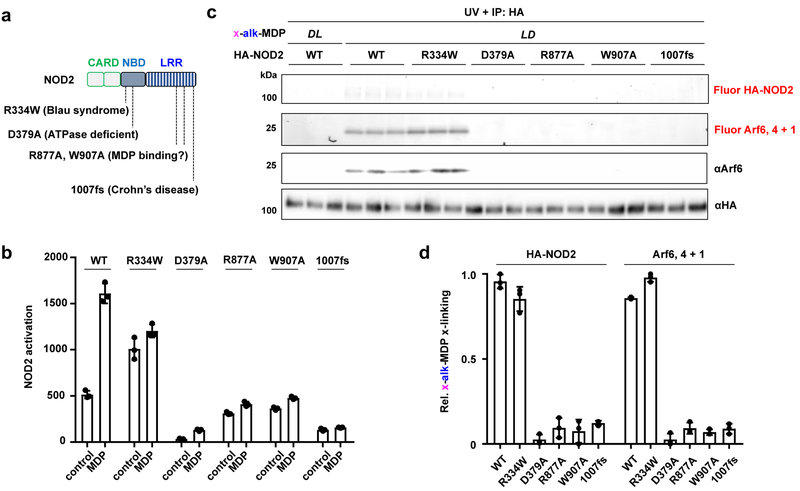
To determine the significance of the NOD2:MDP:GTP-Arf6 interaction, we evaluated disease mutations and candidate MDP binding mutants of NOD2 (Figure 7a). For these experiments, we focused on the constitutively active R334W mutant associated with the Blau syndrome,15 and the most prominent loss-of-function Crohn’s disease mutant 1007fs,12 which encodes a frameshift mutation in the LRR domain. Based on the X-ray structure of monomeric NOD2, we also evaluated R877A and W907A mutants in the LRR domain that were proposed to mediate MDP binding.45 The analysis of these NOD2 variants showed that x-alk-MDP crosslinked to the active NOD2 R334W Blau syndrome mutant and Arf6 at similar levels as wild-type NOD2 (Figure 7b–d). In contrast, x-alk-MDP crosslinking to Arf6 was abrogated with the inactive NOD2 LRR R877A, W907A (potential MDP binding) and 1007fs Crohn’s disease mutants, similar to ATPase- and oligomerization-deficient D379A mutant (Figure 7b–d). These results demonstrate x-alk-MDP crosslinking with Arf6 only occurs with active NOD2 variants (wild-type and R334W Blau syndrome mutant) and suggests that MDP and Arf6 binding is impaired in loss-of-function NOD2 LRR mutants associated with Crohn’s disease in humans.
Figure 7. MDP reporter crosslinking to the NOD2:Arf6 complex is associated gain- and loss-of-function human NOD2 mutations.
(a) Human NOD2 is 1040 amino acids in length composed of tandem-CARD, NBD and LRR domains. Key mutants are highlighted. (b) MDP-dependent NF-κB activation activity of wt and NOD2 variants in HEK293T cells. (c) In-gel fluorescence profile of x-alk-MDP (20 μM) crosslinking and expression of immunoprecipitated HA-NOD2 mutants as well as endogenous Arf6, 4 and 1. Each HA-NOD2 variant was analyzed in triplicate. (d) Quantitation of x-alk-MDP crosslinking of HA-NOD2 mutants by ImageJ analysis. Data represent average and standard deviation of independent replicates shown in (c).
CONCLUSIONS
Peptidoglycan metabolites are sensed and remodeled by diverse protein receptors and enzymes in animals to activate and regulate host immunity.2, 3 Nonetheless, the direct biochemical interactions of these microbial-specific metabolites with their proposed receptors and enzymes have been difficult to observe due to potential transient and non-covalent interactions. Indeed, iE-DAP and MDP were discovered to activate the intracellular pattern recognition receptors NOD1 and NOD2, respectively, over a decade ago,4, 5 but their direct binding to these intracellular pattern recognition receptors in cells has not been demonstrated. To address this key question, we synthesized active photoaffinity reporters of iE-DAP and MDP and showed that they can directly and specifically crosslink active NOD1 and NOD2 in mammalian cells, respectively (Figures 2 and 3). Unexpectedly, we discovered that MDP also binds to Arf GTPases, predominantly Arf6 in mammalian cell lines and primary macrophages (Figure 4 and 5). The analysis of Arf6 variants showed MDP and NOD2 preferentially binds to the N-myristoylated GTP-bound form of Arf6 (Figure 4).
Our discovery of direct MDP binding to both NOD2 and GTP-Arf6 reveals a novel and direct biochemical mechanism for regulating innate immune receptor activation. Previous studies have shown that Rho-family GTPases can modulate NOD1 and NOD2 activity,43 but these proteins do not appear to directly interact with peptidoglycan metabolites, as we did not observe crosslinking to Rac1, RhoA or Cdc42 with x-alk-MDP or x-alk-iE-DAP (Figures 4a and S8). Alternatively, both GTP- and GDP-variants of Arf6 have been suggested to inhibit the endocytosis and signaling of TLR4 and 9 indirectly.39–42 In contrast, we observed that only GDP-bound Arf6 via Arf6-T27N or ACAP1 expression inhibits MDP:NOD2:GTP-Arf6 interactions (Figure 4). Moreover, we observed direct x-alk-MDP crosslinking as well as MDP-induced co-immunoprecipitation of GTP-Arf6 and NOD2 (Figure 4c,d and 5), which suggests that these Arf GTPases may directly regulate NOD2 localization, oligomerization and/or recruitment of downstream signaling components. In the absence of Arf6, MDP-induced NOD2 binding to Arf4 and 1 (Figure 6), which suggests NOD2 interaction with Arf GTPases is crucial and retains activity in Arf6 deficient cell-types. GTPase regulation of cytosolic pattern recognition receptors could be a general mechanism, as interferon-induced GTPases such as guanylate binding protein 5 (GBP5) have also been suggested to regulate the assembly of NLRP3 inflammasomes.49
Notably, formation of the NOD2:MDP:Arf GTPase complex is abrogated with loss-of-function NOD2 LRR and Crohn’s disease mutants (Figure 7) and also disrupted by GDP-Arf6 (Figure 5), suggesting this complex is important for NOD2 activation in human disease and is regulated by GTP loading of Arf GTPases. These results are consistent with previous studies that also demonstrate ACAP1 expression inhibited NOD2 activity,46 which we now demonstrate is associated with MDP and NOD2 binding of Arf (6, 4 and 1) GTPases. Interestingly, patients with inflammatory bowel disease (IBD) were found to harbor inactivating mutations in C1orf106/INAVA, an upstream regulator of GTP-Arf6 levels.50 C1orf106/INAVA is proposed to inhibit the activity of cytohesin-1, an Arf6 guanidine exchange factor (GEF). C1orf106 mutant cells thus exhibit higher levels of GTP-Arf6, which enhanced the endocytosis of tight junction factors such as cadherin that may result in compromised intestinal barrier.50 Our discovery of MDP:NOD2:GTP-Arf6 complex formation suggests loss-of-function mutants in GTP-Arf6 regulators such as C1orf106/INAVA and cytohesin-1 may also affect NOD2 activity. Indeed, recent additional studies have shown that expression of C1orf106/INAVA modulates ARNO (GTP-Arf6 GEF) regulation of MDP-induced NOD2 activation.51 The cellular mechanisms and structural basis by which MDP directly binds NOD2 and GTP-Arf6 is unknown and ongoing in our laboratory, which should provide important insights into innate immune activation and intestinal barrier homeostasis. As bacterial pathogens such as Salmonella enterica serovar Typhimurium47 and enterohaemorrhagic Escherichia coli48 inject proteins into host cells that modulate or directly bind Arf GTPases, these bacterial virulence factors may also suppress innate immune detection by interfering with MDP binding to NOD2 and GTP-Arf6, which should also be an interesting area of future investigation in pathogen immune evasion. In summary, this study highlights the utility of peptidoglycan metabolite photoaffinity reporters for characterizing direct receptor binding in cells and discovering unpredicted regulatory factors, which should afford useful tools for exploring additional peptidoglycan metabolite-protein interactions in other cell types, tissues, whole animals and microbes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank H. Molina for assistance with proteomic analysis (The Rockefeller University Proteomics Resource Center). Y.C.W. was a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. H.C.H. acknowledges support from the National Institutes of Health (NIGMS R01-GM103593) and the Robertson Therapeutic Fund.
Footnotes
Accession Codes
The proteome data sets are available via ProteomeXchange with identifier PXD012350.
The authors declare no competing financial interests.
REFERENCES
- [1].Vollmer W, Blanot D, and de Pedro MA (2008) Peptidoglycan structure and architecture, FEMS Microbiol Rev 32, 149–167. [DOI] [PubMed] [Google Scholar]
- [2].Wolf AJ, and Underhill DM (2018) Peptidoglycan recognition by the innate immune system, Nat Rev Immunol 18, 243–254. [DOI] [PubMed] [Google Scholar]
- [3].Royet J, Gupta D, and Dziarski R (2011) Peptidoglycan recognition proteins: modulators of the microbiome and inflammation, Nat Rev Immunol 11, 837–851. [DOI] [PubMed] [Google Scholar]
- [4].Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, and Girardin SE (2014) NOD proteins: regulators of inflammation in health and disease, Nat Rev Immunol 14, 9–23. [DOI] [PubMed] [Google Scholar]
- [5].Caruso R, Warner N, Inohara N, and Nunez G (2014) NOD1 and NOD2: signaling, host defense, and inflammatory disease, Immunity 41, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fujimoto Y, Inamura S, Kawasaki A, Shiokawa Z, Shimoyama A, Hashimoto T, Kusumoto S, and Fukase K (2007) Chemical synthesis of peptidoglycan fragments for elucidation of the immunostimulating mechanism, J Endotoxin Res 13, 189–196. [DOI] [PubMed] [Google Scholar]
- [7].Ellouz F, Adam A, Ciorbaru R, and Lederer E (1974) Minimal structural requirements for adjuvant activity of bacterial peptidoglycan derivatives, Biochem Biophys Res Commun 59, 1317–1325. [DOI] [PubMed] [Google Scholar]
- [8].Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, and Flavell RA (2005) Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract, Science 307, 731–734. [DOI] [PubMed] [Google Scholar]
- [9].Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, and Kobayashi KS (2009) Nod2 is required for the regulation of commensal microbiota in the intestine, Proc Natl Acad Sci U S A 106, 15813–15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, Mucida D, and Hang HC (2016) A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens, Science 353, 1434–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, and Mucida D (2016) Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance, Sci Immunol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, and Cho JH (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease, Nature 411, 603–606. [DOI] [PubMed] [Google Scholar]
- [13].Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, and Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease, Nature 411, 599–603. [DOI] [PubMed] [Google Scholar]
- [14].Al Nabhani Z, Dietrich G, Hugot JP, and Barreau F (2017) Nod2: The intestinal gate keeper, PLoS Pathog 13, e1006177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, and Hugot JP (2001) CARD15 mutations in Blau syndrome, Nat Genet 29, 19–20. [DOI] [PubMed] [Google Scholar]
- [16].Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, and Mengin-Lecreulx D (2003) Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2, J Biol Chem 278, 41702–41708. [DOI] [PubMed] [Google Scholar]
- [17].Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, and Sansonetti PJ (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection, J Biol Chem 278, 8869–8872. [DOI] [PubMed] [Google Scholar]
- [18].Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, and Philpott DJ (2003) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan, Science 300, 1584–1587. [DOI] [PubMed] [Google Scholar]
- [19].Hisamatsu T, Suzuki M, and Podolsky DK (2003) Interferon-gamma augments CARD4/NOD1 gene and protein expression through interferon regulatory factor-1 in intestinal epithelial cells, J Biol Chem 278, 32962–32968. [DOI] [PubMed] [Google Scholar]
- [20].Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, and Inohara N (2003) An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid, Nat Immunol 4, 702–707. [DOI] [PubMed] [Google Scholar]
- [21].Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, and Flavell RA (2002) RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems, Nature 416, 194–199. [DOI] [PubMed] [Google Scholar]
- [22].Mo J, Boyle JP, Howard CB, Monie TP, Davis BK, and Duncan JA (2012) Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP, J Biol Chem 287, 23057–23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grimes CL, Ariyananda Lde Z, Melnyk JE, and O’Shea EK (2012) The innate immune protein Nod2 binds directly to MDP, a bacterial cell wall fragment, J Am Chem Soc 134, 13535–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Melnyk JE, Mohanan V, Schaefer AK, Hou CW, and Grimes CL (2015) Peptidoglycan Modifications Tune the Stability and Function of the Innate Immune Receptor Nod2, J Am Chem Soc 137, 6987–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lauro ML, D’Ambrosio EA, Bahnson BJ, and Grimes CL (2017) Molecular Recognition of Muramyl Dipeptide Occurs in the Leucine-rich Repeat Domain of Nod2, ACS infectious diseases 3, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schaefer AK, Melnyk JE, Baksh MM, Lazor KM, Finn MG, and Grimes CL (2017) Membrane Association Dictates Ligand Specificity for the Innate Immune Receptor NOD2, ACS Chem Biol 12, 2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Laroui H, Yan Y, Narui Y, Ingersoll SA, Ayyadurai S, Charania MA, Zhou F, Wang B, Salaita K, Sitaraman SV, and Merlin D (2011) L-Ala-gamma-D-Glu-meso-diaminopimelic acid (DAP) interacts directly with leucine-rich region domain of nucleotide-binding oligomerization domain 1, increasing phosphorylation activity of receptor-interacting serine/threonine-protein kinase 2 and its interaction with nucleotide-binding oligomerization domain 1, J Biol Chem 286, 31003–31013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Peng T, and Hang HC (2015) Bifunctional Fatty Acid chemical reporter for analyzing s-palmitoylated membrane protein-protein interactions in Mammalian cells, J Am Chem Soc 137, 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peng T, Yuan X, and Hang HC (2014) Turning the spotlight on protein-lipid interactions in cells, Curr Opin Chem Biol 21, 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pham ND, Parker RB, and Kohler JJ (2013) Photocrosslinking approaches to interactome mapping, Curr Opin Chem Biol 17, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Niphakis MJ, Lum KM, Cognetta AB 3rd, Correia BE, Ichu TA, Olucha J, Brown SJ, Kundu S, Piscitelli F, Rosen H, and Cravatt BF (2015) A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells, Cell 161, 1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hatanaka Y (2015) Development and leading-edge application of innovative photoaffinity labeling, Chem Pharm Bull (Tokyo) 63, 1–12. [DOI] [PubMed] [Google Scholar]
- [33].Agnihotri G, Ukani R, Malladi SS, Warshakoon HJ, Balakrishna R, Wang X, and David SA (2011) Structure-activity relationships in nucleotide oligomerization domain 1 (Nod1) agonistic gamma-glutamyldiaminopimelic acid derivatives, J Med Chem 54, 1490–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, and Nunez G (1999) Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB, J Biol Chem 274, 14560–14567. [DOI] [PubMed] [Google Scholar]
- [35].D’Souza-Schorey C, and Chavrier P (2006) ARF proteins: roles in membrane traffic and beyond, Nat Rev Mol Cell Biol 7, 347–358. [DOI] [PubMed] [Google Scholar]
- [36].Cherfils J (2014) Arf GTPases and their effectors: assembling multivalent membrane-binding platforms, Curr Opin Struct Biol 29, 67–76. [DOI] [PubMed] [Google Scholar]
- [37].van Endert P (2016) Intracellular recycling and cross-presentation by MHC class I molecules, Immunol Rev 272, 80–96. [DOI] [PubMed] [Google Scholar]
- [38].Jimenez A, Chen D, and Alto NM (2016) How Bacteria Subvert Animal Cell Structure and Function, Annu Rev Cell Dev Biol 32, 373–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Van Acker T, Eyckerman S, Vande Walle L, Gerlo S, Goethals M, Lamkanfi M, Bovijn C, Tavernier J, and Peelman F (2014) The small GTPase Arf6 is essential for the Tram/Trif pathway in TLR4 signaling, J Biol Chem 289, 1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wan T, Liu T, Zhang H, Tang S, and Min W (2010) AIP1 functions as Arf6-GAP to negatively regulate TLR4 signaling, J Biol Chem 285, 3750–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kagan JC, and Medzhitov R (2006) Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling, Cell 125, 943–955. [DOI] [PubMed] [Google Scholar]
- [42].Wu JY, and Kuo CC (2012) Pivotal role of ADP-ribosylation factor 6 in Toll-like receptor 9-mediated immune signaling, J Biol Chem 287, 4323–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Keestra-Gounder AM, and Tsolis RM (2017) NOD1 and NOD2: Beyond Peptidoglycan Sensing, Trends Immunol 38, 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, and Randazzo PA (2000) ACAPs are arf6 GTPase-activating proteins that function in the cell periphery, J Cell Biol 151, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Maekawa S, Ohto U, Shibata T, Miyake K, and Shimizu T (2016) Crystal structure of NOD2 and its implications in human disease, Nat Commun 7, 11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, and Podolsky DK (2006) Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation, J Biol Chem 281, 36060–36070. [DOI] [PubMed] [Google Scholar]
- [47].Davidson AC, Humphreys D, Brooks AB, Hume PJ, and Koronakis V (2015) The Arf GTPase-activating protein family is exploited by Salmonella enterica serovar Typhimurium to invade nonphagocytic host cells, MBio 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, and Alto NM (2011) The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold, Nature 469, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, and MacMicking JD (2012) GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals, Science 336, 481–485. [DOI] [PubMed] [Google Scholar]
- [50].Mohanan V, Nakata T, Desch AN, Levesque C, Boroughs A, Guzman G, Cao Z, Creasey E, Yao J, Boucher G, Charron G, Bhan AK, Schenone M, Carr SA, Reinecker HC, Daly MJ, Rioux JD, Lassen KG, and Xavier RJ (2018) C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions, Science 359, 1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Luong PH, Hedl M, Yan J, Zuo T, Fu TM, Jiang X, Thiagarajah JR, Hansen SH, Lesser CF, Wu H, Abraham C, and Lencer WI (2018) INAVA-ARNO complexes bridge mucosal barrier function with inflammatory signaling, eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.