Abstract
The aim of the study was to comprehensively evaluate the clinical value of miR-125b-5p in hepatocellular carcinoma (HCC) and its potential molecular mechanisms. MiR-125b-5p expression was remarkably lower as examined by real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR) in 95 paired HCC and nonmalignant liver tissues in house (P<0.001), which was in accord with the results from miRNA-sequencing data with 371 cases of HCC. miRNA-chips from Gene Expression Omnibus (GEO) and ArrayExpress were screened. Among the seven included miRNA-chips, the relative expression of miR-125b-5p expression levels showed decreasing trends in HCC tissue samples compared with non-cancerous liver tissue samples. Altogether, A total of 655 cases of HCC tissues and 334 non-HCC liver tissues were included in the final meta-analysis. We observed that the expression of miR-125b-5p indeed decreased markedly in HCC tissues compared with the non-HCC tissues (SMD: -1.414, 95% CI: -1.894 to -0.935, P<0.001). The area under the SROC curve of lower expression of miR-125b-5p was 0.91 (95% CI: 0.89 to 0.94). A Kaplan-Meier survival analysis indicated that the lower expression or the absence of miR-125b-5p may be a risk factor for the poor outcome of HCC patients. Furthermore, the potential target genes of miR-125b-5p from 11 miRNA target prediction databases were intersected with 1,486 differentially expressed genes (DEGs) as calculated by RNA-sequencing data. Finally, a total of 330 GEGs were collected and enriched in the pathways of lysosome, focal adhesion, and pathways in cancer. In conclusion, this study utilizes a variety of research methods to confirm the lower level of miR-125b-5p in HCC tissues. This lower expression level of miR-125b-5p is closely related to increased disease progression in HCC patients.
Keywords: miR-125b-5p, hepatocellular carcinoma (HCC), RT-qPCR, miRNA-sequencing, miRNA-chip, pathways
Introduction
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death in solid tumors worldwide, with a particularly poor prognosis in patients in the advanced stages [1-4]. However, because of the low diagnostic rate in the early stages of the disease, many patients unfortunately lose their chance to receive initial surgical treatment [5-8]. Improving the accuracy of diagnosis for HCC patients is therefore an urgent issue. Meanwhile, finding a way to monitor the progress of HCC and reduce its recurrence is also an urgent need [9-12]. Novel signaling pathways and reliable biomarkers, which are involved in the occurrence of tumorigenesis and the progression of the disease, therefore need to be identified.
MicroRNAs (miRNAs) are a class of small non-coding RNAs endogenously expressed; they can suppress or degrade their target messenger RNAs (mRNAs) by binding to them [13-17]. Through the mechanisms of post-transcriptional regulation, certain miRNAs have been found to exert critical functions as tumor suppressors or oncogenes across diverse biological processes in the initiation and progression of malignancies [18-25]. One of these miRNAs, miR-125b-5p (previous name: miR-125b), which has the sequence of ucccugagacccuaacuuguga, has been studied in the tumorigenesis and progression of HCC. The expression levels of miR-125b-5p have been measured by several groups but with small sample sizes, and the results have been inconsistent [26-30]. A study with a large sample size, combined with the use of various detection methods (RT-qPCR, miRNA sequencing and miRNA-chip, etc.) to confirm the clinical implication of miR-125b-5p in HCC, has not been conducted. Furthermore, only several target genes of miR-125b-5p have been determined thus far, including sirtuin6 (SIRT6) [24], eva-1 homolog A, a regulator of programmed cell death (EVA1A) [29], ETS proto-oncogene 1, transcription factor (Ets1) [30], angiopoietin 2 (Angpt2) [25], sirtuin7 (SIRT7) [27], and transcriptional coactivator with PDZ-binding motif (TAZ) [28], to name a few. As miRNAs can have diverse target genes through sequence-complementary relationships, many other target genes may be unidentified for miR-125b-5p in HCC. Currently, the development of in-silico research has provided the possibility of identifying potential miR-125b-5p target genes with the consideration of differentially expressed genes (DEGs) of HCC. Studies that explore comprehensive miR-125b-5p target genes have not yet been conducted either.
In this study, we therefore evaluated the expression of miR-125b-5p based on evidence from three sources, which are in-house data by quantitative reverse transcription-polymerase chain reaction (RT-qPCR), miRNA sequencing data from The Cancer Genome Atlas (TCGA), and public miRNA-chip data from Gene Expression Omnibus (GEO) and ArrayExpress. Meanwhile, the potential targets of miR-125b-5p were obtained by overlapping the predicted candidates and DEGs of HCC. The potential signaling pathways of miR-125b-5p were explored via in-silico methods, such as gene ontology (GO) analysis, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation, and protein-protein interaction (PPI) analysis, thus providing a guide to acquire insights into the potential molecular mechanisms by which miR-125b-5p mediates the tumorigenesis and progression of HCC.
Materials and methods
Clinical significance of miR-125b-5p in clinical HCC samples
This study utilized a retrospective group of HCC patients receiving initial surgical resection without any chemotherapy or radiotherapy in the First Affiliated Hospital of Guangxi Medical University from March 2010 to December 2011. Formalin-fixed paraffin-embedded (FFPE) tissue samples of HCC and paired nonmalignant liver tissues from 95 cases of HCC patients were randomly selected. All patients were aged 29 to 82 years (mean age: 52 years), with tumor sizes ranging from 1 to 11 cm (mean size: 6.4 cm). Three pathologists contributed to the pathological diagnoses independently. The study was legally authorized by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University.
To detect the expression of miR-125b-5p in FFPE tissues, we conducted RT-qPCR, as previously reported in June 2012 [31-38]. The average level of RUN6B and RUN48 was utilized as the internal reference in this study. The primers of miR-125b-5p and the stable internal controls were included in the TaqMan® MicroRNA Assays (4427975, Applied Biosystems, Life Technologies, Grand Island, NY, USA). The expression of miR-125b-5p was then computed with formula 2-Δcq in this experiment.
MiR-125b-5p expression originating from miRNA sequencing data
To extend the scope of the study, we downloaded level 3 miRNA sequencing data from the TCGA database and focused on the expressions and associations between the clinical parameters of miR-125b-5p. The data downloaded included 49 adjacent non-cancerous tissues and 371 HCC tissues, and they were log2 transformed for further analysis.
MiR-125b-5p expression from miRNA-chip data
To inquire into the profiling expression of miR-125b-5p in HCC from microarray studies, we searched the GEO and ArrayExpress databases. The keywords used in the search strategies were as follows: miR*, microRNA, non-coding RNA; liver, hepatic, hepatocellular, HCC; malignan*, neoplas*, cancer, carcinoma, tumor, or tumour. All included studies should be designed with a control group of human non-cancerous liver tissues and a case group of human HCC tissues. Only studies with proper groups and available or calculable expression data of miRNA were included. Finally, we obtained seven eligible miRNA microarray profiles in this study, which were GSE21362 [39], GSE10694 [40], GSE12717 [41], GSE31383 [42], GSE54751 [43], GSE67882, and GSE69580.
Comprehensive meta-analyses of the clinical characteristics of miR-125b-5p in HCC
To further evaluate the veracity of data from the three resources (in-house RT-qPCR, miRNA-seq, and miRNA chips), we performed meta-analyses using Stata software version 15.1 (StataCorp, College Station, TX, USA) in order to calculate both the standard mean difference (SMD) and the summary receiver operating characteristic curve (SROC). The continuous outcomes were evaluated with SMD with a 95% confidence interval (95% CI). The heterogeneity of the analysis was assessed with a Q test (chi-squared test) and the I2 statistic value. A random-effect model was used when heterogeneity existed (P<0.05 or I2>50%); otherwise, a fixed-effect model was chosen. Forest plots of SMDs with CIs of miR-125b-5p in each group were calculated and pooled. The publication bias was tested using Begg’s or Egger’s funnel plots. A two-sided P value over 0.05 indicates no publication bias. Forest plots were built to show the results. The published bias was detected with Deeks’ funnel plot. To support the findings shown by SMD, we used SPSS 23.0 software in order to analyze the true positive, false positive, false negative, and true negative of the individual dataset, as well as to describe the receiver operating characteristic (ROC) curve. The SROC curve was generated with STATA software [44-47].
Prognostic value of miR-125b-5p in HCC
We next focused on the prognostic value of miR-125b-5p in HCC, as assessed by the module miRpower for liver cancer in Kaplan-Meier plotter (http://kmplot.com) [48-52]. This newly updated module provides platforms of “miRNA-seq from TCGA” with 412 miRNAs and 421 cases, “CapitalBio miRNA Array” with 119 miRNAs and 156 cases (GSE10694), “Non-commercial spotted” with 525 miRNAs and 166 cases (GSE31384), and “OSU-CCC” with 209 miRNAs and 481 cases (GSE6857). Overall survival (OS) and disease-free survival were used for the survival evaluation.
Probable targets of miR-125b-5p in HCC
In this study, the potential targets of the candidate miRNA consisted of two parallel parts, which are both predicted and are DEGs from HCC tissues. To achieve the predicted target mRNAs of miR-125b-5p, 11 miRNA target prediction databases (DIANA microT-CDS, miRanda, miRWalk, miRDB, miRNAMap, PicTar, PITA, PolymiRTS Database, RNAv22, TargetScan v7.1, and TargetMiner) were used, and the mRNAs that overlapped at least four times were selected. The validated targets were also included through in-silico databases, such as DIANA TarBase v7.0 and miRTarBase. The possible targets of miR-125b-5p predicted by at least three datasets were obtained from miRwalk database. As miR-125b-5p expression was declined in HCC tissues, we extracted up-regulated genes in HCC from the TCGA and GTEx databases; these genes were prepared with a gene expression profiling interactive analysis (GEPIA) [53-61]. A total of 369 cases of HCC tissues and 160 cases of non-HCC liver controls were involved. The |Log2FC| cutoff was 1, and the q-value Cutoff was 0.01. ANOVA was used to select the DEGs.
Gene functional enrichment and network analysis
To assess the latent function and relative signaling pathways of the potential target mRNAs of miR-125b-5p in HCC, we performed the GO analysis and KEGG pathway annotation with the Metascape database [62,63]. The intersection of the two above sources, including GEPIA and miRwalk, was submitted to the clusterProfiler package in R software for functional enrichment analysis [64-69]. The interaction network among the target mRNAs of miR-125b-5p was built with STRING [70-74]. The mRNA and protein expression levels of one hub gene of the target genes of miR-125b-5p were shown with GEPIA and The Human Protein Atlas project (THPA) databases [75-77].
Statistical analysis
In this study, SPSS software v 23.0 was utilized to conduct most of our statistical analyses. The outcomes were presented as means and standard deviations. The parametric statistics of the different groups were examined with Student’s t-test or one-way ANOVA. The degree of difference in the microarray studies was indicated by the fold change on a log2 scale. The area under curve (AUC) of the ROC curves was used to assess the distinguishing capacity of miR-125b-5p for HCC. A p value less than 0.05 was considered statistically significant.
Results
Expression and clinicopathological significance of miR-125b-5p based on in-house RT-qPCR in HCC
In this study, the relative expression of miR-125b-5p was remarkably lower in HCC tissues than in adjacent non-cancerous liver tissues (P<0.001, Figure 1). Furthermore, the AUC of miR-125b-5p was 0.9293 (95% CI: 0.894-0.9647, P<0.001). The expression of miR-125b-5p was also found to be correlated with several clinical features. It was largely lower in patients with the following clinicopathological characteristics: occurrence of advanced tumor (T)-lymph nodes (N)- and metastasis (M, TNM) stages and a tumor with incomplete capsular and multi-tumor nodes (P<0.05, Figure 1).
Figure 1.
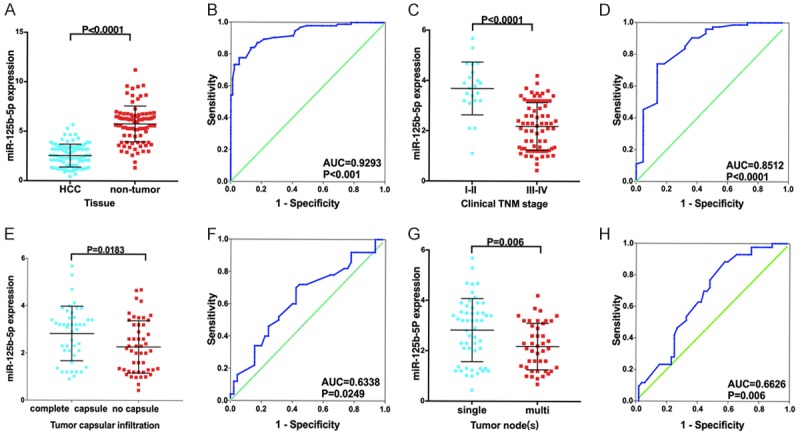
Expression and clinicopathological significance of miR-125b-5p based on in-house RT-qPCR in HCC. A, B: HCC vs. non-tumor; C, D: Clinical TNM stages I-II vs. III-IV; E, F: Complete capsule vs. no capsule; G, H: Single tumor node vs. multiple tumor nodes.
Expression and clinicopathological value of miR-125b-5p in the TCGA database in HCC
Consistent with our in-house findings, the expression of miR-125b-5p was pronouncedly lower in HCC tissues than in non-HCC liver tissues (P<0.001, Figure 2). Additionally, the AUC of miR-125b-5p was 0.9133 (95% CI: 0.8848-0.9417, P<0.001, Figure 2). Regarding the clinical performance of miR-125b-5p, significant differences exist between different groups, as classified by the clinicopathological parameters. The expression levels of miR-125b-5p were notably lower in patients with poorly differentiated grades, clinical T stage, advanced stages (III-IV), and vascular invasion (P<0.05, Figure 2).
Figure 2.
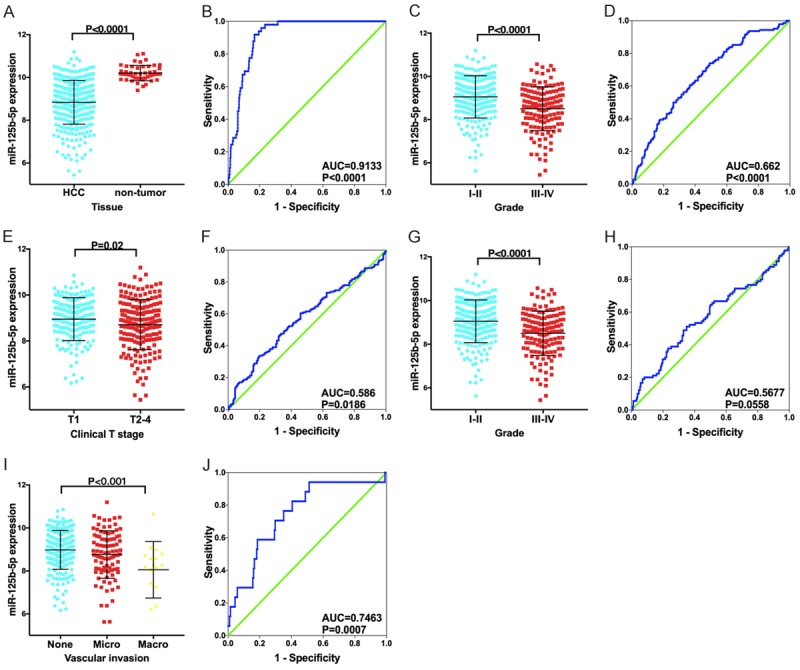
Expression and clinicopathological value of miR-125b-5p in miRNA sequencing data. A, B: HCC vs. non-tumor, C, D: Histology grades I-II vs. III-IV; E, F: Clinical T stage T1 vs. T2-4; G, H: Clinical TNM stages I-II vs. III-IV; I, J: None vascular invasion vs. micro-invasion vs. macro-invasion.
miR-125b-5p expression levels in microarray data in HCC
Among the seven included microarrays, the relative expression of miR-125b-5p expression levels showed decreasing trends in the HCC tissue samples compared with the non-cancerous liver tissue samples (Figure 3).
Figure 3.
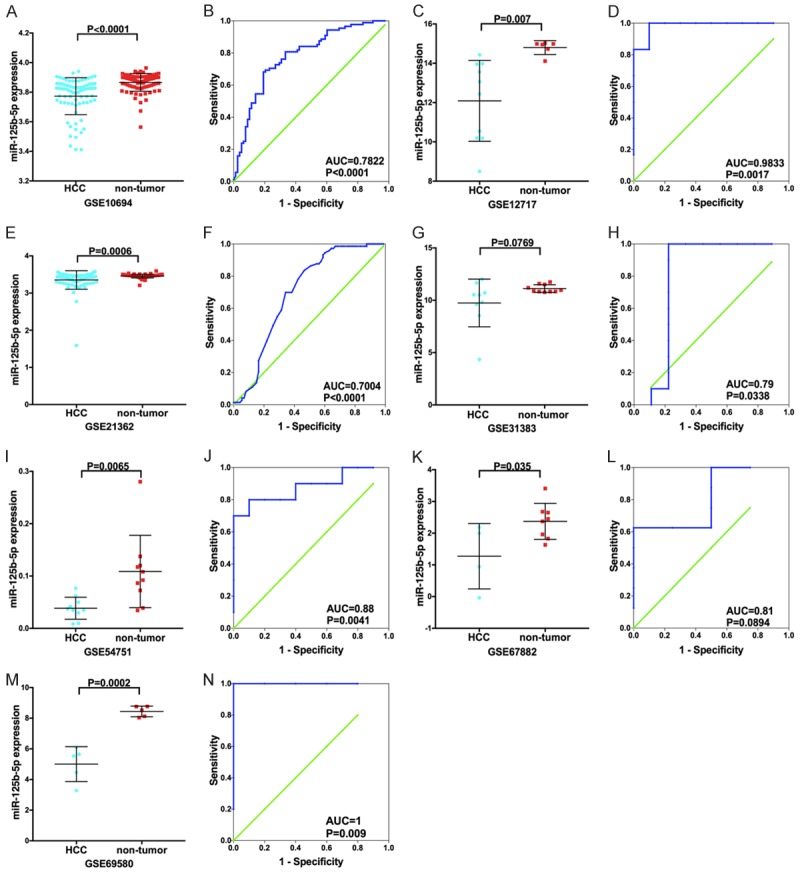
miR-125b-5p expression levels in the miRNA chip data in HCC. Scatter plots (A, C, E, G, I, K, M) and receiver operating characteristic (ROC) curves (B, D, F, H, J, L, N) were used to show the 125b-5p expression levels in HCC samples.
Comprehensive meta-analyses from RT-qPCR, miRNA-seq, and miRNA-microarrays
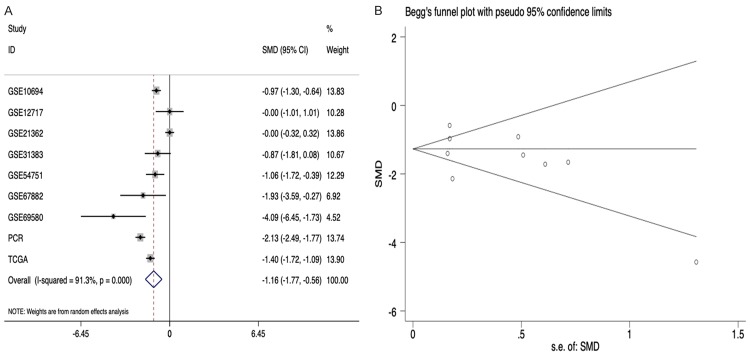
To further confirm the expression of miR-125b-5p in HCC, we conducted two types of meta-analyses by integrating the information from our RT-qPCR, TCGA program, and GEO databases. A total of 655 cases of HCC tissues and 334 non-HCC liver tissues were included. The random-effect model was used because of the presence of heterogeneity. We observed that the expression of miR-125b-5p decreased markedly in HCC tissues compared with non-HCC tissues (SMD=-1.414; 95% CI: -1.894 to -0.935, P<0.001) (Figure 4). However, Begg’s test showed publication bias in the current meta-analysis (z=0.251, P=0.464) (Figure 4).
Figure 4.
Meta-analyses of miR-125b-5p expression levels in HCC. A: The forest plot of the standard mean difference (SMD) indicated that miR-125b-5p was significantly down-regulated in HCC samples compared with non-tumor tissues; B: Begg’s funnel plot indicated that there was no publication bias. CI: confidence interval.
The area under the SROC curve was 0.91 (95% CI: 0.89 to 0.94, Figure 5). The combined sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic score, and odds ratio were 0.83, 0.86, 6.04, 0.20, 3.40, and 29.92, respectively (Figure 6). Furthermore, no publication bias was detected in Deeks’ funnel plot (Figure 5).
Figure 5.
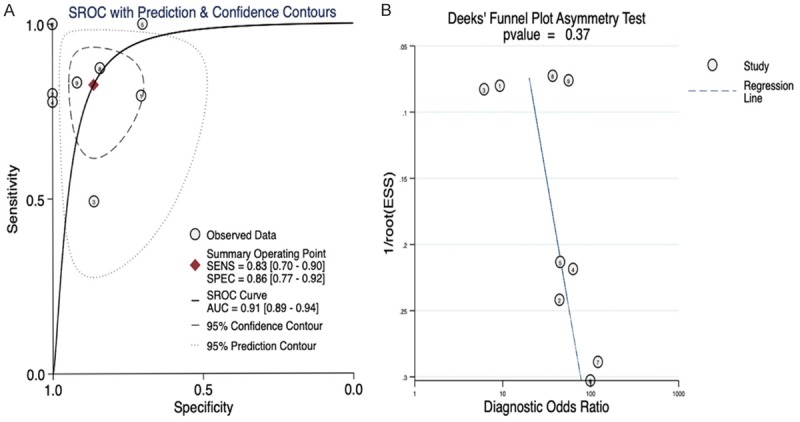
Summary receiver operating characteristic (SROC) curves for the assessment of miR-125b-5p expression in HCC. A. SROC curves for the assessment of miR-125b-5p expression in HCC; the area under the curve was 0.91; B. Deeks’ funnel plot to detect publication bias. No publication bias was found.
Figure 6.
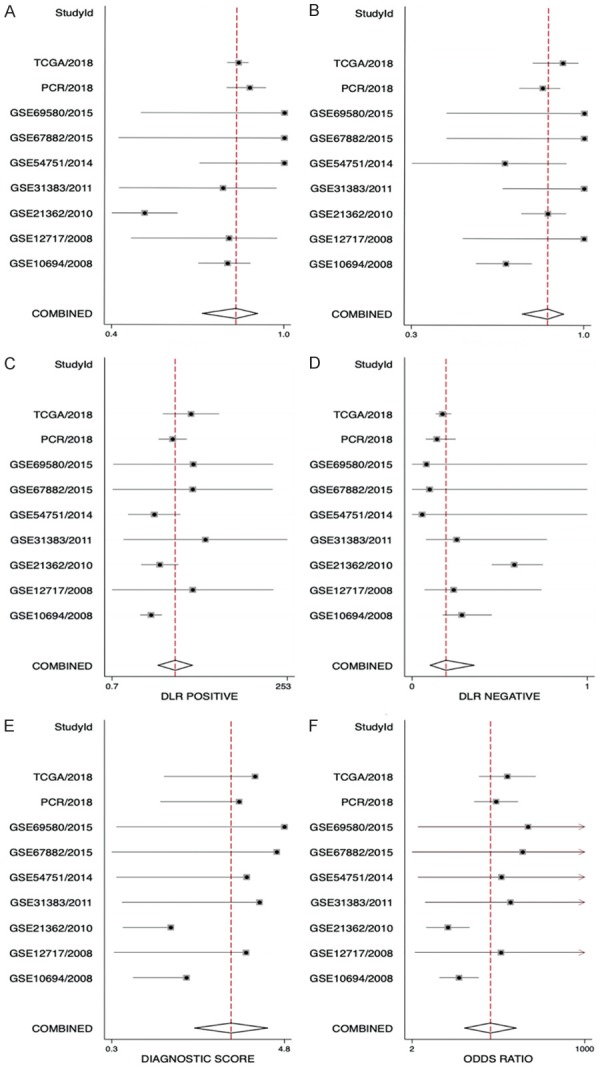
Forest plots showing the sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic score, and odds ratio of miR-125b-5p in HCC. The combined sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic score, and odds ratio were 0.83, 0.86, 6.04, 0.20, 3.40, and 29.92, respectively.
Prognostic consequence of miR-125b-5p in HCC
From the KM plot, in both RNA-seq and CapitalBio miRNA array, patients with higher miR-125b-5p levels tended to have a more favorable OS, with a hazard ratio (HR) of 0.54 and 0.55, respectively (Figure 7), indicating that the lower expression or the absence of miR-125b-5p may be a risk factor for the poor outcome of HCC patients. No significant relationships between miR-125b-5p and survival were noted from the cohort of “Non-commercial spotted”. No data were available from the other cohorts.
Figure 7.
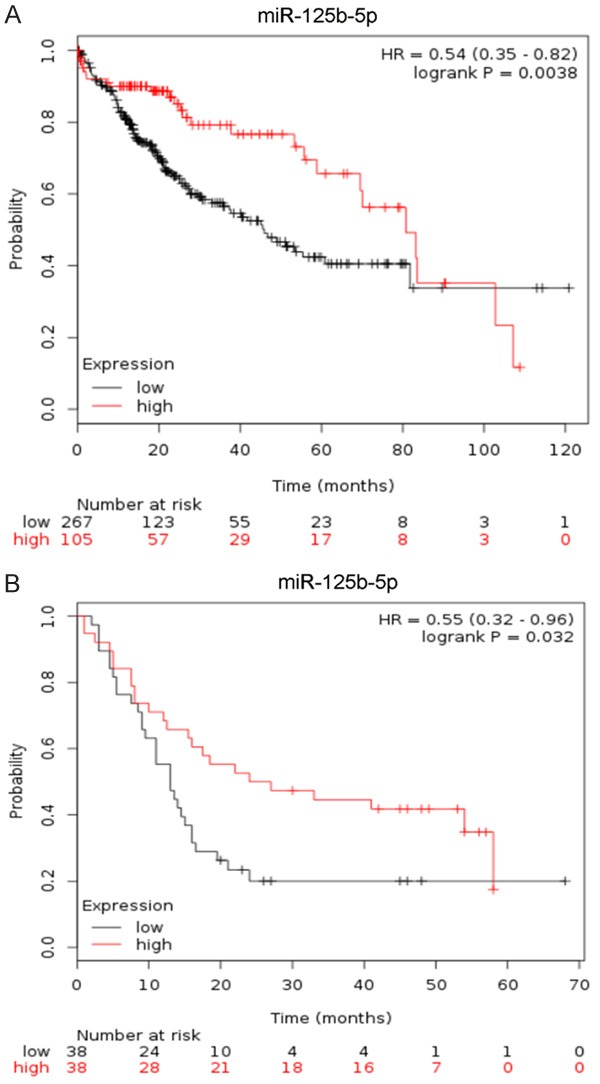
Prognostic role of miR-125b-5p in HCC. A: miRNA-seq data from TCGA; B: CapitalBio miRNA Array from GSE10694.
Potential targets of miR-125b-5p in HCC
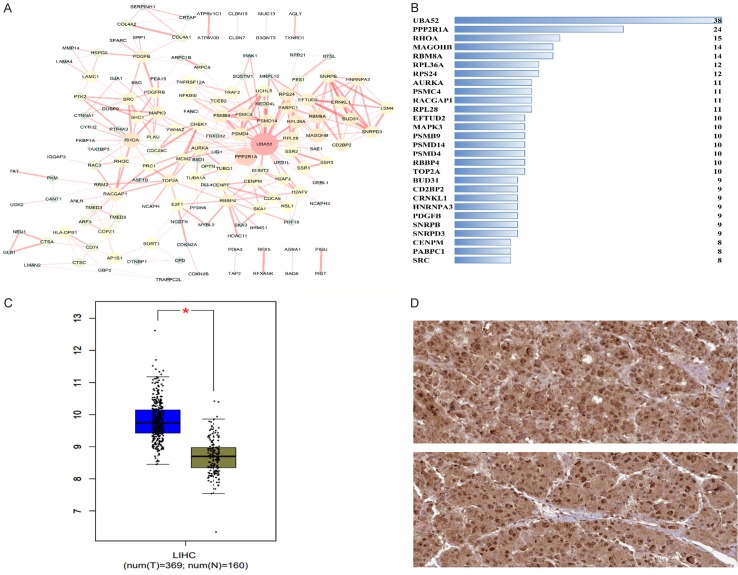
In this study, we obtained the predicted targets of miR-125b-5p via 11 computational algorithms. We achieved a total of 16,000 mRNAs selected by their present frequencies during the process to increase the reliability of our study; only 6,388 mRNAs were selected after the duplicates were excluded. In addition, 1,486 HCC-related over-expressed genes were identified with GEPIA. Finally, the potential targets of miR-125b-5p in HCC were extracted by combining the predicted or validated targets and the ones specifically expressed in HCC. A total of 330 mRNAs were extracted for GO term annotation and KEGG pathway analysis in the next step (Figure 8).
Figure 8.
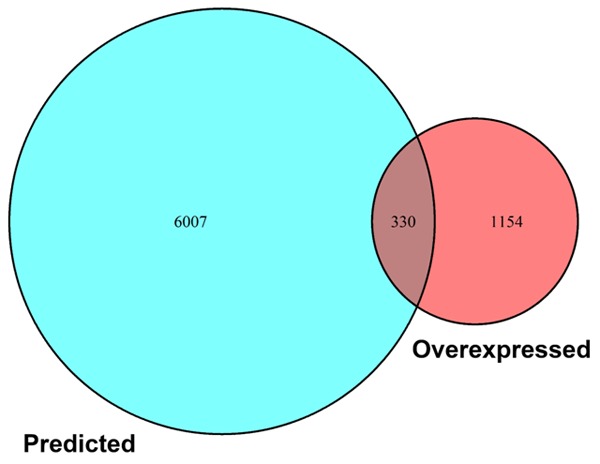
Overlapping miR-125b-5p target genes in HCC. A total of 330 potential targets were achieved.
GO functional enrichment analysis and KEGG pathway annotation of the chosen targets
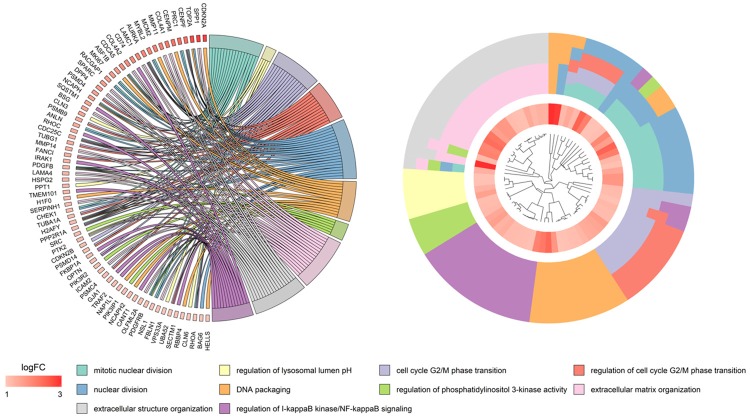
To further describe the potential molecular mechanisms of the miR-125b-5p function in the development of HCC, we conducted the GO analysis and KEGG pathway annotation by using the chosen mRNAs in Metascape. As for the biological process in Figure 9, 333 GO terms were statistically significant, as both P values and FDR q values were less than 0.01, of which the prospective targets of miR-125b-5p were remarkably enriched in mitotic nuclear division (n=17), extracellular matrix organization (n=18), regulation of protein serine/threonine kinase activity (n=22), extracellular structure organization (n=19), and response to endoplasmic reticulum stress (n=15). For the cellular component in Figure 10, 102 terms were significant, such as vacuole (n=36), lytic vacuole (n=32), lysosome (n=32), vacuolar part (n=30), Golgi membrane (n=27), anchoring junction (n=27), and adherens junctions (n=23).
Figure 9.
Biological process (BP) annotations of potential miR-125b-5p targets in HCC in gene ontology (GO) analysis. P value <0.05.
Figure 10.
Cellular component (CC) annotations of potential miR-125b-5p targets in HCC in GO analysis. P value <0.05.
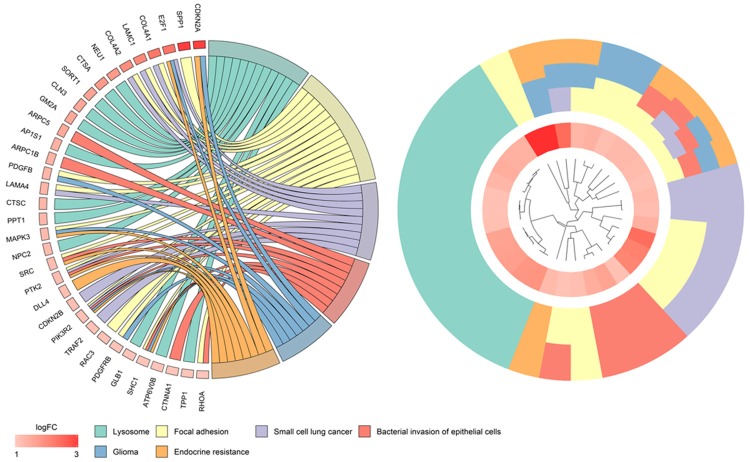
To further explore the potential signaling pathways during the carcinogenesis of HCC, we performed KEGG pathway annotation, and 42 significant pathways were identified accordingly. Among these were lysosome (n=12), focal adhesion (n=14), pathways in cancer (n=20), small cell lung cancer (n=9), bacterial invasion of epithelial cells (n=8), glioma (n=7), endocrine resistance (n=8), spliceosome (n=9), and viral carcinogenesis (n=11). The visualization of KEGG annotations is shown in Figure 11.
Figure 11.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of potential miR-125b-5p targets in HCC. P value <0.05.
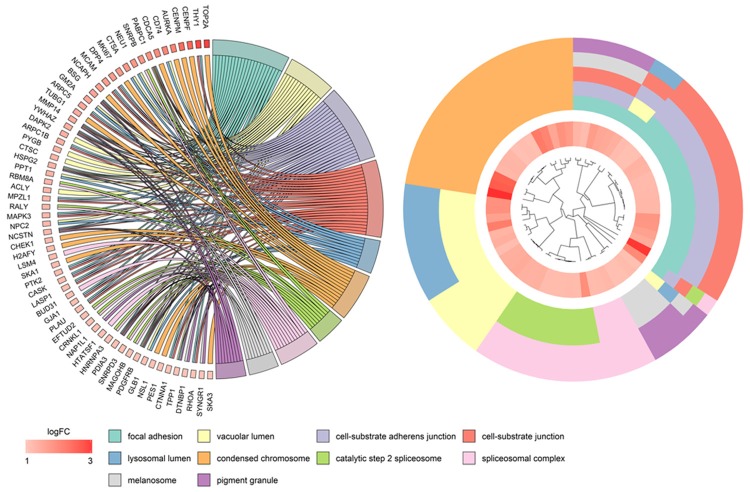
Furthermore, the interaction network among the targets of miR-125b-5p was built and visualized in Figure 10. We selected the top target, UBA52, as an example to show its mRNA and protein level. The UBA52 mRNA level was indeed significantly up-regulated in HCC tissues, and we could only observe a high expression level of UBA52 based on the THPA database because of the limited number of cases available (Figure 12).
Figure 12.
Protein-protein interaction network of the potential miR-125b-5p targets in HCC. The String program was used to construct the PPI network. The PPI network displayed the interacting relationships among these genes. A: PPI network; B: Bar plots showing the top hub genes; C: The mRNA level of the top hub gene, UBA52, based on TCGA and GTEx RNA-seq data; D: Protein level of UBA52, as assessed by immunohistochemistry with the antibody HPA049132 (×400).
Discussion
Although the expression level and target genes of miR-125b-5p in HCC have been documented by several groups, its clinical role has been investigated using a small sample size with a single detecting method, and only a single target was identified in each study. The novelty of the current study is that we combined multiple detecting methods, such as in-house RT-qPCR, miRNA-seq, and miRNA chips, to examine the clinical role of miR-125b-5p in HCC. The larger sample size also led to more convincing findings that the down-regulation of miR-125b-5p may play a vital role in the carcinogenesis and progression of HCC. Furthermore, with the advantages of in-silico tools, we constructed a network of the prospective target genes of miR-125b-5p in HCC. More unconfirmed targets of miR-125b-5p were shown and are worthy of further in-depth investigation.
As a novel biomarker, miR-125b-5p has been studied in several types of malignancies, such as nasopharyngeal carcinoma [78,79], melanoma [80], laryngeal squamous cell carcinoma [81], colorectal cancer [82], gallbladder cancer [83], breast cancer [84], acute myeloid leukemia [85], acute lymphoblastic leukemia [86], osteosarcoma [87], and gastric cancer [88]. The expression of miR-125b-5p was found to be decreased in melanoma [80], laryngeal squamous cell carcinoma [81], and gallbladder cancer [83], but it was increased in nasopharyngeal carcinoma [78,79], gastric cancer [88], colorectal cancer [82], breast cancer [84], acute myeloid leukemia [85], and acute lymphoblastic leukemia [86]. The clinical role and molecular mechanism of miR-125b-5p may be disease specific.
The expression level of miR-125b-5p was found by several groups to be reduced in HCC tissues compared with non-cancerous liver tissues based on the small size of cases [24,26-28]. In the current study, we first investigated the expression level of miR-125b-5p, as detected by a single method using multiple statistical approaches, including the performance of the t test and the drawing of ROCs. Interestingly, all the RT-qPCR, miRNA-seq, and miRNA-chip data demonstrated a consistent decreasing trend for miR-125b-5p in HCC tissues. More convincingly, the subsequent meta-analyses also supported the finding, as the SMD was -1.16 and the AUC of the SROC was 0.91 for miR-125b-5p in HCC, indicating that the loss of miR-125b-5p is closely related to the tumorigenesis of HCC. Recently, the level of plasma miR-125b-5p was also documented to be markedly down-regulated in HCC cases compared with healthy controls. An AUC of 0.891 was achieved for plasma miR-125b-5p to diagnose HCC [89]. The detection of circulating miR-125b-5p has the potential to be a non-invasive marker for the screening of HCC, but this hypothesis needs to be verified.
The loss of miR-125b-5p also influences the development of HCC after the tumor is formed. Several publications have shown that a lower miR-125b-5p level leads to early recurrence and worse five-year HCC survival based on a small sample size of patients [24,90]. In the current study, we also observed that the decreased level of miR-125b-5p was closely related with clinical TNM stages and vascular invasion by both in-house detection and miRNA-seq data from TCGA. The prognostic significance of the low level of miR-125b-5p was also verified with the miRNA-seq data and miRNA-chip data, as evidenced by K-M plots. Therefore, the feature of miR-125b-5p to inhibit tumor cells continues throughout the progression of the tumor, and miR-125b-5p may play a constant role in the process of tumor growth. Such characteristics of miR-125b-5p also make it a potential indicator for HCC prognosis prediction. Non-invasive detection is more assessable in the clinic, and interestingly, the exosomal miR-125b-5p level could also predict the recurrence and survival of HCC patients with an AUC of 0.739 [91]. A larger sample size will be required to test the prognostic value of exosomal miR-125b-5p in the near future.
MiR-125b-5p may exert its tumor suppressive role by modulating different targets. Thus far, only a couple of verified target genes of miR-125b-5p have been confirmed in HCC, including SIRT6 [24], EVA1A [29], Ets1 [30] Angpt2 [25], SIRT7 [27], and TAZ [28]. In fact, there could be many possible targets of miR-125b-5p in HCC that have not been discovered. In this study, we also aimed to discover new targets of miR-125b-5p in HCC. We overlapped the predicted target genes with the highly expressed genes in HCC tissues to obtain a more specific target gene group that influences the development of HCC. The expression levels of some target genes may, of course, change only at the protein level and may not be altered at the mRNA level. Unfortunately, this study could not obtain the expression data of HCC differential proteins, which is also a limitation of bioinformatics research. Nevertheless, the 330 potential target genes we eventually obtained could narrow the scope of future research. Unsurprisingly, these target genes are concentrated in those pathways that have a classical role in tumorigenesis, such as the lysosome, focal adhesion, and pathways in cancer. We also selected one hub gene, UBA52, from the PPI analysis to show its expression levels. The expression levels of UCA52 are the opposite of miR-125b-5p, which has an over-expression trend in both mRNA and protein levels. Our group will also choose UCA52 for the next step in vivo and in vitro studies, as this has never been examined in HCC.
In conclusion, this study utilized a variety of research methods to confirm the lower level of miR-125b-5p expression in HCC tissues. This lower expression level of miR-125b-5p is closely related to the more progressive condition of HCC patients. Furthermore, the low expression level of miR-125b-5p is also an independent prognosticator of poor prognosis in patients with HCC. However, the specific target genes and molecular mechanisms of miR-125b-5p remain to be further studied.
Acknowledgements
The present study was supported by the Fund of Future Academic Star of Guangxi Medical University (WLXSZX18107). The authors thank TCGA, GTEx, GEO, ArrayExpress and all other bioinformatics tools used in the current study.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.de Jesus VH, Dettino AL. Update on hepatocellular carcinoma from the 2018 Gastrointestinal Cancer Symposium (ASCO GI) J Hepatocell Carcinoma. 2018;5:87–90. doi: 10.2147/JHC.S171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y, Xu X, Huang D, Cui D, Liu L, Liu J, He Z, Liu J, Zheng S, Luo Y. Plasma heat shock protein 90alpha as a biomarker for the diagnosis of liver cancer: an official, large-scale, and multicenter clinical trial. EBioMedicine. 2017;24:56–63. doi: 10.1016/j.ebiom.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai J, Li B, Zhu Y, Fang X, Zhu M, Wang M, Liu S, Jiang X, Zheng J, Zhang X, Chen P. Prognostic biomarker identification through integrating the gene signatures of hepatocellular carcinoma properties. EBioMedicine. 2017;19:18–30. doi: 10.1016/j.ebiom.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP Jr. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giovanardi F, Lai Q, Bertacco A, Vitale A. Resection for hepatocellular cancer: overpassing old barriers. Transl Gastroenterol Hepatol. 2018;3:64. doi: 10.21037/tgh.2018.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao P, Qu WK, Wang CY, Tian Y, Ye ML, Sun DG, Sui JD, Wang LM, Fan R, Gao ZM. MicroRNA-205-5p regulates the chemotherapeutic resistance of hepatocellular carcinoma cells by targeting PTEN/JNK/ANXA3 pathway. Am J Transl Res. 2017;9:4300–4307. [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng K, Chen Z, Liu L, Zhao Y, Zhang S, Wang Q, Deng Z, Tan S, Ye Q. ZNF667 serves as a putative oncogene in human hepatocellular carcinoma. Cell Physiol Biochem. 2017;41:2523–2533. doi: 10.1159/000475971. [DOI] [PubMed] [Google Scholar]
- 9.Toesca DAS, Barry A, Sapisochin G, Beecroft R, Dawson L, Owen D, Mouli S, Lewandowski R, Salem R, Chang DT. Clinical case panel: treatment alternatives for inoperable hepatocellular carcinoma. Semin Radiat Oncol. 2018;28:295–308. doi: 10.1016/j.semradonc.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Ding ZB, Fu XT, Shi YH, Zhou J, Peng YF, Liu WR, Shi GM, Gao Q, Wang XY, Song K, Jin L, Tian MX, Shen YH, Fan J. Lamp2a is required for tumor growth and promotes tumor recurrence of hepatocellular carcinoma. Int J Oncol. 2016;49:2367–2376. doi: 10.3892/ijo.2016.3754. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: evidence from bioinformatic analysis. Oncol Rep. 2017;38:2607–2618. doi: 10.3892/or.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi H, Kuroda K, Sugitani M, Takayama T, Hasegawa K, Esumi M. Transglutaminase 2 is upregulated in primary hepatocellular carcinoma with early recurrence as determined by proteomic profiles. Int J Oncol. 2017;50:1749–1759. doi: 10.3892/ijo.2017.3917. [DOI] [PubMed] [Google Scholar]
- 13.Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775–8784. doi: 10.18632/oncotarget.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Liu S, Jia L, Chu F, Zhou Y, He Z, Guo M, Chen C, Xu L. Nanostructured lipid carriers for MicroRNA delivery in tumor gene therapy. Cancer Cell Int. 2018;18:101. doi: 10.1186/s12935-018-0596-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen E, Xu X, Liu R, Liu T. Small but heavy role: microRNAs in hepatocellular carcinoma progression. Biomed Res Int. 2018;2018:6784607. doi: 10.1155/2018/6784607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szymczyk A, Macheta A, Podhorecka M. Abnormal microRNA expression in the course of hematological malignancies. Cancer Manag Res. 2018;10:4267–4277. doi: 10.2147/CMAR.S174476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard EW, Yang X. microRNA regulation in estrogen receptor-positive breast cancer and endocrine therapy. Biol Proced Online. 2018;20:17. doi: 10.1186/s12575-018-0082-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He R, Yang L, Lin X, Chen X, Lin X, Wei F, Liang X, Luo Y, Wu Y, Gan T, Dang Y, Chen G. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8:15632–15641. [PMC free article] [PubMed] [Google Scholar]
- 19.Guan J, Liu Z, Xiao M, Hao F, Wang C, Chen Y, Lu Y, Liang J. MicroRNA-199a-3p inhibits tumorigenesis of hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am J Transl Res. 2017;9:2457–2465. [PMC free article] [PubMed] [Google Scholar]
- 20.Shao Y, Gu W, Ning Z, Song X, Pei H, Jiang J. Evaluating the prognostic value of microRNA-203 in solid tumors based on a meta-analysis and The Cancer Genome Atlas (TCGA) datasets. Cell Physiol Biochem. 2017;41:1468–1480. doi: 10.1159/000470649. [DOI] [PubMed] [Google Scholar]
- 21.He RQ, Wu PR, Xiang XL, Yang X, Liang HW, Qiu XH, Yang LH, Peng ZG, Chen G. Downregulated miR-23b-3p expression acts as a predictor of hepatocellular carcinoma progression: a study based on public data and RT-qPCR verification. Int J Mol Med. 2018;41:2813–2831. doi: 10.3892/ijmm.2018.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang L, Gao L, Zou XP, Huang ML, Chen G, Li JJ, Cai XY. Diagnostic significance and potential function of miR-338-5p in hepatocellular carcinoma: a bioinformatics study with microarray and RNA sequencing data. Mol Med Rep. 2018;17:2297–2312. doi: 10.3892/mmr.2017.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Lin H, Li G, Sun Y, Chen J, Shi L, Cai X, Chang C. The miR-367-3p increases sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine. 2016;12:55–67. doi: 10.1016/j.ebiom.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song S, Yang Y, Liu M, Liu B, Yang X, Yu M, Qi H, Ren M, Wang Z, Zou J, Li F, Du X, Zhang H, Luo J. MiR-125b attenuates human hepatocellular carcinoma malignancy through targeting SIRT6. Am J Cancer Res. 2018;8:993–1007. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu HR, Huang RZ, Yu XN, Shi X, Bilegsaikhan E, Guo HY, Song GQ, Weng SQ, Dong L, Janssen HL, Shen XZ, Zhu JM. Microarray expression profiling of microRNAs reveals potential biomarkers for hepatocellular crcinoma. Tohoku J Exp Med. 2018;245:89–98. doi: 10.1620/tjem.245.89. [DOI] [PubMed] [Google Scholar]
- 26.Zhou HC, Fang JH, Shang LR, Zhang ZJ, Sang Y, Xu L, Yuan Y, Chen MS, Zheng L, Zhang Y, Zhuang SM. MicroRNAs miR-125b and miR-100 suppress metastasis of hepatocellular carcinoma by disrupting the formation of vessels that encapsulate tumour clusters. J Pathol. 2016;240:450–460. doi: 10.1002/path.4804. [DOI] [PubMed] [Google Scholar]
- 27.Zhao L, Wang W. miR-125b suppresses the proliferation of hepatocellular carcinoma cells by targeting Sirtuin7. Int J Clin Exp Med. 2015;8:18469–18475. [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Fang L, Yu W, Wang Y. MicroRNA-125b suppresses the migration and invasion of hepatocellular carcinoma cells by targeting transcriptional coactivator with PDZ-binding motif. Oncol Lett. 2015;9:1971–1975. doi: 10.3892/ol.2015.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren WW, Li DD, Chen X, Li XL, He YP, Guo LH, Liu LN, Sun LP, Zhang XP. MicroRNA-125b reverses oxaliplatin resistance in hepatocellular carcinoma by negatively regulating EVA1A mediated autophagy. Cell Death Dis. 2018;9:547. doi: 10.1038/s41419-018-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng JF, Zeng ZL, Zhang K, Zhao Y, Liu YM, Chen JJ, Tong H, Wei DH, Jiang ZS, Wang Z. miR-23b-3p and miR-125b-5p downregulate apo(a) expression by targeting Ets1 in HepG2 cells. Cell Biol Int. 2018;42:313–323. doi: 10.1002/cbin.10896. [DOI] [PubMed] [Google Scholar]
- 31.Ding H, Ye ZH, Wen DY, Huang XL, Zeng CM, Mo J, Jiang YQ, Li JJ, Cai XY, Yang H, Chen G. Downregulation of miR1365p in hepatocellular carcinoma and its clinicopathological significance. Mol Med Rep. 2017;16:5393–5405. doi: 10.3892/mmr.2017.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan TQ, Tang RX, He RQ, Dang YW, Xie Y, Chen G. Upregulated MiR-1269 in hepatocellular carcinoma and its clinical significance. Int J Clin Exp Med. 2015;8:714–721. [PMC free article] [PubMed] [Google Scholar]
- 33.Huang WT, Chen ZX, He RQ, Wu YZ, Yin SY, Liang XN, Chen G, Yang H, Peng ZG, Yang LH. Clinicopathological role of miR-30a-5p in hepatocellular carcinoma tissues and prediction of its function with bioinformatics analysis. Onco Targets Ther. 2016;9:5061–5071. doi: 10.2147/OTT.S111431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang WT, Wang HL, Yang H, Ren FH, Luo YH, Huang CQ, Liang YY, Liang HW, Chen G, Dang YW. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: a clinicopathological and in silico study. Onco Targets Ther. 2016;9:5163–5180. doi: 10.2147/OTT.S108828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang HW, Ye ZH, Yin SY, Mo WJ, Wang HL, Zhao JC, Liang GM, Feng ZB, Chen G, Luo DZ. A comprehensive insight into the clinicopathologic significance of miR-144-3p in hepatocellular carcinoma. Onco Targets Ther. 2017;10:3405–3419. doi: 10.2147/OTT.S138143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Ren F, Luo Y, Rong M, Chen G, Dang Y. Down-regulation of MiR-193a-3p dictates deterioration of HCC: a clinical real-time qRT-PCR study. Med Sci Monit. 2015;21:2352–2360. doi: 10.12659/MSM.894077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna-203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int. 2015;15:62. doi: 10.1186/s12935-015-0214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Tang W, Li R, He R, Gan T, Luo Y, Chen G, Rong M. Downregulation of microRNA-132 indicates progression in hepatocellular carcinoma. Exp Ther Med. 2016;12:2095–2101. doi: 10.3892/etm.2016.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S, Shimizu K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6:e16435. doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P, Pan Z, Hu X, Zhao Y, Xie H, Jiang G, Chen T, Wang J, Zheng S, Cheng J, Wan D, Yang S, Li Y, Gu J. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 41.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 42.Wang PR, Xu M, Toffanin S, Li Y, Llovet JM, Russell DW. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc Natl Acad Sci U S A. 2012;109:11264–11269. doi: 10.1073/pnas.1117032109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen J, LeFave C, Sirosh I, Siegel AB, Tycko B, Santella RM. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med Genomics. 2015;8:28. doi: 10.1186/s12920-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gan BL, He RQ, Zhang Y, Wei DM, Hu XH, Chen G. Downregulation of HOXA3 in lung adenocarcinoma and its relevant molecular mechanism analysed by RT-qPCR, TCGA and in silico analysis. Int J Oncol. 2018;53:1557–1579. doi: 10.3892/ijo.2018.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng Y, He R, Zhang R, Gan B, Zhang Y, Chen G, Hu X. The expression of HOXA13 in lung adenocarcinoma and its clinical significance: a study based on The Cancer Genome Atlas, Oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol Lett. 2018;15:8556–8572. doi: 10.3892/ol.2018.8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang YY, Huang JC, Tang RX, Chen WJ, Chen P, Cen WL, Shi K, Gao L, Gao X, Liu AG, Peng XT, Chen G, Huang SN, Fang YY, Gu YY. Clinical value of miR-198-5p in lung squamous cell carcinoma assessed using microarray and RT-qPCR. World J Surg Oncol. 2018;16:22. doi: 10.1186/s12957-018-1320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biaoxue R, Hua L, Wenlong G, Shuanying Y. Increased serum amyloid A as potential diagnostic marker for lung cancer: a meta-analysis based on nine studies. BMC Cancer. 2016;16:836. doi: 10.1186/s12885-016-2882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, Gyorffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 49.Gao F, Yu X, Meng R, Wang J, Jia L. STARD13 is positively correlated with good prognosis and enhances 5-FU sensitivity via suppressing cancer stemness in hepatocellular carcinoma cells. Onco Targets Ther. 2018;11:5371–5381. doi: 10.2147/OTT.S170775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aiderus A, Black MA, Dunbier AK. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer. 2018;18:805. doi: 10.1186/s12885-018-4626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Wang X, Hu B, He Y, Qian X, Wang W. Candidate genes in gastric cancer identified by constructing a weighted gene co-expression network. Peer J. 2018;6:e4692. doi: 10.7717/peerj.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu LM, Xiong DD, Lin P, Yang H, Dang YW, Chen G. DNA topoisomerase 1 and 2A function as oncogenes in liver cancer and may be direct targets of nitidine chloride. Int J Oncol. 2018;53:1897–1912. doi: 10.3892/ijo.2018.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen YT, Yao JN, Qin YT, Hu K, Wu F, Fang YY. Biological role and clinical value of miR-99a-5p in head and neck squamous cell carcinoma (HNSCC): a bioinformatics-based study. FEBS Open Bio. 2018;8:1280–1298. doi: 10.1002/2211-5463.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Mo WJ, Wang X, Zhang TT, Qin Y, Wang HL, Chen G, Wei DM, Dang YW. Microarraybased bioinformatics analysis of the prospective target gene network of key miRNAs influenced by long noncoding RNA PVT1 in HCC. Oncol Rep. 2018;40:226–240. doi: 10.3892/or.2018.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wen DY, Lin P, Pang YY, Chen G, He Y, Dang YW, Yang H. Expression of the long intergenic non-protein coding RNA 665 (LINC00665) gene and the cell cycle in hepatocellular carcinoma using The Cancer Genome Atlas, the Gene Expression Omnibus, and quantitative real-time polymerase chain reaction. Med Sci Monit. 2018;24:2786–2808. doi: 10.12659/MSM.907389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Li XJ, He RQ, Wang X, Zhang TT, Qin Y, Zhang R, Deng Y, Wang HL, Luo DZ, Chen G. Upregulation of HOXA1 promotes tumorigenesis and development of nonsmall cell lung cancer: a comprehensive investigation based on reverse transcription-quantitative polymerase chain reaction and bioinformatics analysis. Int J Oncol. 2018;53:73–86. doi: 10.3892/ijo.2018.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie J, Zhu XY, Liu LM, Meng ZQ. Solute carrier transporters: potential targets for digestive system neoplasms. Cancer Manag Res. 2018;10:153–166. doi: 10.2147/CMAR.S152951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang C, Yuan N, Wu L, Wang X, Dai J, Song P, Li F, Xu C, Zhao X. An integrated analysis for long noncoding RNAs and microRNAs with the mediated competing endogenous RNA network in papillary renal cell carcinoma. Onco Targets Ther. 2017;10:4037–4050. doi: 10.2147/OTT.S141951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao L, Li SH, Tian YX, Zhu QQ, Chen G, Pang YY, Hu XH. Role of downregulated miR-133a-3p expression in bladder cancer: a bioinformatics study. Onco Targets Ther. 2017;10:3667–3683. doi: 10.2147/OTT.S137433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian T, Gong Z, Wang M, Hao R, Lin S, Liu K, Guan F, Xu P, Deng Y, Song D, Li N, Wu Y, Dai Z. Identification of long non-coding RNA signatures in triple-negative breast cancer. Cancer Cell Int. 2018;18:103. doi: 10.1186/s12935-018-0598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shuaichen L, Guangyi W. Bioinformatic analysis reveals CYP2C9 as a potential prognostic marker for HCC and liver cancer cell lines suitable for its mechanism study. Cell Mol Biol (Noisy-le-grand) 2018;64:70–74. [PubMed] [Google Scholar]
- 64.Wang Y, Wang Y, Liu F. A 44-gene set constructed for predicting the prognosis of clear cell renal cell carcinoma. Int J Mol Med. 2018;42:3105–3114. doi: 10.3892/ijmm.2018.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu L, Yu J, Wang Q, Xu B, Ji L, Yu L, Zhang X, Cai H. Identification of a 5lncRNA signaturebased risk scoring system for survival prediction in colorectal cancer. Mol Med Rep. 2018;18:279–291. doi: 10.3892/mmr.2018.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S, Wang Y, Zhang L, Su Y, Zhang M, Wang J, Zhang X. Exploration of the mechanism of colorectal cancer metastasis using microarray analysis. Oncol Lett. 2017;14:6671–6677. doi: 10.3892/ol.2017.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng B, Peng J, Mollayup A, Bakri A, Guo L, Zheng J, Xu H. Construction of a prognostic prediction system for pancreatic ductal adenocarcinoma to investigate the key prognostic genes. Mol Med Rep. 2018;17:216–224. doi: 10.3892/mmr.2017.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li XT. Identification of key genes for laryngeal squamous cell carcinoma using weighted co-expression network analysis. Oncol Lett. 2016;11:3327–3331. doi: 10.3892/ol.2016.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Foj L, Filella X. Identification of potential miRNA biomarkers for high-grade prostate cancer by integrated bioinformatics analysis. Pathol Oncol Res. 2018 doi: 10.1007/s12253-018-0508-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 71.Cui W, Gu Z, Liu H, Zhang C, Liu J. Identification modules of gastric cancer based on protein-protein interaction networks and gene expression data. J BUON. 2018;23:1013–1019. [PubMed] [Google Scholar]
- 72.Pan Y, Zhang Y, Liu J. Text miningbased drug discovery in cutaneous squamous cell carcinoma. Oncol Rep. 2018;40:3830–3842. doi: 10.3892/or.2018.6746. [DOI] [PubMed] [Google Scholar]
- 73.Lu Y, Li A, Lai X, Jiang J, Zhang L, Zhong Z, Zhao W, Tang P, Zhao H, Ren X. Identification of differentially expressed genes and signaling pathways using bioinformatics in interstitial lung disease due to tyrosine kinase inhibitors targeting the epidermal growth factor receptor. Invest New Drugs. 2018 doi: 10.1007/s10637-018-0664-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 74.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstrom H, Glimelius B, Sjoblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 76.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, Backstrom A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjostedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Ponten F, von Feilitzen K, Lilley KS, Uhlen M, Lundberg E. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 77.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 78.Li LN, Xiao T, Yi HM, Zheng Z, Qu JQ, Huang W, Ye X, Yi H, Lu SS, Li XH, Xiao ZQ. MiR-125b increases nasopharyngeal carcinoma radioresistance by targeting A20/NF-kappaB signaling pathway. Mol Cancer Ther. 2017;16:2094–2106. doi: 10.1158/1535-7163.MCT-17-0385. [DOI] [PubMed] [Google Scholar]
- 79.Zheng Z, Qu JQ, Yi HM, Ye X, Huang W, Xiao T, Li JY, Wang YY, Feng J, Zhu JF, Lu SS, Yi H, Xiao ZQ. MiR-125b regulates proliferation and apoptosis of nasopharyngeal carcinoma by targeting A20/NF-kappaB signaling pathway. Cell Death Dis. 2017;8:e2855. doi: 10.1038/cddis.2017.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pei G, Lan Y, Chen D, Ji L, Hua ZC. FAK regulates E-cadherin expression via p-SrcY416/p-ERK1/2/p-Stat3Y705 and PPARgamma/miR-125b/Stat3 signaling pathway in B16F10 melanoma cells. Oncotarget. 2017;8:13898–13908. doi: 10.18632/oncotarget.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng J, Fan Y, Ayiheng Q, Zhang H, Yong J, Hu B. MicroRNA-125b targeted STAT3 to inhibit laryngeal squamous cell carcinoma cell growth and motility. Oncol Lett. 2017;14:480–486. doi: 10.3892/ol.2017.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu X, Shi W, Zhang Y, Wang X, Sun S, Song Z, Liu M, Zeng Q, Cui S, Qu X. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci Rep. 2017;7:42226. doi: 10.1038/srep42226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang D, Zhan M, Chen T, Chen W, Zhang Y, Xu S, Yan J, Huang Q, Wang J. miR-125b-5p enhances chemotherapy sensitivity to cisplatin by down-regulating Bcl2 in gallbladder cancer. Sci Rep. 2017;7:43109. doi: 10.1038/srep43109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu B, Su F, Chen M, Li Y, Qi X, Xiao J, Li X, Liu X, Liang W, Zhang Y, Zhang J. Serum miR-21 and miR-125b as markers predicting neoadjuvant chemotherapy response and prognosis in stage II/III breast cancer. Hum Pathol. 2017;64:44–52. doi: 10.1016/j.humpath.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 85.Hu J, Zheng L, Shen X, Zhang Y, Li C, Xi T. MicroRNA-125b inhibits AML cells differentiation by directly targeting Fes. Gene. 2017;620:1–9. doi: 10.1016/j.gene.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Piatopoulou D, Avgeris M, Marmarinos A, Xagorari M, Baka M, Doganis D, Kossiva L, Scorilas A, Gourgiotis D. miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br J Cancer. 2017;117:801–812. doi: 10.1038/bjc.2017.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li JF, Song YZ. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. 2017;39:1010428317709991. doi: 10.1177/1010428317709991. [DOI] [PubMed] [Google Scholar]
- 88.Sui M, Jiao A, Zhai H, Wang Y, Wang Y, Sun D, Li P. Upregulation of miR-125b is associated with poor prognosis and trastuzumab resistance in HER2-positive gastric cancer. Exp Ther Med. 2017;14:657–663. doi: 10.3892/etm.2017.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen S, Chen H, Gao S, Qiu S, Zhou H, Yu M, Tu J. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol Res. 2017;47:312–320. doi: 10.1111/hepr.12739. [DOI] [PubMed] [Google Scholar]
- 90.Shimagaki T, Yoshizumi T, Harimoto N, Yoshio S, Naito Y, Yamamoto Y, Ochiya T, Yoshida Y, Kanto T, Maehara Y. MicroRNA-125b expression and intrahepatic metastasis are predictors for early recurrence after hepatocellular carcinoma resection. Hepatol Res. 2018;48:313–321. doi: 10.1111/hepr.12990. [DOI] [PubMed] [Google Scholar]
- 91.Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–3851. doi: 10.2147/OTT.S140062. [DOI] [PMC free article] [PubMed] [Google Scholar]