Abstract
Microsporidia have the leanest genomes among eukaryotes, and their physiological and genomic simplicity has been attributed to their intracellular, obligate parasitic life-style. However, not all microsporidia genomes are small or lean, with the largest dwarfing the smallest ones by at least an order of magnitude. To better understand the evolutionary mechanisms behind this genomic diversification, we explore here two clades of microsporidia with distinct life histories, Ordospora and Hamiltosporidium, parasitizing the same host species, Daphnia magna. Based on seven newly assembled genomes, we show that mixed-mode transmission (the combination of horizontal and vertical transmission), which occurs in Hamiltosporidium, is found to be associated with larger and AT-biased genomes, more genes, and longer intergenic regions, as compared with the exclusively horizontally transmitted Ordospora. Furthermore, the Hamiltosporidium genome assemblies contain a variety of repetitive elements and long segmental duplications. We show that there is an excess of nonsynonymous substitutions in the microsporidia with mixed-mode transmission, which cannot be solely attributed to the lack of recombination, suggesting that bursts of genome size in these microsporidia result primarily from genetic drift. Overall, these findings suggest that the switch from a horizontal-only to a mixed mode of transmission likely produces population bottlenecks in Hamiltosporidium species, therefore reducing the effectiveness of natural selection, and allowing their genomic features to be largely shaped by nonadaptive processes.
Keywords: microsporidia, genome evolution, population genomics, transmission modes, neutral evolution, genetic drift
Introduction
Streamlining in the context of microbial evolution refers to the minimization of cell and genome size and complexity (Novichkov et al. 2009; Giovannoni et al. 2014). In populations with large effective population sizes, Ne, natural selection enables even mutations with small positive selection coefficients, that is, “slightly” beneficial mutations to go to fixation. Likewise, mutations with small negative selection coefficients (mildly deleterious mutations) can rapidly be eliminated. Considering that the maintenance of any genomic segment involves a cost, and that the efficiency of selection depends on Ne, theory predicts a negative correlation between population and genome sizes (Lynch and Conery 2003; Lynch 2007b). With huge Ne at the order of 109 and compact genomes tightly packed with protein-encoding genes, free-living bacteria are the primary examples for genome streamlining due to natural selection (Bobay and Ochman 2017).
In contrast, natural selection is less effective in populations with low Ne, where there is greater influence of genetic drift (Lynch 2007a). An example for this is bacterial lineages that turned endosymbionts. They are believed to suffer a strong reduction in population size due to host restriction (Mira and Moran 2002; Moran and Plague 2004), and for obligate symbionts, their transmission is exclusively vertical. The evolution of the small genomes of many endosymbionts is believed to be mostly driven by genetic drift at low Ne—not natural selection—with the random fixation of mildly deleterious mutations. Although loss of function by natural selection can occur if selection coefficients are greater than about 1/2Ne (Hottes et al. 2013), metabolic degeneration of mutualistic symbiotic bacteria is caused by a process of genomic erosion that reflects both a mutational bias toward deletions and the reduced efficacy of natural selection in maintaining gene functionality (Mira et al. 2001; Kuo and Ochman 2009; Nowack and Weber 2018). Bacterial populations that have been serially passaged to simulate recurrent bottlenecks revealed that extensive genome reduction can occur on a short evolutionary time scale (Nilsson et al. 2005). This reduction is often accompanied by an overall degeneration of the DNA repair machinery, leading to increased mutations rates and a decrease in GC content, as DNA damage such as cytosine deamination and guanine oxidation bias mutations toward A and T bases (Moran 1996; McCutcheon and Moran 2012).
In eukaryotes, the most extreme genomic reduction is found in some microsporidia, obligate intracellular parasites related to fungi. Because most microsporidia have no mitochondria (but see Haag et al. [2014] for an exception), they rely heavily on their hosts for their energetic requirements. Microsporidian genomes are poor in genes involved in resource-producing metabolic pathways—such as ATP synthesis—and rich in genes that enhance transport mechanisms and enable the hijacking of resources from the host (Keeling et al. 2010; Nakjang et al. 2013; Boakye et al. 2017). This adaptation has been associated with their intracellular life-style (Peyretaillade et al. 2011; Corradi and Slamovits 2011). With genomes smaller than 3 Mb and coding for about 2,000 proteins, microsporidian species from the genus Encephalitozoon are considered paragons of eukaryote streamlining (Katinka et al. 2001; Corradi et al. 2010; Pombert et al. 2012). However, this streamlining did not occur in all microsporidians and several species harbor genomes larger by an order of magnitude (Williams et al. 2008). Genome size variability in microsporidia has been attributed in part to the differential accumulation of transposable elements (Xu et al. 2006; Parisot et al. 2014) but this accumulation cannot, by itself, explain such large differences, suggesting that different evolutionary forces are at play.
An often-overlooked aspect of microsporidian evolution is the diversity present in their life cycles, with some species even relying on more than one host (Becnel and Andreadis 2014). Transmission in microsporidian species can be horizontal, vertical, or both (mixed mode), and their capacity to perform standard meiosis is questionable (Lee et al. 2014). In a few species with well-characterized life cycles, abortive meiosis is observed (Canning et al. 1999), and population genetic studies suggest that some microsporidia are clonal (Haag, Traunecker, et al. 2013). Daphniamagna is the host of a diversity of microsporidia with divergent life-styles and genomic features (Ebert 2005; Corradi et al. 2009; Haag et al. 2014; Pombert et al. 2015) offering the unique opportunity to investigate the influence of microsporidian life history strategies on genome evolution while keeping the host factor constant. Here, to investigate the impact of transmission modes on the evolution of genomic architectures of microsporidia, we sequenced seven genomes from two clades of microsporidia that specifically parasitize D. magna with contrasting life histories: Hamiltosporidium and Ordospora. The two Hamiltosporidium species analyzed in our study, H. tvaerminnensis and H. magnivora, are diploid (Haag, Traunecker, et al. 2013). Hamiltosporidiumtvaerminnensis is asexual and transmitted both horizontally and vertically, whereas H. magnivora is possibly sexual but only transmitted vertically from mother to offspring (Haag et al. 2011; Haag, Traunecker, et al. 2013) and possibly horizontally to a second, yet unknown, host. In contrast, vertical transmission was never observed for Ordospora colligata, which belongs to a derived group of microsporidia with highly reduced genomes (Pombert et al. 2015). Its ploidy and sexuality are unclear, but its closest known relatives have been found diploid, albeit with very low levels of heterozygosity, and postulated to feature a sexual cycle (Selman et al. 2013). In our comparisons, we focus on the relative roles of natural selection and genetic drift in genome evolution of microsporidian parasites with known modes of transmission.
Materials and Methods
Host and Parasite Collection
Daphnia magna are planktonic Crustaceans inhabiting standing fresh- and brackish-water bodies with a Holarctic distribution. In the course of diverse sampling efforts (e.g., Roulin et al. 2013; Yampolsky et al. 2014; Fields et al. 2018), Daphnia were brought to the laboratory either in form of planktonic samples or in form of resting eggs (=ephippia), collected from the sediment surface of the water body. Field collected planktonic females were cloned, that is, individual females were allowed to reproduce asexually. Resting eggs were washed and stimulated to hatch by exposure to continuous light under room temperature in well-oxygenated medium. Hatchlings were isolated and were allowed to produce clonal lines by asexual reproduction. All clones were kept in the laboratory under conditions of continuous asexual reproduction with standard laboratory conditions (20 °C, 16:8 h light:dark cycles, green algae Scenedesmus sp. as food in artificial Daphnia medium (Ebert et al. 1998). Animals from each clone were tested for microsporidian infections. Clones with infections of O.colligata, H.magnivora, and H. tvaerminnensis were mass propagated to produce enough parasite material for DNA isolation using standard protocols.
DNA Isolation and Sequencing
Reduction of nonfocal DNA, mainly from host microbiota and food items, followed the protocol of Dukić et al. (2016). In short, infected D. magna were treated for 72 h with three antibiotics (streptomycin, tetracycline, and ampicillin). At the same time, all animals were fed with dextran beads (Sephadex beads, 50-µm diameter, Sigma Aldrich) to aid gut evacuation. Animals were then placed in 1.5-ml Eppendorf tubes and excess fluids were removed. We added extraction buffer (Qiagen GenePure DNA Isolation Kit) to the tubes and disrupted the tissue using sterile and DNA-free plastic pestles. The sample was incubated overnight with Proteinase K at 55 °C, followed by RNA degradation using RNAse treatment (1 h, 37 °C). Protein removal and DNA precipitation, including the addition of glycogen (Qiagen) to aid DNA precipitation, were done using the Qiagen GenePure DNA Isolation Kit instructions. Resultant DNA was suspended in 40 µl of Qiagen DNA hydration solution and subsequently tested for purity and concentration using a Nanodrop and Qubit 2.0, respectively. Libraries were prepared using Kapa PCR-free kits and sequenced by the Quantitative Genomics Facility service platform at the Department of Biosystem Science and Engineering (D-BSSE, ETH), in Basel, Switzerland, on an Illumina HiSeq 2000 (upgraded to 2500) with the HiSeq 2500 v4 kit in paired ends mode (2× 126 bp [i.e., 125 bp + 1 bp for final quality score assessment]; data sets FI-SK-17-1, GB-EP-1, NO-V-7, IL-G-3) or the HiSeq 2500 Rapid Run v2 kit in paired ends mode (2× 301 bp [300 bp + 1 bp]; data sets BE-OM-2, IL-BN-2, FI-OER-3-3).
Genome Assembly
Ordospora
Daphnia magna clones collected from populations in Finland, England, and Norway were found to be infected with O.colligata (sample codes: FI-SK-17-1, GB-EP-1, and NO-V-7). Low quality bases in the data sets were discarded with Trimmomatic 0.27 (Bolger et al. 2014) using default parameters. Host sequences were filtered out from the Ordospora data sets before the initial assemblies by read mapping with Bowtie2 2.2 against the D.magna (dmagna-v2.4-20100422-assembly.fna; http://arthropods.eugenes.org/EvidentialGene/daphnia/daphnia_magna/Genome/? M=A; last accessed December 18, 2019) and Daphnia pulex (dpulex_jgi060905_evenline.fna; http://wfleabase.org/genome/Daphnia_pulex/dpulex_jgi060905/genome-assembly; last accessed December 18, 2019) draft genomes. Host-filtered Ordospora data sets were assembled iteratively with Ray 2.3.2 (Boisvert et al. 2010) using distinct kmers (31, 41, 51, 61, 71, 81, and 91) and with SPAdes 3.5.0 (Bankevich et al. 2012). For each data set, the best Ray assembly was selected with QUAST 4.4 (Gurevich et al. 2013) and merged with the corresponding SPAdes assembly with CONSED 30 (Gordon et al. 1998). Potential contaminants not belonging to Ordospora in the merged contigs were searched for using BLAST (Altschul et al. 1990) against the NCBI nucleotide database (NT), and discarded when found. The filtered Ordospora contigs were further assembled and polished using the seed-extension approach described in Pombert et al. (2013), using the Ordospora OC4 genome (Pombert et al. 2015) as reference and preliminary annotations derived from PROKKA 1.12 (Seemann 2014) to help guide the assemblies.
Hamiltosporidium
Contaminants from the host were filtered out postassembly of two H. magnivora isolates from Belgium and Israel (BE-OM-2, IL-BN-2) and two H. tvaerminnensis isolates from Finland and Israel (FI-OER-3-3, IL-G-3). The Hamiltosporidium spp. genomes were assembled iteratively with Ray 2.3.2 using kmers 31, 41, 51, 61, 71, 81, 91, 101, and 111, and the best contiguous assemblies were selected with QUAST 4.4. Contaminants were then filtered out in successive steps. Contigs from Daphnia were first filtered by mapping the Hamiltosporidium-free BE-OM-1 data set containing sequencing reads from the host and Ordospora (in very low proportion) using Bowtie2 2.2.9 as implemented in the SSRG pipeline (https://github.com/PombertLab; last accessed December 18, 2019). Contigs showing an average sequencing depth of at least 1× in the resulting coverage files were discarded with the filter_by_coverage.pl custom Perl script (all custom scripts are available on the Pombert lab GitHub page). Contigs were then filtered by GC content with filter_by_GC.pl using minimum and maximum binning values set to 35 and 60, respectively, and the contigs averaging <35% GC kept thereafter. Potential contaminants remaining in these contigs were identified by taxonomized BLASTN searches with the megablastn algorithm against the NCBI nucleotide (NT) and the Taxonomy databases, and then discarded from the final assemblies. To ensure that the read mapping approach and the 35% GC criterion above did not end up removing true Hamiltosporidium contigs, taxonomized BLASTN searches were also performed independently on the full BE-OM-2 assembly.
Genome Annotation
Ordospora
Annotations from the Ordospora OC4 reference genome were transferred to the final FI-SK-17-1, GB-EP-1, and NO-V-7 assemblies with Geneious 9.1.2 (Biomatters, Auckland, New Zealand) using the “Annotate & Predict -> Transfer annotations” built-in function. Transferred annotations were manually curated with Artemis 16.0.0 (Rutherford et al. 2000) to correct incomplete or missing features. Curated EMBL annotations were converted to TBL format with the custom Perl script EMBLtoTBL.pl, and GenBank-compatible annotations were generated from the TBL files with the NCBI tool TBL2ASN.
Hamiltosporidium
Initial gene prediction in the Hamiltosporidium assemblies were performed with the eukaryotic gene predictors GeneMark-ES 4.33 (Ter-Hovhannisyan et al. 2008) and Augustus 3.2.3 (Keller et al. 2011) as implemented in MAKER 2.31 (Holt and Yandell 2011), with Augustus independently using the built-in Encephalitozoon cuniculi gene model and Rozella allomycis gene model from James et al. (2013). Additional intron-less putative open reading frames were also positioned on the contigs with Prodigal 2.6.3 (Hyatt et al. 2010). In parallel, microsporidia proteins from Rozella allomycis (James et al. 2013) and from MicrosporidiaDB (Aurrecoechea et al. 2017) data sets EcuniculiGBM1, AalgeraePRA109, AalgeraePRA339, EaedisUSNM41457, MdaphniaeUGP3, NausubeliERTm2, NausubeliERTm6, NbombycisCQ1, NceranaeBRL01, NparisiiERTm1, NparisiiERTm3, OcolligataOC4, PneurophiliaMK1, Slophii42_110, ThominisUnknown, VcorneaeATCC50505, and Vculicisfloridensis were queried against the Hamiltosporidium spp. assemblies using TBLASTN homology searches. The Hamiltosporidium spp. assemblies, gene predictions, and TBLASTN homology searches were loaded in a local WebApollo 2.0.5 browser (Lee et al. 2013), and the annotations were curated manually from the sum of the independent searches. The curated annotations were exported in GFF3 format using WebApollo’s built-in tools, the gff3 files were split per contig with splitGFF3.pl and then converted to EMBL format with WebApolloGFF3toEMBL.pl, and the predicted protein sequences were exported to FASTA format with EMBLtoPROT.pl. Protein functions were predicted using InterProScan 5.26-65.0 (Jones et al. 2014) searches and BLAST homology searches against the SwissProt, TREMBL, and UniProt databases. The functions inferred from these predictions were compared with the parse_annotators.pl and curate_annotations.pl Perl scripts, and the predicted functions were annotated based on the consensus with ambiguous functions further checked using NCBI’s CDD searches (Marchler-Bauer et al. 2015). EMBL files were converted to TBL format with EMBLtoTBL.pl, and the ASN files generated with NCBI’s TBL2ASN. Miscellaneous annotation issues caused by the gene predictors (e.g., peptides shorter than 50 aa) not automatically fixed by WebApolloGFF3toEMBL.pl and detected in the TBL2ASN validations (.val) files were corrected in the EMBL files by manual curation using the check_errors_1.sh, check_errors_2.sh and check_errors_3.sh Bash scripts and Artemis 16.0.0 (Rutherford et al. 2000). Corrected EMBL files were then converted to TBL and ASN, as described above. Intron/exon junctions in Figure SX and SY were annotated and corrected manually using Artemis 16.0.0; introns (nucleotide sequence) and exons (amino acid sequence) were aligned using MAFFT v7.407 (Katoh and Standley 2013).
Duplication Analyses
During the manual curation phase of the Hamiltosporidium spp. genomes, we noticed that some of the contigs appeared duplicated to some extent in the assemblies. The extent of these duplications was evaluated by performing BLASTN homology searches of each assembly against itself, then by parsing the results with parse_BLAST_selftest.pl with thresholds of 80% and 90% nucleotide identify between candidate duplicates. Hamiltosporidium is known to be diploid (Haag, Traunecker, et al. 2013); therefore in order to test whether these redundant contigs were real duplicates or instead arose from allelic differences representing assembly artifacts, the sequencing reads were mapped against the assemblies with Bowtie 2.2.9 in pair-ends mode as implemented in the SSRG pipeline (Pombert lab, GitHub), then the relative sequencing depth of each contig (Cdepth) was compared with the average depth (Adepth) of the assembly with seq_depth.pl. Statistics on duplicated contigs were further evaluated by t-tests using StatPlus for Excel (AnalystSoft, Walnut, CA) assuming unequal variances.
Repetitive Elements Analyses
A de novo identification of repetitive elements was performed with RepeatModeler 1.0.11 as implemented in RepeatMasker (Smit et al. 2013) with default parameters. RepeatModeler created genome-specific libraries for the seven de novo assemblies generated in this study, as well as ten other assemblies downloaded from NCBI (MdaphniaeUGP3 GCA_000760515.1, Edha_aedisV4b GCA_000230595.3, NosBomCQ1v01 GCA_000383075.1, NapisBRLv01 GCA_000447185.1, NceranaeBRL01 GCA_000182985.1, OcolligataOC4 GCA_000803265.1, EcuniculiGBM1 GCF_000091225.1, EintestinalisATCC50506 GCA_000146465.1, EhellemATCC50504 GCA_000277815.3, and EromalaeSJ2008 GCA_000280035.2). Each genomic-specific library was combined with representative transposable elements (TEs) from fungi obtained from Repbase (Bao et al. 2015). Assemblies were screened for the repetitive elements contained in the genome-specific libraries using BLAST as search engine, as implemented in RepeatMasker, with default parameters. Low complexity DNA sequences and simple repeats were not masked. Kimura distances between genome copies and TE consensus from the library were determined using RepeatLandscape (Caballero 2012) on alignments included in *.align files after genome masking with RepeatMasker. The rates of transitions and transversions were calculated on alignments and transformed to Kimura distance (Kimura 1980).
Phylogenetic Analysis
Orthologous sequences of 27 proteins were extracted from the above-mentioned genome assemblies (supplementary file 1, Supplementary Material online). Orthologs (E-value cutoff 1e-10) were confirmed by manual inspection of postalignment similarity. Each individual protein was aligned using MAFFT (Katoh and Standley 2013), concatenated, and the ambiguously aligned regions removed with Geneious 11.0.4 (Biomatters) using the “Tools -> Mask Alignment” command. The final alignment contained 13,409 sites. The maximum likelihood phylogeny was estimated using the LG model in RAxML version 8 (Stamatakis 2014) with empirically determined amino acid frequencies, and support estimated by 500 bootstrap pseudo-replicates.
dN/dS Ratios
The ratios of nonsynonymous versus synonymous sites (dN/dS) were calculated with SNAP 2.1.1 (Korber 2000) for two distinct data sets: Ordospora/Encephalitozoon spp. (OE) and Hamiltosporidium/Nosema spp. (HN). The OE data set included the three Ordospora isolates from this study (FI-SK-17-1, GB-EP-1, and NO-V-7) and data sets OcolligataOC4, EcuniculiEC1, EcuniculiEC2, EcuniculiEC3, EcuniculiGBM1, EhellemATCC50504, EhellemSwiss, EintestinalisATCC50506, and EromaleaeSJ2008/ from MicrosporidiaDB. The HN data set included the four Hamiltosporidium spp. from this study (BE-OM-2, FI-OER-3-3, IL-BN-2, and IL-G-3), data sets NbombycisCQ1 and NceranaeBRL01 from MicrosporidiaDB, and data set NapisBRLv01 (GCA_000447185.1) from NCBI. Orthologs within each data set were identified with OrthoFinder 2.2.6 (Emms and Kelly 2015) using default parameters and single-copy orthologs were separated from multicopy genes in the OrthoFinder output with split_csv.pl. Ortholog data sets were generated with make_datasets.pl from the corresponding predicted mRNAs, and the nucleotide data sets were aligned at the amino acid level with MACSE 2.01 (Ranwez et al. 2011) using the multithreaded run_macse.pl Perl script. dN/dS values were calculated with SNAP, whose Perl script was modified to generate outputs with filenames derived from input sequences rather than from process IDs. The analyses were performed using this modified Perl script (SNAP_mod.pl) and the automation script run_SNAP.pl. Results were then concatenated into tab-delimited files with cat_summaries.pl. Statistics on dN/dS were further evaluated by t-tests and linear regression analyses using StatPlus for Excel (AnalystSoft) assuming unequal variances.
Recombination Analyses
The population mutation (Θ) and recombination (ρ) rates were assessed directly with mlRho 2.9 (Haubold et al. 2010) as follows (see MlRho.sh on GitHub for the full commands). Briefly, reads were filtered using Trimmomatic 0.39 (Bolger et al. 2014) with the ILLUMINACLIP and SLIDINGWINDOW:4:28 command line switches to clip out Illumina adapters and to discard low quality bases, respectively. Reads passing the quality filters were concatenated in a single FASTQ file, then mapped against the corresponding genomes with minimap2, as implemented in get_SNPs.pl from the SSRG pipeline (https://github.com/PombertLab; last accessed December 18, 2019) with the -bam command line switch to keep the resulting BAM alignments files. The BAM alignments were indexed with samtools 1.9-50, and used as input for formatPro 0.5 and mlRho 2.9, iteratively using minimum coverage values (-c) of 4, 8, and 16.
The effect of recombination in the Hamiltosporidium and Ordospora genomes was further assessed indirectly by calculating GC-biased gene conversion (gBGC) values with phastBias from the PHAST 1.5 package (Capra et al. 2013). Orthologs within each data set were identified with OrthoFinder 2.2.6 using default parameters and single-copy orthologs were separated from multicopy genes in the OrthoFinder output with split_csv.pl. Single-copy ortholog data sets were generated with make_phast_datasets.pl from the corresponding predicted mRNAs, and the nucleotide data sets were aligned at the amino acid level with MACSE 2.01 using the multithreaded run_macse.pl Perl script. Neutral models for each alignment and gBGC values were generated and calculated with the phyloFit and phastBias programs from the PHAST package, respectively, as implemented in the Perl scripts run_phyloFit_ORDOSPORA.pl and run_phyloFit_HAMIL.pl tailored for each data set. These analyses were repeated using each isolate/species iteratively as outgroup for the gBGC calculations.
Data Availability
All customs scripts and software are available on the Pombert lab GitHub page (https://github.com/PombertLab; last accessed December 18, 2019). This project was deposited in NCBI under BioProject accession number PRJNA419750. Sequencing data sets were deposited in the NCBI Sequence Read Archive under the same accession number (PRJNA419750). The Ordospora genomes were deposited in GenBank under accession numbers PITH00000000 (FI-SK-17-1), PITG00000000 (GB-EP-1), and PITF00000000 (NO-V-7). The H.magnivora genomes were deposited under accession numbers PITI00000000 (BE-OM-2) and PIXR00000000 (IL-BN-2). The H.tvaerminnensis genomes were deposited under accession numbers PITJ00000000 (FI-OER-3-3) and PITK00000000 (IL-G-3).
Results
Contrasting Genome Architectures of Ordospora and Hamiltosporidium
We de novo assembled and annotated seven new genomes of microsporidian parasites of D. magna with different modes of transmission belonging to two clades: Ordospora and Hamiltosporidium. Table 1 shows that their genomes differ by one order of magnitude in size. Ordospora—an exclusively horizontally transmitted parasite of the gut epithelium—bears a compact genome with less than half of the coding capacity of Hamiltosporidium—a parasite with horizontal and vertical transmission (mixed-mode transmission) infecting the fat tissue and ovaries of its host. Predicted proteins, on average, were not found to be larger in Hamiltosporidium genomes compared with those found in Ordospora isolates (table 1) and we identified only one clear expansion in the family of genes encoding a minichromosome maintenance (MCM) protein (data not shown). In Hamiltosporidium, these MCM proteins feature in-frame expansions rich in asparagine residues that split conserved MCM domain motifs. However, we could not ascertain if those expansions are inteins that are spliced post-translationally—inteins often interrupt MCM proteins in archaea and bacteria—or simply expansions in variable regions that do not disrupt the function of the protein. Although ab initio predictions with GeneMark suggest the presence of numerous and longer introns in Hamiltosporidium compared with Ordospora, manual investigation of introns inserted at cognate and ectopic sites (supplementary file 2, Supplementary Material online) rather suggests that the introns that are present in Hamiltosporidium spp. are similarly small. In Trachipleistophora hominis (Heinz et al. 2012) for which RNAseq data are available, further investigation of the GeneMark predicted introns revealed that these are not genuine introns (Whelan et al. 2019), and we believe that it is also the case here. The main difference we observed in the genomes of the two clades is that the Hamiltosporidium genomes contain much longer intergenic regions, a large proportion of repetitive elements, and long segmental duplications, which are absent in Ordospora. Interestingly, the Hamiltosporidium genomes presented here vary regarding their accumulation patterns of repetitive elements. Differently from H. magnivora (H.m.) BE-OM-2, H.m. IL-BN-2 and H. tvaerminnensis (H.t.) FI-OER-3-3, which show a unimodal distribution of TE age classes, H.t. IL-G-3 is unique in having a slightly bimodal distribution, suggesting two instances of genomic bursts caused by TE expansion in the latter (supplementary file 3, Supplementary Material online). Furthermore, Hamiltosporidium genomes are strongly AT biased (26% GC, on average) as compared with O. colligata (38% GC, on average).
Table 1.
Contrasting Biologic and Genomic Features of Microsporidia That Parasitize D. magna
|
H. tvaerminnensis
|
H. magnivora
|
O. colligate
|
||||||
|---|---|---|---|---|---|---|---|---|
| Daphnia clone | FI-OER-3-3 | IL-G-3 | BE-OM-2 | IL-BN-2 | FI-SK-17-1 | NO-V-7 | GB-EP-1 | OC4a |
| Geographic origin | Finland | Israel | Belgium | Israel | Finland | Norway | UK | UK |
| Parasitized tissueb | FT, OV | FT, OV | FT, OV | FT, OV | Gut | Gut | Gut | Gut |
| Mode of transmissionc | H, V | H, V | H(?), V | H(?), V | H | H | H | H |
| Genome assembly length (Mb) | 18.34 | 25.20 | 20.73 | 17.18 | 2.26 | 2.32 | 2.30 | 2.29 |
| Duplicated segments (%) | 8.50 | 15.66 | 14.72 | 5.00 | — | — | — | — |
| GC content (%) | 25.82 | 26.14 | 25.80 | 25.74 | 38.51 | 38.39 | 38.44 | 38.52 |
| Number of contigs | 2,915 | 2,738 | 3,550 | 3,833 | 26 | 21 | 18 | 15 |
| N50 (bp) | 9,580 | 12,459 | 10,347 | 6,803 | 174,687 | 151,370 | 217,670 | 228,601 |
| Largest contig (bp) | 60,231 | 84,002 | 68,063 | 52,330 | 299,661 | 301,630 | 299,656 | 299,546 |
| Annotated genes | 4,121 | 6,203 | 4,658 | 4,180 | 1,849 | 1,862 | 1,857 | 1,879 |
| Predicted introns | 890 | 5,708 | 1,891 | 1,491 | 28 | 28 | 28 | 28 |
| Average intron length (bp)d | 302 | 195 | 310 | 264 | 31 | 31 | 31 | 31 |
| Average intergenic region length (bp) | 2,467.05 | 2,431.22 | 2,486.85 | 2,171.20 | 172.71 | 180.61 | 176.60 | 175.48 |
| Average protein length (aa) | 364 | 355 | 350 | 340 | 355 | 356 | 356 | 355 |
From Pombert et al. (2015).
FT, fat tissue; OV, ovaries.
H, horizontal; V, vertical; ?, horizontal transmission does not occur in culture.
Values for Hamiltosporidium spp. are derived from GeneMark ab initio predictions.
To investigate whether assembly artifacts due to allelic divergence generated segmental duplications, the four Hamiltosporidium assemblies were searched for contigs showing regions of at least 80% of sequence identity. Such highly similar segments range from about 0.9 to 59 kb (supplementary file 4, Supplementary Material online) and correspond to 5–15% of the assembled genomes (table 1). Because Hamiltosporidium is known to be diploid (Haag, Traunecker, et al. 2013), the duplicated segments may represent allelic versions of the same genomic regions, or genuine duplications. Therefore, the ratio of contig sequencing depth to average sequencing depth (Cdepth/Adepth) from the two H. tvaerminnensis assemblies showing Adepth larger than 300× (Adepth = 389× for H.t. FI-OER-3-3, and 645× for H.t. IL-G-3) was used as a proxy for their “ploidy.” Sequencing depths from genuine genomic duplications should match that of the assembly average; allelic contigs should yield Cdepth/Adepth ratios near 0.5. Duplicated contigs show a skewed distribution toward a lower Cdepth/Adepth ratios (supplementary file 5, Supplementary Material online); their mean ratio significantly differs from nonduplicated contigs (unequal variance t-test, P = 0) by about 0.3 in both assemblies (supplementary file 3, Supplementary Material online), suggesting that at least some segmental duplications may in fact represent alleles, not duplications.
Genomic Features Associated with the Mode of Transmission of Microsporidia
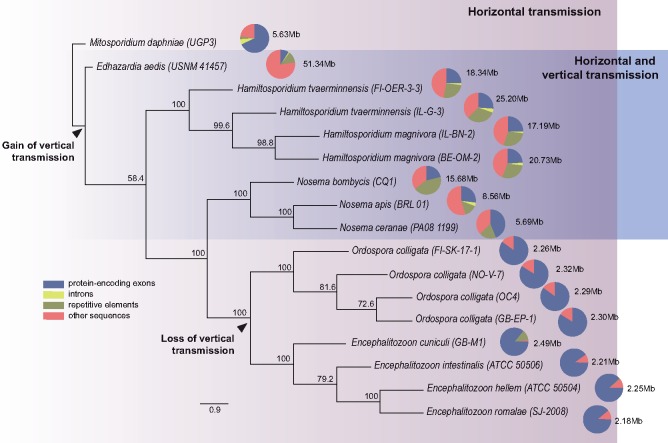
Our hypothesis to explain the divergent genome architectures in these two D. magna parasite taxa is that vertical transmission causes population bottlenecks and thus reduces Ne, and consequently the power of natural selection. If this hypothesis is correct, phylogenetically unrelated microsporidia that exploit similar routes of transmission should evolve similar genome architectures. Therefore, a phylogeny was built including Hamiltosporidium spp., O. colligata, as well as nine other microsporidian species with known genomes and modes of transmission (fig. 1). The tree contains a subset of some highly divergent microsporidia and is in agreement with previously published microsporidian phylogenies built with a larger number of species using different molecular markers (Vossbrinck and Debrunner-Vossbrinck 2005; Pombert et al. 2015). Horizontal transmission is an ancestral feature in our phylogeny and associated with a high proportion of protein-coding sequences within genomes. In contrast, the distantly related microsporidia with vertical transmission (in addition to horizontal transmission) show enlarged, less dense genomes with a reduced proportion of protein-encoding sequences (fig. 1). Nosema and Hamiltosporidium, in particular, are both characterized by the accumulation of repetitive elements. The compact genomes of the exclusively horizontally transmitted microsporidia, on the other hand, are largely devoid of repetitive sequences.
Fig. 1.
—Phylogenetic reconstruction of the history of 12 microsporidia with well-known transmission modes, based on 27 orthologous protein sequences. Mitosporidium daphniae, another horizontally transmitted gut parasite of Daphnia magna, is used as the root. Edhazardia aedis is a parasite of the mosquito Aedis aegypti transmitted vertically from adult hosts to their larval progeny, and horizontally among larvae (Desjardins et al. 2015). Nosema bombycis, a microsporidian parasite of the silkworm, as well as Nosema ceranae and Nosema apis, which are found in the honey bee, are vertically and horizontally transmitted; N. bombycis is transmitted through the host eggs (Han and Watanabe 1988), N. ceranae and N. apis, are sexually transmitted through the sperm of drones (Roberts et al. 2015). Among the Encephalitozoon species, Encephalitozoon romalae is found specifically in grasshoppers, whereas the other three species infect a broad range of mammals including humans. Although transplacental transmission is reported for Encephalitozoon cuniculi, horizontal transmission is the predominant transmission route of parasites from this genus (Cali and Takvorian 2014). Proportions of sequences within assemblies are indicated by pie charts with assembly sizes indicated next to the pie charts.
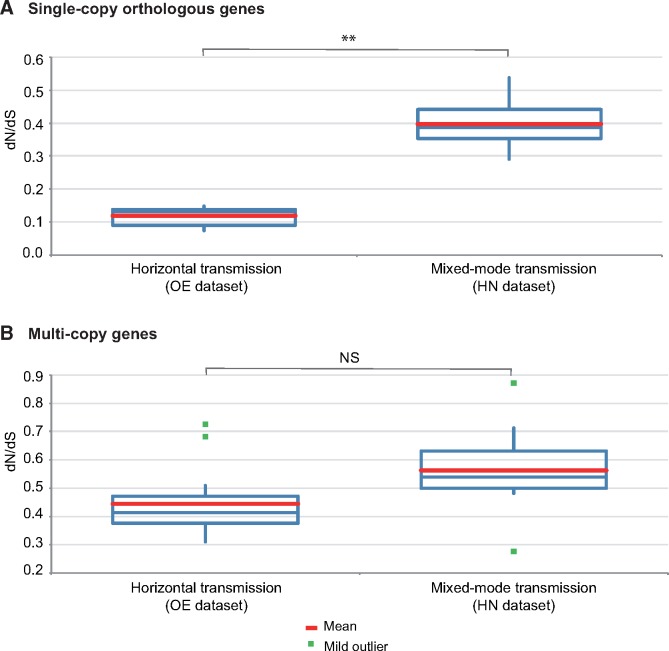
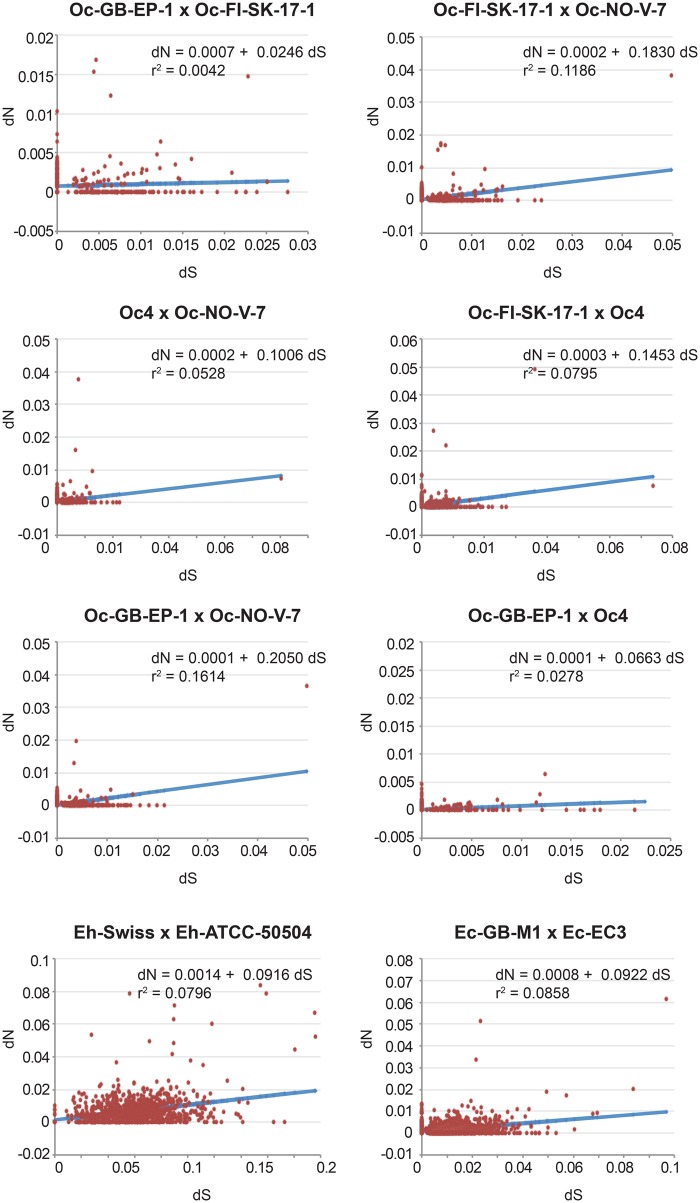
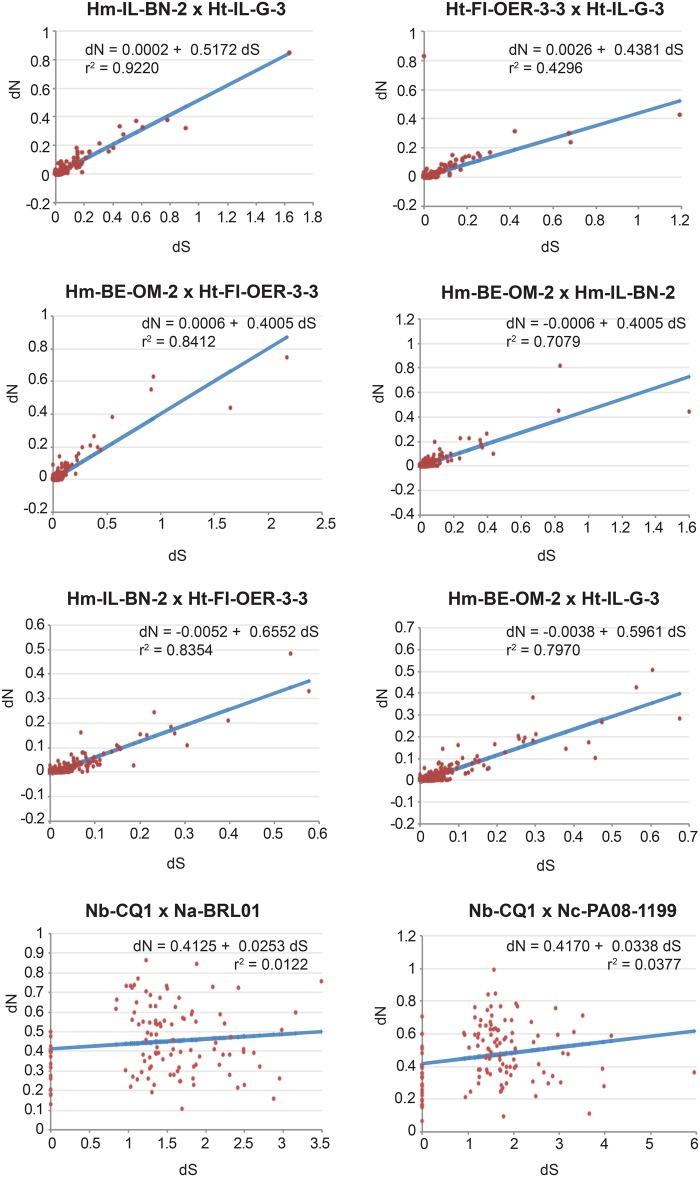
If the accumulation of noncoding and repetitive elements reflects the lack of power of purifying selection due to genetic drift, then vertically transmitted species are expected to show the accumulation of mildly deleterious mutations in their protein-encoding genes. To test for the differential strength of purifying selection in microsporidia with distinct modes of transmission, genome-wide estimates of the ratio of nonsynonymous versus synonymous substitutions (dN/dS) were obtained. We found a 4-fold increase in average dN/dS ratios of single-copy orthologous genes from species with mixed-mode transmission (HN data set) in relation to species with horizontal transmission only (OE data set; fig. 2A and supplementary file 6, Supplementary Material online). The difference is less clear, and not statistically significant, for multicopy genes (fig. 2B and supplementary file 6, Supplementary Material online). However, these multicopy genes include cases where “paralogous” copies might represent alleles. There is large variation in dN/dS estimates of genes encoding isoforms of similar proteins within genomes, reaching extreme values in the HN group with mixed-mode transmission (see supplementary file 6, Supplementary Material online). In order to control for the large variation in evolutionary distances between taxa and to account for differences in behavior of subsets of genes within our data set, we plotted the distribution of dN versus dS (supplementary file 7, Supplementary Material online) for a sample of microsporidian genomes from the OE and HN data sets with comparable dS estimates. Because synonymous substitutions are considered neutral, dS can be used as a proxy for their evolutionary distance. For microsporidia relying exclusively on horizontal transmission (OE data set), dN distributions are flat, independently of the evolutionary distance (fig. 3), with regression slopes of 0.02–0.2 and intercept around 0.0 (fig. 3), whereas for those with mixed-mode transmission (HN data set), the regression slopes are much larger (0.4–0.65, and intercepts around 0.0), but only at shorter evolutionary distances (top graphs in fig. 4). For the highly divergent genomes of Nosema spp. (bottom graphs in fig. 4), the regression slopes are equivalent to those obtained for the OE data set, but the intercepts are much larger, suggesting that dN might saturate at high dS. Furthermore, the large dN versus dS correlation coefficients of the Hamiltosporidium spp. comparisons indicate that the excess of nonsynonymous substitutions in vertically transmitted microsporidia are not biased by small subset of genes with extremely high dN, but instead represent a genomic pattern.
Fig. 2.
—Box plots of mean dN/dS estimates (A) for all orthologous genes identified in OE and HN data sets, based on 112,055 and 8,846 pairwise comparisons, respectively and (B) for all copies of similar genes within genomes based on 7,790 comparisons of data set OE and 64,437 of data set HN. The statistical significance of the difference between dN/dS means was assessed by unequal variance t-tests (**P < 0.01; NS = not significant; supplementary file 4, Supplementary Material online).
Fig. 3.
—Linear regression analyses of dN versus dS estimates for all orthologous genes identified in a subset of comparisons from the OE data set (see supplementary file 7, Supplementary Material online, for details). The regression equations describing the best fit of the dN versus dS distributions are given. Correlation coefficients are indicated. Oc = O. colligata; Eh = E. hellem; Ec = E. cuniculi.
Fig. 4.
—Linear regression analyses of dN versus dS estimates for all orthologous genes identified in a subset of comparisons from the HN data set (see supplementary file 7, Supplementary Material online, for details). The regression equations describing the best fit of the dN versus dS distributions are given. Correlation coefficients are indicated. Hm = H. magnivora; Ht = H. tvaerminnensis; Nb = N. bombycis; Na = N. apis; Nc = N. ceranae.
Reduced Levels of Recombination in Hamiltosporidium spp.
Accumulation of nonsynonymous substitutions as well as assembly artifacts caused by allelic divergence within Hamiltosporidium genomes could result from the lack of recombination (Muller 1964; Welch and Meselson 2000). Assuming that each Hamiltosporidium isolate corresponds to a single diploid individual, we calculated the recombination rate ρ for genomic regions considering three different levels of coverage (c = 4.8 and 16; supplementary file 8, Supplementary Material online). Recombination estimates are slightly above the sequencing error rate, averaging 158 events per 100 kb. Although we also applied the same approach to Ordospora spp., the zygosity correlation (δ) could not be calculated—in line with our current expectations that these species are either haploids or show very low levels of heterozygosity like their Encephalitozoon relatives (Selman et al. 2013)—and as such the values reported for Ordospora spp. in supplementary file 8, Supplementary Material online (1,114 events per 100 kb on average) should only be considered with caution. To enable a comparison between Hamiltosporidium and Ordospora, we assessed recombination indirectly, searching for genomic regions with increased GC content (gBGC tracts). In the absence of recombination, gBGC should occur at greatly reduced rates, eliminating such regions. We employed a Bayesian approach to identify gBGC tracts within all orthologous single-copy genes of the seven new genome assemblies plus the previously published Ordospora genome; 1,227 genes for Hamiltosporidium spp. and 1,748 for O. colligata). Overall, we found 141 gBGC tracts with probability >0.5, ranging from 10 to 2,168 bp in the four Hamiltosporidium genomes, but none for O. colligata (supplementary file 9, Supplementary Material online). The vast majority (111) of those tracts were found in H.m. BE-OM-2, whereas only 12, 10, and 8 tracts were found in H.t. FI-OER-3-3, H.t. IL-G-3, and H.m. IL-BN-2, respectively. Overall, these results suggest that some form of recombination (either mitotic or meiotic) occurs in Hamiltosporidium, favoring the hypothesis that the excess of nonsynonymous substitutions observed in the vertically transmitted microsporidia results from genetic drift, and not from the lack of recombination.
Discussion
Exploring the genomic landscape of seven new microsporidian genomes, we found several differences between Hamiltosporidium and Ordospora taxa: The former have more protein-coding genes, a larger proportion of repetitive sequences relative to the total assembled sequence, longer intergenic regions, and reduced GC content. These differences are associated with a 10-fold larger genome size of Hamiltosporidium compared with Ordospora, suggesting a possible link between the genomic features and genome size. Microsporidian genome size variation has remained elusive due to insufficient genomic data from species with sufficiently well described life histories. The two taxa sequenced here are good models for studying the mechanisms shaping genome evolution, given that our knowledge about their ecology, evolution, and epidemiology is better than for other microsporidia (Ebert et al. 2000, 2001; Ebert 2005; Lass and Ebert 2006; Sheikh-Jabbari et al. 2014; Urca and Ben-Ami 2018; Kirk et al. 2019).
Natural Selection for Smaller or Larger Coding Capacity
It is suggested that the divergence of microsporidia from their fungal relatives coincided with a dramatic bottleneck that resulted in the evolutionary loss of gene families, and proceeded with the gain and subsequent adaptive enlargement of other gene families, resulting in an expanded core proteome (Nakjang et al. 2013). Sharp contrasts in microsporidian genomic architectures are proposed to be derived from divergent factors causing differential selection and being associated with variable degrees of host specificity (Desjardins et al. 2015), such that genomes from microsporidia that use a range of different host species would be selected for having a larger and more flexible protein repertoire than microsporidia restricted to a single host species. However, neither gene family expansions nor host adaptation seems satisfying explanations for their genome size variation, as much of the variation remains unexplained. For example, all encephalitozoans have extremely reduced genomes of about 2.5 Mb, but their host specificities vary. Furthermore, the increase in genome size in microsporidia is associated with a proportional increase in noncoding sequence, whereas the associated increase of the proteome is much smaller. The here studied microsporidia are highly host specific, with D. magna being the only known host (Green 1974; Ebert et al. 2001; Ebert 2005). Still Hamiltosporidium spp. and O. colligata genomes differ by a factor of 10, whereas their proteomes only differ by a factor of 2–3 (table 1). Perhaps a valid generalization on microsporidian genomes is that there are those that we might call large and “gene sparse,” whereas others are smaller and “gene dense” (Keeling et al. 2014). Taking into account that the “gene sparse” genomes are scattered across the microsporidian phylogeny (see Pombert et al. 2015), and if natural selection would be the only mechanism shaping genome architecture, then the accumulation of noncoding and potentially deleterious sequences would remain unexplained.
Accumulation of Selfish TEs
It is believed that the wide variation in genome size observed among eukaryotic species is more closely correlated with the amount of repetitive DNA than with the number of coding genes (Kidwell and Lisch 2000). In microsporidia, the smallest encephalitozoan genomes are devoid of TEs, and large genomes, such as from Anncaliia algerae (23 Mb), contain hundreds of copies of different families of TEs (Parisot et al. 2014). Similarly, the Hamiltosporidium genomes (17–25 Mb) that we have sequenced contain a large proportion of repetitive elements, some of which identified as known TE families. The age-distribution of different TEs in Hamiltosporidium genomes suggests the occurrence of cyclical TE expansions; the largest genome of strain H.t. IL-G-3 shows two peaks in the abundance of TEs from distinct age classes (supplementary file 3, Supplementary Material online). However, the largest known microsporidian genome, from the mosquito parasite Edhazardia aedis (assembly size of 51.34 Mb; fig. 1), is rather poor in TEs, but enriched with intergenic AT-biased sequences (Desjardins et al. 2015). This finding is not in direct support of a simple relationship between total TE-derived DNA and microsporidian genome size.
Reduced Recombination and the Accumulation of TEs
Reduced recombination has been associated with TE accumulation and larger genomes (Tiley and Burleigh 2015). Two potential mechanisms for a negative association between recombination rate and genome size are that recombination either deletes TEs by chance or facilitates selection against TE insertions (Langley et al. 1988). However, recent evidence suggests that TEs might actively contribute to the spread of recombination suppression (Kent et al. 2017). Nevertheless, the absence of recombination reduces effectiveness of natural selection on short and long evolutionary timescales and is predicted to be associated with reduced GC content because gBGC does not occur (Capra et al. 2013). Searching our assemblies for regions suggestive of gBGC and by estimating recombination rates directly, we detected recombination in Hamiltosporidium, though at very low rates (0.0006–0.00158; supplementary file 8, Supplementary Material online). These results are lower than recombination estimates obtained for other asexual fungi such as Candida glabrata (0.003–0.008, Carreté et al. 2018), which may engage in parasexual processes that allow them to reshuffle DNA, and which have been postulated to achieve gBGC by mitotic recombination (Marsolier-Kergoat 2013). In contrast, recombination rates calculated for Ordospora spp. were found higher by a 10-fold, congruent with the idea that the Hamiltosporidium spp. underwent fewer recombination events. Overall, the apparent lower recombination rates in Hamiltosporidium spp. are compatible with the idea that these genomes acquired TE at least in part due to reduced recombination pressure.
Genetic Drift Caused by Vertical Transmission
A general model of genome evolution must account for the accumulation of neutral and maladaptive sequences, as well as their elimination. The two microsporidian taxa that we compared are specific to D. magna but differ in their modes of transmission. Mixed-mode transmitted Hamiltosporidium spp. show larger and “gene sparse” genomes containing duplicated segments and other repetitive elements. These are common features of eukaryotic genomes that have been affected by genetic drift. Mixed-mode transmission is a strategy used by many microsporidia (Dunn et al. 2001), enabling them to achieve very high prevalence at equilibrium (Lipsitch et al. 1995; Lass and Ebert 2006). Vertical transmission in addition to horizontal transmission involves a fitness tradeoff (Vizoso and Ebert 2005) but is probably an important survival strategy when opportunities to transmit horizontally are temporarily reduced or absent, for example, under low host density and during a phase of adverse environmental conditions (Lucarotti and Andreadis 1995; Ebert 2013; Sheikh-Jabbari et al. 2014). Vertical transmission under such conditions may cause transmission bottlenecks and thus reduces Ne. As a consequence, the strength of natural selection is reduced, because genetic drift adds stochastic noise to the fate of variants.
In microsporidia with mixed-mode transmission, two morphologically different spore types are often observed, and vertically transmitted spores resemble those at early stages of development, with a shorter polar filament and a thinner endospore wall (Dunn et al. 2001). Horizontally transmitted spores, on the other hand, show a thick cell wall and are produced in large numbers, creating conditions for multiple infections of a single host with a diversity of parasite genotypes (Dunn and Smith 2001). This is less likely with vertical transmission, because only a small proportion of spores translocate into reproductive cells. Hamiltosporidiumtvaerminnensis produces two types of spores with different shapes, but they do not seem to differ with regard to their roles in horizontal transmission (Urca and Ben-Ami 2018). Although the ratio of vertical to horizontal transmission is not known for microsporidia with mixed-mode transmission, studies using microsatellite polymorphisms in Hamiltosporidium suggest that multiple infections are rare (Haag, Sheikh-Jabbari, et al. 2013). Once the Daphnia host is born vertically infected, the chances of becoming horizontally reinfected by a different Hamiltosporidium genotype may be low. Theory predicts that the tradeoff between horizontal and vertical transmission is controlled by the proportion of available uninfected hosts, with vertical transmission being selectively favored when uninfected hosts are rare (Turner et al. 1998). In Finland, where H. tvaerminnensis is common in a D. magna metapopulation, parasite prevalence is cyclical, reaching up to 100% during summer, and decreasing every year after winter and summer host diapauses (Lass and Ebert 2006). Hence, it is reasonable to assume that vertical transmission is favored when H. tvaerminnensis prevalence is high, thus reducing Ne and facilitating the accumulation of neutral and mildly deleterious mutations by genetic drift.
Accumulation of Mildly Deleterious Mutations
To further investigate the hypothesis that microsporidian genomes expand through a reduced effectiveness of purifying selection, we compared the ratio of nonsynonymous versus synonymous substitutions in the microsporidian genomes. We found that mixed-mode transmission, and larger genomes, are associated with significantly larger dN/dS ratios between single-copy orthologous genes of different genomes (fig. 2A). We speculate that the excess of nonsynonymous substitutions in orthologous genes results from the accumulation of mildly deleterious mutations due to genetic drift. Variable dN/dS ratios among multiple copy sequences (fig. 2B) indicate variable functional constraints (Ohta 1992). Thus, it seems that the reduced power of natural selection associated with the addition of vertical transmission, may contribute to the accumulation of noncoding sequences, TEs, and duplicated genes, leading to microsporidian genome expansions. In the long term, the opposite outcome occurs in exclusively vertically transmitted bacterial endosymbionts, which have extremely miniaturized genomes (Mira and Moran 2002; Moran and Bennett 2014). Genome miniaturization associated with genetic drift in endosymbiotic bacteria is believed to be caused by a mutational bias toward deletions (Mira et al. 2001; Kuo and Ochman 2009).
Relative Roles of Reduced Recombination and Vertical Transmission
Accumulation of mildly deleterious mutations might be generated both by reduced recombination via Muller’s ratchet, or by reduced Ne due to recurrent population bottlenecks. Sex has been lost multiple times in the evolutionary history of microsporidia, including Nosema and Hamiltosporidium (Ironside 2007; Haag, Traunecker, et al. 2013; Lee et al. 2014). Indeed, although most microsporidia seem to contain a conserved set of meiosis genes, key meiosis genes apparently were lost in some microsporidian lineages (Lee et al. 2014), such as Encephalitozooncuniculi (Pelin et al. 2016), an icon of microsporidian genome streamlining. One of the few microsporidia known to perform regular meiosis is Amblyospora, a mosquito parasite with mixed-mode transmission (Hazard and Brookbank 1984). In this group, horizontal transmission happens between the mosquito and the second host, a copepod, that becomes infected by the haploid meiospores produced in the vertically infected mosquito larvae (Becnel and Andreadis 2014). Unfortunately, Amblyospora does not have a sequenced genome, but a closely related species, Ehazhardia aedis, with mixed-mode transmission (fig. 1), and supposed to perform meiosis as well (Becnel et al. 1989), is known for having one of the most “gene sparse” genomes among microsporidia (Williams et al. 2008). It was suggested that H. magnivora from Belgium (here represented by H.m. BE-OM-2) undergoes the same form of a two host life cycle with recombination occurring in the second—still unknown—host (Haag, Traunecker, et al. 2013). Overall, the available data on microsporidian life histories consistently implicate the occurrence of vertical transmission in genome expansions, whereas the lack of meiosis does not.
In summary, our results suggest that microsporidia relying mostly on horizontal transmission, such as O. colligata and Encephalitozoon spp., probably maintain large population sizes that are required for genome streamlining. On the other hand, those that are mixed mode transmitted, such as Nosema spp. and Hamiltosporidium spp., are likely to experience recurrent population bottlenecks with vertical transmission, which would decrease Ne, and consequently the efficiency of purifying selection. Increased levels of genetic drift are correlated with genome expansions in microsporidia, fitting the expectations that nonadaptive forces play a greater role in shaping small sized populations. Thus, the intracellular mode of life is not the only factor playing a role in shaping microsporidian genome architectures, and population genetic structure, strongly influenced by the parasite’s mode of transmission, might explain why their genomes differ so much in size.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (grants 31003A_146462 and 310030B_166677) to D.E., Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant PQ 302121/2017-0) to K.L.H, and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award number R15AI128627) to J.-F.P. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Urs Stiefel and Jürgen Hottinger for support in the laboratory. We especially thank to Sidia Maria Callegari Jacques for her help with the statistical analyses.
Data deposition: This project has been deposited at GenBank under the accession PRJNA419750.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, et al. 2017. EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 45(D1):D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel JJ, Andreadis TG. 2014. Microsporidia in insects In: Weiss LM, Becnel JJ, editors. Microsporidia: pathogens of opportunity. Ames (IA: ): John Wiley & Sons; p. 521–570. [Google Scholar]
- Becnel JJ, Sprague V, Fukuda T, Hazard EI. 1989. Development of Edhazardia aedis (Kudo, 1930) N. G., N. Comb. (Microsporida: Amblyosporidae) in the mosquito Aedes aegypti (L.) (Diptera: Culicidae). J Protozool. 36(2):119–130. [DOI] [PubMed] [Google Scholar]
- Boakye DW, et al. 2017. Decay of the glycolytic pathway and adaptation to intranuclear parasitism within Enterocytozoonidae microsporidia. Environ Microbiol. 19:2077–2089. [DOI] [PubMed] [Google Scholar]
- Bobay L-M, Ochman H. 2017. The evolution of bacterial genome architecture. Front Genet. 8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17(11):1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero J. 2012. RepeatLandscape Institute for Systems Biology, Seattle. Available from: https://github.com/caballero/RepeatLandscape; last accessed December 18, 2019.
- Cali A, Takvorian PM. 2014. Developmental morphology and life cycles of the Microsporidia In: Weiss LM, Becnel JJ, editors. Microsporidia: pathogens of opportunity. Ames (IA: ): John Wiley & Sons; p. 71–133. [Google Scholar]
- Canning EU, Curry A, Cheney S, Lafranchi-Tristem NJ, Haque MA. 1999. Vairimorpha imperfecta n.sp., a microsporidian exhibiting an abortive octosporous sporogony in Plutella xylostella L. (Lepidoptera: Yponomeutidae). Parasitology 119(3):273–286. [DOI] [PubMed] [Google Scholar]
- Capra JA, Hubisz MJ, Kostka D, Pollard KS, Siepel A. 2013. A model-based analysis of GC-biased gene conversion in the human and chimpanzee genomes. PLoS Genet. 9(8):e1003684.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreté L, et al. 2018. Patterns of genomic variation in the opportunistic pathogen Candida glabrata suggest the existence of mating and a secondary association with humans. Curr Biol. 28(1):15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Haag KL, Pombert J-F, Ebert D, Keeling PJ. 2009. Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host–parasite interactions. Genome Biol. 10(10):R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Pombert J-F, Farinelli L, Didier ES, Keeling PJ. 2010. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat Commun. 1:77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Slamovits CH. 2011. The intriguing nature of microsporidian genomes. Brief Funct Genomics. 10(3):115–124. [DOI] [PubMed] [Google Scholar]
- Desjardins CA, et al. 2015. Contrasting host–pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes. Nat Commun. 6(1):7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukić M, Berner D, Roesti M, Haag CR, Ebert D. 2016. A high-density genetic map reveals variation in recombination rate across the genome of Daphnia magna. BMC Genet. 17(1):137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AM, Smith JE. 2001. Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microbes Infect. 3(5):381–388. [DOI] [PubMed] [Google Scholar]
- Dunn AM, Terry RS, Smith JE. 2001. Transovarial transmission in the microsporidia. Adv Parasitol. 48:57–100. [DOI] [PubMed] [Google Scholar]
- Ebert D. 2005. Ecology, epidemiology, and evolution of parasitism in Daphnia [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available from: http://www.ncbi.nlm.nih.gov/books/NBK2036/; last accessed December 18, 2019.
- Ebert D. 2013. The epidemiology and evolution of symbionts with mixed-mode transmission. Annu Rev Ecol Evol Syst. 44(1):623–643. [Google Scholar]
- Ebert D, Hottinger JW, Pajunen VI. 2001. Temporal and spatial dynamics of parasite richness in a Daphnia metapopulation. Ecology 82:3417–3434. [Google Scholar]
- Ebert D, Lipsitch M, Mangin KL. 2000. The effect of parasites on host population density and extinction: experimental epidemiology with Daphnia and six microparasites. Am Nat. 156(5):459–477. [DOI] [PubMed] [Google Scholar]
- Ebert D, Zschokke-Rohringer CD, Carius HJ. 1998. Within- and between-population variation for resistance of Daphnia magna to the bacterial endoparasite Pasteuria ramosa. Proc R Soc Lond B 265(1410):2127–2134. [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PD, et al. 2018. Mitogenome phylogeographic analysis of a planktonic crustacean. Mol Phylogenet Evol. 129:138–148. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Cameron Thrash J, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J. 8(8):1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8(3):195–202. [DOI] [PubMed] [Google Scholar]
- Green J. 1974. Parasites and epibionts of Cladocera. Trans Zool Soc Lond. 32(6):417–515. [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag KL, Larsson JIR, Refardt D, Ebert D. 2011. Cytological and molecular description of Hamiltosporidium tvaerminnensis gen. et sp. nov., a microsporidian parasite of Daphnia magna, and establishment of Hamiltosporidium magnivora comb. nov. Parasitology 138(4):447–462. [DOI] [PubMed] [Google Scholar]
- Haag KL, Sheikh-Jabbari E, Ben-Ami F, Ebert D. 2013. Microsatellite and single-nucleotide polymorphisms indicate recurrent transitions to asexuality in a microsporidian parasite. J Evol Biol. 26(5):1117–1128. [DOI] [PubMed] [Google Scholar]
- Haag KL, Traunecker E, Ebert D. 2013. Single-nucleotide polymorphisms of two closely related microsporidian parasites suggest a clonal population expansion after the last glaciation. Mol Ecol. 22(2):314–326. [DOI] [PubMed] [Google Scholar]
- Haag KL, et al. 2014. Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc Natl Acad Sci U S A. 111(43):15480–15485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M-S, Watanabe H. 1988. Transovarial transmission of two microsporidia in the silkworm, Bombyx mori, and disease occurrence in the progeny population. J Invertebr Pathol. 51(1):41–45. [Google Scholar]
- Haubold B, Pfaffelhuber P, Lynch M. 2010. mlRho—a program for estimating the population mutation and recombination rates from shotgun-sequenced diploid genomes. Mol Ecol. 19:277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazard EI, Brookbank JW. 1984. Karyogamy and meiosis in an Amblyospora sp. (Microspora) in the mosquito Culex salinarius. J Invertebr Pathol. 44(1):3–11. [Google Scholar]
- Heinz E, et al. 2012. The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 8(10):e1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1):491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes AK, et al. 2013. Bacterial adaptation through loss of function. PLoS Genet. 9(7):e1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside JE. 2007. Multiple losses of sex within a single genus of Microsporidia. BMC Evol Biol. 7(1):48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, et al. 2013. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol. 23(16):1548–1553. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414(6862):450–453. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P, Fast NM, Corradi N. 2014. Microsporidian genome structure and function In: Weiss LM, Becnel JJ, editors. Microsporidia: pathogens of opportunity. Ames (IA: ): John Wiley & Sons; p. 221–229. [Google Scholar]
- Keeling PJ, et al. 2010. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol Evol. 2:304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27(6):757–763. [DOI] [PubMed] [Google Scholar]
- Kent TV, Uzunović J, Wright SI. 2017. Coevolution between transposable elements and recombination. Philos Trans R Soc B 372(1736):20160458.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 2000. Transposable elements and host genome evolution. Trends Ecol Evol. 15(3):95–99. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120. [DOI] [PubMed] [Google Scholar]
- Kirk D, Luijckx P, Stanic A, Krkošek M. 2019. Predicting the thermal and allometric dependencies of disease transmission via the metabolic theory of ecology. Am Nat. 193(5):661–676. [DOI] [PubMed] [Google Scholar]
- Korber B. 2000. HIV signature and sequence variation analysis In: Rodrigo AG, Learn GH, editors. Computational analysis of HIV molecular sequences. Dordrecht (the Netherlands: ): Kluwer Academic Publishers; p. 55–72. [Google Scholar]
- Kuo C-H, Ochman H. 2009. Deletional bias across the three domains of life. Genome Biol Evol. 1:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 52(3):223–235. [DOI] [PubMed] [Google Scholar]
- Lass S, Ebert D. 2006. Apparent seasonality of parasite dynamics: analysis of cyclic prevalence patterns. Proc R Soc B 273(1583):199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, et al. 2013. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 14(8):R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Heitman J, Ironside JE. 2014. Sex and the Microsporidia In: Weiss LM, Becnel JJ, editors. Microsporidia: pathogens of opportunity. Ames (IA: ): John Wiley & Sons; p. 231–243. [Google Scholar]
- Lipsitch M, Nowak MA, Ebert D, May RM. 1995. The population dynamics of vertically and horizontally transmitted parasites. Proc R Soc Lond B Biol. 260:321–327. [DOI] [PubMed] [Google Scholar]
- Lucarotti CJ, Andreadis TG. 1995. Reproductive strategies and adaptations for survival among obligatory microsporidian and fungal parasites of mosquitos—a comparative analysis of Ambylospora and Coelomomyces. J Am Mosquito Contr. 11:111–121. [PubMed] [Google Scholar]
- Lynch M. 2007a. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A. 104(Suppl 1):8597–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007b. The origins of genome architecture. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302(5649):1401–1404. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43(D1):D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier-Kergoat M-C. 2013. Models for the evolution of GC content in asexual fungi Candida albicans and C. dubliniensis. Genome Biol Evol. 5(11):2205–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Mira A, Moran NA. 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol. 44(2):137–143. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17(10):589–596. [DOI] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93(7):2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68(1):195–215. [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 14(6):627–633. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1964. The relation of recombination to mutational advance. Mutat Res. 106:2–9. [DOI] [PubMed] [Google Scholar]
- Nakjang S, et al. 2013. Reduction and expansion in microsporidian genome evolution: new insights from comparative genomics. Genome Biol Evol. 5(12):2285–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AI, et al. 2005. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA. 102:12112–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Wolf YI, Dubchak I, Koonin EV. 2009. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J. Bacteriol. 191(1):65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack ECM, Weber A. 2018. Genomics-informed insights into endosymbiotic organelle evolution in photosynthetic eukaryotes. Annu Rev Plant Biol. 69(1):51–84. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 23(1):263–286. [Google Scholar]
- Parisot N, et al. 2014. Microsporidian genomes harbor a diverse array of transposable elements that demonstrate an ancestry of horizontal exchange with metazoans. Genome Biol Evol. 6(9):2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelin A, et al. 2016. The genome of an Encephalitozoon cuniculi type III strain reveals insights into the genetic diversity and mode of reproduction of a ubiquitous vertebrate pathogen. Heredity 116(5):458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyretaillade E, et al. 2011. Extreme reduction and compaction of microsporidian genomes. Res Microbiol. 162(6):598–606. [DOI] [PubMed] [Google Scholar]
- Pombert J-F, Haag KL, Beidas S, Ebert D, Keeling PJ. 2015. The Ordospora colligata genome: evolution of extreme reduction in microsporidia and host-to-parasite horizontal gene transfer. mBio 6(1):e02400-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombert J-F, et al. 2012. Gain and loss of multiple functionally related, horizontally transferred genes in the reduced genomes of two microsporidian parasites. Proc Natl Acad Sci U S A. 109(31):12638–12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombert J-F, et al. 2013. Complete genome sequences from three genetically distinct strains reveal high intraspecies genetic diversity in the microsporidian Encephalitozoon cuniculi. Eukaryot Cell 12(4):503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery E. 2011. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One 6(9):e22594.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KE, Evison SEF, Baer B, Hughes WOH. 2015. The cost of promiscuity: sexual transmission of Nosema microsporidian parasites in polyandrous honey bees. Scientific Reports. 5:10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin AC, et al. 2013. Local adaptation of sex induction in a facultative sexual crustacean: insights from QTL mapping and natural populations of Daphnia magna. Mol Ecol. 22(13):3567–3579. [DOI] [PubMed] [Google Scholar]
- Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069.24642063 [Google Scholar]
- Selman M, et al. 2013. Extremely reduced levels of heterozygosity in the vertebrate pathogen Encephalitozoon cuniculi. Eukaryot Cell 12(4):496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh-Jabbari E, Hall MD, Ben-Ami F, Ebert D. 2014. The expression of virulence for a mixed-mode transmitted parasite in a diapausing host. Parasitology 141(8):1097–1107. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013. RepeatMasker Institute for Systems Biology, Seattle. Available from: http://www.repeatmasker.org/RepeatModeler/; last accessed December 18, 2019.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18(12):1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley GP, Burleigh JG. 2015. The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. BMC Evol Biol. 15:194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Cooper VS, Lenski RE. 1998. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52(2):315–329. [DOI] [PubMed] [Google Scholar]
- Urca H, Ben-Ami F. 2018. The role of spore morphology in horizontal transmission of a microsporidium of Daphnia. Parasitology 145(11):1452–1457. [DOI] [PubMed] [Google Scholar]
- Vizoso DB, Ebert D. 2005. Phenotypic plasticity of host–parasite interactions in response to the route of infection. J Evol Biol. 18(4):911–921. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Debrunner-Vossbrinck B. A. 2005. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitol. 52(1–2):131–142. [DOI] [PubMed] [Google Scholar]
- Welch DBM, Meselson M. 2000. Evidence for the evolution of Bdelloid rotifers without sexual reproduction or genetic exchange. Science 288(5469):1211–1215. [DOI] [PubMed] [Google Scholar]
- Whelan TA, Lee NT, Lee RCH, Fast NM. 2019. Microsporidian introns retained against a background of genome reduction: characterization of an unusual set of introns. Genome Biol Evol. 11(1):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, et al. 2008. Genome sequence surveys of Brachiola algerae and Edhazardia aedis reveal microsporidia with low gene densities. BMC Genomics. 9(1):200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. 2006. The varying microsporidian genome: existence of long-terminal repeat retrotransposon in domesticated silkworm parasite Nosema bombycis. Int J Parasitol. 36(9):1049–1056. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Schaer TMM, Ebert D. 2014. Adaptive phenotypic plasticity and local adaptation for temperature tolerance in freshwater zooplankton. Proc R Soc B 281(1776):20132744.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All customs scripts and software are available on the Pombert lab GitHub page (https://github.com/PombertLab; last accessed December 18, 2019). This project was deposited in NCBI under BioProject accession number PRJNA419750. Sequencing data sets were deposited in the NCBI Sequence Read Archive under the same accession number (PRJNA419750). The Ordospora genomes were deposited in GenBank under accession numbers PITH00000000 (FI-SK-17-1), PITG00000000 (GB-EP-1), and PITF00000000 (NO-V-7). The H.magnivora genomes were deposited under accession numbers PITI00000000 (BE-OM-2) and PIXR00000000 (IL-BN-2). The H.tvaerminnensis genomes were deposited under accession numbers PITJ00000000 (FI-OER-3-3) and PITK00000000 (IL-G-3).