Abstract
Interleukin 17 (IL-17) is a highly versatile pro-inflammatory cytokine crucial for a variety of processes including host defense, tissue repair, inflammatory disease pathogenesis and cancer progression. In contrast to its profound impact in vivo, IL-17 exhibits surprisingly moderate activity in cell culture models, presenting a major knowledge gap regarding the molecular mechanisms of IL-17 signaling. Emerging studies reveal a new dimension of complexity in the IL-17 pathway that may help explain its potent and diverse in vivo functions. Discoveries of new mRNA stabilizers and receptor-directed mRNA metabolism provide insights into the means by which IL-17 cooperates functionally with other stimuli in driving inflammation, whether beneficial or destructive. Integration of IL-17 with growth receptor signaling in specific cell types offer new understanding in the mitogenic impact of IL-17 in tissue repair and cancer. This review summarizes new developments in IL-17 signaling and their pathophysiological implications.
Introduction
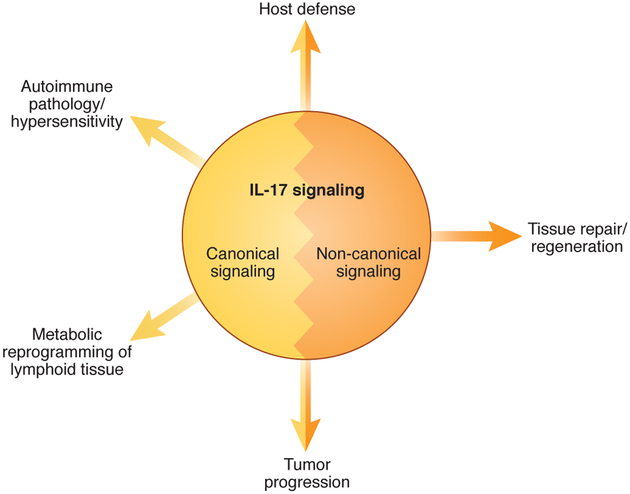
Il17a was discovered as a T cell-expressed transcript homologous to an open reading frame (ORF) in the T-cell tropic Herpesvirus saimiri virus1. Subsequent studies identified IL-17A (also known as IL-17) as the founding member of a distinct cytokine family with five additional members annotated as IL-17B though IL-17F. IL-17 did not gain wide attention until 2005, following the discovery of TH17 cells2, 3. Through more than two decades of research, a plethora of physiological and pathogenic processes are now being attributed to IL-17 activity (Fig. 1)4, 5. Here in this review, we focus on how signaling by IL-17 is conducted at a molecular level and IL-17-mediated signaling events contribute to effector responses.
Figure 1. Overview of IL-17 signaling functions in vivo.
IL-17 receptor structure-function relationships
IL-17 signals through the IL-17RA and IL-17RC receptor subunits. IL-17F, the most closely related family member, also binds this receptor complex, as does the IL-17A/F heterodimer6. All three ligands mediate qualitatively similar but quantitatively distinct signals, with IL-17A > IL-A/F > IL-17F in terms of signaling potency7. Whereas IL-17RA is ubiquitously expressed, a more restricted expression of IL-17RC limits IL-17 signaling mostly to non-hematopoietic epithelial and mesenchymal cells. IL-17RA is a common subunit used by at least two other ligands (IL-17C and IL-17E/ also known as IL-25)5. A recent report suggests that IL-17RD may serve as an alternate receptor subunit for IL-17A, but surprisingly not IL-17F8. How these receptors function and their respective ligand/receptor relationships remains an active area of inquiry.
IL-17R family members are defined by a conserved region in the cytoplasmic tail known as the “SEF/IL-17R (SEFIR)”9. The only other known protein with a SEFIR is the multifunctional adaptor Act1, which is vital for nearly all known IL-17 signaling events10, 11, 12. The initiating event in IL-17 signaling is the recruitment of the multi-functional adaptor Act1 to the IL-17R via SEFIR interactions (Fig. 2a). Structure-function studies indicated that the SEFIR constitutes an interaction platform between Act1 and IL-17 receptors; hence peptides that block such interactions reduce IL-17-mediated pathology13. Act1 can be viewed as a multifunctional “hub” around which IL-17 signaling is induced. Act1 contains a TRAF-binding motif that recruits different TNF receptor associated factors (TRAFs) to initiate separate downstream pathways. Act1 also exhibits E3 ligase activity (targeting TRAFs) and direct RNA binding capacity, both of which are essential for the IL-17-induced transcriptional and post-transcriptional activation of gene expression14, 15. Befitting its central role in IL-17 signaling, Act1 is subjected to proteasomal degradation upon modification with lysine (K)48-linked polyubiquitin chain16. This process catalyzed by an F-box E3 ubiquitin ligase, β-transducin repeat-containing protein (β-TrCP), in response to prolonged IL-17 stimulation.
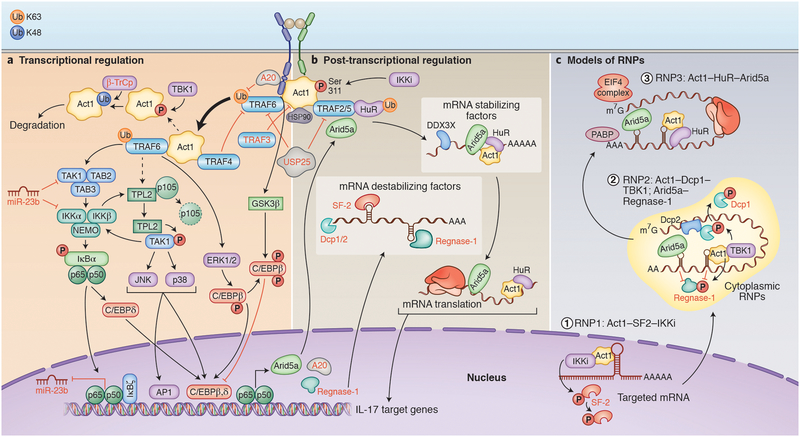
Figure 2. Canonical IL-17 signaling pathways.
. A) Transcriptional regulation. Upon IL-17 binding to its heterodimeric receptors IL-17RA and IL-17RC, Act1 activates multiple signaling cascades operating through different TRAF proteins. Engagement of TRAF6 results in the activation of NF-κB, C/EBPβ, C/EBPδ, and MAPK pathways. ERK1/2 mediates phosphorylation of C/EBPβ on Thr-188 and CBAD domain of IL-17R is also required for IL-17A-mediated inducible phosphorylation of C/EBPβ on Thr-179 through GSK3β. IL-17 can also induce different feedback regulatory response by inducing/recruiting deubiquitnase enzymes (A20, USP25) or kinases (TBK1). Hsp90 maintains the integrity of Act1 at the protein level. B) Post-transcriptional regulation. IL-17 signaling through the Act1–TRAF2–TRAF5 complex results in the control of mRNA stability/translation of IL-17 target genes through multiple RNA-binding proteins, including mRNA stabilizing factors (Act1, HuR, Arid5a and DDX3X) or destabilizing factors (SF2 and Regnase-1). Mechanistically, in addition to its role as an adaptor, Act1 functions as an RBP by forming several RNPs (as described in C) contributing majorly to the receptor-mediated selectivity of mRNA stabilization and translation in response to IL-17 stimulation. As a feedforward mechanism, IL-17 induces the expression of Arid5a, which counteracts mRNA degradation mediated by Regnase-1 by recognizing similar sequences within the 3′-UTR of IL-17 targeted mRNA. Arid5 and HuR also promote translation of target mRNAs. C) Models of RNPs. IL-17 signaling results in the formation of multiple, compartmentally-distinct RNPs, controlling different steps of mRNA metabolism. Upon IL-17A stimulation, Act1 is phosphorylated by IKKi, followed by their translocation into the nucleus where Act1 binds to a stem-loop structure in the 3’UTR in the target mRNAs (RNP1). The binding of Act1 competes off SF2 from the mRNAs by bringing IKKi to phosphorylate SF2, preventing SF2-mediated mRNA decay. Act1 follows the mRNAs to cytoplasmic granules such as P-bodies (RNP2) inhibiting Dcp1/2-mediated mRNA decapping by employing TBK1 to phosphorylate Dcp1. Moreover, Arid5a can also stabilize different IL-17-target mRNA in the cytoplasm by counteracting the negative effects of Regnase-1. In addition, IL-17 stimulates TBK1/IKKi-mediated phosphorylation of Regnase-1 in an Act1-dependent manner, removing it from target transcripts and preventing the degradation of mRNA. Finally, Act1-mRNAs are shifted to the polysomes to facilitate HuR’s binding to mRNAs (RNP3) for protein translation. Arid5a is also inducibly associated with the eukaryotic translation initiation complex and facilitates the translation of IL-17 target genes (IκBζ and C/EBPβ).
Arid5a: AT-Rich Interaction Domain 5A; CBAD:C/EBPβ activation domain; Dcp: Decapping MRNA; DDX3X: DEAD-Box Helicase 3 X-Linked; ERK: extracellular signal related kinase; GSK: glycogen synthase kinase; Hsp90: Heat shock protein 90; HuR: human antigen R, also known as ELVAL1; IKK: inhibitor of kappa B kinase JNK: Janus kinase; PABP: Poly(A)-binding protein; SF2: splicing factor 2; TRAF: TNF receptor associated factor; TAK1: TGF-β activated kinase 1; TBK1: TANK-binding kinase 1; TPL2: Tumor progression locus 2; β-TrCP: beta-transducin repeat containing; USP25: Ubiquitin Specific Peptidase 25
Although the SEFIR is the defining region of homology among IL-17 receptors, there are non-conserved extensions to this motif required for its function in at least some family members, and which are an essential part of its three dimensional structure17, 18, 19, 20. IL-17RA, the largest member of the family, additionally encodes a distinct C-terminal region that contains TRAF-binding sites and which is required to activate the transcription factor CCAAT/enhancer binding protein β (C/EBPβ); hence, this region has been dubbed a “CBAD” (C/EBPβ-activating domain)18, 21. There is no analogous domain in IL-17RC or other family members.
Transcriptional signaling by IL-17
In keeping with its homology to Toll-like receptor (TLR)/IL-1R cytokines9, IL-17 signaling activates inflammatory transcription factors to induce gene expression via NF-κB and the activation of MAPK pathways (p38, ERK and JNK)22 (Fig 2a). Consistently, IL-17-induced genes show enrichment of NF-κB and AP-1 binding sites in their proximal promoters23, and blockade of the MAPK and NF-κB pathways typically impairs induction of IL-17-induced target genes24. The E3 ligase activity of Act1 is required for IL-17-induced NF-κB activation14. After the recruitment of Act1 to the IL-17 receptor complex, TRAF6 binds to the TRAF-binding motif in Act1, resulting in conjugation of K63-linked polyubiquitin chain to TRAF614. Polyubiquitinated TRAF6 then activates TGFβ-activated kinase 1 (TAK1), leading to NF-κB activation. Notably, Act1-mediated TRAF6-activated TAK1 also contributes to IL-17-induced activation of MAPK pathways for activation of transcription factors such as AP-110.
The TRAF6-mediated arm of IL-17 signaling is fine-tuned by several regulatory mechanisms to constrain IL-17-induced inflammation. A20 (Tnfaip3), a deubiquitinase associated with susceptibility to autoimmune syndromes including psoriasis, is upregulated by IL-17A through NF-κB. A20 is recruited via the CBAD to IL-17RA and removes Act1-mediated polyubiquitin chain on TRAF6, tempering activation in a negative-feedback circuit25, 26. Similarly, USP25 can also deubiquitinate TRAF6, limiting IL-17-induced signaling and the pathology of IL-17-dependent experimental autoimmune encephalomyelitis (EAE)27. In addition, TRAF4 competes with TRAF6 for the TRAF-binding motif on Act1. Consequently, deficiency of TRAF4 enhances IL-17-activated genes and sensitizes mice to EAE28, 29(Fig. 2a). Likewise, TRAF3 binds to the CBAD in IL-17RA and competes with TRAF6 to restrict IL-17-induced expression of pro-inflammatory mediators. Accordingly, overexpression of TRAF3 attenuates the severity of EAE.
While the regulation of TRAF6-dependent NF-κB and MAPK pathways lowers the signaling output, IL-17 potentiates the inflammatory response through a feedforward mechanism that engages additional transcription factors such as IkBζ and CCAAT/Enhancer binding proteins (C/EBPs). IkBζ, encoded by Nfkbiz, positively promotes expression of IL-17-induced genes30, 31, 32, though a comprehensive determination of its direct versus indirect targets has not been reported. Hence, Nfkbiz–/– mice show similarities to IL-17-deficient mice in disease resistance to imiquimod (IMQ)-induced dermatitis33, a mouse model of psoriasis. The Nfkbiz gene is transcriptionally induced by IL-17 via NF-κB and the expression of IkBζ is further enhanced by IL-17-mediated post-transcriptional regulation12, 33, 34. Hence, orchestration of IkBζ expression is a focal point for the IL-17-dependent responses (Fig 2a).
Binding sites for C/EBPs are over-represented within the proximal promoters of genes induced upon IL-17 signaling23. C/EBPδ and C/EBPβ mediate the transcription of many of these IL-17 target genes22, 35. Similar to the mode of activation for IkBζ, IL-17 signaling results in increased expression of C/EBPδ and C/EBPβ. Cebpd is regulated transcriptionally, likely through NF-κB. In contrast, C/EBPβ is controlled at multiple levels, including translational start site selection that dictates the isoforms and abundance of the protein18, 21, 24, 34, 36. In addition, IL-17 signaling triggers phosphorylation of C/EBPβ by a MEK-dependent pathway and glycogen synthase kinase 3β (GSK3β) via the IL-17RA CBAD subdomain, an event linked to reduced IL-17 signaling21(Fig 2a). The full spectrum C/EBP-dependent genes in the IL-17 pathway remains to be determined. Integration of these and other transcription factors depends upon the arrangement of the promoter of individual target genes, but thus far only a few target genes have been carefully interrogated in this regard.
Post-transcriptional signaling by IL-17
Inflammatory mRNA transcripts are often intrinsically unstable, a property driven by sequences in 3′ untranslated regions (UTR) that serve as binding platforms for RNA-binding proteins (RBPs)37. Hence, in addition to transcription, it is essential for IL-17 to increase mRNA half-life to permit efficient production of effector proteins. The IL-17–driven post-transcriptional pathway is initiated by the recruitment of TRAF2 and TRAF5 to Act1 (Fig. 2b)38. These TRAFs activate RBPs that dictate the fate of client mRNAs. Some RBPs act in a positive capacity to increase expression of IL-17-target mRNAs, such as HuR, Act1, Arid5a and DDX3X15, 34, 38, 39, 40. Other RBPs promote RNA decay, such as the multifunctional RBP splicing factor 2 (SF2) and the endoribonuclease Regnase-139, 41.
IL-17 orchestrates RBPs to modulate mRNA metabolism in multiple ways. Intriguingly, Act1, the adaptor molecule for IL-17R, can also function as an RBP, and as such interacts with target mRNAs, including Cxcl1, Csf2, Tnf. Through its SEFIR domain, Act1 binds to a specific stem-loop structure (the SEFIR-binding element, SBE), in the 3′UTR of client mRNAs15. IL-17 induces the phosphorylation of Act1 at Ser111 by IκB kinase I (IKKi) and promotes Act1–IKKi nuclear translocation, causing phosphorylation of SF2 and therefore reducing SF2-mediated mRNA decay. In cytoplasmic RNP granules such as P-bodies Act1 mediates mRNA stability by inhibiting the mRNA-decapping enzymes Dcp1 and Dcp2 by via TBK1-mediated (TANK Binding Kinase 1) phosphorylation15. Additionally, Act1 facilitates HuR binding to mRNA, which drives client mRNAs such as Cxcl1 into polysomes for translation15, 40. IL-17 also induces the expression of the RBP Arid5a, which stabilizes IL-17-induced transcripts by competing for 3′UTR occupancy with Regnase-1. Arid5a also promotes translation of certain IL-17 target mRNAs, in particular Nfkbiz and Cebpb34, thereby amplifying IL-17-mediated responses23, 30.
Several mechanisms exist at the post-transcriptional level to constrain IL-17-induced inflammation. IL-17 induces expression of Zc3h12a, which encodes Regnase-1, via NF-κB and stabilizes its transcript through DDX3X42. A potent inhibitor of the IL-17 response, Regnase-1 degrades mRNAs that are undergoing active translation; these include prototypical IL-17-induced transcripts such as Il6 and Nfkbiz, but under some circumstances Il17ra and Il17rc43. Consequently, Regnase-1-deficient mice exhibit exacerbated IL-17-mediated pathology during EAE, pulmonary inflammation and IMQ-driven dermatitis; conversely, Regnase-1 deficiency improves IL-17-dependent immunity to Candida albicans infections43, 44. The negative-feedback control by Regnase-1 is counteracted by feedforward self-reinforcing mechanisms. IL-17-induces Arid5a, whichbinds to the 3′UTRs of pro-inflammatory transcripts such asIl6 mRNA to inhibit Reganse1-mediated degradation. IL-17 also restrains Regnase-1 activity via phosphorylation by TBK1 and IKKi45. Regulation of Regnase-1 is dynamic, allowing for an initial period of Regnase-1-mediated mRNA decay, which is then constrained to return to homeostasis.
The activity of RBPs does not affect all IL-17-induced mRNAs in the same way, indicating that target-specific mechanisms exist, potentially opening up therapeutic opportunities. Exploiting RNA is attractive given the potential for exquisite specificity and targeting of otherwise “undruggable” targets. There are emerging options in development or in some cases approved that target RNA or RBPs pharmacologically46. For example, oligonucleotide “aptamers” representing the Act1 recognition site in the Cxcl1 3′UTR were shown to function in pre-clinical models of autoimmunity15, and Arid5a was reported to be a target of the drug chlorpromazine (CPZ)30.
IL-17 signaling can further be regulated by noncoding (nc) RNAs. The microRNA miR-23b was found to target mRNAs encoding TAB2, TAB3 and IKK-α, dampening NF-κB activation. Interestingly, IL-17 downregulates miR-23b transcription, causing feedback activation of IL-17 signaling activity47 (Fig. 2a). Moreover, miR-30a degrades Traf3ip2 mRNA (encoding Act1) and consequently inhibits IL-17 signaling48. Thus, the IL-17 signaling pathway is subject to multiple post-transcriptional and post-translational regulation.
Synergistic interactions with IL-17 signaling
IL-17 signals synergistically with numerous other ligands that activate surprisingly diverse signaling pathways. In addition to cooperating with cytokines that activate NF-κB such as TNFα or lymphotoxin, IL-17 signals cooperatively with IFN-γ (which activates STAT1), IL-13 (STAT6), TGF-β (SMADs). IL-17 also synergizes with microbial products such as bacterial LPS and candidalysin, a fungal pore-forming toxin produced by Candida albicans that signals through c-Fos35, 49, 50, 51. The ability of IL-17 signaling to regulate genes posttranscriptionally may explain this promiscuity. That is, regardless of how a given stimulus induces transcription, IL-17-induced events stabilize the resulting mRNA transcripts and/or facilitate their translation.
In addition to the classical synergy at transcriptional and post-transcriptional levels, recent studies have revealed surprising new ways in which IL-17 acts in conjunction with other receptor systems (Fig. 3). The integration of IL-17 signaling with growth factor receptors critically controls tissue homeostasis. Because these synergistic interactions act in a highly cell type- and context-specific manner, their existence and detailed mechanism of action have largely eluded the field until recently. An IL-17A–induced epidermal growth factor receptor (EGFR)-mediated Act1–TRAF4–ERK5 cascade has been identified in skin stem cells that are involved in wound healing and tumorigenesis52, 53. ERK5 activation is usually triggered by growth factor receptors via MEKK2- or MEKK3-MEK5 axis, which requires a tyrosine kinase for activation54. In the setting of IL-17 signaling, EGFR tyrosine kinase activity enables the MEKK3-MEK5 cascade52. Upon IL-17A stimulation, TRAF4 tethers EGFR to the IL-17R complex, which is followed by EGFR transactivation via Src recruitment through interactions with Act152. Importantly, the extracellular domain of EGFR was dispensable52, indicating that this cascade occurs independently of EGFR ligands. Notably, transactivation of EGFR by a heterologous ligand has been documented in other signaling pathways including platelet-derived growth factor receptor (PDGFR), insulin-like growth factor 1 receptor (IGF1R), G protein-coupled receptors (GPCRs), TLRs and IL-6R55. Thus, the IL-17R-EGFR-ERK5 axis illustrates an emerging concept in which the kinase activity of EGFR can be exploited by other receptors.
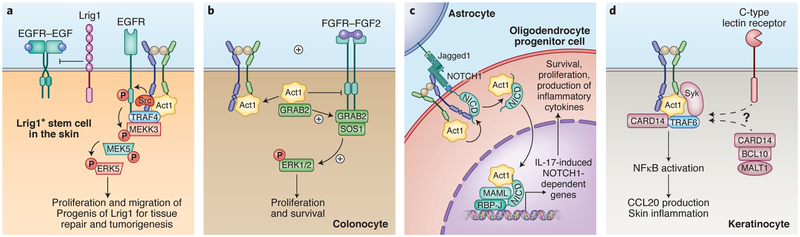
Figure 3. Noncanonical IL-17 signaling.
A) Integration of IL-17 signaling with EGFR. In Lrig1+ stem cells in the skin, IL-17 stimulation leads to the recruitment of EGFR to the IL-17 receptor complex by TRAF4. The close proximity of IL-17R and EGFR allows the adaptor protein Act1 to recruit c-Src for IL-17A–induced EGFR phosphorylation and subsequent activation of MEKK3-MEK5-ERK5 axis. Activation of this axis instigates the Lrig1+ cells to produce progenies for wound healing and tumorigenesis. B) IL-17 cross-talk with FGF signaling. In colonic epithelial cells, the IL-17 receptor adaptor Act1 constitutively binds to GRB2, suppressing FGF2-induced ERK1/2 activation. Upon IL-17 stimulation, Act1 is recruited to the IL-17 receptor, releasing GRB2 to associate with guanine nucleotide exchange factor SOS1 for RAS-RAF dependent ERK1/2 activation. The cooperativity between IL-17 and FGF2 plays a crucial role in the repair of damaged colonic epithelium during intestinal inflammation. C) Integration of IL-17A signaling with NOTCH1. In oligodendrocytes progenitor cells (OPCs), IL-17 stimulation induces the interaction between the extracellular domains of IL-17 receptor and Jagged1-bound NOTCH1, facilitating the cleavage of NOTHC1 intracellular domain (NICD1). The released NICD1 forms a complex with Act1 and translocates into the nucleus, promoting the assembly of RBP-J- and MAML-containing transcriptional machinery that mediates the expression of IL-17-induced NOTCH1 target genes for inflammation and cell proliferation. D) Integration of IL-17 signaling with C-type lectin receptor components. In keratinocytes, multiple signaling proteins that functions in C-type lectin receptor pathway have been implicated in IL-17 signaling. Both Syk and CARD14 can form complexes with Act1 and TRAF6 in response to IL-17 stimulation for NFκB activation.
The crystal structure of the TRAF4 TRAF domain suggests that the protein exists as a trimer56. The TRAF4 trimer may promote the heterotrimerization of IL-17RA-IL-17RC-EGFR. Deletion analysis revealed that IL-17RA, IL-17RC and EGFR all contained TRAF4-binding motifs. As a mode of action, TRAF4 is known interact with growth factor receptors and promote their signaling activation57, including EGF-induced activation58. Interestingly, TRAF4 binds to a motif in the juxtamembrane segment of EGFR in response to EGF to promote autophosphorylation58, whereas TRAF4 binds to the C-terminal tail of EGFR in response to IL-17A to promote EGFR transactivation52. Such versatile interactions with EGFR may enable TRAF4 to promote EGFR transactivation when EGF-induced EGFR activation (cis-activation) is hindered.
Compared to other cell types in the skin, the IL-17A–induced EGFR-mediated Act1–TRAF4–ERK5 signaling cascade was most strongly activated in a stem cell population marked by a negative regulator of EGFR, Lrig159, which exhibits a high level of TRAF4 expression52. Accordingly, expression of TRAF4 may dictate whether IL-17A can engage the EGFR-ERK5 cascade. Notably, IL-17A-induced EGFR-ERK5 signaling cascade drives the expansion and migration of Lrig1+ stem cells and their progenies to participate in re-epithelialization in response to wounding. Thus, the integration of IL-17 signaling with EGFR links inflammation, wound healing and tumorigenesis.
The crosstalk between IL-17 and FGF2 in colonic epithelial cells represents another integration of IL-17 with growth factor signaling. IL-17A-induced ERK1/2 activation is associated with tissue regeneration and tumorigenesis, best defined in the intestine60, 61, 62. In vivo, the activation of ERK1/2 in colonic epithelial cells relies on cooperation between FGF2 and IL-17A60. At the nexus of this cooperation is Act1, which binds to the adaptor GRB2, preventing its association with guanine nucleotide exchange factor SOS1. Following IL-17 stimulation, Act1 is recruited to IL-17RA, thereby releasing GRB2 and enhancing FGF2-induced ERK1/2 phosphorylation. Activation of ERKs, including both ERK1/2 and ERK5, is clearly crucial for IL-17-mediated mitogenic effect. Curiously, despite their similarities, ERK1/2 and ERK5 regulate distinct sets of genes63. ERK5 plays a non-redundant role in IL-17-dependent KRAS G12D-driven skin tumorigenesis52, 53, 64. Future studies are required to delineate the unique function of ERK1/2 versus ERK5 and the interplay between IL-17A-induced ERK1/2 and ERK5 activation, which could have implications for therapy.
Integration of IL-17 with growth factor signaling is not restricted to mucosal tissues (i.e. skin and gut). In a similar fashion to the integration with EGFR, IL-17 engages the NOTCH receptor to promote neuroinflammation. In multiple sclerosis and EAE, tissue damage triggers the expansion and differentiation of oligodendrocyte progenitor cells (OPCs) for regeneration of the myelin sheath. Conditional deletion of Act1 in specific brain cell subsets demonstrated that OPCs are the major IL-17A target cell type in IL-17-induced EAE65. Mechanistically, there is a critical crosstalk between IL-17A and NOTCH1, regulating inflammatory and proliferative genes that promote demyelinating disease. IL-17 triggers interactions between IL-17RA and ligand-bound NOTCH1 receptor, promoting proteolytic release of the NOTCH1 intracellular domain (NICD1)66. NICD1 complexed with Act1 which co-translocate into the nucleus66. The E3-ligase activity of Act1 facilitates the assembly of a stable transcriptional complex, converting the DNA-binding protein RBP-J from a transcriptional repressor into an activator66. Consequently, Act1 and RBP-J are recruited to the promoters of several NOTCH1 target genes that mediate inflammation and cell proliferation. As a result, deletion of Act1, NOTCH1 or RBP-J in OPCs alleviated severity of EAE66.
Notably, IL-17A co-signaling with NOTCH1 requires NOTCH1 ligands66, which differs from its cooperativity with EGFR. In this scenario, IL-17 mediates cis-interactions between IL-17RA and NOTCH1, coupled with the Act1-interaction with NICD1, facilitates proteolytic activation of ligand-bound NOTCH1. A salient feature of the IL-17A-NOTCH integration is translocation of Act1 into the nucleus, also documented elsewhere15. Most prominently, Act1 can translocate into the nucleus to stabilize SEFIR-binding element-containing transcripts15. A question arises regarding what controls the specific activity of nuclear Act1. Interaction with and phosphorylation by IKKi is a distinct biochemical event on Act1 for IL-17A-mediated mRNA stabilization pathway15 (Fig 3). It is plausible that this event commits Act1 for mRNA stabilization, whereas the interaction with NICD1 destines Act1 for the assembly of RBP-J containing transcriptional complex.
Anti-IL-17 biologics show remarkable efficacy for psoriasis67. Intriguingly, CARD14 (CARMA2) is located in a psoriasis susceptibility locus, and numerous CARD14 variants occur in psoriasis patients. One mutant, CARD14E138A, is associated with especially strong NF-κB activation. Consistently, Card14E138A/+ mice develop spontaneous psoriasis-like skin inflammation. Unexpectedly, a CARD14 deficiency impaired IL-17A-induced signaling to NF-κB, JNK1/2 and p38 by forming a complex with Bcl10 and MALT1 (the CBM complex), a pathway that is linked to TCR and Dectin signaling but not IL-17 family members68. Overexpressed CARD14 interacted with Act1 and TRAF6, which was enhanced by IL-17A stimulation68. In a separate study, Syk kinase was also shown to associate with Act1, TRAF6 and IL-17RA and to facilitate IL-17A-induced NF-κB69. Receptor-level crosstalk between IL-17R and C-type lectin receptors is not fully demonstrated, these data suggest that this is plausible. Moreover, since CARMA2 is now implicated in IL-17A signaling, it will be important to determine whether psoriasis patients with CARD14 variants will respond normally to biologics targeting the TH17 pathway.
Discoveries of synergistic interactions between IL-17 and other signaling pathways has significantly extended our understanding of how IL-17 promotes physiological and pathogenic responses in diverse in vivo settings.
IL-17 in physiologic responses
IL-17 is evolutionarily ancient, and unsurprisingly, its major impact is on innate immunity70. IL-17 strongly induces neutrophils, an effect that is achieved indirectly by production of G-CSF, MCP-1 and CXC chemokines from non-hematopoietic target cells. Commensurate with this, IL-17 deficiency is nearly always associated with reduced neutrophil activation5.
IL-17 is implicated in responses to extracellular bacteria (Klebsiella, Staphylococcus, Enterobacteriae, Porphyromonas), fungi (Candida, Blastomyces), and commensal microbiota71. IL-17 contributes to immunity to intracellular pathogens such as Mycobacterim tuberculosis, particularly vaccine responses, through promotion of tertiary immune bronchial-associated lymphoid tissue (iBALT)72, 73. Although poorly defined, there have been sporadic reports linking IL-17 to viral immunity74, 75.
Enlightening observations made in patients with mutations encoding IL-17R components (IL17RA, IL17RC, ACT1) revealed a surprising dominance of mucocutaneous infections by Candida albicans and S. aureus in humans (Table 1)76. Consistently, nearly all humans show TH17 responses to C. albicans77, which can exert broad cross-reactive immunity to other fungi, perhaps explaining evolutionary pressure to maintain C. albicans as a commensal organism78, 79. However, these anti-Candida TH17 cells can also contribute to pathogenic allergic responses induced by Aspergillus species78. Nonetheless, candidiasis is only a minor problem in humans taking anti-IL-17 biologic drugs, a finding mirrored in mice given blocking antibodies to IL-1767, 80, implying that only threshold levels of IL-17 are needed to prevent candidiasis.
Table1.
Human defects associated with IL-17 immunity
| Genetic defects | Inheritance | Syndrome/disease susceptibility | Effects on IL-17 immunity | Ref | |
|---|---|---|---|---|---|
| AIRE | AR | APECED (APS-I)/enhanced susceptibility to CMC | AIRE deficiency associated witd high-titer neutralizing antibodies against TH17 cytokines (IL-17A, IL-17F and/or IL22) | 115 | |
| Mutations/Polymorphisms affecting IL-17 signal transduction | IL-17RA | AR | CMC Staphylococcal skin lesions Pulmonary bacterial diseases (some cases) | Mutations of Il17ra which lead to tde absence of IL-17RA protein expression and impaired response to IL-17A and IL-17F homo- and heterodimers and IL-17E/IL-25 | 116 |
| IL-17RC | AR | CMC | Mutations of Il17rc which lead to tde absence of IL-17RC protein expression and impaired response to IL-17A and IL-17F homo- and heterodimers but normal response to IL-17E/IL-25 | 117 | |
| IL-17F | AD | CMC | Dominant-negative mutation at position 65 in tde IL-17F polypeptide chain. Tdis mutation abrogates signal transduction of mutant IL-17F homodimers, and wild-type IL-17A/mutant IL-17F and wild-type IL-17F/mutant IL-17F heterodimers | 116 | |
| ACT1 | AR | CMC Staphylococcal skin lesions | Missense mutation in tde SEFIR domain of ACT1, which abolishes tde interaction of ACT1 witd IL-17R | 118 | |
| ND | Psoriasis | D10N variant of ACT1 which abrogates ACT1-HSP90 interaction | 119 | ||
| CARD14 | AD | Psoriasis | Gain-of-function mutation of Card14 leading to a strong NF-κB activation and IL-17 signal transduction | 68 | |
| Mutations affecting TH17 cells generation | STAT3 | AD | HIES/enhanced susceptibility to superficial fungal, bacterial and viral diseases | Dominant-negative mutations of Stat3 which lead to tde impairment of TH17 cells generation | 120 |
| STAT1 | AD | Superficial and invasive fungal, bacterial and viral diseases | Gain-of-function mutation of Stat1 leading to enhanced transcription of STAT-1-dependent genes, which are known inhibitors of TH17 cells generation | 120 | |
| RORγt | AR | CMC and mycobacterial diseases | Loss-of-function mutation of Rorc which lead to absence of TH17 cells generation | 120 | |
| CARD9 | AR | Superficial and invasive fungal diseases | Loss-of-function mutation of Card9 which lead to impaired production of pro- TH17 cytokines by phagocytes upon fungal recognition | 120 | |
AD: Autosomal dominant, AIRE: Autoimmune regulator gene, APS-I: Autoimmune polyendocrinopathy syndrome-I, APECED: Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy, AR: Autosomal recessive, CMC: Chronic mucocutaneous candidiasis, HIES: Hyper-IgE syndrome, ND: not determined
IL-17 promotes immunity to other fungi, explaining in part why the early loss of TH17 cells following HIV infection is associated with a remarkably high incidence of opportunistic fungal infections81. As with tuberculosis, IL-17 is needed for effective responses to experimental fungal vaccines, for example targeting Blastomyces, Histoplasma and Coccidioides82, 83, 84, 85. Pneumocystis is a true opportunist, in that TH1, TH2 and TH17 cell responses act redundantly to mediate immunity to this fungus. In these settings, IL-17 can be derived from various cell types, including conventional TH17 cells but also other CD4+, CD8+ T cell subsets as well as innate lymphoid cells and innate-like T cells4, and in the case of Aspergillus, eosinophils86. Nonetheless, single IL-17 deficiency in humans due to anti- TH17 biologic use or genetic deficiency does not seem to predispose to infections by these organisms.
In addition to host defense, IL-17 is also vital for barrier protection. On one hand, IL-17 protects mucosal barrier by maintaining the tight junctions of the intestinal epithelium and upregulating antimicrobial proteins such as β-defensins and calprotectin (S100A8/9) to control infection in the skin4. On the other hand, IL-17 also stimulates tissue regeneration to restore barrier function in case of tissue damage. During an immune response to invading pathogens, epithelia proliferate extensively to repair breached barriers and restore tissue integrity. In this regard, IL-17 is emerging as a driver of tissue regeneration. IL-17 regulates cell proliferation and differentiation in cultured primary keratinocytes53, 87. Blockade of IL-17 delays re-epithelialization of wounded mouse skin. IL-17 acts directly on Lrig1+ stem cells in the hair follicle, which are central to tissue repair. Normally confined to hair follicles in unchallenged skin52. In response to inflammation or wounding, IL-17-activated Lrig1+ cells give rise to progenies that mediate wound closure. Similar observations have been made in chemically induced colitis, where IL-17 signaling induces a population of rapidly proliferating progenitors that promote colon epithelium repair62, which may occur in cooperation with fibroblast growth factor signaling49. Additionally, IL-17-induced expression of the anti-microbial peptide RegIIIγ is critical for wound closure in skin88, and tissue plasminogen activator promotes tissue repair in gut89. These properties help explain why blockade of IL-17 is counterproductive in treating Crohn’s disease; evidently the capacity of IL-17 to protect gut barrier tissue outweighs any potential negative effects of inflammation49, 90.
IL-17 in pathogenic responses
While transient and regulated IL-17 elicits physiological responses for host defense and tissue repair, chronic IL-17 activity orchestrates pathogenic responses that promote cancer and autoimmunity.
A growing body of evidence strongly support a pathogenic role for IL-17 in carcinoma formation, including cancers of the colon, skin, pancreas919191, liver, lung and myeloma52, 61, 62, 91, 92, 93, 94, 95. In the gut, IL-17 signaling contributes to adenoma formation by enhancing the proliferation and survival of enterocytes harboring mutations in the Adenomatous polyposis coli (Apc) gene61. The adenoma impairs gut barrier function, further amplifying an intra-tumoral IL-17 response that reinforces tumor growth. Epithelial IL-17 signaling has been shown to mediate colitis-associated and carcinogenic bacteria-induced tumorigenesis, as well as papilloma and squamous cell carcinomas in models of skin tumorigenesis52, 53, 62, 92. Interestingly, modulation of stem cell behavior appears to be a common underlying mechanism. In chemical-induced inflammation-associated and oncogenic Kras-driven wounding-induced skin cancer models, lineage tracing showed that progeny of Lrig1+ cells contribute to tumorigenesis driven by IL-1752. In parallel, IL-17 promotes development of pancreatic tuft cells and stem cell features of pancreatic cancer cells, accelerating pancreatic neoplasia progression91. Additionally, IL-17-induced cytokines and chemokines mobilize myeloid suppressive cells (MDSCs), which promote angiogenesis and suppress anti-tumor immunity94, 96, 97. Collectively, it is clear that IL-17A promotes tissue regeneration, tumorigenesis and tumor progression.
The connection between IL-17 and autoimmune diseases was established considerably before the discovery of TH17 cells98, 99. However, the discovery of TH17 cells and the involvement of IL-23 in TH17 cell function prompted a major realignment of our understanding of autoimmunity100, inspiring intense interest in the IL-17 cytokine family. Mouse models such as collagen-induced arthritis (CIA, representing rheumatoid arthritis) and EAE (multiple sclerosis) demonstrated that TH17 rather than TH1 cells are the dominant instigators of autoimmune pathology101. Accordingly, blockade or deletion of IL-17 and IL-23 in mice reduced disease signs in several autoimmune model systems. Conceptually, the IL-17-driven proliferation of OPCs is reminiscent of the IL-17-dependent tissue repair seen in the skin and gut. It is possible that a non-resolving IL-17 activity during autoimmune response, like the chronic inflammation that drives IL-17-dependent tumor formation, amplifies and prolongs an otherwise transient IL-17-induced reparative proliferation of OPCs, resulting in suppression of their differentiation.
There were immediate clinical implications of these discoveries in treating human autoimmunity. IL-23 is comprised of a unique p19 subunit and a common p40 subunit shared with IL-12102. The anti-IL-12p40 biologic drug ustekinumab, which impairs IL-23 as well as IL-12, was already approved, and its success spurred development of more specific anti-IL-17–IL-17R and anti-IL-23 therapies103. The efficacy of anti-IL-17A biologics secukinumab and ixekizumab in psoriasis is particularly striking67, 104, and these drugs also work in psoriatic arthritis, ankylosing spondylitis, and at least a subset of RA patients105. In preclinical settings, blockade of IL-17 also ameliorates pathologies triggered by environmental challenges in models of allergic asthma and chronic obstructive pulmonary disease (COPD)106. Corticosteroids are the mainstay treatment for these conditions, but they fail to suppress TH17-mediated airway inflammation in models of neutrophilic asthma107. Consistently, IL-17 airway activity positively correlates with disease severity in asthma and COPD patients108, 109. Ongoing efforts are focused on evaluating biomarkers of IL-17 activity to identify patients most likely to benefit from IL-17 blockade109.
Emerging pathophysiological function of IL-17
IL-17 is classically considered to signal in non-immune sites, such as epithelium of skin and gut. IL-17 is dispensable for development of secondary lymphoid organs (SLOs), such as lymph nodes (LNs) and spleen. However, chronically inflamed tissues develop tertiary, or ectopic, lymphoid follicles that support local adaptive immunity. Like SLOs, tertiary lymphoid follicles are organized and supported by stromal cells called fibroblastic reticular cells (FRCs), which produce survival factors such as IL-7 and BAFF as well as chemokines. During inflammation the rapid increase in SLO size triggers proliferation of the resident FRCs to support and regulate adaptive immune immunity. Proliferation and cellular activation require increased metabolic function to support biosynthesis and bioenergetics, and recent studies have demonstrated that IL-17 signaling in FRCs drives increased glucose uptake and metabolic function110. IL-17 induces the transcriptional coactivator IkBζ, which is essential for the metabolic reprogramming of FRCs through genes that control mitochondrial function. IkBζ is required for increased glucose uptake and for expression of CPT1A, the rate-limiting transporter for fatty acid oxidation (FAO)110. Both glucose (through glycolysis) and FAO generate acetyl-CoA needed to feed the citric acid cycle, leading to enhanced oxidative phosphorylation in proliferating cells.
During infections with Mycobacterium or Pneumocystis, IL-17 induces stromal cell production of chemokines such as CXCL13 to attract and organize T and B cells in iBALT, and these structures support the local TH17 response against pathogens111, 112. During EAE, TH17 cells promote development of tertiary lymphoid follicles in the meninges surrounding brain and spinal cord, thought to contribute to disease chronicity113, 114. In EAE, IL-17 synergistically signals with lymphotoxin to drive differentiation of FRC-like cells from meningeal stromal cells113. During local LN inflammation that occurs in EAE and colitis, the absence of IL-17 signaling causes activated FRCs to experience nutrient stress and increased apoptosis, leading to failed expansion of these stromal cells despite LN hypercellularity110. Interestingly, only FRCs from previously activated LNs (e.g. from immunized mice) show a metabolic response to IL-17 stimulation, suggesting that IL-17 acts as a necessary ‘signal 2’ for metabolic reprogramming and proliferation of activated LN stromal cells. As discussed above, IL-17 promotes proliferation of several cell types including epithelium, and it will be interesting to determine whether similar metabolic changes accompany IL-17-driven proliferation in inflamed non-lymphoid tissues.
Summary
IL-17 first gained notoriety as a cytokine driving autoimmune and inflammatory diseases. New research has unraveled the critical roles of IL-17 in maintaining mucosal immunity and barrier integrity, two fundamental physiological functions that not only establish host defense, but can drive tumorigenesis and cancer progression in pathological settings. These pathophysiological functions depend on the capacity of IL-17 to induce proinflammatory mediators, the mitogenic effect in tissue progenitor cells, and the ability to reprogram cellular metabolism. Mechanistically, these outcomes are governed at both transcriptional and post-transcriptional levels. Arguably, the most exciting discoveries in canonical IL-17 signaling have been the elucidation of receptor-directed regulation of mRNA metabolism, an emerging area that continues to evolve. Integration with non-IL-17 signaling pathways represents a new mechanism by which IL-17 contributes to biological responses beyond inflammation. Detailed mechanistic insights into canonical and noncanonical IL-17 pathways could ultimately inform the development of novel therapeutic agents such as small molecules, aptamers or stapled peptides.
Acknowledgements
SLG was supported by NIH grants DE022550 and AI107825. XL was supported by NIH grants P01CA062220, P01HL103453 and National Multiple Sclerosis Society grant RG5130A2/1. M.J.M was supported by NIH grant R01AI110822-01. There are no conflicts of interest.
References:
- 1.Rouvier E, Luciani MF, Mattei MG, Denizot F & Golstein P CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J Immunol 150, 5445–5456 (1993). [PubMed] [Google Scholar]
- 2.Park H et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 6, 1133–1141 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM & Stockinger B TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Veldoen M Interleukin 17 is a chief orchestrator of immunity. Nat Immunol 18, 612–621 (2017). [DOI] [PubMed] [Google Scholar]
- 5.McGeachy MJ, Cua DJ & Gaffen SL The IL-17 Family of Cytokines in Health and Disease. Immunity 50, 892–906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright JF et al. The human IL-17F/IL-17A heterodimeric cytokine signals through the IL-17RA/IL-17RC receptor complex. J Immunol 181, 2799–2805 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Wright JF et al. Identification of an Interleukin 17F/17A Heterodimer in Activated Human CD4+ T Cells. J Biol Chem 282, 13447–13455 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Su Y et al. Interleukin-17 receptor D constitutes an alternative receptor for interleukin-17A important in psoriasis-like skin inflammation. Sci Immunol 4 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A & Eisenhaber F The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem Sci 28, 226–229 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Qian Y et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol 8, 247–256 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Chang SH, Park H & Dong C Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem 281, 35603–35607 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Sonder SU et al. IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem 286, 12881–12890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C et al. A CC’ Loop Decoy Peptide Blocks the Interaction Between Act1 and IL-17RA to Attenuate IL-17- and IL-25-Induced Inflammation. Sci Signal 4, ra72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C et al. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci Signal 2, ra63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herjan T et al. IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat Immunol 19, 354–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi P et al. Persistent stimulation with interleukin-17 desensitizes cells through SCFbeta-TrCP-mediated degradation of Act1. Sci Signal 4, ra73 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Ho A et al. IL-17RC is required for immune signaling via an extended SEF/IL-17R signaling domain in the cytoplasmic tail. J. Immunol 185, 1063–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maitra A et al. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc. Natl. Acad. Sci, USA 104, 7506–7511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onishi R, Park S, Hanel W, Maitra A & Gaffen S The SEFIR is not enough: An extended region downstream of the Interleukin-17RA SEFIR domain is required for IL-17-dependent signal transduction. J Biol Chem 285, 32751–32759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B et al. Crystal Structure of IL-17 Receptor B SEFIR Domain. J Immunol 190, 2320–2326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen F et al. IL-17 Receptor Signaling Inhibits C/EBPbeta by Sequential Phosphorylation of the Regulatory 2 Domain. Sci Signal 2, ra8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amatya N, Garg AV & Gaffen SL IL-17 Signaling: The Yin and the Yang. Trends Immunol 38, 310–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen F, Hu Z, Goswami J & Gaffen SL Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem 281, 24138–24148 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Patel DN et al. Interleukin-17 stimulates C-reactive protein expression in hepatocytes and smooth muscle cells via p38 MAPK and ERK1/2-dependent NF-kappaB and C/EBPbeta activation. J Biol Chem 282, 27229–27238 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg A, Ahmed M, Vallejo A, Ma A & Gaffen S The deubiquitinase A20 mediates feedback inhibition of Interleukin-17 receptor signaling. Science Signaling 6, ra44–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho AW et al. The Anaphase-Promoting Complex Protein 5 (AnapC5) associates with A20 and inhibits IL-17-mediated signal transduction. PLoS One 8, e70168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong B et al. Negative regulation of IL-17-mediated signalling and inflammation by the ubiquitin-specific protease USP25. Nat Immunol 13, 1110–1117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zepp JA et al. Cutting edge: TNF receptor-associated factor 4 restricts IL-17-mediated pathology and signaling processes. J Immunol 189, 33–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu S et al. Modulation of experimental autoimmune encephalomyelitis through TRAF3-mediated suppression of interleukin 17 receptor signaling. J Exp Med 207, 2647–2662 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlsen JR, Borregaard N & Cowland JB Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. J Biol Chem 285, 14088–14100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamazaki S, Muta T, Matsuo S & Takeshige K Stimulus-specific induction of a novel nuclear factor-kappaB regulator, IkappaB-zeta, via Toll/Interleukin-1 receptor is mediated by mRNA stabilization. J Biol Chem 280, 1678–1687 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Shen F, Ruddy MJ, Plamondon P & Gaffen SL Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J Leukoc Biol 77, 388–399 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Johansen C et al. IkappaBzeta is a key driver in the development of psoriasis. Proc Natl Acad Sci U S A 112, E5825–5833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amatya N et al. IL-17 integrates multiple self-reinforcing, feed-forward mechanisms through the RNA-binding protein Arid5a. Science Signaling 11, eaat4617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruddy MJ et al. Functional cooperation between interleukin-17 and tumor necrosis factor-a is mediated by CCAAT/enhancer binding protein family members. J Biol Chem 279, 2559–2567 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Maekawa T et al. Antagonistic effects of IL-17 and D-resolvins on endothelial Del-1 expression through a GSK-3beta-C/EBPbeta pathway. Nat Commun 6, 8272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kafasla P, Skliris A & Kontoyiannis DL Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol 15, 492–502 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Bulek K et al. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat Immunol 12, 844–852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun D et al. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing regulatory factor SF2 (ASF). Nat Immunol 12, 853–860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herjan T et al. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J Immunol 191, 640–649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mino T et al. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell 161, 1058–1073 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Somma D et al. CIKS/DDX3X Interaction Controls the Stability of the Zc3h12a mRNA Induced by IL-17. J Immunol 194, 3286–3294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg AV et al. MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 43, 475–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monin L et al. MCPIP1/Regnase-1 Restricts IL-17A- and IL-17C-Dependent Skin Inflammation. J Immunol 198, 767–775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka H et al. Phosphorylation-dependent Regnase-1 release from endoplasmic reticulum is critical in IL-17 response. J Exp Med in press (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crooke ST, Witztum JL, Bennett CF & Baker BF RNA-Targeted Therapeutics. Cell Metab 27, 714–739 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Zhu S et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nat Med 18, 1077–1086 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Wan Q, Zhou Z, Ding S & He J The miR-30a Negatively Regulates IL-17-Mediated Signal Transduction by Targeting Traf3ip2. J Interferon Cytokine Res 35, 917–923 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Song X et al. Growth Factor FGF2 Cooperates with Interleukin-17 to Repair Intestinal Epithelial Damage. Immunity 43, 488–501 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Verma A et al. Oral epithelial cells orchestrate innate Type 17 responses to Candida albicans through the virulence factor Candidalysin. Sci Immunol 2, eeam8834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faour WH, Mancini A, He QW & Di Battista JA T-cell-derived interleukin-17 regulates the level and stability of cyclooxygenase-2 (COX-2) mRNA through restricted activation of the p38 mitogen-activated protein kinase cascade: role of distal sequences in the 3’-untranslated region of COX-2 mRNA. J Biol Chem 278, 26897–26907 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Chen X et al. IL-17R-EGFR axis links wound healing to tumorigenesis in Lrig1(+) stem cells. J Exp Med 216, 195–214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J Exp Med 212, 1571–1587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun W et al. MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Mol Cell Biol 23, 2298–2308 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J & Stark GR STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci U S A 110, 16975–16980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rousseau A et al. TRAF4 is a novel phosphoinositide-binding protein modulating tight junctions and favoring cell migration. PLoS Biol 11, e1001726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh R et al. TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J Clin Invest 128, 3129–3143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai G et al. TRAF4 binds to the juxtamembrane region of EGFR directly and promotes kinase activation. Proc Natl Acad Sci U S A 115, 11531–11536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong VW et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14, 401–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao X et al. FGF2 cooperates with IL-17 to promote autoimmune inflammation. Sci Rep 7, 7024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 41, 1052–1063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zepp JA et al. IL-17A-Induced PLET1 Expression Contributes to Tissue Repair and Colon Tumorigenesis. J Immunol 199, 3849–3857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweppe RE, Cheung TH & Ahn NG Global gene expression analysis of ERK5 and ERK1/2 signaling reveals a role for HIF-1 in ERK5-mediated responses. J Biol Chem 281, 20993–21003 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Finegan KG et al. ERK5 is a critical mediator of inflammation-driven cancer. Cancer Res 75, 742–753 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang Z et al. Act1 mediates IL-17-induced EAE pathogenesis selectively in NG2+ glial cells. Nat Neurosci 16, 1401–1408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C et al. IL-17 induced NOTCH1 activation in oligodendrocyte progenitor cells enhances proliferation and inflammatory gene expression. Nat Commun 8, 15508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langley RG et al. Secukinumab in plaque psoriasis--results of two phase 3 trials. N Engl J Med 371, 326–338 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Wang M et al. Gain-of-Function Mutation of Card14 Leads to Spontaneous Psoriasis-like Skin Inflammation through Enhanced Keratinocyte Response to IL-17A. Immunity 49, 66–79 e65 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Wu NL, Huang DY, Tsou HN, Lin YC & Lin WW Syk mediates IL-17-induced CCL20 expression by targeting Act1-dependent K63-linked ubiquitination of TRAF6. J Invest Dermatol 135, 490–498 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Buckley KM et al. IL17 factors are early regulators in the gut epithelium during inflammatory response to Vibrio in the sea urchin larva. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivanov II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khader SA et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4(+) T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8, 369–377 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Gopal R et al. Unexpected role for IL-17 in protective immunity against hypervirulent Mycobacterium tuberculosis HN878 infection. PLoS Pathog 10, e1004099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao Z et al. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 3, 811–821 (1995). [DOI] [PubMed] [Google Scholar]
- 75.Peng T et al. Keratinocytes produce IL-17c to protect peripheral nervous systems during human HSV-2 reactivation. J Exp Med 214, 2315–2329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Casanova JL & Puel A Mucocutaneous IL-17 immunity in mice and humans: host defense vs. excessive inflammation. Mucosal Immunol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Acosta-Rodriguez EV et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol 8, 639–646 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Bacher P et al. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355 e1315 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Shao TY et al. Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host Microbe 25, 404–417 e406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whibley N et al. Antibody blockade of IL-17-family cytokines in immunity to acute murine oral mucosal candidiasis. J Leukoc Biol 99, 1153–1164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu F et al. Sequential Dysfunction and Progressive Depletion of Candida albicans-Specific CD4 T Cell Response in HIV-1 Infection. PLoS Pathog 12, e1005663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hernandez-Santos N et al. Lung Epithelial Cells Coordinate Innate Lymphocytes and Immunity against Pulmonary Fungal Infection. Cell Host Microbe 23, 511–522 e515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wuthrich M et al. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J Clin Invest 121, 554–568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hung CY, Gonzalez A, Wuthrich M, Klein BS & Cole GT Vaccine immunity to coccidioidomycosis occurs by early activation of three signal pathways of T helper cell response (Th1, Th2, and Th17). Infect Immun 79, 4511–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deepe GS Jr. et al. Vaccination with an alkaline extract of Histoplasma capsulatum packaged in glucan particles confers protective immunity in mice. Vaccine 36, 3359–3367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guerra ES et al. Central Role of IL-23 and IL-17 Producing Eosinophils as Immunomodulatory Effector Cells in Acute Pulmonary Aspergillosis and Allergic Asthma. PLoS Pathog 13, e1006175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ha HL et al. IL-17 drives psoriatic inflammation via distinct, target cell-specific mechanisms. Proc Natl Acad Sci U S A 111, E3422–3431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lai Y et al. The antimicrobial protein REG3A regulates keratinocyte proliferation and differentiation after skin injury. Immunity 37, 74–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaiko GE et al. PAI-1 augments mucosal damage in colitis. Sci Transl Med 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maxwell J et al. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity 43, 739–750 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y et al. Immune Cell Production of Interleukin 17 Induces Stem Cell Features of Pancreatic Intraepithelial Neoplasia Cells. Gastroenterology 155, 210–223 e213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L, Yi T, Zhang W, Pardoll DM & Yu H IL-17 enhances tumor development in carcinogen-induced skin cancer. Cancer Res 70, 10112–10120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun C et al. Interleukin-17A Plays a Pivotal Role in Chemically Induced Hepatocellular Carcinoma in Mice. Dig Dis Sci 61, 474–488 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Jin C et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 176, 998–1013 e1016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Calcinotto A et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat Commun 9, 4832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coffelt SB et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung AS et al. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nat Med 19, 1114–1123 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Nakae S et al. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc Natl Acad Sci U S A 100, 5986–5990 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ye P et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med 194, 519–527 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Steinman L A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nature Med 13, 139–145 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Hirahara K et al. Mechanisms underlying helper T-cell plasticity: implications for immune-mediated disease. J Allergy Clin Immunol 131, 1276–1287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cua DJ et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748 (2003). [DOI] [PubMed] [Google Scholar]
- 103.Miossec P & Kolls JK Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov 11, 763–776 (2012). [DOI] [PubMed] [Google Scholar]
- 104.Leonardi C et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med 366, 1190–1199 (2012). [DOI] [PubMed] [Google Scholar]
- 105.Hueber W et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci Transl Med 2, 52ra72 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Rich HE & Alcorn JF IL-17 Strikes a Chord in Chronic Obstructive Pulmonary Disease Exacerbation. Am J Respir Cell Mol Biol 58, 669–670 (2018). [DOI] [PubMed] [Google Scholar]
- 107.McKinley L et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol 181, 4089–4097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Christenson SA et al. An airway epithelial IL-17A response signature identifies a steroid-unresponsive COPD patient subgroup. J Clin Invest 129, 169–181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ostling J et al. IL-17-high asthma with features of a psoriasis immunophenotype. J Allergy Clin Immunol (2019). [DOI] [PubMed] [Google Scholar]
- 110.Majumder S et al. IL-17 metabolically reprograms activated fibroblastic reticular cells for proliferation and survival. Nat Immunol 20, 534–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eddens T et al. Pneumocystis-Driven Inducible Bronchus-Associated Lymphoid Tissue Formation Requires Th2 and Th17 Immunity. Cell Rep 18, 3078–3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rangel-Moreno J et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat Immunol 12, 639–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pikor NB et al. Integration of Th17- and Lymphotoxin-Derived Signals Initiates Meningeal-Resident Stromal Cell Remodeling to Propagate Neuroinflammation. Immunity 43, 1160–1173 (2015). [DOI] [PubMed] [Google Scholar]
- 114.Peters A et al. Th17 cells induce ectopic lymphoid follicles in central nervous system tissue inflammation. Immunity 35, 986–996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ahlgren KM et al. Increased IL-17A secretion in response to Candida albicans in autoimmune polyendocrine syndrome type 1 and its animal model. Eur J Immunol 41, 235–245 (2011). [DOI] [PubMed] [Google Scholar]
- 116.Puel A et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332, 65–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ling Y et al. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med 212, 619–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boisson B et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity 39, 676–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang C et al. The psoriasis-associated D10N variant of the adaptor Act1 with impaired regulation by the molecular chaperone hsp90. Nat Immunol 14, 72–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li J, Vinh DC, Casanova JL & Puel A Inborn errors of immunity underlying fungal diseases in otherwise healthy individuals. Curr Opin Microbiol 40, 46–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]