Abstract
Superoxide dismutases (SODs) are highly efficient enzymes for superoxide dismutation and the first line of defense against oxidative stress. These metalloproteins contain a redox active metal ion in their active site (Mn, Cu, Fe, Ni) with a tightly controlled reduction potential found in a close range around the optimal value of 0.36 V vs. NHE. Rationally designed proteins with well-defined three-dimensional structures offer new opportunities for obtaining functional SOD mimics. Here we explore four different copper binding scaffolds: H3 (His3), H4 (His4), H2DH (His3Asp with 2 His and 1 Asp in the same plane) and H3D (His3Asp with 3 His in the same plane) using the scaffold of the de novo protein GRα3D. EPR and XAS analysis of the resulting copper complexes demonstrates that they are good Cu(II) bound structural mimics of Cu-only SODs. Furthermore, all the complexes exhibit SOD activity, though three orders of magnitude slower than the native enzyme, making them the first de novo copper SOD mimics.
Keywords: bioinorganic chemistry, enzyme models, metalloenzymes, metalloproteins, protein design
Introduction
Superoxide dismutases (SODs) are highly conserved metalloenzymes that have evolved to protect organisms from oxidative stress.1, 2 Four types of SOD have been identified, but only three have been extensively studied (Table 1).1, 3 The first well characterized class of SODs is the Fe/MnSOD, which can function with iron only, manganese only, or either metal in the case of cambialistic enzymes. These proteins have high sequence identity and the same His3Asp metal binding site regardless of the active metal.4 The second is the NiSOD, which contains a His2Cys binding site.5 The third type of well characterized SODs are Cu/ZnSODs. General features of Cu/ZnSODs include a Greek key β-barrel backbone, an electrostatic loop, a disulfide bond, and a conserved active site.1–3 In its reduced state, the catalytic copper is bound by three histidines in a trigonal plane. Upon oxidation, the Cu(II) is bound in a distorted square pyramid by an additional, bridging histidine and a water molecule. The bridging histidine also binds to the structural zinc ion, which is further coordinated by two other histidines and an aspartate.1 Interestingly, a fourth class of SOD, Cu-only SOD, has recently been described containing a single copper metal ion in the active site.6–10 In these enzymes, the two histidines chelating the zinc ion in Cu/Zn SOD are missing, either due to substitution or deletion. Thus, the active site contains only four histidines capable of chelating copper. All four His residues coordinate Cu(II) in a pseudotrigonal pyramid, while Cu(I) is coordinated by only three His residues in a pseudotrigonal planar arrangement with the fourth His at a longer distance.11 In C. albicans Cu-only SOD5, the role of the zinc ion in promoting pH-independent catalysis is adopted by a glutamate residue (Glu110) that interacts through H-bonding with the bridging histidine.10
Table 1.
Summary of SOD activities.
Despite their differences in structure, active site, and metal center, all SODs catalyze superoxide dismutation at diffusion limited rates between pH 4 and 10.1 Their reduction potentials are tightly controlled and fall in a close range around the optimal value of 0.36 V vs. NHE at physiological pH, corresponding to the midpoint potential between the oxidation (−0.18 V vs. NHE) and the reduction (+0.91 vs. NHE) of superoxide.1
Low-molecular weight complexes mimicking SOD with a manganese, iron, or copper metal ion have been thoroughly described in the literature.2, 16–24 The challenges faced in their design include stability, flexibility to adapt to coordination of different metal redox states, and tuning the reduction potential to enable superoxide dismutation. For Cu/Zn SOD mimics, cyclodextrin22, 25 and bisdioxocyclan derivatives26, 27 are among the most efficient SOD mimics reported, with kcat only ten fold lower than that of the native enzyme under similar conditions. Peptidic Cu/Zn SOD mimics have also been studied,15, 28–36 with the aim to reproduce the active site of the enzyme using short sequences of amino acids (3 to 10 residues) that contain two to four histidine moieties. One of the most active peptidic mimics of Cu/ZnSOD was reported by Árus et al.15 These unstructured peptide sequences contain three histidines (HADHDHKK) and bind copper in a 1:1 ratio. At pH 7.0 its kcat is 2.7 107 M−1s−1, which is only two orders of magnitude lower than that of the native Cu/Zn SOD. As many of these peptidic models are mononuclear Cu catalysts, they can provide some amount of insight into the Cu-only SOD system, but no models of Cu-only SOD have previously been described.
Widening possibilities beyond low-molecular weight complexes, rationally designed self-assembling peptidic scaffolds with well-defined secondary and tertiary structures are tools of choice to mimic the structure and activity of an enzyme.37–45 Only one manganese SOD mimic has been reported using this type of construct with modest SOD activity (kcat = 3.7 105 M−1s−1 at pH 7.4).46 Using protein redesign Benson et al. have obtained a functional iron SOD mimic by introducing a His3 metal binding site and a pocket for O2 binding into E. coli thioredoxin (kcat = 6.4 106 M−1s−1 at pH 8).47, 48
Herein, the GRα3D de novo protein scaffold was used to design a functional mimic of Cu-only SOD. The α3D family of proteins, originally designed by DeGrado et al, consists of seven amino acid repeats in which the first and fourth residue of each heptad is hydrophobic.49 In solution these hydrophobic residues collapse to form the core of an antiparallel three helix bundle.50, 51 Metal binding residues can then be introduced into this core to construct metalloproteins, though a loss of stability is incurred.52–54 This scaffold has previously been used to study both electron transfer proteins, such as cupredoxins and rubredoxins, and catalytic proteins, such as carbonic anhydrase.52, 54–57 An elongated version of this peptide, GRα3D, was designed with an additional heptad for improved thermodynamic stability.58 Four binding sites were introduced into the protein; H3 (His3), H4 (His4), H2DH (His3Asp with two His and one Asp in the same plane), and H3D (His3Asp with three His in the same plane) (Table 2). The stability of the apo- and Cu(II)-protein was studied by thermal denaturation and the complexes were characterized by EPR, EXAFS, and CV experiments. Their SOD activity was assessed by the indirect assay of McCord-Fridovich.59–61
Table 2.
Designed protein amino acid sequences with mutations from GRα3D in bold and active site residues in blue. Electrochemical studies were performed with derivatives having a terminal Cys rather than Ala (A98C) indicated by A/C.
| Peptide | Sequence |
|---|---|
| GRα3D H3 | MGSWAEFKQRLAAIKTRLAAIKSRHDALGGSEAELAAHEKEIAAFESEIAAFESELQAYKGKGNPEVEALRKEAAAIRDEAAAIRDEHQAYRLNGSGA/C |
| GRα3D H4 | MGSWAEFKQRLAAIKTRLAAIKSRHDALGGSEAEHAAHEKEIAAFESEIAAFESELQAYKGKGNPEVEALRKEAAAIRDEAAAIRDEHQAYRLNGSGA/C |
| GRα3D H2DH | MGSWAEFKQRLAAIKTRLAAIKSRHDALGGSEAEHAADEKEIAAFESEIAAFESELQAYKGKGNPEVEALRKEAAAIRDEAAAIRDEHQAYRLNGSGA/C |
| GRα3D H3D | MGSWAEFKQRLAAIKTRLAAIKSRHDALGGSEAEDAAHEKEIAAFESEIAAFESELQAYKGKGNPEVEALRKEAAAIRDEAAAIRDEHQAYRLNGSGA/C |
Results
Protein Design
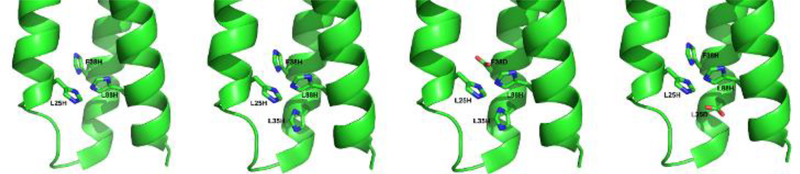
Four constructs were built within the GRα3D de novo protein scaffold with varying active sites meant to recapitulate that of two different classes of SODs, Cu only SOD and Fe/Mn SOD. Their sequences are listed in Table 2. Constructs with an A98C mutation were used for electrochemical experiments. The four different active sites designed within GRα3D for this study are represented in Figure 1. GRα3D H3 contains three histidines that replace leucine or phenylalanine (L25H, F38H, L88H) in positions analogous to the carbonic anhydrase mimic α3DH3, which also models the Cu(I) binding site of Cu only SOD.52 GRα3D H4 contains a fourth histidine in position 35 (L35H) to mimic the Cu(II) binding site of Cu-only SOD.6, 7 Constructs in which the fourth His residue was substituted by an Asp were created to test the effects of modulating the reduction potential of the bound Cu. To this end GRα3D H2DH and GRα3D H3D, in which the Asp is either positioned in the same plane as two other His (L38D), or below a plane constituted by the three His (L35D), respectively, were designed. These constructs also model the active site of Fe/Mn SOD, providing interesting insight into the selectivity and catalytic efficiency of substituting copper into this system.
Figure 1.
PyMol models of the designed His3 (H3), His4 (H4), His2AspHis (H2DH), and His3Asp (H3D) active sites within GRα3D based on the crystal structure of GRα3D (PDB: 6DS9).
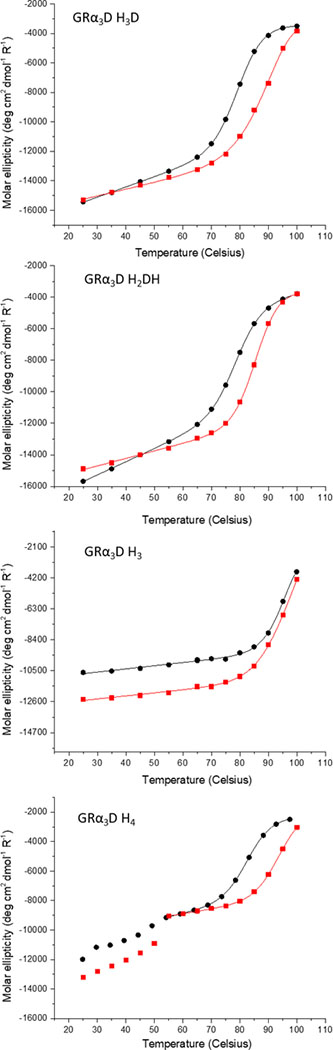
Thermal stability of the apo- and Cu(II)-proteins
CD spectra of the apo-proteins were obtained to test whether GRα3D could properly fold with four large hydrophilic metal binding residues mutated into the hydrophobic core (Figure S1). The double minima bands at 208 and 222 nm are representative of α-helical secondary structure and indicate a well folded three helical bundle peptide.62, 63 We next sought to compare the destabilization effects of the differing active sites. Thermal denaturation of apo- and Cu(II)-proteins were studied and compared to that of GRα3D by following the ellipticity of the proteins at 222 nm at varying temperatures. The midpoint of unfolding (TM) was determined by fitting the data to a two state unfolding model using the program CDpal.63–65 These values are summarized in Table 3.
Table 3.
Thermodynamic parameters of unfolding for apo- and Cu(II)-GRα3D three-helix bundles determined by thermal denaturation.
| Peptide | Apo peptide TM (°C) | Cu peptide TM (°C) |
|---|---|---|
| GRα3D | >95 | N/A |
| GRα3D H2DH | 79.8 ± 0.7 | 85.1 ± 0.9 |
| GRα3D H3D | 80.6 ± 0.6 | 88 ± 6 |
| GRα3D H3 | >95 | >95 |
| GRα3D H4 | 83.21 ±0.01 | 93.8 ± 0.5 |
GRα3D is structurally stable in the range of temperature used with a melting temperature above 95 °C, which precludes calculating the aforementioned thermodynamic parameters. Three of the four constructs reported within this study, however, have melting temperatures below 95° C, allowing for their direct measurement. Comparing these four constructs, we find that addition of a fourth residue destabilizes the protein as the TM decreases over 10°C between GRα3D H3 (TM >95 °C), and GRα3D H2DH, H3D, and H4 (TM ~80°C). Cu(II) bound peptide is more thermodynamically stable than the apo peptide, with TM increasing by 5 to 10 °C depending on the construct. Overall, the data assess that the apo-proteins and the Cu(II) complexes are well folded at room temperature.
Interestingly, GRα3D H4 shows two steps in the unfolding process in both the apo and Cu(II) bound forms (Figure 2). The first transition to an intermediate state occurs at 55 °C. The second step occurs between 70 °C and 90 °C, similar to the other constructs. Upon cooling, the first state is no longer observed and reheating a sample results in a similar denaturation profile to the renaturation profile (Figure S2).
Figure 2.
Thermal denaturation circular dichroism fits of GRα3D H3D, H2DH, H3, and H4 (top to bottom). Apo spectra are shown with black circles and Cu(II) bound spectra with red squares. Only the second denaturation step was fit for GRα3D H4.
Cu(II)-protein XAS and EPR characterization
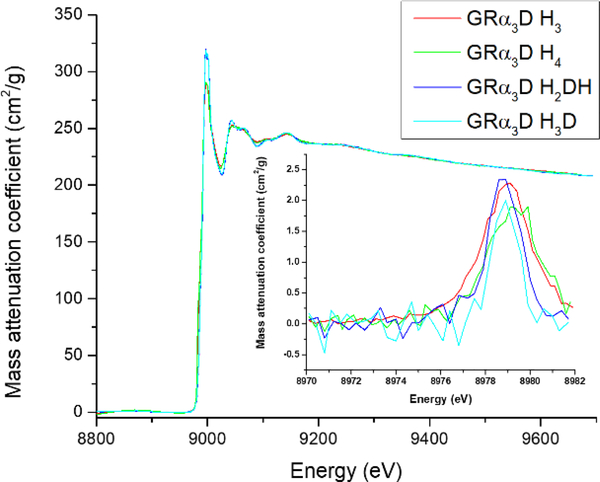
X-ray Absorption Spectroscopy (XAS), consisting of both X-ray absoprtion near edge structure (XANES) and extended X-ray absorption fine structure (EXAFS), was done on Cu(II)-GRα3D H3, Cu(II)-GRα3D H4, Cu(II)-GRα3D H2DH, and Cu(II)-GRα3D H3D and analyzed to investigate the structural differences between these four SOD mimics (Figure 3). The average nearest neighbor bond distance was between 1.94 and 1.95 Å for all constructs analyzed, consistent with 4-coordinate N or O bound Cu(II) (Table 4).
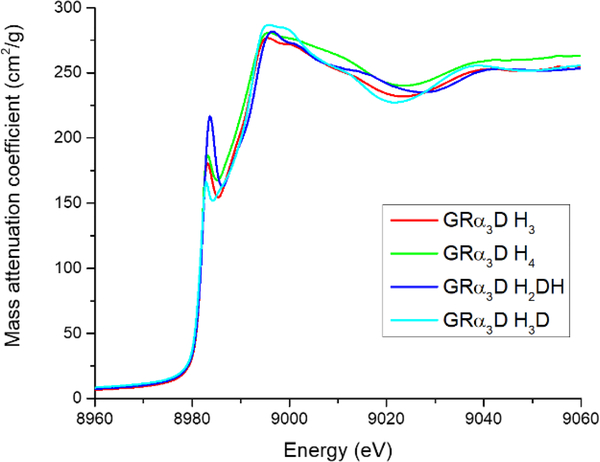
Figure 3.
1s → 3d region of Cu(II) XANES at pH 7.5 for every construct reported.
Table 4.
Cu(II) EXAFS and XANES fitting parameters at pH 7.5
| Construct | Model | Cu-O/N* R Å | Cu-O/N* σ2 10−3 Å2 | Cu-Imid R Å | Cu-Imid σ2 10−3 Å2 | Avg. bond length Å | 1s→3d area |
|---|---|---|---|---|---|---|---|
| GRα3D H3 | His3O1 | 1.966 | 5.7 | 1.935 | 13.28 | 1.943 | 8.27 |
| GRα3D H4 | His3 N1 | 1.881 | 4.4 | 1.972 | 6.23 | 1.949 | 6.89 |
| GRα3D H2DH | His3O1 | 1.981 | 1.5 | 1.933 | 8.06 | 1.945 | 5.41 |
| GRα3D H3D | His3O1 | 1.989 | 2.0 | 1.933 | 7.86 | 1.947 | 3.09 |
All constructs exhibit long distance backscatterers, which best fit to 3 His ligands. All fits attempted are included within the supporting information. This apparent similarity between the four constructs’ structures also extended to XANES analysis, where the 1s → 3d transitions (peak at 8979 eV) for all four constructs were of similar height indicating an equivalent degree of tetrahedral character to their geometry.66, 67
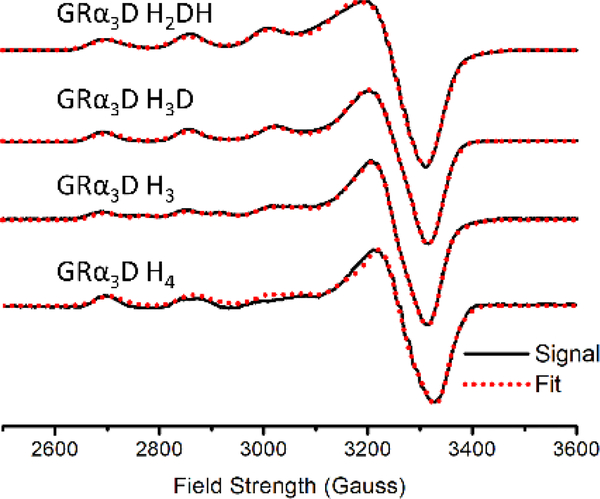
EPR spectra of Cu(II)-proteins were collected in 50 mM HEPES buffer, pH 7.5 at 100 K with a 2:1 protein/Cu(II) ratio to ensure that all copper is bound (Figure 4). The g values and hyperfine coupling constants of the Cu(II) complexes were determined by fitting with the SpinCount software and are listed in Table 5.68 Each complex has anisotropic g values with gx, gy < gz, characteristic of a dx2-y2 SOMO.69 Together with EXAFS data, the results suggest 4-coordinate copper complexes in a distorted square planar geometry.69 Two distinct species (A and B in Table 5) are observed in the EPR spectra of Cu(II)-GRα3D H3 and Cu(II)-GRα3D H4. Simulation of the EPR spectrum of GRα3D H3 shows that form A is dominant and accounts for 75% of the signal observed, whereas form B accounts for 25%. For GRα3D H4 the two species are present in the same ratio. The EPR parameters of a Cu-only SOD found in Mycobacterium tuberculosis (His4 active site) and a copper-substituted FeSOD from the archaeon Acidianus ambivalens (His2AspHis) are given in Table 5.7, 70 The Cu(II)-GRα3D derivatives have g values similar to these native enzymes and slightly higher AII values.
Figure 4.
Electron Paramagnetic Resonance spectra of the constructs presented in this study recorded at pH 7.5. Fits were done using the program SpinCount.
Table 5.
EPR parameters obtained from simulation of EPR spectra of Cu(II)-protein solutions.
| gx | gy | gz | A∥ (10−4 cm−1) | f=gz/ A∥ (cm) | ||
|---|---|---|---|---|---|---|
| [a] GRα3D H3 | 75% A | 2.05 | 2.04 | 2.27 | 166 | 136.8 |
| 25% B | 2.05 | 2.03 | 2.22 | 149 | 149.0 | |
| [a] GRα3D H4 | 50% A | 2.012 | 2.04 | 2.26 | 158 | 143.0 |
| 50% B | 2.04 | 2.03 | 2.23 | 188 | 119.6 | |
| [a] GRα3D H2DH | N/A | 2.06 | 2.03 | 2.27 | 162 | 140.2 |
| [a] GRα3D H3D | N/A | 2.04 | 2.04 | 2.26 | 168 | 134.5 |
| [b] Cu-only SOD | N/A | 2.05 | 2.05 | 2.26 | ∼140 | 161.4 |
| [c] Cu-(Fe) SOD | N/A | 2.05 | 2.05 | 2.37 | 136 | 174.3 |
X-band EPR spectra recorded at 100 K on solutions of 1 mM CuCl2 and 2 mM protein in 50 mM HEPES buffer containing 30% glycerol at pH 7.5, 9.3 GHz, 20.5 mW, 1 G.
Spagnolo et al., J. Biol. Chem., 2004, 33447–33455. X-band EPR spectra of a Cu-only SOD from Mycobacterium tuberculosis
Kardinahl et al., Biol. Chem., 2000, 381, 1089–1101. X-band EPR spectra recorded at 10 K of a copper-substituted FeSOD from the archaeon Acidianus ambivalens in Tris/HCl buffer at pH 7, 9.6452 GHz, 20 mW, and 10.0 G.
One may also attempt to correlate the hyperfine coupling constant observed in EPR to variation in activity between the constructs reported. The empirical factor f = gz/A‖ correlates with tetrahedral distortions where values between 105 to 135 cm are indicative of square planar geometry and higher values indicate distortion towards tetrahedral structures.71, 72 According to the values in table 5, our constructs are more tetragonal than native examples, which may be correlated to the decreased activity compared to native enzymes. This is bolstered by the case of GRα3D H4 in which two species are evident at a 1:1 ratio. The GRα3D H4 species B has an f factor of 119.6 cm, indicating that this species is more tetragonal than GRα3D H4 species A at 143 cm or any other construct reported in this manuscript. The activity of GRα3D H4 is also about half that of any of the other de novo constructs which one could explain by species B being an inactive form. However, the differences in activity observed are too minor to make definitive claims about the requirement of tetrahedral distortion for CuSOD activity. Future studies with other designed proteins may allow us to elucidate this relationship more clearly.
Cu(I)-protein XANES characterization
The XANES region of all four constructs were analyzed to investigate the coordination geometry. The Cu(I) 1s➔4p transition at 8982–8985 eV, was analyzed to determine geometry differences between constructs. The intensity of this peak is indicative of coordination number with higher peak intensities correlating with lower coordination number.73 GRα3D H3D has the lowest 1s➔4p transition signal, indicative of a higher coordination number (Figure 5). GRα3D H3 and H4 have similar intermediate transition signals. GRα3D H2DH has the highest transition signal, indicative of a lower coordination number, likely more 2-coordinate than 3-coordinate.
Figure 5.
1s → 4p region of Cu(I) XANES at pH 7.5 for every construct reported.
Reduction potentials, affinity, and SOD activity
The apparent standard potentials of Cu(II)-complexes containing a C-terminal Cys (Ala98Cys) and grafted on a gold electrode are listed in Table 6 with cyclic voltammograms in Figure S3. All four constructs share similar first coordination sphere His3N/O around the metal center as demonstrated by EXAFS analysis, however, the potential of GRα3D H3 is notably higher than the other three at 550 mV vs. NHE compared to 420–470 mV vs. NHE. The reduction potentials of the four constructs lie in between the potentials for oxidation of superoxide to peroxide (−0.18 V vs. NHE) and the reduction of superoxide to dioxygen (+0.91 V vs. NHE), which should enable catalysis of superoxide dismutation. The Cu(II) affinities for each of these four constructs were determined in an effort to account for these differences in reduction potential (Table 6). Interestingly, though GRα3D H3 has a much higher reduction potential, it does not have a Cu(II) affinity that is significantly different from that of GRα3D H2DH and GRα3D H3D. GRα3D H4 has the weakest Cu(II) affinity with a Kd of 5.7×10−10 M, indicating that a fourth His residue does not play a significant role in the binding of Cu(II).
Table 6.
Apparent standard potentials at pH 7.5, affinity constants and SOD activity of Cu(II)-protein complexes.
| E1/2 [a] (mV vs. NHE) | Cu(II) Kd (10−10 M) | Cu(I) Kd (10−16 M) | IC50[b] (μM) | kMcF[b] (106 M−1s−1) | |
|---|---|---|---|---|---|
| GRα3D H3 | 550 ± 10 | 5.00 ± 0.17 | 1.22 ± 0.04 | 2.9 ± 0.6 | 3.0 ± 0.6 |
| GRα3D H4 | 463 ± 10 | 5.7 ± 0.3 | 41 ± 2 | 8.0 ± 1.7 | 1.1 ± 0.2 |
| GRα3D H2DH | 420 ± 10 | 1.4 ± 0.4 | 54 ± 15 | 3.5 ± 1.1 | 2.6 ± 0.8 |
| GRα3D H3D | 470 ± 10 | 1.6 ± 0.2 | 8.8 ± 1.1 | 3.3 ± 0.3 | 2.6 ± 0.2 |
The Cu(II)-proteins bearing a GSGC tail were grafted on an Au electrode and CV were recorded at v = 0.05 V/s in an electrochemical working cell containing 50 mM HEPES buffer, pH 7.5 at room temperature. CE: Pt wire, ref: SCE.
IC50 (μM) and kMcF (106 M−1s−1) were determined from triplicate experiments with Cu(II)-protein (4:1 L/M) in 50 mM HEPES buffer, pH 7.5 with 100 μM XTT, 200 μM xanthine, and xanthine oxidase at 25 °C. kXTT = 8.6 104 M−1s−1 (in PBS 50 mM, pH 7.8). Note that the rate reported for GRα3D H3 is a composite of two species as observed by EPR.
The calculated Cu(I) affinities vary by two orders of magnitude across the four constructs. Cu(I) binds tightest to GRα3D H3, with a Kd of 1.22×10−16 M. GRα3D H3D has an intermediate Cu(I) affinity (8.8×10−16 M), while both GRα3D H4 and H2DH have a weaker affinity at 4.1×10−15 and 5.4×10−15 M, respectively (Table 6). Thus, adding a fourth peptide ligand decreases the Cu(I) affinity of the protein but to different extents.
The SOD activities of the Cu-proteins were measured by the McCord-Fridovich assay,59, 60 in which a secondary probe, XTT, is used to determine the amount of superoxide removed from solution by the SOD mimic.61 This assay is an indirect method to measure kcat, but its reliability has been validated by direct methods (stopped-flow and pulse radiolysis).74–78 The assay was performed using an excess of ligand (4:1 protein/Cu(II) ratio) to ensure no free copper is present. The IC50 is the concentration of SOD mimics at which 50% of the superoxide produced is dismutated by the SOD mimics. From this value, the kMcF, which can be compared to a catalytic rate constant, is calculated (Table 6).2, 59, 60, 74–77 GRα3D H3, GRα3D H2DH, and GRα3D H3D have similar IC50 (2.9–3.5 μM) and kMcF (3 106 M−1s−1) values, whereas GRα3D H4 has a lower SOD activity with the highest IC50 (8.0 μM) and lowest kMcF (1.1 106 M−1s−1). The apo proteins showed no SOD activity in the same concentration range. Additional controls were performed to check that no reaction occurs between the complex and formazan, and that the complex did not inhibit xanthine oxidase.
Discussion
The Cu-only SOD models presented here demonstrate that an exact reproduction of the active site is not necessary for modest SOD activity, but that the native residues play an important role in mediating this activity. Previous work with Cu/Zn SODs has shown that loss of even a single histidine in the active site results in a loss of SOD activity, typically through a loss of copper binding.79–81 While similar work in Cu only SODs has not been performed, the present work suggests that all four histidine residues are not necessary for copper binding or SOD activity.
Studies of the Cu(II) bound forms indicate that all four models are structurally similar to Cu only SOD. EPR experiments show that all four constructs are similar to both Cu(II) bound Cu only SOD and Cu(II) substituted FeSOD.7, 82 The fourth protein ligand is not vital in modulating the structure of the Cu(II) binding environment as observed by EPR, but does determine how many species are present in solution. Both constructs with an Asp ligand contain a single species, while a fourth His ligand or lack of a fourth protein ligand results in two species. This is confirmed by CD for GRα3D H4 as two unfolding steps are observed. Of these two species, only the more stable is observed upon cooling the sample. The fourth ligand, therefore, is necessary to restrict Cu(II) binding to only a single confirmation. A fourth aspartate ligand, regardless of position, may both coordinate the copper and orient a histidine residue in a single geometry. In both Cu/Zn and Cu only SODs, an aspartate ligand orients the histidine residue that is bound only in the Cu(II) form and loss of this residue results in a decrease in SOD activity.83, 84 The second species present may simply be a result of different histidine coordination in these systems.
This is corroborated by the XAS data. While these experiments cannot distinguish between multiple coordination states, the average indicates that all three histidines are bound in all four constructs. The fourth residue is likely an additional histidine in GRα3D H4 or an oxygen in the remaining three constructs. This oxygen may be from either the Asp ligand that is present in GRα3D H3D or H2DH or a solvent residue. Thus, the Asp may be binding the copper as a fourth residue or the Cu(II) coordination sphere is completed by water and the Asp acts solely to orient a histidine residue.
The Cu(II) affinity is also affected by this fourth ligand. GRα3D H4 has the weakest Cu(II) affinity, though only slightly weaker than GRα3D H3. If, however, the fourth ligand is instead an Asp residue, Cu(II) affinity increases by a factor of 3. The position of this Asp residue does not have a significant effect on the affinity of the cupric ion. This indicates that a fourth oxygen ligand lends to tighter Cu(II) binding than an imidazole ligand.
The fourth ligand is also important in modulating the copper reduction potential. Though all four constructs are catalytically active, GRα3D H3 has a much higher reduction potential than the constructs with a fourth amino acid ligand. Simply adding a fourth ligand decreases the midpoint potential by 80–130 mV, depending on the construct. This may be by removing solvent from the active site or by limiting the amount of rearrangement necessary to convert between the Cu(I) and Cu(II) bound forms.85 None of the constructs are close to the desired 360 mV vs NHE, the midpoint between the reduction and oxidation of superoxide.2, 3 Disrupting the His3 plane decreases the reduction potential by 50 mV from ~470 to 420 mV vs NHE, even though the Cu(II) affinity remains unchanged between GRα3D H3D and H2DH. The rearrangement of the Asp residue does more to stabilize the Cu(II) bound enzyme outside of increasing the affinity for Cu(II). The axial His in GRα3D H2DH is positioned in a more open, solvent exposed cavity, which may allow for easier conversion between the Cu(I) and Cu(II) forms than in GRα3D H3D. In that construct the three histidines are located in a more spatially confined position within the peptide, preventing such easy rearrangement.
Calculated Cu(I) affinities also provide insight into the copper binding environment. Unsurprisingly given the relatively weak Cu(II) affinity and high reduction potential, GRα3D H3 has the tightest Cu(I) affinity. GRα3D H2DH has the weakest Cu(I) affinity, correlated with the low reduction potential and has the most two-coordinate character measured by XANES. Again, the identity and placement of the fourth ligand significantly affects cuprous binding. The identity of the axial ligand may account for this difference. With no axial peptide ligand (GRα3D H3), Cu(I) binds with the highest affinity. In the Cu(II) bound form, the coordination sphere is completed with an axial solvent residue. This is most similar to GRα3D H3D, containing an axial Asp residue in addition to the H3 plane which results in an 8-fold loss in Cu(I) affinity. Maintaining the H3 plane and adding an axial His residue (GRα3D H4) further decreases the Cu(I) affinity (40x weaker than GRα3D H3). Disruption of the His3 plane results in the greatest loss of Cu(I) affinity and corresponds to the lowest reduction potential and highest Cu(II) affinity. Clearly, GRα3D H2DH most favors the oxidized species.
All four constructs exhibit measurable SOD activity, though still three orders of magnitude slower than the native enzyme. They are more efficient than the previously reported manganese de novo mimic (kMcF = 3.7 105 M−1s−1), and have a similar activity to the iron SOD mimic reported by Benson et al. (kcat 6.4 106 M−1s−1).46–48 The slowest construct, GRα3D H4, also has the weakest overall copper affinity which is known to decrease activity.79–81 The remaining three constructs are 2.5–3x faster but do not have statistically different rates. Thus, the trends in Cu(I/II) affinity and reduction potential do not correspond to trends in SOD activity. Previously, trends in reduction potential and SOD activity were observed for low-molecular weight manganese SOD mimics which show a better SOD activity when the reduction potential is closer to 0.36 V vs. NHE, the midpoint potential between the oxidation and reduction of superoxide.1–3, 19, 20, 23 This observation applies for other copper SOD mimics.1–3, 19, 20, 22
At this point we are unable to determine if these peptides are rate-limited by product release or conversion between the oxidized and reduced metal species. There may be a measurable difference in substrate conversion among the three more active peptides following the reduction potential trend that is unobservable due to slow product release.
To conclude, this study is the first example of the design of functional Cu only SOD mimics in de novo proteins and shows that all four His residues from the native active site are not required for SOD activity within a de novo construct. The activity of these constructs is much slower than that of the native enzyme and further studies will be done to address this. The Cu(I) environment and dynamics between the Cu(I) and Cu(II) bound species will be characterized to provide insight into this difference. These states could provide insight into the rate of catalysis and may highlight the importance of the fourth copper ligand. By modifying this fourth ligand, both in ligand type and position, we aim to improve the rate of catalysis and determine the most efficient coordination environment for SOD activity in de novo protein models.
Experimental Section
Protein expression and purification
pET15B recombinant DNA plasmid (Celtek Genes) containing the gene for the GRα3D constructs were transformed and expressed in E. coli BL21(DE3) competent cells (Life Technologies). The Ala98Cys derivatives of GRα3D constructs were prepared for electrochemical studies.
Colonies were inoculated in 30 mL of LB medium and 100 μg.mL−1 of ampicillin before being incubated at 37 °C and 175 rpm for 6–7 h. Autoincubation medium was inoculated with 10 mL/1L culture flask at 25 °C and 250 rpm for 18 h to overexpress the proteins. Cells were pelleted, resuspended in 1.0 mM phosphate buffer saline solution containing 2 mM dithiothreitol and 2 mM EDTA and lysed with a microfluidizer. The soluble protein was isolated after heat denaturation at 55 °C and acidification to pH 2 to remove contaminant proteins. The supernatant was syringe-filtered (0.2 μm) and purified on a reversed-phase C18 HPLC using a flow rate of 20 mL/min and a linear gradient of 0.1% TFA in 3:7 CH3CN:H2O to 0.1% TFA in 7:3 CH3CN:H2O over 35 min. The molecular weight of the purified peptide was confirmed by ESI-MS (collected on a Micromass LCT Time-of-Flight Mass Spectrometer) and found to correspond to the expected protein mass after deletion of the N-terminal methionine residue. Protein concentrations were determined by measuring the absorbance with ε280 = 8480 M−1 cm−1.
Circular Dichroism (CD) Spectroscopy
CD spectra were collected on a Jasco J-1500 CD spectropolarimeter at 25 °C using 1 cm path length quartz cuvettes. Samples contained 10 mM phosphate buffer, pH 7.0, and 10 μM protein. Cu(II) containing samples also included 11 μM CuCl2. Thermal denaturation samples were heated at at 5 °C/min and ellipticity was recorded every 5 °C from 25 to 100 degrees after stabilization within ± 0.1 °C of the desired temperature and an additional equilibration time of 5 minutes. Molar ellipticities ([θ]) were calculated using previously reported procedures.63, 86, 87
Cu(II) binding affinity
The binding affinity of Cu(II) to each peptide was determined using the competitive chelator 3,4-bis(oxamato)benzoic acid (baba). Solutions containing 50 μM peptide and 45 μM CuCl2 in 50 mM HEPES pH 7.5 were titrated with 0.1 equivalents of baba with 10 minutes of equilibration time between additions. The reverse titration was also performed with 50 μM baba and 45 μM CuCl2 in 50 mM HEPES pH 7.5. The absorbance of the Cu(II)baba complex was monitored at 330 nm and fit using previously described methods.88
Cu(I) binding affinity
The binding affinity of Cu(I) was calculated at pH 7.5 based on the Cu(II) binding affinity and the reduction potentials of each peptide (Equation 1). A value of 0.159 V vs. NHE was used for E°(CuII/I, aq).89, 90
| (1) |
Equation 1. Calculation of Cu(I) affinity from Cu(II) affinity and CuII/I standard potential
X-ray Absorption Spectroscopy
Cu(II) samples were prepared with 1.0 mM Cu(II)acetate and 1.5 mM peptide in a 50 mM HEPES buffer at pH 7.5 in aerobic conditions. Samples were then lyophilized before transferring to sample cells as a dried powder. During collection the Cu edge energy and 1s → 4p transition of Cu(I)peptide were monitored. We estimate that no more than 10% of the sample was photoreduced in the scans included in fits.
Cu(I) samples were prepared with 1.0 mM tetrakis(acetonitrile)Cu(I)hexafluorophosphate and 1.5 mM peptide in a 50 mM HEPES buffer at pH 7.5 in anaerobic conditions. 50% glycerol was added to the solution as a glassing agent before sample solutions were loaded into an XAS sample cell and frozen in liquid nitrogen.
Measurements were carried out at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7–3 or 9–3 with a Si(220) double-crystal monochromator and a flat Rh-coated harmonic rejection mirror. Samples were maintained below 10 K with an Oxford Instruments liquid helium cryostat. Data were measured as fluorescence excitation spectra using a 30- or 100-element Ge detector array (for beamlines 7–3 or 9–3 respectively) normalized to incident intensity measured with a N2 filled ion chamber. Data were measured with steps of 0.25 eV in the XANES region (1 sec integration time) and 0.05 Å−1 in the EXAFS region to k = 13.5 Å−1 (1~20 sec integration, k3 weighted). Energies were calibrated by assigning the lowest energy inflection point of a copper metal foil as 8980.3 eV. An initial E0 value of 9000 eV was used to convert data to k-space, and the background was removed using a 3-region cubic spline. EXAFS data were analysed using EXAFSPAK91 and FEFF 9.0.92 XANES data were normalized using MBACK.93 For analysis of the 1s→3d transitions, data were fitted with an arctan background with a pseudo-Voigt peak to model the rising edge and the 1s→3d peak, and this fitted background was then subtracted from the data. Cu(I) spectra were analyzed in this way to determine the degree of oxidation of XAS samples. The absence of any peak above the noise in these spectra indicate that oxidation was minimal.
Single- and multiple-scattering fitting of EXAFS data were performed using EXAFSPAK91 with ab initio amplitude and phase parameters calculated using FEFF 9.0.92 An initial model of Cu-imidazole coordination was built based on the averaged bond distances determined by single-scattering fitting of EXAFS data. An initial model of Cu-imidazole coordination was built based on the average Cu-N bond distances determined by single-scattering fitting of EXAFS data, with the imidazole bond-lengths and angles taken as the average of all Cu-imidazole structures contained in the Cambridge Structural Database. All significant non-H paths, defined as those having an amplitude greater than 4% of the Cu-N amplitude, from this model were then loaded into EXAFSPAK and modeled as a rigid ligand. Initial estimates of the Debye-Waller factors for each Cu-imidazole shell were taken from calculations by Dimakis and Bunker.94 The Cu-N distance and Debye-Waller factor were allowed to vary, with the distance and Debye-Waller factor for the other paths calculated based on the Cu-N values. Thus, the long distance scattering from the Cu-Imid was modeled while only varying two independent variables.
Electron Paramagnetic Resonance (EPR) spectroscopy
X-band EPR spectra were collected on a Bruker EMX electron spin resonance spectrometer with a Varian liquid nitrogen cryostat at 100 K. EPR samples contained 1 mM Cu(II)Cl2, 1.5–2 mM peptide, 50 mM HEPES pH 7.4, and 30% glycerol. Each sample was flash frozen in liquid N2 before measurement. To obtain Cu(II) EPR parameters, each spectrum was simulated on SpinCount.68
Electrochemistry
Cyclic voltammetry measurements were obtained on a Metrohm AUTOLAB potentiostat (PGSTAT302N).
The electrochemical apparatus contained a gold (Au) disk working electrode (1 mm diameter), a platinum wire counter electrode, and an aqueous saturated calomel electrode (SCE) as the reference electrode (0.241 V + SCE = normal hydrogen electrode). The gold surface was polished with diamond slurries having decreasing particle sizes in the following order: 6−3−1 μm. Au electrodes were conditioned in an electrochemical cell containing 0.5 M H2SO4, by scanning from −300 mV to +1500 mV vs. SCE at 500 mV/s until the cyclic voltammograms (CVs) overlaid to indicate a homogeneous surface. After each electrode had been polished and conditioned, 50 μL of a 0.5 mM CuII-protein Ala98Cys solution in 50 mM HEPES buffer pH 7.5 was chemically adsorbed on the Au surface for 2–3 h. CVs were collected in 50 mM HEPES buffer, pH 7.5 at varying scan rates at room temperature (22–28 °C).
SOD activity
The SOD activity of the copper complexes was determined using the indirect assay of McCord-Fridovich observing XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) reduction.59–61 Superoxide anions were generated by a xanthine-xanthine oxidase system and detected by monitoring the formation of formazan at 470 nm. The reactions were performed in 50 mM HEPES buffer pH 7.4 with 100 μM XTT and 200 μM xanthine. An appropriate amount of xanthine oxidase was added to start the reaction and generate a change in absorbance of 0.025–0.030 min−1. The absorbance at 470 nm was monitored for 1.5 minutes (slope P1) before the addition of the SOD mimic, and for another 1.5 minutes after the addition (slope P2). Plot of the ratio (P1-P2)/P1 as a function of SOD mimic concentration is used to calculate the inhibition concentration (IC50) at which the reduction of XTT to formazan is inhibited by 50% (P2 = ½ P1). A pseudo-catalytic rate constant, kMcF, is deduced from the IC50 value using the relation: kXTT × [XTT] = (kMcF)(IC50), with kXTT = 5.94 × 104 M−1s−1 (pH 7.8).2 In order to ensure that no free copper is present in solution the experiments were performed with an excess of protein (4:1 protein/metal ratio). The measurements were performed in triplicate for each compound. Controls with the apo-proteins show no SOD activity. Controls were performed to determine that no inhibition of the xanthine-xanthine oxidase system resulted from the addition of the peptides. The rate of conversion of xanthine to urate was monitored at 290 nm in the presence and absence of peptide and no inhibition was observed. Possible formazan complexation was monitored at 490 nm after the addition of peptide and no decrease in absorbance was detected, indicating no inhibition.95–97
Supplementary Material
Acknowledgements
VP, KK, and AT thank NIH grant R01 ES012236 for funding. At additionally acknowledges the Chateabriand Fellowship for funding. CP and EM acknowledge the network FrenchBIC (http://frenchbic.cnrs.fr/) for EM’s fellowship for her stay at Michigan. They also wish to thank ANR (ANR-15-CE07-0027) and Fondation pour la recherche médicale (DIE20151234413) for funding. OI acknowledges the financial support from Fundação para a Ciência e a Tecnologia [PTDC/QUI-BIQ/098406/2008) and the Marie Curie Actions (FP7-PEOPLE-IRG-230896).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller A-F, Teixeira M and Valentine JS, Chem. Rev, 2014, 114, 3854–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Policar C, in Redox Active Therapeutics, eds. Reboucas JS, Batinic-Haberle I, Spasojevic I, Warner DS and St D Clair, Springer, 2016, ch. Chapter 17, pp. 125–164. [Google Scholar]
- 3.Abreu IA and Cabelli DE, Biochim. Biophys. Acta, 2010, 1804, 263–274. [DOI] [PubMed] [Google Scholar]
- 4.Aguirre JD and Culotta VC, J. Biol. Chem, 2012, 287, 13541–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA and Getzoff ED, Biochemistry, 2004, 43, 8038–8047. [DOI] [PubMed] [Google Scholar]
- 6.Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, Cormack BP, Cabelli DE, Hart PJ and Culotta VC, Proceedings of the National Academy of Sciences, 2014, 111, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spagnolo L, Törö I, D’Orazio M, O’Neill P, Pedersen JZ, Carugo O, Rotilio G, Battistoni A and Djinović-Carugo K, J. Biol. Chem, 2004, 279, 33447–33455. [DOI] [PubMed] [Google Scholar]
- 8.Li CX, Gleason JE, Zhang SX, Bruno VM, Cormack BP and Culotta VC, Proceedings of the National Academy of Sciences, 2015, 112, E5336–E5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinett NG, Peterson RL and Culotta VC, Journal of Biological Chemistry, 2018, 293, 4636–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson RL, Galaleldeen A, Villarreal J, Taylor AB, Cabelli DE, Hart PJ and Culotta VC, Journal of Biological Chemistry, 2016, 291, 20911–20923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, Cormack BP, Cabelli DE, Hart PJ and Culotta VC, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forman HJ and Fridovich I, Arch. Biochem. Biophys, 1973, 158, 396–400. [DOI] [PubMed] [Google Scholar]
- 13.Choudhury SB, Lee J-W, Davidson G, Yim Y-I, Bose K, Sharma ML, Kang S-O, Cabelli DE and Maroney MJ, Biochemistry, 1999, 38, 3744–3752. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y-H, Fu H, Zhao W-X, Chen W-L, Su C-Y, Sun H, Ji L-N and Mao Z-W, Inorg. Chem. (Washington, DC, U. S.), 2007, 46, 734–739. [DOI] [PubMed] [Google Scholar]
- 15.Árus D, Jancsó A, Szunyogh D, Matyuska F, Nagy NV, Hoffmann E, Körtvélyesi T and Gajda T, J. Inorg. Biochem, 2012, 106, 10–18. [DOI] [PubMed] [Google Scholar]
- 16.Riley DP, Chem. Rev, 1999, 99, 2573–2588. [DOI] [PubMed] [Google Scholar]
- 17.Salvemini D, Muscoli C, Riley DP and Cuzzocrea S, Pulm. Pharmacol. Ther, 2002, 15, 439–447. [DOI] [PubMed] [Google Scholar]
- 18.Iranzo O, Bioorg. Chem, 2011, 39, 73–87. [DOI] [PubMed] [Google Scholar]
- 19.Batinic-Haberle I, Tovmasyan A, Roberts ER, Vujaskovic Z, Leong KW and Spasojevic I, Antioxid Redox Signal, 2014, 20, 2372–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batinić-Haberle I, Rebouças JS and Spasojević I, Antioxid. Redox Signaling, 2010, 13, 877–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salvemini D, Muscoli C, Riley DP and Cuzzocrea S, Pulm. Pharmacol. Ther, 2002, 15, 439–447. [DOI] [PubMed] [Google Scholar]
- 22.Bonomo RP, Impellizzeri G, La Mendola D, Maccarrone G, Pappalardo G, Santoro A, Tabbi G, Vecchio G and Rizzarelli E, in Metal-ligand interactions. Molecular, nano-, micro- and macro-systems in complex environments, eds. Russo N, Salahub DR and Witko M, Kluwer Academic Publishers, Dordrecht; Boston; London, 2003, pp. 41–63. [Google Scholar]
- 23.Batinic-Haberle I, Tovmasyan A and Spasojevic I, Antioxid. Redox Signal, 2018, 29, 1691–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batinic-Haberle I and Tome ME, Redox Biol, 2019, DOI: 10.1016/j.redox.2019.101139, 101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y-H, Fu H, Zhao W-X, Chen W-L, Su C-Y, Sun H, Ji L-N and Mao Z-W, Inorganic Chemistry, 2007, 46, 734–739. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J-J, Luo Q-H, Long D-L, Chen J-T, Li F-M and Liu A-D, Journal of the Chemical Society, Dalton Transactions, 2000, DOI: 10.1039/a909018e, 1893–1900. [DOI] [Google Scholar]
- 27.Feng C-J, Luo Q-H, Wang Z-L, Shen M-C, Wang H-W and Zhao M-H, J. Inorg. Biochem, 1999, 75, 1–6. [Google Scholar]
- 28.Kotynia A, Janek T, Czyżnikowska Ż, Bielińska S, Kamysz W and Brasuń J, Int. J. Pept. Res. Ther, 2017, 23, 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Csire G, Timári S, Asztalos J, Király JM, Kiss M and Várnagy K, J. Inorg. Biochem, 2017, 177, 198–210. [DOI] [PubMed] [Google Scholar]
- 30.Timári S, Cerea R and Várnagy K, J. Inorg. Biochem, 2011, 105, 1009–1017. [DOI] [PubMed] [Google Scholar]
- 31.Jakab IN, Lőrincz O, Jancsó A, Gajda T and Gyurcsik B, Dalton Trans, 2008, DOI: 10.1039/b811452h, 6987. [DOI] [PubMed] [Google Scholar]
- 32.Jancsó A, Paksi Z, Jakab N, Gyurcsik B, Rockenbauer A and Gajda T, Dalton Trans, 2005, DOI: 10.1039/b507655b, 3187. [DOI] [PubMed] [Google Scholar]
- 33.Bóka B, Myari A, Sóvágó I and Hadjiliadis N, J. Inorg. Biochem, 2004, 98, 113–122. [DOI] [PubMed] [Google Scholar]
- 34.Costanzo LL, Guidi GD, Giuftida S, Rizzarelli E and Vecchio G, J. Inorg. Biochem, 1993, 50, 273–281. [DOI] [PubMed] [Google Scholar]
- 35.Kubota S and Yang JT, Proceedings of the National Academy of Sciences, 1984, 81, 3283–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brigelius R, Spöttl R, Bors W, Lengfelder E, Saran M and Weser U, FEBS Lett, 1974, 47, 72–75. [DOI] [PubMed] [Google Scholar]
- 37.Huang P-S, Boyken SE and Baker D, Nature, 2016, 537, 320–327. [DOI] [PubMed] [Google Scholar]
- 38.Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HX, O’Neil KT and DeGrado WF, Science, 1995, 270, 935–941. [DOI] [PubMed] [Google Scholar]
- 39.Bryson JW, Desjarlais JR, Handel TM and Degrado WF, Protein Science, 1998, 7, 1404–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeGrado WF, Summa a. C. M., Pavone V, Nastri F and Lombardi a. A., Annu. Rev. Biochem, 1999, 68, 779–819. [DOI] [PubMed] [Google Scholar]
- 41.Plegaria JS and Pecoraro VL, Israel Journal of Chemistry, 2015, 55, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tegoni M, Eur. J. Inorg. Chem, 2014, 2014, 2177–2193. [Google Scholar]
- 43.Ghosh D and Pecoraro VL, Curr. Opin. Chem. Biol, 2005, 9, 97–103. [DOI] [PubMed] [Google Scholar]
- 44.Yu F, Cangelosi VM, Zastrow ML, Tegoni M, Plegaria JS, Tebo AG, Mocny CS, Ruckthong L, Qayyum H and Pecoraro VL, Chemical Reviews, 2014, 114, 3495–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zastrow ML and Pecoraro VL, Coordination Chemistry Reviews, 2013, 257, 2565–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh UP, Singh RK, Isogai Y and Shiro Y, Int. J. Pept. Res. Ther, 2006, 12, 379–385. [Google Scholar]
- 47.Pinto AL, Hellinga HW and Caradonna JP, Proceedings of the National Academy of Sciences, 1997, 94, 5562–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benson DE, Wisz MS and Hellinga HW, Proceedings of the National Academy of Sciences, 2000, 97, 6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryson JW, Desjarlais JR, Handel TM and Degrado WF, Protein Sci, 1998, 7, 1404–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walsh STR, Cheng H, Bryson JW, Roder H and DeGrado WF, Proc. Natl. Acad. Sci. U. S. A, 1999, 96, 5486–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HX, O’Neil KT and DeGrado WF, Science, 1995, 270, 935–941. [DOI] [PubMed] [Google Scholar]
- 52.Cangelosi VM, Deb A, Penner-Hahn JE and Pecoraro VL, Angewandte Chemie International Edition, 2014, 53, 7900–7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plegaria JS, Duca M, Tard C, Friedlander TJ, Deb A, Penner-Hahn JE and Pecoraro VL, Inorg. Chem, 2015, 54, 9470–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plegaria JS, Dzul SP, Zuiderweg ERP, Stemmler TL and Pecoraro VL, Biochemistry, 2015, 54, 2858–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tebo AG, Quaranta A, Herrero C, Pecoraro VL and Aukauloo A, ChemPhotoChem, 2017, 1, 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tebo AG, Pinter TBJ, Garcia-Serres R, Speelman AL, Tard C, Seneque O, Blondin G, Latour JM, Penner-Hahn J, Lehnert N and Pecoraro VL, Biochemistry, 2018, 57, 2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koebke KJ, Yu F, Salerno E, Van Stappen C, Tebo AG, Penner-Hahn JE and Pecoraro VL, Angew. Chem. Int. Ed. Engl, 2018, DOI: 10.1002/anie.201712757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koebke KJ, Ruckthong L, Meagher JL, Mathieu E, Harland J, Deb A, Lehnert N, Policar C, Tard C, Penner-Hahn JE, Stuckey JA and Pecoraro VL, Inorg. Chem, 2018, 57, 12291–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fridovich I, J. Biol. Chem, 1970, 245, 4053–4057. [PubMed] [Google Scholar]
- 60.McCord JM and Fridovich I, J. Biol. Chem, 1969, 244, 6049–6055. [PubMed] [Google Scholar]
- 61.Sutherland MW and Learmonth BA, Free Radical Res, 1997, 27, 283–289. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y-H, Yang JT and Chau KH, Biochemistry, 1974, 13, 3350–3359. [DOI] [PubMed] [Google Scholar]
- 63.Creighton TE, Protein structure. A practical approach, Oxford University Press, New York, 1997. [Google Scholar]
- 64.Greenfield NJ, Nat. Protoc, 2007, 1, 2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niklasson M, Andresen C, Helander S, Roth MG, Zimdahl Kahlin A, Lindqvist Appell M, Martensson LG and Lundstrom P, Protein Sci, 2015, 24, 2055–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn JE, Scott RA, Hodgson KO, Doniach S, Desjardins SR and Solomon EI, Chem. Phys. Lett, 1982, 88, 595–598. [Google Scholar]
- 67.Frank de G, György V and Pieter G, J. Phys.: Condens. Matter, 2009, 21, 104207.21817427 [Google Scholar]
- 68.Golombek AP and Hendrich MP, J. Magn. Reson, 2003, 165, 33–48. [DOI] [PubMed] [Google Scholar]
- 69.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG and Tian L, Chem. Rev, 2014, 114, 3659–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kardinahl S, Anemüller S and Schäfer G, Biol. Chem, 2000, 381. [DOI] [PubMed] [Google Scholar]
- 71.Sakaguchi U and Addison AW, J. Chem. Soc., Dalton Trans, 1979, DOI: 10.1039/DT9790000600, 600–608. [DOI] [Google Scholar]
- 72.Diaz A, Pogni R, Cao R and Basosi R, Inorg. Chim. Acta, 1998, 275–276, 552–556. [Google Scholar]
- 73.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO and Solomon EI, J. Am. Chem. Soc, 1987, 109, 6433–6442. [Google Scholar]
- 74.Eckshtain M, Zilbermann I, Mahammed A, Saltsman I, Okun Z, Maimon E, Cohen H, Meyerstein D and Gross Z, Dalton Trans, 2009, DOI: 10.1039/B911278B, 7879–7882. [DOI] [PubMed] [Google Scholar]
- 75.Spasojevic I, Batinic-Haberle I, Stevens RD, Hambright P, Thorpe AN, Grodkowski J, Neta P and Fridovich I, Inorg. Chem, 2001, 40, 726–739. [DOI] [PubMed] [Google Scholar]
- 76.Durot S, Policar C, Cisnetti F, Lambert F, Renault J-P, Pelosi G, Blain G, Korri-Youssoufi H and Mahy J-P, Eur. J. Inorg. Chem, 2005, 3513–3523. [Google Scholar]
- 77.Durot S, Lambert F, Renault J-P and Policar C, Eur. J. Inorg. Chem, 2005, 2789–2793. [Google Scholar]
- 78.Friedel FC, Lieb D and Ivanovic-Burmazovic I, Journal of Inorganic Biochemistry, 2012, 109, 26–32. [DOI] [PubMed] [Google Scholar]
- 79.Carri MT, Battistonia A, Polizio F, Desideri A and Rotilio G, FEBS J, 1994, 356, 314–316. [DOI] [PubMed] [Google Scholar]
- 80.Wang J, Slunt H, Gonzales V, Fromholt D, Coonfield M, Copeland NG, Jenkins NA and Borchelt DR, Hum. Mol. Genet, 2003, 12, 2753–2764. [DOI] [PubMed] [Google Scholar]
- 81.Wang J, Caruano-Yzermans A, Rodriguez A, Scheurmann JP, Slunt HH, Cao X, Gitlin J, Hart PJ and Borchelt DR, J. Biol. Chem, 2007, 282, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kardinahl S, Anemuller S and Schafer G, Biol. Chem, 2000, 381, 1089–1101. [DOI] [PubMed] [Google Scholar]
- 83.Peterson RL, Galaleldeen A, Villarreal J, Taylor AB, Cabelli DE, Hart PJ and Culotta VC, J. Biol. Chem, 2016, 291, 20911–20923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fisher CL, Cabelli DE, Tainer JA, Hallewell RA and Getzoff ED, Proteins Struct. Funct. Bioinformat, 1994, 19, 24–34. [DOI] [PubMed] [Google Scholar]
- 85.Moore GR, Pettigrew GW and Rogers NK, Proceedings of the National Academy of Sciences, 1986, 83, 4998–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rohl CA and Baldwin RL, Biochemistry, 1997, 36, 8435–8442. [DOI] [PubMed] [Google Scholar]
- 87.Luo P and Baldwin RL, Biochemistry, 1997, 36, 8413–8421. [DOI] [PubMed] [Google Scholar]
- 88.Conte-Daban A, Borghesani V, Sayen S, Guillon E, Journaux Y, Gontard G, Lisnard L and Hureau C, Anal. Chem, 2017, 89, 2155–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tegoni M, Yu F, Bersellini M, Penner-Hahn JE and Pecoraro VL, Proceedings of the National Academy of Sciences, 2012, 109, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Atkins P, Overton T, Rourke J, Weller M, Armstrong F and Hagerman M, Inorganic Chemistry, Oxford University Press, W. H. Freeman and Company, New York, Fifth Edition edn., 2010. [Google Scholar]
- 91.George GN and Pickering IJ, EXAFSPAK, http://ssrl.slac.stanford.edu/~george/exafspak/exafs.htm).
- 92.Ankudinov AL and Rehr JJ, Physical Review B, 1997, 56, R1712–R1716. [Google Scholar]
- 93.Weng T-C, Waldo GS and Penner-Hahn JE, Journal of Synchrotron Radiation, 2005, 12, 506–510. [DOI] [PubMed] [Google Scholar]
- 94.Dimakis N and Bunker G, Physical Review B, 2004, 70, 195114. [Google Scholar]
- 95.Friedel FC, Lieb D and Ivanović-Burmazović I, Journal of Inorganic Biochemistry, 2012, 109, 26–32. [DOI] [PubMed] [Google Scholar]
- 96.Faulkner KM, Liochev SI and Fridovich I, J. Biol. Chem, 1994, 269, 23471–23476. [PubMed] [Google Scholar]
- 97.Weiss RH, Flickinger AG, Rivers WJ, Hardy MM, Aston KW, Ryan US and Riley DP, J. Biol. Chem, 1993, 268, 23049–23054. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.