Abstract
MiR-381 has been reported to be deregulated in many different types of human cancers. However, the clinical significance and function of miR-381 in gastric cancer (GC) remains unclear. Based on real-time quantitative PCR (RTq-PCR) analysis, we found that miR-381 was frequently lost or downregulated in GC tissues and cell lines, and decreased miR-381 was correlated with GC tumor size (P=0.012), TNM stage (P=0.007), invasion depth (P=0.008) and lymphatic metastasis (P=0.019). Cellular function assays revealed that miR-381 restoration inhibited cell proliferation, migration and invasion of GC cells. In addition, we validated that ROCK2 was as a direct target of miR-381, and knockdown of ROCK2 had similar with effects of miR-381 restoration in GC cells, while overexpression of ROCK2 attenuated the inhibitory effects of miR-381 on GC cells. These data suggest that miR-381 serves as a tumor suppressor by targeting ROCK2, which may provide a potential therapeutic tool for GC therapy.
Keywords: miR-381, ROCK2, gastric cancer, cell proliferation, cell migration, cell invasion
Introduction
Gastric cancer (GC) is one of the most common cancers and cause of cancer-related deaths worldwide, with a 5-year survival rate of 25% or less, which is mainly due to the non-specific symptoms during the early stages of GC [1-4]. Although extensive studies have investigated the etiology of gastric cancer, the molecular mechanisms underlying the pathogenesis and progression of GC remain undefined.
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNAs and act as important regulators of more than half of genes by binding to specific target mRNAs and leading to direct mRNA degradation or translational repression [5-7]. Numerous studies have revealed that miRNAs play crucial roles in various biological processes and diseases including human cancers [8-10]. Emerging evidence demonstrates that miRNA alteration is highly associated with the initiation and progression of all types of human cancers, which may provide new therapeutic targets to deal with human cancers [11]. MiR-381 has been reported to be deregulated in several human cancers, such as colorectal [12,13], ovarian [14], breast [15], glioma [16-18], lung [19], hepatocellular [20] and prostate cancer [21]. These reports also revealed that aberrant regulation of miR-381 is involved in many processes of tumor progression including cell proliferation, invasion and metastasis. However, the clinical significance and function of miR-381 in GC progression and the underlying mechanism remain unknown.
In the present study, we aimed to elucidate the involvement of miR-381 in GC. Our study indicated that miR-381 was frequently downregulated in primary GC and correlated with GC progression. In vitro functional study revealed that enhanced expression of miR-381 in GC cell line BGC823 inhibited cell proliferation, migration and invasion. Furthermore, we found that miR-381 directly targeted and inhibited ROCK2, and ROCK2 overexpression significantly attenuated the inhibitory effects of miR-381 on BGC823 cells. In conclusion, we demonstrated that miR-381 might function as a tumor suppressor by directly targeting ROCK2 in GC cells.
Materials and methods
Clinical samples and cell lines
Primary GC tissue samples were collected from Xiangya Hospital between Apr, 2013 to Oct, 2015, and the clinicopathologic information of the GC patients was summarized in Table 1. Written informed consent was obtained from all participants, and the use of these samples was approved by the ethical review committees of the Xiangya Hospital Ethic Committee of Central South University. Five GC cell lines (SGC7901, MGC803, BGC823, AGS and MKN45) and the gastric epithelial cell line GES-1 were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) and maintained in 90% RPMI-1640 (BioTeke Corporation, Beijing, China) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Table 1.
Correlation between miR-381 expression and clinicopathological features in 134 gastric cancer patients
| Parameters | No. of cases | High miR-381 expression | Low miR-381 expression | P value |
|---|---|---|---|---|
| Gender | 0.36 | |||
| Male | 69 | 36 | 33 | |
| Female | 65 | 31 | 34 | |
| Age | 0.431 | |||
| ≥60 | 74 | 38 | 36 | |
| <60 | 60 | 29 | 31 | |
| Tumor size | 0.012 | |||
| <5 cm | 74 | 44 | 30 | |
| ≥5 cm | 60 | 23 | 37 | |
| Differentiation | 0.191 | |||
| Well/Moderate | 56 | 31 | 25 | |
| Poor | 78 | 36 | 42 | |
| TNM stage | 0.007 | |||
| I+II | 79 | 47 | 32 | |
| III+IV | 55 | 20 | 35 | |
| Invasion depth | 0.008 | |||
| T1, T2 | 71 | 43 | 28 | |
| T3, T4 | 63 | 24 | 39 | |
| Lymphatic metastasis | 0.019 | |||
| Negative | 69 | 41 | 28 | |
| Positive | 65 | 26 | 39 |
Real-time quantitative PCR (RTq-PCR)
Total RNA was isolated from GC tissues and cell lines by Trizol reagent (Life Technologies, Gaithersburg, MD). First strand cDNA was synthesized according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Reverse transcription and RTq-PCR were performed by using a qSYBR-green PCR kit (Qiagen, Germantown, USA). U6 snRNA and β-actin were used as endogenous controls for miR381 and ROCK2 relative expression, respectively. The primers used are as follow: ROCK2-F: 5’-CAGTGGCATTGGGATAACATAA-3’, ROCK2R: 5’-GGAATTGGGAAGGTTTCTACAT3’; β-actin-F: 5’-AGGGGCCGGACTCGTCATACT-3’, β-actin-R: 5’-GGCGGCACCACCATGTACCCT-3’. The specific primers for miR-381 and U6 snRNA were purchased from GeneCopoeia. The relative levels of miR-381 and ROCK2 of each sample were measured in triplicates using the 2-ΔΔCt method.
Western blot
Proteins were separated on 10% SDS-PAGE and then transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). The membrane was blocked with 5% nonfat milk and incubated with indicated antibodies, followed by the HRP-conjugated secondary antibody. Antibodies of ROCK2, MMP2, MMP9 and β-actin were obtained from Abcam (Abcam, MA, USA). The Signals were detected with enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA).
Luciferase reporter assay
A fragment of the 3’UTR of ROCK2 containing predicted miR-381 target seed sequence was amplified and cloned into the psiCHECK-2 vector (Promega, Shanghai, China). The mutation of the predicted seed regions in the ROCK2 3’UTR sequence was performed using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Relative luciferase activity (a ratio of firefly luciferase to renilla luciferase) after cotransfection of miR-381 mimics or scramble mimics with wild type or mutant type of psiCHECK-2 vector into BGC823 cells was measured by the Dual Luciferase Reporter Assay system (Promega, Shanghai, China).
Plasmid construction, siRNA and cell transfection
ROCK2 coding region was amplified with the following primers (ROCK2-F, 5’-GAGCCTGGATGGACAAGAGGAA-3’; ROCK2-R, 5’-CTAGATTTTCGAGCTAATCATC-3’) and cloned into pcDNA3.1 vector to generate ROCK2 expressing vectors. The empty pCDNA3.1 vector was used as control. MiR-381 mimics, miR-381 inhibitor mimics and scramble mimics were purchased from RiboBio (Guangzhou, China). ROCK2 specific small interfering RNAs (siRNA-ROCK2, sense, 5’-GGCACGACUAGCAGAUAAAUU-3’; antisense, 5’-UUUAUCUGCUAGUCGUGCCUU-3’) and the negative control siRNA (siRNA-NC) were purchased from GeneChem (Shanghai, China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacture’s instruction.
Cell proliferation, invasion and migration assays
BGC823 cells were plated in 96-well plates (5,000 per well), and allowed to grow for 24, 48 and 72 h, then cell proliferation ability was assessed by a colorimetric assay using MTT solution (5 mg/ml) at 570 nm. Cell invasion ability was assessed using transwell invasion chambers coated with mgatrigel (BD Biosciences, Franklin Lakes, NJ, USA). Cells suspended in 200 µl of FBS-free medium were added into the upper chamber. 500 µl of medium with 10% FBS as the nutritional attractant was added into the lower chamber. After 24 h, the cells that had invaded through the membrane were fixed with 20% methanol, stained with 0.1% crystal violet, imaged and counted using an inverted microscope (Olympus, Tokyo, Japan). Cell migration ability was assessed by wound healing assays. An artificial “wound” was created using a pipette tip on a confluent cell monolayer. Then, the cells were then cultured for 24 hours and the wound closure was assessed by Scion Image Software (Scion Corporation, Frederick, MD).
Statistical analysis
SPSS 19.0 software (IBM, NY, USA) was used for all statistical analysis. Student’s t-test was used to analyze the differences in the experiments. Chi-squared test was used to analyze differences of miR-381 expression with clinicopathological features. P<0.05 was considered statistically significant.
Results
MiR-381 is frequently downregulated in gastric cancer
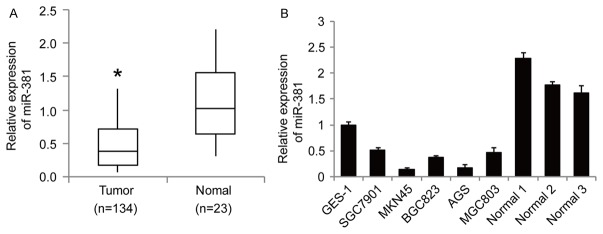
To examine the expression signature of miR-381 in GC progression, we performed RTq-PCR analysis in 134 cases of primary GC and 23 cases of normal gastric mucosa. We found that miR-381 expression levels were significantly decreased in GC cases compared to normal gastric mucosa (Figure 1A). We then extended to GC cell lines, and found that the expression of miR-381 was significantly downregulated in GC cell lines (SGC7901, MKN45, BGC823, AGS and MGC803) than that of the gastric epithelial cell line GES-1 and normal gastric mucosa (Figure 1B). Using the median value of the relative expression levels of miR-381 in all 134 samples as a cutoff, we classified 67 samples as the low expression group. Correlation analysis between miR-381 expression levels and clinicopathologic parameters revealed that the downregulation of miR-381 was significantly correlated with tumor size (P=0.012), TNM stage (P=0.007), invasion depth (P=0.008) and lymphatic metastasis (P=0.019) (Table 1). Taken together, these data suggest that decreased expression level of miR-381 may be associated with GC carcinogenesis.
Figure 1.
MiR-381 is frequently downregulated in GC tissues and cell lines RTq-PCR were performed to examine the expression levels of miR-381 in 134 primary GC and 23 normal gastric mucosa (A) and six GC cell lines (GES-1, SGC7901, MKN45, BGC823, AGS and MGC803) (B). *P<0.05 versus normal tissues, data shown are means ± SD.
MiR-381 inhibits the proliferation, invasion and migration of BGC823 cells
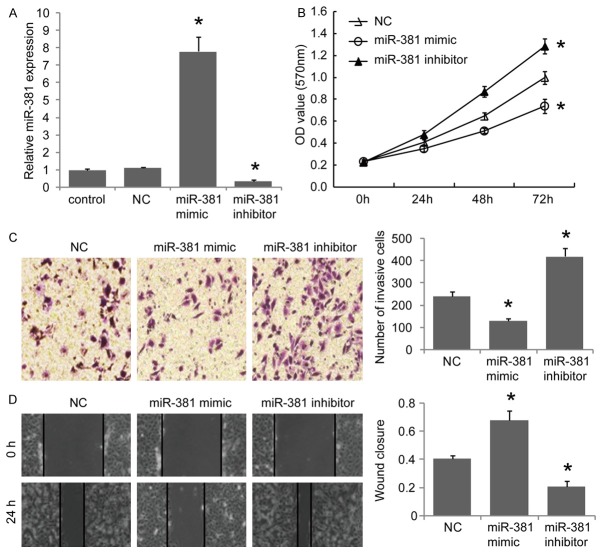
We then assessed the biological role of miR-381 in GC cells. We restored miR-381 expression in BGC823 cells (Figure 2A). MTT assays revealed that restoration of miR-381 inhibited the proliferation of BGC823 cells (Figure 2B). Transwell assays with matrigel (Figure 2C) and wound healing assays (Figure 2D) also showed that miR-381 had inhibitory effect on cell invasion and migration ability, respectively. In contrast, when endogenous miR-381 was silenced with anti-miR-381 mimics in BGC823 cells, the cell proliferation (Figure 2B), invasion (Figure 2C) and migration (Figure 2D) were increased. These observations suggested that miR-381 might act as a tumor suppressor miRNA in GC cells.
Figure 2.
MiR-381 inhibits the proliferation, invasion and migration of BGC823 cells. (A) BGC823 cells were transfected with miR-381 mimic, miR-381 inhibitor mimic or scramble mimic as negative control (NC), then miR-381 expression levels were determined by RTq-PCR. The effects of miR-381 on cell proliferation, invasion and migration were determined using MTT assays (B), transwell assays with matrigel (C) and wound healing assay (D), respectively. *P<0.05 versus the control, data shown are means ± S.D.
MiR-381 directly inhibits ROCK2 by targeting its 3’UTR
To elucidate the molecular mechanism by which miR-381 exerts its inhibitory effects on GC cells, miRNA target analysis tools PicTar and TargetScan were used to explore potential target of miR-381. Among hundreds of potential targets, we focused on ROCK1 and ROCK2 in the study, considering the involvement of them in the pathogenesis of many human cancers including gastric cancer. To validate targeting of ROCK1 and ROCK2 by miR-381, we performed luciferase reporter assays. The wild type 3’UTR luciferase reporter plasmid (Wt-3’UTR) or the mutated (Mut-3’UTR) was cotransfected with miR-381 expressing plasmid or the control vector into BGC823 cells. The cotransfection of miR-381 expression plasmid with ROCK2 Wt-3’UTR (but not ROCK1 Wt-3’UTR, data not shown) showed significant decreased luciferase activity compared with the control vector group, while ROCK2 Mut-3’UTR luciferase activity was not changed (Figure 3A, 3B). In addition, western blot analysis revealed that miR-381 mimic significantly reduced the expression of ROCK2, while miR-381 inhibitor increased ROCK2 expression in BGC823 cells (Figure 3C). We did not found that ROCK1 protein expression was significantly changed by miR-381 mimic or inhibitor treatment (data not shown). These results revealed that miR-381 directly inhibited ROCK2 expression.
Figure 3.
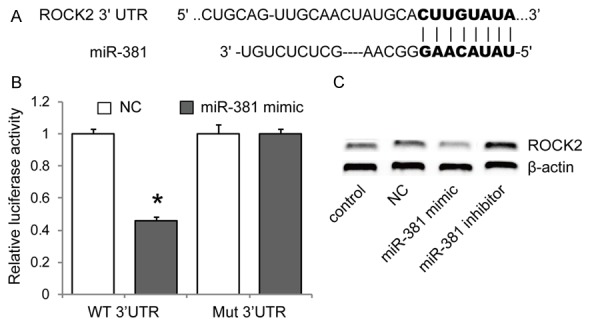
MiR-381 directly inhibits ROCK2 by targeting its 3’UTR. A. Predicted binding sequences of miR-381 in the 3’UTR of ROCK2. B. The ROCK2 3’UTR containing wild-type (WT) or mutant (Mut) 3’UTR of ROCK2 was subcloned into the luciferase reporter vector, then luciferase assay was performed in BGC823 cells cotransfected with miR-381 mimic and a luciferase reporter containing the wild-type (WT) or mutant (Mut) 3’UTR of ROCK2. Luciferase activity was determined 48 h after transfection. C. The expression levels of ROCK2 protein in BGC823 cells after 48 h transfection with miR-381 mimic, miR-381 inhibitor or scramble mimic (NC) were measured western blot. *P<0.05 versus the negative control (NC), data shown are means ± S.D.
Silencing of ROCK2 inhibits the proliferation, invasion and migration of BGC823 cells
To further validate that cell proliferation and migration is induced by ROCK2, siRNA knockdown of ROCK2 was performed and validated by western blot (Figure 4A). Similar to miR-381 restoration, silencing ROCK2 inhibited cell proliferation, invasion and migration ability (Figure 4B-D). To further validate whether miR-381 exerts its inhibitory effect through downregulation of ROCK2 in GC cells, we overexpressed ROCK2 in miR-381 mimic transfected BGC823 cells by transfecting ROCK2 expression plasmid. As expected, ROCK2 overexpression significantly attenuated the inhibitory effects of miR-381 mimics on cellular proliferation, invasion and migration of BGC823 cells (Figure 4F-H). Collectively, our results revealed that ROCK2 participates in cell growth and migration of GC cells. In addition, we examined the effect of ROCK2 on the expression levels of MMP2 and MMP9, which are well known important regulators of cellular invasion and migration. Western blot analysis revealed that silencing of ROCK2 significantly inhibited MMP2 and MMP9 expression while ROCK2 overexpression significantly increased MMP2 and MMP9 in BGC823 cells (Figure 5), suggesting that ROCK2 might promote cell invasion and migration, at least partially by upregulating MMP2 and MMP9 in GC cells.
Figure 4.
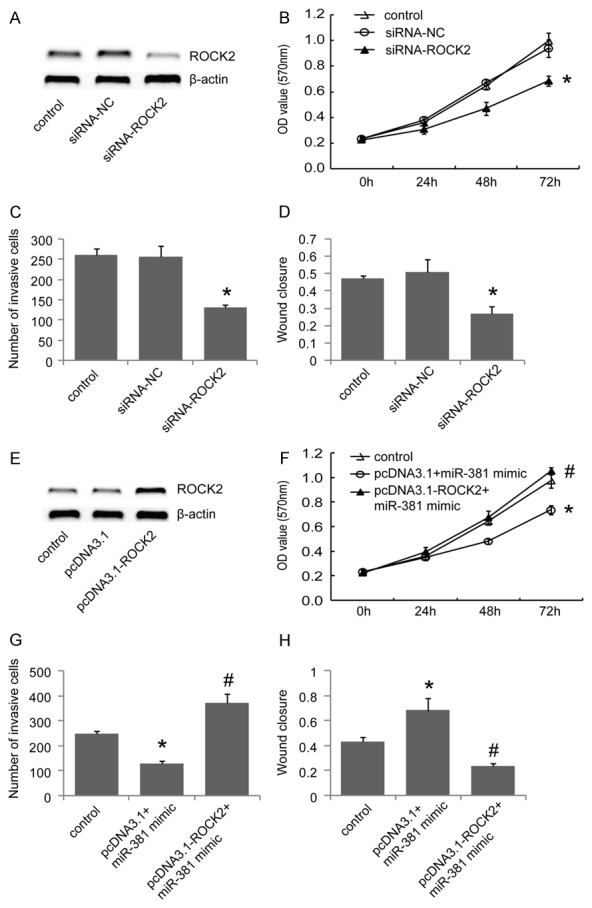
Silencing of ROCK2 inhibits the proliferation, invasion and migration of BGC823 cells. (A) BGC823 cells were transfected with ROCK2 specific siRNA (siRNA-ROCK2) or negative control siRNA (siRNA-NC). The knockdown of ROCK2 was validated by western blot. The effects of ROCK2 knockdown on cell proliferation, invasion and migration were determined using MTT assays (B), transwell assays with matrigel (C) and wound healing assay (D), respectively. (E) BGC823 cells were transfected with ROCK2 expression plasmids lacking its 3’UTR (pcDNA3.1-ROCK2) or empty pCDNA3.1 vector, and the expression of ROCK2 was examined by western blot. The effects of ROCK2 overexpression in miR-381 mimic transfected BGC823 cells on the proliferation, invasion and migration were assessed by MTT assays (F), transwell assays with matrigel (G) and wound healing assay (H), respectively. *P<0.05 versus the control, #P<0.05 versus the pcDNA3.1+miR-381 mimic group, data shown are means ± S.D.
Figure 5.
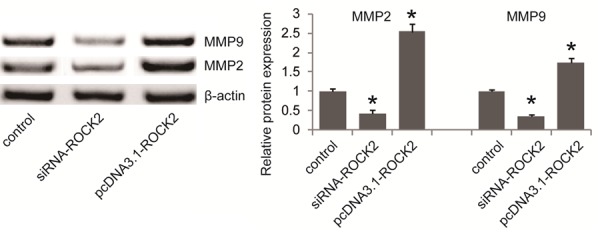
ROCK2 promoted MMP2 and MMP9 expression in BGC823 cells. Western blot was carried out to examine the effect of ROCK2 on the expression levels of MMP2 and MMP9 in BGC823 cells transfected with ROCK2 siRNA (siRNA-ROCK2) and ROCK2 expression plasmids (pcDNA3.1-ROCK2). *P<0.05 versus the control.
Discussion
Emerging evidence implied that miRNAs play crucial roles in the regulation of cellular functions and altered miRNA levels have been found in the initiation or development process of human cancers [10]. MiR-381 has been reported to be downregulated in several human cancers [12,14,15,19,20], but increased in glioma [16-18], suggesting that miR-381 may act as a tumor suppressor or oncogene in different types of cancers. In the present study, based on RTq-PCR analysis in 134 cases of primary GC and 23 cases of normal gastric mucosa, we found that miR-381 was frequently downregulated or lost in GC clinical samples. In addition, decreased miR-381 was correlated with GC tumor size, TNM stage, invasion depth and lymphatic metastasis (Table 1), suggesting that miR-381 may act as a tumor suppressor in GC cells.
To validate our hypothesis, we restored and inhibited miR-381 in BGC823 cells, respectively. MTT, transwell and wound healing assays demonstrated that miR-381 restoration inhibited BGC823 cell proliferation, invasion and migration ability, respectively, and inhibiting the endogenous expression of miR-381 in BGC823 cells resulted in opposite effects. These observations were consistent with previous findings that miR-381 has an inhibitory effect on cellular growth and migration in many types of cancers. For example, miR-381 mimics significantly inhibited cell proliferation, invasion and migration ability of HT29 and SW480 cells [12]. In hepatocellular carcinoma (HCC), miR-381 overexpression significantly inhibited HCC cell proliferation and colony formation and suppressed cell invasion [20]. Inconsistently, Tang et al reported that miR-381 increased the in vitro and in vivo proliferation of glioma cells, and inhibiting the endogenous expression of miR-381 decreased cell proliferation and tumor growth, suggesting that increased miR-381 is involved in the pathogenesis of glioma [16]. These data suggest that the roles of miR-381 might vary in different types of cancers. Here, we demonstrated that miR-381 functions as a tumor suppressor by inhibiting the proliferation and migration and invasion of GC cells.
Generally, a specific microRNA exerts its function during a certain cancer development through post-transcriptional regulating specific target genes [22,23]. Thus, a specific miRNA may function as a tumor suppressor by negatively regulating oncogenes, or an oncogene by downregulating tumor suppressor genes [11,24]. In the present study, we predicted and validated that ROCK2 was a novel direct target of miR-381 in GC cells. Luciferase reporter assay and western blot analysis revealed that miR-381 directly targeted and inhibited ROCK2 expression in BGC823 cells. ROCK2 plays a central role in the organization of the actin cytoskeleton [25-27] and is involved in a wide range of fundamental processes such as cell adhesion, proliferation, migration and apoptosis [28,29]. Emerging evidence revealed that ROCK2 level is elevated and involved in the growth and metastasis of many kinds of cancers including GC [30,31]. In the present study, we firstly conformed that targeting ROCK2 by siRNA inhibited the proliferation, invasion and migration ability of BGC823 cells. Matrix metalloproteinases (MMPs) are modulators of cancer invasion and metastasis in many kinds of cancers including GC [32,33]. We found that silencing of ROCK2 significantly inhibited MMP2 and MMP9 expression while ROCK2 overexpression significantly increased MMP2 and MMP9 in BGC823 cells. Our observation was consistent with another report that knockdown of ROCK2 significantly decreased MMP2 expression in hepatocellular carcinoma [34]. To further validate whether miR-381 exerts its inhibitory effect through targeting ROCK2 in GC cells, we rescued ROCK2 in BGC823 cells transfected with miR-381 mimics, and found ROCK2 overexpression attenuated the inhibitory effects of miR-381 on BGC823 cells. Collectively, our data suggest a functional miR-381/ROCK2 interaction in GC cells.
In summary, our current results demonstrated that miR-381 might act as a tumor suppressor in GC cells via inhibiting ROCK2. Therefore, restoration of miR-381 might provide a therapeutic strategy for GC treatment.
Disclosure of conflict of interest
None.
References
- 1.Duraes C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464:367–378. doi: 10.1007/s00428-013-1533-y. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 4.Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 7.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 8.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 9.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 10.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 12.He X, Wei Y, Wang Y, Liu L, Wang W, Li N. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther. 2016;9:1231–1239. doi: 10.2147/OTT.S99228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y, Zhao Q, Fan L, Zhang Z, Tan B, Liu Y, Li Y. Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomed Pharmacother. 2015;75:137–141. doi: 10.1016/j.biopha.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Xia B, Li H, Yang S, Liu T, Lou G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol. 2016;37:9157–9167. doi: 10.1007/s13277-016-4805-8. [DOI] [PubMed] [Google Scholar]
- 15.Ming J, Zhou Y, Du J, Fan S, Pan B, Wang Y, Fan L, Jiang J. miR-381 suppresses C/EBPalpha-dependent Cx43 expression in breast cancer cells. Biosci Rep. 2015;35 doi: 10.1042/BSR20150167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang H, Liu X, Wang Z, She X, Zeng X, Deng M, Liao Q, Guo X, Wang R, Li X, Zeng F, Wu M, Li G. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 2011;1390:21–32. doi: 10.1016/j.brainres.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Wang Z, Liu Q, Liu X, Wu M, Li G. Disturbing miR-182 and -381 inhibits BRD7 transcription and glioma growth by directly targeting LRRC4. PLoS One. 2014;9:e84146. doi: 10.1371/journal.pone.0084146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Yang J, Xu G, Wang W, Liu C, Yang H, Yu Z, Lei Q, Xiao L, Xiong J, Zeng L, Xiang J, Ma J, Li G, Wu M. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6:3147–3164. doi: 10.18632/oncotarget.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thorac Oncol. 2012;7:1069–1077. doi: 10.1097/JTO.0b013e31824fe976. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Zhao S, Pang X, Chi B. MicroRNA-381 suppresses cell growth and invasion by targeting the liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep. 2016;35:1831–1840. doi: 10.3892/or.2015.4491. [DOI] [PubMed] [Google Scholar]
- 21.Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 22.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain CK, Gupta A, Dogra N, Kumar VS, Wadhwa G, Sharma SK. MicroRNA therapeutics: the emerging anticancer strategies. Recent Pat Anticancer Drug Discov. 2014;9:286–296. doi: 10.2174/1574892809666140307101519. [DOI] [PubMed] [Google Scholar]
- 24.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 25.Newell-Litwa KA, Badoual M, Asmussen H, Patel H, Whitmore L, Horwitz AR. ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J Cell Biol. 2015;210:225–242. doi: 10.1083/jcb.201504046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schofield AV, Steel R, Bernard O. Rho-associated coiled-coil kinase (ROCK) protein controls microtubule dynamics in a novel signaling pathway that regulates cell migration. J Biol Chem. 2012;287:43620–43629. doi: 10.1074/jbc.M112.394965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J, Wu X, Surma M, Vemula S, Zhang L, Yang Y, Kapur R, Wei L. Distinct roles for ROCK1 and ROCK2 in the regulation of cell detachment. Cell Death Dis. 2013;4:e483. doi: 10.1038/cddis.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei L, Surma M, Shi S, Lambert-Cheatham N, Shi J. Novel insights into the roles of Rho Kinase in cancer. Arch Immunol Ther Exp (Warsz) 2016;64:259–278. doi: 10.1007/s00005-015-0382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinsenkamp I, Schulz S, Roscher M, Suhr AM, Meyer B, Munteanu B, Fuchser J, Schoenberg SO, Ebert MP, Wangler B, Hopf C, Burgermeister E. Inhibition of Rho-associated Kinase 1/2 Attenuates tumor growth in murine gastric cancer. Neoplasia. 2016;18:500–511. doi: 10.1016/j.neo.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia S, Jia Y, Weeks HP, Ruge F, Feng X, Ma R, Ji J, Ren J, Jiang WG. Down-regulation of WAVE2, WASP family verprolin-homologous protein 2, in gastric cancer indicates lymph node metastasis and cell migration. Anticancer Res. 2014;34:2185–2194. [PubMed] [Google Scholar]
- 32.Chen SX, Yin JF, Lin BC, Su HF, Zheng Z, Xie CY, Fei ZH. Upregulated expression of long noncoding RNA SNHG15 promotes cell proliferation and invasion through regulates MMP2/MMP9 in patients with GC. Tumour Biol. 2016;37:6801–6812. doi: 10.1007/s13277-015-4404-0. [DOI] [PubMed] [Google Scholar]
- 33.Wang HL, Zhou PY, Zhang Y, Liu P. Relationships between abnormal MMP2 expression and prognosis in gastric cancer: a meta-analysis of cohort studies. Cancer Biother Radiopharm. 2014;29:166–172. doi: 10.1089/cbr.2014.1608. [DOI] [PubMed] [Google Scholar]
- 34.Huang D, Du X, Yuan R, Chen L, Liu T, Wen C, Huang M, Li M, Hao L, Shao J. Rock2 promotes the invasion and metastasis of hepatocellular carcinoma by modifying MMP2 ubiquitination and degradation. Biochem Biophys Res Commun. 2014;453:49–56. doi: 10.1016/j.bbrc.2014.09.061. [DOI] [PubMed] [Google Scholar]