Exploratory myocardial and plasma proteomics screens in patients with hypertrophic cardiomyopathy identified new biomarkers that were developed into a multiplexed targeted liquid chromatography-tandem/mass spectrometry-based assay for validation across a larger cohort of patients with this disease. Using quantitative proteomics and supervised machine learning, this six-biomarker panel shows a relationship with myocardial substrate changes in patients with hypertrophic cardiomyopathy and with their estimated sudden cardiac death risk.
Keywords: Cardiovascular Disease, Cardiovascular Function or Biology, Diagnostic, Mass Spectrometry, Multiple Reaction Monitoring
Graphical Abstract
Highlights
Quantitative proteomics and machine learning to study plasma biomarkers in HCM.
Six peptides are increased in plasma of LVH+ HCM compared to controls.
Peptide biomarkers correlate with imaging markers of phenotype severity.
Peptide biomarkers correlate with the estimated sudden cardiac death risk.
Abstract
Hypertrophic cardiomyopathy (HCM) is defined by pathological left ventricular hypertrophy (LVH). It is the commonest inherited cardiac condition and a significant number of high risk cases still go undetected until a sudden cardiac death (SCD) event. Plasma biomarkers do not currently feature in the assessment of HCM disease progression, which is tracked by serial imaging, or in SCD risk stratification, which is based on imaging parameters and patient/family history. There is a need for new HCM plasma biomarkers to refine disease monitoring and improve patient risk stratification. To identify new plasma biomarkers for patients with HCM, we performed exploratory myocardial and plasma proteomics screens and subsequently developed a multiplexed targeted liquid chromatography-tandem/mass spectrometry-based assay to validate the 26 peptide biomarkers that were identified. The association of discovered biomarkers with clinical phenotypes was prospectively tested in plasma from 110 HCM patients with LVH (LVH+ HCM), 97 controls, and 16 HCM sarcomere gene mutation carriers before the development of LVH (subclinical HCM). Six peptides (aldolase fructose-bisphosphate A, complement C3, glutathione S-transferase omega 1, Ras suppressor protein 1, talin 1, and thrombospondin 1) were increased significantly in the plasma of LVH+ HCM compared with controls and correlated with imaging markers of phenotype severity: LV wall thickness, mass, and percentage myocardial scar on cardiovascular magnetic resonance imaging. Using supervised machine learning (ML), this six-biomarker panel differentiated between LVH+ HCM and controls, with an area under the curve of ≥ 0.87. Five of these peptides were also significantly increased in subclinical HCM compared with controls. In LVH+ HCM, the six-marker panel correlated with the presence of nonsustained ventricular tachycardia and the estimated five-year risk of sudden cardiac death. Using quantitative proteomic approaches, we have discovered six potentially useful circulating plasma biomarkers related to myocardial substrate changes in HCM, which correlate with the estimated sudden cardiac death risk.
Hypertrophic cardiomyopathy (HCM) is a common myocardial disorder characterized by left ventricular hypertrophy (LVH) caused predominantly by mutations in cardiac sarcomere protein genes (1, 2). Disease manifestations, including symptoms, are highly variable among patients with HCM and occasionally first presentation is with a major adverse sudden cardiac event, including death. Although the genetic architecture of the disease has been substantially resolved, the impact of this knowledge on therapy has been limited largely due to a lack of understanding about the determinants of disease progression. In HCM, genetic testing is used to identify known pathogenic disease-causing mutations, while electrocardiographic (EKG) and cardiac imaging tests are used to elaborate the overall phenotype and monitor disease progression. New biomarkers are needed to better guide the intensity of imaging surveillance, refine current risk stratification algorithms, and track disease progression so the impact of existing and novel therapies can be assessed.
Global proteome studies have provided mechanistic insights into many cardiovascular diseases, but there are few such studies in human cardiomyopathies. Recently, we examined myocardial tissue removed from patients with HCM at the time of cardiac surgery using an unbiased label-free proteomic characterization of myocardium and demonstrated that the tissue proteome of the disease is characterized by dysregulation of metabolic and structural proteins (3).
The aim of the present study was to combine, into one targeted multiple reaction monitoring (MRM) liquid chromatography mass spectrometry-based (LC-MS/MS) assay, the peptides identified in the previous myocardial study, with peptides identified in a proteomics plasma screen. We then applied this “tier 2” (4) targeted proteomic assay on a prospective cohort of patients with LVH+ HCM and controls to validate if any of these biomarkers circulating in the blood have potential clinical utility in terms of disease monitoring and risk stratification.
There is evidence that cardiac structural changes (crypts (5), anterior mitral valve leaflet elongation, increased apical trabecular complexity (6)), myocardial disarray (7), and aberrant mechanics (8) precede the establishment of LVH in at least some patients with HCM. We therefore additionally studied biomarker levels in the plasma of HCM sarcomere gene mutation carriers before the development of LVH (subclinical HCM).
MATERIALS AND METHODS
Experimental Design and Statistical Rationale
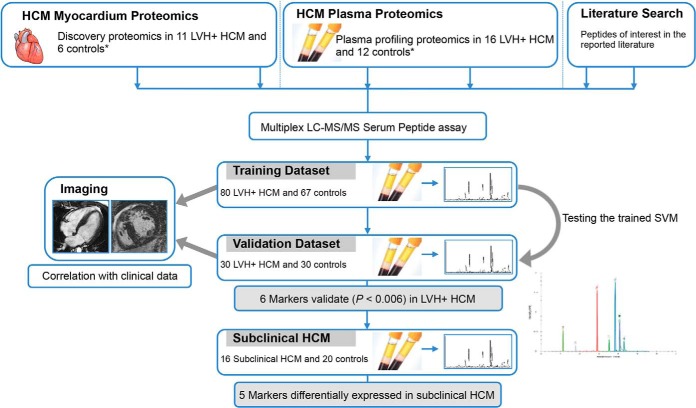
Fig. 1 outlines the experimental design. This observational, prospective, single-center study recruited 110 LVH+ HCM patients and 97 healthy volunteers as controls, randomly split into training and validation cohorts, in order to identify and subsequently verify, differentially expressed proteomics biomarkers and develop them into a preliminary plasma assay. The assay was then prospectively applied to 16 patients with subclinical HCM and biomarker levels compared with those from 20 age, sex, and body surface area-matched healthy controls (from within the original control cohort of 97).
Fig. 1.
Experimental design and workflow. Previous published work identified differentially expressed proteotypic peptides in the myocardium of patients with HCM and LVH+ (*). Plasma from another set of LVH+ HCM patients was proteomically profiled to identify differentially expressed candidate peptides (*). A panel of 26 quantatypic peptide biomarkers was created from candidates identified in these profiling experiments. This panel was multiplexed into a 10-min LC-MS/MS assay and first applied to plasma samples from LVH+ HCMs and controls in a training dataset using ML, where six proteolytic peptide biomarkers were found to be differentially expressed. These differences were confirmed in the validation dataset. Correlation analysis with clinical and imaging information and with the five-year HCM sudden cardiac risk score was subsequently performed in LVH+ HCM patients to explore the assay's potential clinical utility. Five of the six biomarkers were also elevated in the plasma of a smaller group of patients with five subclinical HCM compared with controls. SVM, support vector machine.
HCM patients were recruited from a dedicated cardiomyopathy clinic at The Heart Hospital, University College London Hospital, London, UK, and given written informed consent conforming to the Declaration of Helsinki (fifth revision, 2000). Diagnosis of LVH+ HCM was based on demonstration by cardiovascular magnetic resonance (CMR) or echocardiography of a hypertrophied LV (maximal wall thickness [MWT] ≥15 mm) in the absence of loading conditions that could produce the same magnitude of hypertrophy. Inclusion criteria for subclinical HCM patients were as follows: (1) confirmed pathogenic or likely pathogenic HCM sarcomere gene mutation; (2) maximal LV wall thickness <13 mm by CMR and mass within the normal range relative to body surface area, age, and sex; (3) sinus rhythm, no LVH, and no pathological Q waves/T-wave inversion on 12-lead electrocardiography; and (4) no causes of secondary LVH (valve disease, hypertension). For study inclusion, healthy controls recruited from staff at University College London and The Heart Hospital were required to have no personal or family history of cardiac disease. Exclusion criteria for all participants were needle-phobia and a recent history (<1 month) of blood transfusion or hemodialysis. Estimates for five-year risk of sudden cardiac death (SCD) were calculated using the European Society of Cardiology online clinical tool (9, 10). Patients were categorized as low (< 4%), intermediate (≥ 4–<6%) or high (≥ 6%) five-year risk.
The UK National Research Ethics Service approved the generic analysis of anonymized clinical scans. The Local Research Ethics Committee gave approval for both proteomic method development on plasma specimens and plasma collection and sampling for this study: REC Ref. no. 04/0035 and REC 11/LO/0913.
Plasma Sample Preparation
Whole blood collected from individual participants was centrifuged on-site. Aliquoted plasma samples were stored in a freezer at −80°C until use. All patients with HCM donated blood samples for measurement of N-terminal prohormone of brain natriuretic peptide (NT-proBNP) levels, serum creatinine, and for genetic analysis.
Genetic Analysis
Genotyping of all patients with HCM (LVH+ and subclinical groups) was approved by the University College London/University College London Hospital Trust Joint Research Ethics Committee. Blood samples were collected at initial evaluation, and genomic DNA was isolated from peripheral blood lymphocytes using standard methodology. Patients with LVH+ and subclinical HCM were screened using a targeted high-throughput sequencing methodology, and sequencing data were subjected to bioinformatics analysis as previously described (11). Briefly, 2.1 Mb of genomic DNA sequence was screened per patient, covering coding, intronic, and selected regulatory regions of 41 cardiovascular genes. Solution-based sequence capture was used followed by massive parallel resequencing on Illumina GAIIx. Average read depth in the 2.1-Mb target region was 120. For identified variants, nonsynonymous pathogenic and likely pathogenic variants were selected on frequency (12) and putative functional consequence (13): either missense variants previously published to be associated with the disease or splicing, nonsense, and frameshift variants (11). Variants affecting the sarcomere genes were classified as either thick-filament (myosin-binding protein C [MYBPC3], β-myosin heavy chain [MYH7], myosin regulatory light chain [MYL2]) or thin-filament variants (troponin T [TNNT2], troponin I [TNNI3], tropomyosin [TPM1], cardiac α-actin [ACTC1]).
Label-free Proteomics to Identify Potential Biomarkers for HCM Present in Plasma
HCM Plasma Profiling Experiment
To identify candidate plasma biomarkers for HCM (Table S1) 200 μl of plasma were pooled from 16 LVH+ HCM patients and compared with that from 12 age-matched healthy controls. Pooled plasma samples were depleted for high abundance proteins using a Proteominer Protein Enrichment Kit (Bio-Rad UK). Eluted depleted protein concentration was determined using a protein bicinchoninic acid assay (14). Eighty micrograms (80 μg) were loaded onto a BioRad Any kDaTM gradient gel and Coomassie® stained. Nine gel bands from each lane were excised and in-gel digested with trypsin (Promega, UK). Fractions were then analyzed using MSE label-free quantitation. Analyses were performed as previously described (15, 16).
MSE Label-free Quantitation
All analyses were performed using a nanoAcquity high-performance liquid chromatography and quadrupole time of flight Premier mass spectrometer (Waters Corporation, Manchester, UK) as previously described (3). Briefly, peptides were trapped and desalted prior to reverse phase separation using a Symmetry C18 5-μm, 5 mm × 300 μm precolumn. Peptides were then separated prior to mass spectral analysis using a 15 cm × 75 μm C18 reverse phase analytical column. Peptides were loaded onto the precolumn at a flow rate of 4 μl/min in 0.1% formic acid for a total time of 4 min. Peptides were eluted off the precolumn and separated on the analytical column using a gradient of 3–40% acetonitrile [0.1% formic acid] over a period of 90 min and at a flow rate of 300 nl/min. The column was washed and regenerated at 300 nl/min for 10 min using a 99% acetonitrile [0.1% formic acid] rinse. After all nonpolar and nonpeptide material was removed the column was re-equilibrated at the initial starting conditions for 20 min. All column temperatures were maintained at 35 °C. Mass accuracy was maintained during the run using a lock spray of the peptide [glu1]-fibrinopeptide B delivered through the auxiliary pump of the nanoAcquity at a concentration of 300 fmol/l and at a flow rate of 300 nl/min, a collision energy of 25 V and over the mass range 50–2000 m/z. Peptides were analyzed in positive ion mode and operated in v-mode with a typical resolving power of 10,000 fwhm. Post calibration of data files was corrected using the doubly charged precursor ion of [glu1]-fibrinopeptide B (785.8426 m/z) with a sampling frequency of 30 s. Accurate mass LC-MS data were collected in MSe mode with data independent and alternating low and high collision energy mode (MSE). Each low/high acquisition was 1.5 s with 0.1-s interscan delay. Low energy data collections were performed at a constant collision energy of 4 V, high collision energy acquisitions were performed using a 15–40 V ramp over a 1.5-s time period and a complete low/high energy acquisition achieved every 3.2 s.
Protein identifications were obtained by ProteinLynx Global server (version 2.5). Data were searched against a combined target and reversed (decoy) human sequences to determine a 4% false discovery rate using a UniprotKB database and list of common laboratory contaminants (version 2013_06) with 50,901 entries. Trypsin was set as the protease, and two missed cleavages were allowed. Protein identification from the low/high collision spectra for each sample was processed using a hierarchical approach where more than three fragment ions per peptide, five fragment ions per protein, and more than two peptides per protein had to be matched. Protein identification parameters used in the database search included a <10-ppm mass accuracy tolerance, fixed modification of carboamidomethylation of cysteines and dynamic modifications of deamidation of asparagine/glutamine, and oxidation of methionine. Relative protein abundance was calculated using the Hi-3 method (17).
Proteins with >95% confidence identification were exported for comparative analysis using Non Linear Dynamics Progenesis software. Proteins of interest were identified from this experiment on the basis of confidence score, fold change, and clinical relevance.
Targeted Proteomics Analysis
Development of a Tandem Mass Spectrometry UPLC-MS/MS Assay for Validation
Potential biomarkers identified through plasma profiling were combined with those identified through a separate HCM myocardial tissue proteomic profiling experiment (3). In total, 26 proteotypic peptides (Table S2) demonstrated potential as HCM biomarkers and were developed into a multiplexed and targeted proteomic plasma test (Table S3). On the basis of these aforementioned label-free proteomics analyses, the proteotypic peptides specific to proteins of interest were determined from label-free proteomics data using either one of the top three most abundant peptides and selecting the optimum daughter spectra for quantitation or the open source online global proteome machine MRM database at www.thegpm.org (18). Custom synthesized peptides (Genscript, UK) were used to optimize the detection of the peptides in plasma digest matrix. Only peptides that gave good quantitative data by assessment using a standard curve spiked in plasma, and good signal to noise ratio of the endogenous peptide, were used in the assay. Two transitions per peptide were selected for the assay: one for quantitation and one for confirmation. The most abundant clean transitions without interfering nonspecific peaks were selected using the synthetic peptides spiked into matrix.
MRM-LC-MS/MS Assay Sample Preparation
Ten microliters (10 μl) of plasma were precipitated with 40 μl of ice cold 10% trichloroacetic acid in acetone. Samples were vortexed and incubated on ice for 1–2 h then centrifuged for 10 min at 500 g. Supernatant was discarded, and the pellet washed in 1 ml of ice cold acetone. Pellets were centrifuged, supernatant removed, and freeze-dried overnight. Freeze-dried pellets were resuspended in 20 μl of 100-mm Tris, 1% amidosulfobetaine-14 (ASB-14), pH 7.8, containing 6 m urea, 5 pmol heavy-labeled peptide internal standard and agitated at room temperature for 60 min. Disulfide bridges were reduced by the addition of 3 μl of 100-mm tris-hydrochloride, pH 7.8, containing 20-mm 1,4-dithioerythritol and incubated at room temperature for 60 min. Free thiol groups were carboamidomethylated by incubation with 6 μl of 100-mm tris-hydrochloride, pH 7.8, containing 20 mm iodoacetamide and incubated at room temperature for 45 min. The reaction mixture was then diluted with 155 μl of water and vortexed, and 150 ng of sequence grade trypsin (Promega, UK) were added to the solution. Samples were incubated overnight (12–16 h) at 37 °C in a water bath. Digested peptides were cleaned and desalted using C18 Bond Elute (Agilent, UK) as described previously (19).
MRM LC-MS/MS Analysis
Dried peptides were resuspended in 100 μl 3% acetonitrile 0.1% trifluoroacetic acid. Ten microliters (10 μl) of each sample were injected into a Xevo TQ-S triple quadrupole mass spectrometer coupled to a Waters standard Acquity ultra-performance liquid chromatography (UPLC) system (Waters PLC). The instrument was operated in positive ion mode. The capillary voltage was maintained at 3.7 kV with the source temperature held constant at 150 °C. A Waters Acquity UPLC Cortecs C18+ column 1.7-μm 2.1 × 100 mm attached to a C18+ VanGuard precolumn was used for separation with solution A (99.9% LC-MS-grade water with 0.1% formic acid) and solution B (LC-MS-grade 99.9% acetonitrile with 0.1% formic acid). The flow rate was set to 0.8 ml/min and a linear gradient of 0 to 97% solution A over 7 min. The total run time was 10 min. Pooled plasma digest was used as a quality control, which was run every 10 injections. The peptide and transitions of the final biomarker panel are given in Table S3. Quality controls were monitored throughout the run, and a coefficient of variation of ± 15% was considered acceptable. A standard curve 0–40 pmol was run at the start and end of the run. Data were analyzed using Target Lynx software (Waters). Integrated peak areas were expressed as a ratio to internal standard, and pmol were extracted from the standard curve.
Protein Interaction Network Analysis
A human protein interaction network was constructed for the six promising HCM biomarkers using interaction data gathered from the publicly available database IntAct (20) (11–2018) and filtered to remove non-human nodes and interactions, interactions with chemicals, self-loops, and duplicated edges. Cytoscape (21) (version 3.0.2) was used as a visualization tool. Data presented in Fig. S2 is from an advanced IntAct search limited to species Homo sapiens, with interaction confidence MiScores ranging between 0.27–0.74. The final interactome topology was cross-checked with that generated in Interactome3D (22). A second protein interaction network for the six biomarkers to include non-human nodes and edges is presented in Fig. S3. The complete list of proteins and interaction metadata is provided in Data file S1.
Cardiac Imaging
Transthoracic Echocardiography
All patients with LVH+ and subclinical HCM were evaluated by standard two-dimensional and Doppler transthoracic echocardiography in accordance with previously published methods (23). Recorded variables included: LA diameter on parasternal long axis view, LV end-systolic dimension, LV end-diastolic dimension, MWT, and LV outflow tract gradient at rest and with Valsalva. MWT was defined as the greatest thickness in any single LV segment measured in the parasternal short-axis plane at either the level of the mitral valve, mid-ventricle, or apex at end-diastole. LV volumes and ejection fraction were assessed by biplane Simpson's equation using the apical four- and two-chamber views. Pulsed Doppler sample volume (1–3 mm) was placed on the edge of the mitral leaflets to determine mitral diastolic flow properties in the apical four-chamber view. The peak flow velocity of early diastolic E-wave and late diastolic A-wave, E wave velocity/A wave velocity (E/A) ratio, and E velocity deceleration times were recorded (24). Tissue Doppler imaging in the apical four-chamber view, with sample volume at the septal and lateral segment of the mitral annulus provided e′ values.
Cardiovascular Magnetic Resonance
All controls and patients with LVH+ and subclinical HCM free of cardiac implantable electronic devices, permanent atrial fibrillation, and breath-holding difficulties at baseline, underwent CMR scanning. Standard clinical scans were performed using a 1.5 Tesla magnet (Avanto VB17, Siemens Medical Solutions, Erlangen, Germany). CMR short-axis volumetric studies (25) were acquired as previously described (26). Native T1 mapping was performed on a basal and mid LV short-axis slice in diastole using the shortened modified Look-Locker inversion recovery (ShMOLLI) sequence (5b(1b)1b(1b)1b, WIP# 448) (27). Standard late gadolinium enhancement (LGE) images were acquired using a fast low angle single-shot inversion recovery sequence following a contrast bolus of 0.1 mmol/kg of gadoterate meglumine (Dotarem; Guerbet, Paris, France). For septal T1 measurements, a region of interest of standard size in the mid septum (segment 8) was manually drawn to avoid the blood-myocardial boundary with a 20% offset. Standard LGE images were analyzed by two observers each with six years of experience in CMR (G.C., S.R.) using the same software (cvi42, Circle CVI, Calgary, Canada) (28). Total LGE volume was quantified using the signal threshold versus reference mean (STRM) semi-automated technique with an STRM-based threshold of > 3 standard deviations (SD) above the mean signal intensity of reference myocardium as previously described (28).
Statistics
Statistical analysis was performed in R (29) (version 3.0.1). Distribution of data was assessed on histograms and using the Shapiro-Wilk test. Continuous variables are expressed as mean ± 1 SD; categorical variables, as counts and percentages. Unpaired t test was used for the comparison of normally distributed data between HCM patients and controls and χ2 or Fisher's exact test for noncontinuous variables. Chromatograms were analyzed using Waters Targetlynx software. Peak integration was processed by manual inspection to correct for false assignments. Data were exported to Microsoft Excel (Microsoft, Redmond, WA). Levels of peptides in the form of nonparametric continuous data were compared between LVH+/subclinical HCM cases and controls using nonparametric Mann-Whitney-Wilcoxon test with p value adjustment for multiple comparisons by the Bonferroni method. Correlations were calculated using Spearman's rho or point biserial correlation as appropriate. The best biomarker panel for LVH+ HCM was built using supervised ML with the support vector machine (SVM) classification as previously described (30). SVMs are a set of effective, supervised nonparametric ML techniques that analyze data and recognize patterns. They are increasingly being applied to proteomic datasets for classification and regression analysis (31, 32) and are especially suited to two-group separation challenges like the one presented in the current work. The goal of our SVM model was to use the proteomics biomarker panel to predict which phenotypic category a participant belonged to (LVH+ HCM or control), based on an initial training set example. Performance of the tuned SVM was then verified in the validation dataset and overfitting avoided through the implementation of a 10-fold cross validation (R package 'E1071′). We constructed the SVM with a radial kernel tuned to cost 2 and gamma 1 to derive ML prediction scores per participant. For the optimal model, area under the receiver operating characteristics curve was calculated using package ROCR. p values are two sided and considered significant when < 0.05. Datasets used in this analysis are provided in Data file S2.
RESULTS
Characteristics of Study Participants
Clinical and demographic characteristics of the cohorts are provided in Table I. The training and validation LVH+ HCM cohorts were matched to controls except for age in the validation cohort. In this regard, we show that individual proteomics biomarker levels as well as the ML prediction score exhibit no correlation with age (Table II). Genetic variants identified in LVH+ and subclinical HCM patients are summarized in Table S4.
Table I. Clinical and imaging characteristics of study participants.
| Variable† | All Samples |
Training Set |
P‡ | Validation Set |
P‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 20) | Subclinical HCM (n = 16) | P‡ | Control (n = 97) | LVH+ HCM (n = 110) | P‡ | Control (n = 67) | LVH+ HCM (n = 80) | Control (n = 30) | LVH+ HCM (n = 30) | |||
| At recruitment | ||||||||||||
| Age (yrs) | 41.4 ± 9.7 | 38.93 ± 15.4 | 0.714 | 49.6 ± 13.4 | 50.1 ± 15.0 | 0.834 | 53.2 ± 13.1 | 48.8 ± 15.2 | 0.060 | 41.5 ± 10.2 | 53.5 ± 14.2 | 0.001 |
| Sex (M/F) | 12/8 | 5/11 | 0.086 | 56/41 | 77/33 | 0.066 | 41/26 | 56/24 | 0.171 | 15/15 | 21/9 | 0.114 |
| BSA (m2) | 1.71 ± 0.65 | 1.79 ± 0.54 | 0.622 | 1.88 ± 0.23 | 1.95 ± 0.21 | 0.03 | 1.90 ± 0.23 | 1.95 ± 0.22 | 0.130 | 1.83 ± 0.23 | 1.95 ± 0.20 | 0.088 |
| BMI (kg/m2) | 25.29 ± 4.42 | 24.01 ± 3.55 | 0.074 | 24.66 ± 3.63 | 27.93 ± 4.71 | <0.001 | 24.55 ± 3.56 | 27.68 ± 4.60 | <0.001 | 24.97 ± 3.90 | 28.64 ± 5.06 | 0.007 |
| FH of HCM [%] | 0 [0] | 0 [0] | – | 0 [0] | 44 [40] | – | 0 [0] | 35 [44] | <0.001 | 0 [0] | 9 [30] | <0.001 |
| FH of S.D. [%] | 0 [0] | 0 [0] | – | 0 [0] | 35 [32] | <0.001 | 0 [0] | 26 [33] | <0.001 | 0 [0] | 9 [30] | <0.001 |
| CIED in situ [%] | 0 [0] | 0 [0] | – | 0 [0] | 36 [33] | <0.001 | 0 [0] | 21 [26] | <0.001 | 0 [0] | 15 [50] | <0.001 |
| NYHA class | I = 20 | I = 16 | – | I = 97 | I = 47 | I = 67 | I = 33 | I = 30 | I = 14 | |||
| II = 0 | II = 0 | – | II = 0 | II = 46 | II = 0 | II = 33 | II = 0 | II = 13 | ||||
| III = 0 | III = 0 | – | III = 0 | III = 17 | <0.001 | III = 0 | III = 14 | <0.001 | III = 0 | III = 3 | <0.001 | |
| IV = 0 | IV = 0 | – | IV = 0 | IV = 0 | IV = 0 | IV = 0 | IV = 0 | IV = 0 | ||||
| AF [%] | 0 [0] | 0 [0] | – | 0 [0] | 28 [25] | <0.001 | 0 [0] | 20 [25] | <0.001 | 0 [0] | 8 [27] | <0.001 |
| NT-proBNP (pg/ml) | – | 27.0 ± 53.97 | – | – | 147.7 ± 179.0 | – | – | 148.0 ± 187.1 | – | – | 146.9 ± 159.0 | – |
| Creatinine (ml/min) | – | 70.8 ± 17.8 | – | – | 84.0 ± 16.2 | – | – | 85.1 ± 15.9 | – | – | 81.0 ± 16.9 | – |
| Drug therapy | ||||||||||||
| Antiplatelet [%] | 0 [0] | 0 [0] | – | 0 [0] | 39 [35] | <0.001 | 0 [0] | 27 [34] | <0.001 | 0 [0] | 12 [40] | <0.001 |
| Anticoagulation [%] | 0 [0] | 0 [0] | – | 0 [0] | 23 [21] | <0.001 | 0 [0] | 15 [19] | <0.001 | 0 [0] | 8 [27] | <0.001 |
| Beta-blocker [%] | 0 [0] | 0 [0] | – | 0 [0] | 61 [55] | <0.001 | 0 [0] | 43 [54] | <0.001 | 0 [0] | 18 [60] | <0.001 |
| ACE-i/ARB [%] | 0 [0] | 0 [0] | – | 0 [0] | 12 [11] | <0.001 | 0 [0] | 7 [9] | <0.001 | 0 [0] | 5 [17] | <0.001 |
| Ca2+ channel blocker [%] | 0 [0] | 0 [0] | – | 0 [0] | 27 [25] | <0.001 | 0 [0] | 17 [21] | <0.001 | 0 [0] | 10 [33] | <0.001 |
| Diuretic [%] | 0 [0] | 0 [0] | – | 0 [0] | 21 [19] | <0.001 | 0 [0] | 16 [20] | <0.001 | 0 [0] | 5 [17] | <0.001 |
| Other antiarrhythmic [%] | 0 [0] | 0 [0] | – | 0 [0] | 24 [22] | <0.001 | 0 [0] | 18 [23] | <0.001 | 0 [0] | 6 [20] | <0.001 |
| Genetic Variants* | ||||||||||||
| MYBPC3 [%] | – | 4 [25] | – | – | 18 [16] | – | – | 14 [18] | – | – | 4 [13] | – |
| MYH7 [%] | – | 5 [31] | – | – | 18 [16] | – | – | 12 [15] | – | – | 6 [20] | – |
| TNNT2 [%] | – | 3 [19] | – | – | 2 [2] | – | – | 1 [1] | – | – | 1 [3] | – |
| TNNI3 [%] | – | 2 [13] | – | – | 2 [2] | – | – | 2 [3] | – | – | 0 [0] | – |
| TPM1 [%] | – | 1 [6] | – | – | 2 [2] | – | – | 2 [3] | – | – | 0 [0] | – |
| MYL2 [%] | – | 0 [0] | – | – | 3 [3] | – | – | 2 [3] | – | – | 1 [3] | – |
| Other [%] | – | 1 [6] | – | – | 2 [2] | – | – | 1 [1] | – | – | 1 [3] | – |
| CMR features | ||||||||||||
| MWT (mm) | 8.9 ± 1.3 | 9.0 ± 1.5 | 0.832 | 9.4 ± 1.7 | 21.0 ± 4.2 | <0.001 | 9.7 ± 1.6 | 21.0 ± 42 | <0.001 | 8.7 ± 1.6 | 21.5 ± 6.1 | 0.206 |
| LV EDV (ml) | 146 ± 33 | 141 ± 29 | 0.762 | 136 ± 30 | 135 ± 31 | 0.901 | 137 ± 31 | 135 ± 31 | 0.796 | 134 ± 31 | 139 ± 21 | 0.797 |
| LV ESV (ml) | 52 ± 15 | 50 ± 15 | 0.338 | 45 ± 13 | 37 ± 16 | 0.002 | 45 ± 12 | 36 ± 16 | 0.002 | 46 ± 15 | 50 ± 16 | 0.806 |
| EF (%) | 64 ± 5 | 65 ± 7 | 0.252 | 67 ± 5 | 73 ± 9 | <0.001 | 67 ± 4 | 74 ± 9 | <0.001 | 66 ± 6 | 65 ± 7 | 0.829 |
| LV mass (g) | 135 ± 41 | 138 ± 36 | 0.601 | 127 ± 34 | 223 ± 81 | <0.001 | 129 ± 35 | 221 ± 80 | <0.001 | 120 ± 31 | 281 ± 117 | 0.301 |
| Native T1 ShMOLLI (ms) | 957 ± 36 | 965 ± 30 | 0.752 | 955 ± 28 | 1002 ± 47 | <0.001 | 952 ± 28 | 1001 ± 47 | <0.001 | 963 ± 30 | 1000 ± 52 | 0.251 |
| LGE mass (g) | 2.67 ± 2.38 | 2.37 ± 1.99 | 0.612 | 2.47 ± 1.97 | 53.2 ± 43.1 | <0.001 | 2.23 ± 1.75 | 54.5 ± 43.4 | <0.001 | 3.09 ± 2.41 | 20.6 ± 2.29 | 0.037 |
| LGE volume (ml) | 2.55 ± 2.27 | 2.11 ± 1.75 | 0.840 | 2.35 ± 1.88 | 50.7 ± 41.0 | <0.001 | 2.13 ± 1.67 | 51.9 ± 41.4 | <0.001 | 2.95 ± 2.29 | 19.6 ± 2.18 | 0.037 |
| TTE features | ||||||||||||
| MWT (mm) | – | 8.6 ± 1.7 | – | – | 17.8 ± 3.9 | – | – | 18.3 ± 4.2 | – | – | 17.2 ± 3.1 | – |
| LV EDD (mm) | – | 46.5 ± 4.3 | – | – | 46.9 ± 5.9 | – | – | 46.5 ± 6.0 | – | – | 47.7 ± 5.3 | – |
| LV EF (%) | – | 65.1 ± 4.1 | – | – | 66 ± 7 | – | – | 67 ± 6 | – | – | 65 ± 9 | – |
| LAd (mm) | – | 34.9 ± 6.4 | – | – | 44.8 ± 7.6 | – | – | 44.0 ± 7.3 | – | – | 47.0 ± 8.0 | – |
| Resting LVOT gradient (mmHg) | – | 4 ± 1 | – | – | 24 ± 33 | – | – | 28 ± 36 | – | – | 14 ± 21 | – |
| E/A ratio | – | 1.7 ± 1.0 | – | – | 1.3 ± 0.6 | – | – | 1.4 ± 0.6 | – | – | 1.0 ± 0.5 | – |
| EdecT (ms) | – | 185 ± 32 | – | – | 229 ± 70 | – | – | 231 ± 72 | – | – | 226 ± 66 | – |
| E/e' ratio | – | 5.5 ± 1.6 | – | – | 10.5 ± 7.1 | – | – | 10.3 ± 5.8 | – | – | 11.5 ± 10.9 | – |
| 5-year HCM SCD Risk (ESC 2014) | – | – | – | – | 3.61 ± 2.98 | – | – | 3.96 ± 3.24 | – | – | 2.69 ± 1.93 | – |
| ML Prediction Score | – | – | – | 0.65 ± 0.57 | 2.04 ± 0.61 | <0.0001 | 0.59 ± 0.47 | 2.08 ± 0.64 | <0.0001 | 0.79 ± 0.73 | 1.94 ± 0.54 | <0.0001 |
† Categorical data are presented as counts/number of participants [%], and continuous data as mean ± S.D. except where otherwise stated.
*The full list of genetic variants is provided in Table S4.
‡Significant p values are highlighted in bold for differences between HCM patients and controls in the respective datasets. Differences were calculated using unpaired t-test, χ2 or Fisher's exact test as appropriate.
ACE-i, angiotensin converting enzyme inhibitor; AF, atrial fibrillation; ARB, angiotensin II receptor blocker; BMI, body mass index; BSA, body surface area; Ca2+, calcium; CIED, cardiac implantable electronic device; CMR, cardiovascular magnetic resonance; EDD, end diastolic dimension; EdecT, E wave deceleration time; EDV, end diastolic volume; EF, ejection fraction; ESC, European Society of Cardiology; ESV, end systolic volume; FH, family history; HCM, hypertrophic cardiomyopathy; LAd, left atrial diameter; LGE, late gadolinium enhancement; LVOTO, left ventricular outflow tract obstruction; M/F, male/female; ML, machine learning; MWT, maximal wall thickness in diastole; MYBPC3, myosin-binding protein C; MYH7, β-myosin heavy chain; MYL2, myosin regulatory light chain; NT-proBNP, n-terminal pro-brain natriuretic peptide; NYHA, New York Heart Association Functional Class; S.D., sudden death; ShMOLLI, shortened modified Look-Locker inversion recovery; TNNI3, troponin I; TNNT2, troponin T; TPM, tropomyosin; yrs, years.
Table II. Correlations of proteomic biomarkers with baseline demographic characteristics in healthy controls and with biohumoral, clinical, genetic and imaging characteristics in LVH+ HCM.
| Variable | ALDOA rs/pb [p value] | C3 rs/pb [p value] | GST01 rs/pb [p value] | RSU1 rs/pb [p value] | TLN1rs/pb [p value] | THBS1 rs/pb [p value] | ML Score* rs/pb [p value] |
|---|---|---|---|---|---|---|---|
| Demographic [Controls only] | |||||||
| Age (yrs) | 0.03 | 0.17 | 0.03 | 0.04 | –0.13 | 0.11 | 0.01 |
| [0.764] | [0.097] | [0.797] | [0.692] | [0.204] | [0.311] | [0.990] | |
| BSA (m2) | –0.04 | –0.11 | –0.06 | –0.02 | 0.06 | 0.05 | 0.08 |
| [0.745] | [0.311] | [0.583] | [0.869] | [0.617] | [0.677] | [0.268] | |
| Biohumoral | |||||||
| NT-proBNP (pmol/L) | –0.17 | 0.03 | –0.16 | 0.21 | –0.16 | 0.10 | –0.12 |
| [0.071] | [0.790] | [0.220] | [0.028] | [0.095] | [0.311] | [0.225] | |
| Creatinine (ml/min) | –0.27 | –0.12 | –0.10 | –0.03 | –0.27 | 0.06 | –0.02 |
| [0.193] | [0.577] | [0.632] | [0.904] | [0.188] | [0.773] | [0.907] | |
| Sarcomere gene variants | |||||||
| Thin vs. Thick filament | –0.23 | 0.19 | –0.18 | 0.27 | –0.01 | 0.07 | 0.240 |
| [0.254] | [0.509] | [<0.001] | [0.607] | [0.175] | [0.051] | [0.045] | |
| TTE | |||||||
| LAd (mm) | –0.02 | –0.09 | 0.08 | –0.08 | –0.05 | 0.05 | –0.07 |
| [0.865] | [0.360] | [0.395] | [0.411] | [0.569] | [0.569] | [0.441] | |
| LVOT gradient (mmHg) | –0.08 | –0.01 | –0.04 | 0.01 | –0.05 | –0.16 | 0.01 |
| [0.393] | [0.958] | [0.704] | [0.883] | [0.636] | [0.098] | [0.956] | |
| E/A ratio | 0.10 | –0.13 | 0.08 | –0.02 | 0.10 | –0.06 | 0.06 |
| [0.324] | [0.201] | [0.429] | [0.973] | [0.337] | [0.537] | [0.578] | |
| E/e' | –0.17 | –0.02 | –0.12 | 0.09 | –0.07 | –0.01 | –0.04 |
| [0.097] | [0.887] | [0.245] | [0.524] | [0.520] | [0.927] | [0.710] | |
| CMR | |||||||
| MWT (mm) | 0.28 | 0.33 | 0.27 | 0.27 | 0.15 | 0.31 | 0.61 |
| [0.001] | [<0.001] | [0.002] | [0.002] | [0.084] | [0.002] | [<0.0001] | |
| LV EDV (ml) | 0.05 | –0.07 | 0.04 | 0.05 | 0.14 | –0.06 | –0.08 |
| [0.548] | [0.409] | [0.673] | [0.606] | [0.117] | [0.489] | [0.386] | |
| LV ESV (ml) | 0.02 | –0.16 | 0.01 | –0.05 | 0.06 | –0.15 | –0.25 |
| [0.845] | [0.073] | [0.905] | [0.543] | [0.519] | [0.076] | [0.004] | |
| EF (%) | –0.03 | 0.17 | –0.01 | 0.12 | 0.02 | 0.15 | 0.28 |
| [0.713] | [0.048] | [0.924] | [0.180] | [0.861] | [0.082] | [0.001] | |
| LV mass (g) | 0.26 | 0.19 | 0.28 | 0.23 | 0.21 | 0.22 | 0.40 |
| [0.003] | [0.029] | [0.001] | [0.008] | [0.017] | [0.012] | [<0.0001] | |
| Native T1 ShMOLLI (ms) | 0.09 | 0.11 | 0.07 | 0.10 | 0.10 | 0.17 | 0.25 |
| [0.351] | [0.301] | [0.511] | [0.331] | [0.344] | [0.087] | [0.011] | |
| LGE mass (g) | 0.27 | 0.28 | 0.25 | 0.23 | 0.20 | 0.35 | 0.62 |
| [0.003] | [<0.001] | [0.005] | [0.011] | [0.030] | [<0.001] | [<0.001] | |
| NSVT | |||||||
| Y/N | 0.09 | –0.16 | 0.14 | –0.04 | 0.22 | 0.11 | 0.39 |
| [<0.0001] | [0.341] | [<0.0001] | [0.170] | [<0.0001] | [0.019] | [<0.0001] | |
| 5-Year HCM SCD Risk | |||||||
| Intermediate & High (≥ 4) | 0.03 | –0.16 | 0.07 | –0.15 | 0.12 | 0.04 | 0.23 |
| [<0.0001] | [0.701] | [<0.0001] | [0.212] | [<0.0001] | [0.017] | [<0.0001] | |
| High (≥ 6%) | 0.06 | –0.14 | 0.06 | –0.19 | 0.20 | 0.14 | 0.23 |
| [<0.0001] | [0.763] | [<0.0001] | [0.210] | [<0.0001] | [0.014] | [<0.0001] |
*Machine learning prediction scores of the combined six marker assay calculated by a support vector machine supervised machine learning method in the study population.
Reporting Spearman's correlations (rs) for continuous variables (proteomic analyte distributions are non-parametric) or point biserial correlations (rpb) for binary variables.
Significant correlations are highlighted in bold.
NSVT, non-sustained ventricular tachycardia; SCD, sudden cardiac death; TTE, transthoracic echocardiography; Y/N, yes/no.
Other abbreviations as in Table I.
Overall Workflow and Quantitative Reliability of Plasma MRM Analysis
Targeted proteomic MRM analysis of 26 candidate peptides (Table S2) was performed blindly for 223 individual plasma samples over three days. Eleven of the 26 peptides were filtered out from further analysis because their levels were below the limits of reliable detection in plasma. Of the remaining peptides that could be reliably detected, six (Fig. 2) showed significant differential expression between LVH+ HCM cases and controls in the training dataset (Table III, Fig. 3). A mean standard curve linearity of R2 0.95 ± 0.04 was achieved across the calibration curves for the six candidate peptides of interest (Table S5 and Data file S3). Mean coefficients of variation for target peptides between replicate quality control samples was 9.8 ± 3.3% (Table S5 and Data file S4). The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (33) partner repository with the dataset identifier PXD009859.
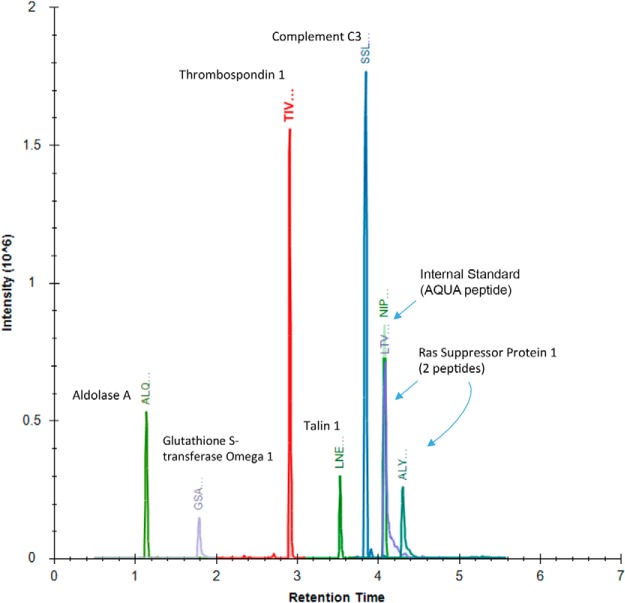
Fig. 2.
Overlaid chromatogram of the six marker peptides that validated in the multiplexed targeted proteomic assay. Individual chromatograms are provided in Fig. S1.
Table III. Candidate plasma biomarker levels in LVH+ HCM and controls (training dataset) and in subclinical HCM and controls.
| UniProt ID | Accession [QC CoV %] | Peptide Description | Symbol | Control n = 67 (pmol/ml) | LVH+ HCM n = 67 (pmol/ml) | Fold Change | Direction‡ | P* | Control n = 20 (pmol/ml) | Subclinical HCM n = 16 (pmol/ml) | Fold Change | Direction‡ | P* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P04075 | ALDOA_HUMAN [8.92%] | Aldolase, Fructose-Bisphosphate A-peptide | ALDOA | 50.59 ± 49.52 | 132.05 ± 220.21 | 2.59 | Up | 0.021 | 21.91 ± 25.93 | 130.01 ± 140.02 | 5.9 | Up | 0.019 |
| P01024 | CO3_HUMAN [10.04%] | Complement C3-peptide | C3 | 1988.69 ± 2502.66 | 2974.76 ± 2553.83 | 1.50 | Up | 0.014 | 939.35 ± 1636.96 | 2540.91 ± 1801.76 | 2.7 | Up | 0.006 |
| P78417 | GSTO1_HUMAN [6.39%] | Glutathione S-Transferase, Omega 1-peptide | GST01 | 1.18 ± 4.61 | 10.50 ± 20.16 | 8.90 | Up | 0.007 | 5.45 ± 5.08 | 12.08 ± 9.76 | 2.2 | Up | 0.007 |
| Q15404 | RSU1_HUMAN | Ras Suppressor Protein 1 | RSU1 | 18.06 ± 32.13 | 34.70 ± 56.99 | 1.76 | Up | 0.042 | 25.69 ± 14.90 | 70.18 ± 46.47 | 2.7 | Up | 0.001 |
| [12.42%] | ALY–Peptide 1 | 95.65 ± 96.50 | 142.34 ± 106.56 | 1.39 | Up | 0.035 | 100.62 ± 110.24 | 188.80 ± 88.37 | 1.9 | Up | 0.004 | ||
| [13.57%] | LTV–Peptide 2 | ||||||||||||
| Q9Y490 | TLN1_HUMAN [8.63%] | Talin 1-peptide | TLN1 | 22.61 ± 24.04 | 63.47 ± 113.05 | 2.81 | Up | 0.007 | 26.72 ± 16.73 | 85.14 ± 38.13 | 3.2 | Up | 0.529 |
| P07996 | TSP1_HUMAN [8.2%] | Thrombospondin 1-peptide | THBS1 | 36.03 ± 35.66 | 58.47 ± 56.35 | 1.62 | Up | 0.007 | 39.19 ± 54.54 | 67.78 ± 61.55 | 1.7 | Up | 0.010 |
Plasma protein analytes selected for the biomarker assay were quantified by label-free mass spectrometry in HCM patients and controls. The six proteins showing differential expression in LVH+ HCM and controls were combined into a multimarker assay using machine learning.
‡Direction of the fold change comparing LVH+ HCM vs. controls and subclinical HCM vs. controls.
*Significant p values are highlighted in bold. Significance levels were calculated using the non-parametric Mann-Whitney-Wilcoxon test with p value adjustment for multiple comparisons by the Bonferroni method.
CoV, coefficient of variation; QC, quality control (see Supplementary Table S5). Other abbreviations as in Table I.
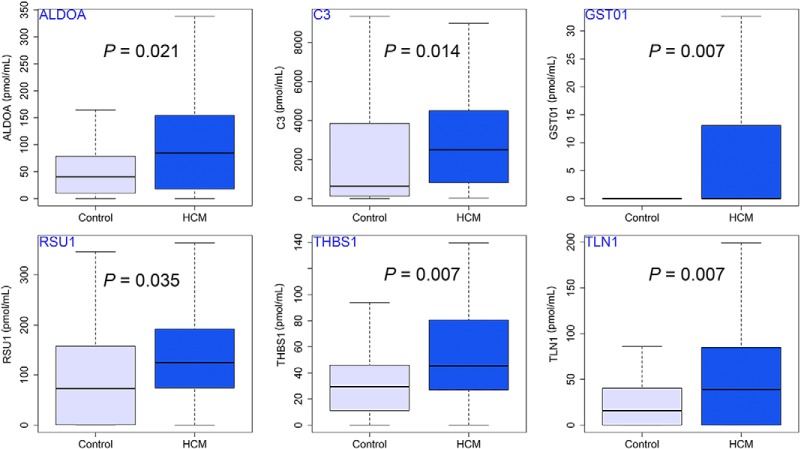
Fig. 3.
Box and whisker plots showing the six differentially expressed plasma peptides identified in training dataset consisting of LVH+ HCM and controls by the targeted proteomic multiplexed assay (using the Mann-Whitney-Wilcoxon test with p value adjustment for multiple comparisons by the Bonferroni method).
Biomarkers Differentially Expressed in the Plasma of HCM Patients Compared with Controls
In the LVH+ HCM versus controls training dataset, the levels of six proteotypic peptides (aldolase fructose-bisphosphate A-peptide [ALDOA-peptide], complement C3-peptide [C3-peptide], glutathione S-transferase omega 1-peptide [GSTO1-peptide], Ras suppressor protein 1-peptide [RSU1-peptide], talin 1-peptide [TLN1-peptide], and thrombospondin 1-peptide [THBS1-peptide]) were elevated significantly in LVH+ HCM plasma samples compared with controls. We found no significant sex-related differences for any of the studied peptides across LVH+ HCM participants (all p > 0.05). As atrial fibrillation has been reported to influence the levels of some of our candidate biomarkers (C3, TLN1, and THBS1) (34–36), we repeated the analysis after excluding 20 LVH+ HCM patients in the training dataset with a history of atrial fibrillation; differences persisted for all markers except TLN1 (Table S6). In a subanalysis limited to LVH+ HCM patients with pathogenic sarcomere gene mutations (n = 41, 47.3 ± 14.3 years, 27 male), similar differences relative to controls persisted for THBS1 (51.92 ± 45.85 pmol/ml versus 34.69 ± 35.03 pmol/ml; p = 0.013) and C3 (2969.39 ± 2776.19 pmol/ml versus 1691.42 ± 2297.59 pmol/ml; p = 0.002). Though myocardial lumican concentration was recently shown to be elevated in LVH+ HCM compared with controls (3), plasma differences, albeit present, did not achieve statistical significance in the present work (plasma lumican in LVH+ HCM versus controls: 262.87 ± 131.87 versus 227.14 ± 153.84, p = 0.06). In patients with subclinical HCM, five of the proteotypic peptides were significantly elevated (Table III) compared with controls.
Diagnostic Performance of the Plasma Biomarkers
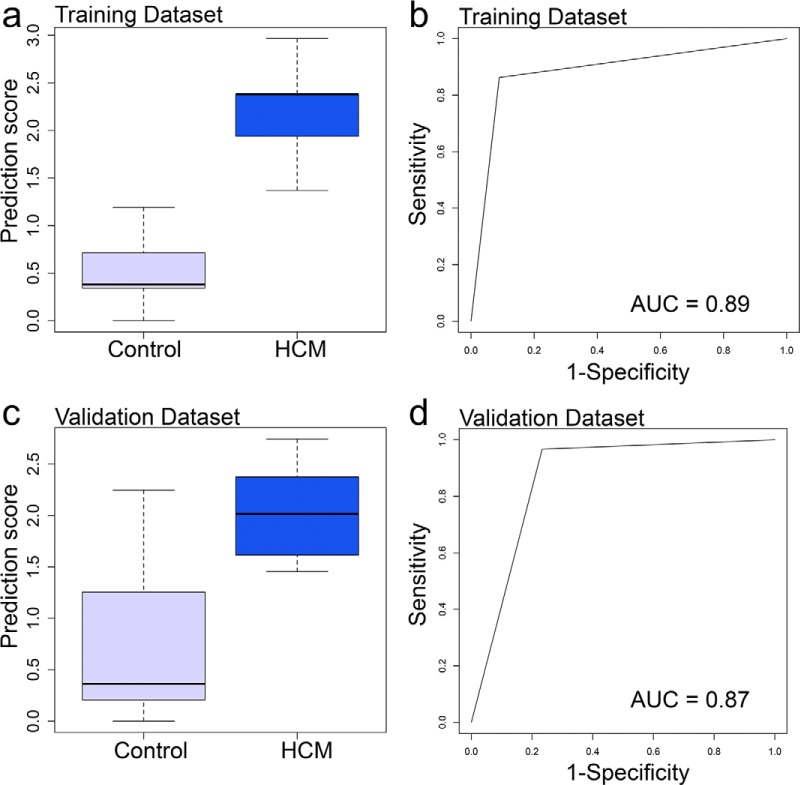
The six circulating peptide biomarkers in combination (ALDOA-peptide, C3-peptide, GSTO1-peptide, RSU1-peptide, TLN1-peptide, and THBS1-peptide) identified an LVH+ HCM phenotype compared with controls, with an area under the curve of 0.89 in the training dataset (sensitivity 96%, 95% confidence interval [CI] 77–93; specificity 87%, 95% CI 77–94) and 0.87 in the validation dataset (sensitivity 97%, 95% CI 83–100; specificity 77%, 95% CI 58–90, Fig. 4).
Fig. 4.
Box plots showing performance of prediction scores calculated by a support vector machine supervised ML method in the training (A) and validation (C) datasets made up of LVH+ HCM patients and controls (whiskers indicate variability outside the third and first quartiles [75th and 25th percentiles] represented as hinges around the median [bold midline]). Receiver operating characteristics (ROC) curves (B and D) show performance of the ML prediction score in training and validation datasets.
Correlation of Biomarker Levels With Clinical, Humoral, Genetic, and Imaging Variables
Correlations are reported in Table II. None of the six biomarkers demonstrated a correlation with age, body surface area, or serum creatinine levels, echocardiographic measures of left atrial size, diastolic function, or LV outflow tract obstruction. RSU1-peptide weakly correlated with NT-proBNP levels (rs = 0.21; p = 0.028).
Considering individual biomarker correlations with thin (14%) versus thick filament (86%) sarcomere gene variants, only GST01-peptide showed a significant association (higher in LVH+ HCM patients with thick filament variants), while the composite ML prediction score was significantly higher in those with thin filament mutations.
The majority of biomarkers correlated with LV MWT, LV mass and myocardial scar burden by CMR. THBS1-peptide and C3-peptide showed the strongest correlation with MWT (rs = 0.31 and 0.33; p = 0.002 and <0.001, respectively) and percentage myocardial scar (rs = 0.35 and 0.28; p < 0.001 both, respectively). GST01-peptide and ALDOA-peptide showed the strongest correlation with LV mass (rs = 0.28 and 0.26; p = 0.001 and 0.003, respectively). In LVH+ HCM, four of the biomarkers (ALDOA-peptide, GST01-peptide, TLN1-peptide, and THBS1-peptide) as well as the composite ML prediction score, correlated with the svtpresence of nonsustained ventricular tachycardia (NSVT) and with the five-year HCM SCD risk score (37). The composite ML score, was independently predictive of NSVT after adjusting for MWT (odds ratio [OR] 4.06; 95% confidence intervals [CI] 1.57–13.70, p = 0.010) but not with the addition of myocardial scar (OR 1.49; 95% CI 0.00–6.48, p = 0.541).
Interaction Network Analysis
Human protein interaction network analysis of the six biomarkers (Figs. S2 and S3) highlights their predominant connections to HCM pathophysiology (38). The complementary interactome enriched with non-human nodes and edges (Fig. S3) implicates the enzyme glutamine gamma-glutamyltransferase 2 (TGM2, UniProtKB: P21980) as the interacting node between C3 and THBS1.
DISCUSSION
Using a pipeline that combined potential biomarkers discovered through proteomic profiling of heart tissue and plasma with others identified in the literature (39), we assembled a list of 26 possible biomarkers that were put forward for verification. From these, we were able to accurately quantify and confirm six biomarkers that were elevated in the plasma of patients with LVH+ HCM compared with controls. These six multiplexed plasma markers (one extracellular, two intracellular, two enzymes, one complement component) were related to myocardial substrate changes in HCM as suggested by their correlation with LV wall thickness, LV mass, percentage myocardial scar, and presence of NSVT and with the five-year risk estimate for SCD (37). Adverse clinical outcomes in patients with HCM, such as cardiac arrhythmias, heart failure, and SCD, are thought to be related to myocardial substrate changes including myoarchitectural disarray, fibrosis, and small vessel disease. We found that five of the biomarkers identified in LVH+ HCM patients were also elevated in the plasma of a limited number of patients with subclinical HCM where, despite the absence of LVH by standard imaging methods, myocardial substrate changes are already presumed to exist (40, 41). There is a growing body of evidence to suggest that such myocardial substrate changes in subclinical HCM may be prognostically relevant. For example, in a post-mortem histopathological study of subclinical HCM, the hearts of four related SCD victims with apparently normal LV mass and wall thickness demonstrated widespread myoarchitectural disarray (42), and a pathogenic HCM-causing sarcomere gene mutation was later implicated. In another UK regional post-mortem registry, nine hearts from athletes who had succumbed to SCD, again showing normal wall thickness and mass, were discovered to have myoarchitectural disarray consistent with HCM (7). These preliminary biomarker findings described for subclinical HCM suggest that the plasma proteome of individuals with subclinical HCM merits further exploration at scale to better understand its potential clinical utility in terms of tracking pathophysiological myocardial substrate changes ahead of manifest LVH.
Description and Function of the Parent Proteins Linked to Peptides Identified in the Study
Interactome data indicate that five of the proteins are connected in a network related to hypertrophy and fibrosis with potential relevance to the known myocardial substrate changes driving SCD in HCM, while C3 participates more distinctly in the inflammation network with inflammation increasingly gaining traction as a key pathophysiological player in HCM (38).
Extracellular Protein: Thrombospondin 1
(THBS1-peptide) is a nonstructural extracellular matrix component with anti-angiogenic activity that is able to activate transforming growth factor-β, a potent profibrotic and anti-inflammatory factor (43). THBS1 has been described as a crucial regulator of cardiac matrix integrity, enabling the myocardium to adapt to increased pressure loading (44). It is minimally expressed in the normal heart but markedly up-regulated following cardiac injury (45). The THBS family has five members, of which THBS1 is one of the best studied. In a mouse model of pulmonary hypertension, overexpression of THBS1 was demonstrated in the hypertrophied right ventricle (46). It was also overexpressed in LV myocardium from mouse models of cardiac hypertrophy (44, 47) and from patients with LVH secondary to aortic stenosis (44).
Intracellular Proteins
The ubiquitously expressed single-copy RSU1 (RSU1-peptide) gene is expressed specifically in the human heart (48) where it encodes a leucine-rich repeat protein. RSU1 interacts with PINCH and integrin-linked kinase (ILK) serving as a molecular scaffold for cellular focal adhesion (49). A marked increase in myocardial levels of ILK proteins has been reported in patients with congenital and acquired outflow tract obstruction (50) causing ventricular hypertrophy. Our study is the first to describe RSU1-peptide elevated in HCM and the first study to describe its potential as a plasma biomarker.
TLN1 (TLN1-peptide) is a large dimeric cytoskeletal protein that activates integrins and mediates their adhesion to the actin cytoskeleton. Integrins are key mechanotransducers in cardiomyocytes and are intimately involved in the process of cardiac hypertrophy (51). During embryogenesis, cardiomyocytes exhibit high TLN1 levels, but these decline in the mature heart (52). TLN1 has been found to be up-regulated in the costameres, both in a HCM mouse model and in the myocardium of adult humans with heart failure (52).
Enzymes
We observed elevated levels of GSTO-1 (GSTO1-peptide) in the myocardium (3) and plasma of HCM patients but reduced levels of another glutathione S-transferase (Kappa-1, GSTK1) has been reported in murine HCM models (53). This may reflect the fact that the two anti-oxidant enzymes, GSTO1 and GSTK1, are functionally distinct with the former localizing to the cytosol and the latter to the mitochondria and peroxisomes (54) thus their expression may be differentially impacted by HCM. Another study of a cor pulmonale mouse model has shown high levels of GSTO1 in the hypertrophied right ventricular myocardium (55) confirming our observation in human tissue and plasma and indicating its potential as a marker to monitor cardiac oxidative stress.
ALDOA
(ALDOA-peptide) was found altered in the previously published myocardial proteomics analysis (3). It is a glycolytic enzyme found in skeletal as well as cardiac muscle where it functions as a scaffolding protein binding to actin or actin-tropomyosin (56). Serum aldolase concentrations have been shown to be elevated in patients with Danon disease, a genetic disorder causing a weakening of the heart (excluded from this study) (57). ALDOA activity has been observed to be up-regulated in response to hypoxia (58), which could indicate this protein could be an indirect marker of hypoxic stress in HCM.
Complement Component
The anaphylatoxin C3 (C3-peptide) is generated by activation of the innate immune system and has been shown to be elevated in a limited number of patients with LVH (five HCM, three hypertensives, one athlete) (59) and in a larger cohort of hypertensive patients (60). The human protein-protein interaction network for our six biomarkers (Fig. S2) illustrates how the C3 cluster representing inflammation appears distinct from the remaining five biomarkers that more generally reflect myocyte hypertrophy and fibrosis. Fig. S3 illustrates the glutamine gamma-glutamyltransferase 2 (TGM2) as the interacting node putatively linking mechanisms of myocyte hypertrophy and inflammation. Indeed, a mouse model with increased cardiomyocyte TGM2 (61) has been shown to have up-regulated COX2 expression leading to LVH, fibrosis, and eventual cardiomyocyte apoptosis.
It is known that myocyte hypertrophy and disarray lead to augmented protein synthesis in the myocardium, partly from the reactivation of fetal transcription (62). This may partly explain the high circulating levels of some of our biomarkers, particularly for those involved in intracellular hypertrophic signal transduction. The presence in the plasma of HCM patients of some of these proteotypic peptides may reflect hypertrophy-related microparticle secretion by cardiomyocytes or the liberation of intracellular components following cell death.
Clinical Correlations
We applied the assay to a randomly selected cohort of patients with LVH+ HCM and controls and replicated the findings in a validation cohort. The emergence of a common biomarker panel in both the training and validation LVH+ HCM datasets and similar profiles in subclinical HCM, could suggest that the six biomarkers reflect aspects of HCM disease pathophysiology. Peptide biomarkers identified in the plasma of our HCM patients could be related to mechanisms of myocyte hypertrophy and disarray, but they may also reflect myocardial fibrosis or other downstream effects observed in heart failure.
Our data suggest that LVH+ HCM patients with high ML proteomics prediction scores (upper quartile) are more likely to be at high risk of SCD, with projected SCD rates that are 1.4-fold higher than in patients with ML prediction scores in the lower quartile. Apart from an association with NSVT (9, 10), the ML prediction score also related to CMR markers of adverse disease progression in HCM, including extensive LGE, LV mass, and diffuse myocardial fibrosis by native T1 mapping. If our findings are confirmed in larger cohorts, the proteotypic peptide biomarkers we describe in this study may be a valuable additional factor for future risk stratification schemes.
We developed the entire proteomics method on a triple quadrupole MS-based platform to make the validation of multiple biomarkers high throughput for large numbers of samples. This approach and development of this assay makes potential further validation and signature refinement possible on other center cohorts in the future, as well as its application to disease models for novel therapy studies. Our assay uses plasma—a patient biofluid—that, compared with myocardial tissue, is less limited in sample access, obtained less invasively, and easier to sample repeatedly and to process for storage and analysis in a more standardized and less complex manner.
Elevated levels of NT-proBNP (a biomarker of ventricular wall stress (63)) associate with a higher risk of heart failure, death, or transplantation in LVH+ HCM (64). In this study, there was no correlation between NT-proBNP and our proteomics assay (with the exception of RSU1, rs = 0.21; p = 0.028), suggesting that the majority of our candidate peptides may be tracking myocyte hypertrophy (LV mass and MWT) and fibrosis (native T1 and LGE) rather than myocardial stress. Though results from enzyme-linked immunosorbent assays appraising collagen synthesis biomarkers in venous blood samples of HCM patients have previously been conflicting (40, 64), recent data in Fabry disease (a genetic lysosomal storage disorder resulting in pathological cardiac hypertrophy) demonstrated a correlation between levels of type I collagen synthesis and degradation biomarkers in blood, and LV mass and scar by CMR (65). This would fit with our findings where a different set of proteomics plasma markers of fibrosis similarly correlate with LV wall thickness, mass, and scar by CMR in LVH+ HCM. We recently reported exploratory myocardial proteomics profiling experiments in which lumican was up-regulated in LVH+ HCM compared with controls (3). A similar trend was observed in the current plasma experiments, but the differences between LVH+ HCM and controls were not significant to justify inclusion of lumican in the final multiplex panel. Factors potentially explaining the mitigated lumican trends in HCM plasma compared with myocardium include the relatively small number of control samples used in the myocardial experiments (n = 7) when compared with the current work (n = 97), that patients in the myocardial study exhibited more advanced drug refractory disease (given they all underwent myectomy to treat LV outflow tract obstruction), and that peptide biomarkers discovered in whole tissue myocardial homogenates are not necessarily liberated or secreted into the circulating plasma at sufficiently high concentrations to impact the HCM plasma proteome.
Study Limitations
Though assay results were promising, this was a single center study. Results merit validation in a larger, multicenter study of both LVH+ and subclinical HCM. Other diseases with hypertrophy and other cardiomyopathies were not explored in the current work, thus the detected biomarkers may not be unique to HCM. The utility of candidate biomarkers was assessed at a single time point in subclinical and established HCM versus healthy volunteers and against a limited panel of clinically relevant parameters thought to be significant (e.g. BNP, LGE, MWT). Due to the high prevalence of cardiac implantable electronic devices at the time of this study, not all participants underwent CMR. Targeted validation of the identified peptides by enzyme-linked immunosorbent assay approaches was not undertaken in this study.
CONCLUSIONS
Using a combination of targeted and nontargeted proteomic approaches, we have discovered six potentially useful circulating plasma biomarkers related to myocardial substrate changes in HCM, which correlate with the estimated sudden cardiac death risk.
DATA AVAILABILITY
ProteomeXchange Accession Details - Project accession: PXD009859 (http://www.ebi.ac.uk/pride).
Supplementary Material
Footnotes
* This work was funded by the Translational Mass Spectrometry Research Group, kind donations from the Peto Foundation and the National Institute for Health Research (NIHR) Great Ormond Street Hospital Biomedical Research Center. The views expressed are those of the author(s) and not necessarily those of the National Health Service, the NIHR, or the Department of Health. G.C. is supported by the NIHR Rare Diseases Translational Research Collaboration. G.C., J.C.M., and B.O.B. are supported by the Barts Charity HeartOME1000 grant MGU0427. K.M./W.H. and N.P. are supported by the NIHR Biomedical Research Center at Great Ormond Street Hospital for Children National Health Service Foundation Trust and UCL. P.M.E. is supported by University College London Hospitals (UCLH) NIHR Biomedical Research Center. J.C.M. is directly and indirectly supported by UCLH NIHR Biomedical Research Center and the Biomedical Research Unit at Barts Hospital, respectively. P.S. is funded by the UCLH NIHR Biomedical Research Center, and his research is supported by the Foundation Leducq. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material Tables S1–S6 and Figs. S1–S3.
This article contains supplemental material Tables S1–S6 and Figs. S1–S3.
1 The abbreviations used are:
- ALDOA
- aldolase fructose-bisphosphate A
- ASB-14
- amidosulfobetaine-14
- CMR
- cardiovascular magnetic resonance
- C3
- complement C
- GSTO1
- glutathione S-transferase omega-1
- HCM
- hypertrophic cardiomyopathy
- LGE
- late gadolinium enhancement
- LVH
- left ventricular hypertrophy
- ML
- machine learning
- MRM
- multiple reaction monitoring
- MWT
- maximal wall thickness
- NSVT
- nonsustained ventricular tachycardia
- NT-proBNP
- N-terminal prohormone of brain natriuretic peptide
- RSU1
- Ras suppressor protein 1
- SCD
- sudden cardiac death
- SD
- standard deviation
- ShMOLLI
- shortened modified look-locker inversion recovery
- STRM
- signal threshold versus reference mean
- SVM
- support vector machine
- TLN1
- talin 1
- THBS1
- thrombospondin 1.
REFERENCES
- 1. Ho C. Y., Charron P., Richard P., Girolami F., Van Spaendonck-Zwarts K. Y., and Pinto Y. (2015) Genetic advances in sarcomeric cardiomyopathies: State of the art. Cardiovasc. Res. 105, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carrier L., Mearini G., Stathopoulou K., and Cuello F. (2015) Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 573, 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coats C., Heywood W., Virasami A., Syrris P., Dos Remedios C., Treibel T., Moon J., McKenna W., McGregor C., Sebire N., Ashworth M., Mills K., and Elliott P. (2018) Proteomic analysis of the myocardium in hypertrophic obstructive cardiomyopathy. Circ. Cardiovasc. Genet. 11, e001974. [DOI] [PubMed] [Google Scholar]
- 4. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Hüttenhain R., Koomen J. M., Liebler D. C., Liu T., MacLean B., Mani D. R., Mansfield E., Neubert H., Paulovich A. G., Reiter L., Vitek O., Aebersold R., Anderson L., Bethem R., Blonder J., Boja E., Botelho J., Boyne M., Bradshaw R. A., Burlingame A. L., Chan D., Keshishian H., Kuhn E., Kinsinger C., Lee J. S., Lee S.-W., Moritz R., Oses-Prieto J., Rifai N., Ritchie J., Rodriguez H., Srinivas P. R., Townsend R. R., Van Eyk J., Whiteley G., Wiita A., and Weintraub S. (2014) Targeted peptide measurements in biology and medicine: Best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Captur G., Ho C. Y., Schlossarek S., Kerwin J., Mirabel M., Wilson R., Rosmini S., Obianyo C., Reant R., Bassett P., Cook A. C., Lindsay S., McKenna W. J., Mills K., Elliott P. M., Mohun T. J., Carrier L., and Moon J. C. (2016) The embryological basis of subclinical hypertrophic cardiomyopathy. Sci. Rep. 8, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Captur G., Lopes L. R., Mohun T. J., Patel V., Li C., Bassett P., Finocchiaro G., Ferreira V. M., Esteban M. T., Muthurangu V., Sherrid M. V., Day S. M., Canter C. E., McKenna W. J., Seidman C. E., Bluemke D. A., Elliott P. M., Ho C. Y., and Moon J. C. (2014) Prediction of sarcomere mutations in subclinical hypertrophic cardiomyopathy. Circ. Cardiovasc. Imaging 7, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finocchiaro G., Papadakis M., Robertus J.-L., Dhutia H., Steriotis A. K., Tome M., Mellor G., Merghani A., Malhotra A., Behr E., Sharma S., and Sheppard M. N. (2016) Etiology of sudden death in sports: Insights from a United Kingdom regional registry. J. Am. Coll. Cardiol. 67, 2108–2115 [DOI] [PubMed] [Google Scholar]
- 8. Williams L. K., Misurka J., Ho C. Y., Chan W.-X., Agmon Y., Seidman C., Rakowski H., and Carasso S. (2018) Multilayer myocardial mechanics in genotype-positive left ventricular hypertrophy-negative patients with hypertrophic cardiomyopathy. Am. J. Cardiol. 122, 1754–1760 [DOI] [PubMed] [Google Scholar]
- 9. O'Mahony C., Jichi F., Pavlou M., Monserrat L., Anastasakis A., Rapezzi C., Biagini E., Gimeno J. R., Limongelli G., McKenna W. J., Omar R. Z., Elliott P. M., and Investigators H. C. O. (2014) A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM risk-SCD). Eur. Heart J. 35, 2010–2020 [DOI] [PubMed] [Google Scholar]
- 10. O'Mahony C., Jichi F., Ommen S. R., Christiaans I., Arbustini E., Garcia-Pavia P., Cecchi F., Olivotto I., Kitaoka H., Gotsman I., Carr-White G., Mogensen J., Antoniades L., Mohiddin S., Maurer M. S., Tang H. C., Geske J. B., Siontis K. C., Mahmoud K. D., Vermeer A., Wilde A., Favalli V., Guttmann O. P., Gallego-Delgado M., Dominguez F., Tanini I., Kubo T., Keren A., Bueser T., Waters S., Issa I. F., Malcolmson J., Burns T., Sekhri N., Hoeger C. W., Omar R. Z., and Elliott P. M. (2018) International external validation study of the 2014 European Society of Cardiology guidelines on sudden cardiac death prevention in hypertrophic cardiomyopathy (EVIDENCE-HCM). Circulation 137, 1015–1023 [DOI] [PubMed] [Google Scholar]
- 11. Lopes L. R., Zekavati A., Syrris P., Hubank M., Giambartolomei C., Dalageorgou C., Jenkins S., McKenna W., Plagnol V., and Elliott P. M. (2013) Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J. Med. Genet. 50, 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T., and McVean G. A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W. W., Hegde M., Lyon E., Spector E., Voelkerding K., and Rehm H. L.; ACMG Laboratory Quality Assurance Committee. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17, 405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., and Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 15. Heywood W., Mills K., Wang D., Hogg J., Madgett T. E., Avent N. D., and Chitty L. S. (2012) Identification of new biomarkers for Down's syndrome in maternal plasma. J. Proteomics 75, 2621–2628 [DOI] [PubMed] [Google Scholar]
- 16. Manwaring V., Heywood W. E., Clayton R., Lachmann R. H., Keutzer J., Hindmarsh P., Winchester B., Heales S., and Mills K. (2013) The identification of new biomarkers for identifying and monitoring kidney disease and their translation into a rapid mass spectrometry-based test: evidence of presymptomatic kidney disease in pediatric Fabry and type-I diabetic patients. J. Proteome Res. 12, 2013–2021 [DOI] [PubMed] [Google Scholar]
- 17. Silva J. C., Gorenstein M. V., Li G.-Z., Vissers J. P. C., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 18. Craig R., Cortens J. P., and Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 19. Heywood W. E., Baud A., Bliss E., Sirka E., Schott J. M., Zetterberg H., Galimberti D., Sebire N. J., and Mills K. (2016) A high throughput, multiplexed and targeted proteomic CSF assay to quantify neurodegenerative biomarkers and apolipoprotein e isoforms status. J. Vis. Exp. 116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N. H., Chavali G., Chen C., Del-Toro N., Duesbury M., Dumousseau M., Galeota E., Hinz U., Iannuccelli M., Jagannathan S., Jimenez R., Khadake J., Lagreid A., Licata L., Lovering R. C., Meldal B., Melidoni A. N., Milagros M., Peluso D., Perfetto L., Porras P., Raghunath A., Ricard-Blum S., Roechert B., Stutz A., Tognolli M., Van Roey K., Cesareni G., and Hermjakob H. (2014) The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, 358–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mosca R., Céol A., and Aloy P. (2013) Interactome3D: Adding structural details to protein networks. Nat. Methods 10, 47–53 [DOI] [PubMed] [Google Scholar]
- 23. Lang R. M., Bierig M., Devereux R. B., Flachskampf F. A., Foster E., Pellikka P. A., Picard M. H., Roman M. J., Seward J., Shanewise J., Solomon S., Spencer K. T., St John Sutton M., and Stewart W. (2006) Recommendations for chamber quantification. Eur. J. Echocardiogr. 7, 79–108 [DOI] [PubMed] [Google Scholar]
- 24. Quiñones M. A., Otto C. M., Stoddard M., Waggoner A., and Zoghbi W. A. (2002) Recommendations for quantification of Doppler echocardiography: A report from the Doppler Quantification Task Force of the Nomenclature and Standards Committee of the American Society of Echocardiography. J. Am. Soc. Echocardiogr. 15, 167–184 [DOI] [PubMed] [Google Scholar]
- 25. Kramer C. M., Barkhausen J., Flamm S. D., Kim R. J., and Nagel E. (2008) Standardized cardiovascular magnetic resonance imaging protocols, Society for Cardiovascular Magnetic Resonance: Board of trustees task force on standardized protocols. J. Cardiovasc. Magn. Reson. 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Captur G., Lopes L. R., Patel V., Li C., Bassett P., Syrris P., Sado D. M., Maestrini V., Mohun T. J., McKenna W. J., Muthurangu V., Elliott P. M., and Moon J. C. (2014) Abnormal cardiac formation in hypertrophic cardiomyopathy fractal analysis of trabeculae and preclinical gene expression. Circ. Cardiovasc. Genet. 7, 241–248 [DOI] [PubMed] [Google Scholar]
- 27. Piechnik S. K., Ferreira V. M., Dall'Armellina E., Cochlin L. E., Greiser A., Neubauer S., and Robson M. D. (2010) Shortened modified look-locker inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J. Cardiovasc. Magn. Reson. 12, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mikami Y., Kolman L., Joncas S. X., Stirrat J., Scholl D., Rajchl M., Lydell C. P., Weeks S. G., Howarth A. G., and White J. A. (2014) Accuracy and reproducibility of semi-automated late gadolinium enhancement quantification techniques in patients with hypertrophic cardiomyopathy. J. Cardiovasc. Magn. Reson. 16, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. RCore Team. (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/ [Google Scholar]
- 30. Hollander Z., Dai D. L., Putko B. N., Yogasundaram H., Wilson-McManus Thompson J. E., Khan A., West M. L., McManus B. M., and Oudit G. Y. (2015) Gender-specific plasma proteomic biomarkers in patients with Anderson–Fabry disease. Eur. J. Heart Fail. 17, 291–300 [DOI] [PubMed] [Google Scholar]
- 31. Varanasi H. (2015) in Practical Predictive Analytics and Decisioning Systems for Medicine. 1st ed 2014, pp. 850–865, Academic Press [Google Scholar]
- 32. Swan A. L., Mobasheri A., Allaway D., Liddell S., and Bacardit J. (2013) Application of machine learning to proteomics data: Classification and biomarker identification in postgenomics biology. OMICS 17, 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dernellis J., and Panaretou M. (2006) Effects of C-reactive protein and the third and fourth components of complement (C3 and C4) on incidence of atrial fibrillation. Am. J. Cardiol. 97, 245–248 [DOI] [PubMed] [Google Scholar]
- 35. Zemljic-Harpf A. E., Miller J. C., Henderson S. A., Wright A. T., Manso A. M., Elsherif L., Dalton N. D., Thor A. K., Perkins G. A., McCulloch A. D., and Ross R. S. (2007) Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol. Cell. Biol. 27, 7522–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Procter N. E., Ball J., Liu S., Hurst N., Nooney V. B., Goh V., Stafford I., Heresztyn T., Carrington M., Ngo D. T., Hylek E. M., Isenberg J. S., Chirkov Y. Y., Stewart S., and Horowitz J. D. (2015) Impaired platelet nitric oxide response in patients with new onset atrial fibrillation. Int. J. Cardiol. 179, 160–165 [DOI] [PubMed] [Google Scholar]
- 37. Elliott P. M., Anastasakis A., Borger M. A., Borggrefe M., Cecchi F., Charron P., Hagege A. A., Lafont A., Limongelli G., Mahrholdt H., McKenna W. J., Mogensen J., Nihoyannopoulos P., Nistri S., Pieper P. G., Pieske B., Rapezzi C., Rutten F. H., Tillmanns C., Watkins H., O'Mahony C., Zamorano J. L., Achenbach S., Baumgartner H., Bax J. J., Bueno H., Dean V., Deaton C., Erol C., Fagard R., Ferrari R., Hasdai D., Hoes A. W., Kirchhof P., Knuuti J., Kolh P., Lancellotti P., Linhart A., Piepoli M. F., Ponikowski P., Sirnes P. A., Tamargo J. L., Tendera M., Torbicki A., Wijns W., Windecker S., Alfonso F., Basso C., Cardim N. M., Gimeno J. R., Heymans S., Holm P. J., Keren A., Lionis C., Muneretto C., Priori S., Salvador M. J., and Wolpert C. (2014) 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy. Eur. Heart J. 35, 2733–2779 [DOI] [PubMed] [Google Scholar]
- 38. Fang L., Ellims A., Beale A., Taylor A., Murphy A., and Dart A. (2017) Relationships between systemic inflammation and myocardial fibrosis, diastolic dysfunction, and cardiac hypertrophy in patients with hypertrophic cardiomyopathy. Hear. Lung Circ. 26, S110. [PMC free article] [PubMed] [Google Scholar]
- 39. Rehulkova H., Rehulka P., Myslivcova Fucikova A., Stulik J., and Pudil R. (2016) Identification of novel biomarker candidates for hypertrophic cardiomyopathy and other cardiovascular diseases leading to heart failure. Physiol. Res. 65, 751–762 [DOI] [PubMed] [Google Scholar]
- 40. Ho C. Y., López B., Coelho-Filho O. R., Lakdawala N. K., Cirino A. L., Jarolim P., Kwong R., González A., Colan S. D., Seidman J. G., Díez J., and Seidman C. E. (2010) Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N. Engl. J. Med. 363, 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ho C. Y. (2009) Hypertrophic cardiomyopathy: Preclinical and early phenotype. J. Cardiovasc. Transl. Res. 2, 462–470 [DOI] [PubMed] [Google Scholar]
- 42. McKenna W. J., Stewart J. T., Nihoyannopoulos P., McGinty F., and Davies M. J. (1990) Hypertrophic cardiomyopathy without hypertrophy: Two families with myocardial disarray in the absence of increased myocardial mass. Br. Heart J. 63, 287–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chistiakov D. A., Melnichenko A. A., Myasoedova V. A., Grechko A. V., and Orekhov A. N. (2017) Thrombospondins: A role in cardiovascular disease. Int. J. Mol. Sci. 18, e1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schroen B., Heymans S., Sharma U., Blankesteijn W. M., Pokharel S., Cleutjens J. P., Porter J. G., Evelo C. T., Duisters R., van Leeuwen R. E., Janssen B. J., Debets J. J., Smits J. F., Daemen M. J., Crijns H. J., Bornstein P., and Pinto Y. M. (2004) Thrombospondin-2 is essential for myocardial matrix integrity: Increased expression identifies failure-prone cardiac hypertrophy. Circ. Res. 95, 515–522 [DOI] [PubMed] [Google Scholar]
- 45. Frangogiannis N. G. (2012) Matricellular proteins in cardiac adaptation and disease. Physiol. Rev. 92, 635–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imoto K., Okada M., and Yamawaki H. (2017) Expression profile of matricellular proteins in hypertrophied right ventricle of monocrotaline-induced pulmonary hypertensive rats. J. Vet. Med. Sci. 79, 1096–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Belmadani S., Bernal J., Wei C.-C., Pallero M. A., Dell'italia L., Murphy-Ullrich J. E., and Berecek K. H. (2007) A thrombospondin-1 antagonist of transforming growth factor-beta activation blocks cardiomyopathy in rats with diabetes and elevated angiotensin II. Am. J. Pathol. 171, 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mégy K., Audic S., and Claverie J.-M. (2002) Heart-specific genes revealed by expressed sequence tag (EST) sampling. Genome Biol. 3, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elias M. C., Pronovost S. M., Cahill K. J., Beckerle M. C., and Kadrmas J. L. (2012) A crucial role for Ras suppressor-1 (RSU-1) revealed when PINCH and ILK binding is disrupted. J. Cell Sci. 125, 3185–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu H., Fedak P. W. M., Dai X., Du C., Zhou Y.-Q., Henkelman M., Mongroo P. S., Lau A., Yamabi H., Hinek A., Husain M., Hannigan G., and Coles J. G. (2006) Integrin-linked kinase expression is elevated in human cardiac hypertrophy and induces hypertrophy in transgenic mice. Circulation 114, 2271–2279 [DOI] [PubMed] [Google Scholar]
- 51. Brancaccio M., Hirsch E., Notte A., Selvetella G., Lembo G., and Tarone G. (2006) Integrin signalling: The tug-of-war in heart hypertrophy. Cardiovasc. Res. 70, 422–433 [DOI] [PubMed] [Google Scholar]
- 52. Manso A. M., Li R., Monkley S. J., Cruz N. M., Ong S., Lao D. H., Koshman Y. E., Gu Y., Peterson K. L., Chen J., Abel E. D., Samarel A. M., Critchley D. R., and Ross R. S. (2013) Talin1 has unique expression versus talin 2 in the heart and modifies the hypertrophic response to pressure overload. J. Biol. Chem. 288, 4252–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sasagawa S., Nishimura Y., Okabe S., Murakami S., Ashikawa Y., Yuge M., Kawaguchi K., Kawase R., Okamoto R., Ito M., and Tanaka T. (2016) Downregulation of GSTK1 is a common mechanism underlying hypertrophic cardiomyopathy. Front. Pharmacol. 7, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petit E., Michelet X., Rauch C., Bertrand-Michel J., Terce F., Legouis R., and Morel F. (2009) Glutathione transferases kappa 1 and kappa 2 localize in peroxisomes and mitochondria, respectively, and are involved in lipid metabolism and respiration in Caenorhabditis elegans. FEBS J. 276, 5030–5040 [DOI] [PubMed] [Google Scholar]
- 55. Souza-Rabbo M. P., Silva L. F., Auzani J. A., Picoral M., Khaper N., and Belló-Klein A. (2008) Effects of a chronic exercise training protocol on oxidative stress and right ventricular hypertrophy in monocrotaline-treated rats. Clin. Exp. Pharmacol. Physiol. 35, 944–948 [DOI] [PubMed] [Google Scholar]
- 56. Clarke F. M., and Morton D. J. (1976) Aldolase binding to actin-containing filaments. Formation of paracrystals. Biochem. J. 159, 797–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Di Mauro S., Tanji K., and Hirano M. (2007) LAMP-2 deficiency (Danon disease). Acta Myol. 26, 79–82 [PMC free article] [PubMed] [Google Scholar]
- 58. Dawson N. J., Biggar K. K., and Storey K. B. (2013) Characterization of fructose-1,6-bisphosphate aldolase during anoxia in the tolerant turtle, Trachemys scripta elegans: An assessment of enzyme activity, expression and structure. PLoS ONE 8, e68830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boudonas G., Boura P., Lefkos N., Zacharioydaki E., and Efthymiadis A. (1994) A possible role for autoantibodies in left ventricular hypertrophy. Cardiology 84, 278–283 [DOI] [PubMed] [Google Scholar]
- 60. Lefkos N., Boura P., Boudonas G., Zacharioudaki E., Efthimiadis A., Tsougas M., and Epivatianos P. (1995) Immunopathogenic mechanisms in hypertension. Am. J. Hypertens. 8, 1141–1145 [DOI] [PubMed] [Google Scholar]
- 61. Zhang Z., Vezza R., Plappert T., McNamara P., Lawson J. A., Austin S., Praticò D., Sutton M. S., and FitzGerald G. A. (2003) COX-2-dependent cardiac failure in Gh/tTG transgenic mice. Circ. Res. 92, 1153–1161 [DOI] [PubMed] [Google Scholar]
- 62. Sack M. N., Disch D. L., Rockman H. A., and Kelly D. P. (1997) A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc. Natl. Acad. Sci. U.S.A. 94, 6438–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rajtar-Salwa R., Hladij R., and Dimitrow P. P. (2014) Elevated level of troponin but not n-terminal probrain natriuretic peptide is associated with increased risk of sudden cardiac death in hypertrophic cardiomyopathy calculated according to the ESC Guidelines. Dis. Markers 417908, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ho J. E., Shi L., Day S. M., Colan S. D., Russell M. W., Towbin J. A., Sherrid M. V., Canter C. E., Jefferies J. L., Murphy A., Taylor M., Mestroni L., Cirino A. L., Sleeper L. A., Jarolim P., Lopez B., Gonzalez A., Diez J., Orav E. J., and Ho C. Y. (2017) Biomarkers of cardiovascular stress and fibrosis in preclinical hypertrophic cardiomyopathy. Open Heart 4, e000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aguiar P., Azevedo O., Pinto R., Marino J., Cardoso C., Sousa N., Cunha D., Hughes D., and Soares J. L. (2018) Biomarkers of myocardial fibrosis: Revealing the natural history of fibrogenesis in Fabry disease cardiomyopathy. J. Am. Heart Assoc. 7, e007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ProteomeXchange Accession Details - Project accession: PXD009859 (http://www.ebi.ac.uk/pride).