Abstract
Background
Toothache often occurs with pulpitis. Lipopolysaccharide (LPS) is produced by gram-negative bacteria, and its accumulation is related to clinical symptoms of pain. MicroRNAs (miRNAs) display anti-inflammatory potential due to their direct regulation of cellular protein expression, which can promote inflammatory changes in dental pulp tissues. However, the mechanism of LPS-induced pulpitis is still unclear.
Material/Methods
In this study, dental pulp stem cells (DPSCs) were separated and cultured from rat dental pulp tissues; then, LPS was administered to induce inflammation and activate the TLR4 pathway.
Results
It was found that miR-506 was upregulated following LPS treatment in DPSCs. The inhibition of miR-506 in LPS-treated DPSCs led to attenuated inflammation and deactivation of the TLR4 pathway. Furthermore, the bioinformatic analysis and dual-luciferase reporter gene assay indicated that miR-506 could target the 3′-UTR of sirtuin 1 (SIRT1). Additionally, SIRT1 decreased in LPS-treated DPSCs, and miR-506 transfection resulted in SIRT1 upregulation. SIRT1 overexpression showed a similar inhibitory effect as that of miR-506 downregulation on inflammation and TLR4 activation in DPSCs.
Conclusions
In brief, miR-506 can protect dental pulp in LPS-induced inflammation by inhibiting the SIRT1-mediated TLR4 pathway.
MeSH Keywords: Inflammation, Lipopolysaccharides, Pulpitis
Background
Toothache during or after treatment is the primary cause of irregular visits in a dental clinic. Therefore, it is necessary to properly treat toothaches before, during, and after the treatment of dental diseases. However, toothache treatment is often challenging. As the neurobiological mechanism that causes toothaches is unclear, it is therefore difficult to develop mechanism-based therapies.
Recently, studies on microbial infection in dental pulp tissues have demonstrated that microRNAs (miRNAs) are an important regulator of inflammatory response in dental pulp stem cells (DPSC) [1,2]. First discovered in 1993, miRNA is a type of non-coding small RNA [3]. Recent studies have shown that ectopic miRNA expression is related to the progression of various malignant tumors. miRNA is a target for molecular therapy of malignant tumors and other diseases because it can be used as both an oncogene and tumor inhibitor [4]. A previous study has confirmed that multiple miRNA members can be differentially expressed in inflamed human pulps [4], including upregulated miRNAs related to the development of pulpitis and downregulated miRNAs with the potential to treat pulpitis.
As an NAD+-dependent lysine deacetylase, sirtuin 1 (SIRT1) plays a key role in metabolism, inflammation, and aging [5]. At the same time, SIRT6 is associated with the treatment of LPS-induced pulpitis [6]. The expression of SIRT6 in human DPSCs was downregulated using LPS treatment. MTT and LDH assays have shown that LPS-induced cell death was decreased by SIRT6 overexpression in human DPSCs. SIRT6 can protect human DPSCs from apoptosis, which is consistent with our results. SIRT1 may be mediated by Berberine to inhibit LPS-stimulated RAW264.7 macrophage inflammation [7]. However, a comprehensive study has yet to be conducted on the impact of SIRT1 on pulpitis or LPS-induced inflammation in DPSCs. In the present study, an in vitro pulpitis model was constructed to demonstrate the influence of SIRT1 on the inflammation of DPSCs, which was induced by injection of LPS. The roles of miR-506 on DPSC inflammation and inflammation-related TLR4 signaling activity were evaluated.
Material and Methods
Cell culture
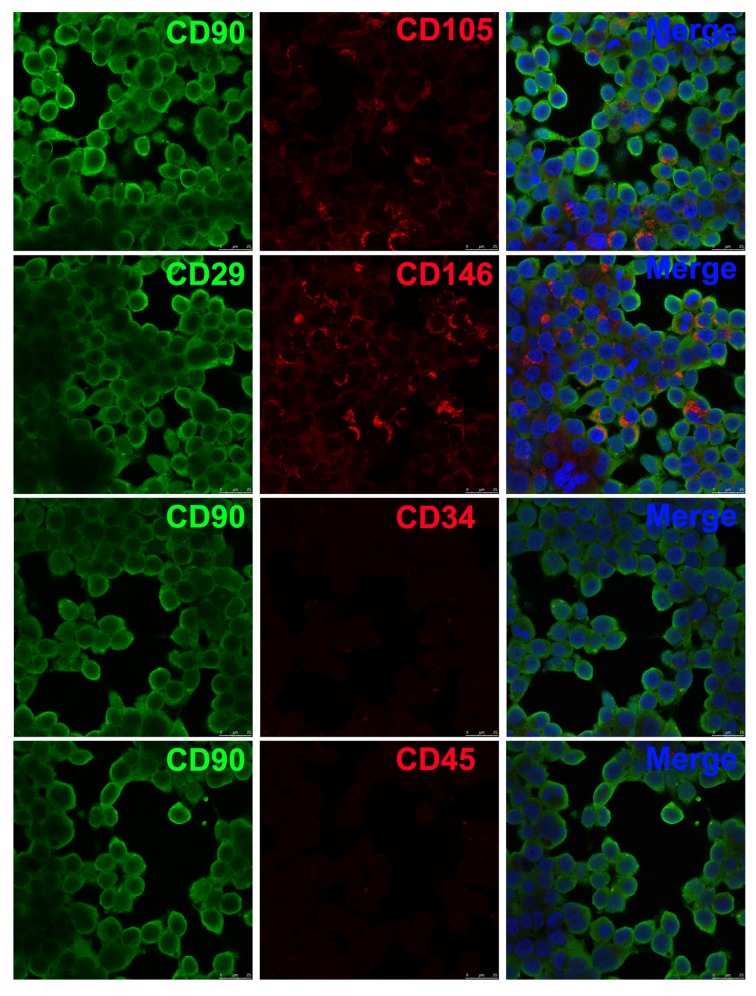
SD rats aged 8 weeks old were killed by intravenous injection of excessive pentobarbital sodium, and dental tissues were collected. After removal of soft tissues, dentinal cartilage was incised to expose dental pulp tissues. Collagenase I (3 mg/mL) and neutral protease (3 mg/mL) were used for the lysis and culture of pulp tissues for 20 min at 37°C. Tissues were centrifuged for 7 min at 309×g and then filtered. The procedure was repeated until tissues were completely digested. Cells were re-suspended and kept at 37°C with 5% CO2. The expressions of CD29, CD90, CD146, CD105, CD34, and CD45 [8–10] were determined with immunofluorescence method. All animal experiments were carried out based on the Animal Ethics Committee and Guidelines for Care and Use of Laboratory Animals, the Second People’s Hospital of Lanzhou.
Cell administration
To induce inflammatory response, LPS (10 μg/mL) was used to incubate cells for 24 h. To validate the role of SIRT1 and miR-506 on DPSC inflammation, the DPSCs were further transfected with pcDNA3-SIRT1 and miR-506 inhibitor for 24 h following LPS administration.
Immunofluorescence assay
DPSCs were inoculated into a 24-well plate and fixed with 4% poly-formaldehyde for 15 min. DPSCs were permeated for 30 min with 0.1% Triton X-100, cultured with 10% goat serum for 15 min at room temperature, and treated with the first antibody overnight at 4°C. DPSCs were rinsed 3 times with PBS and then incubated with secondary Cy3-labeled antibody in the dark for 1 h at room temperature. Then, the cells were dyed with DAPI for 15 min. The images were magnified 400 times by a fluorescent microscope.
Western blotting (WB)
DPSCs were lysed with cell lysis buffer. Protein was determined by a bicinchoninic analysis kit, separated by 10% SDS-PAGE, and then transferred to a PVDF membrane. Tween 20 was added to bovine serum albumin (BSA; 5%) phosphate buffer to block non-binding sites on the membrane for 1 h. Protein was cultured with the first antibody overnight at 4°C, and the secondary antibody was bound to peroxidase (Amersham ECL). The protein bands were stained, and the gray values were measured on a C-DiGit Blot Scanner.
RNA extraction and Q-PCR
Total RNA was extracted, and GAPDH was applied as an internal reference. Then, qRT-PCR was conducted using a SYBR-Green Kit to detect RNA in a 20 μL system under the following conditions: pre-denaturation (95°C, 10 min), denaturation (95°C, 15 s, 40 cycles), annealing (60°C, 30 s), and extension (72°C, 30 s). Quantitative analysis was based on the 2−ΔΔCT method and normalized to GAPDH.
Dual-luciferase reporter gene assay (DLRGA)
The target gene of miR-506 was detected using DLRGA in the 3′-UTR of wild-type (WT) and mutant-type (MU) SIRT1. The Renilla and firefly luciferase sequences were used for luciferase reporter luminescence (Rluc) and calibration luminescence (Luc) analyses, respectively. The cells were cultured with vectors and miRNA mimic for 24 h.
Statistical analysis
Results are expressed as the mean±SD. Comparisons were analyzed by one-way analysis of variance (ANOVA) or 2-tailed t test. P<0.05 represents a significant difference.
Results
Purification and feature of DPSCs
DPSCs were extracted and purified. Immunofluorescence assay suggested that some cells (CD90, CD105, CD29, and CD146) were positive and some (CD34 and CD45) were negative. DPSCs possessed surface antigenic features (Figure 1) [11].
Figure 1.
DPSC identification using immunofluorescence. DPSCs were separated from dental tissues of SD rats, and immunofluorescence assay was applied for analysis of the molecular surface antigen markers. The confocal results showed that some cells (CD90, CD105, CD29, and CD146) were positive and some (CD34 and CD45) were negative. Magnification: 400×.
LPS triggered pro-inflammatory cytokine expression and TLR4-NFκB p65 activation in DPSCs
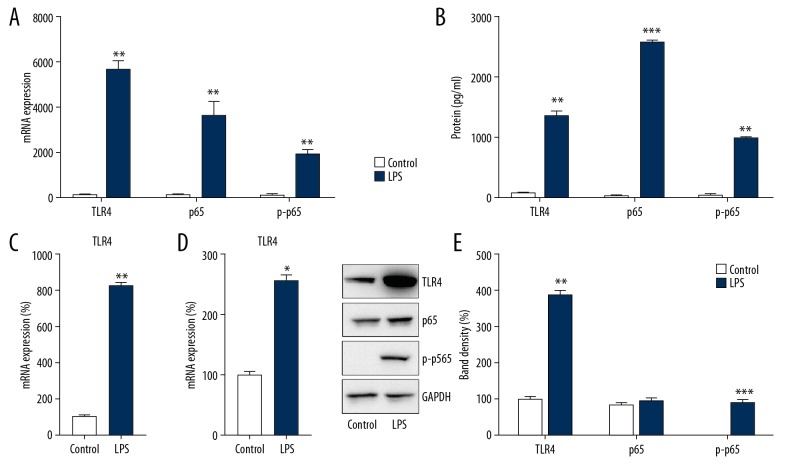
To explore DPSC inflammation following LPS incubation, pro-inflammatory cytokine expression was assessed in DPSCs. The results of Q-PCR and ELISA showed that after pretreatment, LPS could significantly increase the mRNA and protein expressions of IL-1β, IL-6, and TNF-α in DPSCs (p<0.01) (Figure 2A, 2B).
Figure 2.
LPS triggered inflammation and activation of TLR4-NFκB p65 signaling in DPSCs. The experimental group and the control group were treated with LPS and PBS for 8 h. After lysis, the expressions of IL-1β, IL-6, and TNF-α were assessed by (A) Q-PCR and (B) ELISA. (C, D) Cell lysates were subjected to Q-PCR to assess TLR4 and NFκB p65 expression. (E) WB analysis was performed to probe with antibodies for phosphorylated p65, total p65, and TLR4 in cell lysates. Data are represented as means±SD. * P<0.05; ** P<0.01; *** P<0.001.
As activation of the TLR4-NFκB p65 pathway is crucial for triggering cytokine expression to counteract LPS treatment [12], the expressions of TLR4 and p65 after LPS treatment were studied. Q-PCR results showed that LPS could significantly upregulate the mRNA expressions of TLR4 and p65 (p<0.05) (Figure 2C, 2D). Moreover, the WB results indicated that protein levels of TLR4 and p65 were increased following LPS incubation. Meanwhile, the phosphorylation level of p65, a key signal in the NFκB pathway, was also augmented (Figure 2E). These results suggest that LPS treatment triggered inflammatory reactions in DPSCs by stimulating the TLR4-NFκB p65 pathway.
In LPS-stimulated DPSCs, miR-506 was upregulated while SIRT1 was reduced
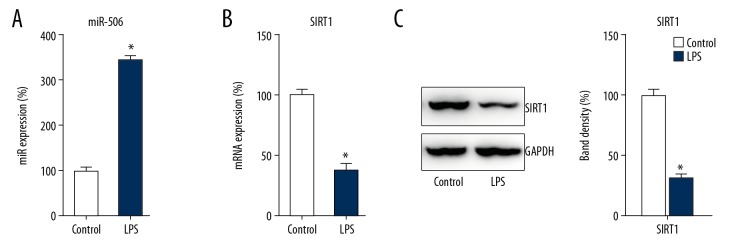
Previous research showed that miR-506 and SIRT1 are involved in LPS-induced host defense gene expression [7,13]. Therefore, Q-PCR and WB were utilized to detect the expressions of miRNA and SIRT1 after LPS stimulation. It was revealed that miR-506 was significantly upregulated in DPSCs at 8 h after LPS treatment (p<0.05) (Figure 3A). Meanwhile, Q-PCR and WB results showed that SIRT1 was downregulated in DPSCs due to LPS stimulation (p<0.05) (Figure 3B, 3C), suggesting that these 2 signals serve a potential role in LPS-treated DPSCs.
Figure 3.
LPS triggered upregulation of miR-506 and downregulation of SIRT1. DPSCs and the control group were treated with LPS or PBS for 8 h. The expressions of miR-506 and SIRT1 at 8 h after LPS treatment were detected by (A, B) Q-PCR and (C) WB. Data are represented as means±SD. * P<0.05.
LPS-induced inflammation of DPSCs required miR-506
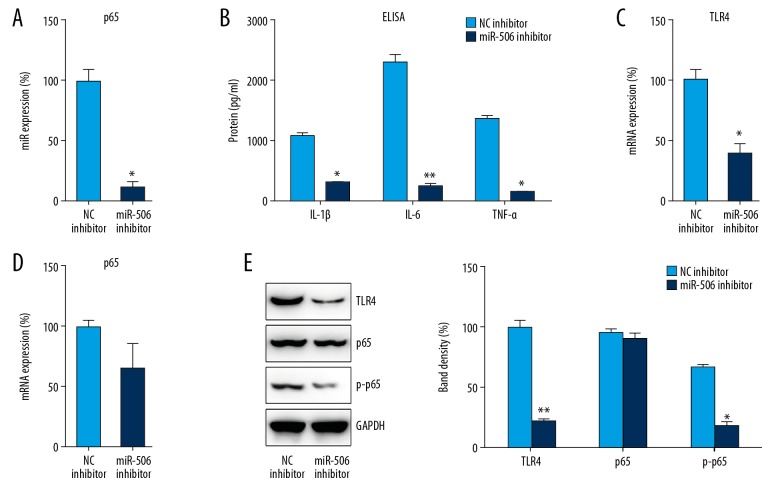
In LPS-treated DPSCs, the effect of miR-506 on LPS-induced inflammation was evaluated via the transfection of miR-506 inhibitor (p<0.05) (Figure 4A). ELISA analysis revealed that, compared with the NC group, cytokine production was obviously decreased in the miR-506 inhibition group (p<0.05) (Figure 4B). We then assessed the expression of TLR4 and p65 in DPSCs after transfection and stimulation. Q-PCR and WB indicated that TLR4 and p65 expression, as well as p65 phosphorylation, were suppressed after transfection with miR-506 inhibitor, especially for TLR4 expression (p<0.05) (Figure 4C–4E), indicating that miR-506 confers a protective effect in the inflammatory reactions of DPSCs.
Figure 4.
The inhibition of miR-506 repressed LPS-induced inflammation in DPSCs. DPSCs were transfected with miR-506 inhibitor or NC inhibitor for 24 h followed by LPS treatment for 8 h. (A) Q-PCR was used to examine miR-506 levels in cell lysates. (B) ELISA was conducted to assess IL-1β, IL-6, and TNF-α expression. (C, D) Cell lysates were subjected to Q-PCR to assess TLR4 and NFκB p65 expression. (E) WB analysis was performed to probe with antibodies for phosphorylated p65, total p65, and TLR4 in cell lysates. Data are represented as means±SD. * P<0.05; ** P<0.01.
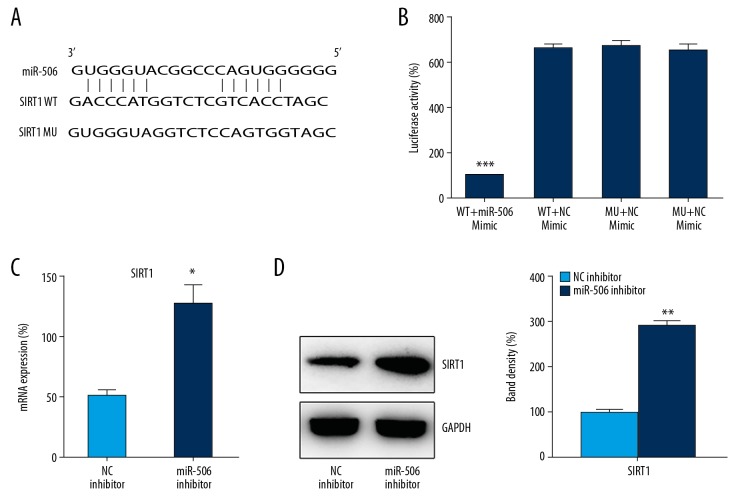
The 3′-UTR of SIRT1 was targeted by miR-506
Bioinformatic analysis suggested that miR-506 can target the 3′-UTR of SIRT1 (Figure 5A). DLRGA was then applied to analyze the relationship between miR-506 and the 3′-UTR of SIRT1 (Figure 5B). Compared with other control groups, the activity of luciferase binding to the 3′-UTR of SIRT1 was inhibited (70%) by transfection with miR-506 mimic (p<0.001). To further validate the relationship between SIRT1 expression and miR-506 in LPS-treated DPSCs, we evaluated SIRT1 expression in cells inhibited with miR-506. SIRT1 expression was significantly upregulated in LPS-treated DPSCs containing transfected miR-506 inhibitor, as determined by Q-PCR and WB (p<0.05) (Figure 5C, 5D). These results demonstrated that miR-506 potentially targeted the 3′-UTR of SIRT1.
Figure 5.
The 3′-UTR of SIRT1 is a target of miR-506. (A) Illustration of the conserved binding motifs of miR-506 in the 3′-UTR of WT SIRT1. (B) DLRGA contained the wild-type (WT) and mutant-type (MU) 3′-UTR of human SIRT1 after miR-506 mimic transfection. Luciferase activity was normalized to β-galactosidase activity. (C) Q-PCR and (D) WB analyses determined the expression of SIRT1 mRNA in LPS-treated DPSCs transfected with miR-506 or NC inhibitor. Data are represented as means±SD. * P<0.05; *** P<0.001.
Effects of SIRT1 on LPS-induced inflammation and TLR4 pathway activation in DPSCs
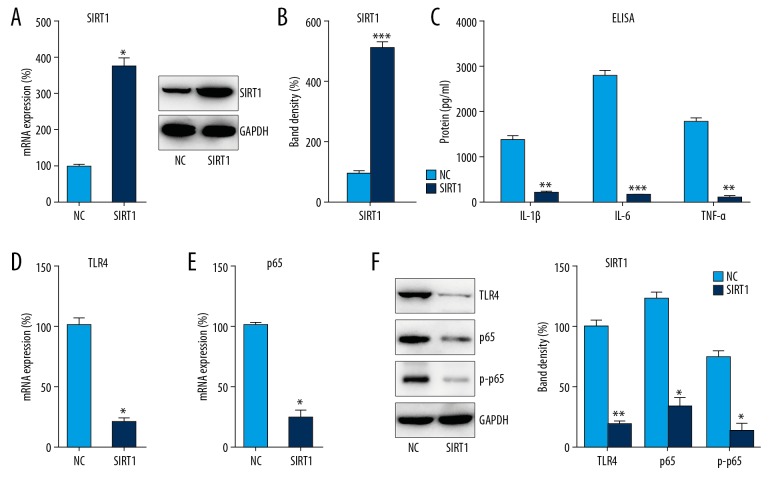
To further examine the influence of SIRT1 on LPS-induced inflammation in DPSCs, cells were transfected with pcDNA3-SIRT1, which expresses SIRT1, or the NC plasmid, showing that the mRNA and protein expressions of SIRT1 were obviously increased after transfection (p<0.05) (Figure 6A, 6B). We also found that pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) were markedly decreased after DPSCs transfected with pcDNA3-SIRT1 according to the results of ELISA. In addition, downregulated expression of TLR4 and NFκB p65 were observed with SIRT1 overexpression (p<0.05) (Figure 6C–6F), while p65 phosphorylation was also decreased, suggesting that SIRT1 overexpression counteracts the effect of LPS on DPSC inflammation or pulpitis.
Figure 6.
SIRT1 overexpression repressed LPS-induced inflammation in DPSCs. DPSCs were transfected with pcDNA3-SIRT1 or NC plasmid for 24 h followed by LPS treatment for 8 h. (A) Q-PCR and (B) WB analyses were used to examine SIRT1 levels in cell lysates. (C) ELISA was conducted to assess IL-1β, IL-6, and TNF-α expression. (D, E) Cell lysates were subjected to Q-PCR to assess TLR4 and NFκB p65 expression. (F) WB analysis was performed to probe with antibodies for phosphorylated p65, total p65, and TLR4 in cell lysates. Data are represented as means±SD. * P<0.05; ** P<0.01; *** P<0.001.
Discussion
As a single miRNA has different targets, the mechanism of miRNAs regulating inflammation is complex. Several studies have explored the relationship between miRNAs and inflammation-related genes in dental pulp tissues. Our results showed that miR-506, a differentially expressed miRNA, was a pro-inflammatory factor during the development of pulpitis. We further explored the specific mechanism of miR-506 in protecting pulp tissue from inflammation, and concluded that miR-506 inhibited inflammation induced by LPS in dental pulp cells in vitro, and the specific mechanism was that miR-506 could inhibit the activation of TLR4-NFκB signaling through SIRT1 inhibition.
The regulation of mRNA transcription and expression is complex and is affected by many factors [14]. This study focused on upstream mRNA regulators of SIRT1, and bioinformatics analysis was used to predict upstream genes regulating SIRT1. Previous research demonstrated the possibility that SIRT1 interacts with the TLR4-NFκB pathway. Lin et al. reported that SIRT1 could regulate LPS-induced CD40 expression in renal medullary collecting duct cells. LPS can downregulate SIRT1 expression and upregulate the expression of CD40, TLR4, and phosphorylated NFκB p65 in time- and dose-dependent manners. The expression levels of CD40, TLR4, and p-NFκB p65 were decreased by SIRT1720-induced SIRT1 overexpression, but were increased by SIRT1 overexpression induced by SIRT1 siRNA [15]. A study by Li et al. explored the effects of SIRT1 on periodontal ligament fibroblasts (PDLFs) injured by LPS [16] and found that cell viability was increased, cell apoptosis was decreased, and pro-inflammatory cytokine levels (IL-1α, IL-6, IL-8, and TNF-α) were decreased by SIRT1 overexpression in LPS-injured PDLFs. The TLR4 and JNK/NFκB pathways were downregulated by SIRT1 overexpression. SIRT1 can downregulate the HMGB 1/TLR4 pathway to alleviate murine allergic rhinitis [17]. These previous studies have shown that miR-506 can regulate the upstream region of SIRT1. The miRNAs can cut mRNA and act as a translation inhibitor, thus producing a negative feedback effect on their target RNA. Therefore, miRNAs are the primary regulator of human physiology, development, and disease. It has been confirmed that miRNA quantity is a biomarker of specific diseases [18, 19]. Previous studies showed that miR-506 may be involved in many activities, such as inflammation [13], cardiomyocyte dysfunction [20], malignant tumor development [21–25], stem cell differentiation [26], and epithelial-mesenchymal transition [27], and have effects on primary biliary cirrhosis [28], rheumatoid arthritis [29], and blood vessel regeneration [30]. Therefore, miR-506 is associated with various cell biological processes, and this is consistent with the results of the present study. In the pulpitis cell model, miR-506 and SIRT1 were significantly imbalanced, indicating that there was an antagonistic relationship between them in inflammation. The hypothesis that LPS can upregulate miR-506 expression and downregulate SIRT1 expression in vitro was confirmed by our study. LPS-induced DPSC inflammation can be alleviated by miR-506 overexpression. On the molecular level, LPS-induced activation of TLR4-NFκB signaling can be inhibited by miR-506 overexpression, which inhibited SIRT1 expression and hence suppressed the phosphorylation of NFκB p65. These results confirmed the pro-inflammatory function of miR-506 and the anti-inflammatory function of SIRT1.
Conclusions
Our results suggest that miR-506 is a positive regulator of pulpitis. The expression of miR-506 was high while that of SIRT1 was low in LPS-treated DPSCs, which were closely associated with pro-inflammatory cytokines and activation of the TLR4-NFκB pathway.
Footnotes
Conflicts of interests
None.
Source of support: Departmental sources
References
- 1.Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: Novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–40. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284:4667–78. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong S, Zhang S, Bair E, et al. Differential expression of microRNAs in normal and inflamed human pulps. J Endod. 2012;38:746–52. doi: 10.1016/j.joen.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Zhong S, Zhang S, Bair E, et al. Differential expression of microRNAs in normal and inflamed human pulps. J Endod. 2012;38:746–52. doi: 10.1016/j.joen.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Sebastian C, Satterstrom FK, Haigis MC, Mostoslavsky R. From sirtuin biology to human diseases: An update. J Biol Chem. 2012;287:42444–52. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Bai L, Ren Q, et al. Protective effects of SIRT6 against lipopolysaccharide (LPS) are mediated by deacetylation of Ku70. Mol Immunol. 2018;101:312–18. doi: 10.1016/j.molimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Shan Y, Wu Y, et al. Berberine suppresses LPS-induced inflammation through modulating Sirt1/NF-kappaB signaling pathway in RAW264.7 cells. Int Immunopharmacol. 2017;52:93–100. doi: 10.1016/j.intimp.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 8.Matsui M, Kobayashi T, Tsutsui TW. CD146 positive human dental pulp stem cells promote regeneration of dentin/pulp-like structures. Hum Cell. 2018;31:127–38. doi: 10.1007/s13577-017-0198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledesma-Martínez E, Mendoza-Núñez VM, Santiago-Osorio E. Mesenchymal stem cells derived from dental pulp: A review. Stem Cells Int. 2016;2016 doi: 10.1155/2016/4709572. 4709572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan H, Zhang H, Hong L, et al. MicroRNA let-7c-5p suppressed lipopolysaccharide-induced dental pulp inflammation by inhibiting dentin matrix protein-1-mediated nuclear factor kappa B (NF-κB) pathway in vitro and in vivo. Med Sci Monit. 2018;24:6656–66. doi: 10.12659/MSM.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Jiang W, Luo Z, et al. IFN-γ regulates human dental pulp stem cells behavior via NF-κB and MAPK signaling. Sci Rep. 2017;7:40681. doi: 10.1038/srep40681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Ann Rev Immunol. 2014;32:461–88. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 13.Erice O, Munoz-Garrido P, Vaquero J, et al. MicroRNA-506 promotes primary biliary cholangitis-like features in cholangiocytes and immune activation. Hepatology. 2018;67:1420–40. doi: 10.1002/hep.29533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Yao X, Wang G. ‘Mediator-ing’ messenger RNA processing. Wiley Interdiscip Rev RNA. 2015;6:257–69. doi: 10.1002/wrna.1273. [DOI] [PubMed] [Google Scholar]
- 15.Lin QQ, Geng YW, Jiang ZW, Tian ZJ. SIRT1 regulates lipopolysaccharide-induced CD40 expression in renal medullary collecting duct cells by suppressing the TLR4-NF-kappaB signaling pathway. Life Sci. 2017;170:100–7. doi: 10.1016/j.lfs.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Li K, Lv G, Pan L. Sirt1 alleviates LPS induced inflammation of periodontal ligament fibroblasts via downregulation of TLR4. Int J Biol Macromol. 2018;119:249–54. doi: 10.1016/j.ijbiomac.2018.07.099. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y, Liu Q, Zhao J, et al. SIRT1 attenuates murine allergic rhinitis by downregulated HMGB 1/TLR4 pathway. Scand J Immunol. 2018;87:e12667. doi: 10.1111/sji.12667. [DOI] [PubMed] [Google Scholar]
- 18.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–76. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Varshney J, Subramanian S. MicroRNAs as potential target in human bone and soft tissue sarcoma therapeutics. Front Mol Biosci. 2015;2:31. doi: 10.3389/fmolb.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Liu F, Wang Q, Geng Y. Overexpressed microRNA-506 and microRNA-124 alleviate H2O2-induced human cardiomyocyte dysfunction by targeting kruppel-like factor 4/5. Mol Med Rep. 2017;16:5363–69. doi: 10.3892/mmr.2017.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zu C, Liu T, Zhang G. MicroRNA-506 inhibits malignancy of colorectal carcinoma cells by targeting LAMC1. Ann Clin Lab Sci. 2016;46:666–74. [PubMed] [Google Scholar]
- 22.Cheng R-F, Wang J, Zhang J-Y, et al. MicroRNA-506 is up-regulated in the development of pancreatic ductal adenocarcinoma and is associated with attenuated disease progression. Chin J Cancer. 2016;35:64. doi: 10.1186/s40880-016-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G, Yang D, Rupaimoole R, et al. Augmentation of response to chemotherapy by microRNA-506 through regulation of RAD51 in serous ovarian cancers. J Natl Cancer Inst. 2015;107(7) doi: 10.1093/jnci/djv108. pii: djv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao WJ, Wang YL, Lu JG, et al. MicroRNA-506 inhibits esophageal cancer cell proliferation via targeting CREB1. Int J Clin Exp Pathol. 2015;8:10868–74. [PMC free article] [PubMed] [Google Scholar]
- 25.Deng J, Lei W, Xiang X, et al. MicroRNA-506 inhibits gastric cancer proliferation and invasion by directly targeting Yap1. Tumour Biol. 2015;36:6823–31. doi: 10.1007/s13277-015-3364-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Jiaqi C, Zhaoying C, Huimin C. MicroRNA-506-3p regulates neural stem cell proliferation and differentiation through targeting TCF3. Gene. 2016;593:193–200. doi: 10.1016/j.gene.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, Xu Q, Yan S, et al. Suppression of forkhead box Q1 by microRNA-506 represses the proliferation and epithelial-mesenchymal transition of cervical cancer cells. Oncol Rep. 2016;35:3106–14. doi: 10.3892/or.2016.4651. [DOI] [PubMed] [Google Scholar]
- 28.Banales JM, Saez E, Uriz M, et al. Up-regulation of microRNA 506 leads to decreased Cl−/HCO3− anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology. 2012;56:687–97. doi: 10.1002/hep.25691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Zhou Q, Hu G, Wang G. MicroRNA-506 inhibits rheumatoid arthritis fibroblast-like synoviocytes proliferation and induces apoptosis by targeting TLR4. Biosci Rep. 2019;39(5) doi: 10.1042/BSR20182500. pii: BSR20182500. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Yi F, Hao Y, Chong X, Zhong W. Overexpression of microRNA-506-3p aggravates the injury of vascular endothelial cells in patients with hypertension by downregulating Beclin1 expression. Exp Ther Med. 2018;15:2844–50. doi: 10.3892/etm.2018.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]