Editor: Despite the success of antiretroviral therapy (ART), HIV persists and viremia rebounds if ART is interrupted.1 Characterizing proviruses in infected cells that persist on ART is critical to developing strategies to eliminate them. Important studies from a limited number of donors have revealed that the vast majority of proviruses that persist on ART are defective, leaving only a small fraction that are potentially replication competent or “intact”.2–4 Despite the importance of characterizing the proviral landscape, there is no centralized public database dedicated to proviral sequences, nor are there bioinformatics tools available to differentiate defective from sequence intact proviruses. To facilitate our understanding of the genetic structure and dynamics of the HIV reservoir, we developed the Proviral Sequence Database (PSD; https://psd.cancer.gov) for the storage and meta-analyses of HIV sequences before ART, that persist on ART, or that rebounded after ART was interrupted (Fig. 1). The PSD described in this study only includes HIV sequences from donors who provided informed consent in studies with institutional review board approval.
FIG. 1.
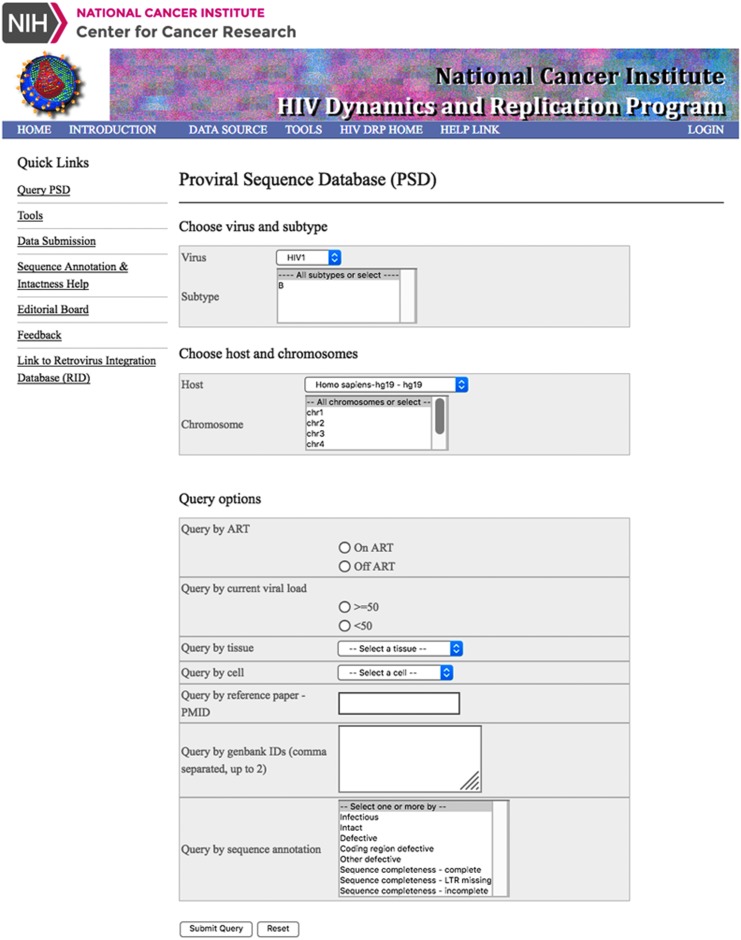
HIV PSD home page. It contains tabs for DATA SOURCE that lists all the articles from which the provirus sequences are collected, and TOOLS tab that provide the link to Pro-Seq IT. The home page also lists query options (Reprinted with permission from National Cancer Institute—Center for Cancer Research). PSD, Proviral Sequence Database.
The PSD was constructed with MySql (https://www.mysql.com), an open source relational database management system. It stores near full-length DNA and RNA HIV sequences from publications along with characteristics of the donor and sample type. HIV sequences are obtained by single-genome techniques and thus are derived from a single template. Proviral data can be queried in the PSD by donor characteristics (e.g., on or off ART and plasma HIV RNA levels), sample type (e.g., PBMC, lymph node, or other tissue or cell type), or sequence characteristics (e.g., intact, infectious, and defective). Query results include downloadable sequence FASTA files with extensive annotation of each sequence, including insertions, deletions, frameshifts, premature stop codons in coding regions, and mutations likely to disrupt function in LTR regulatory elements.5 A built-in tool called the Proviral Sequence Annotation & Intactness Test (Pro-Seq IT) allows users to submit their sequences, obtain the sequence annotations, and computationally infer proviral intactness. Host integration sites of proviruses are included when available.
In summary, the HIV PSD and accompanying tools are available for sequence storage and meta-analyses of HIV RNA and DNA genomes at (https://psd.cancer.gov). This resource and the analyses derived from it should contribute to the understanding of HIV persistence on ART and may reveal new curative strategies. PSD will be regularly updated and extended to include other retroviruses and tools as they become available.
Authors' Contributions
All authors participated in the design of the database. W.S., J.W.M., J.M.C., and M.F.K. involved in data interpretation. W.S. designed and constructed the database structure, wrote Proviral Sequence Annotation & Intactness Test (Pro-Seq IT) tool, and participated in the web application design. J.S. participated in the database structure design and constructed the web application of the database and Pro-Seq IT. E.K.S. provided testing sequences and analysis. W.S.H. contributed to the development of the intactness tool.
Author Disclosure Statement
J.W.M. is a consultant to Gilead Sciences and a shareholder of Co-Crystal, Inc. The remaining authors have no potential conflicts.
Funding Information
We acknowledge the funding sources for this study from NCI CCR, the Office of AIDS Research, NIH, NCI intramural funding and OAR funding to M.F.K., and NCI contract no. HHSN261200800001E to Leidos Biomedical Research, Inc. J.W.M. is supported by NCI contract no. HHSN261200800001E and by grants from NIAID (UM1AI126603) and the Bill and Melinda Gates Foundation (OPP1115715). J.M.C. was a research professor of the American Cancer Society, and was supported by Leidos contract l3XS110.
References
- 1. Chun TW, Stuyver L, Mizell SB, et al. : Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997;94:13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ho YC, Shan L, Hosmane NN, et al. : Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013;155:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruner KM, Murray AJ, Pollack RA, et al. : Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016;22:1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hiener B, Horsburgh BA, Eden JS, et al. : Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep 2017;21:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuzembayeva M, Dilley K, Sardo L, Hu WS: Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 2014;454–455:362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]