Abstract
Eukaryotic chromosomes are composed of chromatin, in which regularly spaced nucleosomes containing ~147 bp of DNA are separated by linker DNA. Most eukaryotic cells have a characteristic average nucleosome spacing of ~190 bp, corresponding to a ~45 bp linker. However, cortical neurons have a shorter average spacing of ~165 bp. The significance of this atypical global chromatin organization is unclear. We have compared the chromatin structures of purified mouse dorsal root ganglia (DRG) neurons, cortical oligodendrocyte precursor cells (OPCs) and cortical astrocytes. DRG neurons have short average spacing (~165 bp), whereas OPCs (~182 bp) and astrocytes (~183 bp) have longer spacing. We measured nucleosome positions by MNase-seq and gene expression by RNA-seq. Most genes in all three cell types have a promoter chromatin organization typical of active genes: a nucleosome-depleted region at the promoter flanked by regularly spaced nucleosomes phased relative to the transcription start site. In DRG neurons, the spacing of phased nucleosomes downstream of promoters (~182 bp) is longer than expected from the genomic average for DRG neurons, whereas phased nucleosome spacing in OPCs and astrocytes is similar to the global average for these cells (~183 bp). Thus, the atypical nucleosome spacing of neuronal chromatin does not extend to promoter-proximal regions.
Keywords: chromatin, nucleosome, transcription, dorsal root ganglia neurons, oligodendrocyte progenitor cells, astrocytes
1. Introduction
Eukaryotic chromosomes are composed of chromatin, the basic subunit of which is the nucleosome. Chromatin can be visualized in the electron microscope as regularly spaced ‘beads-on-a-string’, in which the beads represent nucleosome cores and the string is the intervening linker DNA [1]. The nucleosome contains ~147 bp of DNA coiled almost twice around a central octamer of core histones, composed of two molecules each of histones H3, H4, H2A and H2B [2]. The DNA entry-exit points of the nucleosome are sealed by a molecule of H1, the linker histone, which binds both to the nucleosome core and linker DNA. H1 is required for full compaction of chromatin [1,3].
Most eukaryotic cells have a characteristic spacing in the range of 185-195 bp, corresponding to 147 bp of nucleosomal DNA and ~45 bp of linker DNA. However, there are notable exceptions, including mammalian cortical neurons, which have a much shorter genomic spacing of 160-170 bp, corresponding to a linker of only ~15-20 bp [4,5]. Interestingly, cerebellar neurons have a more typical nucleosome spacing [4,5] and spacing in both cortical and cerebellar neurons increases with age [6]. The chromatin of cortical neurons is deficient in H1, but still forms compact fibers [7,8]. In contrast, glial cells, which interact with neurons, have a typical nucleosome spacing and normal H1 levels [4,5] and their average spacing does not change with age [6]. The significance of tissue-specific differences in nucleosome spacing is uncertain, but they are likely to be important because cells contain ATP-dependent chromatin remodeling enzymes that alter spacing [9–11].
Previous reports of the difference in nucleosome spacing between cortical neurons and glial cells involved separation of neuronal and glial nuclei by size [4,5,12]. However, these glial cells were probably a mixture of astrocytes, oligodendroglia and microglia. To address this issue and to extend the original observations using both classical and modern genomic methods, we analyzed nucleosome spacing and positioning in purified primary cultures of mouse dorsal root ganglia (DRG) neurons, astrocytes and oligodendrocyte progenitor cells (OPCs). OPCs are precursors of oligodendrocytes, which synthesize the myelin responsible for insulating neuronal axons. We have previously shown direct interactions between OPCs and DRG neurons in co-culture [13]. Astrocytes are the most abundant non-neuronal cell population in the mammalian brain [14]. They are involved in many essential brain functions [14–16]. Our results demonstrate a significant difference in genomic chromatin structure between neurons and glial cells.
2. Materials and methods
2.1. Cell culture
All research involving animals was approved by the NICHD Institutional Animal Care and Use Committee. OPCs were purified from the cortices of postnatal mice (P6-P9) of either sex and cultured for 6-7 days [17]. OPC purity was determined by olig2 immunoreactivity (routinely >90% Olig2-positive cells). Astrocytes were isolated from the cortices of postnatal mice (P1-P3) of either sex and cultured for ~21 days until confluent. After 6-7 days of growth, flasks were shaken overnight to reduce contamination by OPCs and microglia [14,18]. Astrocyte purity was determined by GFAP immunoreactivity (routinely >90% GFAP-positive cells). DRG neurons were isolated from the dorsal root ganglia of embryonic mice (E13.5) [19]. Cells were plated onto coverslips and cultured for ~2 weeks. DRG purity was verified by NeuN immunoreactivity; the numbers of non-DRG fibroblast-like cells on coverslips before and after removal of DRG neurons were compared after fixation and staining. Cultures were >70% NeuN-positive cells, with no astrocytes or OPCs [14]. Cell purity for all cell types was confirmed by RNA-seq (Fig. S1). The purity of our cultures is also shown by a cross comparison with an established database [20]. There is no correlation between the RNA seq data from our OPCs and their astrocytes (R = 0.15-0.18), or between our DRG neurons and their astrocytes (R = 0.14-0.16), or between our astrocytes and their OPCs (0.07-0.08).
2.2. MNase digestion
Frozen cells were thawed on ice and gently resuspended in 500 μl Buffer A with 0.5% NP40 (0.34 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl, 0.5 mM spermidine, 0.15 mM spermine, 1 mM EDTA, 15 mM 2-mercaptoethanol and protease inhibitors (Roche 11666975001)). Nuclei were pelleted in a microfuge (21,000g, 4°C, 1 min), washed with 500 μl Buffer A and resuspended in 250 μl Buffer A. The DNA concentration was measured in a fluorimeter. Nuclei were divided into 100 μl aliquots at 0.5 to 9 μg/ml and digested for 3 min at ~25°C with 0 to 128 units of MNase (Worthington LS004798). Digestion was stopped with 1 μl 0.5 M EDTA and 5 μl 20% SDS. Purified DNA was dissolved in 25 μl 10 mM Tris.HCl pH 8.0, 0.1 mM EDTA, 0.1 mg/ml RNase and incubated at 37°C for 30 min. Samples were analyzed in a 1.2% (w/v) agarose-LE gel stained with SYBR-Gold. Repeat lengths were measured by analysis in a 1.5% agarose gel (containing 1% 3:1 agarose and 0.5% agarose-LE to maximize separation).
2.3. MNase-seq and RNA-seq
Samples containing predominantly mono-nucleosomal DNA were treated with repair enzymes to seal any MNase-induced nicks using the PreCR Repair kit (New England Biolabs M0309) according to the manufacturer’s instructions. Repaired DNA was purified using a Qiagen PCR column and the DNA concentration was measured. Paired-end libraries were constructed using the NEBNext Ultra DNA Library Prep Kit for illumina (New England Biolabs E7370) according to the manufacturer’s instructions for < 100 ng input DNA. Adaptor-ligated DNA was purified using an equal volume of AMPure XP beads (Beckman A63880) and then amplified by PCR (11 cycles). Libraries were purified using AMPure XP beads and checked in a gel prior to sequencing (Illumina HiSeq 2500). RNA was extracted from cells using standard protocols. RNA was sequenced by ACGT Inc. (Replicate 1) or the NICHD Molecular Genomics Core Facility (Replicates 2 and 3) (Fig. S2).
2.4. Bioinformatics
MNase-seq reads were aligned to the mouse genome (mm10) using Bowtie 2 [21] with parameters -X 1000 --very-sensitive. Nucleosome positions were defined by the centers of sequenced DNA fragments of 120 - 180 bp. RefSeq [22] annotations for transcription start sites were obtained using the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables). Heat maps representing nucleosome distributions near gene promoters were generated in MATLAB using the heatmap plotting function (http://www.mathworks.com/matlabcentral/fileexchange/24253-customizable-heat-maps). RNA-seq reads were aligned using TopHat2 [23] using default parameters. Transcript levels were quantified using HTSeq [24]. The data have been deposited in the GEO database: GSE133966 (MNase-seq); GSE133745 (RNA-seq).
3. Results
3.1. Mouse DRG neurons have short genomic nucleosome spacing
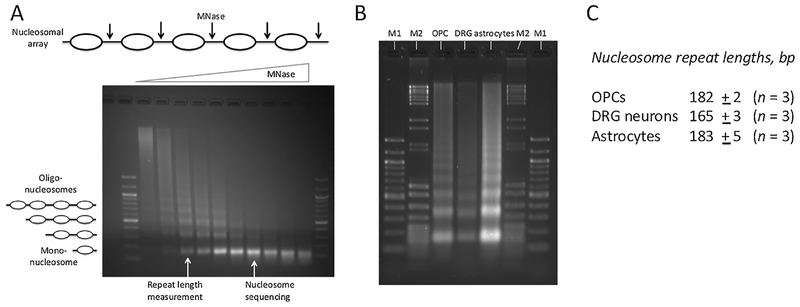
We measured the average nucleosome spacing (or “repeat length”) in DRG neurons, OPCs and astrocytes (Fig. 1). Nuclei were digested with increasing concentrations of micrococcal nuclease (MNase), which cuts the linker DNA much faster than the nucleosome core, giving rise to a typical “ladder” of DNA fragments, corresponding to mono-, di-, tri-nucleosomes etc. (Fig. 1A). To estimate the repeat length, DNA samples at the appropriate level of digestion were compared side-by-side in an agarose gel (Fig. 1B). As expected, the mono-nucleosome band is about the same size in all three cell types. However, the di-, tri- and tetra-nucleosome bands from DRG neurons are clearly shorter than those from OPCs and astrocytes, indicating that the linker DNA averages shorter in neuronal chromatin. The average spacings derived from three independent experiments are 165±3 bp for DRG neurons, 183±5 bp for OPCs and 182±2 bp for astrocytes (Fig. 1C). Thus, DRG neurons resemble cortical neurons in having atypically short nucleosome spacing.
Fig. 1.
DRG neurons have shorter nucleosome spacing than OPCs and astrocytes. (A) Gel electrophoresis of DNA purified from a typical MNase titration (example: astrocytes). The oligo-nucleosomes become progressively shorter as more linkers are cut by MNase, eventually resulting in mono-nucleosomes. The repeat length is measured using samples with multiple nucleosome bands (as indicated). Nucleosome positioning (MNase-seq) data are obtained using samples containing predominantly mono-nucleosomes, avoiding over-digestion (indicated by the smearing below the mono-nucleosome band in the last 3 samples). 100-bp marker. (B) Typical gel used for repeat length measurement. Specific samples from titrations like that in A are analyzed side-by-side. M1: 100-bp marker; M2: mixture of pBR322 MspI and λ-DNA BstEII digests. (C) Repeat length data (average of 3 independent experiments with standard deviation). The lengths of the mono-, di-, tri- and tetra-nucleosomes were determined using the markers for calibration. The repeat length is given by the slope of the regression line in a plot of DNA length vs. nucleosome number (Fig. S5).
3.2. Gene expression patterns
We determined the gene expression patterns for all three cell types by RNA-seq. Histograms of the number of genes with a given mRNA count indicated a wide range of gene expression in each cell type (Fig. S3). All three cell types exhibited a peak corresponding to highly expressed genes, representing roughly half of the 23,997 genes. The remaining genes had little or no activity in each cell type. As expected, pairwise comparisons of these gene expression patterns showed that a large fraction of genes had similar expression levels in all three cell types (Fig. S3). Also as expected, many genes showed much greater differential expression, corresponding to transcripts characteristic of each cell type.
3.3. Most gene promoters have a nucleosome-depleted region and phased nucleosomes in all three cell types
The measured repeat lengths (Fig. 1C) represent the global average nucleosome spacing for the entire genome in each cell type. More locally, the average spacing in the vicinity of promoters can be estimated using MNase-seq data, because nucleosomes are phased with respect to the transcription start site (TSS). The promoters of active genes are typically associated with a nucleosome-depleted region (NDR) about 200 bp wide; the TSS is usually located just upstream of the first (“+1”) nucleosome on the gene. Regularly spaced nucleosomal arrays form on either side of the promoter NDR and are said to be phased because they adopt similar positions relative to the DNA sequence in every cell. Nucleosome phasing upstream of the promoter NDR is generally weaker than downstream nucleosome phasing. In contrast, the promoters of inactive genes typically lack both an NDR and nucleosome phasing [25].
We obtained genome-wide nucleosome positioning data for all three cell types using MNase-seq. Accurate and relatively unbiased nucleosome mapping with MNase requires extensive digestion in order to isolate most of the genome in mono-nucleosomes (Fig. 1A). Two biological replicate experiments with excellent coverage were performed for each cell type (Table S1). The distance in base pairs between the ends of each pair of 50 nt reads after alignment to the mouse genome gives the length of the sequenced nucleosome, which is expected to be ~147 bp. The central nucleotide in each nucleosome sequence marks the genomic position (dyad) of the nucleosome, subject to the caveat that some nucleosomal DNA sequences are longer or shorter than the expected 147 bp, due to incomplete removal of linker DNA or to internal digestion of the nucleosome by MNase, respectively.
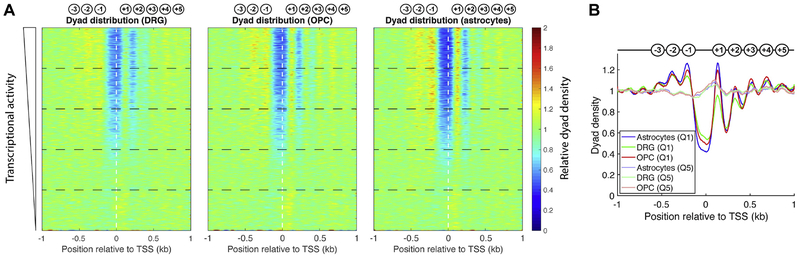
We sorted all 23,997 genes according to their transcriptional activity using our RNA-seq data for the three cell types (Fig. S3A) and constructed heat maps representing our nucleosome positioning (MNase-seq) data aligned on the TSS, indicated by the vertical dashed line (Fig. 2A). Each row in the heat map represents a gene, with the most active gene at the top and the least active gene at the bottom. As expected, we observed that the more active genes are associated with a prominent NDR at the promoter, which includes the TSS. These promoters are also associated with an array of well-phased nucleosomes downstream of the TSS and weaker upstream phasing. The genes in the bottom half of each heat map have much less prominent NDRs and weaker nucleosome phasing, both of which decrease with transcriptional activity. The least active genes, at the bottom of each heat map, have very weak NDRs and very poor nucleosome phasing. Overall, more than half of the genes in each cell type have the active chromatin configuration.
Fig. 2.
Transcriptionally active promoters have an NDR and phased nucleosomes in all three cell types. (A) Nucleosome dyad densities are shown for each gene according to the scale at right (MNase-seq data). The genomic average density is set at 1. Genes are sorted according to transcriptional level for each cell type using the RNA-seq data for that cell type (Fig. S3) and divided into quintiles (horizontal dashed lines); quintile 1 corresponds to the most active genes (at top). Genes are aligned on the TSS (vertical dashed line). (B) Nucleosome phasing relative to the TSS for the most active and least active genes in all three cell types (quintiles 1 and 5, respectively). The spacing of phased nucleosomes near active gene promoters (quintile 1) is similar in DRG neurons, OPCs and astrocytes. Nucleosomes on inactive genes (quintile 5) are not phased and there is no NDR. See Fig. S4 for analysis of all quintiles and Fig. S5 for regression analysis.
3.4. Similar nucleosome spacing on active genes In all three cell types
Since chromatin structure is related to transcriptional activity, we measured the nucleosome spacing of the most active genes in each cell type. The genes were first divided into quintiles (Fig. 2A). The distribution of nucleosomes relative to the TSS on the most active genes (quintile 1) was compared with that on the least active genes (quintile 5) by plotting the average nucleosome dyad density against distance from the TSS (Fig. 2B; Fig. S4). In all three cell types, the active genes exhibited a deep NDR at the promoter and phased nucleosomes upstream and downstream of the TSS, whereas the least active genes had no NDR and no nucleosome phasing (the approximately flat line indicates that nucleosomes have a similar probability of formation at any location relative to the promoter). The average spacing between the phased nucleosomes at the 5’-ends of the most active genes (quintile 1) in each cell type was estimated from the slope of a regression line in a plot of the distance between nucleosome peaks (for the +1 to +4 nucleosomes) against the nucleosome number (Fig. S5). The results for two biological replicate experiments were as follows: DRG neurons: 183 and 181 bp; OPCs: 183 and 184 bp; astrocytes: 190 and 184 bp. The nucleosome spacing at active promoters in OPCs and astrocytes is therefore very similar to the global spacing measured in gels (182 and 183 bp, respectively; Fig. 1C). Surprisingly, nucleosome spacing at promoters in DRG neurons (~182 bp) is clearly higher than the global spacing in the same cells (165 bp).
4. Discussion
We report here that DRG neurons have short nucleosome spacing at the global level (~165 bp). OPCs and astrocytes have more typical global nucleosome spacing (~182 bp and ~183 bp, respectively), although these spacings are shorter than reported for nuclei isolated from heterogeneous glial cell populations (~200 bp) and on the lower edge of the range for many other cells and tissue types [26]. The significance of the atypical global chromatin organization of DRG neurons is unclear. DRG neurons are non-dividing, terminally differentiated cells, OPCs are dividing, non-differentiated progenitor cells, whereas astrocytes are dividing, differentiated cells. Short spacing is not a simple consequence of terminal differentiation, because cerebellar neurons are also terminally differentiated, but have relatively long nucleosome spacing.
It has been proposed that shorter nucleosome spacing in cortical neurons may reflect higher overall transcriptional activity relative to other cells [4,5,12]. This hypothesis is supported by the similarity with budding yeast, which is very active in transcription and has similarly short spacing (~165 bp) [27]. Furthermore, the most highly transcribed yeast genes generally have the shortest nucleosome spacing [9,28,29]. However, our observation that the most active genes in DRG neurons have longer nucleosome spacing than the genomic average, at least near their promoters, is inconsistent with this hypothesis.
An alternative hypothesis is that nucleosome spacing affects the compaction of the chromatin fiber. A short linker is likely to reduce the overall affinity of H1 for chromatin, because the interaction of the long C-terminal domain of H1 with linker DNA may be limited if the linker is too short [9]. This model is supported by the direct correlation between the amount of bound H1 and nucleosome spacing both in vitro [30,31] and in vivo [32,33]. The chromatin of cortical neurons contains only about half as much H1 as glial chromatin or chromatin from other cells with long nucleosome spacing [4]. Similarly, yeast chromatin has short spacing and low levels of H1 [34]. Mice lacking three of a total of about eight linker histone genes have reduced H1 in their chromatin and reduced nucleosome spacing relative to wild type mice [32]. However, low H1 does not result in full chromatin decompaction because both neuronal and yeast chromatin fragments form condensed fibers in vitro [7,8,35]. Instead, the key point may be that chromatin with long spacing can decondense if H1 levels are low, whereas chromatin with short spacing would be much less affected. Thus, H1 binding may regulate the degree of compaction of long-spaced chromatin but not of short-spaced chromatin. Our data indicate that active genes have relatively long spacing, suggesting that they would be decondensed at low H1 levels, presumably facilitating transcription. In DRG and cortical neurons, the bulk of the chromatin has short spacing and its level of compaction would be predicted to be relatively insensitive to H1 levels. More work needs to be done to test this hypothesis.
Supplementary Material
Highlights.
Mouse dorsal root ganglia (DRG) neurons have atypically short nucleosome spacing
Oligodendrocyte progenitor cells and astrocytes have more typical nucleosome spacing
Nucleosome spacing at active gene promoters is similar in all three cell types
In DRG neurons, nucleosomes are spaced farther apart at active genes than elsewhere
Acknowledgements
We thank William Huffman for culturing DRG neurons and OPCs. We thank the NICHD Molecular Genomics Core Facility (Tianwei Li and Steve Coon) for paired-end sequencing. This study utilized the computational resources of the NIH HPC Biowulf cluster.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health (NICHD) (grants ZIAHD000713 to R.D.F. and ZIAHD008775 to D.J.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests: None.
References
- [1].Thoma F, Koller T, Klug A, Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin., J. Cell Biol 83 (1979) 403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution., Nature. 389 (1997) 251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [3].Clark DJ, Kimura T, Electrostatic mechanism of chromatin folding., J. Mol. Biol 211 (1990) 883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- [4].Pearson EC, Bates DL, Prospero TD, Thomas JO, Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents., Eur. J. Biochem 144 (1984) 353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- [5].Thomas JO, Thompson RJ, Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons., Cell. 10 (1977) 633–640. [DOI] [PubMed] [Google Scholar]
- [6].Berkowitz EM, Sanborn AC, Vaughan DW, Chromatin structure in neuronal and neuroglial cell nuclei as a function of age., J. Neurochem 41 (1983) 516–523. doi: 10.1111/j.1471-4159.1983.tb04769.x. [DOI] [PubMed] [Google Scholar]
- [7].Pearson EC, Butler PJ, Thomas JO, Higher-order structure of nucleosome oligomers from short-repeat chromatin., EMBO J. 2 (1983) 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Allan J, Rau DC, Harborne N, Gould H, Higher order structure in a short repeat length chromatin., J. Cell Biol 98 (1984) 1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ocampo J, Chereji RV, Eriksson PR, Clark DJ, The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo., Nucleic Acids Res. 44 (2016) 4625–4635. doi: 10.1093/nar/gkw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Corona DFV, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, et al. , ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo., PLoS Biol. 5 (2007) e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT, Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo., Genes Dev. 18 (2004) 170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Thompson RJ, Studies on RNA synthesis in two populations of nuclei from the mammalian cerebral cortex., J. Neurochem 21 (1973) 19–40. [DOI] [PubMed] [Google Scholar]
- [13].Stevens B, Porta S, Haak LL, Gallo V, Fields RD, Adenosine: a neuron-glial transmitter promoting myelination in the CNS in response to action potentials., Neuron. 36 (2002) 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ishibashi T, Dakin KA, Stevens B, Lee PR, Kozlov SV, Stewart CL, et al. , Astrocytes promote myelination in response to electrical impulses., Neuron. 49 (2006) 823–832. doi: 10.1016/j.neuron.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Allen NJ, Astrocyte regulation of synaptic behavior., Annu. Rev. Cell Dev. Biol 30 (2014) 439–463. doi: 10.1146/annurev-cellbio-100913-013053. [DOI] [PubMed] [Google Scholar]
- [16].Dutta DJ, Woo DH, Lee PR, Pajevic S, Bukalo O, Huffman WC, et al. , Regulation of myelin structure and conduction velocity by perinodal astrocytes., Proc Natl Acad Sci USA. 115 (2018) 11832–11837. doi: 10.1073/pnas.1811013115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu J, Magri L, Zhang F, Marsh NO, Albrecht S, Huynh JL, et al. , Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation., J. Neurosci 35 (2015) 352–365. doi: 10.1523/JNEUROSCI.2606-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McCarthy KD, de Vellis J, Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue., J. Cell Biol 85 (1980) 890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee PR, Cohen JE, lacobas DA, lacobas S, Fields RD, Gene networks activated by specific patterns of action potentials in dorsal root ganglia neurons., Sci. Rep 7 (2017) 43765. doi: 10.1038/srep43765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex., J. Neurosci 34 (2014) 11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2., Nat. Methods 9 (2012) 357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pruitt KD, Tatusova T, Brown GR, Maglott DR, NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy., Nucleic Acids Res. 40 (2012) D130–5. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions., Genome Biol. 14 (2013) R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Anders S, Pyl PT, Huber W, HTSeq — a Python framework to work with high-throughput sequencing data., Bioinformatics. 31 (2015) 166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chereji RV, Clark DJ, Major determinants of nucleosome positioning., Biophys. J 114 (2018) 2279–2289. doi: 10.1016/j.bpj.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van Holde KE, Chromatin, Springer New York, New York, NY, 1989. doi: 10.1007/978-1-4612-3490-6. [DOI] [Google Scholar]
- [27].Thomas JO, Furber V, Yeast chromatin structure., FEBS Lett. 66 (1976) 274–280. [DOI] [PubMed] [Google Scholar]
- [28].Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ, Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases., Nucleic Acids Res. 42 (2014) 12512–12522. doi: 10.1093/nar/gku1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N, High-resolution nucleosome mapping reveals transcription-dependent promoter packaging., Genome Res. 20 (2010) 90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Blank TA, Becker PB, Electrostatic mechanism of nucleosome spacing., J. Mol. Biol 252 (1995) 305–313. doi: 10.1006/jmbi.1995.0498. [DOI] [PubMed] [Google Scholar]
- [31].Lusser A, Urwin DL, Kadonaga JT, Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly., Nat. Struct. Mol. Biol 12 (2005) 160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- [32].Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, et al. , Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation., Cell. 123 (2005) 1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- [33].Woodcock CL, Skoultchi AI, Fan Y, Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length., Chromosome Res. 14 (2006) 17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- [34].Freidkin I, Katcoff DJ, Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin., Nucleic Acids Res. 29 (2001) 4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lowary PT, Widom J, Higher-order structure of Saccharomyces cerevisiae chromatin., Proc Natl Acad Sci USA. 86 (1989) 8266–8270. doi: 10.1073/pnas.86.21.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.