Abstract
PURPOSE
The identification of a heritable tumor predisposition often leads to changes in management and increased surveillance of individuals who are at risk; however, for many rare entities, our knowledge of heritable predisposition is incomplete.
METHODS
Families with childhood medulloblastoma, one of the most prevalent childhood malignant brain tumors, were investigated to identify predisposing germline mutations. Initial findings were extended to genomes and epigenomes of 1,044 medulloblastoma cases from international multicenter cohorts, including retrospective and prospective clinical studies and patient series.
RESULTS
We identified heterozygous germline mutations in the G protein-coupled receptor 161 (GPR161) gene in six patients with infant-onset medulloblastoma (median age, 1.5 years). GPR161 mutations were exclusively associated with the sonic hedgehog medulloblastoma (MBSHH) subgroup and accounted for 5% of infant MBSHH cases in our cohorts. Molecular tumor profiling revealed a loss of heterozygosity at GPR161 in all affected MBSHH tumors, atypical somatic copy number landscapes, and no additional somatic driver events. Analysis of 226 MBSHH tumors revealed somatic copy-neutral loss of heterozygosity of chromosome 1q as the hallmark characteristic of GPR161 deficiency and the primary mechanism for biallelic inactivation of GPR161 in affected MBSHH tumors.
CONCLUSION
Here, we describe a novel brain tumor predisposition syndrome that is caused by germline GPR161 mutations and characterized by MBSHH in infants. Additional studies are needed to identify a potential broader tumor spectrum associated with germline GPR161 mutations.
INTRODUCTION
Cancer predisposition syndromes are defined by germline mutations that result in a highly or moderately increased tumor risk in affected individuals. Knowledge of these syndromes has a profound impact on patient care and the prevention of malignant disease through the provision of genetic counseling and careful surveillance of families who are at risk.1 Many of the genes mutated in cancer predisposition syndromes are also somatically mutated in sporadic types of cancer. Thus, understanding the molecular function of these genes can lead to new therapeutic concepts in both sporadic and heritable tumors. In children and adolescents with cancer, tumor predisposition as a result of germline mutations is found in at least 7% to 10%.2-4 Medulloblastoma (MB), a tumor that originates in the cerebellum and dorsal brainstem, has a peak incidence in childhood and makes up a large proportion of embryonal brain tumors. Consensus molecular subgroups of MB include WNT, sonic hedgehog (SHH), group 3, and group 4, each showing distinct transcriptional and epigenetic profiles.5 Recent advances in deep molecular phenotyping suggest additional MB subtypes with distinct somatic driver mutations and epigenetic signatures.6-8 The SHH subgroup (MBSHH) is characterized by a constitutive transcriptional and genomic activation of the SHH pathway9; driver mutations in PTCH1, SUFU, SMO, TERT, DDX3X, TP53, and KMT2D; and structural aberrations that affect most frequently chromosomes 9, 10, and 17. Most MBs are thought to develop sporadically, but inherited forms also exist, most often in children with MBSHH.4 Heritable predisposition to MB is observed in Li-Fraumeni syndrome,10 APC-associated polyposis,11 subtypes of Fanconi anemia,12,13 and Gorlin syndrome (nevoid basal-cell carcinoma syndrome). The latter is associated with germline mutations in SUFU14-16 and PTCH1,17,18 which play crucial roles in the SHH pathway. Active SHH signaling is a key element of embryonic development and cell differentiation.19 Here, we describe the SHH regulator G protein-coupled receptor 161 (GPR161) as a novel brain tumor predisposition gene in children.
METHODS
Patients
Written informed consent was obtained from study participants after approval from the institutional review boards at the participating institutions (Uniklinik RWTH Aachen: EK302-16). Consent was obtained according to the Declaration of Helsinki. The study had access to international multicenter MB studies, including retrospective cohorts (International Cancer Genome Consortium [ICGC] PedBrain, Medulloblastoma Advanced Genomics International Consortium, CEFALO series, and clinic of pediatric oncogenetics from Gustave Roussy), and prospective cohorts from clinical studies or patient series (SJMB03, SJMB12, SJYC07, and I-HIT-MED).4,20
Germline and Tumor Sequencing
Whole-exome sequencing (WES) of the index patient (M20769) was done at the Institute of Human Genetics, Uniklinik RWTH Aachen, using the Nextera Rapid Capture Exome kit (version 1.2; Illumina, San Diego, CA). The library was sequenced on a NextSeq500 Sequencer with 2 × 75 cycles on a high-output flow cell. For sample MB11_06, WES data for germline and tumor samples were generated at the German Cancer Research Centre (Heidelberg, Germany) using Agilent SureSelect Human All Exon V5 (Agilent Technologies, Santa Clara, CA) without untranslated regions and sequenced on an Illumina HiSEq 4000 System (paired-end mode). For sample MB13_03, whole-genome sequencing data for germline and tumor samples were generated at the German Cancer Research Centre and sequenced on the Illumina HiSeq X Ten System (paired-end mode). For samples SJMB335, SJMB303, and SJMB054, WES data for germline and tumor samples were generated at St Jude Children’s Research Hospital (Memphis, TN) using the Agilent SureSelect Human All Exon V5 kit. For the detection of single-nucleotide variations (SNVs), small insertions and deletions (indels), fusions, and copy number aberrations, the customized enrichment/hybrid capture–based next-generation sequencing gene panel analysis covering 130 genes of particular relevance in brain tumors was applied, as previously described.21
Somatic and Germline Variant Calling
Sequencing data were processed using a standardized alignment and somatic variant calling pipeline, which was developed in the context of the ICGC Pan-Cancer Analysis of Whole Genomes (PCAWG) project (https://dockstore.org/containers/quay.io/pancancer/pcawg-dkfz-workflow). In brief, reads were aligned to the phase II reference human genome assembly of the 1000 Genomes Project, including decoy sequences (hs37d5), using BWA-MEM (version 0.7.15). Somatic SNVs were called with the previously described SAMtools-based German Cancer Research Centre pipeline adjusted for ICGC PCAWG settings, and short somatic indels were called using Platypus.22,23 The CNVkit library24 and ACEseq25 were used to infer and visualize somatic genome copy number from the WES and whole-genome sequencing data. Loss of heterozygosity (LOH) analysis for tumor-only panel sequencing data was based on known germline variants (1000 Genomes Project) in target and off-target regions, 20× sequencing coverage, and one or more read support for the reference and alternative allele. Germline genomes and exomes were analyzed using the freebayes (version 1.1.0) pipeline with ICGC PCAWG settings26 and deleterious germline variants—SNVs, multi-nucleotide variants, indels, and complex variants—were inferred using automated pipelines and manually curated. For sequencing data of the index case, demultiplexing and fastq file generation were performed using bcl2fastq2 (version 2.2; Illumina) and read alignment and variant calling using the automated SeqMule pipeline (v1.2.6).27 For germline variant detection in the index patient, different variant callers were used (GATKLite UnifiedGenotyper, SAMtools, freebayes consensus) and variants shared by at least one pair of variant callers were written to the final variant file. Variant annotation and bioinformatics prioritization were performed using KGGSeq (version 1.0; 20 June 2018).28 Synonymous variants and variants with a minor allele frequency greater than 0.75% in public databases, that is, gnomAD, ExAC, 1000 Genomes Project, and National Heart, Lung, and Blood Institute Exome Sequencing Project, were excluded.
DNA Methylation Array Processing
DNA from tumor tissue with tumor cell content greater than 80% by histopathologic evaluation was extracted from formalin-fixed, paraffin-embedded tissue using the automated Maxwell system with the Maxwell 16 formalin-fixed, paraffin-embedded Plus LEV DNA Purification Kit (Promega, Madison, WI). We performed DNA methylation profiling of all samples using the Infinium MethylationEPIC (850k) BeadChip (Illumina) or Infinium HumanMethylation450 (450k) BeadChip (Illumina) array, as previously described.29 Filtering and genome-wide copy number analyses were performed using the conumee package in R (http://www.bioconductor.org) as previously described.29
RESULTS
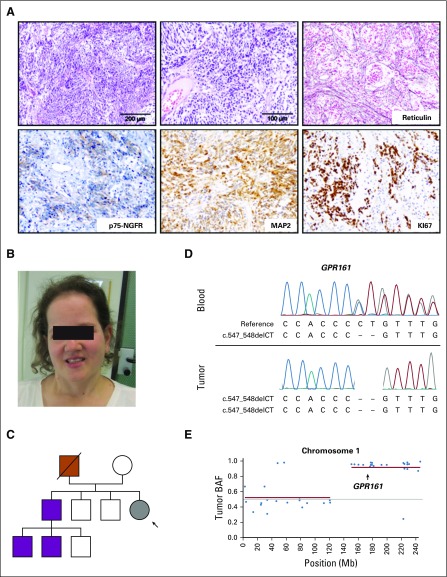
Our index patient was a female (M20769) with an extensive history of neoplasia that began with the diagnosis of MB at the age of 12 months. Histologic evaluation showed a highly cellular, undifferentiated small-cell neoplasm with increased mitotic activity. Cells had hyperchromatic nuclei and a scant cytoplasm. Silver impregnation demonstrated an increased density of argyric fibers and fiber-rich areas of ensheathed fiber-free islands of tumor cells. Cells expressed the neural marker MAP2 and the SHH target protein p75-NGFR (Fig 1A). Consistent with SHH activation, OTX2 was negative, and TP53 was not accumulated (not shown). DNA methylation profiling classified the tumor into the MBSHH subgroup. According to the revised WHO classification of tumors of the CNS 2016, the tumor classifies as desmoplastic/nodular, SHH-activated, TP53 wild-type MB (WHO grade IV).30 At age 16 years, the patient developed her first basal-cell carcinoma (BCC) and has subsequently gone on to develop 10 more BCCs, all within the radiation field and all amenable to surgical removal. By age 18, the patient underwent a total thyroidectomy for a multinodular goiter, and at 23 years old, the patient underwent the excision of a rectal tubular adenoma with low-grade dysplasia, a low-grade intraepithelial neoplasia in the stomach, and several hyperplastic serrated polyps. A meningioma in the right temporal region was removed at age 24. The patient, currently age 29 years, has a microcephaly and a mild frontal bossing and in summary fulfills some, but not all, criteria of Gorlin syndrome (Fig 1B).
FIG 1.
Index patient with medulloblastoma and germline GPR161 mutation. (A) Desmoplastic/nodular medulloblastoma, SHH-activated, TP53 wild type (WHO grade IV) in the index case (M20769). Highly cellular, undifferentiated small-cell neoplasm with increased mitotic activity. Silver impregnation shows an increased density of argyric fibers. The cells express the neural marker MAP2 and the SHH target protein p75-NGFR. (B) Index patient (27 years old). (C) Pedigree of the index patient (gray) with MB (black arrow). Father (red) was carrier of the GPR161 germline mutation and died from colorectal cancer. Asymptomatic GPR161 mutation carriers are indicated in purple. (D) Sanger sequencing-based validation of the germline GPR161 frameshift mutation (c.547_548delCT) in peripheral blood (upper panel) and medulloblastoma (lower panel) of the index case. (E) Loss of heterozygosity analysis of chromosome 1 based on targeted gene panel sequencing of tumor DNA. GPR161 location is highlighted with an arrow.
Matched WES was performed using DNA extracted from the patient’s MB tumor and blood. Analysis revealed no candidate germline mutation in consensus MB predisposition genes4; no somatic copy-number alterations affecting MBSHH hallmark chromosomes 9, 10, and 17; and no somatic mutations in MBSHH driver genes (eg, PTCH1, SUFU, TP53; Fig 2B). However, exome-wide analysis for rare damaging germline mutations revealed a frameshift mutation in GPR161 on chromosome 1q24.2 (Figs 1D and 2A), and subsequent analysis of tumor DNA showed a somatic copy-neutral loss of heterozygosity (cnLOH) event on 1q (Fig 1E and Data Supplement). Of note, five additional rare germline protein-truncating variants were identified, but none of them were located in regions that showed focal deletions and/or LOH. Sanger sequencing confirmed that the 1q cnLOH event affected the wild-type GPR161 allele and consequently led to somatic biallelic inactivation of GPR161 (Fig 1D). These results provide the first clue that GPR161 might be a novel MB predisposition gene.
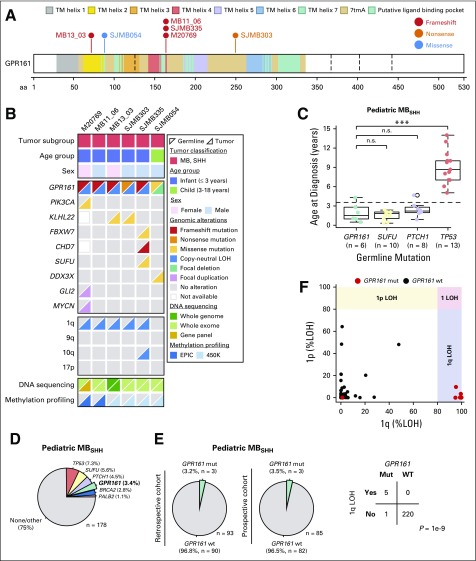
FIG 2.
Genomic landscape of GPR161-associated medulloblastoma. (A) Germline GPR161 mutations in patients with MB. (B) Demographic and molecular characteristics of GPR161-associated MB. (C) Age at diagnosis across pediatric patients with pathogenic germline mutations in MBSHH predisposition genes. (D) Frequency of pathogenic germline mutations in pediatric MBSHH. (E) Frequency of germline GPR161 mutations in retrospective and prospective pediatric MBSHH cohorts. (F) Frequency of somatic 1p/1q LOH events in MBSHH tumors and association between 1q LOH and GPR161 mutation status. LOH, loss of heterozygosity; MB, medulloblastoma; Mut, mutant; SHH, sonic hedgehog; WT, wild type.
To further corroborate the role of GPR161 in predisposition to MB, we specifically analyzed the genomic data of 1,044 patients with MB who were enrolled in previous sequencing studies.4,20 These analyses revealed five additional patients with rare and damaging germline GPR161 mutations (Figs 2A and 2B). Clinical details on these patients are summarized in the Data Supplement. Four patients were found to harbor GPR161 protein-truncating variants and one patient was a carrier of a predicted damaging missense variant (SJMB054). Two patients (MB11_06 and SJMB335) shared the same frameshift mutation as that of the index patient; kinship analysis demonstrated that all three patients were unrelated.
Classification of 872 human MBs into four consensus MB subgroups4,20 revealed that all six patients with GPR161 mutations developed MBs that belonged to the SHH subgroup (P < 5×10−4, Fishers’ exact test; Fig 2B). Moreover, all patients with GPR161 mutations developed MBSHH at a young age (median age, 1.5 years; P = .025, MWU test; Fig 2C), which is similar to the age at which patients with germline PTCH1 and SUFU mutations develop MBSHH (Fig 2C). Overall prevalence of germline GPR161 mutations among pediatric (age < 18 years) and infant (age < 4 years) patients with MBSHH was 3.4% (six of 178) and 5.5% (five of 91), respectively (Fig 2D). GPR161 mutation carriers were also observed at a similar frequency in retrospective (3.2%; three of 93) and prospective (3.5%; three of 85) pediatric MBSHH cohorts (Fig 2E). Of note, the frequency of germline mutations in GPR161 (3.4%), PTCH1 (4.5%), and SUFU (5.6%) was comparable among pediatric patients with MBSHH.
The latest edition of the WHO CNS tumor classification divides MBSHH into TP53 mutated and wild type, respectively.30,31 We assessed TP53 mutation status across all MBSHH and observed that all GPR161-associated MBSHH were TP53 wild type and represented 4.1% (six of 147) of pediatric patients with MBSHH, TP53-wt. Recently, MBSHH tumors have been further classified into four molecular subtypes.8 These include two infant subtypes SHH-γ (low risk) and SHH-β (high risk), which are also equivalent to infant-SHH-I and infant-SHH-II in another study.32 Classification of these infant GPR161-associated MBSHH tumors into molecular subtypes demonstrated that the majority (five of six) belonged to the MBSHH-β subtype; however, this was not statistically significant when considering all infant patients with MBSHH in our series (P = .09). Finally, germline GPR161 protein-truncating variants were significantly enriched in pediatric patients with MBSHH compared with the general population (one in 30 patients v one in 1,664 controls in gnomAD; P = 2×10−9, Fisher’s exact test).
Analysis of tumor DNA in GPR161 germline mutation carriers revealed LOH at the GPR161 locus, unexceptionally leading to loss of the wild-type allele in all affected patients (P = .03, n = 6, binomial test; Fig 2B and Data Supplement). Other than this, no shared somatic driver event was detected, which further substantiates a role for GPR161 deficiency as a driver event for MB development. Of note, patient SJMB054 had a somatic chromosome 6 monosomy, an alteration associated with MBWNT; however, a somatic CTNNB1 mutation was not detected and the brain tumor methylation classifier score was highest for MBSHH (MBSHH:0.9897 v MBWNT:0.0003). Patient SJMB335 had a somatic SUFU missense mutation, but clonal evolution analysis did not provide evidence for an SUFU-associated tumor as a result of low mutation frequency. Finally, the MB of the index patient showed low-level copy number gain at the GLI2 locus, but this was not a true gene amplification and the tumor lacked a loss of 17p and a TP53 mutation that usually accompanies a prototypical MB with GLI2 amplification. In summary, biallelic GPR161 inactivation occurred in all six patients with MB that otherwise did not harbor any other driver events that affect the SHH pathway (Fig 2B).
Remarkably, five of six patients with MB demonstrated an inactivation of GPR161 as a result of cnLOH of 1q and in one tumor a focal approximate 425-kb deletion spanned all exons of GPR161. Recurrent cnLOH of 1q so far has not been reported in the MB literature and we therefore assessed the overall frequency of 1q cnLOH events among pediatric patients with MBSHH and its association with GPR161 mutation status (Fig 2B). This analysis revealed that 1q cnLOH is exclusively present in tumors with germline GPR161 mutations (GPR161MUT = five of six v GPR161WT = zero of 220; P = 8.8×10−8, Fisher’s exact test), which suggests that 1q cnLOH is a hallmark for GPR161 deficiency and a molecular marker for the identification of patients with GPR161 mutations.
DISCUSSION
Recognition of tumor predisposition syndromes helps to optimize tumor surveillance programs and to improve clinical outcome. Recent progress in high-throughput genomics and access to larger patient cohorts enables the identification of novel tumor predisposition syndromes. In the current study, we have identified a novel tumor predisposition syndrome with SHH-activated and early-onset MB as the primary clinical manifestation. Carriers of a protein-truncating germline mutation in GPR161 reported here developed MBSHH before the age of 3 years. Overall, we estimate that GPR161 mutations are responsible for approximately 5% of infant MBSHH cases. All GPR161 mutations that were identified within our patients with MBSHH were also found to be present at a low carrier frequency in the general population (approximately one in 42,000 to one in 125,000 individuals), and analysis of two available parent-offspring trios (M20769 and MB13_03) demonstrated parental transmission in both affected patients. Of note, the frequency of germline GPR161 protein-truncating variants in the general population is six in 10,000 individuals, and extrapolated across the global population one would anticipate approximately four million people worldwide to be affected (Data Supplement).
Patients with truncating germline GPR161 mutations (genomic location: 1q24.2) showed recurrent LOH of GPR161 as a second hit in the tumor. This primarily resulted from segmental cnLOH of 1q, an event that is completely absent among GPR161 wild-type MBSHH and can thus be considered a hallmark characteristic of GPR161-associated MB. cnLOH and the unmasking of germline mutations have previously been shown to be of importance in the inactivation of tumor suppressor genes and are described in a variety of different tumor types.33 The 1q cnLOH event most likely originates from mitotic recombination as a result of DNA double-strand breaks that necessitate the second, homologous chromosome as a template for DNA repair. Such double-strand breaks tend to be more frequent at common fragile sites, which are prone to breakage upon DNA replication stress.34 A common fragile site (FRA1F) is also present at the 1q cnLOH breakpoints in GPR161-associated MB (1q21.1 to 1q21.3). Moreover, the lack of 1q loss events among GPR161 wild-type MBSHH tumors and the preferential mode of GPR161 inactivation, either focal or via cnLOH, suggests that 1q harbors dose-sensitive genes that might restrict MBSHH tumor development. Future studies could therefore explore whether genes on 1q might serve as a substrate for novel treatment options based on synthetic lethal interactions. Of note, analysis of somatic mutations in known and putative MB driver genes revealed biallelic inactivation of GPR161 as the sole shared event across the six patients, indicating that loss of GPR161 is sufficient for tumor growth, which is consistent with results of recent Gpr161 knockout studies in mice.35
GPR161 function is essential for several aspects of embryonic development and, among other things, is also relevant for granule cell (GC) proliferation.36-38 GCs of the cerebellum are by far the most numerous neurons in the brain and MBSHH originates from their progenitors (GC progenitors [GCPs]).39 Proliferation of GCPs is critically regulated by the mitogenic activity of the SHH ligand. SHH secretion from cerebellar Purkinje cells at late stages of embryonic development boosts the postnatal mitotic rate of GCPs before differentiated cells exit the cell cycle to become postmitotic neurons.40 Consequently, constitutive activation of SHH signaling increases the proliferation rate and neoplastic transformation of GCPs.41 As Gpr161 acts as a negative regulator of the SHH pathway37 and prevents GCP overproduction, a loss of GPR161 activity is consistent with MB pathogenesis, which was recently demonstrated in a mouse model with a neural stem cell–specific Gpr161 deletion.35 The timing of the GPR161–SHH interplay at the stage of GPC differentiation is in line with a vulnerable phase for MB development in early childhood. Similar to PTCH1- and SUFU-associated MBSHH in Gorlin syndrome, patients in this study were typically diagnosed with MB during infancy.
In our study, patients were mainly children and other tumors could potentially occur at later stages of life. The oldest patient of the study (index patient, M20769, age 29 years) developed recurrent BCCs and a meningioma at age 24 in the radiation field. She also developed multiple hyperplastic GI polyps and a tubular adenoma with low-grade dysplasia. Her father, likewise a carrier of the GPR161 germline mutation, has died of an adenocarcinoma of the colon at age 55 years. In a second family (SJMB335), both the maternal aunt and grandmother had ovarian cancer, but were not available for additional genetic testing. Other potential manifestations of GPR161-associated tumors are clearly an avenue for additional studies, not least because of their fundamental importance for genetic screening programs. In summary, the GPR161 disorder defines a novel MB predisposition syndrome.
ACKNOWLEDGMENT
The authors acknowledge the Next Generation Sequencing Diagnostic Center Aachen and the Comprehensive Diagnostic Center Aachen. The authors thank Christina Backhaus, Elvira Golz-Staggemeyer, Hai Yen Nguyen, and Laura Dörner for excellent technical assistance. The authors also thank the Genomics and Proteomics Core Facility at the German Cancer Research Centre (Stephan Wolf and team) for performing whole-exome sequencing and the Microarray Core Facility at the German Cancer Research Centre (Melanie Bewerunge-Hudler and team) for performing DNA methylation analysis.
Footnotes
Supported by Deutsche Forschungsgemeinschaft Grants No. EG110/15-1 and 948/32-1 FUGG; partially funded by the PedBrain Tumor Project contributing to the International Cancer Genome Consortium, Grant No. 109252 from German Cancer Aid, and Grants No. 01KU1201A and 01KU1201C from the German Federal Ministry of Education and Research. Additional funding through German Federal Ministry of Education and Research BioTop Grants No. 01EK1502A and 01EK1502B and International Cancer Genome Consortium Data Mining (Grant No. 01KU1505F), Grant No. 111234 from the German Cancer Aid, and Grant No. A2013/46 DKS2014.12 from the German Childhood Cancer Foundation (Deutsche Kinderkrebsstiftung). The Hirntumor-Studiengruppe für Medulloblastome study group and the Deutsche Gesellschaft für Neuropathologie und Neuroanatomie Brain Tumor Reference Center are supported by the German Childhood Cancer Foundation (Deutsche Kinderkrebsstiftung). S.M.W. received funding through a Swiss National Science Foundation early postdoctoral mobility fellowship (P2ELP3_155365) and a European Molecular Biology Organization Long-Term Fellowship (ALTF 755‐2014). J.O.K. is a European Research Council investigator. F.S. is a fellow of the Else Kröner Excellence Program of the Else Kröner-Fresenius Stiftung (2017_EKES.24).
AUTHOR CONTRIBUTIONS
Conception and design: Matthias Begemann, Sebastian M. Waszak, Miriam Elbracht, Stefan M. Pfister, Udo Kontny, Ingo Kurth
Provision of study materials or patients: Matthias Begemann, Giles W. Robinson, Cordula Knopp, Florian Kraft, Olga Moser, Martin Mynarek, Lea Guerrini-Rousseau, Laurence Brugieres, Pascale Varlet, Torsten Pietsch, Daniel C. Bowers, Murali Chintagumpala, Stefan Rutkowski, Amar Gajjar, Miriam Elbracht, Stefan M. Pfister, Udo Kontny, Ingo Kurth
Collection and assembly of data: Matthias Begemann, Sebastian M. Waszak, Natalie Jäger, Tanvi Sharma, Martin Mynarek, Lea Guerrini-Rousseau, Laurence Brugieres, Pascale Varlet, Daniel C. Bowers, Murali Chintagumpala, Felix Sahm, Jan O. Korbel, Thomas Eggermann, Paul Northcott, Miriam Elbracht, Stefan M. Pfister, Udo Kontny, Ingo Kurth
Data analysis and interpretation: Matthias Begemann, Sebastian M. Waszak, Giles W. Robinson, Natalie Jäger, Tanvi Sharma, Cordula Knopp, Florian Kraft, Olga Moser, Torsten Pietsch, Felix Sahm, Jan O. Korbel, Stefan Rutkowski, Thomas Eggermann, Amar Gajjar, Paul Northcott, Stefan M. Pfister, Udo Kontny, Ingo Kurth
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Germline GPR161 Mutations Predispose to Pediatric Medulloblastoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/journal/jco/site/ifc.
Giles W. Robinson
Consulting or Advisory Role: Eli Lilly
Research Funding: Novartis (Inst), Genentech (Inst), Novartis (Inst)
Martin Mynarek
Employment: Medac (I)
Felix Sahm
Consulting or Advisory Role: AbbVie
Speakers' Bureau: Agilent Technologies, Illumina, Medac
Travel, Accommodations, Expenses: AbbVie, Agilent Technologies
Amar Gajjar
Research Funding: Genentech (Inst), Kazia Pharmaceutical (Inst)
Stefan Rutkowski
Research Funding: Riemser Pharma (Inst)
Stefan M. Pfister
Research Funding: Eli Lilly (Inst), Bayer (Inst), Roche (Inst), PharmaMar (Inst), Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Patent for using DNA methylation profiling for tumor classification
Udo Kontny
Consulting or Advisory Role: Eisai
No other potential conflicts of interest were reported.
REFERENCES
- 1.Rahman N.Realizing the promise of cancer predisposition genes Nature 505302–308.2014[Erratum: Nature 510:176, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gröbner SN, Worst BC, Weischenfeldt J, et al. The landscape of genomic alterations across childhood cancers Nature 555321–327.2018[ErratumNature559:E10, 2018] [DOI] [PubMed] [Google Scholar]
- 3.Ripperger T, Bielack SS, Borkhardt A, et al. Childhood cancer predisposition syndromes: A concise review and recommendations by the Cancer Predisposition Working Group of the Society for Pediatric Oncology and Hematology. Am J Med Genet A. 2017;173:1017–1037. doi: 10.1002/ajmg.a.38142. [DOI] [PubMed] [Google Scholar]
- 4.Waszak SM, Northcott PA, Buchhalter I, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: A retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018;19:785–798. doi: 10.1016/S1470-2045(18)30242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Northcott PA, Jones DT, Kool M, et al. Medulloblastomics: The end of the beginning. Nat Rev Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwalbe EC, Lindsey JC, Nakjang S, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol. 2017;18:958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–754.e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kool M, Jones DT, Jäger N, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleihues P, Schäuble B, zur Hausen A, et al. Tumors associated with p53 germline mutations: A synopsis of 91 families. Am J Pathol. 1997;150:1–13. [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton SR, Liu B, Parsons RE, et al. The molecular basis of Turcot’s syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 12.Offit K, Levran O, Mullaney B, et al. Shared genetic susceptibility to breast cancer, brain tumors, and Fanconi anemia. J Natl Cancer Inst. 2003;95:1548–1551. doi: 10.1093/jnci/djg072. [DOI] [PubMed] [Google Scholar]
- 13.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 14.Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 15.Smith MJ, Beetz C, Williams SG, et al. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol. 2014;32:4155–4161. doi: 10.1200/JCO.2014.58.2569. [DOI] [PubMed] [Google Scholar]
- 16.Brugières L, Remenieras A, Pierron G, et al. High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol. 2012;30:2087–2093. doi: 10.1200/JCO.2011.38.7258. [DOI] [PubMed] [Google Scholar]
- 17.Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 18.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 19.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 20.Forget A, Martignetti L, Puget S, et al. Aberrant ERBB4-SRC signaling as a hallmark of group 4 medulloblastoma revealed by integrative phosphoproteomic profiling. Cancer Cell. 2018;34:379–395.e7. doi: 10.1016/j.ccell.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Sahm F, Schrimpf D, Jones DT, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131:903–910. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
- 22.Jones DT, Jäger N, Kool M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimmer A, Phan H, Mathieson I, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talevich E, Shain AH, Botton T, et al. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLOS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinheinz K, Bludau I, Hübschmann D, et al. ACEseq: Allele specific copy number estimation from whole genome sequencing. bioRxiv. 2017 [Google Scholar]
- 26.Waszak SM, Tiao G, Zhu B, et al. Germline determinants of the somatic mutation landscape in 2,642 cancer genomes. bioRxiv. 2017 [Google Scholar]
- 27.Guo Y, Ding X, Shen Y, et al. SeqMule: Automated pipeline for analysis of human exome/genome sequencing data. Sci Rep. 2015;5:14283. doi: 10.1038/srep14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Li J, Li MJ, et al. Robust and rapid algorithms facilitate large-scale whole genome sequencing downstream analysis in an integrative framework. Nucleic Acids Res. 2017;45:e75. doi: 10.1093/nar/gkx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturm D, Orr BA, Toprak UH, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietsch T, Wiestler OD, Ellison DW, et al. Desmoplastic/nodular medulloblastomainLouis DN, Ohgaki H, Wiestler OD, et al.WHO Classification of Tumours of the Central Nervous System ed 4Lyon, France: IARC Press; 2016. pp195–210. [Google Scholar]
- 31.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 32.Robinson GW, Rudneva VA, Buchhalter I, et al. Risk-adapted therapy for young children with medulloblastoma (SJYC07): Therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncol. 2018;19:768–784. doi: 10.1016/S1470-2045(18)30204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lapunzina P, Monk D. The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. Biol Cell. 2011;103:303–317. doi: 10.1042/BC20110013. [DOI] [PubMed] [Google Scholar]
- 34.LaFave MC, Sekelsky J. Mitotic recombination: Why? When? How? Where? PLoS Genet. 2009;5:e1000411. doi: 10.1371/journal.pgen.1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada IS, Hwang SH, Somatilaka BN, et al. Basal suppression of the sonic hedgehog pathway by the G-protein-coupled receptor Gpr161 restricts medulloblastoma pathogenesis. Cell Reports. 2018;22:1169–1184. doi: 10.1016/j.celrep.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung T, Humbert JE, Stauffer AM, et al. The orphan G protein-coupled receptor 161 is required for left-right patterning. Dev Biol. 2008;323:31–40. doi: 10.1016/j.ydbio.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay S, Wen X, Ratti N, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 38.Hwang SH, White KA, Somatilaka BN, et al. The G protein-coupled receptor Gpr161 regulates forelimb formation, limb patterning and skeletal morphogenesis in a primary cilium-dependent manner. Development. 2018;145:145. doi: 10.1242/dev.154054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuccillo M, Joyner AL, Fishell G.Morphogen to mitogen: The multiple roles of hedgehog signalling in vertebrate neural development Nat Rev Neurosci 7772–783.2006[Erratum: Nat Rev Neurosci 7:902, 2006] [DOI] [PubMed] [Google Scholar]
- 41.Yang ZJ, Ellis T, Markant SL, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]