Abstract
Higher order tasks in development for brain-computer interfacing applications require the invasiveness of intracortical microelectrodes. Unfortunately, the resulting inflammatory response contributes to the decline of detectable neural signal. The major components of the neuroinflammatory response to microelectrodes have been well-documented with histological imaging, leading to the identification of broad pathways of interest for its inhibition such as oxidative stress and innate immunity. To understand how to mitigate the neuroinflammatory response, a more precise understanding is required. Advancements in genotyping have led the development of new tools for developing temporal gene expression profiles. Therefore, we have meticulously characterized the gene expression profiles of the neuroinflammatory response to mice implanted with non-functional intracortical probes. A time course of differential acute expression of genes of the innate immune response were compared to naïve sham mice, identifying significant changes following implantation. Differential gene expression analysis revealed 22 genes that could inform future therapeutic targets. Particular emphasis is placed on the largest changes in gene expression occurring 24 h post-implantation, and in genes that are involved in multiple innate immune sets including Itgam, Cd14, and Irak4.
Keywords: Pattern recognition receptors, Intracortical microelectrodes, Gene expression, Brain, Innate immune response
1. Introduction
Intracortical microelectrodes have been explored as the interface with the brain for brain computer interfacing (BCI) since the 1990’s. Neural signals captured using intracortical microelectrodes can be used to inform systems that can restore movement and function to patients with sensorimotor disabilities and or other neurological disease. To do so, intracortical microelectrodes acquire single unit neuronal signals that are sent to a computer which decodes neural activity into a patient’s intent to move. Algorithms translate desire to move to a signal instructing a cursor, prosthetic, or a patient’s own limb to move accordingly [1–3].
Due to the level of invasiveness of the implanted BCI technology, the benefits need to outweigh the risks. Specifically, intracortical microelectrodes need to be able to sustain reliable and consistent recorded signals throughout the duration of device implantation [4]. Currently, fluctuations and steady declines in the quality of detected neural signals over time limit the utility for long-term human use [5–7]. Multimodal failure mechanisms contribute to the stability [8], one of the most significant challenges being the self-destructive inflammatory response [9–11]. Upon implantation, the immune system quickly responds resulting in an immediate and imminent neuronal death at the probe interface, ultimately yielding poor neural signal acquisition. The immediate portion of the neuroinflammatory response is facilitated by the innate immune response. The innate immune response is part of the body’s first line of defense against foreign invaders such as pathogens or foreign bodies.
The innate immune response senses and processes external and internal danger signals, initiates a response to clear the danger, and prompts other aspects of the immune system to respond further if necessary [12]. The main components of the innate immune response include physical barriers (i.e. epithelial surfaces), the complement system, effector cells, and subsequent cytokine and chemokine response. Broadly, effector cells such as astrocytes, microglia, and blood-derived macrophages have been implicated in neuronal death at the tissue-probe interface resulting in poor electrode performance [13, 14]. Effector cells have pattern recognition receptors (PRRs), which are used to detect damage and initiate intracellular signaling cascades that coordinate the inflammatory response through cytokine and chemokine production.
There is increasing emphasis in further understanding the intricacies of the biological response at the tissue-probe interface to better inform more targeted therapeutic approaches to decrease neuroinflammation [15]. To accomplish this, one can elucidate the expression of genes involved in the inflammatory response. A temporal delineation of a gene expression profile can detail the activity of a large number of relevant genes providing benefit over traditional means of assessing the inflammatory response, such as immunohistochemistry. High information content can advance the identification of key mechanisms to target the reduction of neuroinflammation leading to improvements of intracortical microelectrode performance.
We have established that two specific pathways are critical to the connection between microelectrode-induced neuroinflammation and microelectrode performance: oxidative stress and innate immunity [16–22]. Oxidative stress is a result of the intense or prolonged inflammatory response associated with microelectrode implantation [21, 22]. We, and others, have previously looked at genes involved in oxidative stress [13, 22–25]. Since oxidative stress is downstream from the initiation of the innate immune response, a profile of the specific innate immune system-related genes could provide a more robust approach to attenuation of microelectrode-induced neuroinflammation. Therefore, here we investigated molecular changes of different subsets of the acute innate immune response driven by damage resulting from the intracortical probe implantation. Using gene expression profiling, changes in innate immune gene expression was measured over time and compared to no-surgery shams. Understanding how gene expression profiles change immediately following probe implantation provides valuable insights into inflammatory mechanisms at the tissue-probe interface. By looking at key players within each subset of the innate immune response and determining if there are any commonalities, we can find key targets to decrease the neuroinflammatory response.
2. Material and methods
2.1. Probes
“Dummy”, non-recording, single shank, uncoated, Michigan-style silicon probes (2 mm × 15 μm × 123 μm) were used as intracortical probes as previously described [26]. Probes were washed in 95% ethanol, three times for five minutes each wash, and sterilized via a cold ethylene oxide gas cycle as previously described in Ravikumar et al. (2014) [27].
2.2. Animals
Prior to surgery, mice were housed in groups of 3–5 with food and water provided ad libitium, while maintained on a 12-hour light/dark cycle. At the time of surgery, all mice were 8 to 10 weeks of age (Male C57/BL6, strain #000,664, Jackson Laboratories). After surgery, mice were individually housed to avoid injury to the implant site. All animal handling was performed in a class II sterile hood using microisolator techniques. There were a total of 15 animals utilized in this study, designated as three animals used per time point (6 h, 24 h, 72 h, and 2 weeks) and three animals used as sham controls. Sham animals are defined as naïve non-surgery controls. Sham animals were all male, age matched, and did not have any pre-, post- or surgical procedures. All procedures and animal care practices were performed in accordance with a protocol approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
2.3. Surgical implantation of probes
Animals were prepped for surgery using established methods [17]. Briefly, after mice reached surgical plane using isoflurane (3% in 1.0 L/min O2), mice were mounted onto a stereotaxic frame for the remainder of the surgical preparations and the surgery itself. A single dose of 0.2 ml of 0.25% Marcaine was administered subcutaneously (SQ) around the surgical site as a topical anesthetic, and Meloxicam (2 mg/kg) and Buprenorphine (0.05 mg/kg) were administered subcutaneously as analgesics. The surgical site was shaved and sterilized with alternating wipes of betadine and isopropanol. Following which, a midline incision was used to expose the skull and four craniotomies were made over the motor and sensorimotor cortex in each brain hemisphere (1.5 mm lateral and 1 mm anterior and posterior to bregma respectively) [28] were created using a high-speed dental drill with a 0.45 mm size bit. Adequate breaks in the drilling were taken to prevent thermal damage [29]. In order to ensure repeatability, the same surgeon manually inserted one dummy probe per craniotomy by hand, for a total of four probes implanted per animal. Insertion speed has been shown to be a contributor to electrode recording quality, likely due to surgical trauma during electrode insertion [30]. Therefore, our method of implantation is a potential limitation to the study. However, the same-trained surgeon, limiting variability, performed all implantations. Nonetheless, precautions were made to ensure repeatable implantations with minimal surgical trauma. Probes were held at the wide tab segment using fine tip forceps and slowly implanted into the cortex at a rate of ~2–3 mm/s until the 2 cm long shank was implanted. The wide tab ensured that the same depth of probe shank was implanted per craniotomy/animal, because it is too wide to be implanted and always remained above the cortical tissue. Following which, the probe was tethered to the skull with silicone elastomer and dental acrylic. Lastly, the incision site was sutured closed using 5–0 monofilment polypropylene suture. Post-operative care and monitoring was provided the week following surgery, with administration of Meloxicam (2 mg/kg) analgesia for three days following surgery.
2.4. Tissue processing
Sham and implanted animals were euthanized via cardiac perfusions in order to prepare tissue extracts for analysis. Briefly, deeply anesthetized animals, were transcardially perfused with cold 1X phosphate-buffered saline (PBS) followed by 30% (w/v) sucrose. Brains were immediately extracted and probes (if implanted) were explanted. Perfusion and explantation was done quickly to prevent excessive degradation of RNA. Brain tissue was flash frozen in optimal cutting temperature compound (OCT) on dry ice. Using a cryostat, the cortical brain tissue surrounding the neural probes was sectioned at 150 μm thick frozen slices and collected onto glass microscope slides. A biopsy punch (1 mm deep and 500 μm diameter) centered directly over the implant site of the collected tissue on the glass slide, was used to immediately excise the tissue of the frozen tissue slices, resulting in tissue samples each 150 μm thick and with 250 μm radii from the implant site. This step was repeated for a total of 6–7 tissue slices, resulting in a collection of 900 – 1050 μm total thickness of cortical tissue down the electrode shaft. Tissue collection started at ~150 μm depth, continuing down the length of the device, spanning most of the cortex. Tissue was collected from all four implant sites per animal, but only two implant sites per animal were utilized for further downstream RNA isolation and gene expression analysis to account for technical replicates within each biological animal replicate.
2.5. RNA isolation
Excised brain tissue samples were directly placed in microcentrifuge tubes containing Qiazol (RNA extraction lysate). Extracted tissue was homogenized in Qiazol using 1.5 mm zirconium beads using the Bead Bug Homogenizer at high speed. RNA was immediately extracted and purified using the RNeasy Plus Universal Mini Kit in accordance with manufacturer’s protocol. RNA was stored at −80 °C until further processing. RNA purity and concentrations were determined using Qubit Flourometer. Samples were shipped on dry ice to the Nanostring processing facility (Seattle, WA) for further quality control and quantification.
2.6. RNA quantification
For each sample, 50 ng of total RNA was hybridized to 777 capture probes provided by Nanostring at 96 °C overnight with the nCounter® Mouse Neuroinflammation Plus panel. The nCounter® Mouse Neuroinflammation Plus panel (with custom add-ins genes of interest) interrogated 777 neuro-immune-related mouse genes, associated controls, and 13 housekeeping genes. Hybridized samples were digitally analyzed for frequency of each gene using the nCounter® MAX Analyzer.
2.7. Statistical analysis
Raw mRNA abundance frequency data were imported into nSolver® software Version 4.0. Default quality control checks were run to assess technical assay performance. Raw counts were normalized to internal controls and housekeeping genes to remove any technical variance. Internal controls include positive and negative controls to assess success of hybridization. Background noise was filtered out using thresholding which utilized the mean count of negative controls to substitute all raw counts at or below estimated background to the threshold value. The geometric mean of the positive controls was used to compute the positive control normalization factor. After 2 housekeeping genes were removed due to low average counts, the geometric mean of the housekeeping genes was used to calculate the CodeSet Content Normalization. Low count genes were removed by setting the Threshold Count Value to 25. Observation frequency of the Threshold Count Value was also set to 0.9. The Threshold Count Value was chosen to achieve balance between false positives and false negatives in the data.
Normalized data was Log transformed by base 2 before analysis to achieve a normal distribution of the data. From here on in, log2-fold changes will just be referred to as “fold change” for simplicity. Fold-change in gene expression levels was calculated by dividing individual expression values (normalized reads) for each time point by the mean of values from shams. Each gene was tested for differential expression in response to ‘time’ (covariate). For each gene, a negative binomial regression without dispersion was fit using ‘time’ to predict expression.
Functional analyses were performed using Advanced Analysis nSolver® software Version 2.0.115. Gene sets were curated by NanoString and informed by KEGG analysis pathways. Gene Set Analysis Scores were output by the software. Gene Set Analysis Scores summarize differential expression testing at the gene set level. Gene Set Analysis Scores are calculated as the square root of the mean squared t-statistic for the genes in a gene set (Eq. (1)), with t-statistics coming from the linear regression underlying the differential expression analysis. A correction for multiple comparisons due to the multiplicity given by the many genes was included in the nSolver software analysis. Statistical significance was performed in R 3.6 using the MASS package. The average fold change from the technical replicates within each animal was first calculated and then used in a generalized linear model (GLM) with a Poisson distribution (n = 3). For the GLM, each time point was evaluated as a categorical variable. Genes regulated differentially by more than 1 Log 2 fold from sham condition with a p < 0.05 were considered significant.
Eq. (1). Calculation of Gene Set Analysis Scores for each gene set. ti: t-statistic from ith pathway gene
3. Results
The investigation of gene expression profiles from mice implanted with neural probes (n = 3 animals with a technical duplicate per animals) at various acute time points was compared to sham controls (n = 3 animals with a technical duplicate per animal), resulting in 6 samples tested for each time point. One sham sample was removed due to high technical variance, resulting in 5 sham samples from 3 different animals. Gene expression profiles were assessed using a digital molecular multiplex barcoding array to allow for 777 genes to be analyzed from each sample. Out of the 777 genes of the panel, 590 genes met the Threshold Count requirements and were used for analysis. The present study only examined the genes related to gene sets curated by NanoString involving the innate immune system (101 genes) (Table 1). All genes examined exhibited an up-regulation compared to sham at least one time point due to intracortical probe implantation. None of the genes demonstrated significant down-regulation compared to sham (data not shown).
Table 1.
Genes from the innate immune response included in the gene sets used for analysis: complement cascade, cytokine response, chemokine response, toll-like receptor (TLR) cascade, and pattern recognition receptors (PRRs).
| Gene symbol | Gene name | Role in the Immune Response |
|---|---|---|
| AIM2 | Interferon Inducible protein | Pattern recognition receptors |
| Atm | ataxia telangiectasia mutated | NFκB pathway |
| Bcl10 | B cell leukemia/lymphoma 10 | NFκB pathway |
| Bcl2 | B cell leukemia/lymphoma 2 | NFκB pathway |
| Bcl2a1a | B cell leukemia/lymphoma 2 related protein A1a | NFκB pathway |
| Bcl2l1 | BCL2-like 1 | NFκB pathway |
| Birc2 | baculoviral IAP repeat-containing 2 | Cytokine response, NFκB pathway, TLR pathway |
| Birc3 | baculoviral IAP repeat-containing 3 | Cytokine response |
| Blnk | B cell linker | NFκB pathway |
| Btk | Bruton agammaglobulinemia tyrosine kinase | NFκB pathway |
| C1qa | complement component 1, q subcomponent, α polypeptide | Complement and coagulation |
| C1qb | complement component 1, q subcomponent, β polypeptide | Complement and coagulation |
| C1qc | complement component 1, q subcomponent, C chain | Complement and coagulation |
| C3 | complement component 3 | Complement and coagulation |
| C3ar1 | complement component 3a receptor 1 | Complement and coagulation |
| C5ar1 | complement component 5a receptor 1 | Complement and coagulation |
| Casp1 | caspase 1 | Cytokine response |
| Casp3 | caspase 3 | Cytokine response |
| Casp8 | caspase 8 | TLR pathway |
| Cd14 | CD14 antigen | Pattern recognition receptors, TLR pathway, NFκB pathway |
| Cd36 | CD36 molecule | TLR pathway |
| Cd45 | CD45 antigen | Cellular markers |
| Cd68 | CD68 antigen | Cellular markers |
| Cflar | CASP8 and FADD-like apoptosis regulator | NFκB pathway |
| Chuk | conserved helix-loop-helix ubiquitous kinase | Cytokine response, NFκB pathway, TLR pathway |
| Csf1 | colony stimulating factor 1 (macrophage) | Cytokine response |
| Csf1r | colony stimulating factor 1 receptor | Cytokine response |
| Csf2rb | colony stimulating factor 2 receptor, β, low-affinity (granulocyte-macrophage) | Cytokine response |
| Ctss | cathepsin S | TLR pathway |
| Ddx58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | Cytokine response, NFκB pathway |
| Dusp7 | dual specificity phosphatase 7 | Cytokine response, TLR pathway |
| F3 | coagulation factor III | Complement and coagulation |
| Fos | FBJ osteosarcoma oncogene | Cytokine response, TLR pathway |
| GFAP | glial fibrillary acidic protein | Cellular markers |
| Fyn | Fyn proto-oncogene | Cytokine response |
| Hmgb1 | high mobility group box 1 | Cytokine response, TLR pathway |
| Ifnar1 | interferon (α and β) receptor 1 | Cytokine response |
| Ifnar2 | interferon (α and β) receptor 2 | Cytokine response |
| Ikbkb | inhibitor of kappaB kinase β | Cytokine response, NFκB pathway, TLR pathway |
| Ikbkg | inhibitor of kappaB kinase gamma | NFκB pathway |
| Il10rb | interleukin 10 receptor, β | Cytokine response |
| Il1a | interleukin 1 α | Cytokine response |
| Il1b | interleukin 1 β | NFκB pathway |
| Il1r1 | interleukin 1 receptor, type I | NFκB pathway |
| Il1rap | interleukin 1 receptor accessory protein | Cytokine response |
| Il2rg | interleukin 2 receptor, gamma chain | Cytokine response |
| Il6ra | interleukin 6 receptor, a | Cytokine response |
| Inpp5d | inositol polyphosphate-5-phosphatase D | Cytokine response |
| Irak1 | interleukin-1 receptor-associated kinase 1 | Cytokine response, NFκB pathway, TLR pathway |
| Irak2 | interleukin-1 receptor-associated kinase 2 | Cytokine response, TLR Pathway |
| Irak4 | interleukin-1 receptor-associated kinase 4 | Cytokine response, NFκB pathway, TLR pathway |
| Irf3 | interferon regulatory factor 3 | Cytokine response, TLR pathway |
| Irf7 | interferon regulatory factor 7 | TLR pathway |
| Itgam | integrin α M | Pattern recognition receptors, Complement and coagulation, TLR pathway |
| Jun | jun proto-oncogene | Cytokine response, TLR pathway |
| Lgmn | legumain | TLR pathway |
| Ltbr | lymphotoxin B receptor | Cytokine response, NFκB pathway |
| Lyn | LYN proto-oncogene, Src family tyrosine kinase | NFκB pathway |
| Map2k4 | mitogen-activated protein kinase 4 | Cytokine response, TLR pathway |
| Map3k1 | mitogen-activated protein kinase 1 | TLR pathway |
| Map3k14 | mitogen-activated protein kinase 14 | Cytokine response |
| Mapk10 | mitogen-activated protein kinase 10 | Cytokine response, TLR pathway |
| Mapk14 | mitogen-activated protein kinase 14 | Cytokine response, TLR pathway |
| Myd88 | myeloid differentiation primary response gene 88 | Cytokine response, NFκB pathway, TLR pathway |
| NFκB1 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 | Cytokine response, NFκB pathway, TLR pathway |
| NFκB2 | nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 | Cytokine response |
| NFκBia | nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, α | Cytokine response, NFκB pathway, TLR pathway |
| Nod1 | nucleotide-binding oligomerization domain containing 1 | Cytokine response, Pattern recognition receptors, TLR pathway |
| Osmr | oncostatin M receptor | Cytokine response |
| Parp1 | poly (ADP-ribose) polymerase family, member 1 | NFκB pathway |
| Pik3ca | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α | Cytokine response |
| Pik3cb | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β | Cytokine response |
| Pik3r2 | phosphoinositide-3-kinase regulatory subunit 2 | Cytokine response |
| Plcg2 | phospholipase C, gamma 2 | NFκB pathway, TLR pathway |
| Prkaca | protein kinase, cAMP dependent, catalytic, α | Cytokine response |
| Prkcq | protein kinase C, theta | NFκB pathway |
| Pros1 | protein S (α) | Complement and coagulation |
| Psmb8 | proteasome (prosome, macropain) subunit, β type 8 (large multifunctional peptidase 7) | Cytokine response |
| Ptgs2 | prostaglandin-endoperoxide synthase 2 | NFκB pathway |
| Ptpn6 | protein tyrosine phosphatase, non-receptor type 6 | Cytokine response |
| RAGE | receptor for advanced glycation endproducts | Pattern recognition receptors |
| Rela | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Cytokine response, NFκB pathway, TLR pathway |
| Relb | avian reticuloendotheliosis viral (v-rel) oncogene related B | Cytokine response, NFκB pathway |
| Rig-1 | retinoic acid-inducible gene I | Pattern recognition receptors |
| Ripk1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | NFκB pathway, TLR pathway |
| Ripk2 | receptor (TNFRSF)-interacting serine-threonine kinase 2 | Cytokine response, TLR pathway |
| S100b | S100 protein, β polypeptide, neural | Cytokine response, TLR pathway |
| Serpine1 | serine (or cysteine) peptidase inhibitor, clade E, member 1 | Complement and coagulation |
| Smarca4 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Cytokine response |
| Sqstm1 | sequestosome 1 | Cytokine response |
| Stat1 | signal transducer and activator of transcription 1 | Cytokine response |
| Sumo1 | small ubiquitin-like modifier 1 | Cytokine response |
| Syk | spleen tyrosine kinase | Cytokine response, NFκB pathway |
| Tlr2 | toll-like receptor 2 | Pattern recognition receptors, TLR pathway |
| Tnfrsf12a | tumor necrosis factor receptor superfamily, member 12a | Cytokine response |
| Tnfrsf1a | tumor necrosis factor receptor superfamily, member 1a | Cytokine response, NFκB pathway |
| Tnfrsf1b | tumor necrosis factor receptor superfamily, member 1b | Cytokine response |
| Tnfrsf25 | tumor necrosis factor receptor superfamily, member 25 | Cytokine response |
| Tnfsf12 | tumor necrosis factor (ligand) superfamily, member 12 | Cytokine response |
| Traf2 | TNF receptor-associated factor 2 | Cytokine response, NFκB pathway |
| Traf3 | TNF receptor-associated factor 3 | Cytokine response, NFκB pathway, TLR pathway |
| Traf6 | TNF receptor-associated factor 6 | Cytokine response, NFκB pathway, TLR pathway |
| Vamp7 | vesicle-associated membrane protein 7 | Cytokine response |
| Vav1 | vav 1 oncogene | Cytokine response |
| Xiap | X-linked inhibitor of apoptosis | NFκB pathway |
3.1. Cellular response to implanted microelectrode
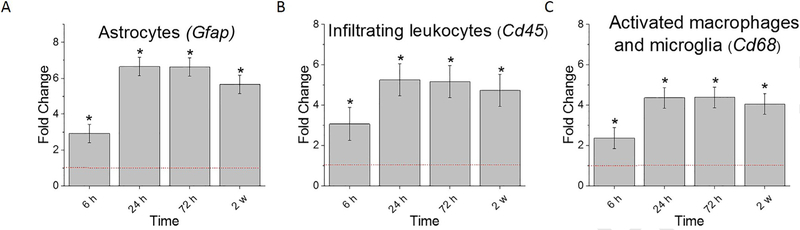
We have previously shown that the inhibition of specific innate immunity pathways reduces the glial scar and neuronal death and the probe/tissue interface, and improves recording quality [16–18]. To connect our previous studies that utilized traditional immunohistochemistry to the gene expression profiles measured here, we first looked at the genes commonly used to code for proteins with immunohistochemistry: astrocytes (Gfap), infiltrating leukocytes (Cd45), and activated microglia/macrophages (Cd68) [13, 31, 32]. It was found that all three genes were significantly upregulated at all time points examined (6 h, 24 h, 72 h, and 2 w post-implantation) compared to sham controls (Fig. 1, Table 2). Furthermore, all three cellular genes were also significantly upregulated at 24 h, 72 h, 2 w compared to 6 h post-implantation, suggesting that transcriptional changes due to the implantation of the probe are not maximized until at least after 6 h post-implantation (Fig. 1, Table 2).
Fig. 1.
Differential expression of genes for commonly used markers of astrocyte and microglial/macrophage activity compared to sham. A. Gfap, commonly used as anastroglial marker B. Cd45, an infiltrating leukocyte marker C. Cd68, commonly used as a marker for microglia/macrophage activation; Fold change is a log2-fold changecompared to sham for each time point post-implantation of dummy probe. * indicates (log2-fold change > 1and p < 0.05).
Table 2. Statistical comparisons between time points of genes commonly used as markers of astrocyte and microglial/macrophage activity.
Bold values represent conditions in which p value is <0.05, indicating statistical significance. n = 3.
| Sham v 6 h | Sham v 24 h | Sham v 72 h | Sham v 2 w | 6 h v 24 h | 6 h v 72 h | 6 h v 2 w | 24 h v 72 h | 24 h v 2w | 72 h v 2 w | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gfap | 2.33E-04 | 8.50E-27 | 4.07E-41 | 3.68E-24 | 9.74E-13 | 4.46E-23 | 5.85E-11 | 5.49E-03 | 5.55E-01 | 7.62E-04 |
| Cd45 | 6.05E-03 | 8.37E-08 | 4.79E-13 | 2.46E-08 | 6.30E-05 | 2.00E-13 | 1.21E-05 | 8.05E-06 | 6.49E-01 | 5.01E-05 |
| Cd68 | 1.14E-02 | 1.28E-12 | 9.99E-31 | 2.28E-16 | 1.24E-06 | 2.58E-22 | 1.37E-09 | 1.85E-07 | 2.03E-01 | 7.40E-05 |
3.2. The complement cascade
The complement system is a component of the innate immune system comprised of plasma proteins which can opsonize foreign substances for clearance and destruction by phagocytes, such as microglia and macrophages, and induce inflammation. As with any injury, the complement system is activated when an intracortical probe is implanted. There has been minimal research investigating the role of the complement system in the response to implanted probes, thus leaving a gap of knowledge that is necessary to be addressed.
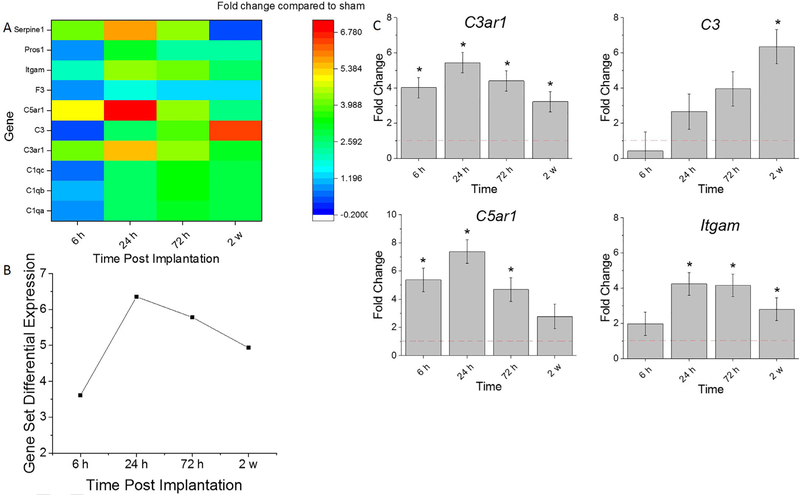
A heatmap depicting the genes expressed in the complement cascade is shown in Fig. 2A. The Complement Cascade Gene Set Differential Expression Score (Eq. (1)), a summary score that describes the collective expression of complement associated genes compared to sham, fluctuates throughout the time course of the study (Fig. 2A, B). At 6 h, there is a collective upregulation in complement factors, followed by a further increase at 24 h post-implantation. By 72 h post-implantation, the extent of upregulation slightly subsides compared to 24 h post-implantation, remaining fairly consistent through the 2 w time point (Fig. 2A, B).
Fig. 2.
Differential expression of gene set involved in complement cascade compared to sham. A. Heat map of differential expression of each gene of this set significantly upregulated at least one time point (log2-fold change > 1and p < 0.05) compared to sham. B. Gene Set Differential Expression Score, a composite score for gene set. C.Top differentially expressed genes from this gene set. Fold change is a log2-fold change and compared to sham for each time point post-implantation of dummy probe. * indicates (log2-fold change > 1 and p < 0.05).
It was found that the genes that had the highest differential expression within the complement cascade included: C3ar1, C3, C5ar1, and Itgam (Fig. 2A, C). There was a significant fold increase for C3ar1 at all time points assessed compared to sham (Fig. 2C, Table 3). Additionally, there was a significant increase of Itgam upregulation from 6 h to 24 h post-implantation which decreased by 2 weeks (Fig. 2C, Table 3). C3 gene expression was significantly upregulated at 2 weeks compared to sham 6, 24 and 72 hrs. (Fig. 2C, Table 3). Of note, by 2 weeks post-implantation, there was a significantly less up-regulation of C3ar1, C5ar1, and Itgam compared to 24 h post-implantation detected (Fig. 2C, Table 3). Thus, the complement cascade is a subset of the innate immune response that warrants further investigation at early time points.
Table 3. Statistical comparisons between time points for genes for complement cascade.
Bold values represent conditions in which p value is <0.05, indicating statistical significance. n = 3.
| Sham v 6 h | Sham v 24 h | Sham v 72 h | Sham v 2 w | 6 h v 24 h | 6 h v 72 h | 6 h v 2 w | 24 h v 72 h | 24 h v 2w | 72 h v 2 w | |
|---|---|---|---|---|---|---|---|---|---|---|
| C3ar1 | 4.65E-09 | 4.51E-15 | 1.29E-16 | 7.15E-06 | 1.84E-02 | 3.91E-03 | 1.19E-01 | 5.94E-01 | 1.01E-04 | 1.04E-05 |
| C3 | 2.98E-01 | 1.53E-01 | 6.15E-02 | 2.45E-02 | 9.24E-02 | 1.16E-04 | 1.08E-08 | 6.32E-03 | 1.91E-07 | 6.06E-03 |
| C5ar1 | 1.32E-07 | 1.09E-12 | 1.18E-06 | 8.50E-02 | 1.75E-03 | 5.10E-01 | 7.35E-06 | 1.67E-04 | 2.90E-12 | 8.64E-05 |
| Itgam | 1.97E-01 | 1.56E-08 | 9.46E-19 | 1.11E-04 | 2.42E-06 | 7.39E-17 | 6.29E-03 | 7.55E-06 | 2.94E-02 | 1.30E-10 |
3.3. Cytokine response
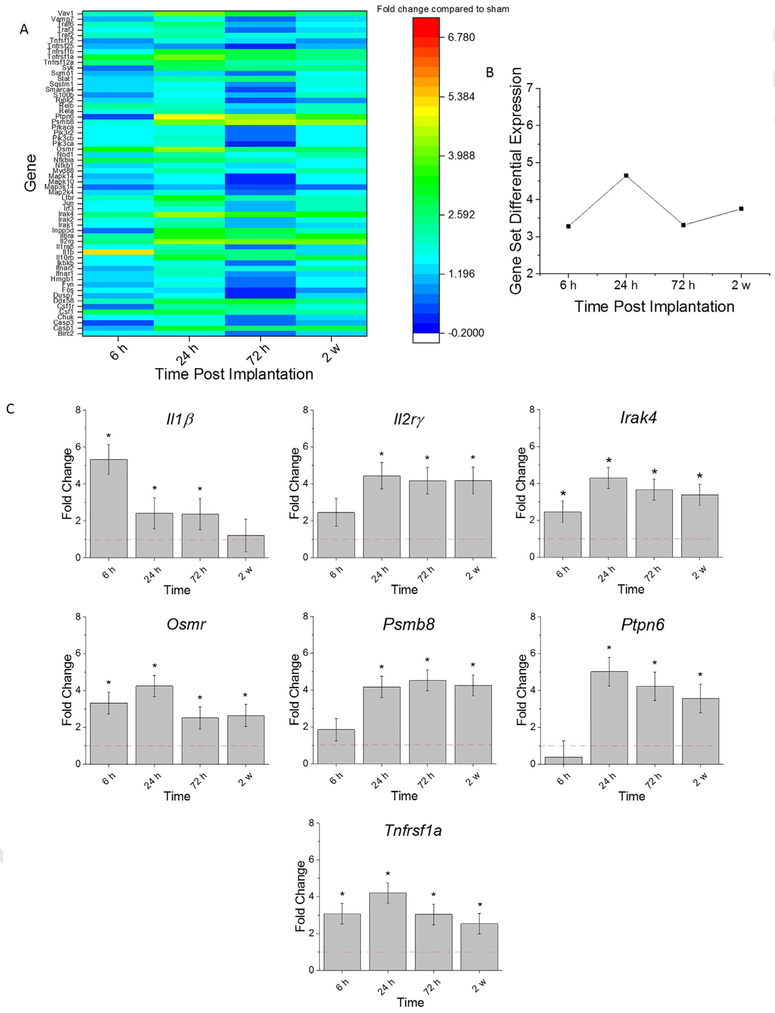
Cells involved in the innate immune response, post intracortical probe implantation, release proinflammatory mediators such as cytokines and chemokines. Cytokines are pleiotropic polypeptides (8–26 kDa) that are chemical messengers of the innate immune response. Cytokines orchestrate cellular events and are the key players in the dynamic cross-talk among all the cell types involved in the innate immune response to intracortical probe implantation [33]. Chemokines are discussed in the following section. The Gene Set Analysis scores summarize differential expression testing compared to sham at the gene set level. At acute time points, the genes associated with cytokine expression were at their highest upregulated levels at 24 h post-implantation (Fig. 3A, B). Interestingly, there was a decline in the collective cytokine response gene expression from 24 h to 72 hr, which was followed by an increase at 2 w post-implantation (Fig. 3A, B).
Fig. 3.
Differential expression of gene set involved in cytokine response compared to sham. A. Heat map of differential expression of each gene of this set significantly upregulated at least one time (log2-fold change > 1and p < 0.05) compared to sham. B. Gene Set Differential Expression Score, a composite score for gene set. C. Top differentially expressed genes from this gene set. Fold change is a log2-fold change and compared to sham for each time point post-implantation of dummy probe.*indicates (log2-fold change > 1and p < 0.05).
Notably, the genes that had the highest differential expression within the cytokine response pathway, compared to sham, included: Il1β, Il2rγ, Irak4, Osmr, Psmb8, Ptpn6, and Tnfrsf1a (Fig. 3A, C). There was a significant –fold increase of Irak4, Osmr, and Tnfrsf1a at all time points assessed, compared to sham (Fig. 3C, Table 4). Moreover, there was a significant upregulation of Il2rγ, Irak4, and Psmb8 at 24 h, 72 h, and 2 w post-implantation compared to 6 h post-implantation (Fig. 3C, Table 4). Inversely, it was observed that Il1β was at the highest upregulation at 6 h post-implantation (Fig. 3C, Table 4). Furthermore, Ptpn6 was significantly upregulated at 24 h, 72 h, 2 w post-implantation compared to sham (Fig. 3C, Table 4). By 2 weeks post-implantation, there was significantly less up-regulation of Osmr, Ptpn6, and Tnfrsf1a compared to 24 h post-implantation (Fig. 3C, Table 4). Many signaling molecules involved in the cytokine response are strongly upregulated early in the neuroinflammatory response to probes, suggesting that targeting strategies need to be present near or before the time of implantation.
Table 4. Statistical comparisons between time points for genes in cytokine response.
Bold values represent conditions in which p value is <0.05, indicating statistical significance. n = 3.
| Sham v 6 h | Sham v 24 h | Sham v 72 h | Sham v 2 w | 6 h v 24 h | 6 h v 72 h | 6 h v 2 w | 24 h v 72 h | 24 h v 2w | 72 h v 2 w | |
|---|---|---|---|---|---|---|---|---|---|---|
| Il1β | 1.96E-05 | 4.03E-02 | 1.03E-02 | 7.64E-02 | 1.71E-09 | 2.19E-08 | 1.44E-09 | 2.40E-01 | 5.92E-01 | 9.58E-02 |
| Il2rγ | 5.79E-02 | 9.75E-06 | 6.50E-10 | 1.63E-07 | 2.10E-03 | 5.28E-08 | 3.22E-05 | 4.39E-03 | 2.08E-01 | 1.04E-01 |
| Irak4 | 2.08E-02 | 3.13E-06 | 1.02E-08 | 3.59E-05 | 5.83E-03 | 4.18E-05 | 3.50E-02 | 1.44E-01 | 4.95E-01 | 3.31E-02 |
| Osmr | 2.08E-07 | 1.41E-08 | 1.61E-05 | 9.08E-04 | 5.80E-01 | 3.22E-01 | 3.82E-02 | 1.23E-01 | 8.83E-03 | 2.76E-01 |
| Psmb8 | 4.25E-01 | 8.40E-05 | 1.88E-11 | 1.03E-08 | 1.08E-03 | 4.16E-10 | 2.17E-07 | 8.16E-04 | 3.31E-02 | 2.16E-01 |
| Ptpn6 | 2.82E-01 | 4.58E-07 | 4.55E-09 | 5.82E-05 | 1.85E-07 | 5.22E-09 | 9.73E-06 | 1.99E-01 | 1.45E-01 | 6.68E-03 |
| Tnfrsf1a | 5.07E-06 | 3.42E-09 | 2.29E-09 | 1.18E-03 | 1.02E-01 | 8.63E-02 | 1.35E-01 | 9.34E-01 | 1.96E-03 | 1.49E-03 |
3.4. Chemokine response
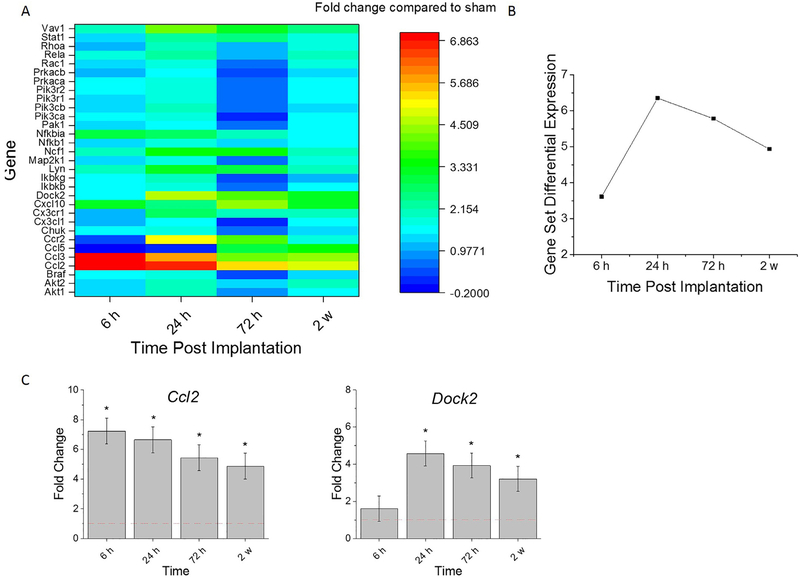
Chemokines are a class of cytokines which specifically mediate immune cell activation and migration to and from injured tissue. There are three main groups of chemokines based on the location of cysteine (C) residues: the CXC, CC, and CX3C [34]. The Gene Set Analysis scores summarize the expression compared to sham at the gene set level. As a set, the genes involved in the chemokine signaling are at their highest upregulation at 24 h post-implantation (Fig. 4A, B). Similar to the cytokine response gene expression trends, we observed a continual decline in the extent of up-regulation of the collective chemokine response genes from 24 h to 72 h and from 72 h to 2 w post-implantation (Fig. 4A, B).
Fig. 4.
Differential expression of gene set involved in chemokine response compared to sham. A. Heat map of differential expression of each gene of this set significantly upregulated at least one time (log2-fold change > 1and p < 0.05) compared to sham. B. Gene Set Differential Expression Score, a composite score for gene set. C. Top differentially expressed genes from this gene set. Fold change is a log2-fold change and compared to sham for each time point post-implantation of dummy probe.* indicates (log2-fold change > 1and p < 0.05).
The top differentially expressed chemokine response genes compared to sham were: Ccl2 and Dock2. There was a significant fold increase of Ccl2 at all time points assessed compared to sham (Fig. 4C, Table 5). Dock2 was significantly upregulated at 24 h, 72 h, and 2 weeks compared to sham (Fig. 4C, Table 5). Compared to the 6 h time point, Ccl2 was significantly less up-regulated at 24 h, 72 h, and 2 weeks (Fig. 4C, Table 5). Conversely, Dock2 had a significant fold increase at 24 h, 72 h, and 2 weeks compared to 6 h (Fig. 4C, Table 5). Similar to the cytokine response, chemokines were strongly upregulated within a day post implantation indicating chemokine targeting strategies need to be present at the site of implantation immediately following implantation.
Table 5. Statistical comparisons between time points for genes in chemokine response.
Bold values represent conditions in which p value is <0.05, indicating statistical significance. n = 3.
| Sham v 6 h | Sham v 24 h | Sham v 72 h | Sham v 2 w | 6 h v 24 h | 6 h v 72 h | 6 h v 2 w | 24 h v 72 h | 24 h v 2w | 72 h v 2 w | |
|---|---|---|---|---|---|---|---|---|---|---|
| Ccl2 | 4.94E-09 | 1.13E-06 | 6.91E-07 | 8.95E-06 | 2.85E-02 | 4.78E-02 | 1.88E-03 | 8.32E-01 | 3.53E-01 | 2.54E-01 |
| Dock2 | 6.27E-01 | 2.16E-08 | 1.03E-11 | 7.27E-06 | 1.59E-07 | 8.42E-11 | 4.34E-05 | 1.40E-01 | 1.92E-01 | 5.68E-03 |
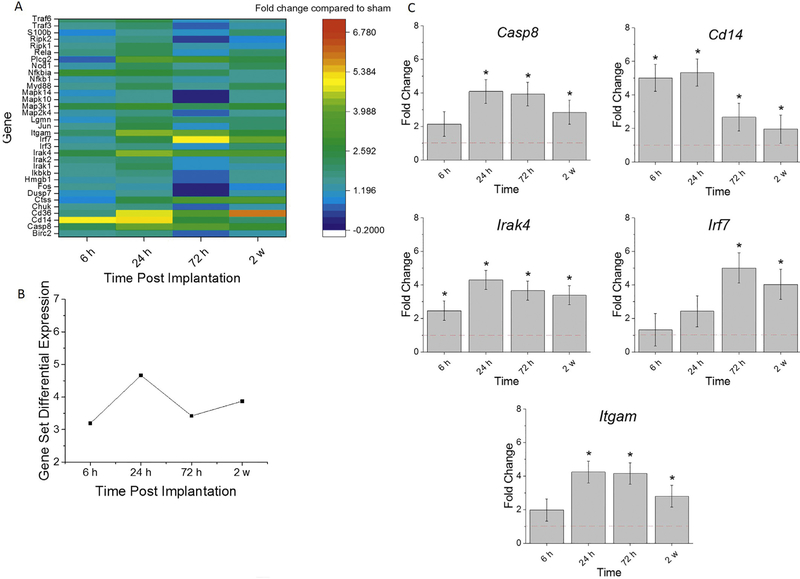
3.5. Toll-like receptor (TLR) signaling cascade
TLRs are some of the most well characterized among PRRs that detect damage due to injury, such as that occurring during intracortical probe implantation. This class of receptors recognize molecular patterns on exogenous or endogenous damaged moieties. Ligand binding to TLRs result in necrosis factor κB (NF-κB) activation leading to inflammatory mechanisms [35]. When activated, the TLR signaling cascade leads to the secretion of proinflammatory cytokines and chemokines such as TNF, IL1β, IL1R, IL6, IL8, IL10, IL23, MIP-1 α/1β, and IFNβ [36, 37]. After probe implantation, some of these cytokines and chemokines have been implicated in the facilitation of the entry of blood-derived inflammatory cells to the brain, increases of blood brain barrier (BBB) permeability, and activation of glial cells [33]. Gene Set Analysis scores in Fig. 5A are a summary score that describes the collective expression of TLR-associated genes compared to sham. The TLR-associated gene set was elevated at all time point examined, and collectively at their highest expression at 24 h post-implantation (Fig. 5A, B). The collective expression of genes involved in the TLR signaling cascade was less up-regulated at 72 h than at 24 h, followed by a slight increase in up-regulation at 2 w, from the 72 h time point (Fig. 5A, B).
Fig. 5.
Differential expression of gene set involved in TLR cascade compared to sham. A. Heat map of differential expression of each gene of this set significantly upregulated at least one time (log2-fold change > 1and p < 0.05) compared to sham. B. Gene Set Differential Expression Score, a composite score for gene set. C. Top differentially expressed genes from this gene set. Fold change is a log2-fold change and compared to sham for each time point post-implantation of dummy probe. * indicates (log2-fold change > 1and p < 0.05).
The top differentially expressed genes from the TLR-associated gene set were: Casp8, Cd14, Irak4, Irf7, and Itgam. Itgam and Irak4 were previously discussed in Sections 3.2 and 3.3, respectively. There was a significant fold increase of Cd14 at all time points assessed compared to sham (Fig. 5C, Table 6). Whereas Irf7 was significantly up-regulated at 72 h and 2 weeks compared to sham (Fig. 5C, Table 6). Upregulation of Cd14 was significantly higher at 6 h and 24 h compared to 72 h and 2 w post-implantation (Fig. 5C, Table 6). There was a significant fold increase of Casp8 at 24 h, 72 h, and 2 weeks compared to 6 h (Fig. 5C, Table 6). By 2 w post-implantation, Cd14 was significantly less up-regulated, and Irf7 was significantly up-regulated, compared to 24 h post-implantation (Fig. 5C, Table 6). Overall, the TLR pathway is upregulated immediately following probe implantation suggesting that TLR associated genes need to be further explored for potential therapeutic targeting immediately after implantation.
Table 6. Statistical comparisons between time points for genes in TLR cascade.
Bold values represent conditions in which p value is <0.05, indicating statistical significance. n = 3.
| Sham v 6 h | Sham v 24 h | Sham v 72 h | Sham v 2 w | 6 h v 24 h | 6 h v 72 h | 6 h v 2 w | 24 h v 72 h | 24 h v 2w | 72 h v 2 w | |
|---|---|---|---|---|---|---|---|---|---|---|
| Casp8 | 1.79E-01 | 1.37E-04 | 1.86E-08 | 1.95E-03 | 4.27E-03 | 2.61E-07 | 4.66E-02 | 4.22E-03 | 3.36E-01 | 2.04E-04 |
| Cd14 | 4.04E-09 | 1.49E-07 | 1.04E-03 | 2.07E-02 | 2.73E-01 | 5.68E-05 | 4.64E-07 | 2.70E-03 | 4.21E-05 | 2.09E-01 |
| Irak4 | 2.08E-02 | 3.13E-06 | 1.02E-08 | 3.59E-05 | 5.83E-03 | 4.18E-05 | 3.50E-02 | 1.44E-01 | 4.95E-01 | 3.31E-02 |
| Irf7 | 7.17E-01 | 3.98E-01 | 9.41E-05 | 7.34E-04 | 6.25E-01 | 2.51E-04 | 1.87E-03 | 1.07E-03 | 7.00E-03 | 5.40E-01 |
| Itgam | 1.97E-01 | 1.56E-08 | 9.46E-19 | 1.11E-04 | 2.42E-06 | 7.39E-17 | 6.29E-03 | 7.55E-06 | 2.94E-02 | 1.30E-10 |
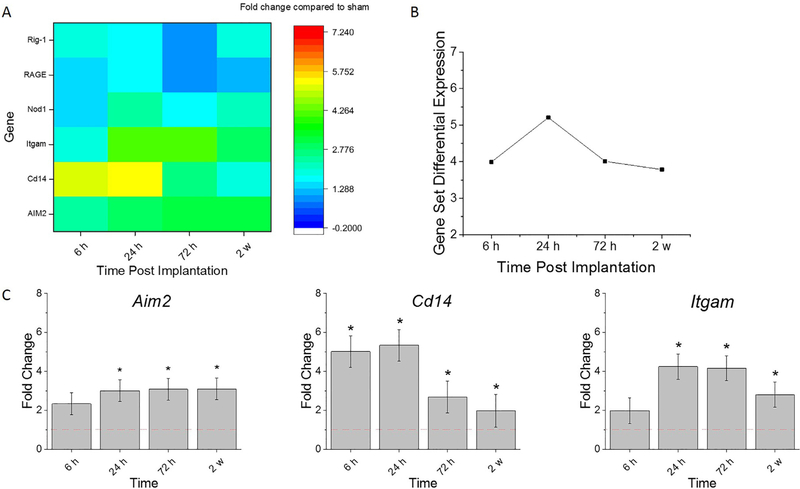
3.6. Pattern recognition receptors
Pattern recognition receptors (PRRs) are the molecular links between tissue damage and innate immunity. When certain molecular patterns or structures, including both PAMPs and DAMPs from serum proteins or damaged cells resulting from implantation of the probe, bind to PRRs, signaling of inflammatory processes ensues. Thus, the gene expression of PRRs was of particular interest to investigate in this study. Of specific interest were the PRRs residing on the cell surface, as they are released after probe implantation. Because of the role of receptor binding in cell phenotype, it is hypothesized that PRRs will be feasible future therapeutic target which can be used to block downstream cellular processes.
Fig. 6 A provides Gene Set Analysis scores, a score summarizing the collective expression of PRRs compared to sham. Genes associated with PRRs are upregulated at all time points included in this study. Collectively, the PRRs gene expression was at the highest upregulation at 24 h post-implantation, and return to approximately the same level of up-regulation at both 72 h and 2 w, as to that detected at 6 h post-implantation (Fig. 6 A, B).
Fig. 6.
Differential expression of gene set of pattern recognition receptors compared to sham. A. Heat map of differential expression of each gene of this set significantly upregulated at least one time point (log2-fold change > 1and p < 0.05) compared to sham. B. Gene Set Differential Expression Score, a composite score for gene set. C. Top differentially expressed genes from this gene set. Fold change is a log2-fold change and compared to sham for each time point post-implantation of dummy probe. * indicates (log2-fold change > 1and p < 0.05).
Aim2, Cd14, and Itgam were the top differentially expressed genes compared to sham. Because of the biological redundancies in pathways, Itgam and Cd14 were previously discussed in Sections 3.2 and 3.5, respectively. There was a significant fold increase of Aim2 at 24 h, 72 h, and 2 w post-implantation compared to sham (Fig. 6 C, Table 7). Moreover, Aim2 expression levels were significantly higher at 72 h and 2 weeks compared to 6 h (Fig. 6 C, Table 7). Since PRRs were upregulated post intracortical probe implantation and remain to be upregulated up for 2 weeks, methods to reduce PRR signaling should continue to be explored to reduce inflammation.
Table 7. Statistical comparisons between time points for genes of PRR.
Bold values represent conditions in which p value is <0.05, indicating statistical significance. n = 3.
| Sham v 6 h | Sham v 24 h | Sham v 72 h | Sham v 2 w | 6 h v 24 h | 6 h v 72 h | 6 h v 2 w | 24 h v 72 h | 24 h v 2w | 72 h v 2 w | |
|---|---|---|---|---|---|---|---|---|---|---|
| Aim2 | 1.49E-01 | 1.61E-02 | 4.71E-08 | 4.41E-05 | 3.10E-01 | 9.60E-06 | 4.27E-03 | 3.71E-04 | 5.76E-02 | 8.01E-02 |
| Cd14 | 4.04E-09 | 1.49E-07 | 1.04E-03 | 2.07E-02 | 2.73E-01 | 5.68E-05 | 4.64E-07 | 2.70E-03 | 4.21E-05 | 2.09E-01 |
| Itgam | 1.97E-01 | 1.56E-08 | 9.46E-19 | 1.11E-04 | 2.42E-06 | 7.39E-17 | 6.29E-03 | 7.55E-06 | 2.94E-02 | 1.30E-10 |
4. Discussion
There has been widespread interest investigating the neuroinflammatory response after intracortical probe implantation [10, 31, 38, 39] Recent progress has utilized gene expression analysis to evaluate the molecular pathways following implantation [22, 40, 41]. However, a comprehensive assessment of the molecular sequelae directed at the acute innate immune response post probe implantation, was yet to be performed. Therefore, this study quantitatively analyzed gene expression profiles from implanted mice at four time points during the acute inflammatory response and compared them to a non-surgical sham control. In order to have a comprehensive molecular analysis of the acute innate immune response, genes involved in the inflammatory cellular markers, the complement response, the cytokine response and the chemokine response were investigated. Since our lab has previously found targeting the TLR/CD14 pathway yields promising results in both improving intracortical microelectrode recording and neuronal density [16, 17], it was critical to continue investigation of this pathway. Furthermore, we explored PRRs that could also be potential therapeutic targets, as we have previously shown CD14 to be [18].
There were 101 different genes examined from the innate immune response. As expected, our findings indicate that genes associated with the innate immune response are significantly upregulated in the neuroinflammatory response following probe implantation. This is consistent with previous studies demonstrating the inflammatory response to intracortical probe implantation [31, 42, 43]. Within the subsets of the innate immune response, we identified 22 genes that were significantly upregulated during the first two weeks post intracortical probe implantation. The Gene Set Analysis score for all the gene within those five pathways demonstrated a robust upregulation of genes involved in the innate immune response 24 h post-implantation (Figs. 2B, 3B, 4B, 5B, 6B), suggesting that the innate immune response should be targeted at this time point.
Some highly upregulated genes of this study were involved in multiple subsets of the innate immune response. Itgam, Cd14, and Irak4 genes had the highest upregulation compared to sham, and are involved in multiple innate immunity pathways. Itgam is involved in the complement cascade, TLR signaling, and is a PRR (Figs. 2, 5, 6). Cd14 is involved in TLR signaling and functions as a PRR (Figs. 5 and 6). Irak4 is involved with both cytokine and TLR signaling (Figs. 3 and 5).
ITGAM [44], CD14 [45], and IRAK4 [46] are all involved in NF-κB signaling. ITGAM (CD11b) is a subunit for the Mac-1 (complement receptor 3 (CR3)) integrin. ITGAM is expressed on both blood-derived cells and microglia, playing a role in neutrophil and macrophage adhesion, activation, and fusion [44, 47, 48]. Activated by TLR pathway [49], ITGAM is also involved in the phagocytosis of complement coated particles [50]. The integrin is a fibrinogen [51], fibronectin [52], and collagen [53] receptor which mediates cell adhesion to these proteins, all of which can adsorb onto the probe surface [54]. Notably, Davalos et al. demonstrated that targeting ITGAM can decrease neuroinflammation. They demonstrated that blocking the fibrinogen binding motif recognized by ITGAM decreases microglial response and axonal damage in experimental autoimmune encephalomyelitis (EAE) [55]. Monoclonal antibodies to ITGAM have been developed and future studies could test the ability of this antibody to reduce intracortical probe induced inflammation [56].
Another of the strongly upregulated genes common to multiple gene sets in this study is Cd14. CD14 is a PRR involved in the recognition of TLR2 and TLR4 by its ligands. The CD14/TLR4 complex recognize DAMPs originating from cells or the extracellular matrix [37] and CD14 is mandatory for microglial reactions to DAMPs [57]. Some DAMPs CD14 is a co-receptor for include hsp70, a common DAMP released by necrotic cells, and S100A9, another DAMP released by neutrophils (blood derived cells present in the few days post-implantation) during inflammation [58, 59]. We have previously demonstrated that mice lacking Cd14 had decreases in both neuronal death and astroglial scar around the probe interface [17]. We have also shown that targeting CD14 can improve intracortical microelectrode performance [16, 18]. Since Cd14 is at its highest upregulation at 6 h and 24 h post-implantation, the best therapeutic window to target CD14 is suggested prior to 24 h post-implantation (Figs. 5C, 6C). Future studies should evaluate this recommended dosing regimen to avoid unnecessary treatment.
Irak4 is also highly upregulated at acute time points post intracortical probe implantation (Figs. 3C and 5C). Irak4 is involved in intracellular signaling cascades downstream of ligand binding to TLRs [60]. This molecule transduces pro-inflammatory downstream signals through both physical protein-protein interaction and kinase activity [61]. There are several IRAK4 inhibitors currently being developed through small-molecule screening of the IRAK4 kinase which were reviewed by Wang et al. [62]. Additionally, Nyrada is developing an IRAK4 inhibitor which is able to cross the BBB which may show promise to mitigate intracortical probe induced inflammation [63].
The following sections highlight in detail the specific genes within each innate immunity pathway that were significantly upregulated.
4.1. The complement system
The complement system is integral to the innate immune system and has not been explored with regards to intracortical probe implantation. Complement induces phagosome mediated loss of neurons in the substatnia nigra as well as microglial associated synapse loss, suggesting that the complement cascade could lead to these same results in the cortex post intracortical probe implantation [64]. The complement system can have deleterious effects [65]; however, there are studies which demonstrate that the complement system can be neuroprotective through its clearance of damage [66]. Conversely, even though the complement pathway can promote the inflammatory response, there is support for its beneficial role in promoting the health of neurons
To enhance the understanding of the complement system, genes involved with this system were investigated temporally (Fig. 2). Interestingly, the complement cascade had a slightly different time course than the other four pathways evaluated in this study. Although the genes involved in the complement cascade were similarly at the highest upregulation at 24 h, the complement cascade experienced a much slower decrease in the extent of up-regulation from 24 h to 2 w post-implantation compared to the other four pathways analyzed (Figs. 2B, 3B, 4B, 5B, 6B). Moreover, C3 and C3ar1 had the highest upregulation compared to the other genes involved in the complement response (Fig. 2A, C). C3 is a downstream effector of NF-κB, found primarily in astrocytes [67] and involved in phagocytosis [68], especially of the elimination of unwanted synapses [69]. Upregulation of C3 around the implant site suggests that further experiments are needed to determine whether synapses are being removed around the probe via C3 opsonization. C3 can also mediate macrophage adhesion to different implanted surfaces, suggesting that C3 is a protein which adsorbs onto the implanted probe, inducing a cellular response through detection by its receptor, C3ar [70]. C3 could potentially be involved with the initial stages of the innate immune response around the probe to clean up cellular debris resulting from implantation. The chemotactic receptor, C3ar, has been shown to be upregulated following CNS injury potentially promoting the migration of microglia, macrophages, and astrocytes towards the probe [71, 72].
As can be seen by the roles the upregulated genes from the complement cascade have in neuroinflammation, this pathway is a major factor that influences the outcome to intracortical probe implantation. Unfortunately, due to controversial reports, it is still unclear whether the complement system’s role in neuroinflammation are beneficial or detrimental. Therefore, before approaches can be developed to “target” any of the genes identified in this grouping, a more mechanistic study must be completed to further elucidate the function in response to this “injury” model.
4.2. Cytokine response
There were numerous molecules associated with cytokine signaling significantly upregulated post neural probe implantation compared to sham. Specifically, Il1β, Il2rγ, Irak4, Osmr, Psmb8, Ptpn6, and Tnfrsf1a (Fig. 3) were all expressed at higher levels in implanted animals than in sham. Irak4 was discussed above as it is involved with multiple gene sets examined in this study. IL1β is a hallmark cytokine involved in the acute inflammatory response and produced by both blood-derived cells and microglia. [73]. IL1β is commonly thought of as a first response cytokine [74], correspondingly, it was found that the highest upregulation of IL1β in this study was at the 6 h post-implantation (Fig. 3C). In the brain, IL1β activates endothelial cells of the BBB, enabling the trafficking of blood-derived cells into the parenchyma [11, 75]. We have previously demonstrated that monocyte trafficking into the brain following probe implantation dominates the cell population at the probe tissue interface [13], ultimately perpetuating the inflammatory response. Similarly, Karumbaiah et al. examined the gene response of several cytokines for multiple probe designs 72 h post-implantation. They found that Il1β was significantly upregulated for all probe designs examined, as demonstrated in our study for single-shank 15 μm Michigan style probes (Fig. 3C) [33]. Targeting IL1β immediately after probe implantation might be neuroprotective and lead to more robust recordings as targeting IL1B has been found to reduce neural ischemic damage [76, 77].
Other notable cytokines that were upregulated include, Il2rγ and Tnfrsf1a, which comprise portions of the IL-2 receptor and TNF cell surface receptor, respectively. The IL-2 ligand/receptor complex is involved in the signaling of other cellular players of the immune response [78]. Both the IL-2 and the TNF complex are known to lead to downstream NF-κB activation and are viable targets for as therapeutic methods to reduce intracortical probe induced inflammation [79, 80]. Conversely, Osmr and Ptpn6 negatively regulate the inflammatory response [81]. OSMR signaling inhibited both recruitment of monocytes and NF-κB signaling in a peritoneal model of acute inflammation. Furthermore, the ligand for OSMR, oncostatin M, regulates neuronal function and survival [82, 83]. PTPN6 is expressed both in astrocytes and neurons. PTPN6 negatively regulates astrocyte proliferation post ischemic injury [84] supporting functional recovery after brain injury [85]. Thus, controlled modulation of OSMR and PTPN6 signaling could be of therapeutic benefit after probe implantation.
4.3. Chemokine signaling
Chemokines mainly attract blood-derived cells and microglia to the area of the probe after implantation; however, they orchestrate other cell to cell interactions as well. The top differentially expressed genes compared to sham within the chemokine signaling pathway include: Ccl2 and Dock2. CCR2/CCL2 is a ligand-receptor pair that mediates macrophage infiltration into the brain resulting in neuronal loss [86, 87]. In fact, Hsieh et.al. implicated CCR2 in macrophage induced neuronal loss after traumatic brain injury (TBI), which is in accordance with other models of CNS injury, including experimental autoimmune encephalomyelitis (EAE) and hemorrhagic stroke [88–90]. The main ligand for CCR2 is CCL2 which was highly upregulated as early as 6 h post-implantation (Fig. 4C). Since it has been shown that the amount of blood-derived cells at the tissue-probe interface temporally correlate with neuronal loss [13], targeting the CCL2/CCR2 pathway to mitigate blood-derived cell infiltration might be a potent therapeutic to reduce neuronal loss following probe implantation. Sawyer et al. demonstrated that targeting the CCL2/CCR2 pathway, both via a knock-model and CCR2 antagonist, improves the neuroinflammatory response to intracortical probes. Targeting the CCL2/CCR2 pathway reduced BBB permeability at 2 weeks. Furthermore, targeting CCR2 resulted in higher neuronal density within 100 μm of the probe [91]. Next steps should include the administration of small-molecule drugs available to target the CCR2/CCL2 ligand-receptor pair, to see if effective dosing can be determined to improve microelectrode performance.
4.4. TLR cascade
The TLR family detects endogenous tissue damage and elicits innate immune responses through NF-κB. The highest upregulated genes involved in this cascade are: Casp8, Cd14, Irak4, Irf7, and Itgam. Itgam, CD14, and Irak4 were already discussed above.
Caspase 8 is a protease involved in several cell death mechanisms and in microglial activation [92]. Caspase 8 is expressed in neurons [93], macrophages, and microglia [94] and has been found to directly regulate IL1β in response to TLR3 and TLR4 activation [95]. Caspase 8 can prevent necroptosis [96], regulate cytokine transcription, and is a negative regulator of inflammasomes [97]. Furthermore, neuron specific casp8 deletion in mice resulted in decreased neurodegneration and neuronal cell death in a TBI model [93]. Thus, caspase 8 could be a key mediator of neuron death in intracortical probe induced inflammation. Considering that neuronal death by the probe limits the functionality of the device, future studies should explore caspase 8 and other potential neuronal death mediators in more detail.
4.5. Pattern recognition receptors
PRRs play an essential role in coordinating the innate immune response to clear damage. Given that cell-surface receptors are a popular target for therapeutic agents, we explored the families of PRRs (regardless or not if they are cell-surface receptors). The different families of PRRs converge with similar signaling cascades which induce the activation of pro-inflammatory transcription factors such as NF-κB. As mentioned above, the activation of NF-κB mediates the production of cytokines and chemokines which promote blood-borne cell infiltration and cellular activation.
In addition to Cd14 and Itgam (discussed in detail above), Aim2 was the highest upregulated gene of the PRRs analyzed. AIM2 is part of the AIM2 inflammasome, a cytosolic sensor which detects mislocalized self-DNA, suggesting a loss of cellular integrity [98]. It is unclear what DNA AIM2 is detecting post intracortical probe implantation. However, we speculate that cellular damage which occurs due to the implantation trauma could cause mislocalized DNA. Hyperactivation of AIM2 has been shown to mediate pyroptosis, a mechanism of proinflammatory cell death in neurons [99]. Thus, pyroptosis could be a possible neuronal death mechanism at the neural interface that needs explored.
5. Conclusion
A crucial step to mitigate the inflammatory response is to identify the molecular mediators involved. This study comprises an important stride towards understanding temporal changes to the biology at the tissue-probe interface. Here, the use of gene expression analysis was utilized to better understand the molecular players in the innate immune response to intracortical microelectrodes and discover new targets to mitigate neuroinflammation to intracortical microelectrodes. Since the innate immune system is fast responding, we first focused on acute time points post-implantation. Our findings corroborate studies that demonstrate the critical role the innate immune response plays in the neuroinflammatory response to intracortical probes. An increased temporal understanding of the innate immune response can inform more targeted intervention strategies to mitigate inflammation at the tissue-probe interface. This work identified Cd14, Irak4 and Itgam, among others, to be promising candidates to reduce the intracortical microelectrode mediated inflammatory response, and to potentially improve the function and stability of the device. CD14 has already been demonstrated to be a key player in the inflammatory response to intracortical microelectrodes, however future studies need to be conducted to confirm of biological relevance of IRAK4 and ITGAM.
Upregulated genes could be further explored using knock-out models or RNA interference (RNAi) to further validate ITGAM and IRAK4 as viable, potent targets to reduce neuroinflammation. Future studies also need to explore the effects of knock out models such as Itgam−/−, and Irak4−/− on electrophysiology in order to correlate the gene expressions with the decline of detectable neural signals commonly observed in control animals. Further analysis deciphering the contribution of specific cell types, including peripheral immune, on the changes of gene expression and protein levels, is a next step towards uncovering the neuroinflammatory response to implanted microelectrodes. Additional analysis could also warrant the collection of appropriate data for future modeling to fit kinetic or spatial profiles of the inflammatory response over time. The findings of this study can inform local delivery of immunomodulatory agents, such as dissolvable probe coatings, to reduce inflammation at the probe interface. Collectively, this study provides an important resource for future studies exploring the role innate immunity plays in intracortical microelectrode failure.
Statement of Significance.
Current understanding of the cellular response contributing to the failure of intracortical microelectrodes has been limited to the evaluation of cellular presence around the electrode. Minimal research investigating gene expression profiles of these cells has left a knowledge gap identifying their phenotype. This manuscript represents the first robust investigation of the changes in gene expression levels specific to the innate immune response following intracortical microelectrode implantation. To understand the role of the complement system in response to implanted probes, we performed gene expression profiling over acute time points from implanted subjects and compared them to no-surgery controls. This manuscript provides valuable insights into inflammatory mechanisms at the tissue-probe interface, thus having a high impact on those using intracortical microelectrodes to study and treat neurological diseases and injuries.
Acknowledgements
The authors thank Dr. Martina Veigl and Mr. Vai Pathak for guidance and assistance with the RT-PCR and scientific discussions. The authors also thank Mr. George Hoeferlin for surgical assistance and his support in processing the samples. Finally, the authors acknowledge Ms. Jennifer Kerbo of the Cleveland Advanced Platform Technology (APT) Center for exceptional artistry of the graphical abstract.
6. Funding
This work was supported in part by the National Institute of Health, National Institute of Neurological Disorders and Stroke, (Grant # 1R01NS082404-01A1, Capadona), the NIH Neuroengineering Training Grant (5T- 32EB004314-16, Kirsch), and NIH National Institute of Dental and Craniofacial Research Grant (2T32DE007057-41, Schaub). This study was further supported by United States (US) Department of Veterans Affairs Rehabilitation Research and Development Service Career Development Awards #RX001664-01A1 (CDA-1, Ereifej) and #RX002628-01A1 (CDA-2, Ereifej), Merit Review Awards #B1495-R and A2611-R (Capadona), and Presidential Early Career Award for Scientist and Engineers (PECASE, Capadona), from the United States (US) Department of Veterans Affairs Rehabilitation Research and Development Service. This publication was also supported by the Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. The contents do not represent the views of the U.S. Department of Veterans Affairs, the National Institute of Health, or the United States Government.
Footnotes
Data Availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations. Also, the raw/processed data required to reproduce these findings cannot be shared at this time as the data is also from a part of an ongoing study.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.actbio.2019.11.017.
References
- [1].Ajiboye AB, Bolu Ajiboye A, Willett FR, Young DR, Memberg WD, Murphy BA, Miller JP, Walter BL, Sweet JA, Hoyen HA, Keith MW, Hunter Peckham P, Simeral JD, Donoghue JP, Hochberg LR, Kirsch RF, Restoration of reaching and grasping movements through brain-controlled muscle stimulation in a person with tetraplegia: a proof-of-concept demonstration, Lancet 389 (10081) (2017) 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Collinger JL, Wodlinger B, Downey JE, Wang W, Tyler-Kabara EC, Weber DJ, McMorland AJ, Velliste M, Boninger ML, Schwartz AB, High-performance neuroprosthetic control by an individual with tetraplegia, The Lancet 381 (9866) (2013) 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Donati ARC, Shokur S, Morya E, Campos DSF, Moioli RC, Gitti CM, Augusto PB, Tripodi S, Pires CG, Pereira GA, Brasil FL, Gallo S, Lin AA, Takigami AK, Aratanha MA, Joshi S, Bleuler H, Cheng G, Rudolph A, Nicolelis MAL, Long-Term training with a brain-machine interface-based gait protocol induces partial neurological recovery in paraplegic patients, Sci. Rep 6 (2016) 30383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shih JJ, Krusienski DJ, Wolpaw JR, Brain-computer interfaces in medicine, Mayo Clinic Proceedings, Elsevier, 2012, pp. 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Burns SP, Xing D, Shapley RM, Comparisons of the dynamics of local field potential and multiunit activity signals in macaque visual cortex, J. Neurosci 30 (41) (2010) 13739–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jackson A, Fetz EE, Compact movable microwire array for long-term chronic unit recording in cerebral cortex of primates, J. Neurophysiol 98 (5) (2007) 3109–3118. [DOI] [PubMed] [Google Scholar]
- [7].Liu B, Kim E, Meggo A, Gandhi S, Luo H, Kallakuri S, Xu Y, Zhang J, Enhanced biocompatibility of neural probes by integrating microstructures and delivering anti-inflammatory agents via microfluidic channels, J. Neural Eng. 14 (2) (2017) 026008. [DOI] [PubMed] [Google Scholar]
- [8].Jorfi M, Skousen JL, Weder C, Capadona JR, Progress towards biocompatible intracortical microelectrodes for neural interfacing applications, J. Neural Eng. 12 (1) (2015) 011001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rennaker R, Miller J, Tang H, Wilson D, Minocycline increases quality and longevity of chronic neural recordings, J. Neural Eng 4 (2) (2007) L1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kozai TD, Li X, Bodily LM, Caparosa EM, Zenonos GA, Carlisle DL, Friedlander RM, Cui XT, Effects of caspase-1 knockout on chronic neural recording quality and longevity: insight into cellular and molecular mechanisms of the reactive tissue response, Biomaterials 35 (36) (2014) 9620–9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Saxena T, Karumbaiah L, Gaupp EA, Patkar R, Patil K, Betancur M, Stanley GB, Bellamkonda RV, The impact of chronic blood-brain barrier breach on intracortical electrode function, Biomaterials 34 (20) (2013) 4703–4713. [DOI] [PubMed] [Google Scholar]
- [12].Bedell H, Capadona J, Anti-inflammatory approaches to mitigate the neuroinflammatory response to brain-dwelling intracortical microelectrodes, J. Immunol. Sci 2–4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ravikumar M, Sunil S, Black J, Barkauskas DS, Haung AY, Miller RH, Selkirk SM, Capadona JR, The roles of blood-derived macrophages and resident microglia in the neuroinflammatory response to implanted intracortical microelectrodes, Biomaterials 35 (28) (2014) 8049–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Barrese JC, Rao N, Paroo K, Triebwasser C, Vargas-Irwin C, Franquemont L, Donoghue JP, Failure mode analysis of silicon-based intracortical microelectrode arrays in non-human primates, J. Neural Eng. 10 (6) (2013) 066014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Salatino JW, Ludwig KA, Kozai TDY, Purcell EK, Glial responses to implanted electrodes in the brain, Nature Biomed. Eng 1 (11) (2017) 862–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bedell HW, Hermann JK, Ravikumar M, Lin S, Rein A, Li X, Molinich E, Smith PD, Selkirk SM, Miller RH, Targeting CD14 on blood derived cells improves intracortical microelectrode performance, Biomaterials (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bedell HW, Song S, Li X, Molinich E, Lin S, Stiller A, Danda VR, Ecker M, Voit WE, Pancrazio JJ, Understanding the effects of both CD14-meditated innate immunity and device/tissue mechanical mismatch in the neuroinflammatory response to intracortical microelectrodes, Front Neurosci. 12 (2018) 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hermann JK, Ravikumar M, Shoffstall AJ, Ereifej ES, Kovach KM, Chang J, Soffer A, Wong C, Srivastava V, Smith P, Inhibition of the cluster of differentiation 14 innate immunity pathway with IAXO-101 improves chronic micro-electrode performance, J. Neural Eng. 15 (2) (2018) 025002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hermann JK, Lin S, Soffer A, Wong C, Srivastava V, Chang J, Sunil S, Sudhakar S, Tomaszewski WH, Protasiewicz G, The role of toll-like receptor 2 and 4 innate immunity pathways in intracortical microelectrode-induced neuroinflammation, Front Bioeng Biotechnol 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Potter-Baker KA, Nguyen JK, Kovach KM, Gitomer MM, Srail TW, Stewart WG, Skousen JL, Capadona JR, Development of superoxide dismutase mimetic surfaces to reduce accumulation of reactive oxygen species for neural interfacing applications, J. Mater. Chem B 2 (16) (2014) 2248–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Potter KA, Buck AC, Self WK, Callanan ME, Sunil S, Capadona JR, The effect of resveratrol on neurodegeneration and blood brain barrier stability surrounding intracortical microelectrodes, Biomaterials 34 (2013) 7001–7015. [DOI] [PubMed] [Google Scholar]
- [22].Ereifej ES, Rial GM, Hermann JK, Smith CS, Meade SM, Rayyan JM, Chen K, Feng H, Capadona JR, Implantation of neural probes in the brain elicits oxidative stress, Front Bioeng Biotechnol 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Streit WJ, Walter SA, Pennell NA, Reactive microgliosis, Prog. Neurobiol 57 (6) (1999) 563–581. [DOI] [PubMed] [Google Scholar]
- [24].Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A, Physiology of microglia, Physiol. Rev 91 (2) (2011) 461–553. [DOI] [PubMed] [Google Scholar]
- [25].Bennett C, Mohammed F, Álvarez-Ciara A, Nguyen MA, Dietrich WD, Rajguru SM, Streit WJ, Prasad A, Neuroinflammation, oxidative stress, and blood-brain barrier (BBB) disruption in acute utah electrode array implants and the effect of deferoxamine as an iron chelator on acute foreign body response, Biomaterials 188 (2019) 144–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ereifej ES, Smith CS, Meade SM, Chen K, Feng H, Capadona JR, The neuroinflammatory response to nanopatterning parallel grooves into the surface structure of intracortical microelectrodes, Adv Funct Mater 28 (12) (2018) 1704420. [Google Scholar]
- [27].Ravikumar M, Hageman DJ, Tomaszewski WH, Chandra GM, Skousen JL, Capadona JR, The effect of residual endotoxin contamination on the neuroinflammatory response to sterilized intracortical microelectrodes, J. Mater. Chem B 2 (17) (2014) 2517–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tennant KA, Adkins DL, Donlan NA, Asay AL, Thomas N, Kleim JA, Jones TA, The organization of the forelimb representation of the C57BL/6 mouse motor cortex as defined by intracortical microstimulation and cytoarchitecture, Cereb Cortex 21 (4) (2011) 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shoffstall AJ, Paiz JE, Miller DM, Rial GM, Willis MT, Menendez DM, Hostler SR, Capadona JR, Potential for thermal damage to the blood–brain barrier during craniotomy: implications for intracortical recording microelectrodes, J Neural Eng 15 (3) (2018) 034001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fiáth R, Márton AL, Mátyás F, Pinke D, Márton G, Tóth K, Ulbert I, Slow insertion of silicon probes improves the quality of acute neuronal recordings, Sci Rep 9 (1) (2019) 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Potter KA, Buck AC, Self WK, Capadona JR, Stab injury and device implantation within the brain results in inversely multiphasic neuroinflammatory and neurodegenerative responses, J. Neural Eng 9 (4) (2012) 046020. [DOI] [PubMed] [Google Scholar]
- [32].Song L, Lee C, Schindler C, Deletion of the murine scavenger receptor CD68, J. Lipid Res. 52 (8) (2011) 1542–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Karumbaiah L, Saxena T, Carlson D, Patil K, Patkar R, Gaupp EA, Betancur M, Stanley GB, Carin L, Bellamkonda RV, Relationship between intracortical electrode design and chronic recording function, Biomaterials 34 (33) (2013) 8061–8074. [DOI] [PubMed] [Google Scholar]
- [34].Zlotnik A, Yoshie O, Chemokines: a new classification system and their role in immunity, Immunity 12 (2) (2000) 121–127. [DOI] [PubMed] [Google Scholar]
- [35].Arroyo DS, Soria JA, Gaviglio EA, Rodriguez-Galan MC, Iribarren P, Toll-like receptors are key players in neurodegeneration, Int. Immunopharmacol 11 (10) (2011) 1415–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee MS, Kim Y-J, Signaling pathways downstream of pattern-recognition receptors and their cross talk, Annu. Rev. Biochem 76 (1) (2007) 447–480. [DOI] [PubMed] [Google Scholar]
- [37].Trotta T, Porro C, Calvello R, Panaro MA, Biological role of toll-like receptor-4 in the brain, J. Neuroimmunol 268 (1–2) (2014) 1–12. [DOI] [PubMed] [Google Scholar]
- [38].Oakes RS, Polei MD, Skousen JL, Tresco PA, An astrocyte derived extracellular matrix coating reduces astrogliosis surrounding chronically implanted microelectrode arrays in rat cortex, Biomaterials 154 (2018) 1–11. [DOI] [PubMed] [Google Scholar]
- [39].McConnell GC, Rees HD, Levey AI, Gutekunst C-A, Gross RE, Bellamkonda RV, Implanted neural electrodes cause chronic, local inflammation that is correlated with local neurodegeneration, J. Neural Eng 6 (5) (2009) 056003. [DOI] [PubMed] [Google Scholar]
- [40].Falcone JD, Carroll SL, Saxena T, Mandavia D, Clark A, Yarabarla V, Bellamkonda RV, Correlation of mRNA expression and signal variability in chronic intracortical electrodes, Front Bioeng Biotechnol 6 (2018) 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bennett C, Samikkannu M, Mohammed F, Dietrich WD, Rajguru SM, Prasad A, Blood brain barrier (BBB)-disruption in intracortical silicon microelectrode implants, Biomaterials 164 (2018) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Polikov VS, Tresco PA, Reichert WM, Response of brain tissue to chronically implanted neural electrodes, J. Neurosci. Methods 148 (1) (2005) 1–18. [DOI] [PubMed] [Google Scholar]
- [43].Turner J, Shain W, Szarowski D, Andersen M, Martins S, Isaacson M, Craighead H, Cerebral astrocyte response to micromachined silicon implants, Exp. Neurol 156 (1) (1999) 33–49. [DOI] [PubMed] [Google Scholar]
- [44].Sisco M, Chao JD, Kim I, Mogford JE, Mayadas TN, Mustoe TA, Delayed wound healing in mac-1–deficient mice is associated with normal monocyte recruitment, Wound Repair Regenerat. 15 (4) (2007) 566–571. [DOI] [PubMed] [Google Scholar]
- [45].Delude RL, Fenton MJ, Savedra R, Perera P-Y, Vogel SN, Thieringer R, Golenbock DT, CD14-mediated translocation of nuclear factor-kappa b induced by lipopolysaccharide does not require tyrosine kinase activity, J. Biol. Chem 269 (35) (1994) 22253–22260. [PubMed] [Google Scholar]
- [46].Yuan B, Shen H, Lin L, Su T, Zhong L, Yang Z, MicroRNA367 negatively regulates the inflammatory response of microglia by targeting IRAK4 in intracerebral hemorrhage, J Neuroinflammat. 12 (1) (2015) 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Coxon A, Rieu P, Barkalow FJ, Askari S, Sharpe AH, von Andrian UH, Arnaout MA, Mayadas TN, A novel role for the β2 integrin CD11b/CD18 in neutrophil apoptosis: a homeostatic mechanism in inflammation, Immunity 5 (6) (1996) 653–666. [DOI] [PubMed] [Google Scholar]
- [48].Podolnikova NP, Kushchayeva YS, Wu Y, Faust J, Ugarova TP, The role of integrins αMβ2 (Mac-1, CD11b/CD18) and αDβ2 (CD11d/CD18) in macrophage fusion, Am. J. Pathol 186 (8) (2016) 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Han C, Jin J, Xu S, Liu H, Li N, Cao X, Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating syk and promoting degradation of myd88 and Trif via Cbl-b, Nat. Immunol 11 (8) (2010) 734. [DOI] [PubMed] [Google Scholar]
- [50].Reichert F, Rotshenker S, Complement-receptor-3 and scavenger-receptor-AI/II mediated myelin phagocytosis in microglia and macrophages, Neurobiol. Dis 12 (1) (2003) 65–72. [DOI] [PubMed] [Google Scholar]
- [51].Lishko VK, Podolnikova NP, Yakubenko VP, Yakovlev S, Medved L, Yadav SP, Ugarova TP, Multiple binding sites in fibrinogen for integrin αMβ2 (Mac-1), J. Biol. Chem 279 (43) (2004) 44897–44906. [DOI] [PubMed] [Google Scholar]
- [52].Lishko VK, Yakubenko VP, Ugarova TP, The interplay between integrins αMβ2 and α5β1 during cell migration to fibronectin, Exp. Cell Res. 283 (1) (2003) 116–126. [DOI] [PubMed] [Google Scholar]
- [53].Walzog B, Schuppan D, Heimpel C, Hafezi-Moghadam A, Gaehtgens P, Ley K, The leukocyte integrin mac-1 (CD11b/CD18) contributes to binding of human granulocytes to collagen, Exp. Cell Res. 218 (1) (1995) 28–38. [DOI] [PubMed] [Google Scholar]
- [54].Davis GE, The mac-1 and p150, 95 β2 integrins bind denatured proteins to mediate leukocyte cell-substrate adhesion, Exp. Cell Res 200 (2) (1992) 242–252. [DOI] [PubMed] [Google Scholar]
- [55].Davalos D, Ryu JK, Merlini M, Baeten KM, Le Moan N, Petersen MA, Deerinck TJ, Smirnoff DS, Bedard C, Hakozaki H, Fibrinogen-induced perivascular microglial clustering is required for the development of axonal damage in neuroinflammation, Nat Commun. 3 (2012) ncomms2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].PROFILE AR, 23F2G, LeukArrest™, Drugs R D 1 (1999) 25–26. [DOI] [PubMed] [Google Scholar]
- [57].Janova H, Böttcher C, Holtman IR, Regen T, van Rossum D, Götz A, Ernst A-S, Fritsche C, Gertig U, Saiepour N, Gronke K, Wrzos C, Ribes S, Rolfes S, Weinstein J, Ehrenreich H, Pukrop T, Kopatz J, Stadelmann C, Salinas-Riester G, Weber MS, Prinz M, Brück W, Eggen BJL, Boddeke HWGM, Priller J, Hanisch U-K, CD14 is a key organizer of microglial responses to cns infection and injury, Glia 64 (4) (2016) 635–649. [DOI] [PubMed] [Google Scholar]
- [58].Asea A, Kraeft S-K, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK, HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine, Nat. Med 6 (4) (2000) 435–442. [DOI] [PubMed] [Google Scholar]
- [59].He W, Bellamkonda R, A molecular perspective on understanding and modulating the performance of chronic central nervous system (CNS) recording electrodes, Frontiers, Neuroeng. Ser (2007) 151–175. [PubMed] [Google Scholar]
- [60].Flannery S, Bowie AG, The interleukin-1 receptor-associated kinases: critical regulators of innate immune signalling, Biochem. Pharmacol 80 (12) (2010) 1981–1991. [DOI] [PubMed] [Google Scholar]
- [61].Li S, Strelow A, Fontana EJ, Wesche H, IRAK-4: a novel member of the irak family with the properties of an IRAK-kinase, Proc. Natl. Acad. Sci 99 (8) (2002) 5567–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wang Z, Wesche H, Stevens T, Walker N, Yeh W-C, IRAK-4 inhibitors for inflammation, Curr Top Med Chem 9 (8) (2009) 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].N. Limited, Noxopharm Subsidiary Announces Important Drug Discovery. <https://www.prnewswire.com/news-releases/noxopharm-subsidiary-announces-important-drug-discovery-300717396.html>, 2018. (accessed january 25, 20192019).
- [64].Bodea L-G, Wang Y, Linnartz-Gerlach B, Kopatz J, Sinkkonen L, Musgrove R, Kaoma T, Muller A, Vallar L, Di Monte DA, Neurodegeneration by activation of the microglial complement–phagosome pathway, J. Neurosci 34 (25) (2014) 8546–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hammad A, Westacott L, Zaben M, The role of the complement system in traumatic brain injury: a review, J Neuroinflammat. 15 (1) (2018) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Fang S, Parsa AT, Complement and the central nervous system: emerging roles in development, protection and regeneration, Immunol. Cell Biol. 88 (8) (2010) 781. [DOI] [PubMed] [Google Scholar]
- [67].Lian H, Yang L, Cole A, Sun L, Chiang AC-A, Fowler SW, Shim DJ, Rodriguez-Rivera J, Taglialatela G, Jankowsky JL, NF κB-activated astroglial release of complement C3 compromises neuronal morphology and function associated with Alzheimer’s disease, Neuron 85 (1) (2015) 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Fu H, Liu B, Frost JL, Hong S, Jin M, Ostaszewski B, Shankar GM, Costantino IM, Carroll MC, Mayadas TN, Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar aβ by microglia, Glia 60 (6) (2012) 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, The classical complement cascade mediates CNS synapse elimination, Cell 131 (6) (2007) 1164–1178. [DOI] [PubMed] [Google Scholar]
- [70].McNally A, Anderson J, Complement C3 participation in monocyte adhesion to different surfaces, Proc. Natl. Acad. Sci 91 (21) (1994) 10119–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Van Beek J, Bernaudin M, Petit E, Gasque P, Nouvelot A, MacKenzie ET, Fontaine M, Expression of receptors for complement anaphylatoxins C3a and C5a following permanent focal cerebral ischemia in the mouse, Exp. Neurol 161 (1) (2000) 373–382. [DOI] [PubMed] [Google Scholar]
- [72].Sirko S, Irmler M, Gascón S, Bek S, Schneider S, Dimou L, Obermann J, De Souza Paiva D, Poirier F, Beckers J, Astrocyte reactivity after brain injury—: the role of galectins 1 and 3, Glia 63 (12) (2015) 2340–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dinarello CA, Immunological and inflammatory functions of the interleukin-1 family, Annu. Rev. Immunol 27 (2009) 519–550. [DOI] [PubMed] [Google Scholar]
- [74].Kim YJ, Hwang SY, Oh ES, Oh S, Han IO, IL–1β, an immediate early protein secreted by activated microglia, induces iNOS/NO in C6 astrocytoma cells through p38 mapk and nf-κb pathways, J. Neurosci. Res 84 (5) (2006) 1037–1046. [DOI] [PubMed] [Google Scholar]
- [75].Paré A, Mailhot B, Lévesque SA, Juzwik C, Doss PMIA, Lécuyer M-A, Prat A, Rangachari M, Fournier A, Lacroix S, IL-1β enables cns access to CCR2hi monocytes and the generation of pathogenic cells through gm-csf released by cns endothelial cells, Proc. Natl. Acad. Sci ((2018) 201714 94 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang G-Y, Mao Y, Zhou L-F, Ye W, Liu X-H, Gong C, Betz AL, Attenuation of temporary focal cerebral ischemic injury in the mouse following transfection with interleukin-1 receptor antagonist, Molecular brain research 72 (2) (1999) 129–137. [DOI] [PubMed] [Google Scholar]
- [77].Mulcahy NJ, Ross J, Rothwell NJ, Loddick SA, Delayed administration of interleukin-1 receptor antagonist protects against transient cerebral ischaemia in the rat, Br. J. Pharmacol 140 (3) (2003) 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kovanen PE, Leonard WJ, Cytokines and immunodeficiency diseases: critical roles of the γc-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways, Immunol. Rev 202 (1) (2004) 67–83. [DOI] [PubMed] [Google Scholar]
- [79].Arima N, Kuziel W, Grdina T, Greene W, IL-2-induced signal transduction involves the activation of nuclear NF-kappa b expression, J. Immunol 149 (1) (1992) 83–91. [PubMed] [Google Scholar]
- [80].Schütze S, Wiegmann K, Machleidt T, Krönke M, TNF-induced activation of NF-KB, Immunobiology 193 (2–4) (1995) 193–203. [DOI] [PubMed] [Google Scholar]
- [81].Christophi GP, Hudson CA, Gruber RC, Christophi CP, Mihai C, Mejico LJ, Jubelt B, Massa PT, SHP-1 deficiency and increased inflammatory gene expression in PBMCs of multiple sclerosis patients, Laboratory Investigat. 88 (3) (2008) 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Guo S, Li Z-Z, Gong J, Xiang M, Zhang P, Zhao G-N, Li M, Zheng A, Zhu X, Lei H, Oncostatin m confers neuroprotection against ischemic stroke, J. Neurosci 35 (34) (2015) 12047–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hams E, Colmont CS, Dioszeghy V, Hammond VJ, Fielding CA, Williams AS, Tanaka M, Miyajima A, Taylor PR, Topley N, Oncostatin m receptor-β signaling limits monocytic cell recruitment in acute inflammation, J. Immunol 181 (3) (2008) 2174–2180. [DOI] [PubMed] [Google Scholar]
- [84].Lurie DI, Solca F, Fischer EH, Rubel EW, Tyrosine phosphatase SHP1 immunoreactivity increases in a subset of astrocytes following deafferentation of the chicken auditory brainstem, J. Comparat. Neurol 421 (2) (2000) 199–214. [PubMed] [Google Scholar]
- [85].Tanaka T, Fujita Y, Ueno M, Shultz LD, Yamashita T, Suppression of SHP-1 promotes corticospinal tract sprouting and functional recovery after brain injury, Cell Death Dis. 4 (4) (2013) e567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hsieh CL, Niemi EC, Wang SH, Lee CC, Bingham D, Zhang J, Cozen ML, Charo I, Huang EJ, Liu J, CCR2 deficiency impairs macrophage infiltration and improves cognitive function after traumatic brain injury, J. Neurotrauma 31 (20) (2014) 1677–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sherry B, Tekamp-Olson P, Gallegos C, Bauer D, Davatelis G, Wolpe S, Masiarz F, Coit D, Cerami A, Resolution of the two components of macrophage inflammatory protein 1, and cloning and characterization of one of those components, macrophage inflammatory protein 1 beta, J. Exper. Med 168 (6) (1988) 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Dimitrijevic OB, Stamatovic SM, Keep RF, Andjelkovic AV, Absence of the chemokine receptor CCR2 protects against cerebral ischemia/reperfusion injury in mice, Stroke 38 (4) (2007) 1345–1353. [DOI] [PubMed] [Google Scholar]
- [89].Hammond MD, Taylor RA, Mullen MT, Ai Y, Aguila HL, Mack M, Kasner SE, McCullough LD, Sansing LH, CCR2 + Ly6Chi inflammatory monocyte recruitment exacerbates acute disability following intracerebral hemorrhage, J. Neurosci 34 (11) (2014) 3901–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Izikson L, Klein RS, Charo IF, Weiner HL, Luster AD, Resistance to experimental autoimmune encephalomyelitis in mice lacking the cc chemokine receptor (CCR2), J. Exper. Med 192 (7) (2000) 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sawyer AJ, Tian W, Saucier-Sawyer JK, Rizk PJ, Saltzman WM, Bellamkonda RV, Kyriakides TR, The effect of inflammatory cell-derived MCP-1 loss on neuronal survival during chronic neuroinflammation, Biomaterials 35 (25) (2014) 6698–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hou W, Han J, Lu C, Goldstein LA, Rabinowich H, Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis, Autophagy 6 (7) (2010) 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Krajewska M, You Z, Rong J, Kress C, Huang X, Yang J, Kyoda T, Leyva R, Banares S, Hu Y, Neuronal deletion of caspase 8 protects against brain injury in mouse models of controlled cortical impact and kainic acid-induced excito-toxicity, PLoS One 6 (1) (2011) e24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Kavanagh E, Burguillos MA, Carrillo-Jimenez A, Oliva-Martin MJ, Santiago M, Rodhe J, Joseph B, Venero JL, Deletion of caspase-8 in mouse myeloid cells blocks microglia pro-inflammatory activation and confers protection in Mptp neurodegeneration model, Aging (Albany NY) 7 (9) (2015) 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Moriwaki K, Bertin J, Gough PJ, Chan FK-M, A RIPK3–caspase 8 complex mediates atypical pro–IL-1β processing, J. Immunol (2015) 1402167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR, Catalytic activity of the caspase-8–FLIP l complex inhibits RIPK3-dependent necrosis, Nature 471 (7338) (2011) 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Feltham R, Vince JE, Lawlor KE, Caspase-8: not so silently deadly, Clin. Transl. Immunol 6 (1) (2017) e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA, AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC, Nature 458 (7237) (2009) 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Yogarajah T, Ong KC, Perera D, Wong KT, AIM2 inflammasome-mediated pyroptosis in enterovirus A71-infected neuronal cells restricts viral replication, Sci. Rep 7 (1) (2017) 5845. [DOI] [PMC free article] [PubMed] [Google Scholar]