Abstract
Background: Long non-coding RNA (lncRNA) Hox transcript antisense intergenic RNA (HOTAIR) has been reported to play an important role in hepatocellular carcinoma (HCC) progression. Although there is evidence on HOTAIR being associated with HCC progression, the underlying mechanism remains to be further clarified. Methods: HOTAIR, miR-214-3p and FLOT1 expression were measured by quantitative real-time PCR or western blot. Cell proliferation, migration, and invasion were displayed by transwell assay, MTT, and colony formation assay. The relationship between HOTAIR, miR-214-3p, and FLOT1 was investigated by luciferase reporter assay. Tumor suppressive effect of HOTAIR was displayed by HepG2 xenografting to nude mice. Results: HOTAIR and FLOT1 expression were significantly up-regulated, whereas miR-214-3p was obviously down-regulated. Also, down-regulation of HOTAIR and FLOT1 as well as up-regulation of miR-214-3p inhibited cell proliferation, invasion, and migration. Intriguingly, the effects of miR-214-3p overexpression on HCC cells were rescued by high expression of HOTAIR. Otherwise, HOTAIR regulated FLOT1 expression through targeting miR-214-3p in HCC cells. Simultaneously, low HOTAIR expression inhibited FLOT1 expression by up-regulating miR-214-3p, which suppressed tumor growth in vivo. Conclusion: HOTAIR expression indirectly regulated FLOT1 expression through endogenous competition with miR-214-3p. HOTAIR/miR-214-3p/FLOT1 axis affected the cell proliferation, migration, and invasion of HCC.
Keywords: Hepatocellular carcinoma (HCC), lncRNA, HOTAIR, miR-214-3p, FLOT1
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world, of which the incidence has been growing in the past decades accompanied by high mortality and poor prognosis [1,2]. Although much work focused on the progression of HCC, the underlying molecular mechanism is still unclear.
Non-coding RNAs have been demonstrated to be involved in the development, progression, invasion, metastasis, and drug resistance to cancers [3-6]. Long non-coding RNAs (lncRNAs) are one class of ncRNAs with transcripts longer than 200 nucleotides [7]. Studies have found that lncRNAs play an indispensable role in cell growth, drug resistance and tumor stem cell formation of tumors [5,8,9]. Of note, lncRNA HOX transcript antisense RNA (HOTAIR) has been proved to act as an oncogene to affect the cell growth and the prognosis in HCC [10], and is a 2.2-kb nucleotide lncRNA located on chromosome 12 [11].
MicroRNAs (miRNAs) are a class of endogenous small ncRNAs with a length of ~22 nucleotides and target their downstream mRNAs for cleavage or translational repression [12]. Also, miRNAs have been indicated to matter much in the progression of cancers, such as breast cancer, non-small lung cancer, glioma cells, and HCC [3,4,13-16]. miR-214-3p influenced the proliferation and migration of glioma cells [15]. Moreover, miR-214-3p was significantly down-regulated in HCC [4]. In addition, the mechanism of HOTAIR binding to miR-214-3p has been suggested to associate with ovarian cancer cell progression [17]. However, whether HOTAIR could sponge miR-214-3p to influence cell growth and tumorigenesis of HCC has not been clarified.
Flotillin 1 (FLOT1), a member of the flotillin protein family, has been reported as a stable scaffold in recruitment of multiprotein complexes [18]. FLOT1 was discovered to participate in signal transduction as a signal molecule [19]. Recent studies have noted that FLOT1 was a key oncogene in various cellular processes in breast cancer, clear renal cell carcinoma, hepatocellular carcinoma, and non-small cell lung cancer [20-22]. However, hardly any investigations have reported whether HOTAIR, miR-214-3p, and FLOT1 cooperate to affect progression in HCC.
In this study, we found that high expression of HOTAIR was detected in HCC patients and highly related with poor prognosis. Knockdown of HOTAIR in Hep3B and HepG2 could impair the proliferation, migration, and invasion. As a negatively regulated target of HOTAIR, ectopic expression of miR-214-3p can reverse the effects induced by HOTAIR overexpression. Computed analysis and luciferase assay revealed that FTOL1 is a downstream target of miR-214-3p. Meanwhile, FTOL1 knockdown may affect the proliferation, migration, and invasion of Hep3B and HepG2. In conclusion, HOTAIR/miR-214-3p/FTOL1 act as an axis in HCC, and this new pathway may provide a new strategy to treat HCC.
Materials and methods
Tissues samples and cell culture
HCC tissues and matched tumor-adjacent tissues were obtained from 35 patients at Second Hospital of Shanxi Medical University. No patient underwent chemotherapy or radiation therapy. This study has acquired informed consents from the guardians of all patients, and it has been approved by Experiments committee of Second Hospital of Shanxi Medical University.
HCC cell lines (Hep3B and HepG2) and normal hepatic cell line (L-02) were purchased from RiboBio Co. (Guangzhou, China). These cells were cultured in RPMI-1640 medium (GIBCO, Grand Island, NE, USA) with 10% fetal bovine serum (Thermo, Waltham, MA, USA), L-glutamine (Thermo), 1% antibiotics (TargetMol, Boston, MA, USA) in an atmosphere containing 5% CO2 at 37°C without Mycoplasma contamination.
Transient transfection
Small interfering RNA (siRNA) of siNC, siHOTAIR, and siFLOT1 as well as pcDNA3.1 of pcDNA-NC and pcDNA-HOTAIR were synthesized by GenePharma (Shanghai, China). miR-NC, miR-214-3p, inhibitor-NC and miR-214-3p inhibitor were purchased from GenePharma. Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) was used in the transfection assay.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissues and cells by using Trizol reagent (Invitrogen). The NanoDropND-1000 spectrophotometer was used to detect RNA concentration. M-MLV Reverse Transcriptase (Invitrogen) and SYBR® Green (Promega, Madison, WI, USA) were used to determine the expression of mRNA or lncRNA with Tubulin as an internal control. The TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and SYBR® Green were used to determine the expression of miR-214-3p with U6 as an endogenous control. The 2-ΔΔCt method was used to calculate the relative expression of mRNA, miRNA, and lncRNA. The primer of HOTAIR forward, 5’-AAATATGGCGGCGTCTACACGGA-3’ and reverse 5’-TCCAGAACCCTCTGACATTTGCCT-3’; FLOT1 forward, 5’-CCCATCTCAGTCACTGGCATT-3’ and reverse 5’-CCGCCAACATCTCCTTGTTC-3’; miR-214-3p forward, 5’-GAGTGTTGGCCTGTCCTCAA-3’ and reverse 5’-TTGTGCCCAGTTGCCTGTAT-3’; Tubulin forward, 5’-GATGACCATTTCTTGCTTC-3’ and reverse 5’-GTTCTGACATTTGCTACCG-3’; U6 Forward, 5’-CTCGCTTCGGCAGCACA-3’ and reverse 5’-AACGCTTCACGAATTTGCGT-3’.
Western blot
Total proteins were extracted from HCC tissues and cells with PIPA buffer (Pierce, Rockford, IL, US). Total proteins (20 µg) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% dried skimmed milk in TBS for 1 h at room temperature and then were incubated with specific primary antibodies against FLOT1 (1:2000, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or Tubulin (1:2000 dilution, Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight. After washing three times in TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (anti-rabbit IgG, 1:2000 dilution) for 2 h at room temperature. The blots were detected using ECL detection kit (Thermo) and ChemiDoc XRS System (Bio-Rad, Hercules, CA, USA).
Cell migration and invasion assays
Cells with a density of 2 × 104/well containing serum-free medium were seeded into upper chamber for migration assays (8-μm pore size, Millipore) and invasion assays with Matrigel (Sigma, St. Louis, MO, USA). The transwell plates were placed in an atmosphere with 5% CO2 at 37°C for 24 h. Then the migrated cells on the lower surface of the membranes were stained with methanol and crystal violet. The number of migrated and invasive cells was observed and calculated with a microscope (Leica, Wetzlar, Germany).
3-(4, 5-Dimethyl-2-thiazolyl)-2, 5-diphenyl-2Htetrazolium bromide (MTT) assay
Cells were inoculated in 96-well cell culture plates (Corning Inc., Corning, NY, USA) at a density of 2 × 103 cells/well for 48 h. Then 20 μL MTT (5 g/L) were added into each well, followed by incubation for 4 h at 37°C. The dimethyl sulfoxide (DMSO, Sigma) was added to dissolve intracellular formazan crystals after discarding supernatant (150 µL/well). Cell viability was determined by using a spectrophotometric microplate reader (Beyotime Institute of Biotechnology, Haimen, China) at a wavelength of 450 nm.
Luciferase assay
The binding sequences of HOTAIR with miR-214-3p were predicted by starBase (http://starbase.sysu.edu.cn/mirLncRNA.php) and the binding sequences of 3’UTR of FLOT1 with miR-214-3p were predicted by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php). In order to probe whether HOTAIR and FLOT1 are bound to miR-124-3p, we constructed the psiCHECK2 vector (Promega) of wild-type of HOTAIR (HOTAIR WT), mutated type of HOTAIR (HOTAIR MUT), FLOT1 WT and FLOT1 MUT by amplifying and cloning into the luciferase reporter vectors. These vectors were co-transfected with miR-124-3p or miR-NC into Hep3B and HepG2 cells by using Lipofectamine 3000. After 48 h post-transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Colony formation assay
The HCC cell suspension with transfection of siNC, siHOTAIR, miR-NC, miR-214-3p, inhibitor-NC, miR-214-3p inhibitor, pcDNA-NC, pcDNA-HOTAIR or siFLOT1 was inoculated into 6-well plates, and then cultured for 2 weeks in an environment of 37°C, 5% CO2 and saturated humidity. The colonies were washed with PBS, then stained with crystal violet and counted.
Tumor xenografts in vivo
The experiments were approved by the animal care and Experiments committee of Second Hospital of Shanxi Medical University. The nude mice were purchased from the Model Animal Research Center of Nanjing University. 2 × 106 HepG2 cells transfected with siHOTAIR or siNC was subcutaneously injected into nude mice. At 35 days after cell implantation, mice were sacrificed for further analysis.
Statistical analysis
All data were analyzed using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA). Data were shown as the mean ± standard deviation (SD) with triplicates. The survival curve was analyzed and evaluated by Kaplan-Meier analysis. Student’s t-test and Pearson’s correlation analysis was used. Statistical significance was established at p value < 0.05.
Results
HOTAIR was up-regulated in HCC tissues and cells, and related to survival
To verify whether HOTAIR is required for HCC progression, the expression pattern of HOTAIR was investigated in tissues from 35 patients. As shown in Figure 1A, the expression of HOTAIR was much more in tumor tissues than in juxtacancerous tissue. Also, we compared the survival of patients with high-expressed (n = 22) and low-expressed (n = 13) HOTAIR, and low HOTAIR expression showed significant correlation with poor prognosis (Figure 1B). Then we double check the HOTAIR expression in two HCC cell lines, Hep3B and HepG2. As in HCC patients, HOTAIR also was expressed more strongly in HCC cell lines than in the liver normal cell line (L-02) (Figure 1C). The results implied that the HOTAIR might be involved in tumorigenesis and prognostic in HCC.
Figure 1.
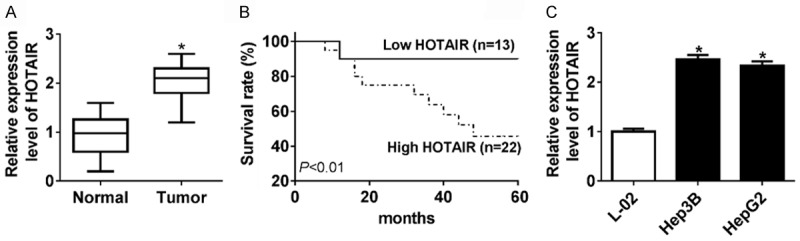
HOTAIR was up-regulated in HCC tissues and cells, and related to survival. A. The expression of HOTAIR was significantly increased in HCC tissues and their adjacent normal tissues by qRT-PCR. B. Correlation between HOTAIR expression and survival of glioma patients by Kaplan-Meier analysis. The survival rates of high expression of HOTAIR were significantly different from low expression of HOTAIR. C. The expression of HOTAIR was evidently enhanced in Hep3B, HepG2 cells and L-02. *P < 0.05.
Knockdown of HOTAIR inhibited proliferation, migration, and invasion in HCC
HOTAIR expression was significantly decreased after transfection of siHOTAIR (Figure 2A). Moreover, cell viability of Hep3B and HepG2 were conspicuously decreased in siHOTAIR group compared with siNC group using MTT assay (Figure 2C and 2D). Meanwhile, the result of cell colony formation assay indicated that low expression of HOTAIR significantly inhibited proliferation in Hep3B and HepG2 cell lines (Figure 2B-D). Otherwise, transwell experiments indicated that knockdown of HOTAIR could significantly decrease cell migration ability and invasion ability of Hep3B and HepG2 cells (Figure 2E and 2F). In summary, these data hinted that knockdown of HOTAIR reduced the capacity of proliferation, migration, and invasion.
Figure 2.
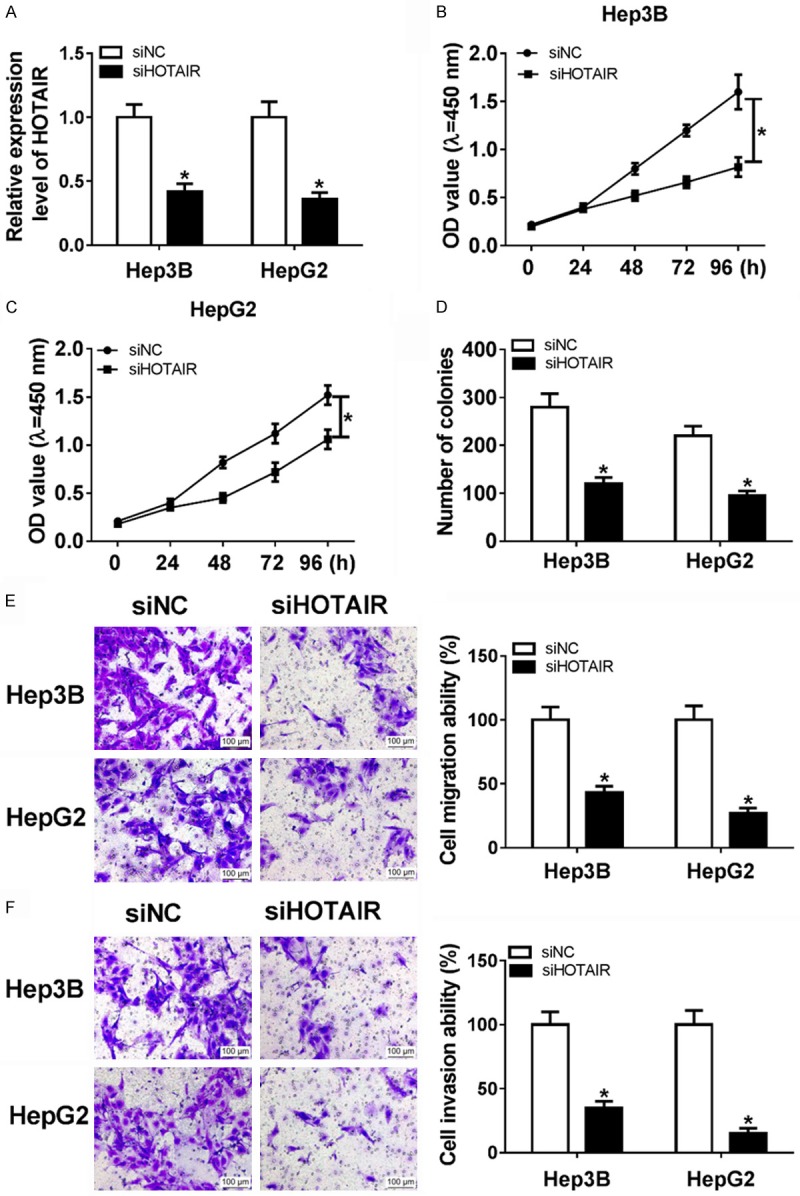
Knockdown of HOTAIR inhibited proliferation, metastasis, and invasion. (A) HOTAIR expression was significantly down-regulated in Hep3B and HepG2 with transfection of siHOTAIR. (B and C) MTT assay was performed to detected cell viability of Hep3B and HepG2 by transfection of siHOTAIR and siNC. (D) Colony formation assay was performed to count the cell proliferation by transfection of siHOTAIR and siNC. (E and F) Cell migration (E) and invasion (F) ability were detected in Hep3B and HepG2 with transfection of siHOTAIR and siNC. *P < 0.05.
miR-214-3p is a target of HOTAIR in HCC
Previous studies reveal that long non-coding RNA as competing endogenous RNAs may play biological functions by competitively binding their target miRNAs [23,24], so to better understand the relationship between HOTAIR and miR-214-3p in HCC, we predicted that microRNA-214-3p was a candidate microRNA target of HOTAIR using starBase. To test the hypothesis, we checked the miRNA-214-3p expression in patients (Figure 3B) and Hep3B and HepG2 cell lines (Figure 3C) and the microRNA expression was indeed down-regulated in HCC. Then, miR-214-3p inhibitor, miR-NC, miR-214-3p and miR-NC were transfected into Hep3B and HepG2 cells. miR-214-3p expression was down-regulated in the miR-214-3p inhibitor group and up-regulated in miR-214-3p (Figure 3D), which could apply to the following tests.
Figure 3.
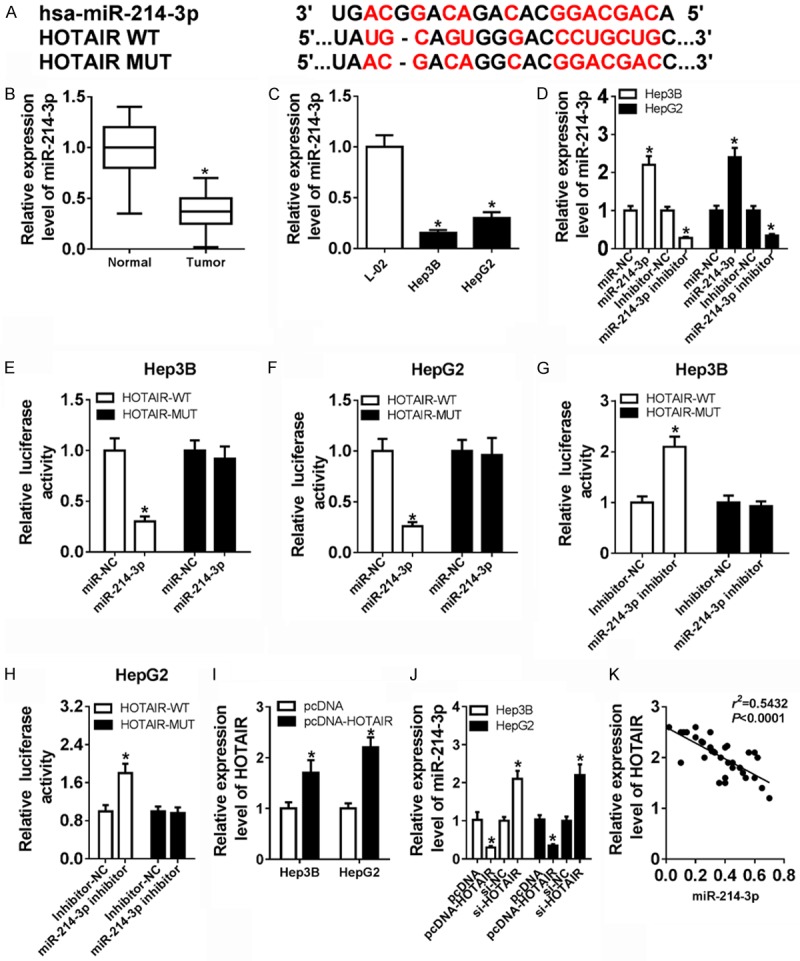
miR-214-3p is a target of HOTAIR in HCC (A) The binding sequence of the HOTAIR 3’UTR with miR-214-3p. (B) The expression of miR-214-3p was significantly decreased in HCC tissues and their adjacent normal tissues by qRT-PCR. (C) The expression of miR-214-3p was evidently enhanced in Hep3B, HepG2 cells. (D) miR-214-3p expression was significantly increased in miR-214-3p group compared with miR-NC group and obviously decreased in miR-214-3p inhibitor group compared with inhibitor-NC. (E and F) The luciferase activities were measured in Hep3B and HepG2 co-transfected with a luciferase reporter containing miR-214-3p mimics, HOTAIR WT and HOTAIR MUT. (G and H) The luciferase activities were measured in Hep3B and HepG2 co-transfected with a luciferase reporter containing miR-214-3p inhibitor, HOTAIR WT, and HOTAIR MUT. (I) HOTAIR expression was markedly up-regulated in Hep3B and HepG2 with transfection of pcDNA-HOTAIR. (J) The expression level of miR-214-3p after transfection of pcDNA, pcDNA-HOTAIR, siNC and siHOTAIR in Hep3B and HepG2. (K) miR-214-3p expression was negatively correlated with the expression of HOTAIR. *P < 0.05.
Next, we constructed wild-type HOTAIR plasmid and a mutant HOTAIR named HOTAIR WT and HOTAIR MUT (Figure 3A) and used the luciferase assay to verify whether lncRNA HOTAIR sponged miR-214-3p. Then, HOTAIR WT and HOTAIR MUT were co-transfected with miR-NC, miR-214-3p, inhibitor-NC or miR-214-3p inhibitor into Hep3B and HepG2, respectively. As expected, the luciferase activity significantly decreased due to the combination miR-214-3p and HOTAIR WT, rather than with HOTAIR MUT in Hep3B and HepG2 (Figure 3E and 3F). However, the luciferase activity was remarkably enhanced due to the combination of miR-214-3p-inhibitor and HOTAIR WT, rather than with HOTAIR MUT in Hep3B and HepG2 (Figure 3G and 3H).
To further explain the relationship between miR-214-3p and HOTAIR, we also constructed the plasmid of pcDNA and pcDNA-HOTAIR. The data showed that HOTAIR expression was remarkably up-regulated in pcDNA-HOTAIR group (Figure 3I). Then, we used qRT-PCR to measure the expression of miR-214-3p in pcDNA, pcDNA-HOTAIR, siNC and siHOTAIR groups. High HOTAIR expression could substantially reduce miR-214-3p expression, while low HOTAIR expression drastically induced miR-214-3p expression in Hep3B and HepG2 (Figure 3J). On the other hand, HOTAIR and miR-214-3p expression showed a dramatic negative correlation in HCC tissues by Pearson’s correlation analysis (Figure 3K). Thus, the results indicated that miR-214-3p was a target of HOTAIR, which was suppressed by HOTAIR expression in HCC.
Overexpression of HOTAIR reversed the effects of miR-214-3p in HCC
To further explore the role of HOTAIR in miR-214-3p-mediated cell growth, overexpression of miR-214-3p was co-transfected with pcDNA-HOTAIR and pcDNA into Hep3B and HepG2, respectively. The MTT assay result displayed that cell viability was sharply lower in miR-214-3p group than miR-NC group (Figure 4A and 4B). Furthermore, overexpression of miR-214-3p remarkably decreased proliferation capacity in Hep3B and HepG2 (Figure 4C). As shown in Figure 4D, 4E, cell migration and invasion ability of HCC were evidently attenuated in miR-214-3p group compared with miR-NC group. Interestingly, cell viability and proliferation capacity were significantly increased in miR-214-3p+pcDNA-HOTAIR group compared with miR-214-3p+pcDNA group (Figure 4A-C). Similar changes were displayed in the ability of cells for migration and invasion (Figure 4D and 4E). Up-regulated HOTAIR could enhance cell growth in miR-214-3p+pcDNA group. From these data, the suppressive effects of miR-214-3p were rescued by high expression of HOTAIR on proliferation, migration, and invasion of HCC cells.
Figure 4.
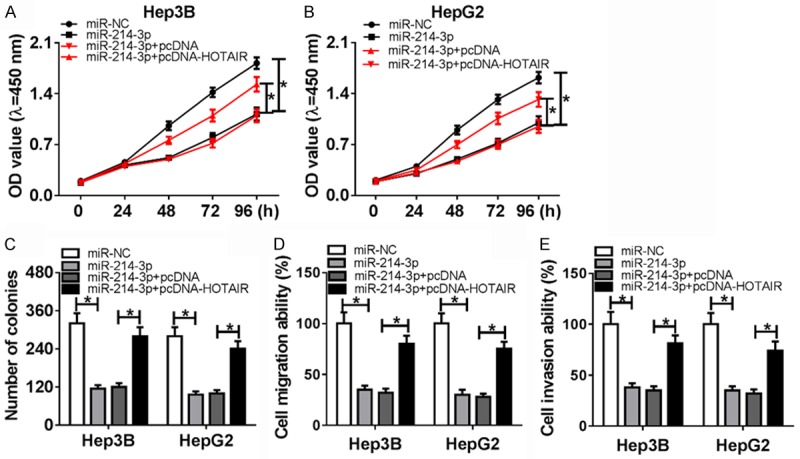
Overexpression of HOTAIR reverses the effects of miR-214-3p in HCC. (A and B) MTT assay was performed to detect cell viability in Hep3B, and HepG2 by transfection of miR-NC, miR-214-3p, miR-214-3p+pcDNA and miR-214-3p+pcDNA-HOTAIR. (C) Colony formation assay was performed to count theproliferation by transfection of miR-NC, miR-214-3p, miR-214-3p+pcDNA, and miR-214-3p+pcDNA-HOTAIR. (D and E) Migration (D) and invasion (E) ability were detected in Hep3B and HepG2 with transfection of miR-NC, miR-214-3p, miR-214-3p+pcDNA and miR-214-3p+pcDNA-HOTAIR. *P < 0.05.
FLOT1 is a downstream target of miR-214-3p in HCC
miRNA regulates its downstream target genes [12]. FLOT1 was predicted by miRTarBase as a potential target gene of miR-214-3p (Figure 5A). First, qRT-PCR and western blot were used to detect FLOT1 expression in HCC tissues and cells and FLOT1 expression was indeed up-regulated (Figure 5B-D). Next, luciferase assay showed that miR-214-3p significantly reduced luciferase activity by binding to the FLOT1 WT 3’UTR, not FLOT1 MUT 3’UTR in Hep3B and HepG2 cells. In comparison, down-regulated miR-214-3p expression obviously enhanced the luciferase activity with binding to FLOT1 WT 3’UTR (Figure 5E-H). Otherwise, FLOT1 expression was obviously decreased in Hep3B and HepG2 cells with transfection of miR-214-3p compared with miR-NC, whereas loss of miR-214-3p expression could strongly reinforce FLOT1 expression in Hep3B and HepG2 cells (Figure 5J). Based on qRT-PCR data with tissues, miR-214-3p expression was negatively correlated with the expression of FLOT1 via Pearson’s analysis (Figure 5I). Hence, these results demonstrated that FLOT1 was a target gene of miR-214-3p and miR-214-3p might affect HCC progression by regulating FLOT1.
Figure 5.
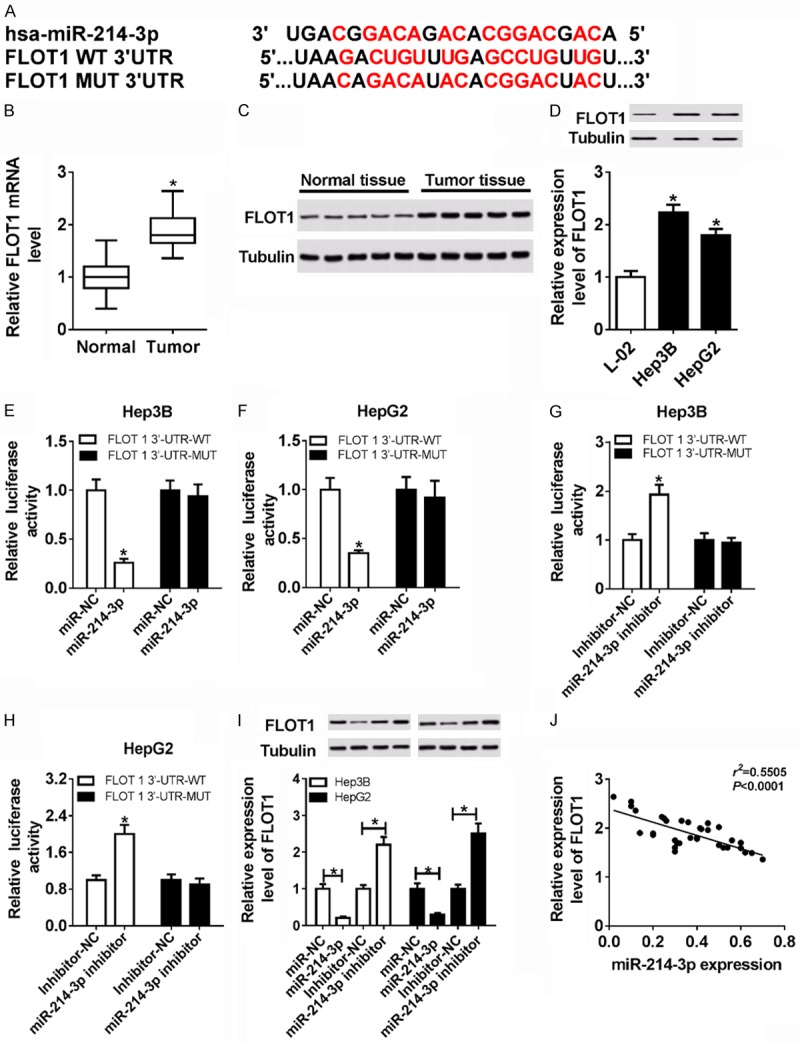
FLOT1 was a downstream target of miR-214-3p in HCC. (A) The binding sequence of the FLOT1 3’UTR with miR-214-3p. (B and C) The expression of FLOT1 was significantly increased in HCC tissues and their adjacent normal tissues by qRT-PCR (B) and western blot (C). (D) The expression of FLOT1was evidently enhanced in Hep3B, HepG2 cells and L-02 via western blot. (E and F) The luciferase activities were measured in Hep3B and HepG2 co-transfected with a luciferase reporter containing miR-214-3p mimics, FLOT1 WT and FLOT1 MUT. (G and H) The luciferase activities were measured in Hep3B and HepG2 co-transfected with a luciferase reporter containing miR-214-3p inhibitor, FLOT1 WT, and FLOT1 MUT. (I) The expression of FLOT1 in miR-NC, miR-214-3p, inhibitor-NC and miR-214-3p inhibitor in Hep3B and HepG2. (J) miR-214-3p expression was negatively correlated with the expression of FLOT1. *P < 0.05.
HOTAIR regulated FLOT1 positively by targeting miR-214-3p
To further investigate the function of FLOT1 in HCC, siFLOT1 was transfected into Hep3B and HepG2 cells. FLOT1 expression was effectively declined at the transcriptional and translational level in siFLOT1 cells by qRT-PCR and western blot (Figure 6A and 6B). As shown in Figure 6C-E, cell proliferative capacity was remarkably decreased in siFLOT group compared with siNC group. Additionally, cell migration and invasive ability was significantly weakened in siFLOT1 group (Figure 6F and 6G). Moreover, to determine the relationship of HOTAIR, miR-214-3p and FLOT1, FLOT1 expression was measured in HCC cells with transfection of siNC, siHOTAIR and siHOTAIR+miR-214-3p inhibitor. The results indicated that FLOT1 expression was drastically decreased in siHOTAIR group, while miR-214-3p inhibitor restored FLOT1 expression with reduction in HCC cells with transfection of siHOTAIR (Figure 6H) Correlation analysis displayed that HOTAIR expression was positively correlated with the expression of FLOT1 (Figure 6I). Accordingly, these data demonstrated that knockdown of FLOT1 inhibited the cell growth in HCC, and HOTAIR regulated FLOT1 expression through sponging miR-214-3p in HCC.
Figure 6.
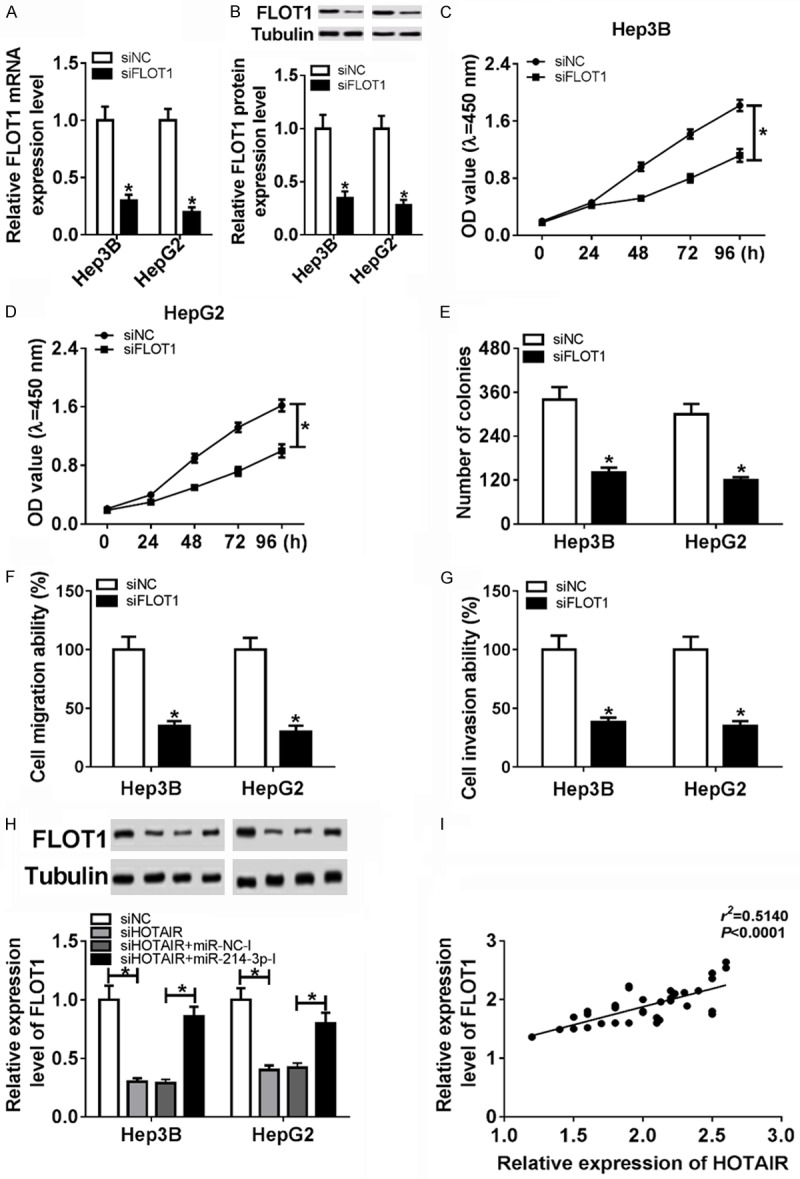
HOTAIR regulated FLOT1 positively by targeting miR-214-3p. (A) The expression of FLOT was decreased significantly in Hep3B and HepG2 with transfection of siFLOT1. (B) The protein level of FLOT was reduced significantly in Hep3B and HepG2 with transfection of siFLOT1. (C and D) MTT assay was performed to detect cell viability of Hep3B and HepG2 by transfection of siNC and siFLOT1. (E) Colony formation assay was performed to count the proliferation by transfection of siNC and siFLOT1. (F and G) Migration (F) and invasion (G) ability were detected in Hep3B and HepG2 with transfection of siNC and siFLOT1. (H) The expression of FLOT1 in siHOTAIR, siNC, siHOTAIR+miR-NC-inhibitor and siHOTAIR+miR-214-3p-inhibitor. (I) HOTAIR expression was negatively correlated with the expression of FLOT1. *P < 0.05.
Knockdown of HOTAIR inhibited tumor growth of HCC in vivo
To evaluate the function of HOTAIR on tumorigenesis, HepG2 cells with transfection of siHOTAIR or siNC were subcutaneously injected into nude mice. Tumor volume in siHOTAIR group was smaller than that in siNC group (Figure 7A). Furthermore, as shown in Figure 7B, down-regulation of HOTAIR significantly decreased the tumor weights. The results showed that low expression of HOTAIR suppressed the tumor growth in vivo. In addition, the HOTAIR (Figure 7C) and FLOT1 expression (Figure 7E, 7F) were obviously diminished and the miR-214-3p expression (Figure 7D) was sharply up-regulated in siHOTAIR compared with siNC via qRT-PCR or western blot. Therefore, knockdown of HOTAIR suppressed tumorigenesis by up-regulating miR-214-3p and down-regulating FLOT1 in HCC.
Figure 7.
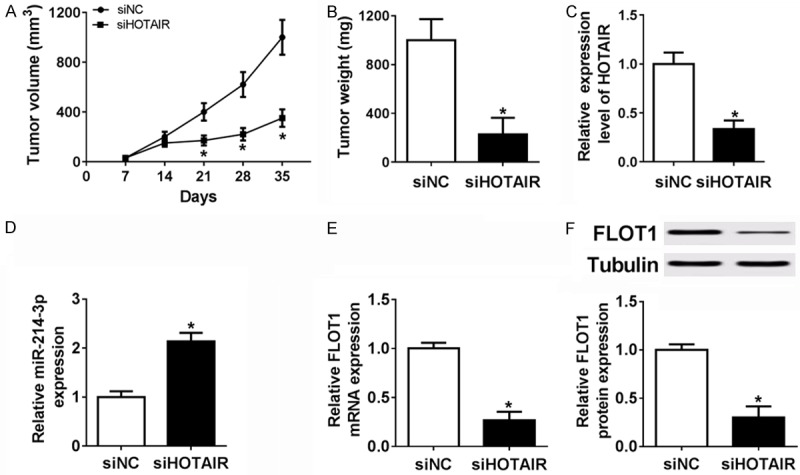
Knockdown of HOTAIR inhibited tumor growth in vivo. A and B. Tumor volume and weight were reduced significantly in siHOTAIR cells that were injected into nude mice. C-F. HOTAIR, miR-214-3p and FLOT1 mRNA or protein level were detected in siHOTAIR cells that were injected into nude mice. *P < 0.05.
Discussion
HCC is the third leading cause of cancer mortality worldwide [25]. With time, more and more investigators showed great promise in HCC [26-28]. Recently, many studies have revealed that lncRNAs plays an essential role in tumor development, including proliferation, invasion, and migration [29-31]. For example, HOTAIR could enhance epithelial-mesenchymal transition to promote the malignancy of HCC by miR-23b from ZEB1 [30]. Xu et al. reported MT1JP inhibited cell proliferation and migration by MT1JP/miR-214-3p/RUNX3 axis [31]. Therefore, lncRNAs act as ceRNAs to interact with miRNAs to regulate target gene expression and provide new drivers of HCC treatment.
HOTAIR has been declared to be up-regulated and participates in various cancers’ progression as a tumor biomarker. Liu et al. noted that HOTAIR promoted gastric cancer cell migration and invasion by sponging miR-331-3p to regulate HER2 expression [24]. Yang et al. pointed out that HOTAIR modulated retinoblastoma cells progression and epithelial-mesenchymal transition by the miR-613/c-met axis [32]. Moreover, overexpression of HOTAIR also influenced cell progression and autophagy of HCC by increasing ATG3 and ATG7 expression [33]. In the present study, HOTAIR expression was up-regulated in HCC tissues and cells lines, which was consistent with the previous study [10]. Knockdown of HOTAIR may suppress Hep3B and HepG2 proliferation and migration and be relevant to prognosis of patients. Consequently, knockdown of HOTAIR can be invoked as a target for the treatment of HCC.
miRNAs serve as important regulatory factors, which affects HCC progression. Ectopic expression of miR-19a-3p and miR-494-3p induced HCC cell metastasis and chemoresistance by regulating PTEN expression [3,34]. Liu et al. indicated that overexpression of miR-141 suppressed HCC migration and invasion by down-regulating Tiam1 [35]. Numerous studies have found that miR-214-3p not only affected cell proliferation, but also was associated with prognosis in cancers, such as HCC, gastric cancer, ulcerative colitis, and ovarian cancer [4,31,36,37]. We found that as a target of HOTAIR, miR-214-3p expression was lower in HCC tissues and cell lines than normal tissues and cells. Also, overexpression of miR-214-3p inhibited cell proliferation, migration, and invasion of HCC. High expression of HOTAIR significantly reversed the suppressive effects of overexpressed miR-214-3p on cell growth in HCC. As shown above, the interaction between HOTAIR and miR-214-3p affects cell progression in HCC cells.
On the basis that miRNA is involved in cellular processes by regulating target mRNA, we identified that FLOT1 was a potential target of miR-214-3p through biological analysis and luciferase assay. In previous studies, FLOT1 was also associated with cellular processes and prognosis in various cancers [20-22,38]. Zhang et al. noted that highly expression of FLOT1 was associated with cell progression and poor prognosis in HCC [20]. SA study by Lin et al. reported that low expression of FLOT1 inhibited cell proliferation and tumorigenicity in breast cancer by up-regulating FOXO3 [21]. Our study demonstrated that FLOT1 is a target of miR-214-3p, and functional analysis showed that knocking down FLOT1 showed less proliferation, migration, and invasion in HCC.
The regulatory mechanism of competing for endogenous RNAs (ceRNA) is that lncRNA competitively sponged miRNA to regulate the expression of target genes [21], which introduces new ideas and methods for the treatment of cancer. For example, Shan et al. reported that lncRNA SNHG7/miR-216b/GALNT1 axis promoted proliferation and liver metastasis of colorectal cancer [5]. Song et al. indicated that CLDN4 promoted proliferation and metastasis in vivo, which could be suppressed by miR-596 and miR-3620-3p and enhanced by lncRNA-KRTAP5-AS1 and TUBB2A [23]. Meanwhile, HOXA11-AS/miR-214-3p/E2H2 axis has been reported to regulate glioma cell migration and invasion [16].
Based on the above results, we speculate that HOTAIR might regulate FLOT1 expression by competing with endogenous miR-214-3p. Subsequent investigations suggested that HOTAIR expression was significantly positively correlated with FLOT1, and obviously negatively correlated with miR-214-3p. Also, miR-214-3p was targeted to HOTAIR, and FLOT1 was a target of miR-214-3p. Of note, up-regulated HOTAIR expression recruited the effects of miR-214-3p on cell growth in HCC. On the other hand, low expression of HOTAIR inhibited tumor growth by up-regulating miR-214-3p to down-regulate FLOT1 in mouse xenograft tumor. Our results indicated that HOTAIR as a ceRNA sponged miR-214-3p to regulate FLOT, which affects cell proliferation, migration, and invasion in HCC.
Conclusions
In this study, we found a novel regulatory network: HOTAIR/mir-214-3p/FLOT1 axis which is involved in HCC cell proliferation, migration, and invasion, providing a new target for accurate treatment of HCC.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T, Aikata H, Kobayashi T, Ohdan H, Arihiro K, Chayama K. Patients with early recurrence of hepatocellular carcinoma have poor prognosis. Hepatobiliary Pancreat Dis Int. 2017;16:279–288. doi: 10.1016/s1499-3872(16)60181-9. [DOI] [PubMed] [Google Scholar]
- 3.Jiang XM, Yu XN, Liu TT, Zhu HR, Shi X, Bilegsaikhan E, Guo HY, Song GQ, Weng SQ, Huang XX, Dong L, Janssen HLA, Shen XZ, Zhu JM. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed Pharmacother. 2018;105:1147–1154. doi: 10.1016/j.biopha.2018.06.097. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Chen Y, Xie Q, Dong N, Gao Y, Deng H, Lu C, Wang S. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int. 2017;17:102. doi: 10.1186/s12935-017-0471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9:722. doi: 10.1038/s41419-018-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu LL, Cai WP, Lei X, Shi KQ, Lin XY, Shi L. NRAL mediates cisplatin resistance in hepatocellular carcinoma via miR-340-5p/Nrf2 axis. J Cell Commun Signal. 2018;136:E359–E386. doi: 10.1007/s12079-018-0479-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, Xu RH. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han L, Zhang HC, Li L, Li CX, Di X, Qu X. Downregulation of long noncoding rna hotair and ezh2 induces apoptosis and inhibits proliferation, invasion, and migration of human breast cancer cells. Cancer Biother Radiopharm. 2018;33:241–251. doi: 10.1089/cbr.2017.2432. [DOI] [PubMed] [Google Scholar]
- 10.Wang LP, Wang JP, Wang XP. HOTAIR contributes to the growth of liver cancer via targeting miR-217. Oncol Lett. 2018;15:7963–7972. doi: 10.3892/ol.2018.8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Guo Z, Li J, Sun J, Sun L, Zhou Y, Yu Z. MiR-346 promotes HCC progression by suppressing Breast cancer metastasis suppressor 1 expression. Oncol Res. 2018;26:1073–1081. doi: 10.3727/096504017X15145088802439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei L, Huang Y, Gong W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30:2897–2902. doi: 10.3892/or.2013.2755. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–1513. doi: 10.1016/j.biopha.2017.08.097. [DOI] [PubMed] [Google Scholar]
- 16.Zhan M, He K, Xiao J, Liu F, Wang H, Xia Z, Duan X, Huang R, Li Y, He X. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13633. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yiwei T, Hua H, Hui G, Mao M, Xiang L. HOTAIR Interacting with MAPK1 regulates ovarian cancer skov3 cell proliferation, migration, and invasion. Med Sci Monit. 2015;21:1856–1863. doi: 10.12659/MSM.893528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langhorst MF, Reuter A, Stuermer CA. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riento K, Frick M, Schafer I, Nichols BJ. Endocytosis of flotillin-1 and flotillin-2 is regulated by fyn kinase. J Cell Sci. 2009;122:912–918. doi: 10.1242/jcs.039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang SH, Wang CJ, Shi L, Li XH, Zhou J, Song LB, Liao WT. High expression of FLOT1 is associated with progression and poor prognosis in hepatocellular carcinoma. PLoS One. 2013;8:e64709. doi: 10.1371/journal.pone.0064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C, Wu Z, Lin X, Yu C, Shi T, Zeng Y, Wang X, Li J, Song L. Knockdown of FLOT1 impairs cell proliferation and tumorigenicity in breast cancer through upregulation of FOXO3a. Clin Cancer Res. 2011;17:3089–3099. doi: 10.1158/1078-0432.CCR-10-3068. [DOI] [PubMed] [Google Scholar]
- 22.Yang FQ, Zhang HM, Chen SJ, Yan Y, Zheng JH. MiR-506 is down-regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT1. PLoS One. 2015;10:e0120258. doi: 10.1371/journal.pone.0120258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, Chen XW, Gao P, Miao ZF, Wang ZN. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527–539. doi: 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, LV SW, Sun S, LI YJ. Studies on molecular mechanism of oleanolic acid on HCC cells. Journal of Harbin University of Commerce. 2014 [Google Scholar]
- 27.Brechot C, Kremsdorf D, Soussan P, Pineau P, Dejean A, Paterlinibrechot P, Tiollais P. Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC): molecular mechanisms and novel paradigms. Pathol Biol. 2010;58:278–287. doi: 10.1016/j.patbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2018 doi: 10.1136/gutjnl-2017-315084. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang X, Cai J. LncRNAs and their role in cancer stem cells. Oncotarget. 2007;8:11065–110692. doi: 10.18632/oncotarget.22161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang T, He X, Chen A, Tan K, Du X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670:114–122. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Zhang G, Zou C, Zhang H, Gong Z, Wang W, Ma G, Jiang P, Zhang W. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46:2445–2459. doi: 10.1159/000489651. [DOI] [PubMed] [Google Scholar]
- 32.Yang G, Fu Y, Lu X, Wang M, Dong H, Li Q. LncRNA HOTAIR/miR-613/c-met axis modulated epithelial-mesenchymal transition of retinoblastoma cells. J Cell Mol Med. 2018;22:5083–5096. doi: 10.1111/jcmm.13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605–2612. doi: 10.1039/c6mb00114a. [DOI] [PubMed] [Google Scholar]
- 34.Lin H, Huang ZP, Liu J, Qiu Y, Tao YP, Wang MC, Yao H, Hou KZ, Gu FM, Xu XF. MiR-494-3p promotes PI3K/AKT pathway hyperactivation and human hepatocellular carcinoma progression by targeting PTEN. Sci Rep. 2018;8:10461. doi: 10.1038/s41598-018-28519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Ding Y, Huang J, Wang S, Ni W, Guan J, Li Q, Zhang Y, Ding Y, Chen B. MiR-141 suppresses the migration and invasion of HCC cells by targeting tiam1. PLoS One. 2014;9:e88393. doi: 10.1371/journal.pone.0088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Wang Y, Wang K, Wang Z, Jia D, Yang B, Xiong C. Downregulation of miR-214-3p may contribute to pathogenesis of ulcerative colitis via targeting STAT6. Biomed Res Int. 2017;2017:8524972. doi: 10.1155/2017/8524972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2016;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 38.Guo AY, Liang XJ, Liu RJ, Li XX, Bi W, Zhou LY, Tang CE, Yan A, Chen ZC, Zhang PF. Flotilin-1 promotes the tumorigenicity and progression of malignant phenotype in human lung adenocarcinoma. Cancer Biol Ther. 2017;18:715–722. doi: 10.1080/15384047.2017.1360445. [DOI] [PMC free article] [PubMed] [Google Scholar]


