Immunopeptidomics is a rapidly evolving field that is catalyzed by neoantigen discovery and cancer immunotherapy. Emulating the path of other omic disciplines, Vizcaíno et al. proposes that technological advances in the mass spectrometry field will lead to large immunopeptidomic cohort studies, and ultimately, the immunopeptidome-wide association study (IWAS) paradigm, which will provide profound insights into disease susceptibility and resistance.
Keywords: Immunology, peptides, mass spectrometry, infectious disease, cancer therapeutics, HLA/MHC, immunopeptidome, IWAS
Graphical Abstract
Highlights
Immunopeptidomics bears the potential to link diseases to antigen representation.
We suggest to achieve this by analyzing the immunopeptidomes of cohorts of patients.
Current mass spectrometry-based techniques to analyze immunopeptidomes are described.
We term the proposed approach “Immunopeptidome-wide association studies” (IWAS).
Abstract
The science that investigates the ensembles of all peptides associated to human leukocyte antigen (HLA) molecules is termed “immunopeptidomics” and is typically driven by mass spectrometry (MS) technologies. Recent advances in MS technologies, neoantigen discovery and cancer immunotherapy have catalyzed the launch of the Human Immunopeptidome Project (HIPP) with the goal of providing a complete map of the human immunopeptidome and making the technology so robust that it will be available in every clinic. Here, we provide a long-term perspective of the field and we use this framework to explore how we think the completion of the HIPP will truly impact the society in the future. In this context, we introduce the concept of immunopeptidome-wide association studies (IWAS). We highlight the importance of large cohort studies for the future and how applying quantitative immunopeptidomics at population scale may provide a new look at individual predisposition to common immune diseases as well as responsiveness to vaccines and immunotherapies. Through this vision, we aim to provide a fresh view of the field to stimulate new discussions within the community, and present what we see as the key challenges for the future for unlocking the full potential of immunopeptidomics in this era of precision medicine.
“Treatment without prevention is simply unsustainable.”
Bill Gates
A RATIONALE FOR THE IWAS PARADIGM
The Human Genome Project was a major milestone in the life sciences (1, 2). First-completed twenty years ago, this mind-shifting project has changed and will continue to change the way we practice medicine. Historically, physicians have largely focused on treating disease already in progress, but modern medicine is now progressively shifting from disease treatment to disease prevention based on an individual's risk (3). The past two decades have seen an enormous success of wide-scale studies in identifying genetic variants that predict an individual's predisposition to common diseases (4). In fact, robust, rapid and inexpensive identification of functional genetic variants in individuals is now enabling predictive, preventive and personalized medicine approaches (5–7). Since the first report of single-nucleotide polymorphisms (SNPs)1 analyzed for association with myocardial infarction by genome-wide association studies (GWAS) in 2002 (8), the GWAS Catalogue resource has now grown to contain tens of thousands of SNPs associated with hundreds of common diseases (9). In this post-GWAS era, the HLA has been established as the region of the genome that is associated with the greatest number of human diseases (10). In fact, population studies of diverse ancestries have identified hundreds of susceptibility loci within the HLA region that predispose individuals to immune diseases (11–18). Most HLA disease associations that were reported over the last 50 years are related to the immune system, but the exact mechanisms driving the associations remain largely elusive.
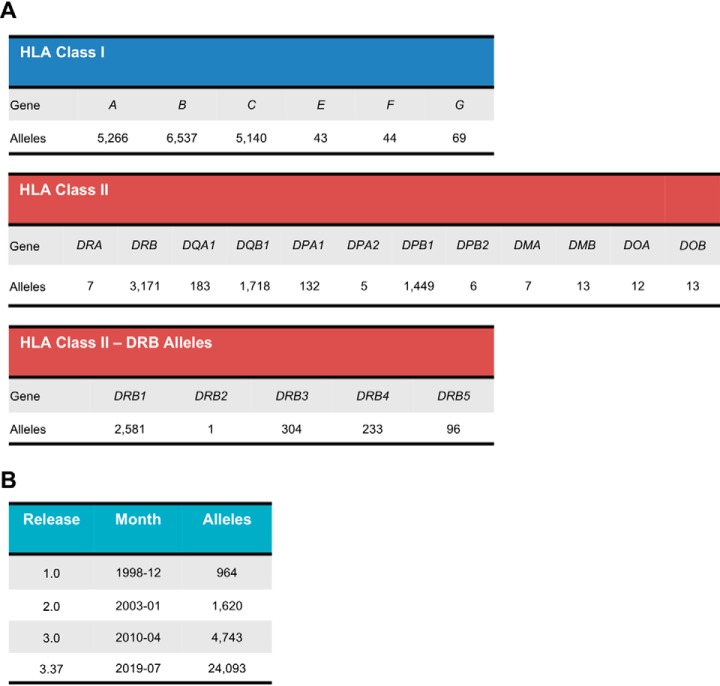
The HLA is divided into two main subclasses: class I, which include the classical HLA-A, HLA-B and HLA-C molecules, as well as the nonclassical HLA-E, HLA-F and HLA-G molecules; and class II, which includes HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DRA, HLA-DRB1, HLA-DRB2, HLA-DRB3, HLA-DRB4, and HLA-DRB5 molecules (Fig. 1A) (19). To date, more than 23,000 different classical HLA alleles have been identified (https://www.ebi.ac.uk/ipd/imgt/hla/stats.html) and this number may keep climbing up to nearly 8–9 million HLA variants (Fig. 1B) (20). The role of HLA molecules is to present a repertoire of peptides at the cell surface for T-cell recognition. The non-random amino acid composition of those peptides is restricted by length, generally between 8 and 12 (can be up to 15) amino acids for class I and between 13 and 25 amino acids for class II, and by the presence of allele-specific binding motifs (21). Therefore, the nature of those peptides and the extreme diversity of HLA alleles at the population level greatly enhances the complexity of the peptide repertoires, which are collectively termed as the human immunopeptidome (22, 23).
Fig. 1.
Number of HLA Class I and Class II alleles from the IPD-IMGT/HLA Database. The IPD-IMGT/HLA is a database for sequences of the human major histocompatibility complex (MHC) and is part of the international ImMunoGeneTics project (IMGT). A, Number of named alleles for each HLA gene. B, History of the database growth, from the first version (1.0) in 1998 to the latest version (3.37) in 2019. The number includes other non-HLA alleles and confidential alleles. Data were extracted from the database on 2019/08/20. Updated numbers can be found online at https://www.ebi.ac.uk/ipd/imgt/hla/stats.html.
T cells scan the HLA immunopeptidome and seek for “abnormal” peptides that originate from metabolic perturbations, pathogenic sources or neoplastic transformations (24–28). Those abnormal cells are then eradicated once engaged by T cells. The extreme diversity of the human immunopeptidome maximizes the probability that at least some individuals within the world population can mount a T-cell attack against an emerging infection and survive (29). This is exemplified by the HIV epidemic and evidences that HLA-B57 individuals are more likely protected against HIV (30, 31). However, such evolutionary advantage comes with the price of disease susceptibility. In fact, individuals expressing specific HLA alleles are more susceptible to suffer from specific auto-inflammatory and autoimmune diseases. For instance, birdshot chorioretinopathy was shown to be associated with HLA-A29, ankylosing spondylitis with HLA-B27, Behçet's disease with HLA-B51 and psoriasis with HLA-C06. Combinations of endoplasmic reticulum aminopeptidases (ERAP) 1 haplotypes, involved in trimming HLA-associated peptides, are also risk factors for these diseases in people that have specific HLAs (e.g. HLA-B27*05) (32–35). Moreover, an increasing number of GWAS results indicate that amino acid polymorphisms associated with immune diseases are likely affecting binding affinities of peptides within the groove of HLA proteins, and thus, affecting the repertoire of peptides presented to T cells that are capable of triggering or perpetuating human diseases (10, 12, 19, 36–45). Interestingly, those evidences obtained from GWAS, meta-analysis and HLA region fine-mapping studies point toward a fundamental role of the immunopeptidome in driving human diseases. We highlight below four cases of such studies: (1) in allergic rhinitis, the most common clinical presentation of allergy affecting 400 million people worldwide, a recent meta-analysis including 59,762 cases and 152,358 controls from European ancestry, confirmed in a replication phase of 60,720 cases and 618,527 controls, indicated that the strongest associated amino acid variants in HLA-DQB1 and HLA-B were both located in the peptide-binding groove (36); (2) in lung cancer, analysis of HLA genetic variation among 26,044 lung cancer patients and 20,836 controls revealed amino acid variants in the HLA-B*08:01 peptide-binding groove (37); (3) in alopecia areata, one of the most prevalent autoimmune diseases, a meta-analysis including 3253 cases and 7543 controls highlighted HLA-DR as the primary risk factor of the disease with amino acid variants located in the peptide-binding groove (39); and (4) in common infections, a meta-analysis including over 200,000 individuals of European ancestry suggested important roles of specific amino acid polymorphisms in the peptide-binding groove, for eight common infectious diseases, in particular: i.e. chicken pox, shingles, cold sores, tuberculosis, scarlet fever, tonsillectomy, pneumonia and plantar warts (38). GWAS studies have also shown the association of Parkinson's disease with HLA-DRB5*01 and DRB1*15:01, which are present in ∼15% of the general population (46). In this study, the authors showed that a defined set of peptides that are derived from alpha-synuclein, a protein aggregated in Parkinson's disease, act as antigenic epitopes displayed by the alleles and drive helper and cytotoxic T-cell response in patients with Parkinson's disease (47). Thus, if further tested and validated for other diseases, cohort studies may ultimately suggest a universal role of the immunopeptidome in disease susceptibility.
In addition to qualitative changes in the peptide repertoire, the level of expression and stability of HLA molecules at the cell surface have been found to be associated with diseases (11, 12, 19). For instance, a high level of expression of HLA-C increases the risk of Crohn's disease but also promotes the control of HIV infection (13, 14); instability of HLA-DQ2 and HLA-DQ8 could increase the risk of type 1 diabetes whereas stability of HLA-DQ6 may confer protection (15–17). In cancer, a recent analysis of more than 1500 melanoma patients treated by immune checkpoint inhibitors (ICIs) showed that HLA-B44 alleles were associated with an improved survival rate whereas HLA-B62 alleles were associated with a decreased effectiveness of ICIs (48, 49). These data suggest that HLA-B44 immunopeptidomes may favor presentation of a broader, and perhaps more stable repertoire of tumor-specific antigens to trigger the function of cytotoxic T cells in response to ICIs. This is consistent with the recent observation that specific HLA genotypes are associated with the appearance of specific oncogenic mutations (50, 51).
GWAS can identify genetic variants at the gene level and can be complemented with genome-wide transcript quantitative trait loci (eQTL) and genome-wide protein quantitative trait loci (pQTL) to better understand genotype-phenotype associations (52–55). However, attempts to understand HLA disease associations by only measuring expression levels of intracellular genes and proteins is unlikely to work very accurately for immune diseases in which HLA-peptide—T-cell interactions play a critical role. In contrast, measuring HLA-bound peptides is highly relevant because those peptides are directly “seen” by T cells and are involved in disease progression (56). Therefore, technologies capable of robust and comprehensive measurements of immunopeptidomes at population-scale (from tens of thousands of samples) would be extremely powerful as they would provide direct physical evidences about the identity and quantity of the peptides that are directly “seen” and engaged by T cells. Such technologies could provide key information to better understand associations between HLA alleles and human diseases, or between HLA alleles and responsiveness to treatment. Thinking forward in this context, we anticipate that further development of “immunopeptidomics” (57) technologies will ultimately lead to the immunopeptidome-wide association study (IWAS) paradigm and will provide a new layer of information about disease susceptibility. Emulating the path of transcriptome-, proteome- and metabolome-wide association studies (TWAS, PWAS and MWAS, respectively) (55, 58–61), the road toward IWAS is technically very challenging, but conceptually straightforward: quantify the immunopeptidome of large human population cohorts and statistically link immunopeptidomic variations to different clinical outcomes (Fig. 2A). IWAS coming from very large immunopeptidomic sample cohorts could be of great value to treat patients but may also provide highly precise information about the predisposition of an individual to get specific immune diseases, hence representing a new look at human disease risk factors.
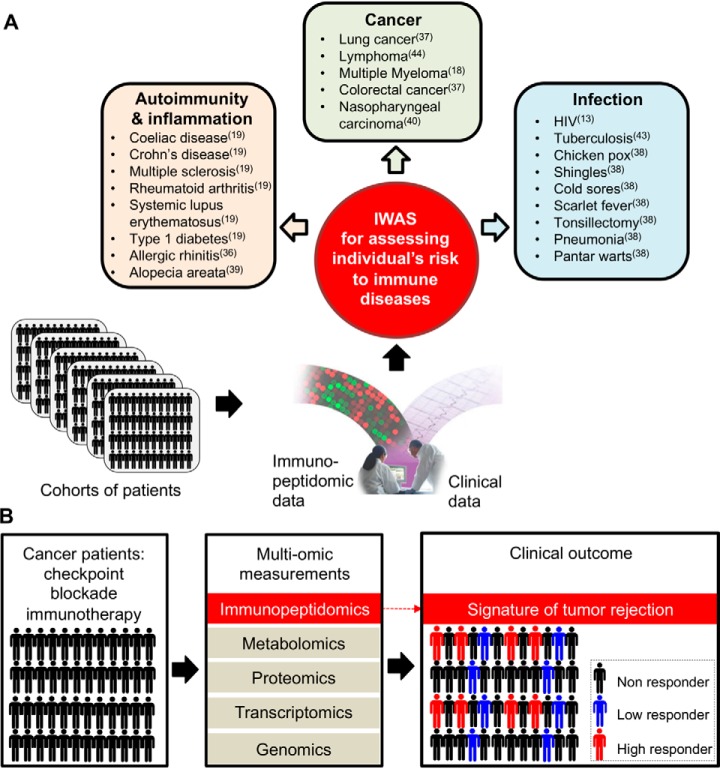
Fig. 2.
Next-generation immunopeptidomics to predict, diagnose, monitor and treat immune diseases. A, Further advances in MS technologies will ultimately enable IWAS in which robust, quantitative and comprehensive analysis of HLA class I and class II immunopeptidomes will be performed from thousands for patients. Such analysis will add a new layer of biological information and will enable the deciphering of immunopeptidome quantitative trait loci to predict disease and responsiveness to vaccines and immunotherapies. Human diseases listed in the boxes were shown to be associated with particular HLA alleles, with amino acid variants affecting either the peptide-binding groove of the HLA or its level of expression. The literature reference showing evidences for the HLA disease association is indicated in parenthesis. B, Integration of immunopeptidomics into a multi-omic biomarker discovery platform for cancer immunotherapy. Patients are treated by checkpoint blockade immunotherapy. In this example, cohort samples will be collected longitudinally and analyzed using an integrative multi-omic approach. HLA-bound peptides will be identified and quantified before, during and after treatment in a robust and comprehensive fashion using advanced immunopeptidomics technologies. Data will be analyzed and correlated with clinical outcomes. Molecular signatures of tumor rejection will be identified, in the immunopeptidome of checkpoint blockade high responders. Signatures (or epitope biomarkers) observed from such cohort studies will then be used prospectively to stratify patients and to treat those that will best respond to checkpoint therapy.
The road toward IWAS is probably very long and it is actually very likely that the feasibility of conducting IWAS from large human population cohorts will face a great deal of skepticism within the immunopeptidomics community as many technical issues can be considered at this point of time. In fact, the immunopeptidome is very different from one tissue to the other of every person and obvious questions will arise. For instance, (1) which tissues and cell types should be used?; (2) which disease should be prioritized while considering sample accessibility?; (3) what is the minimum number of samples (disease and control) that would be required?; and (4) which HLA alleles would be included and would they need to be shared across all samples to enable proper analysis of the IWAS data set? Addressing these questions would require the expertise of biostatisticians and are difficult to answer now. To our knowledge, the largest immunopeptidomic cohort study so far was conducted by the group of Arie Admon through the Glioma Actively Personalized Vaccine Consortium (GAPVAC). In this study, the authors have isolated HLA-associated peptides from 142 plasma samples collected from glioblastoma patients (62). Although impressive, 142 samples are not enough to conduct an IWAS. Akin to other association studies, it is likely that over 10,000 samples sharing specific HLA alleles (e.g. HLA-A02) would be required to enable IWAS. Accessibility to such cohorts of samples would probably need to be done through large multi-institutional networks and biobanking efforts, which may include both academia and industrial partners. Such an endeavor may be viewed as over-optimistic by some experts in the field. Here, our intent is neither to answer all potential questions to make IWAS a reality, nor to provide a detailed roadmap to achieve this goal. Instead, we aim to emphasize the most pressing technical issues to move faster toward this long-term goal, and most importantly, stimulate a mind-shift within the community to move the field to a higher orbit. If the community acknowledges the impact of moving the field from small to large immunopeptidomic cohort studies, we predict that immunopeptidomics will emerge in the future as a central science of immunology to predict, diagnose, monitor and treat immune diseases, and will therefore profoundly impact medicine (Fig. 2).
The Human Immunopeptidome Project
Development of mass spectrometry (MS) technology platforms may pave the way toward the IWAS paradigm. In fact, MS technologies have recently attracted a high level of interest for providing direct measurements of immunopeptidomes given the ongoing revolution in immuno-oncology and the intense interest in identifying therapeutically relevant antigenic peptides that are actionable in the clinic for vaccine development (63–73). More specifically, advances in genomics and MS techniques have enabled the development of proteogenomic methods for the direct detection of immunogenic tumor-specific mutated and non-mutated peptides that originate from both coding and non-coding regions of the genome (74–77). Those studies have provided an important proof-of-concept that MS is an effective approach that can be deployed for direct identification of immunogenic tumor-specific antigens. If further developed and routinely applied in cohort studies, such analytical method would be of great value for biomarker discovery in cancer immunotherapy to provide molecular signatures or epitope biomarkers of tumor rejection to discriminate patients who best respond to ICIs (Fig. 2B) (78). For the time being, imprecise predictions of HLA-associated peptides from genes and mRNA is still preferred over direct HLA-peptide measurements by MS (26, 79). This choice can be explained by the state and accessibility of the respective measurement techniques: whereas essentially complete genome and transcriptome analysis is readily available to immunologists through core facilities, MS-based analysis of immunopeptidomes is most effectively performed by expert labs and cannot easily reach the throughput, sensitivity and reproducibility of genome and transcriptome analysis. To solve this issue, the MS-based immunopeptidomics Human Immunopeptidome Project (HIPP) was launched under the umbrella of the Human Proteome Organization (HUPO) with the long-term goal of 1) mapping the entire composition of the human immunopeptidome and 2) making data and the robust experimental and computational techniques of immunopeptidomics accessible to any researcher and clinical investigators (22, 23). Ultimately, absolute quantification of immunopeptidomes should become as cheap, fast, sensitive and reproducible as it is nowadays for genome and transcriptome analysis. Only then, immunopeptidomics will become widely accessible to the broader immunology community to enable very large immunopeptidomic cohort studies, the development of epitope biomarker discovery platforms, and ultimately, IWAS.
Historical Milestones in MS-based Immunopeptidomics
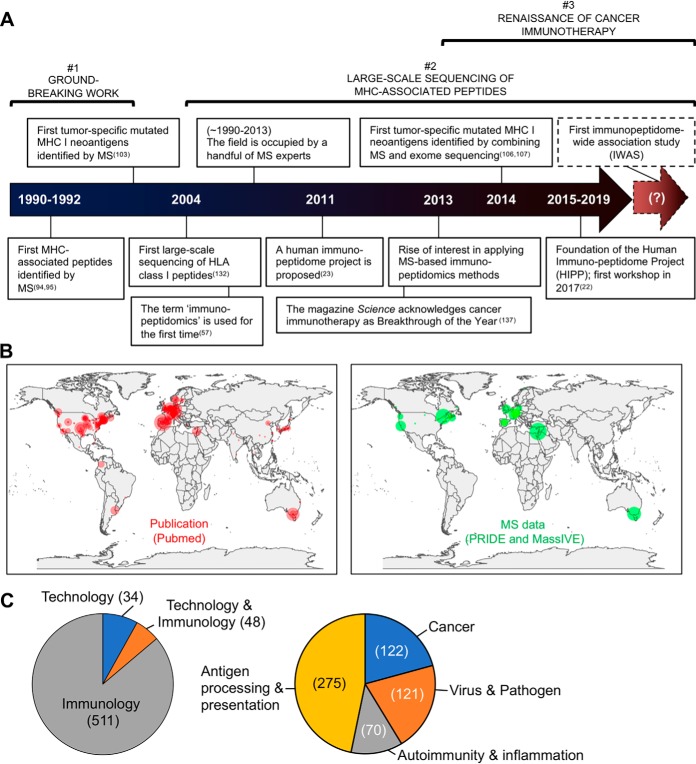
Over the last five years, many excellent reviews have been published regarding the analysis of immunopeptidomes using MS technologies (71, 73, 80–93). To avoid redundancies with those reviews, we focus below on key historical milestones within three different timeframes, which we view as important toward the IWAS paradigm: (1) the groundbreaking work of Rammensee and Hunt/Engelhard, (2) the capability of MS technologies for large-scale sequencing of MHC-associated peptides, and (3) the renaissance of cancer immunotherapy (Fig. 3A).
Fig. 3.
Historical perspective from a systematic review of the literature. A, Technical and cultural milestones in MS-based immunopeptidomics. The dotted arrow to the right indicates a hypothetical future point in time at which the first IWAS will have been successfully conducted. B, Global distribution of research groups that have shown productivity in the field of MS-based immunopeptidomics. (Left panel) A systematic PubMed search (2019/06/24) resulted in a total number of > 500 publications from 381 corresponding authors distributed across 180 cities and 36 countries, since the first peptide was sequenced in 1990. (Right panel) A systematic PRIDE and MassIVE (ProteomeXchange resources) search (2019/07/24) resulted in 94 publicly available MS immunopeptidomic datasets coming from 46 laboratory heads, distributed across 32 cities and 14 countries, since the first dataset was deposited in 2013. The circles indicate the location of the corresponding authors/laboratory heads. The size of the circles is proportional to the number of publications/datasets reported by the corresponding author/laboratory head. C, The pie charts show the proportion of publications from the systematic PubMed search. (Left) The pie chart shows the proportion of publications that have answered an immunology question and/or that have tackled a technological limitation. (Right) The pie chart shows the proportion of studies that have applied the available technology to answer an immunology question in the context of basic research (antigen processing and presentation) or applied research (cancer, infectious diseases and autoimmunity/inflammation). See supplemental Table S1 for details about the PubMed and PRIDE/MassIVE searches.
In the early 1990s, the teams of Hans-Georg Rammensee, Victor Engelhard and Donald Hunt provided truly groundbreaking work (Fig. 3A). They isolated MHC/HLA class I-associated peptides by immunoaffinity purification and applied liquid chromatography (LC)-MS methods to provide the first physical evidence about the nature of MHC class I-bound peptides (94, 95). Specifically, in 1990, Rammensee's group applied LC-MS to analyze naturally processed viral peptides recognized by cytotoxic T cells (95). In 1991, his group reported pooled sequencing of peptides eluted from several MHC class I molecules, with identification of allele-specific peptide binding motifs (21). The year after, the findings by Hunt et al. were consistent with the Rammensee motif (94). In fact, Hunt/Engelhard's group quantified ∼200 peptide species by LC-MS and partially sequenced ∼19 of them, validating the motif concept and providing the first estimate that the total number of different peptides presented by HLA-A2 could easily exceed 1000, with most of the peptides present in 100 or fewer copies per cell, hence suggesting for the first time the existence of a large and complex immunopeptidome (94, 96–101). In 1992, Hunt/Engelhard provided physical evidences for non-canonical long peptides presented by HLA class I molecules (102), and in 1997, sequenced the very first mutated MHC class I peptides (Fig. 3A) (103), largely referred nowadays as tumor-specific neoantigens (26). Further, in 2006, Hunt/Engelhard's group showed evidences for phosphopeptides as potential targets for cancer immunotherapy (104, 105). Since then, MS-based discovery of mutated (75, 76, 80, 106–108), non-canonical (86, 87, 109–111) and post-translationally modified HLA-associated peptides [phosphorylation (112–115), methylation (116), citrullination (117), deamidation (118–121), kynurenine (122), glycosylation (123–125), peptide splicing (126–130)] continues to be a topic of much interest in the field.
A key technical milestone that greatly impacted the immunopeptidomics field was the commercialization of the Orbitrap technology in 2005 (131). In fact, the launch of this instrument has enabled researchers to routinely perform large-scale sequencing of MHC-associated peptides, going from dozens to hundreds, to lately thousands of MHC-associated peptides identified in single experiments (87, 132). Computational algorithms were created and improved (91–93, 133–136) and different MS techniques developed by the proteomics community were used for the analysis of MHC peptides: data-dependent acquisition (DDA) for the discovery of new antigenic peptides, selected/multiple/parallel reaction monitoring (S/M/PRM) for targeted analysis of pre-defined sets of peptides with a high level of sensitivity, reproducibility and quantitative accuracy, and more recently, sequential window acquisition of all theoretical fragment ion spectra/data-independent acquisition (SWATH/DIA). SWATH/DIA makes use of high resolution qTOF instruments (SWATH) or Orbitrap instruments for high-throughput targeted analysis of large fractions of immunopeptidomes and is reviewed in (71). For three decades, a very small community of experts have deployed these MS techniques to generate MS data—generally shared nowadays through MS public data repositories such as PRIDE and MassIVE—in most cases through collaborations with immunologists and clinical investigators (Fig. 3B). Until now, more than 500 original publications related to MS-based sequencing of MHC-associated peptides were reported (supplemental Table S1; see filtered PubMed search result). Overall, only ∼10% were about the development of new methodologies and ∼90% were about the application of the available technology to better understand the basic mechanisms of antigen presentation (46%), followed by application in the fields of cancer (21%), autoimmunity or inflammation (12%), and infectious diseases (21%) (Fig. 3C).
A clear milestone in immunopeptidomics also became apparent in 2013 following the recognition of cancer immunotherapy as breakthrough of the year by the magazine Science (Fig. 3A) (137). In fact, the renaissance of cancer immunotherapy over the last recent years, thanks to the astonishing clinical success of checkpoint blockade inhibitors (138), has significantly increased the level of interest of researchers and clinical oncologists for applying MS-based immunopeptidomics in the quest of tumor-specific antigen discovery (62, 73, 139–141). In 2014, Gubin et al. demonstrated that tumor-specific mutated peptides were targeted by T cells in response to checkpoint blockade therapy (106), thereby providing groundbreaking mechanistic insights about how checkpoint blockade works to eradicate cancer in patients (142–144). In the following years, the capability of MS techniques to directly identify tumor-specific mutated antigens as physical molecules was further demonstrated, collectively providing an important proof-of-concept about the potential utility of MS technologies in the clinic (65, 138, 145, 146). This enthusiasm has created new business opportunities and has resulted in an increasing number of biotech and pharma companies interested in applying MS-based immunopeptidomics in the space of antigen discovery (Table I) (147). Advances in MS technologies and the increasing clinical relevance of the immune system to treat cancer has greatly impacted the culture of the immunopeptidomics field, progressively leading it into a new and exciting era, as highlighted by the recent “Immunopeptidomics” special issue in the journal Proteomics (148).
Table I. Inclusive list of biotech and pharmaceutical companies applying MS-based immunopeptidomics. Note that the list may not be exhaustive.
| Biotech or Pharma company | Headquarters | Founded (year) | Website |
|---|---|---|---|
| Adaptimmune | Abingdon, Oxfordshire, UK | 2007 | https://www.adaptimmune.com/ |
| Adicet Bio | Haifa, Israel | 2014 | https://www.adicetbio.com/ |
| Agenus | Cambridge, UK | 1994 | https://agenusbio.com/ |
| Bristo-Myers Squibb | New York, NY | 1887 | https://www.bms.com/ |
| Caprion | Montreal, Canada | 2000 | http://www.caprion.com/ |
| Ervaxx | Oxford, UK | 2018 | https://www.ervaxx.com/ |
| Evotec A.G. | Hamburg, Germany | 1993 | https://www.evotec.com/ |
| Genentech | San Francisco, CA | 1976 | https://www.gene.com/ |
| Gritstone Oncology | Emeryville, CA | 2015 | https://gritstoneoncology.com/ |
| Immatics | Tuebingen, Germany | 2000 | http://immatics.com/ |
| Immunocore | Abingdon, Oxfordshire, UK | 2008 | https://www.immunocore.com/ |
| Eli Lilly and Company | Indianapolis, IN | 1876 | https://www.lilly.com/ |
| Neon Therapeutics | Cambridge, MA | 2013 | https://neontherapeutics.com/ |
| Pfizer | New York, NY | 1849 | https://www.pfizer.com/ |
| Philochem | Zurich, Switzerland | 2006 | http://www.philochem.ch/ |
| PureMHC | Austin, TX | 2013 | http://puremhc.com/ |
| Roche | Basel, Switzerland | 1896 | https://www.roche.com/ |
Sample Preparation Is The Achilles' Heel For Large Immunopeptidomic Cohort Studies
Next-generation sequencing and MS technologies are revolutionizing the life sciences by enabling robust quantitative analysis of genomes, transcriptomes and proteomes in many laboratories, making multi-omic cohort studies a reality (149). In contrast, the field of immunopeptidomics is lagging behind even though its potential impact in the clinic is well recognized. In immunopeptidomics, the isolation of MHC-associated peptides is currently in our view the “Achilles' heel” of the whole workflow. Below, we argue that the development of new protocols, reagents and consumables will need to be prioritized over the next five years to accelerate the explosion of large-scale cohort studies in immunopeptidomics.
As of today, the field of immunopeptidomics heavily relies on the use of antibodies for the specific isolation of MHC-associated peptides by immunoaffinity purification (see (72) for a list of HLA-specific antibodies). Notably, for the isolation of HLA-ABC-associated peptides, researchers have generally used the exact same antibody (W6/32) over the last 30 years. The method has been refined by a handful of experts but remains fundamentally like the methods used by Donald Hunt and Hans-Georg Rammensee in the early 1990s (72, 150–153). In addition, relatively large quantities of antibodies are required to perform this procedure (i.e. ∼1 mg of antibody on average per sample). As of today, any laboratory aiming at conducting large-scale immunopeptidomic studies must generally produce and purify antibodies from hybridoma cell lines, which is relatively costly and time consuming, and therefore represents a considerable limitation for scaling up the procedure and for rapidly implementing immunopeptidomics workflows into new laboratories. Moreover, very little is known about the yield of the immunoaffinity purification procedure. Until now, the use of isotopically labeled peptide-MHC monomers represents the best strategy for the accurate quantification of stepwise yields of the procedure (154). In this regard, the group of Peter van Veelen has recently synthesized 24 different isotopically peptide-HLA monomers and showed that the yield was sequence-dependant. Specifically, the yield ranged between 2.1 and 15%, and was below 10% for 70% of the peptides tested (17 out of 24), indicating important losses and/or biases during sample preparation (155). Along these lines, other separation techniques were reported to create a bias in the detectable immunopeptidome, which can be explained by the enormous biochemical diversity of those peptides (156). For instance, highly hydrophilic peptides may not be captured by conventional reversed-phase C18 material whereas highly hydrophobic peptides may be preferentially lost during the procedure simply because they stick to plastic materials during the multiple handling steps.
To address the issues mentioned above, exploration of new approaches in digital microfluidics for immunoprecipitation may help increase the sample throughput (157); further development of acoustic technologies for contactless handling may accelerate unbiased analysis of immunopeptidomes (158, 159); and low-cost distribution of antibodies may accelerate the testing of new sample preparation protocols, which may ideally enable all sample processing steps to be carried out in a single tube, thereby enhancing sensitivity, throughput and scalability of immunopeptidomic analyses (160, 161). As a first step, HUPO-HIPP recently proposed to design a multi-laboratory study in which a large library of isotopically labeled peptide-HLA monomers would be distributed across different research groups for the accurate quantification of stepwise yields of the established immunoaffinity purification procedure (22). This type of work has been underappreciated over the years but is nevertheless invaluable to define the physicochemical properties of peptides that are preferentially detected or lost during the isolation procedure. Inspired by landmark studies from other -omic disciplines (162–167), conducting the proposed multi-laboratory study will clarify the uncertainty about the yield of the peptide isolation procedure, will determine the robustness of the methodology used by different groups and will possibly indicate the need to prioritize the development of innovative sample preparation protocols.
In summary, the field has greatly benefited from the traditional immunoaffinity purification method to make groundbreaking discoveries. The method will likely continue to be used for small-scale studies and relatively larger cohort studies in expert laboratories soon (62, 141). However, the development of new sample preparation protocols will become critical to scale up and bring the field to the next level. Isolation of MHC-associated peptides in a cheap, fast, unbiased, reproducible and high-throughput fashion has the potential to transform the field and increase the impact of immunopeptidomics in biomedical research through robust clinical applications, and ultimately, population-scale immunopeptidomic studies.
Disruptive Technologies for Large-Scale Immunopeptidomics
IWAS and population-scale studies will undoubtedly require immunopeptidomic technologies to be widely accessible and commercially well-supported. In genomics, high throughput generation of complete genomic maps became widely accessible only with the development and commercialization of technologies and methods that enabled the sequencing of millions of nucleic acid segments in parallel (168). Reaching this state in the field of immunopeptidomics for high throughput generation of complete digital immunopeptidomic maps is possible but embracing public-private partnerships will need to be prioritized in the coming years to accelerate commercialization of new immunopeptidomics technologies/methods and their wide-spread distribution. It is conceivable that MS technologies, such as SWATH/DIA-MS, will continue to improve toward this goal. However, it is also possible that disruptive new approaches will be developed for massively parallel peptide sequencing, making MS technologies potentially obsolete in the future. In this regard, Swaminathan et al. have recently introduced a groundbreaking peptide fluorosequencing technology which may accelerate the process toward the IWAS paradigm. Akin to massively parallel measurements of DNA using fluorescence as readout they demonstrated that parallel fluorescence sequencing is also achievable for peptides (169). Their method is promising because it paves the way toward single-molecule peptide sequencing at very high throughput from minute amounts of samples (http://www.erisyon.com/) (170).
Their method builds on three well-characterized methods: Edman chemistry, massively parallel DNA sequencing and MS-based computational strategies for sequence database searching (171). Briefly, isolated peptides are first labeled with fluorophores for each amino acid residues; second, labeled peptides are immobilized on a glass surface and imaged by total internal reflection microscopy to monitor decreases in each molecule's fluorescence after consecutive rounds of Edman degradation; third, the pattern of drops in fluorescence intensity is interpreted to provide a sequencing annotation for each peptide, which is matched and scored against a peptide sequence database to infer the most likely set of peptides present in the sample (169, 170, 172). In our opinion, this approach is very promising for proteins soon, but further development of the technology will be essential for sequencing HLA-associated peptides because it is currently impossible to label most of the amino acids with fluorophore. Although this proof of concept approach has limitations at this stage (e.g. dyes used, sample complexity, sequencing yield), it capitalizes on three established techniques that may accelerate maturation from proof of concept to routine application and wide adoption by the community.
If further developed, highly parallel single-molecule peptide sequencing may offer an improvement of more than one million-fold in sensitivity over conventional MS technologies and may allow for millions of distinct peptides to be sequenced in parallel, identified and digitally quantified (169, 171). Given the qualitative and quantitative complexity of the human immunopeptidome, this technology may radically transform the immunopeptidomics field. However, one has to keep in mind that the performance of this approach will always rely on the quality of upstream peptide isolation procedures. Hence, the shortcomings mentioned above related to the isolate of MHC-associated peptides by immunoaffinity purification will need to be overcome for a generally accessible, reliable and truly universal immunopeptidomic technology.
Data Analysis Challenges
The main approaches used currently for the analysis of immunopeptidomics experiments were developed originally for the analysis of MS proteomics datasets. Database searches are the main analysis method of choice for data dependent acquisition workflows. Here, the acquired spectra are compared with generated (theoretical) ones coming from peptide sequences drawn from a given protein sequence database. Several limitations of this type of analyses can apply to immunopeptidomics. First of all, the size of the search database (and the related search space) can become extremely large. For instance, proteasome-generated spliced peptides have been detected by MS in recent studies (126, 129, 173, 174), so the inclusion of spliced variants in the search database could be required (129). However, this strategy has sparked intense discussions in the field over the last two years and remains actively debated (130, 175, 176). Additionally, in most proteomics experiments, trypsin is used as the digestion enzyme. Because “no enzyme specificity” must be used in immunopeptidome searches, the search space grows exponentially. As a result of these two related issues, the time required for performing the searches can increase enormously and even more, if variable post-translational modifications such as phosphorylation have to be considered in the search. More importantly, when making use of the decoy-target approach, it becomes very challenging for search engines to differentiate between true and false matches, considering the huge search space. Consequently, a compromise is often needed, and the accepted PSM false discovery rate can become 5% instead of the standard 1% used in standard MS proteomics experiments.
In parallel, the average percentage of identified spectra can be significantly lower in immunopeptidomics experiments, because of different factors (e.g. shortness of MHC class I peptides, lack of basic amino acid residues, low amount of samples, acquisition parameters, etc), as described previously (71). To address this issue, several approaches have been proposed, like the sequential use of different search engines or the use of Percolator. This tool performs machine learning on high-confidence matches to rescore database search results for lower-confidence peptides (177). Other approaches for performing rescoring have also been successfully used recently in the field (133).
Moreover, alternative analysis approaches to standard database searches can be also used, either alone or in combination. One example is the use of de novo search methods, where the spectra can be sequenced directly, without the need of a protein sequence database (178). The use of de novo algorithms has recently been recommended in immunopeptidomics protocols (72). Spectral searches approaches can also be employed (179). Here, the experimental spectra are compared with existing collections of spectra in the public domain called spectral libraries. However, this approach, although increasingly popular, has had a limited uptake so far. One of the reasons is that the generation of high-quality spectral libraries is not straightforward, and some researchers are reluctant to use publicly available spectral libraries generated by others.
Additionally, the increasing popularity of open modification searches makes them a promising approach. The new tools developed in the last few years (e.g. MSFragger, ANN-SoLo, TagGraph, among others (180–182) have solved the high computational cost of these methods. Additionally, in our opinion, a promising approach for targeting interesting spectra would be the use of clustering of MS/MS spectra (183–185). This approach can be used to select those spectra that remain unidentified and that are commonly found across MS runs in different samples. The hypothesis is that the corresponding peptides are potentially interesting and biologically relevant, because they are abundant. The resulting representatives of these clusters of unidentified spectra could then be subjected to those alternative analysis methods explained above. It is certainly a possibility that a significant fraction of unidentified spectra from such clusters includes contaminating peptides that show up in many samples. They could indeed be actual contaminants usually found in proteomics experiments (e.g. keratins), or not biologically relevant peptides as artifacts derived from the experimental protocol. If the latter case applies, information from such clusters could be used to feed a database of contaminating peptides that are constantly observed in immunopeptidomics. Such database does not exist yet but would be useful for the growing immunopeptidomics community and would be conceptually like the CRAPome, which serves as a repository of protein contaminants in protein-protein interaction studies (186).
Further, artificial intelligence approaches have the potential to improve the analysis dramatically. In MS-based proteomics, deep-learning techniques have been recently deployed using millions of high quality MS2 spectra generated from hundreds of thousands of synthetic tryptic peptides, to build new algorithms to predict fragmentation patterns of peptides, thereby enabling successful generation of in-silico peptide spectral libraries for high throughput targeted analysis of DIA data (187–189). These developments should also greatly benefit the further improvement of de novo search algorithms. Deep-learning was also recently applied for de novo identification of HLA-associated peptides using DIA-MS (190, 191).
Finally, it is worth highlighting that a significant number of challenges would need to be considered in addition with regards to the quantification of immunopeptidomes. This quantitative aspect is much less mature at present. Analogous challenges in quantitative MS proteomics approaches have been described before (192).
The SysteMHC Atlas Project
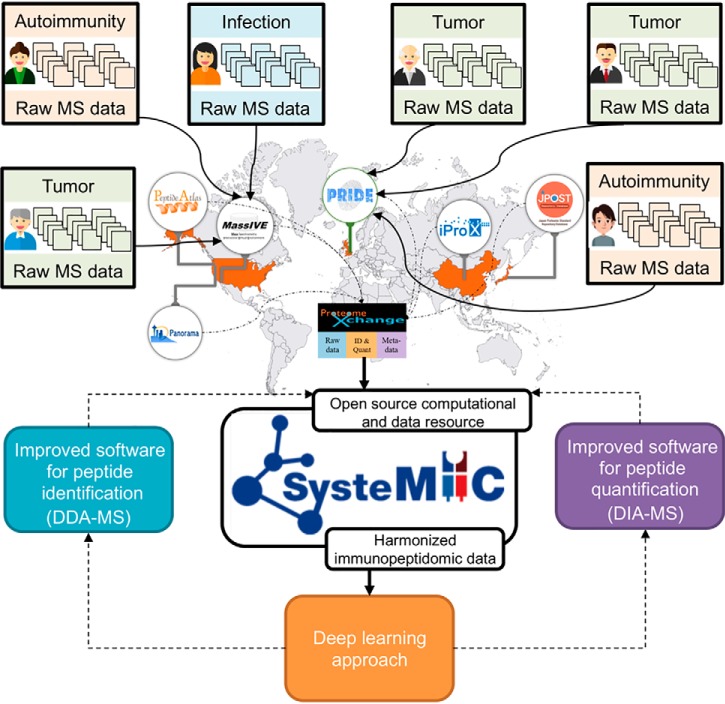
Open and comprehensive reference atlas in life sciences are increasingly beneficial for the scientific community (193–198). Similarly, the creation of a comprehensive atlas of the immunopeptidome in human, mouse, and other species would be of great value for both understanding health and diagnosing, monitoring and treating immune diseases (22, 23, 69). In this context, participants of the HIPP initiative recognized the need for an open immunopeptidomics atlas/repository in which output files of mass spectrometric measurements of immunopeptidome samples would be annotated, stored and shared without restriction. In response to this need, the SysteMHC Atlas project was created—a project fully dedicated to the public dissemination and analysis of immunopeptidomic data generated by MS (https://systemhcatlas.org) (Fig. 4) (199). The SysteMHC Atlas uploads raw immunopeptidomics MS data originally deposited into public proteomics databases (mainly the PRIDE database (200), which is the leading ProteomeXchange repository (201, 202)) along with the metadata associated with the experiment (203). Raw MS data are then processed through the uniform and open-source Trans-Proteomic Pipeline (TPP)-based computational pipeline for HLA peptide identification, annotation (204–206) and statistical validation (205, 206). Lists of HLA peptides as well as allele-specific peptide spectral libraries (207) are generated and presented in the Atlas.
Fig. 4.
The vision of the SysteMHC Atlas project. The SysteMHC Atlas project was launched in 2018 with the main goal of collecting, harmonizing and sharing immunopeptidomic data generated by the community. Over the next years, we anticipate that the growing MS community working on various immunopeptidomic-related research topics (e.g. cancer, autoimmunity, infectious diseases) will contribute to the expansion of the SysteMHC Atlas by uploading data into existing repositories of the ProteomeXchange consortium (mainly PRIDE and MassIVE; the others shown are PeptideAtlas, Panorama, iProX and jPOST). Those data will be reprocessed and harmonized using a uniform and open source computational pipeline developed under the SysteMHC Atlas project. We envision that ProteomeXchange resources will be integrated better with the SysteMHC Atlas in the near future, thereby facilitating the application of deep-learning approaches from quality-controlled immunopeptidomic “Big Data” to improve software tools for MHC peptide identification and quantification by DDA- and DIA-MS techniques.
The SysteMHC Atlas project is still at an early-stage of development. The current version is composed of 23 immunopeptidomic datasets (https://systemhcatlas.org/datasets). In the short-term, we plan to enhance the capabilities of the SysteMHC Atlas project by integrating emerging computational MS workflows (e.g. proteogenomics approaches, ultrafast search engines, identification of post-translationally modified peptides) and expand its content through a closer partnership with the ProteomeXchange consortium (Fig. 4). High-quality harmonized immunopeptidomic data stored in the SysteMHC Atlas will find use over the next years to build and improve software algorithms for the identification and quantification of HLA peptides by DDA- and DIA-MS (Fig. 4). Hence, one can anticipate that the increasing number of high quality MS2 spectra in the SysteMHC Atlas will be of great value for computational scientists to tailor next-generation software for the analysis of immunopeptidomic data. New computational tools will also be used to re-process and re-score data in the Atlas and keep improving the quality of the data. By doing so, the ever-increasing number of HLA peptides in the Atlas will be useful to enhance MS identification and quantification of canonical and non-canonical HLA-associated peptides, including neoantigens that are unpredictable from genomic information as well as those that originate from foreign organisms and the microbiome (208). Moreover, it is our intention to create, in longer-term, the SysteMHC Atlas Data Analysis Center, which will support robust measurements of large immunopeptidomic sample cohorts by SWATH/DIA-MS. In fact, an important utility of the SysteMHC Atlas project is to access reference HLA allele-specific peptide spectral libraries for reproducible, quantitative and comprehensive analysis of immunopeptidomes by SWATH/DIA-MS (71, 209–212). Hence, we envision that the SysteMHC Atlas project is the beginning of an enterprise that will accelerate the design of large-scale immunopeptidomic studies at population scale, and therefore, represents a potential strategy toward the IWAS concept.
CONCLUSION
Cohort studies of the post-GWAS era are increasingly impactful and an important driving force for revolutionizing healthcare (4). Recent GWAS-based evidences indicate that generation of digital immunopeptidome maps from thousands of individuals may unveil a vast array of immunopeptidomic signatures (or epitope biomarkers) that could eventually be used in the clinic to predict the susceptibility or resistance of an individual to immune diseases as well as an individual's predisposition to respond to vaccines and immunotherapies. Inspired by the vision and achievements of pioneers in genomics (213–216) and proteomics (217, 218), we envision that the ongoing enthusiasm in MS profiling of the human immunopeptidome will lead to the development of the IWAS paradigm, in which absolute quantification of immunopeptidomes at population-scale will become a reality. If tested and validated, we envision that IWAS will radically change the way we think about immunopeptidomics and will represent a new milestone in the history of the field. Achieving this goal will require tremendous technological development and community efforts; from highly standardized techniques for high throughput isolation of peptides to advanced knowledge about the baseline and dynamics of the human immunopeptidome, as well as appropriate data and computational resources to handle the analysis of very large immunopeptidomic datasets. With the advent of numerous new “biobank” type of studies, great opportunities will exist to collect samples for immunopeptidomic analyses—together with genomic, transcriptomic, proteomic and metabolomic analyses—that will impact personalised health care management and public health care policy in the future.
Supplementary Material
Acknowledgments
We thank the members of the Human Immunopeptidome Project for insightful discussions.
Footnotes
* This work was supported by funding from the Fonds de recherche du Québec – Santé (FRQS), the Cole Foundation and the CHU Sainte-Justine and the Charles-Bruneau Foundations. JAV wants to acknowledge funding from Wellcome Trust [grant number 208391/Z/17/Z], BBSRC [grant number BB/P024599/1] and the EPIC-XS project (grant number 823839), funded by the Horizon 2020 programme of the European Union. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Tables.
This article contains supplemental Tables.
1 The abbreviations used are:
- SNP
- single-nucleotide polymorphisms
- GWAS
- genome-wide association studies
- eQTL
- genome-wide transcript quantitative trait loci
- IWAS
- immunopeptidome-wide association studies
- DDA
- data-dependent acquisition.
REFERENCES
- 1. Venter C. J., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., Gocayne J. D., Amanatides P., Ballew R. M., Huson D. H., Wortman J., Zhang Q., Kodira C. D., Zheng X. H., Chen L., Skupski M., Subramanian G., Thomas P. D., Zhang J., Miklos G. L., Nelson C., Broder S., Clark A. G., Nadeau J., McKusick V. A., Zinder N., Levine A. J., Roberts R. J., Simon M., Slayman C., Hunkapiller M., Bolanos R., Delcher A., Dew I., Fasulo D., Flanigan M., Florea L., Halpern A., dhar Hannenhalli Kravitz S., Levy S., Mobarry C., Reinert K., Remington K., Abu-Threideh J., Beasley E., Biddick K., Bonazzi V., Brandon R., Cargill M., Chandramouliswaran I., Charlab R., Chaturvedi K., Deng Z., Francesco V., Dunn P., Eilbeck K., Evangelista C., Gabrielian A. E., Gan W., Ge W., Gong F., Gu Z., Guan P., Heiman T. J., Higgins M. E., Ji R.-R., Ke Z., Ketchum K. A., Lai Z., Lei Y., Li Z., Li J., Liang Y., Lin X., Lu F., Merkulov G. V., Milshina N., Moore H. M., Naik A. K., Narayan V. A., Neelam B., Nusskern D., Rusch D. B., Salzberg S., Shao W., Shue B., Sun J., Wang Z., Wang A., Wang X., Wang J., Wei M.-H., Wides R., Xiao C., Yan C., Yao A., Ye J., Zhan M., Zhang W., Zhang H., Zhao Q., Zheng L., Zhong F., Zhong W., Zhu S. C., Zhao S., Gilbert D., Baumhueter S., Spier G., Carter C., Cravchik A., Woodage T., Ali F., An H., Awe A., Baldwin D., Baden H., Barnstead M., Barrow I., Beeson K., Busam D., Carver A., Center A., Cheng M., Curry L., Danaher S., Davenport L., Desilets R., Dietz S., Dodson K., Doup L., Ferriera S., Garg N., Gluecksmann A., Hart B., Haynes J., Haynes C., Heiner C., Hladun S., Hostin D., Houck J., Howland T., Ibegwam C., Johnson J., Kalush F., Kline L., Koduru S., Love A., Mann F., May D., McCawley S., McIntosh T., McMullen I., Moy M., Moy L., Murphy B., Nelson K., Pfannkoch C., Pratts E., Puri V., Qureshi H., Reardon M., Rodriguez R., Rogers Y.-H., Romblad D., Ruhfel B., Scott R., Sitter C., Smallwood M., Stewart E., Strong R., Suh E., Thomas R., Tint N., Tse S., Vech C., Wang G., Wetter J., Williams S., Williams M., Windsor S., Winn-Deen E., Wolfe K., Zaveri J., Zaveri K., Abril J. F., Guigó R., Campbell M. J., Sjolander K. V., Karlak B., Kejariwal A., Mi H., Lazareva B., Hatton T., Narechania A., Diemer K., Muruganujan A., Guo N., Sato S., Bafna V., Istrail S., Lippert R., Schwartz R., Walenz B., Yooseph S., Allen D., Basu A., Baxendale J., Blick L., Caminha M., Carnes-Stine J., Caulk P., Chiang Y.-H., Coyne M., Dahlke C., Mays A., Dombroski M., Donnelly M., Ely D., Esparham S., Fosler C., Gire H., Glanowski S., Glasser K., Glodek A., Gorokhov M., Graham K., Gropman B., Harris M., Heil J., Henderson S., Hoover J., Jennings D., Jordan C., Jordan J., Kasha J., Kagan L., Kraft C., Levitsky A., Lewis M., Liu X., Lopez J., Ma D., Majoros W., niel J., Murphy S., Newman M., Nguyen T., Nguyen N., Nodell M., Pan S., Peck J., Peterson M., Rowe W., Sanders R., Scott J., Simpson M., Smith T., Sprague A., Stockwell T., Turner R., Venter E., Wang M., Wen M., Wu D., Wu M., Xia A., Zandieh A., and Zhu X. (2001) The sequence of the human genome. Science 291, 1304–1351 [DOI] [PubMed] [Google Scholar]
- 2. Lander E., Linton L., Birren B., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., yle, FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R., Wilson R., Hillier L., McPherson J., Marra M., Mardis E., Fulton L., Chinwalla A., Pepin K., Gish W., Chissoe S., Wendl M., Delehaunty K., Miner T., Delehaunty A., Kramer J., Cook L., Fulton R., Johnson D., Minx P., Clifton S., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R., Muzny D., Scherer S., Bouck J., Sodergren E., Worley K., Rives C., Gorrell J., Metzker M., Naylor S., Kucherlapati R., Nelson D., Weinstock G., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R., Federspiel N., Abola A., Proctor M., Myers R., Schmutz J., ckson, Grimwood J., Cox D., Olson M., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G., Athanasiou M., Schultz R., Roe Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J., Bateman A., Batzoglou S., Birney E., Bork P., Brown D., Burge C., Cerutti L., Chen H., Church D., Clamp M., Copley R., Doerks T., Eddy S., Eichler E., Furey T., Galagan J., Gilbert J., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L., Jones T., Kasif S., Kaspryzk A., Kennedy S., Kent W., Kitts P., Koonin E., Korf I., Kulp D., Lancet D., Lowe T., McLysaght A., Mikkelsen T., Moran J., Mulder N., Pollara V., Ponting C., Schuler G., Schultz J., Slater G., Smit A., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y., Wolfe K., Yang S., Yeh R., Collins F., Guyer Peterson J., Felsenfeld A., Wetterstrand K., Patrinos A., Morgan M., de Jong P., Catanese J., Osoegawa K., Shizuya H., Choi S., Chen Y., Szustakowki J., and Consortium I. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 3. Gambhir S., Ge J. T., Vermesh O., and Spitler R. (2018) Toward achieving precision health. Sci. Transl. Med. 10, eaao3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wijmenga C., and Zhernakova A. (2018) The importance of cohort studies in the post-GWAS era. Nat. Genet. 50, 322–328 [DOI] [PubMed] [Google Scholar]
- 5. Tardif J.-C., Rhéaume E., Perreault L.-P., Grégoire J. C., Zada Y., Asselin G., Provost S., Barhdadi A., Rhainds D., L'Allier P. L., Ibrahim R., Upmanyu R., Niesor E. J., Benghozi R., Suchankova G., Laghrissi-Thode F., Guertin M.-C., Olsson A. G., Mongrain I., Schwartz G. G., and Dubé M.-P. (2015) Pharmacogenomic determinants of the cardiovascular effects of Dalcetrapib. Circulation Cardiovasc. Genetics 8, 372–382 [DOI] [PubMed] [Google Scholar]
- 6. Tardif J.-C., Rhainds D., Brodeur M., Zada Y., Fouodjio R., Provost S., Boulé M., Alem S., Grégoire J. C., L'Allier P. L., Ibrahim R., Guertin M.-C., Mongrain I., Olsson A. G., Schwartz G. G., Rhéaume E., and Dubé M.-P. (2018) Genotype-dependent effects of dalcetrapib on cholesterol efflux and inflammation. Circulation Cardiovasc. Genetics 9, 340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayden E. (2017) The rise and fall and rise again of 23andMe. Nat. News 550, 174. [DOI] [PubMed] [Google Scholar]
- 8. Ozaki K., Ohnishi Y., Iida A., Sekine A., Yamada R., Tsunoda T., Sato H., Sato H., Hori M., Nakamura Y., and Tanaka T. (2002) Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat. Genet. 32, 650–654 [DOI] [PubMed] [Google Scholar]
- 9. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., and Parkinson H. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trowsdale J., and Knight J. C. (2012) Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. G 14, 301–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaur G., Gras S., Mobbs J. I., Vivian J. P., Cortes A., Barber T., Kuttikkatte S., Jensen L., Attfield K. E., Dendrou C. A., Carrington M., McVean G., Purcell A. W., Rossjohn J., and Fugger L. (2017) Structural and regulatory diversity shape HLA-C protein expression levels. Nat. Commun. 8, 15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petersdorf E. W., and O'hUigin C. (2018) The MHC in the era of next-generation sequencing: implications for bridging structure with function. Hum. Immunol. 80, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Apps R., Qi Y., Carlson J. M., Chen H., Gao X., Thomas R., Yuki Y., Prete G. Q., Goulder P., Brumme Z. L., Brumme C. J., John M., Mallal S., Nelson G., Bosch R., Heckerman D., Stein J. L., Soderberg K. A., Moody A. M., Denny T. N., Zeng X., Fang J., Moffett A., Lifson J. D., Goedert J. J., Buchbinder S., Kirk G. D., Fellay J., McLaren P., Deeks S. G., Pereyra F., Walker B., Michael N. L., Weintrob A., Wolinsky S., Liao W., and Carrington M. (2013) Influence of HLA-C expression level on HIV control. Science 340, 87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jung E., Cheon J., Lee J., Park S., Jang H., Chung S., Park M., Kim T.-G., Oh H.-B., Yang S.-K., Park S., Han J., Hong S., Kim T., Kim W., and Lee M. (2016) HLA-C*01 is a risk factor for Crohn's disease. Inflamm. Bowel Dis. 22, 796–806 [DOI] [PubMed] [Google Scholar]
- 15. Miyadera H., Ohashi J., Lernmark Å., Kitamura T., and Tokunaga K. (2015) Cell-surface MHC density profiling reveals instability of autoimmunity-associated HLA. J. Clin. Invest. 125, 275–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Z., and Jensen P. E. (2013) Structural characteristics of HLA-DQ that may impact DM editing and susceptibility to type-1 diabetes. Front. Immunol. 4, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu X., Deutsch A. J., Lenz T. L., Onengut-Gumuscu S., Han B., Chen W.-M., Howson J. M., Todd J. A., de Bakker P. I., Rich S. S., and Raychaudhuri S. (2015) Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, ng.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beksac M., Gragert L., Fingerson S., Maiers M., Zhang M.-J., Albrecht M., Zhong X., Cozen W., Dispenzieri A., Lonial S., and Hari P. (2016) HLA polymorphism and risk of multiple myeloma. Leukemia 30, 2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dendrou C. A., Petersen J., Rossjohn J., and Fugger L. (2018) HLA variation and disease. Nat. Rev. Immunol. 8, 193. [DOI] [PubMed] [Google Scholar]
- 20. Robinson J., Guethlein L. A., Cereb N., Yang S., Norman P. J., Marsh S. G., and Parham P. (2017) Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLOS Genet. 13, e1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Falk K., Rötzschke O., Stevanovic S., Jung G., and Rammensee H.-G. (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 [DOI] [PubMed] [Google Scholar]
- 22. Caron E., Aebersold R., Banaei-Esfahani A., Chong C., and Bassani-Sternberg M. (2017) A case for a human Immuno-Peptidome Project Consortium. Immunity 47, 203–208 [DOI] [PubMed] [Google Scholar]
- 23. Admon A., and Bassani-Sternberg M. (2011) The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol. Cell. Proteomics 10, O111.011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Croft N. P., Smith S. A., Pickering J., Sidney J., Peters B., Faridi P., Witney M. J., Sebastian P., Flesch I. E., Heading S. L., Sette A., Gruta N. L., Purcell A. W., and Tscharke D. C. (2019) Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. U.S.A. 73, 3112–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu T., Guan J., Handel A., Tscharke D. C., Sidney J., Sette A., Wakim L. M., Sng X. Y., Thomas P. G., Croft N. P., Purcell A. W., and Gruta N. L. (2019) Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 10, 2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schumacher T. N., Scheper W., and Kvistborg P. (2018) Cancer neoantigens. Annu Rev. Immunol. 37, 173–200 [DOI] [PubMed] [Google Scholar]
- 27. Gilchuk P., Spencer C. T., Conant S. B., Hill T., Gray J. J., Niu X., Zheng M., Erickson J. J., Boyd K. L., McAfee J. K., Oseroff C., Hadrup S. R., Bennink J. R., Hildebrand W., Edwards K. M. Jr., J. E., Williams J. V., Buus S., Sette A., humacher T. N., Link A. J., and Joyce S. (2013) Discovering naturally processed antigenic determinants that confer protective T cell immunity. J. Clin. Invest. 123, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caron E., Vincent K., Fortier M., Laverdure J., Bramoullé A., Hardy M., Voisin G., Roux P. P., Lemieux S., Thibault P., and Perreault C. (2011) The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol. Syst. Biol. 7, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Messaoudi I., Patiño J. A., Dyall R., LeMaoult J., and Nikolich-Žugich J. (2002) Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science 298, 1797–1800 [DOI] [PubMed] [Google Scholar]
- 30. Migueles S. A., Sabbaghian S. M., Shupert L. W., Bettinotti M. P., Marincola F. M., Martino L., Hallahan C. W., Selig S. M., Schwartz D., Sullivan J., and Connors M. (2000) HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U.S.A. 97, 2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaslow R. A., Carrington M., Apple R., Park L., Muñoz A., Saah A. J., Goedert J. J., Winkler C., O'Brien S. J., Rinaldo C., Detels R., Blattner W., Phair J., Erlich H., and Mann D. L. (1996) Influence of combinations of human major histocompatibility complex genes on the course of HIV–1 infection. Nat. Med. 2, 405–411 [DOI] [PubMed] [Google Scholar]
- 32. de Castro J. A. (2018) How ERAP1 and ERAP2 shape the peptidomes of disease-associated MHC-I proteins. Front Immunol 9, 2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matzaraki V., Kumar V., Wijmenga C., and Zhernakova A. (2017) The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 18, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reeves E., and James E. (2018) The role of polymorphic ERAP1 in autoinflammatory disease. Bioscience Rep. 38, BSR20171503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Unanue E. R., Turk V., and Neefjes J. (2015) Variations in MHC class II antigen processing and presentation in health and disease. Annu. Rev. Immunol. 34, 265–297 [DOI] [PubMed] [Google Scholar]
- 36. Team, T., collaborators, A., Waage J., Standl M., Curtin J. A., Jessen L. E., Thorsen J., Tian C., Schoettler N., Flores C., Abdellaoui A., Ahluwalia T. S., Alves A. C., Amaral A. F., Antó J. M., Arnold A., Barreto-Luis A., Baurecht H., van Beijsterveldt C. E., Bleecker E. R., Bonàs-Guarch S., Boomsma D. I., Brix S., Bunyavanich S., Burchard E. G., Chen Z., Curjuric I., Custovic A., den Dekker H. T., Dharmage S. C., Dmitrieva J., Duijts L., Ege M. J., Gauderman J. W., Georges M., Gieger C., Gilliland F., Granell R., Gui H., Hansen T., Heinrich J., Henderson J., Hernandez-Pacheco N., Holt P., Imboden M., Jaddoe V. W., Jarvelin M.-R., Jarvis D. L., Jensen K. K., Jónsdóttir I., Kabesch M., Kaprio J., Kumar A., Lee Y.-A., Levin A. M., Li X., Lorenzo-Diaz F., Melén E., Mercader J. M., Meyers D. A., Myers R., Nicolae D. L., Nohr E. A., Palviainen T., Paternoster L., Pennell C. E., Pershagen G., Pino-Yanes M., Probst-Hensch N. M., Rüschendorf F., Simpson A., Stefansson K., Sunyer J., Sveinbjornsson G., Thiering E., Thompson P. J., Torrent M., Torrents D., Tung J. Y., Wang C. A., Weidinger S., Weiss S., Willemsen G., Williams K. L., Ober C., Hinds D. A., Ferreira M. A., Bisgaard H., Strachan D. P., and Bønnelykke K. (2018) Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis. Nat. Genet. 50, 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferreiro-Iglesias A., Lesseur C., McKay J., Hung R. J., Han Y., Zong X., Christiani D., Johansson M., Xiao X., Li Y., Qian D. C., Ji X., Liu G., Caporaso N., Scelo G., Zaridze D., Mukeriya A., Kontic M., Ognjanovic S., Lissowska J., Szołkowska M., Swiatkowska B., Janout V., Holcatova I., Bolca C., Savic M., Ognjanovic M., Bojesen S., Wu X., Albanes D., Aldrich M. C., Tardon A., Fernandez-Somoano A., Fernandez-Tardon G., Marchand L., Rennert G., Chen C., Doherty J., Goodman G., Bickeböller H., Wichmann H.-E., Risch A., Rosenberger A., Shen H., Dai J., Field J. K., Davies M., Woll P., Teare D. M., Kiemeney L. A., van der Heijden E. H., Yuan J.-M., Hong Y.-C., Haugen A., Zienolddiny S., Lam S., Tsao M.-S., Johansson M., Grankvist K., Schabath M. B., Andrew A., Duell E., Melander O., Brunnström H., Lazarus P., Arnold S., Slone S., Byun J., Kamal A., Zhu D., Landi M., Amos C. I., and Brennan P. (2018) Fine mapping of MHC region in lung cancer highlights independent susceptibility loci by ethnicity. Nat. Commun. 9, 3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian C., Hromatka B. S., Kiefer A. K., Eriksson N., Noble S. M., Tung J. Y., and Hinds D. A. (2017) Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 8, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Betz R. C., Petukhova L., Ripke S., Huang H., Menelaou A., Redler S., Becker T., Heilmann S., Yamany T., Duvic M., Hordinsky M., Norris D., Price V. H., Mackay-Wiggan J., de Jong A., DeStefano G. M., Moebus S., Böhm M., Blume-Peytavi U., Wolff H., Lutz G., Kruse R., Bian L., Amos C. I., Lee A., Gregersen P. K., Blaumeiser B., Altshuler D., Clynes R., de Bakker P. I., Nöthen M. M., Daly M. J., and Christiano A. M. (2015) Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat. Commun. 6, 5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang T., Zhou T., He Y., Xue W., Zhang J., Zheng X., Li X., Zhang S., Zeng Y., and Jia W. (2018) Fine-mapping of HLA class I and class II genes identified two independent novel variants associated with nasopharyngeal carcinoma susceptibility. Cancer Med. 7, 6308–6316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu Y., Kweon S.-S., Tanikawa C., Jia W.-H., Xiang Y.-B., Cai Q., Zeng C., Schmit S. L., Shin A., Matsuo K., Jee S., Kim D.-H., Kim J., Wen W., Shi J., Guo X., Li B., Wang N., Zhang B., Li X., Shin M.-H., Li H.-L., Ren Z., Oh J., Oze I., Ahn Y.-O., Jung K., Conti D. V., Schumacher F. R., Rennert G., Jenkins M. A., Campbell P. T., Hoffmeister M., Casey G., Gruber S. B., Gao J., Gao Y.-T., Pan Z.-Z., Kamatani Y., Zeng Y.-X., Shu X.-O., Long J., Matsuda K., and Zheng W. (2018) Large-scale genome-wide association study of east asians identifies loci associated with risk for colorectal cancer. Gastroenterology 156, 1455–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scepanovic P., Alanio C., Hammer C., Hodel F., Bergstedt J., Patin E., Thorball C. W., Chaturvedi N., Charbit B., Abel L., Quintana-Murci L., Duffy D., Albert M. L., Fellay J., and Consortium M. (2018) Human genetic variants and age are the strongest predictors of humoral immune responses to common pathogens and vaccines. Genome Med. 10, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sveinbjornsson G., Gudbjartsson D. F., Halldorsson B. V., Kristinsson K. G., Gottfredsson M., Barrett J. C., Gudmundsson L. J., Blondal K., Gylfason A., Gudjonsson S., Helgadottir H. T., Jonasdottir A., Jonasdottir A., Karason A., Kardum L., Knežević J., Kristjansson H., Kristjansson M., Love A., Luo Y., Magnusson O. T., Sulem P., Kong A., Masson G., Thorsteinsdottir U., Dembic Z., Nejentsev S., Blondal T., Jonsdottir I., and Stefansson K. (2016) HLA class II sequence variants influence tuberculosis risk in populations of European ancestry. Nat. Genet. 48, 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. consortium, T., Sud A., Thomsen H., Law P. J., Försti A., da Filho M., Holroyd A., Broderick P., Orlando G., Lenive O., Wright L., Cooke R., Easton D., Pharoah P., Dunning A., Peto J., Canzian F., Eeles R., Kote-Jarai Zs., Muir K., Pashayan N., Hoffmann P., Nöthen M. M., Jöckel K-H, von Strandmann E., Lightfoot T., Kane E., Roman E., Lake A., Montgomery D., Jarrett R. F., Swerdlow A. J., Engert A., Orr N., Hemminki K., and Houlston R. S. (2017) Genome-wide association study of classical Hodgkin lymphoma identifies key regulators of disease susceptibility. Nat. Commun. 8, 1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okada Y., Momozawa Y., Sakaue S., Kanai M., Ishigaki K., Akiyama M., Kishikawa T., Arai Y., Sasaki T., Kosaki K., Suematsu M., Matsuda K., Yamamoto K., Kubo M., Hirose N., and Kamatani Y. (2018) Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat. Commun. 9, 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wissemann W. T., Hill-Burns E. M., Zabetian C. P., Factor S. A., Patsopoulos N., Hoglund B., Holcomb C., Donahue R. J., Thomson G., Erlich H., and Payami H. (2013) Association of Parkinson disease with structural and regulatory variants in the HLA region. Am. J. Hum. Genetics 93, 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sulzer D., Alcalay R. N., Garretti F., Cote L., Kanter E., Agin-Liebes J., Liong C., McMurtrey C., Hildebrand W. H., Mao X., Dawson V. L., Dawson T. M., Oseroff C., Pham J., Sidney J., Dillon M. B., Carpenter C., Weiskopf D., Phillips E., Mallal S., Peters B., Frazier A., Arlehamn C. S., and Sette A. (2017) T cells from patients with Parkinson's disease recognize α-synuclein peptides. Nature 546, 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chowell D., Morris L. G., Grigg C. M., Weber J. K., mstein R., Makarov V., Kuo F., Kendall S. M., Requena D., Riaz N., Greenbaum B., Carroll J., Garon E., Hyman D. M., Zehir A., Solit D., Berger M., Zhou R., Rizvi N. A., and Chan T. A. (2018) Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kvistborg P., and Yewdell J. W. (2018) Enhancing responses to cancer immunotherapy. Science 359, 516–517 [DOI] [PubMed] [Google Scholar]
- 50. Marty R., Kaabinejadian S., Rossell D., Slifker M. J., van de Haar J., Engin H., de Prisco N., Ideker T., Hildebrand W. H., Font-Burgada J., and Carter H. (2017) MHC-I genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pyke R., Thompson W., lem R., Font-Burgada J., Zanetti M., and Carter H. (2018) evolutionary Pressure against MHC class II binding cancer mutations. Cell 175, 416–428.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chick J. M., Munger S. C., Simecek P., Huttlin E. L., Choi K., Gatti D. M., Raghupathy N., Svenson K. L., Churchill G. A., and Gygi S. P. (2016) Defining the consequences of genetic variation on a proteome-wide scale. Nature 534, 500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu Y., Williams E. G., Dubuis S., Mottis A., Jovaisaite V., Houten S. M., Argmann C. A., Faridi P., Wolski W., Kutalik Z., Zamboni N., Auwerx J., and Aebersold R. (2014) Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell 158, 1415–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Williams E. G., Wu Y., Jha P., Dubuis S., Blattmann P., Argmann C. A., Houten S. M., Amariuta T., Wolski W., Zamboni N., Aebersold R., and Auwerx J. (2016) Systems proteomics of liver mitochondria function. Science 352, aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A. N., Knowles D. A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K., Björkegren J. L., Im H., Pasaniuc B., Rivas M. A., and Kundaje A. (2019) Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tscharke D. C., Croft N. P., Doherty P. C., and Gruta N. L. (2015) Sizing up the key determinants of the CD8(+) T cell response. Nat. Rev. Immunol. 15, 705–716 [DOI] [PubMed] [Google Scholar]
- 57. Istrail S., Florea L., Halldórsson B. V., Kohlbacher O., Schwartz R. S., Yap V., Yewdell J. W., and Hoffman S. L. (2004) Comparative immunopeptidomics of humans and their pathogens. Proc. Natl. Acad. Sci. U.S.A. 101, 13268–13272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nicholson J. K., Holmes E., and Elliott P. (2008) The metabolome-wide association study: a new look at human disease risk factors. J. Proteome Res. 7, 3637–3638 [DOI] [PubMed] [Google Scholar]
- 59. Holmes E., Loo R., Stamler J., Bictash M., Yap I. K., Chan Q., Ebbels T., Iorio M., Brown I. J., Veselkov K. A., Daviglus M. L., Kesteloot H., Ueshima H., Zhao L., Nicholson J. K., and Elliott P. (2008) Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okada H., Ebhardt A. H., Vonesch S., Aebersold R., and Hafen E. (2016) Proteome-wide association studies identify biochemical modules associated with a wing-size phenotype in Drosophila melanogaster. Nat. Commun. 7, 12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Butter F., Davison L., Viturawong T., Scheibe M., Vermeulen M., Todd J. A., and Mann M. (2012) Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. Plos Genet. 8, e1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shraibman B., Barnea E., Kadosh D., Haimovich Y., Slobodin G., Rosner I., López-Larrea C., Hilf N., Kuttruff S., Song C., Britten C., Castle J., Kreiter S., Frenzel K., Tatagiba M., Tabatabai G., Dietrich P.-Y., Dutoit V., Wick W., Platten M., Winkler F., von Deimling A., Kroep J., Sahuquillo J., Martinez-Ricarte F., Rodon J., Lassen U., Ottensmeier C., van der Burg S. H., Straten P., Poulsen H., Ponsati B., Okada H., Rammensee H.-G., Sahin U., Singh H., and Admon A. (2019) Identification of tumor antigens among the HLA peptidomes of glioblastoma tumors and plasma. Mol. Cell. Proteomics 18, 1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Purcell A. W., McCluskey J., and Rossjohn J. (2007) More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 6, nrd2224. [DOI] [PubMed] [Google Scholar]
- 64. Romero P., Banchereau J., Bhardwaj N., Cockett M., Disis M. L., Dranoff G., Gilboa E., Hammond S. A., Hershberg R., Korman A. J., Kvistborg P., Melief C., Mellman I., Palucka K. A., Redchenko I., Robins H., Sallusto F., Schenkelberg T., Schoenberger S., Sosman J., Türeci Ö., den Eynde B., Koff W., and Coukos G. (2016) The Human Vaccines Project: A roadmap for cancer vaccine development. Sci. Transl. Med. 8, 334ps9–334ps9 [DOI] [PubMed] [Google Scholar]
- 65. Sahin U., and Türeci Ö. (2018) Personalized vaccines for cancer immunotherapy. Science 359, 1355–1360 [DOI] [PubMed] [Google Scholar]
- 66. Hu Z., Ott P. A., and Wu C. J. (2018) Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 18, 168–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Koff W. C., Gust I. D., and Plotkin S. A. (2014) Toward a human vaccines project. Nat. Immunol. 15, 589–592 [DOI] [PubMed] [Google Scholar]
- 68. Schenkelberg T., Kieny M.-P., Bianco A., and Koff W. C. (2015) Building the Human Vaccines Project: strategic management recommendations and summary report of the 15–16 July 2014 business workshop. Expert Rev. Vaccines 14, 629–636 [DOI] [PubMed] [Google Scholar]
- 69. Sette A., Schenkelberg T. R., and Koff W. C. (2015) Deciphering the human antigenome. Expert Rev. Vaccines 15, 167–171 [DOI] [PubMed] [Google Scholar]
- 70. Koff W. C., Burton D. R., Johnson P. R., Walker B. D., King C. R., Nabel G. J., Ahmed R., Bhan M. K., and Plotkin S. A. (2013) Accelerating next-generation vaccine development for global disease prevention. Clin. Infect. Dis. 340, 1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Caron E., Kowalewski D. J., Koh C., Sturm T., Schuster H., and Aebersold R. (2015) Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes using mass spectrometry. Mol. Cell. Proteomics 14, 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Purcell A. W., Ramarathinam H., and Ternette N. (2019) Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 14, 1687–1707 [DOI] [PubMed] [Google Scholar]
- 73. Bassani-Sternberg M., and Coukos G. (2016) Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 41, 9–17 [DOI] [PubMed] [Google Scholar]
- 74. Laumont C. M., Vincent K., Hesnard L., Audemard É., Bonneil É., Laverdure J-P, Gendron P., Courcelles M., Hardy M.-P., Côté C., Durette C., St-Pierre C., Benhammadi M., Lanoix J., Vobecky S., Haddad E., Lemieux S., Thibault P., and Perreault C. (2018) Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 10, eaau5516. [DOI] [PubMed] [Google Scholar]
- 75. Ebrahimi-Nik H., Michaux J., Corwin W. L., Keller G., Shcheglova T., Pak H., Coukos G., Baker B. M., Mandoiu I. I., Bassani-Sternberg M., and vastava P. K. (2019) Mass spectrometry driven exploration reveals nuances of neoepitope-driven tumor rejection. JCI Insight 5, pii: 129152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bassani-Sternberg M., Bräunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., Martignoni M. E., Werner A., Hein R., Busch D. H., Peschel C., Rad R., Cox J., Mann M., and Krackhardt A. M. (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Marijt K. A., Blijleven L., Verdegaal E., Kester M. G., Kowalewski D. J., Rammensee H.-G., Stevanović S., Heemskerk M., van der Burg S. H., and van Hall T. (2018) Identification of non-mutated neoantigens presented by TAP-deficient tumors. J. Exp. Med. 215, 2325–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Havel J. J., Chowell D., and Chan T. A. (2019) The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ehx G., and Perreault C. (2019) Discovery and characterization of actionable tumor antigens. Genome Med. 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wilson E. A., and Anderson K. S. (2018) Lost in the crowd: identifying targetable MHC class I neoepitopes for cancer immunotherapy. Expert Rev. Proteomic 15, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 81. Ramarathinam H., Croft N. P., Illing P. T., Faridi P., and Purcell A. W. (2018) Employing proteomics in the study of antigen presentation: an update. Expert Rev. Proteomic 124, 637–645 [DOI] [PubMed] [Google Scholar]
- 82. Bräunlein E., and Krackhardt A. M. (2017) Tools to define the melanoma-associated immunopeptidome. Immunology 152, 536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Freudenmann L., Marcu A., and Stevanović S. (2018) Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 154, 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schmidt M., and Lill J. R. (2018) MHC class I presented antigens from malignancies: A perspective on analytical characterization & immunogenicity. J. Proteomics 191, 48–57 [DOI] [PubMed] [Google Scholar]
- 85. Granados D. P., Laumont C. M., Thibault P., and Perreault C. (2015) The nature of self for T cells—a systems-level perspective. Curr. Opin. Immunol. 34, 1–8 [DOI] [PubMed] [Google Scholar]
- 86. Laumont C. M., and Perreault C. (2018) Exploiting non-canonical translation to identify new targets for T cell-based cancer immunotherapy. Cell. Mol. Life Sci. 75, 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gfeller D., and Bassani-Sternberg M. (2018) Predicting antigen presentation-what could we learn from a million peptides? Front. Immunol. 9, 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Purcell A. W., Croft N. P., and Tscharke D. C. (2016) Immunology by numbers: quantitation of antigen presentation completes the quantitative milieu of systems immunology!. Curr. Opin. Immunol. 40, 88–95 [DOI] [PubMed] [Google Scholar]
- 89. Croft N. P., Purcell A. W., and Tscharke D. C. (2015) Quantifying epitope presentation using mass spectrometry. Mol. Immunol. 68, 1–4 [DOI] [PubMed] [Google Scholar]
- 90. Stern L. J., and Santambrogio L. (2016) The melting pot of the MHC II peptidome. Curr. Opin. Immunol. 40, 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Faridi P., Purcell A. W., and Croft N. (2018) in immunopeptidomics we need a sniper instead of a shotgun. Proteomics 18, 1700464. [DOI] [PubMed] [Google Scholar]
- 92. Alvarez B., Barra C., Nielsen M., and Andreatta M. (2018) Computational tools for the identification and interpretation of sequence motifs in immunopeptidomes. Proteomics 18, e1700252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Creech A. L., Ting Y. S., Goulding S. P., Sauld J. F., Barthelme D., Rooney M. S., Addona T. A., and Abelin J. G. (2018) The role of mass spectrometry and proteogenomics in the advancement of HLA epitope prediction. Proteomics 18, e1700259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hunt D., Henderson R., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A., Appella E., and Engelhard V. H. (1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 [DOI] [PubMed] [Google Scholar]
- 95. Rötzschke O., Falk K., Deres K., Schild H., Norda M., Metzger J., Jung G., and Rammensee H. (1990) Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 348, 252–254 [DOI] [PubMed] [Google Scholar]
- 96. Joyce S., and Nathenson S. G. (1994) Methods to study peptides associated with MHC class I molecules. Curr. Opin. Immunol. 6, 24–31 [DOI] [PubMed] [Google Scholar]
- 97. Engelhard V. H. (2007) The contributions of mass spectrometry to understanding of immune recognition by T lymphocytes. Int. J. Mass. Spectrom. 259, 32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Engelhard V. H., Appella E., Benjamin D. C., Bodnar W. M., Cox A. L., Chen Y., Henderson R. A., Huczko E. L., Michel H., Sakaguichi K., Shabanowitz J., Sevilir N., Slingluff C. L., and Hunt D. F. (1993) Mass spectrometric analysis of peptides associated with the human class I MHC molecules HLA-A2.1 and HLA-B7 and identification of structural features that determine binding. Chem. Immunol. Allergy 57, 39–62 [PubMed] [Google Scholar]
- 99. Huczko E., Bodnar W., Benjamin D., Sakaguchi K., Zhu N., Shabanowitz J., Henderson R., Appella E., Hunt D., and Engelhard V. H. (1993) Characteristics of endogenous peptides eluted from the class I MHC molecule HLA-B7 determined by mass spectrometry and computer modeling. J. Immunol. 151, 2572–2587 [PubMed] [Google Scholar]
- 100. Henderson R., Cox A., Sakaguchi K., Appella E., Shabanowitz J., Hunt D., and Engelhard V. (1993) Direct identification of an endogenous peptide recognized by multiple HLA-A2.1-specific cytotoxic T cells. Proc. Natl. Acad. Sci. U.S.A. 90, 10275–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Engelhard V. H. (1994) Structure of Peptides Associated with Class I and Class II MHC Molecules. Annu. Rev. Immunol. 12, 181–207 [DOI] [PubMed] [Google Scholar]
- 102. Henderson R., Michel H., Sakaguchi K., Shabanowitz J., Appella E., Hunt D., and Engelhard V. (1992) HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science 255, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 103. Dubey P., Hendrickson R. C., Meredith S. C., Siegel C. T., Shabanowitz J., Skipper J., Engelhard V. H., Hunt D. F., and Schreiber H. (1997) The immunodominant antigen of an ultraviolet-induced regressor tumor is generated by a somatic point mutation in the DEAD box helicase p68. J. Exp. Med. 185, 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zarling A. L., Polefrone J. M., Evans A. M., Mikesh L. M., Shabanowitz J., Lewis S. T., Engelhard V. H., and Hunt D. F. (2006) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 103, 14889 14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mohammed F., Cobbold M., Zarling A. L., Salim M., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Engelhard V. H., and Willcox B. E. (2008) Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat. Immunol. 9, 1236–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gubin M. M., Zhang X., Schuster H., Caron E., Ward J. P., Noguchi T., Ivanova Y., Hundal J., Arthur C. D., Krebber W.-J., Mulder G. E., Toebes M., Vesely M. D., Lam S. S., Korman A. J., Allison J. P., Freeman G. J., Sharpe A. H., Pearce E. L., Schumacher T. N., Aebersold R., Rammensee H.-G., Melief C. J., Mardis E. R., Gillanders W. E., Artyomov M. N., and Schreiber R. D. (2014) Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yadav M., Jhunjhunwala S., Phung Q. T., Lupardus P., Tanguay J., Bumbaca S., Franci C., Cheung T. K., Fritsche J., Weinschenk T., Modrusan Z., Mellman I., Lill J. R., and Delamarre L. (2015) Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 [DOI] [PubMed] [Google Scholar]
- 108. Carreno B. M., Magrini V., Becker-Hapak M., Kaabinejadian S., Hundal J., Petti A. A., Ly A., Lie W.-R., Hildebrand W. H., Mardis E. R., and Linette G. P. (2015) A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pymm P., Illing P. T., Ramarathinam H., O'Connor G. M., Hughes V. A., Hitchen C., Price D. A., Ho B. K., McVicar D. W., Brooks A. G., Purcell A. W., Rossjohn J., and Vivian J. P. (2017) MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat. Struct. Mol. Biol. 24, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. McMurtrey C., Trolle T., Sansom T., Remesh S. G., Kaever T., Bardet W., Jackson K., McLeod R., Sette A., Nielsen M., Zajonc D. M., Blader I. J., Peters B., and Hildebrand W. (2016) Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove. Elife 5, e12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guillaume P., Picaud S., Baumgaertner P., Montandon N., Schmidt J., Speiser D. E., Coukos G., Bassani-Sternberg M., Filippakopoulos P., and Gfeller D. (2018) The C-terminal extension landscape of naturally presented HLA-I ligands. Proc. Natl. Acad. Sci. USA 115, 5083–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Alpízar A., Marino F., Ramos-Fernandez A., Lombardía M., Jeko A., Pazos F., Paradela A., Santiago C., Heck A. J., and Marcilla M. (2017) A molecular basis for the presentation of phosphorylated peptides by HLA-B antigens. Mol. Cell. Proteomics 16, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Marcilla M., Alpízar A., Lombardía M., Ramos-Fernandez A., Ramos M., and Albar J. (2014) Increased diversity of the HLA-B40 ligandome by the presentation of peptides phosphorylated at their main anchor residue. Mol. Cell. Proteomics 13, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Abelin J. G., Trantham P. D., Penny S. A., Patterson A. M., Ward S. T., Hildebrand W. H., Cobbold M., Bai D. L., Shabanowitz J., and Hunt D. F. (2015) Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat. Protoc. 10, 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cobbold M., Peña H., Norris A., Polefrone J. M., Qian J., English A., Cummings K. L., Penny S., Turner J. E., Cottine J., Abelin J. G., Malaker S. A., Zarling A. L., Huang H.-W., Goodyear O., Freeman S. D., Shabanowitz J., Pratt G., Craddock C., Williams M. E., Hunt D. F., and Engelhard V. H. (2013) MHC class I–associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci. Transl. Med. 5, 203ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Marino F., Mommen G. P., Jeko A., Meiring H. D., van den Brink J. A., Scheltema R. A., van Els C. A., and Heck A. J. (2016) Arginine (di)methylated human leukocyte antigen class I peptides are favorably presented by HLA-B*07. J. Proteome Res. 16, 34–44 [DOI] [PubMed] [Google Scholar]
- 117. Sidney J., Becart S., Zhou M., Duffy K., Lindvall M., Moore E. C., Moore E. L., Rao T., Rao N., Nielsen M., Peters B., and Sette A. (2017) Citrullination only infrequently impacts peptide binding to HLA class II MHC. Plos One 12, e0177140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Molberg Ø., Mcadam S. N., Körner R., Quarsten H., Kristiansen C., Madsen L., Fugger L., Scott H., Norén O., Roepstorff P., Lundin K., Sjöström H., and Sollid L. M. (1998) Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4, 713–717 [DOI] [PubMed] [Google Scholar]
- 119. Bodd M., Kim C., Lundin K., and Sollid L. M. (2012) T-cell response to gluten in patients with HLA-DQ2.2 reveals requirement of peptide-MHC stability in celiac disease. Gastroenterology 142, 552–561 [DOI] [PubMed] [Google Scholar]
- 120. Fallang L.-E., Bergseng E., Hotta K., Berg-Larsen A., Kim C.-Y., and llid L. (2009) Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat. Immunol. 10, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 121. Hovhannisyan Z., Weiss A., Martin A., Wiesner M., Tollefsen S., Yoshida K., Ciszewski C., Curran S. A., Murray J. A., David C. S., Sollid L. M., Koning F., Teyton L., and Jabri B. (2008) The role of HLA-DQ8 β57 polymorphism in the anti-gluten T-cell response in coeliac disease. Nature 456, 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ramarathinam H., Gras S., Alcantara S., Yeung A., Mifsud N. A., Sonza S., Illing P. T., Glaros E. N., Center R. J., Thomas S. R., Kent S. J., Ternette N., Purcell D. F., Rossjohn J., and Purcell A. W. (2018) Identification of native and posttranslationally modified HLA-B*57:01-restricted HIV envelope derived epitopes using immunoproteomics. Proteomics 18, 1700253. [DOI] [PubMed] [Google Scholar]
- 123. Malaker S. A., Ferracane M. J., Depontieu F. R., Zarling A. L., Shabanowitz J., Bai D. L., Topalian S. L., Engelhard V. H., and Hunt D. F. (2016) Identification and characterization of complex glycosylated peptides presented by the MHC class II processing pathway in melanoma. J. Proteome Res. 16, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Dengjel J., Rammensee H., and Stevanovic S. (2005) Glycan side chains on naturally presented MHC class II ligands. J. Mass Spectrom. 40, 100–104 [DOI] [PubMed] [Google Scholar]
- 125. Marino F., Bern M., Mommen G. P., Leney A. C., van den Brink J. A., Bonvin A. M., Becker C., van Els C. A., and Heck A. J. (2015) Extended O-GlcNAc on HLA class-I-bound peptides. J. Am. Chem. Soc. 137, 10922–10925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Faridi P., Li C., Ramarathinam H., Vivian J. P., Illing P. T., Mifsud N. A., Ayala R., Song J., Gearing L. J., Hertzog P. J., Ternette N., Rossjohn J., Croft N. P., and Purcell A. W. (2018) A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, eaar3947. [DOI] [PubMed] [Google Scholar]
- 127. Liepe J., Sidney J., Lorenz F. K., Sette A., and Mishto M. (2018) Mapping the MHC class I spliced immunopeptidome of cancer cells. Cancer Immunol. Res. 7, 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Mishto M., and Liepe J. (2017) Post-translational peptide splicing and T cell responses. Trends Immunol. 38, 904–915 [DOI] [PubMed] [Google Scholar]
- 129. Liepe J., Marino F., Sidney J., Jeko A., Bunting D. E., Sette A., Kloetzel P. M., Stumpf M. P., Heck A. J., and Mishto M. (2016) A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 [DOI] [PubMed] [Google Scholar]
- 130. Mylonas R., Beer I., Iseli C., Chong C., Pak H.-S., Gfeller D., Coukos G., Xenarios I., Müller M., and Bassani-Sternberg M. (2018) Estimating the contribution of proteasomal spliced peptides to the HLA-I ligandome. Mol. Cell. Proteomics 17, 2347–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., and Mann M. (2005) Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 132. Hickman H. D., Luis A. D., Buchli R., Few S. R., Sathiamurthy M., VanGundy R. S., Giberson C. F., and Hildebrand W. H. (2004) Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J. Immunol. 172, 2944–2952 [DOI] [PubMed] [Google Scholar]
- 133. Andreatta M., Nicastri A., Peng X., Hancock G., Dorrell L., Ternette N., and Nielsen M. (2019) MS-rescue: a computational pipeline to increase the quality and yield of immunopeptidomics experiments. Proteomics 19, 1800357. [DOI] [PubMed] [Google Scholar]
- 134. Nielsen M., Lundegaard C., Blicher T., Lamberth K., Harndahl M., Justesen S., Røder G., Peters B., Sette A., Lund O., and Buus S. (2007) NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. Plos One 2, e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Dhanda S., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M., Jurtz V., Andreatta M., Greenbaum J. A., Marcatili P., Sette A., Nielsen M., and Peters B. (2019) IEDB-AR: immune epitope database—analysis resource in 2019. Nucleic Acids Res., 47, W502–W506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. O'Donnell T. J., Rubinsteyn A., Bonsack M., Riemer A. B., Laserson U., and Hammerbacher J. (2018) MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 7, 129–132.e4 [DOI] [PubMed] [Google Scholar]
- 137. Couzin-Frankel J. (2013) Breakthrough of the year 2013. Cancer immunotherapy. Science 342, 1432–1433 [DOI] [PubMed] [Google Scholar]
- 138. Ribas A., and Wolchok J. D. (2018) Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Schuster H., Peper J. K., Bösmüller H.-C., Röhle K., Backert L., Bilich T., Ney B., Löffler M. W., Kowalewski D. J., Trautwein N., Rabsteyn A., Engler T., Braun S., Haen S. P., Walz J. S., Schmid-Horch B., Brucker S. Y., Wallwiener D., Kohlbacher O., Fend F., Rammensee H.-G., Stevanovic S., Staebler A., and Wagner P. (2017) The immunopeptidomic landscape of ovarian carcinomas. Proc. Natl. Acad. Sci. U.S.A. 114, E9942–E9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hilf N., Kuttruff-Coqui S., Frenzel K., Bukur V., x00107 S., cile Gouttefangeas C., Platten M., Tabatabai G., Dutoit V., Burg S. H., thor Straten P., nez Ricarte -F., Ponsati B., Okada H., Lassen U., Admon A., Ottensmeier C. H., Ulges A., Kreiter S., Deimling A., Skardelly M., Migliorini D., Kroep J. R., Idorn M., Rodon J., x000F3 J., Poulsen H. S., Shraibman B., McCann K., Mendrzyk R., wer M., Stieglbauer M., Britten C. M., Capper D., Welters M. J., Sahuquillo J., Kiesel K., Derhovanessian E., Rusch E., Bunse L., Song C., Heesch S., Wagner C., ck A., rg Ludwig J., Castle J. C., Schoor O., Tadmor A. D., Green E., Fritsche J., Meyer M., Pawlowski N., Dorner S., Hoffgaard F., ssler B., Maurer D., Weinschenk T., Reinhardt C., Huber C., Rammensee H.-G., Singh-Jasuja H., Sahin U., Dietrich P.-Y., and Wick W. (2018) Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 [DOI] [PubMed] [Google Scholar]
- 141. Consortium H., Löffler M. W., Mohr C., Bichmann L., Freudenmann L., Walzer M., hroeder C., Trautwein N., Hilke F. J., Zinser R. S., Mühlenbruch L., Kowalewski D. J., Schuster H., Sturm M., Matthes J., Riess O., Czemmel S., Nahnsen S., Königsrainer I., Thiel K., Nadalin S., Beckert S., Bösmüller H., Fend F., Velic A., Maček B., Haen S. P., Buonaguro L., Kohlbacher O., Stevanović S., Königsrainer A., and Rammensee H.-G. (2019) Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Le D. T., Durham J. N., Smith K. N., Wang H., Bartlett B. R., Aulakh L. K., Lu S., Kemberling H., Wilt C., Luber B. S., Wong F., Azad N. S., Rucki A. A., Laheru D., Donehower R., Zaheer A., Fisher G. A., Crocenzi T. S., Lee J. J., Greten T. F., Duffy A. G., Ciombor K. K., Eyring A. D., Lam B. H., Joe A., Kang P. S., Holdhoff M., Danilova L., Cope L., Meyer C., Zhou S., Goldberg R. M., Armstrong D. K., Bever K. M., Fader A. N., Taube J., Housseau F., Spetzler D., Xiao N., Pardoll D. M., Papadopoulos N., Kinzler K. W., Eshleman J. R., Vogelstein B., Anders R. A., and Diaz L. A. (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Le D. T., Uram J. N., Wang H., Bartlett B. R., Kemberling H., Eyring A. D., Skora A. D., Luber B. S., Azad N. S., Laheru D., Biedrzycki B., Donehower R. C., Zaheer A., Fisher G. A., Crocenzi T. S., Lee J. J., Duffy S. M., Goldberg R. M., de la Chapelle A., Koshiji M., Bhaijee F., Huebner T., Hruban R. H., Wood L. D., Cuka N., Pardoll D. M., Papadopoulos N., Kinzler K. W., Zhou S., Cornish T. C., Taube J. M., Anders R. A., Eshleman J. R., Vogelstein B., and Diaz L. A. (2015) PD-1 blockade in tumors with mismatch-repair deficiency. New Engl. J. Med. 372, 2509–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Samstein R. M., Lee C.-H., Shoushtari A. N., Hellmann M. D., Shen R., Janjigian Y. Y., Barron D. A., Zehir A., Jordan E. J., Omuro A., Kaley T. J., Kendall S. M., Motzer R. J., Hakimi A. A., Voss M. H., Russo P., Rosenberg J., Iyer G., Bochner B. H., Bajorin D. F., Al-Ahmadie H. A., Chaft J. E., Rudin C. M., Riely G. J., Baxi S., Ho A. L., Wong R. J., Pfister D. G., Wolchok J. D., Barker C. A., Gutin P. H., Brennan C. W., Tabar V., Mellinghoff I. K., DeAngelis L. M., Ariyan C. E., Lee N., Tap W. D., Gounder M. M., D'Angelo S. P., Saltz L., Stadler Z. K., Scher H. I., Baselga J., Razavi P., Klebanoff C. A., Yaeger R., Segal N. H., Ku G. Y., DeMatteo R. P., Ladanyi M., Rizvi N. A., Berger M. F., Riaz N., Solit D. B., Chan T. A., and Morris L. G. (2019) Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. June C. H., O'Connor R. S., Kawalekar O. U., Ghassemi S., and Milone M. C. (2018) CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 [DOI] [PubMed] [Google Scholar]
- 146. Kelly P. N. (2018) The cancer immunotherapy revolution. Science 359, 1344–1345 [DOI] [PubMed] [Google Scholar]
- 147. Bouchie A., and DeFrancesco L. (2016) Nature Biotechnology's academic spinouts of 2015. Nat. Biotechnol. 34, 484. [DOI] [PubMed] [Google Scholar]
- 148. Ternette N., and Purcell A. W. (2018) Immunopeptidomics special issue. Proteomics 18, 1800145. [DOI] [PubMed] [Google Scholar]
- 149. Liu Y., Mi Y., Mueller T., Kreibich S., Williams E. G., Drogen A., Borel C., Frank M., Germain P.-L., Bludau I., Mehnert M., Seifert M., Emmenlauer M., Sorg I., Bezrukov F., Bena F., Zhou H., Dehio C., Testa G., Saez-Rodriguez J., Antonarakis S. E., Hardt W.-D., and Aebersold R. (2019) Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 37, 314–322 [DOI] [PubMed] [Google Scholar]
- 150. Lanoix J., Durette C., Courcelles M., Cossette É., Comtois-Marotte S., Hardy M.-P., Côté C., Perreault C., and Thibault P. (2018) Comparison of the MHC I immunopeptidome repertoire of B-cell lymphoblasts using two isolation methods. Proteomics 18, e1700251. [DOI] [PubMed] [Google Scholar]
- 151. Bassani-Sternberg M. (2018) Mass spectrometry based immunopeptidomics for the discovery of cancer neoantigens. Methods Mol. Biol. 1719, 209–221 [DOI] [PubMed] [Google Scholar]
- 152. Marino F., Chong C., Michaux J., and Bassani-Sternberg M. (2019) High-throughput, fast, and sensitive immunopeptidomics sample processing for mass spectrometry. Methods Mol. Biol. 1913, 67–79 [DOI] [PubMed] [Google Scholar]
- 153. Nelde A., Kowalewski D. J., and Stevanović S. (2019) Antigen processing, methods and protocols. Methods Mol. Biol. 1988, 123–136 [DOI] [PubMed] [Google Scholar]
- 154. Hassan C., Kester M., Oudgenoeg G., de Ru A. H., Janssen G., Drijfhout J. W., Spaapen R. M., Jiménez C. R., Heemskerk M., Falkenburg J. H., and van Veelen P. A. (2014) Accurate quantitation of MHC-bound peptides by application of isotopically labeled peptide MHC complexes. J. Proteomics 109, 240–244 [DOI] [PubMed] [Google Scholar]
- 155. Bijen H. M., Hassan C., Kester M. G., Janssen G. M., Hombrink P., Ru A. H., Drijfhout J., Meiring H. D., Jong A. P., Falkenburg F. J., Jimenez C. R., Heemskerk M. H., and Veelen P. A. (2018) Specific T Cell Responses against minor histocompatibility antigens cannot generally be explained by absence of their allelic counterparts on the cell surface. Proteomics 18, 1700250. [DOI] [PubMed] [Google Scholar]
- 156. Demmers L. C., Heck A. J., and Wu W. (2019) Pre-fractionation extends, but also creates a bias in the detectable HLA class Ι ligandome. J. Proteome Res. 18, 1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Seale B., Lam C., Rackus D. G., Chamberlain M. D., Liu C., and Wheeler A. R. (2016) Digital microfluidics for immunoprecipitation. Anal. Chem. 88, 10223–10230 [DOI] [PubMed] [Google Scholar]
- 158. Sinclair I., Bachman M., Addison D., Rohman M., Murray D. C., Davies G., Mouchet E., Tonge M. E., Stearns R. G., Ghislain L., Datwani S. S., Majlof L., Hall E., Jones G. R., Hoyes E., Olechno J., Ellson R. N., Barran P. E., Pringle S. D., Morris M. R., and Wingfield J. (2019) Acoustic mist ionization platform for direct and contactless ultrahigh-throughput mass spectrometry analysis of liquid samples. Anal. Chem. 91, 3790–3794 [DOI] [PubMed] [Google Scholar]
- 159. Zhang H. (2017) Acoustic dispensing-mass spectrometry: the next high throughput bioanalytical platform for early drug discovery. Bioanalysis 9, 1619–1621 [DOI] [PubMed] [Google Scholar]
- 160. Hughes C. S., Foehr S., Garfield D. A., Furlong E. E., einmetz L., and Krijgsveld J. (2014) Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Kanshin E., and Thibault P. (2014) Efficient sample processing for proteomics applications—Are we there yet? Mol. Syst. Biol. 10, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Iacobucci C., Piotrowski C., Aebersold R., Amaral B. C., Andrews P., Bernfur K., Borchers C., Brodie N. I., Bruce J. E., Cao Y., Chaignepain S., Chavez J. D., Claverol S., Cox J., Davis T., Degliesposti G., Dong M.-Q., Edinger N., Emanuelsson C., Gay M., Götze M., Gomes-Neto F., Gozzo F. C., Gutierrez C., Haupt C., Heck A. J., Herzog F., Huang L., Hoopmann M. R., Kalisman N., Klykov O., Kukačka Z., Liu F., MacCoss M. J., Mechtler K., Mesika R., Moritz R. L., Nagaraj N., Nesati V., Neves-Ferreira A. G., Ninnis R., Novák P., O'Reilly F. J., Pelzing M., Petrotchenko E., Piersimoni L., Plasencia M., Pukala T., Rand K. D., Rappsilber J., Reichmann D., Sailer C., Sarnowski C. P., Scheltema R. A., Schmidt C., Schriemer D. C., Shi Y., Skehel M. J., Slavin M., Sobott F., Solis-Mezarino V., Stephanowitz H., Stengel F., Stieger C. E., Trabjerg E., Trnka M., Vilaseca M., Viner R., Xiang Y., Yilmaz S., Zelter A., Ziemianowicz D., Leitner A., and Sinz A. (2019) First community-wide, comparative cross-linking mass spectrometry study. Anal. Chem. 91, 6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A.-J. L., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander Å., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson L. N., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., and Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring–based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Bell A. W., Deutsch E. W., Au C. E., Kearney R. E., Beavis R., Sechi S., Nilsson T., Bergeron J. J., Beardslee T. A., Chappell T., Meredith G., Sheffield P., Gray P., Hajivandi M., Pope M., Predki P., Kullolli M., Hincapie M., Hancock W. S., Jia W., Song L., Li L., Wei J., Yang B., Wang J., Ying W., Zhang Y., Cai Y., Qian X., He F., Meyer H. E., Stephan C., Eisenacher M., Marcus K., Langenfeld E., May C., Carr S. A., Ahmad R., Zhu W., Smith J. W., Hanash S. M., Struthers J. J., Wang H., Zhang Q., An Y., Goldman R., Carlsohn E., van der Post S., Hung K. E., Sarracino D. A., Parker K., Krastins B., Kucherlapati R., Bourassa S., Poirier G. G., Kapp E., Patsiouras H., Moritz R., Simpson R., Houle B., LaBoissiere S., Metalnikov P., Nguyen V., Pawson T., Wong C. C., Cociorva D. III, J. R., Ellison M. J., Lopez-Campistrous A., Semchuk P., Wang Y., Ping P., Elia G., Dunn M. J., Wynne K., Walker A. K., Strahler J. R., Andrews P. C., Hood B. L., Bigbee W. L., Conrads T. P., Smith D., Borchers C. H., Lajoie G. A., Bendall S. C., Speicher K. D., Speicher D. W., Fujimoto M., Nakamura K., Paik Y.-K., Cho S., Kwon M.-S., Lee H.-J., Jeong S.-K., Chung A., Miller C. A., Grimm R., Williams K., Dorschel C., Falkner J. A., Martens L., and Vizcaíno J. (2009) A HUPO test sample study reveals common problems in mass spectrometry–based proteomics. Nat. Methods 6, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., and Enard W. (2017) Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4 [DOI] [PubMed] [Google Scholar]
- 166. Svensson V., Natarajan K., Ly L.-H., Miragaia R. J., Labalette C., Macaulay I. C., Cvejic A., and Teichmann S. A. (2017) Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Collins B. C., Hunter C. L., Liu Y., Schilling B., Rosenberger G., Bader S. L., Chan D. W., Gibson B. W., Gingras A.-C., Held J. M., Hirayama-Kurogi M., Hou G., Krisp C., Larsen B., Lin L., Liu S., Molloy M. P., Moritz R. L., Ohtsuki S., Schlapbach R., Selevsek N., Thomas S. N., Tzeng S.-C., Zhang H., and Aebersold R. (2017) Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 8, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Goodwin S., McPherson J. D., and McCombie R. W. (2016) Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Swaminathan J., Boulgakov A. A., Hernandez E. T., Bardo A. M., Bachman J. L., Marotta J., Johnson A. M., Anslyn E. V., and Marcotte E. M. (2018) Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 36, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Collins B. C., and Aebersold R. (2018) Proteomics goes parallel. Nat. Biotechnol. 36, 1051. [DOI] [PubMed] [Google Scholar]
- 171. Tang L. (2018) Next-generation peptide sequencing. Nat. Methods 15, 997. [DOI] [PubMed] [Google Scholar]
- 172. Swaminathan J., Boulgakov A. A., and Marcotte E. M. (2015) A theoretical justification for single molecule peptide sequencing. Plos Comput. Biol. 11, e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Platteel A., Liepe J., Textoris-Taube K., Keller C., Henklein P., Schalkwijk H. H., Cardoso R., Kloetzel P. M., Mishto M., and Sijts A. (2017) Multi-level strategy for identifying proteasome-catalyzed spliced epitopes targeted by CD8+ T cells during bacterial infection. Cell Rep. 20, 1242–1253 [DOI] [PubMed] [Google Scholar]
- 174. Delong T., Wiles T. A., Baker R. L., Bradley B., Barbour G., Reisdorph R., Armstrong M., Powell R. L., Reisdorph N., Kumar N., Elso C. M., DeNicola M., Bottino R., Powers A. C., Harlan D. M., Kent S. C., Mannering S. I., and Haskins K. (2016) Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Rolfs Z., Müller M., Shortreed M. R., Smith L. M., and Bassani-Sternberg M. (2019) Comment on “A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.” Sci. Immunol. 4, eaaw1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Faridi P., Li C., Ramarathinam H., Illing P. T., Mifsud N. A., Ayala R., Song J., Gearing L. J., Croft N. P., and Purcell A. W. (2019) Response to Comment on “A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.” Sci. Immunol. 4, eaaw8457. [DOI] [PubMed] [Google Scholar]
- 177. The M., MacCoss M. J., Noble W. S., and Käll L. (2016) Fast and accurate protein false discovery rates on large-scale proteomics data sets with Percolator 3.0. J. Am. Soc. Mass Spectr. 27, 1719–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Devabhaktuni A., and Elias J. E. (2016) Application of de Novo Sequencing to Large-Scale Complex Proteomics Data Sets. J. Proteome Res. 15, 732–742 [DOI] [PubMed] [Google Scholar]
- 179. Griss J. (2016) Spectral library searching in proteomics. Proteomics 16, 729–740 [DOI] [PubMed] [Google Scholar]
- 180. Kong A. T., Leprevost F. V., Avtonomov D. M., Mellacheruvu D., and Nesvizhskii A. I. (2017) MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Bittremieux W., Meysman P., Noble W., and Laukens K. (2018) Fast open modification spectral library searching through approximate nearest neighbor indexing. J. Proteome Res. 17, 3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Devabhaktuni A., Lin S., Zhang L., Swaminathan K., Gonzalez C. G., Olsson N., Pearlman S. M., Rawson K., and Elias J. E. (2019) TagGraph reveals vast protein modification landscapes from large tandem mass spectrometry datasets. Nat. Biotechnol. 37, 469–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Griss J., Perez-Riverol Y., Lewis S., Tabb D. L., Dianes J. A., del-Toro N., Rurik M., Walzer M., Kohlbacher O., Hermjakob H., Wang R., and Vizcaíno J. (2016) Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nat. Methods 13, 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Griss J., Foster J. M., Hermjakob H., and Vizcaíno J. (2013) PRIDE Cluster: building a consensus of proteomics data. Nat. Methods 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Perez-Riverol Y., Vizcaíno J., and Griss J. (2018) Future prospects of spectral clustering approaches in proteomics. Proteomics 18, e1700454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J.-P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T., Halim V. A., Bagshaw R. D., Hubner N. C., al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z.-Y., Badillo B., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A.-C., and Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Röst H. L. (2019) Deep learning adds an extra dimension to peptide fragmentation. Nat. Methods 16, 469. [DOI] [PubMed] [Google Scholar]
- 188. Gessulat S., Schmidt T., Zolg D., Samaras P., Schnatbaum K., Zerweck J., Knaute T., Rechenberger J., Delanghe B., Huhmer A., Reimer U., Ehrlich H.-C., Aiche S., Kuster B., and Wilhelm M. (2019) Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 16, 509–518 [DOI] [PubMed] [Google Scholar]
- 189. Tiwary S., Levy R., Gutenbrunner P., Soto F., Palaniappan K. K., Deming L., Berndl M., Brant A., Cimermancic P., and Cox J. (2019) High-quality MS/MS spectrum prediction for data-dependent and data-independent acquisition data analysis. Nat. Methods 16, 519–525 [DOI] [PubMed] [Google Scholar]
- 190. Tran N., Qiao R., Xin L., Chen X., Liu C., Zhang X., Shan B., Ghodsi A., and Li M. (2018) Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat. Methods 16, 63–66 [DOI] [PubMed] [Google Scholar]
- 191. Bulik-Sullivan B., Busby J., Palmer C. D., Davis M. J., Murphy T., Clark A., Busby M., Duke F., Yang A., Young L., Ojo N. C., Caldwell K., Abhyankar J., Boucher T., Hart M. G., Makarov V., Montpreville V., Mercier O., Chan T. A., Scagliotti G., Bironzo P., Novello S., Karachaliou N., Rosell R., Anderson I., Gabrail N., Hrom J., Limvarapuss C., Choquette K., Spira A., Rousseau R., Voong C., Rizvi N. A., Fadel E., Frattini M., Jooss K., Skoberne M., Francis J., and Yelensky R. (2018) Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol. 37, 1–17 [DOI] [PubMed] [Google Scholar]
- 192. Cappadona S., Baker P. R., Cutillas P. R., Heck A. J., and van Breukelen B. (2012) Current challenges in software solutions for mass spectrometry-based quantitative proteomics. Amino Acids 43, 1087–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Papatheodorou I., Fonseca N. A., Keays M., Tang A. Y., Barrera E., Bazant W., Burke M., Füllgrabe A., Fuentes A., George N., Huerta L., Koskinen S., Mohammed S., Geniza M., Preece J., Jaiswal P., Jarnuczak A. F., Huber W., Stegle O., Vizcaíno J., Brazma A., and Petryszak R. (2018) Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 46, D246–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Wilhelm M., Schlegl J., Hahne H., Gholami A., Lieberenz M., vitski M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J.-H., Bantscheff M., Gerstmair A., Faerber F., and Kuster B. (2015) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 195. Uhlen M., Fagerberg L., Hallstrom B., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C., Odeberg J., Djureinovic D., Takanen J., Hober S., Alm T., Edqvist P., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., hwenk J., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Ponten F. (2015) Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 196. Rosenberger G., Koh C., Guo T., Röst H. L., Kouvonen P., Collins B. C., Heusel M., Liu Y., Caron E., Vichalkovski A., Faini M., Schubert O. T., Faridi P., Ebhardt A. H., Matondo M., Lam H., Bader S. L., Campbell D. S., Deutsch E. W., Moritz R. L., Tate S., and Aebersold R. (2014) A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci. Data 1, 140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Regev A., Teichmann S. A., Lander E. S., Amt I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., Clevers H., Deplancke B., Dunham I., Eberwine J., Elis R., Enard W., Farmer A., Fugger L., Gottgens B., Hacohen N., Haniffa M., Hemberg M., Kim S., Klenerman P., Kriegstein A., Lein E., Linnarsson S., Lundberg E., Lundeberg J., Majumder P., Marioni J. C., Merad M., Mhlanga M., Nawijin M., Netea M., Nolan G., Pe'er D., Phillipakis A., Ponting C. P., Quake S., Reik W., Rozenblatt-Rosen O., Sanes J., Satija R., Schumacher T. N., Shalek A., Shapiro E., Sharma P., Shin J. W., Stegle O., Stratton M., Stubbington M. J., Theis F. J., Uhlen M., van Oudenaarden A., Wagner A., Watt F., Weissman J., Wold B., Xavier R., Yosef N., and MEETING, H. (2017) The human cell atlas. Elife 6, e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Rozenblatt-Rosen O., Stubbington M. J., Regev A., and Teichmann S. A. (2017) The human cell atlas: from vision to reality. Nat. News 550, 451–453 [DOI] [PubMed] [Google Scholar]
- 199. Shao W., Pedrioli P. G. A., Wolski W., Scurtescu C., Schmid E., Vizcaíno J. A., Courcelles M., Schuster H., Kowalewski D., Marino F., Arlehamn C. S. L., Vaughan K., Peters B., Sette A., Ottenhoff T. H. M., Meijgaarden K. E., Nieuwenhuizen N., Kaufmann S. H. E., Schlapbach R., Castle J. C., Nesvizhskii A. I., Nielsen M., Deutsch E. W., Campbell D. S., Moritz R. L., Zubarev R. A., Ytterberg A. J., Purcell A. W., Marcilla M., Paradela A., Wang Q., Costello C. E., Ternette N., van Veelen P. A., van Els C. A. C. M., Heck A. J. R., de Souza G. A., Sollid L. M., Admon A., Stevanovic S., Rammensee H. G., Thibault P., Perreault C., Bassani-Sternberg M., Aebersold R., and Caron E. (2018) The SysteMHC Atlas project. Nucleic Acids Res. 46, D1237–D1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., and Vizcaíno J. (2018) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res., 47, D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P.-A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H.-J., Albar J., Martínez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Deutsch E., Csordas A., and Sun Z. (2016) The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Lill J. R., van Veelen P. A., Tenzer S., Admon A., Caron E., Elias J. E., Heck A. J., Marcilla M., Marino F., Müller M., Peters B., Purcell A., Sette A., Sturm T., Ternette N., Vizcaíno J., and Bassani-Sternberg M. (2018) Minimal information about an immuno-peptidomics experiment (MIAIPE). Proteomics 18, e1800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Karosiene E., Lundegaard C., Lund O., and Nielsen M. (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 [DOI] [PubMed] [Google Scholar]
- 205. Shteynberg D., Nesvizhskii A., Moritz R., and Deutsch E. (2013) Combining Results of Multiple Search Engines in Proteomics. Mol. Cell. Proteomics 12, 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Shteynberg D., Deutsch E., Lam H., Eng J., Sun Z., Tasman N., Mendoza L., Moritz R., Aebersold R., and Nesvizhskii A. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., Stein S. E., and Aebersold R. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Asquith M., Sternes P. R., Costello M., Karstens L., Diamond S., Martin T. M., Li Z., Marshall M. S., Spector T. D., le Cao K., Rosenbaum J. T., and Brown M. A. (2019) HLA alleles associated with risk of ankylosing spondylitis and rheumatoid arthritis influence the gut microbiome. Arthritis Rheumatol. 71, 1642–1650 [DOI] [PubMed] [Google Scholar]
- 209. Caron E., Espona L., Kowalewski D. J., Schuster H., Ternette N., Alpízar A., Schittenhelm R. B., Ramarathinam H., Arlehamn C. S., Koh C., Gillet L. C., Rabsteyn A., Navarro P., Kim S., Lam H., Sturm T., Marcilla M., Sette A., Campbell D. S., Deutsch E. W., Moritz R. L., Purcell A. W., Rammensee H.-G., Stevanovic S., and Aebersold R. (2015) An open-source computational and data resource to analyze digital maps of immunopeptidomes. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Ritz D., Kinzi J., Neri D., and Fugmann T. (2017) Data-independent acquisition of HLA class I peptidomes on the Q Exactive mass spectrometer platform. Proteomics 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Ritz D., Sani E., Debiec H., Ronco P., Neri D., and Fugmann T. (2018) Membranal and blood-soluble HLA class II peptidome analyses using data-dependent and independent acquisition. Proteomics 18, e1700246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Schuster H., Shao W., Weiss T., Pedrioli P. G., Roth P., Weller M., Campbell D. S., Deutsch E. W., Moritz R. L., Planz O., Rammensee H.-G., Aebersold R., and Caron E. (2018) A tissue-based draft map of the murine MHC class I immunopeptidome. Sci. Data 5, 180157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Hood L., and Price N. D. (2014) Demystifying disease, democratizing health care. Sci. Transl. Med. 6, 225ed5. [DOI] [PubMed] [Google Scholar]
- 214. Hood L., and Friend S. H. (2011) Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat. Rev. Clin. Oncol. 8, 184–187 [DOI] [PubMed] [Google Scholar]
- 215. Auffray C., Charron D., and Hood L. (2010) Predictive, preventive, personalized and participatory medicine: back to the future. Genome Med. 2, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Green E. D., Watson J. D., and Collins F. S. (2015) Human Genome Project: twenty-five years of big biology. Nat. News 526, 29–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Aebersold R., and Mann M. (2016) Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 [DOI] [PubMed] [Google Scholar]
- 218. Ahrens C. H., Brunner E., Qeli E., Basler K., and Aebersold R. (2010) Generating and navigating proteome maps using mass spectrometry. Nat. Rev. Mol. Cell. Biol. 11, 789–801 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.