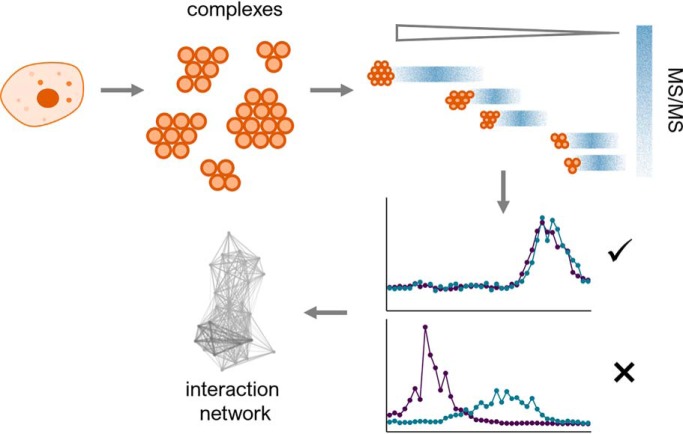
Interactome studies are necessary to understand cellular processes and co-elution methods are well suited for the simultaneous and global exploration of the interactome, as well as the assessment of biological perturbations of the network. These methods rely on the fundamental idea that proteins from the same complex migrate together during fractionation. We review the different separation techniques along with the quantification and bioinformatic approaches used for co-elution methods and provide design considerations to choose between them.
Keywords: Protein-protein interactions, protein complex analysis, separation technologies, mass spectrometry, chromatography, bioinformatics, SILAC, co-elution, co-fractionation, complexes, interactome, protein correlation profiling
Graphical Abstract
Highlights
Co-elution stands out as a global interactome mapping method.
Benefits include all-to-all protein analysis and measurement of interactome perturbations.
Different separation, quantification and bioinformatic strategies are available.
Design considerations depend largely on system under study.
Abstract
Understanding how proteins interact is crucial to understanding cellular processes. Among the available interactome mapping methods, co-elution stands out as a method that is simultaneous in nature and capable of identifying interactions between all the proteins detected in a sample. The general workflow in co-elution methods involves the mild extraction of protein complexes and their separation into several fractions, across which proteins bound together in the same complex will show similar co-elution profiles when analyzed appropriately. In this review we discuss the different separation, quantification and bioinformatic strategies used in co-elution studies, and the important considerations in designing these studies. The benefits of co-elution versus other methods makes it a valuable starting point when asking questions that involve the perturbation of the interactome.
Cellular functions and responses are coordinated by proteins working in concert through networks of protein-protein interactions (PPIs)1, often involving higher-order complexes. Understanding the architecture of this interactome from a dynamic, topological and quantitative perspective is key to discerning biological processes and their involvement in disease (1–4).
There are numerous techniques available for studying PPIs (5–12). These have evolved from the classic yeast two-hybrid (Y2H) method to mass spectrometry (MS) approaches based on the co-purification of interacting proteins. Currently, the most widely used technique is affinity purification (AP-MS), thanks to its simplicity and improvements made in quantification and data analysis (13, 14). BioID is a novel strategy (15) that has rapidly found a niche with important applications, despite the method still evolving (16). For a systems-level analysis, the ideal interactome mapping method should be high-throughput, quantitative, simple, physiologically relevant and give information about stoichiometry, topology and dynamics. However, current techniques show limitations in at least a few of these characteristics so the key is to use complementary methods to corroborate results. One approach particularly useful for exploratory studies are co-elution methods.
Co-elution or co-fractionation methods are collectively a global approach used to simultaneously study the whole interactome (as opposed to piece-by-piece, as in AP-MS) and will be the focus of this review. Co-elution methods all rely on separation of protein complexes under native conditions, with the fundamental idea being that proteins belonging to the same complex co-elute or migrate together during separation, showing the same migration profile (Fig. 1A). Co-elution strategies were originally introduced to assign proteins to the same subcellular localization if these displayed similar profiles across a density gradient (17–19). More recently, this method has been adapted to detect protein interactions, using chromatography (20, 21) or blue-native polyacrylamide gel electrophoresis (22) (BN-PAGE) to generate high-resolution elution profiles for thousands of proteins. The analysis of co-elution data involves plotting the MS1 intensities of proteins across many fractions, matching and scoring those profiles to detect binary protein interactions and provide an interactome map from those interactions (Fig. 1B). Current advances in the analysis of co-elution data include the development of a bioinformatics pipeline (PrInCE) (23) and the software toolkit EPIC (24), both freely available. In this review we discuss the comparable advantages of co-elution, the different separation strategies used and design considerations with an emphasis on the separation method, quantification and data analysis.
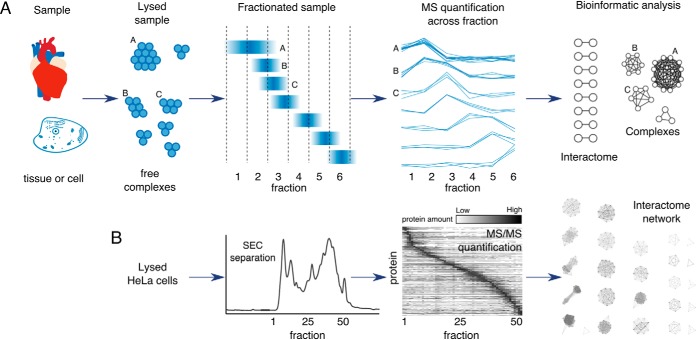
Fig. 1.
Schematic of the steps commonly involved in co-elution methods. A, General workflow of a co-elution experiment. The lysed sample containing protein complexes under native conditions is separated in a set number of fractions. Proteins from the same complex show the same co-elution profile after a bioinformatic analysis to extract an interactome map. B, Co-elution data representative from an experiment where lysed HeLa cells were fractionated by size exclusion chromatography (SEC), quantified by MS/MS, and finally used to construct an interactome network through data analysis.
Existing Co-elution Strategies Are Well-suited for Global Exploratory PPI Studies When Compared with Other Methods
The main comparative benefit of co-elution strategies (Table I) is that hundreds to thousands of protein complexes can be simultaneously and rapidly analyzed in a single experiment (25). Because the primary measurement in these experiments is the abundances of thousands of proteins across the elution gradient, rather than a focus on bait proteins, co-elution studies scale much more easily than Y2H or AP-MS (26). Thus, co-elution can identify all the interactors for many proteins simultaneously, as well as to identify when a single protein participates in multiple complexes (6), which is more difficult to determine by AP-MS. A similar and recently developed complementary approach to co-elution is thermal proteome profiling (TPP), which can provide the proteome-wide detection of protein complexes and their rearrangements (27, 28) but is based on comparing protein melting curves instead of co-elution profiles.
Table I. Comparison between co-elution approaches and other commonly used strategies to detect protein-protein interactions.
| Co-elution | Y2H-based | AP-MS | IP-MS | BioID | |
|---|---|---|---|---|---|
| “All-to-all” interactome | + | − | − | − | − |
| High-throughput | + | + | + | − | − |
| Protein tagging | − | + | + | − | + |
| Study different biological contexts | + | − | + | + | − |
| Type of interactions | |||||
| Direct | + | + | + | + | + |
| Indirect | + | − | + | + | + |
| Proximal | − | − | − | − | + |
| Selectivity | + | − | − | − | − |
| Same protein in multiple complexes | + | − | − | − | − |
| In vivo | − | + | − | − | + |
| Suited for weak/transient interactions | − | + | − | − | + |
An added attraction of co-elution studies is that the generated interactome should be more physiologically relevant than results from studies involving protein tagging because modifying proteins can perturb endogenous interactions by the presence of the tag or overexpression of the bait (5, 10). In this sense, co-elution is similar to immunoprecipitation-type AP-MS studies, where proteins are purified with an antibody against the bait itself, rather than an added epitope, but co-elution is not dependent on the existence of a specific and high affinity antibody (8, 10). In AP-MS, the bait is fused to an affinity tag allowing the purification of this bait and its interacting partners without the need for a specific antibody, but it still relies on the fusion step.
Co-elution studies take considerably less time and resources than an equivalently scaled AP-MS study, so replicates can be conducted more easily. This also means that biological perturbations of the network can be measured. This has so far only been done globally using SILAC (20, 29), but in principle could also be accomplished using label-free quantitation. Improved quantitation in AP-MS has allowed the comparison of proteins that co-purify with a bait protein under normal and perturbed conditions in a quantitative manner (13), but not nearly at the scale enabled by co-elution.
The various interactome methods provide fundamentally different types of information. Co-elution, at its heart, identifies binary interactions, but these interactions do not necessarily represent direct physical connections, and can include proteins that co-elute because they are members of the same complex but not in direct physical contact (Table I, “Indirect” interactions). AP-MS targets only the complexes co-purified with a specific protein. The BioID method is similar in that it focuses on the potential interactors of a specific protein. However, the candidates identified can be direct or indirect interactors, and/or vicinal proteins that do not physically interact with the fusion protein (15).
False positive interactions are a problem for all PPI technologies, to a greater or lesser degree. In co-elution strategies, functionally unrelated complexes can co-elute, leading the user to conclude that all the component proteins interact and thus manifesting as false positives. Therefore, co-elution results should be regarded with caution. Potential novel complexes provide a good seed for follow-up analyses to obtain more detailed and high confidence biochemical information. These types of false positives can be mitigated by using as high resolution separation conditions as possible. The use of multiple orthogonal separation strategies can also decrease the effect of co-elution by chance. Targeted complex quantification should also be helpful for follow-up experiments. In addition, rigorous bioinformatic analyses lower the chances of predicting false positive interactions.
The chromatograms or electropherograms generated in co-elution studies can also be used to quantify the relative distribution of a protein into multiple different protein complexes. That is, if one protein participates in more than one complex, the relative amounts of those different complexes can be derived. This can yield information about the dynamics of PPIs as substoichiometric interactors will, e.g., more likely be dynamic partners in the complex (5, 30).
When compared with other methods, co-elution stands out as a global approach capable of producing vast information of the interactome. It is therefore particularly suited for exploratory studies that can later be validated with complementary approaches.
Separation Strategies Used for Co-elution
In co-elution studies, tissues or cells are lysed to extract protein complexes that are subsequently fractionated under conditions that are designed to preserve the PPIs within the complexes. Different separation techniques have been used for fractionation, including size-exclusion chromatography (SEC), ion-exchange (IEX) and hydrophobic interaction chromatography (HIC), and BN-PAGE (Table II) (7). Protein complexes can also be separated according to their sedimentation rate or isoelectric point by fractionating in sucrose gradients (31) or native capillary isoelectric focusing (IEF) (32). However, considering that sucrose gradients have low resolution and IEF is mostly coupled to native MS and can be challenging for whole lysates, we recommend the use of these for orthogonal separations or complementary experiments.
Table II. Comparison between co-elution separation strategies: SEC, IEX, HIC, BN-PAGE.
| Separation technique | Stationary phase | Separation principle | Mobile phasea | Benefits/Drawbacks | Applications |
|---|---|---|---|---|---|
| SEC | Material with different pore sizes (silica, polymeric or cross-linked agarose) | Hydrodynamic size | Millimolar salt buffers at neutral pH | Less buffer requirements, isocratic/Modest resolution | Soluble complexes |
| IEX | Material with ionic groups (silica-based or polymeric) with SAX, WAX, WCX or mixed-bed (WAX & WCX) properties | Salt gradient | Increasing molar salt gradient (NaCl) at neutral pH | Higher resolution, more chemistries available/Higher salt concentrations required | Soluble complexes |
| HIC | Hydrophobic | Salt gradient | Decreasing molar salt gradient (e.g. (NH4)2SO4) at neutral pH | Higher salt concentrations required | Multiple orthogonal separations |
| BN-PAGE | Polyacrylamide gel | Electrophoretic mobility | Coomassie blue G, salt buffers | Not limited to soluble complexes/Less reproducible | Membrane protein complexes |
aSalts commonly used for buffers: NaCl, HEPES, KCl, MgCl2, Tris, PBS, NaCH3COO, NaN3, (NH4)2SO4. May contain additives like proteases inhibitors, dithiothreitol or glycerol.
An early proof-of-principle study (32) demonstrated how E. coli polypeptides from protein complexes had the same elution profile through multiple orthogonal chromatographic steps (including IEX, HIC and SEC) performed successively. A simpler approach using only SEC (33) provided a biologically relevant map of soluble chloroplast-localized complexes of Arabidopsis thaliana, showing the potential of the approach for interactome study. The use of SEC in global monitoring of protein complexes was limited until the introduction of the first co-elution study using SILAC and SEC (20). The same year, Havugimana et al. (21) used multiple orthogonal separations including weak-anion exchange and mixed-bed ion exchange, sucrose gradient centrifugation and IEF. This strategy was later used to examine complexes among diverse metazoan models, studying eight different organisms in total (34).
Current co-elution studies are mostly based on the two previously mentioned approaches using SILAC-SEC (20) or label-free-IEX (21), with some variations but keeping the basis of co-elution (35–38). E.g., some SEC studies have used label-free quantification approaches instead of SILAC (38), including SWATH-MS (37). Recently, both SEC and IEX were used in parallel to separate the same samples and obtain an overlapping data set to hopefully reduce the confounding effect of chance co-elution (39). A recent study was based on the IEX approach but using SILAC instead of label-free quantification to monitor interactome changes following perturbation assays (40) using a single mixed-bed exchange column rather than two columns in series as originally.
One downside of previous co-elution methods is that they only target soluble complexes and do not focus on membrane complexes (29, 41, 42), as lysis is done under mild, complex-preserving conditions. To allow the study of soluble and membrane-bound complexes of entire mitochondria, Heide et al. (22) used BN-PAGE and large-pore BN-PAGE after digitonin solubilization. With this approach, they also resolved large complexes (up to a molecular mass of 30 MDa) that cannot be resolved by SEC. More recently, Scott et al. (29) also used the BN-PAGE approach for fractionation instead of SEC, as an adaptation of the SILAC-SEC method (20). Other methods have used fractionation after detergent solubilization, including SEC or IEX (43–45). Detergent-free solubilization strategies have been recently introduced to improve the study of membrane proteins, where amphipathic scaffold proteins (46) or bi-helical peptides (41) wrap around the hydrophobic parts of the target membrane protein and shield them from the aqueous solution.
Design Considerations
Choice of Separation Method
Perhaps the first question to ask when designing a co-elution protocol is which type of proteins are the focus of study: soluble or membrane proteins. The mild, detergent-free lysis conditions at neutral pH and physiological salt concentration used to preserve protein complexes are not suitable to solubilize membrane complexes because of their hydrophobicity (47). Beside the soluble cytosolic protein complexes, these conditions extract soluble intra-organellar protein complexes such as nuclear, mitochondrial and lysosomal ones. Thus, for initial and exploratory investigations, the soluble interactome provides a large map of the biological processes of an organism (20, 21). However, membrane proteins are involved in important cell processes and they can be the focus of study. Some studies have used mild and non-denaturing detergents to solubilize membrane complexes, which are then separated by SEC or BN-PAGE (22, 45). However, the use of detergent in SEC or IEX deteriorates separation because detergent micelles can bound proteins (29, 48) or interfere with solvent access to charged proteins. Instead, BN-PAGE has the advantage of being an established method for membrane protein separations and has proved to be well suited for co-elution interactome studies (29). A recent co-elution method used in vivo formaldehyde protein crosslinking with denaturing SEC separation which identified membrane and membrane-associated protein complexes compared with the only-soluble-complexes approach (49). No current method allows the simultaneous study of native soluble and membrane proteins as mild detergents can disrupt soluble PPIs. However, new detergent-free technologies to solubilize membrane proteins might lead to a global method (41). Potentially, the use of crosslinking could also help overcome the limitation of co-elution (shared with other lysis-based methods) of possibly missing important transient and weak interactions. However, this adds a layer of complexity to the bioinformatics analysis involving the identification of crosslinked peptides.
In theory, soluble proteins can be effectively separated in any type of chromatography that allows separations in aqueous conditions with proper column dimensions to accommodate protein complexes. Traditional reversed-phase or hydrophilic interaction LC require the use of organic solvents that denature proteins and disrupt PPIs. The biggest advantage of SEC is precisely that separations can be performed under aqueous and isocratic conditions, as separation only depends on the hydrodynamic volume of the complexes (SEC columns have pores of different sizes where small hydrodynamic volumes equilibrate more often than large ones and therefore smaller complexes elute later (48)). The mobile phase can be the same buffer used for lysis at neutral pH and physiological salt concentration. One downside of SEC is that it has modest resolution and is thus prone to co-elution by chance. One way to increase resolution in SEC separations (applicable to any LC) is to use two long columns (300 mm) in series.
IEX separation is based on the charge attraction between column and protein, which carries surface charges depending on their isoelectric point and buffer pH. Salt concentration is controlled to drive the actual separation by ion displacement of immobilized proteins by mobile phase ions. Compared with SEC, IEX might show enhanced retention and therefore more characteristic profiles. There are also more columns available with different chemistries. However, the increased salt concentration required for separation might disrupt some PPIs. To minimize this, shallow salt gradients are used to not perturb nonionic protein associations and maintain non-denaturing conditions. In HIC, separation is also driven by salt concentration, where high concentrations reduce solvation of proteins, promoting interaction of the protein's hydrophobic parts with the hydrophobic stationary phase. HIC requires higher salt concentrations to promote retention, which is why HIC is less used than other chromatographies (50, 51).
As mentioned before, several studies have combined several of the above techniques in sequence or in parallel to obtain multiple orthogonal fractionation (21, 32, 34, 39). The main advantage of these approaches is that complexes that might be lost by one strategy can be rescued by another one (e.g. salt in IEX may disrupt some complexes that can be rescued by SEC). Multiple separations also further separate protein complexes that might be poorly resolved by a single separation. These methods are however time-consuming, and they still require validation experiments by complementary approaches.
LC stationary phases require a suitable column (typically high resolution, analytical-grade), particle and pore dimensions to separate protein complexes with high efficiency. Large biomolecules require large pore sizes to allow unrestricted diffusion inside the pores and larger columns with smaller particles (e.g. 500Å, 300 mm, ≤5 μm) give narrower peaks, with limits imposed by separation time, column backpressure and material synthesis (52, 53). Material technology for chromatography is constantly introducing advances, which are applied to biomolecules, such as mixed-mode materials or superficially porous particles, and co-elution methods could benefit from them to achieve faster and more efficient separations (54, 55). To achieve faster separations, temperature is also controlled, often set at room or higher temperatures. However, for protein complexes keeping the temperature during separation (and sample handling) lower (e.g. work on ice, LC separations <10 °C) is critical for complex stability. The use of low temperatures also prevents protein aggregation when the sample is concentrated to a suitable volume for LC injection. The absence of large macromolecules eluting at void-volume in SEC are evidence of absence of protein aggregation. Column dimensions and separation conditions will determine overall separation resolution and, in turn, this determines the optimum number of fractions that should be collected to obtain adequate co-elution data. Narrow peaks are desired because it gives characteristic elution profiles that can be more effectively compared for co-migration data. However, narrow peaks can also go undetected if they are only spread across one or two fractions. The solution to this could be to collect a larger number of fractions, but this comes at the cost of more sample preparation and increased MS analysis time.
Once protein complexes are fractionated their stability as complexes is not important and the goal is to digest the proteins adequately for peptide LC-MS/MS analysis. The sample handling considerations for this step are the same as for any MS-based proteomics procedure. Nevertheless, it is important to mention here that digestion procedures free of detergents, salts and contaminants produce clean samples that are key to maximize protein identification.
Quantitative Approaches
Some form of quantitation is required to generate chromatograms or electropherograms from co-elution data and, thus, the choice of the quantitation method is important. The main approaches used to quantify co-elution data are SILAC and label-free methods, both frequently used in normal MS-based proteomic workflows. Much has been written about the comparative advantages of both quantification strategies (56–58) and those apply to co-elution workflows. SILAC provides accuracy and consistency across different samples as the metabolic labels are introduced during cell culture, allowing normalization to be done at an early stage in the sample handling. SILAC also saves a significant amount of sample preparation time as different conditions can be pooled into one sample for simultaneous analysis (58). For co-elution, these benefits are key, as high accuracy across fractions is achieved and the introduction of a third channel allows the study of interactome rearrangements on perturbation.
A common misconception about SILAC is that it is expensive. Although it is true that SILAC reagents add cost to an experiment, the increased accuracy in quantitation means that fewer fractions or samples are required to get equivalent data, and thus much less instrument time (which also has a cost) is needed. One caveat to using SILAC is that there are certain biological systems that cannot be easily labeled metabolically, such as primary cells, clinical samples or most whole organisms. The applications of SILAC are still vast, being compatible with numerous cell lines and, though costly, whole organisms (stable isotope labeling of mammals, SILAM (59)) so long as they are not large or unrealistic.
Global interactome studies have been conducted involving heterologous expression of genetically manipulated cell lines which raises the question of how physiologically relevant the results obtained are. Skinnider et al. recently produced a SILAM mouse for tissue interactome study (60) of seven mouse tissues to map tissue-specific mammalian interactomes. Despite being experimentally challenging, these types of studies yield interactome maps that are more relevant.
SILAC limitations have also been addressed by producing a SILAC-labeled spike-in standard (61), where a SILAC sample is prepared separately in a compatible material and is added as a reference to each of the experimental samples. This method allows a SILAC-like quantification for SILAC incompatible samples and is an alternative to whole-organism labeling. Spike-in SILAC could be applied to co-elution, but the same as with label-free approaches, different physiological conditions cannot be pooled for simultaneous MS analysis.
In theory, other labeling approaches like isobaric labeling (i.e. iTRAQ or tandem mass tag, TMT) could be used in co-elution approaches to minimize MS analysis time. This could be particularly useful for multiple separation approaches (32). However, pooling samples is the only advantage facing several disadvantages, including that normalization is done at a late stage (after protein digestion), sample handling is increased and data analysis becomes more challenging.
As previously mentioned, label-free approaches have also been successfully used to quantify co-elution data sets (21, 34, 35, 37, 38, 62, 63). Both available label-free methods, spectral counting from MS/MS scans or MS1 precursor ion intensities, have been used for this purpose, employing appropriate software (e.g. PepQuant (62) and MaxQuant (57)). Label-free methods are arguably simpler and have no sample limitations. While SILAC can compare up to two conditions in perturbation studies, label-free has virtually no limits. This strategy is therefore quite useful for quantification across larger comparison sets (>2 and up to 10s of biological conditions). In these cases, data-independent acquisition (DIA or SWATH-MS) is another alternative that has already been applied to co-elution studies (37). However, this comes with a significant increase in sample preparation and MS analysis time and, in the case of SWATH, additional computational challenges.
Data Analysis for Co-elution Profiling Studies
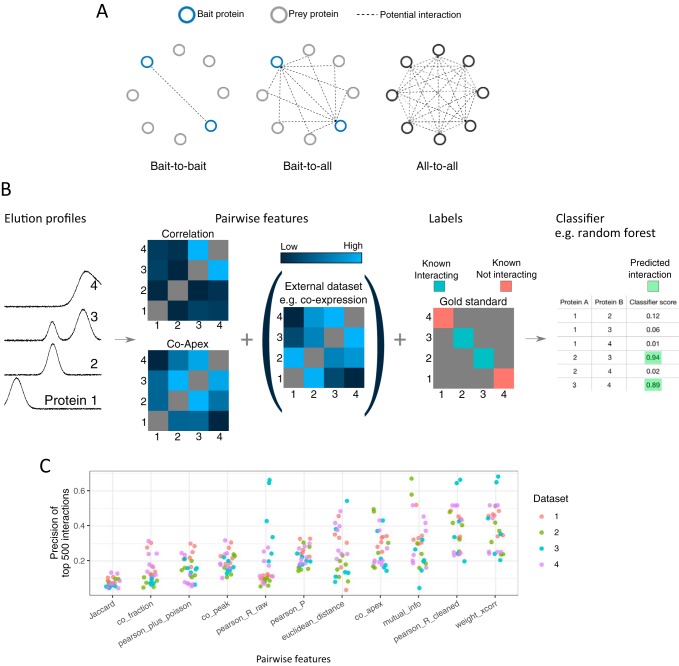
A distinct advantage of co-elution studies over other high-throughput methods is that they can detect PPIs between all proteins identified in a sample (“all-to-all”, also known as the matrix model (64)). Other high-throughput methods are limited to detecting interactions between two tagged or labeled proteins (“bait-to-bait”) such as Y2H, or between a tagged protein and any other (“bait-to-all”, also known as the spoke model) such as BioID (Fig. 2A). This increased number of potential interactions can result in a combinatorial explosion, however. For example, a co-elution data set can contain millions of potential interactions, only thousands of which are likely to be real. Analyzing co-elution data sets, therefore, often involves separating true interactors from a background of spurious false positives through bioinformatic analysis.
Fig. 2.
Bioinformatic analysis of co-elution data. A, Bioinformatic analysis of co-elution data is complicated by the number of potential interactions. In contrast to techniques such as Y2H that find interactions between tagged proteins (“Bait-to-bait”) or BioID (and sometimes AP-MS) that find interactions involving at least one bait protein (“Bait-to-all”), co-elution experiments have the potential to find interactions between all identified proteins in a sample (“All-to-all”). B, Schematic of classifier-based analysis of co-elution data. The strength of co-elution is quantified for every pair of proteins using multiple metrics (“features”). Features derived from external data can be included, such as co-citation or co-expression. Using a gold standard set of known complexes, a subset of the protein pairs are labeled as interacting or not-interacting. Finally, a classifier uses to the features and labels to assign every pair of profiles a classifier score, to which a threshold is applied. C, Performance of single co-elution features. Interactomes were predicted from four data sets using a single co-elution metric. Each dot represents an interactome from one replicate, and the y axis gives the precision of the 500 best-scoring interactions. Interactomes were predicted using PrInCE with default parameters (CORUM gold standard). weighted_xcorr: Weighted cross-correlation, measured with R function wccsom. pearson_R_cleaned: Pearson correlation (cleaned profiles). mutual_info: Mutual information. co_apex: Mininum number of fractions between fitted Gaussian centers. pearson_P: Pearson correlation (raw profiles) p value. pearson_R_raw: Pearson correlation (raw profiles). euclidean_distance: Euclidean distance (cleaned profiles). co_peak: Number of fractions between maximum value. pearson_plus_poisson: Pearson R (raw profiles) plus Poisson noise. co_fraction: 1 if maximum values are in the same fraction, 0 otherwise. Jaccard: Overlapping fractions in which both proteins are quantified, measured with Jaccard. Data sets: 1 Kristensen et al. 2012 (20), 2 Scott et al. 2017 (29), Carlson et al. 2019 (42), Scott et al. 2015 (78).
Although there are many workflows for analyzing co-elution data, it is common to use co-elution data to generate a list of pairwise PPIs (i.e. an interactome), typically done via a machine learning classifier (21, 23, 26, 34, 42, 65, 66). In this analysis, the strength of co-elution is measured for every pair of proteins using a variety of metrics (Fig. 2B). Across published studies, we count eleven metrics used to evaluate the co-elution strength of pairs of proteins (Fig. 2C). These fall into five general categories: correlational metrics, such as weighted cross-correlation and Pearson correlation strength between raw and cleaned elution profiles (23, 34), sometimes with the addition of Poisson noise (21, 23, 34); co-apex measures, that attempt to quantify whether two proteins share an elution peak (21, 23, 34, 67); mutual information (67); the degree to which proteins are quantified in the same fractions, measured with the Jaccard index (67); and Euclidean distance (23, 34). Fig. 2C shows how these metrics perform when predicting interactomes using a single metric (PrInCE, default parameters). In general, we find that correlational metrics such as Pearson R and weighted cross-correlation that use quantified protein amounts are more informative than measures that just detect if proteins are quantified in the same fractions (Jaccard and co_fraction), although each metric differs between data sets. In practice, multiple metrics are used to better differentiate between true interactors and spurious pairs, because truly interacting protein pairs should score highly in most measures.
Using a gold standard reference of known protein complexes (e.g. CORUM (68)) to label a subset of pairs in a data set as known PPIs or known non-interactors, it is possible to estimate the probability that any given protein pair is interacting. That is, combined with a gold standard reference, classifiers assign an interaction score to all protein pairs, with high-scoring pairs more closely resembling known PPIs. Finally, to arrive at an interactome, it is typical to take all protein pairs whose score is greater than a threshold as predicted PPIs. This threshold is typically chosen such that the ratio of true positives to false positives in the interactome, which are derived from the gold standard, satisfies a given FDR. Therefore, the task of finding pairwise PPIs in a co-elution data set can be framed as separating truly interacting protein pairs from a large background of non-interacting pairs. As an optional step, the resulting interactome can be clustered into protein complexes using a network-based clustering algorithm (34, 66), such as ClusterONE (69). Although it can be difficult to assess the quality of clusters, at least in part because metrics for measuring the similarity between clusters have biases and display non-intuitive behavior (70), a number of studies find differences in robustness between algorithms (71, 72), with MCL performing relatively well (73). Additionally, Nepusz et al. (69) show that clustering weighted networks can be more robust than unweighted.
Free classifier-based bioinformatic tools exist for co-elution data (23, 24). These tools can be used as both standalone executable programs, where data is loaded and output files and figures are generated, and as R packages. Parameters to take note of when using these tools are the number of quantified proteins in a data set (ideally greater than 500), the number of missing values in the data set, and, primarily, the width of elution peaks, because elution profiles with poor resolution (“wide” peaks) will be poorly distinguishable and yield more spuriously correlated pairs. For example, we find that in data sets with 50 fractions, elution peaks should have a full width at half maximum of no more than 10 fractions.
Although classifier-based data analysis is common, there are many ways to treat co-elution data. For example, it is also common to cluster co-elution data into groups of similar profiles, as these groups can represent protein complexes (22, 47). Clustering like this does not require a gold standard, although reference complexes can be used to select an optimal number of clusters (47) and to validate the plausibility of the clustered proteins (74). Data analysis methods discussed so far identify novel and known interactions, often focusing on PPIs with complex prediction as a downstream analysis. In contrast, “complex-centric” approaches (37) start with known protein complexes (e.g. CORUM) and assess whether members of a known complex are co-eluting. Although this approach does not detect novel PPIs, it does detect novel subunits of complexes and assembly intermediates. CCProfiler is a free software for complex-centric data analysis (37).
An important consideration for both classifier-based and complex-centric methods is the choice of reference complexes (“gold standards”). Gold standards do not exist for all organisms, and although proteins from non-model organisms can be mapped to model organism proteins, this can introduce errors because orthologs between organisms do not necessarily interact with the same partners. Therefore, co-elution analysis often works best on human data sets, or data sets from other well-studied organisms. A further issue regarding gold standards is that many protein interactions only occur under certain conditions (75). Therefore, it can be beneficial to tweak gold standards so that they more accurately reflect individual experiments (76). Another caveat pertains to including external data as evidence of interaction, such as including a protein pairs' tendency to co-express (21, 34, 35, 49). Although this can help filter out spuriously co-elution proteins, it can also bias results toward highly-studied proteins and away from less-well-studied and/or harder to identify proteins (77).
CONCLUSIONS
Co-elution can investigate all the existing interactions between all the proteins quantified in a given sample whereas other methods focus on a protein's interactions at a time. In addition, it does not use protein tagging, gives quantitative information (including relative amounts of different complexes with a common protein), and, when combined with SILAC, provides interactome rearrangement information on perturbation in record time. Depending on whether soluble or membrane complexes are the focus of study, the separation strategy changes from SEC or IEX to BN-PAGE or mild detergent-based separations, but the introduction of recent membrane protein solubilization strategies might produce global approaches. To a large extent, the system under study defines the quantification strategy to use. SILAC, label-free and other methods are available depending on the cell line or tissue and whether the goal is to find new interactions or study the interactome under different physiological conditions. One important consideration of co-elution experiments is that they typically require sophisticated bioinformatic analyses, because co-elution analyses often compare all pairs of proteins quantified in a sample, and this number is large (millions) for modern data sets. Further, classifier-based analyses of co-elution data require gold standard databases of known protein complexes, a requirement which is not met for all organisms. Co-elution is a powerful tool for uncovering interactomes, and it provides many advantages over existing high-throughput interactome mapping technologies. In the future, we believe co-elution studies should move toward maximizing quantitation accuracy, lowering quantification limits and increasing separation resolution. This would allow the study of the interactome beyond the protein level (e.g. post-translational modifications) and the use of less sample amount, translating in lower costs and sustainable methods. Automatization of sample digestion would also improve the technique greatly, to alleviate the time-consuming analysis of multiple (>2) conditions.
Footnotes
* This work is supported by funding to L.J.F. from NSERC, Genome Canada and Genome British Columbia (project 214PRO). The authors declare that they have no conflicts of interest with the contents of this article.
1 The abbreviations used are:
- PPIs
- protein–protein interactions
- Y2H
- yeast two-hybrid
- MS
- mass spectrometry
- AP
- affinity purification
- BioID
- proximity-dependent biotin identification
- BN-PAGE
- blue-native polyacrylamide gel electrophoresis
- PrInCE
- prediction of interactomes from co-elution
- EPIC
- elution profile-based inference of complexes
- SEC
- size-exclusion chromatography
- IEX
- ion-exchange chromatography
- HIC
- hydrophobic interaction chromatography
- SAX
- strong anion exchange
- WAX
- weak anion exchange
- WCX
- weak cation exchange
- iTRAQ
- isobaric tagging for relative and absolute quantitation
- TOF
- time of flight
- MS/MS
- tandem mass spectrometry
- SILAC
- stable isotope labeling by/with amino acids in cell culture
- SWATH-MS
- sequential windowed acquisition of all theoretical fragment ion mass spectra
- SILAM
- stable isotope labeling of mammals
- CORUM
- comprehensive resource of mammalian protein complexes
- TPP
- thermal proteome profiling
- TMT
- tandem mass tag.
REFERENCES
- 1. Huttlin E. L., Bruckner R. J., Paulo J. A., Cannon J. R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M. P., Parzen H., Szpyt J., Tam S., Zarraga G., Pontano-Vaites L., Swarup S., White A. E., Schweppe D. K., Rad R., Erickson B. K., Obar R. A., Guruharsha K. G., Li K., Artavanis-Tsakonas S., Gygi S. P., and Harper J. W. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hein M. Y., Hubner N. C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I. A., Weisswange I., Mansfeld J., Buchholz F., Hyman A. A., and Mann M. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 [DOI] [PubMed] [Google Scholar]
- 3. Aebersold R., and Mann M. (2016) Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 [DOI] [PubMed] [Google Scholar]
- 4. Altelaar A. F. M., Munoz J., and Heck A. J. R. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 5. Titeca K., Lemmens I., Tavernier J., and Eyckerman S. (2019) Discovering cellular protein-protein interactions: Technological strategies and opportunities. Mass Spectrom. Rev. 38, 79–111 [DOI] [PubMed] [Google Scholar]
- 6. Rattray D. G., and Foster L. J. (2019) Dynamics of protein complex components. Curr. Opinion Chem. Biol. 48, 81–85 [DOI] [PubMed] [Google Scholar]
- 7. Minic Z., Dahms T. E. S., and Babu M. (2018) Chromatographic separation strategies for precision mass spectrometry to study protein-protein interactions and protein phosphorylation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1102–1103, 96–108 [DOI] [PubMed] [Google Scholar]
- 8. Snider J., Kotlyar M., Saraon P., Yao Z., Jurisica I., and Stagljar I. (2015) Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 11, 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yugandhar K., Gupta S., and Yu H. (2019) Inferring protein-protein interaction networks from mass spectrometry-based proteomic approaches: a mini-review. Comput. Struct. Biotechnol. J. 17, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Claire M., Delahunty J. R. Y. I. (2019) in Proteomics for Biological Discovery, Second Edition, ed Timothy D. Veenstra JRYI (Wiley), pp 125–144 [Google Scholar]
- 11. Woodsmith J., and Stelzl U. (2014) Studying post-translational modifications with protein interaction networks. Curr. Opin. Struct. Biol. 24, 34–44 [DOI] [PubMed] [Google Scholar]
- 12. Kristensen A. R., and Foster L. J. (2013) High throughput strategies for probing the different organizational levels of protein interaction networks. Mol. Biosyst. 9, 2201–2212 [DOI] [PubMed] [Google Scholar]
- 13. Meyer K., and Selbach M. (2015) Quantitative affinity purification mass spectrometry: a versatile technology to study protein–protein interactions. Front. Genet. 6, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunham W. H., Mullin M., and Gingras A.-C. (2012) Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics 12, 1576–1590 [DOI] [PubMed] [Google Scholar]
- 15. Varnaitė R., and MacNeill S. A. (2016) Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 16, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim D. I., Jensen S. C., Noble K. A., Kc B., Roux K. H., Motamedchaboki K., and Roux K. J. (2016) An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., and Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 18. Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., and Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 19. Dunkley T. P. J., Watson R., Griffin J. L., Dupree P., and Lilley K. S. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteomics 3, 1128–1134 [DOI] [PubMed] [Google Scholar]
- 20. Kristensen A. R., Gsponer J., and Foster L. J. (2012) A high-throughput approach for measuring temporal changes in the interactome. Nat. Methods 9, 907–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Havugimana P. C., Hart G. T., Nepusz T., Yang H., Turinsky A. L., Li Z., Wang P. I., Boutz D. R., Fong V., Phanse S., Babu M., Craig S. A., Hu P., Wan C., Vlasblom J., Dar V.-U. -N., Bezginov A., Clark G. W., Wu G. C., Wodak S. J., Tillier E. R. M., Paccanaro A., Marcotte E. M., and Emili A. (2012) A census of human soluble protein complexes. Cell 150, 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heide H., Bleier L., Steger M., Ackermann J., Dröse S., Schwamb B., Zörnig M., Reichert A. S., Koch I., Wittig I., and Brandt U. (2012) Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 16, 538–549 [DOI] [PubMed] [Google Scholar]
- 23. Stacey R. G., Skinnider M. A., Scott N. E., and Foster L. J. (2017) A rapid and accurate approach for prediction of interactomes from co-elution data (PrInCE). BMC Bioinformatics 18, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu L. Z., Goebels F., Tan J. H., Wolf E., Kuzmanov U., Wan C., Phanse S., Xu C., Schertzberg M., Fraser A. G., Bader G. D., and Emili A. (2019) EPIC: software toolkit for elution profile-based inference of protein complexes. Nat. Methods 16, 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larance M., and Lamond A. I. (2015) Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 16, 269–280 [DOI] [PubMed] [Google Scholar]
- 26. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., Erickson B. K., Wühr M., Chick J., Zhai B., Kolippakkam D., Mintseris J., Obar R. A., Harris T., Artavanis-Tsakonas S., Sowa M. E., De Camilli P., Paulo J. A., Harper J. W., and Gygi S. P. (2015) The BioPlex Network: A systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai L., Zhao T., Bisteau X., Sun W., Prabhu N., Lim Y. T., Sobota R. M., Kaldis P., and Nordlund P. (2018) Modulation of protein-interaction states through the cell Cycle. Cell 173, 1481–1494.e13 [DOI] [PubMed] [Google Scholar]
- 28. Becher I., Andrés-Pons A., Romanov N., Stein F., Schramm M., Baudin F., Helm D., Kurzawa N., Mateus A., Mackmull M.-T., Typas A., Müller C. W., Bork P., Beck M., and Savitski M. M. (2018) Pervasive protein thermal stability variation during the cell cycle. Cell 173, 1495–1507.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scott N. E., Rogers L. D., Prudova A., Brown N. F., Fortelny N., Overall C. M., and Foster L. J. (2017) Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol. Syst. Biol. 13, 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smits A. H., and Vermeulen M. (2016) Characterizing protein–protein interactions using mass spectrometry: challenges and opportunities. Trends Biotechnol. 34, 825–834 [DOI] [PubMed] [Google Scholar]
- 31. Ramani A. K., Li Z., Hart G. T., Carlson M. W., Boutz D. R., and Marcotte E. M. (2008) A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol. Syst. Biol. 4, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong M., Yang L. L., Williams K., Fisher S. J., Hall S. C., Biggin M. D., Jin J., and Witkowska H. E. (2008) A “tagless” strategy for identification of stable protein complexes genome-wide by multidimensional orthogonal chromatographic separation and iTRAQ reagent tracking. J. Proteome Res. 7, 1836–1849 [DOI] [PubMed] [Google Scholar]
- 33. Olinares P. D. B., Ponnala L., and van Wijk K. J. (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9, 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wan C., Borgeson B., Phanse S., Tu F., Drew K., Clark G., Xiong X., Kagan O., Kwan J., Bezginov A., Chessman K., Pal S., Cromar G., Papoulas O., Ni Z., Boutz D. R., Stoilova S., Havugimana P. C., Guo X., Malty R. H., Sarov M., Greenblatt J., Babu M., Derry W. B., Tillier E. R., Wallingford J. B., Parkinson J., Marcotte E. M., and Emili A. (2015) Panorama of ancient metazoan macromolecular complexes. Nature 525, 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kastritis P. L., O'Reilly F. J., Bock T., Li Y., Rogon M. Z., Buczak K., Romanov N., Betts M. J., Bui K. H., Hagen W. J., Hennrich M. L., Mackmull M.-T., Rappsilber J., Russell R. B., Bork P., Beck M., and Gavin A.-C. (2017) Capturing protein communities by structural proteomics in a thermophilic eukaryote. Mol. Syst. Biol. 13, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gilbert M., and Schulze W. X. (2019) Global identification of protein complexes within the membrane proteome of Arabidopsis roots using a SEC-MS approach. J. Proteome Res. 18, 107–119 [DOI] [PubMed] [Google Scholar]
- 37. Heusel M., Bludau I., Rosenberger G., Hafen R., Frank M., Banaei-Esfahani A., Drogen A., Collins B. C., Gstaiger M., and Aebersold R. (2019) Complex-centric proteome profiling by SEC - SWATH - MS. Molecular Systems Biology 15, e8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aryal U. K., McBride Z., Chen D., Xie J., and Szymanski D. B. (2017) Analysis of protein complexes in Arabidopsis leaves using size exclusion chromatography and label-free protein correlation profiling. J. Proteomics 166, 8–18 [DOI] [PubMed] [Google Scholar]
- 39. McBride Z., Chen D., Lee Y., Aryal U. K., Xie J., and Szymanski D. B. (2019) A label-free mass spectrometry method to predict endogenous protein complex composition. Mol. Cell. Proteomics 18, 1588–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Meara T. R., O'Meara M. J., Polvi E. J., Pourhaghighi M. R., Liston S. D., Lin Z.-Y., Veri A. O., Emili A., Gingras A.-C., and Cowen L. E. (2019) Global proteomic analyses define an environmentally contingent Hsp90 interactome and reveal chaperone-dependent regulation of stress granule proteins and the R2TP complex in a fungal pathogen. PLos Biol. 17, e3000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carlson M. L., Young J. W., Zhao Z., Fabre L., Jun D., Li J., Li J., Dhupar H. S., Wason I., Mills A. T., Beatty J. T., Klassen J. S., Rouiller I., and Duong F. (2018) The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. Elife 7, e34085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carlson M. L., Stacey R. G., Young J. W., Wason I. S., Zhao Z., Rattray D. G., Scott N., Kerr C. H., Babu M., Foster L. J., and Duong F. V. H. (2019) Profiling the E. coli membrane interactome captured in peptidisc libraries. Elife 8, e46615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maddalo G., Stenberg-Bruzell F., Götzke H., Toddo S., Björkholm P., Eriksson H., Chovanec P., Genevaux P., Lehtiö J., Ilag L. L., and Daley D. O. (2011) Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 10, 1848–1859 [DOI] [PubMed] [Google Scholar]
- 44. Walian P. J., Allen S., Shatsky M., Zeng L., Szakal E. D., Liu H., Hall S. C., Fisher S. J., Lam B. R., Singer M. E., Geller J. T., Brenner S. E., Chandonia J.-M., Hazen T. C., Witkowska H. E., Biggin M. D., and Jap B. K. (2012) High-throughput isolation and characterization of untagged membrane protein complexes: outer membrane complexes of Desulfovibrio vulgaris. J. Proteome Res. 11, 5720–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Babu M., Bundalovic-Torma C., Calmettes C., Phanse S., Zhang Q., Jiang Y., Minic Z., Kim S., Mehla J., Gagarinova A., Rodionova I., Kumar A., Guo H., Kagan O., Pogoutse O., Aoki H., Deineko V., Caufield J. H., Holtzapple E., Zhang Z., Vastermark A., Pandya Y., Lai C. C.-L., El Bakkouri M., Hooda Y., Shah M., Burnside D., Hooshyar M., Vlasblom J., Rajagopala S. V., Golshani A., Wuchty S. F., Greenblatt J, Saier M., Uetz P. F, Moraes T, Parkinson J., and Emili A. (2018) Global landscape of cell envelope protein complexes in Escherichia coli. Nat. Biotechnol. 36, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Denisov I. G., and Sligar S. G. (2016) Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 23, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kirkwood K. J., Ahmad Y., Larance M., and Lamond A. I. (2013) Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics. Mol. Cell. Proteomics 12, 3851–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kunji E. R. S., Harding M., Butler P. J. G., and Akamine P. (2008) Determination of the molecular mass and dimensions of membrane proteins by size exclusion chromatography. Methods 46, 62–72 [DOI] [PubMed] [Google Scholar]
- 49. Larance M., Kirkwood K. J., Tinti M., Brenes Murillo A., Ferguson M. A. J., and Lamond A. I. (2016) Global membrane protein interactome analysis using in vivo crosslinking and mass spectrometry-based protein correlation profiling. Mol. Cell. Proteomics 15, 2476–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jungbauer A., and Hahn R. (2009) Ion-exchange chromatography. Methods Enzymol. 463, 349–371 [DOI] [PubMed] [Google Scholar]
- 51. Havugimana P. C., Wong P., and Emili A. (2007) Improved proteomic discovery by sample pre-fractionation using dual-column ion-exchange high performance liquid chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 54–61 [DOI] [PubMed] [Google Scholar]
- 52. Wagner B. M., Schuster S. A., Boyes B. E., and Shields T. J. (2017) Superficially porous particles with 1000 Å pores for large biomolecule high performance liquid chromatography and polymer size exclusion chromatography. J. Chromatogr. A 1489, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goyon A., Beck A., Colas O., Sandra K., Guillarme D., and Fekete S. (2017) Evaluation of size exclusion chromatography columns packed with sub-3μm particles for the analysis of biopharmaceutical proteins. J. Chromatogr. A 1498, 80–89 [DOI] [PubMed] [Google Scholar]
- 54. Chen W., Jiang K., Mack A., Sachok B., Zhu X., Barber W. E., and Wang X. (2015) Synthesis and optimization of wide pore superficially porous particles by a one-step coating process for separation of proteins and monoclonal antibodies. J. Chromatogr. A 1414, 147–157 [DOI] [PubMed] [Google Scholar]
- 55. Zhang K., and Liu X. (2016) Mixed-mode chromatography in pharmaceutical and biopharmaceutical applications. J. Pharm. Biomed. Anal. 128, 73–88 [DOI] [PubMed] [Google Scholar]
- 56. Merl J., Ueffing M., Hauck S. M., and von Toerne C. (2012) Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Müller cells. Proteomics 12, 1902–1911 [DOI] [PubMed] [Google Scholar]
- 57. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 59. Rauniyar N., McClatchy D. B., and Yates J. R. 3rd (2013) Stable isotope labeling of mammals (SILAM) for in vivo quantitative proteomic analysis. Methods 61, 260–268 [DOI] [PubMed] [Google Scholar]
- 60. Skinnider M. A., Scott N. E., Prudova A., Stoynov N., Stacey R. G., Gsponer J., Foster L. J. (2018) An atlas of protein-protein interactions across mammalian tissues. bioRxiv 10.1101/351247 [DOI] [PubMed] [Google Scholar]
- 61. Geiger T., Wisniewski J. R., Cox J., Zanivan S., Kruger M., Ishihama Y., and Mann M. (2011) Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 62. Wan C., Liu J., Fong V., Lugowski A., Stoilova S., Bethune-Waddell D., Borgeson B., Havugimana P. C., Marcotte E. M., and Emili A. (2013) ComplexQuant: high-throughput computational pipeline for the global quantitative analysis of endogenous soluble protein complexes using high resolution protein HPLC and precision label-free LC/MS/MS. J. Proteomics 81, 102–111 [DOI] [PubMed] [Google Scholar]
- 63. Connelly K. E., Hedrick V., Paschoal Sobreira T. J., Dykhuizen E. C., and Aryal U. K. (2018) Analysis of human nuclear protein complexes by quantitative mass spectrometry profiling. Proteomics 18, e1700427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hakes L., Robertson D. L., Oliver S. G., and Lovell S. C. (2007) Protein interactions from complexes: a structural perspective. Comparative Functional Genomics 2007, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shatsky M., Dong M., Liu H., Yang L. L., Choi M., Singer M. E., Geller J. T., Fisher S. J., Hall S. C., Hazen T. C., Brenner S. E., Butland G., Jin J., Witkowska H. E., Chandonia J.-M., and Biggin M. D. (2016) Quantitative tagless copurification: a method to validate and identify protein-protein interactions. Mol. Cell. Proteomics 15, 2186–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Drew K., Lee C., Huizar R. L., Tu F., Borgeson B., McWhite C. D., Ma Y., Wallingford J. B., and Marcotte E. M. (2017) Integration of over 9,000 mass spectrometry experiments builds a global map of human protein complexes. Mol. Syst. Biol. 13, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goebels F., Hu L., Bader G., and Emili A. (2018) Automated computational inference of multi-protein assemblies from biochemical co-purification data. Methods Mol. Biol. 1764, 391–399 [DOI] [PubMed] [Google Scholar]
- 68. Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., and Ruepp A. (2019) CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nepusz T., Yu H., and Paccanaro A. (2012) Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 9, 471–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gates A. J., Wood I. B., Hetrick W. P., and Ahn Y.-Y. (2019) Element-centric clustering comparison unifies overlaps and hierarchy. Sci. Rep. 9, 8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Freytag S., Tian L., Lönnstedt I., Ng M., and Bahlo M. (2018) Comparison of clustering tools in R for medium-sized 10x Genomics single-cell RNA-sequencing data. F1000Research 7, 1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Song J., and Singh M. (2009) How and when should interactome-derived clusters be used to predict functional modules and protein function? Bioinformatics 25, 3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brohée S., and van Helden J. (2006) Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics 7, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hillier C., Pardo M., Yu L., Bushell E., Sanderson T., Metcalf T., Herd C., Anar B., Rayner J. C., Billker O., and Choudhary J. S. (2019) Landscape of the Plasmodium interactome reveals both conserved and species-specific functionality. Cell Rep. 28, 1635–1647.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Celaj A., Schlecht U., Smith J. D., Xu W., Suresh S., Miranda M., Aparicio A. M., Proctor M., Davis R. W., Roth F. P., and St Onge R. P. (2017) Quantitative analysis of protein interaction network dynamics in yeast. Mol. Syst. Biol. 13, 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stacey R. G., Skinnider M. A., Chik J. H. L., and Foster L. J. (2018) Context-specific interactions in literature-curated protein interaction databases. BMC Genomics 19, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Skinnider M. A., Stacey R. G., and Foster L. J. (2018) Genomic data integration systematically biases interactome mapping. PLoS Comput. Biol. 14, e1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Scott N. E., Brown L. M., Kristensen A. R., and Foster L. J. (2015) Development of a computational framework for the analysis of protein correlation profiling and spatial proteomics experiments. J. Proteomics 118, 112–129 [DOI] [PubMed] [Google Scholar]