A broad-based interlaboratory study of glycosylation profiles of a reference and modified IgG antibody involving 103 reports from 76 laboratories.
Keywords: Glycomics, mass spectrometry, fluorescence, glycosylation, glycoproteins, glycan, glycopeptide, interlaboratory study, NISTmAb, reference antibody
Graphical Abstract
Highlights
A broad-based interlaboratory study of the glycosylation of a reference antibody: NISTmAb.
103 reports were received from 76 diverse laboratories worldwide.
Analysis involved two samples, the NISTmAb and an enzymatically modified sample, enabling within-lab separation of random and systematic errors using the “Youden two-sample” method.
Consensus values were derived and similar performance across all experimental methods was noted.
Abstract
Glycosylation is a topic of intense current interest in the development of biopharmaceuticals because it is related to drug safety and efficacy. This work describes results of an interlaboratory study on the glycosylation of the Primary Sample (PS) of NISTmAb, a monoclonal antibody reference material. Seventy-six laboratories from industry, university, research, government, and hospital sectors in Europe, North America, Asia, and Australia submitted a total of 103 reports on glycan distributions. The principal objective of this study was to report and compare results for the full range of analytical methods presently used in the glycosylation analysis of mAbs. Therefore, participation was unrestricted, with laboratories choosing their own measurement techniques. Protein glycosylation was determined in various ways, including at the level of intact mAb, protein fragments, glycopeptides, or released glycans, using a wide variety of methods for derivatization, separation, identification, and quantification. Consequently, the diversity of results was enormous, with the number of glycan compositions identified by each laboratory ranging from 4 to 48. In total, one hundred sixteen glycan compositions were reported, of which 57 compositions could be assigned consensus abundance values. These consensus medians provide community-derived values for NISTmAb PS. Agreement with the consensus medians did not depend on the specific method or laboratory type. The study provides a view of the current state-of-the-art for biologic glycosylation measurement and suggests a clear need for harmonization of glycosylation analysis methods.
Biologics have recently emerged as critically important drugs from health and economic perspectives. Two-thirds of approved biologics are glycoproteins, i.e. proteins containing glycans as post-translational modification. Alteration in glycosylation may impact the safety and efficacy of the drug, including its clearance rates, effector functions, folding, immunogenicity, solubility, and biological activity. In addition to glycomic profiling of new drug candidates, analysis of glycoforms is essential for monitoring production batches of established drugs and comparing biosimilars and biobetters to originator drugs.
This report describes results of a broad interlaboratory study designed to determine both the level of variability in current measurement methods as well as to support consensus measurement values for a reference material. Participation was open to all laboratories, regardless of experience or preferred analytical method. Because specific methods selected by participating laboratories varied greatly, as did their degree of expertise, this study was not designed to determine “best” methods, but to provide a “snapshot” of the currently used methods for biologic glycosylation measurement. Unfortunately, this diversity in experience and objective prevented a deeper analysis of the variability of results, with some highly experienced labs using well-developed standard operating procedures, and with others using novel approaches or exploiting their unique capabilities. The study rationale and design are presented in detail in supplementary Discussion S1.
Glycosylation analysis is inherently challenging because, unlike amino acids in proteins which are encoded by the genome, sequential addition of monosaccharide residues is not template-driven. It is rather dictated by competing enzymatic activities, leading to heterogeneity. Even at the same site of glycosylation, diverse glycans with different linkages, number of antenna, and monosaccharide compositions are possible, giving rise to challenges in separation (chromatography) and isomerization (mass spectrometry).
A common glycosylation in mAbs is N-glycosylation where the glycans are linked to the nitrogen of the Asn residue of the protein with a consensus sequence Asn-X-Ser/Thr or, more rarely, Asn-X-Cys where X is any amino acid except proline. Moreover, N-glycans have a common five-membered trimannosyl chitobiose core, Manα1–6(Manα1–3)Manβ1- 4GlcNAcβ1–4GlcNAcβ1-Asn-X-Ser/Thr. The highly complex nature of N-glycosylation analysis has given rise to a proliferation of different methods (1–14). Currently, N-glycosylation is examined at the level of intact proteins, protein fragments, peptides, glycans, or monosaccharides. Analytes are then analyzed by mass spectrometry (MS)1 (1); liquid chromatography (LC) with fluorescence detection (FD)(2) and/or MS detection; capillary electrophoresis (CE) with MS detection (3); CE-laser-induced fluorescence detection (CE-LIF); high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD); nuclear magnetic resonance (NMR) spectroscopy; or a combination of these techniques (4).
One popular approach is the release of glycans where N-glycans are cleaved from proteins using Peptide-N-Glycosidase F (PNGase F), which hydrolyzes the side-chain amide group of the glycosylated asparagine. Before analysis, glycans may be subjected to permethylation, reduction, or fluorophore labeling to increase sensitivity and specificity. Structure elucidation and isomer separation is possible using the glycan-release approach, but it lacks information on the site of glycosylation because analysis is performed after the glycans are cleaved from the protein.
Analysis of glycopeptides can provide glycosylation site information along with glycan compositions. In this approach, mAbs are digested with proteases such as trypsin (and less commonly used enzymes such as chymotrypsin, LysC, LysN, AspN, GluC, or ArgC) to produce peptides and glycopeptides that are then typically analyzed using MALDI-MS and LC-MS(/MS) methods (and less commonly CE-MS(/MS) methods (15)). The peptide attached to the glycoform gives information on the site of glycosylation. Potential disadvantages include challenges in differentiating isomers and suppression of glycopeptide ions because of peptide ions at the precursor (MS1) level. The latter could be alleviated by (two-dimensional) LC or enrichment methods (16).
Middle-down and top-down approaches characterize the glycosylation by analyzing protein fragments and intact proteins, respectively. In the middle-down approach, mAbs are treated with immunoglobulin G-degrading enzyme (IdeS), an endopeptidase that cleaves heavy chains below the hinge region, resulting in antigen-binding (Fab) and crystallizable (Fc) fragments. These large fragments are then usually analyzed by MS. Protein fragments have a lower molecular mass than the intact protein and could be better resolved in MS compared with the analysis of intact mAbs in the top-down approach. Compared with other techniques, the top-down approach provides the advantage that little-to-no sample preparation steps are needed before the analysis. Typically, only desalting of the intact mAb is necessary, which is normally performed with a desalting column (e.g. C4, C8) followed by the analysis with MS. However, because top-down and middle-down analyses often result in higher masses, fewer glycan compositions can be distinguished because of lack of resolution compared with other MS-based methods.
The diversity of these methods presents a major challenge in the interpretation of N-glycosylation measurements. Unfortunately, only a few multi-laboratory studies have been reported assessing the performance of the different approaches (17–21). In two studies by the Human Proteome Organization (HUPO), relative abundances of N-glycans (in transferrin and IgG) (17) and O-glycans (in IgA1) (18) were analyzed by 20 and 15 laboratories, respectively. They observed that MS-based methods are efficient in identifying and quantifying glycans. However, there were no participants from biopharmaceutical companies.
Here we present the design and results of our interlaboratory study of two materials: primary sample (PS) 8670, commonly referred to as NISTmAb (22), and mod-NISTmAb, a material derived from PS 8670 by modification with galactosidase. PS 8670 is the in-house standard for NIST Reference Material 8671 (23). The rationale for the use of these samples is presented in supplementary Discussion S2. This report is based on 103 reports submitted by 76 laboratories worldwide. It builds on the NIST internal report (NISTIR) 8186 (24).
This interlaboratory study had two goals. The first goal was to determine measurement variability in identifying and quantifying N-glycosylation in monoclonal antibodies across laboratories in the glycomics and glycoproteomics community, including laboratories form biopharmaceutical companies and universities. The second goal was to aid in determining community-based consensus medians for the glycosylation of the PS. The community's consensus values for NISTmAb PS 8670 glycosylation, robustly estimated as medians, represent an unparalleled diversity of approaches applied to the same material and serve as a seminal baseline for comparing glycoanalytical strategies.
Finally, we note two quite different levels of identification - by composition and by structure. Compositions are determined by high mass accuracy mass spectrometry, whereas confident isomer identification often requires reference materials or chromatographic retention matching.
EXPERIMENTAL PROCEDURES
Monoclonal Antibody Sample Preparation
Two materials were used in the study, (1 the Primary sample (PS) for NIST Reference Material 8671, NISTmAb, Humanized IgG1κ Monoclonal Antibody produced in NS0 cells, and (2 a material derived from the PS by treatment with galactosidase, termed “mod-NISTmAb.”
NISTmAb was obtained as a bulk substance prepared using mammalian cell culture and downstream processing. It has one N-glycosylation site at the Fc region of the antibody. mod-NISTmAb was prepared by subjecting a portion of NISTmAb to β-1,4-galactosidase (New England Biolabs, Ipswich, MA) and then adding the resulting solution back to the original NISTmAb (30:70 by mass).
Study Execution
The study was conducted in two stages: Stage 1 involved nine selected laboratories who volunteered to assist in final study design; Stage 2 was widely advertised and open to all laboratories. Two samples were shipped to laboratories on June 2015 and August-September 2015 for Stage 1 and Stage 2, respectively. Laboratories received three vials consisting of two blinded monoclonal antibody samples and one buffer solution in 1.0 ml screw-top tubes (Matrix™ Thermo Fisher Scientific, #3740) as follows:
Sample A: white label, frozen liquid, 0.4 mg, 100 mg/ml mAb
Sample B: blue label, frozen liquid, 0.4 mg, 100 mg/ml mAb
Buffer: yellow label, frozen liquid, 1 ml, 25 mmol/L l-Histidine, pH 6.0
Laboratories were informed that both samples are humanized IgG1k expressed in murine suspension culture and that the samples are “drug-like substances” not for human use. The buffer solution was provided as a diluent.
Participants used their method of choice to determine the glycan content in the two samples. Participants were requested to provide measurement results using NIST-provided data and method reporting templates (24) by July 30, 2015 (Stage 1) and November 6, 2015 (Stage 2). Some laboratories submitted more than one report; each report was assigned a confidential laboratory number (and was treated as a separate laboratory). Participants could enter other glycans or methods in the template; no other post-translational modifications, e.g. lysine glycation, could be reported.
Data were analyzed as reported, i.e. no normalization, using a variety of robust statistical analysis techniques to assess measurement reproducibility and to characterize glycan distributions. Results were compiled and evaluated for determination of community's consensus medians, within-laboratory precision, and concordance within the laboratories. A technical summary (24) of reported and derived values from all laboratories, a table of all identified glycans, and an individualized graphical analysis of their performance for the exercise were sent to the participating laboratories on June 2, 2017.
Shipping
Package shipped to each laboratory consisted of three vials (Sample A, Sample B, and l-Histidine buffer solution) and a welcome packet (24). The three vials were stowed in a rolled, self-sealing bubble wrap bag and placed in an insulated box filled with dry ice. The welcome packet consisted of a cover letter; instructions; packing list/shipment receipt confirmation form; and data, method, and comment reporting sheets. These documents were enclosed in a waterproof sleeve and placed at the top of the shipping box, between the cardboard covering and the foam insulation. A soft copy of the welcome packet was emailed to participants as one spreadsheet workbook with multiple worksheets. Participants were requested to return the filled shipment receipt confirmation form as soon as they received the shipped package.
Analysis Methods
Each laboratory was asked to perform glycosylation analysis of the two samples in triplicate using their own method(s), as summarized in Table I. Briefly, glycans were cleaved by incubating mAbs with PNGase F (74 reports), trypsin/PNGase F (1 report), and Pepsin/PNGase A (1 report). Cleaved glycans were derivatized using fluorescent (54 reports) or non-fluorescent (22 reports) methods. Next, glycans were separated with chromatography (CE (5 reports), HILIC (46 reports), IC (1 report), PGC (6 reports), RP (6 reports)) or without chromatography (12 reports), and then identified by various analytical methods.
Table I. Overview of analytical techniques for mAb glycosylation analysis used in this interlaboratory study.
| Analyte | Derivatization | Analytical method | Chromatography | Identification | Quantification |
|---|---|---|---|---|---|
| Glycan (76) | 2-AB labeling (20) | LC-FD (15) | HILIC (19) | MS mass (5) | PA (19) |
| LC-FD-MS (4) | RP (1) | RT std (8) | MS int (1) | ||
| LC-MS (1) | RT GU (5) | ||||
| RT GU & exo (2) | |||||
| Glycosylamine labeling (18): InstantPC | LC-FD (6) | HILIC (17) | MS mass (8) | PA (16) | |
| InstantAB | LC-FD-MS (8) | RP (1) | MS/MS (2) | MS int (2) | |
| Rapifluor | LC-MS (4) | RT std (6) | |||
| RT GU (1) | |||||
| MS mass & RT GU (1) | |||||
| APTS labeling (6) | CE-LIF (6) | CE (5) | exo (2) | PA (4) | |
| None (1) | MT std (1) | PH (2) | |||
| MT GU (2) | |||||
| MT std & exo (1) | |||||
| Permethylation (6) | MALDI-MS (4) | None (4) | MS mass (4) | MS int (4) | |
| DI-MS (1) | RP (2) | MS/MS (1) | PA (2) | ||
| LC-MS (1) | MSn (1) | ||||
| Procainamide (6) | LC-FD-MS (5) | HILIC (6) | MS mass (3) | PA (6) | |
| LC-FD (1) | RT std (1) | ||||
| RT GU (1) | |||||
| All + MS/MS & exo (1) | |||||
| Reduction (5) | LC-MS (5) | PGC (5) | MS mass (2) | MS int (2) | |
| MS/MS (2) | PA (3) | ||||
| MS mass & MS/MS (1) | |||||
| None (4) | LC-MS (2) | HILIC (1) | MS mass (3) | PA (2) | |
| MALDI-MS (1) | IC (1) | RI (1) | MS int (1) | ||
| HPAEC-PAD (1) | None (1) | sum isotope pks (1) | |||
| PGC (1) | |||||
| Ethyl esterification (3) | MALDI-MS (3) | None (3) | MS mass (3) | sum isotope pks (2) | |
| isotopic dil, (1) | |||||
| 2-AA labeling (2) | LC-FD (1) | HILIC (1) | MS/MS (1) | PA (1) | |
| MALDI-MS (1) | None (1) | RT std (1) | MS int (1) | ||
| 2-AA & permethylation (1) | LC-MS (1) | HILIC (1) | MSn (1) | PA (1) | |
| 2-aminopyridine labeling (1) | LC-FD,MS (1) | RP (1) | MS mass & RT std (1) | PA (1) | |
| 4-AA labeling (1) | LC-FD,MS (1) | HILIC (1) | RT std (1) | PA (1) | |
| INLIGHT (1) | LC-MS (1) | RP (1) | MS mass & MS/MS (1) | PA (1) | |
| Phenylhydrazine (1) | MALDI-MS (1) | None (1) | MS/MS (1) | MS int (1) | |
| p-toluidine (1) | MALDI-MS (1) | None (1) | MS (1) | MS int (1) | |
| Glycopeptide (21) | None (18) | LC-MS (16) | RP (17) | MS/MS (9) | PA (12) |
| CE- MS (1) | None (1) | MS (7) | MS int (4) | ||
| MALDI-MS (1) | MS mass & MS/MS (2) | sum isotope pks (2) | |||
| Dimethylamidation (1) | MALDI-MS (1) | None (1) | MS mass (1) | sum isotope pks (1) | |
| Ludger V-tag (1) | LC-FD (1) | HILIC (1) | RT std (1) | PA (1) | |
| Reduction (1) | CE-MS (1) | CE (1) | MS/MS (1) | MS int (1) | |
| Protein fragment (3) | None (3) | LC-MS (3) | CE (1) | MS mass (3) | MS int (3) |
| RP (1) | |||||
| SEC (1) | |||||
| Intact protein (2) | None (2) | LC-MS (1) | RP (1) | MS mass (2) | MS int (2) |
| DI-MS (1) | None (1) | ||||
| Intact, fragments, glycans (1) | None (1) | LC-MS (1) | PGC (1) | MS mass, MS/MS & exo (1) | MS int (1) |
Number in parenthesis indicates number of laboratories.
Additional abbreviations: std = standard; int = intensity; dil = dilution; sum = summation; pks = peaks.
Glycopeptides were cleaved from mAbs using trypsin (21 reports). Cleaved glycopeptides were left underivatized (18 reports) or subjected to dimethylamidation (1 report), Ludger V-tag (1 report) or reduction (1 report). Glycopeptide separation was performed using RP (17 reports), HILIC (1 report) or CE (1 report) chromatography. MS (20 reports) or FD (1 report) was used for analysis.
To obtain protein fragments, Ides (2 reports) or Endo-S (1 report) enzymes were added to mAbs. No derivatization was performed before chromatography using CE (1 report), RP (1 report), or SEC (1 report). Analysis was performed using LC-MS. Intact mAbs were analyzed with (1 report) or without PNGase F (1 report) and with (1 report) or without RP (1 report) chromatography. Intact mAbs analysis was performed using MS.
Laboratories recorded in a provided template a) methods used and b) percent abundances of glycans. Laboratories were asked to create separate reports for each method of analysis. If a value obtained was below their limit of detection or quantification, participants were asked to indicate this result as “ND” (not detected) or “NQ” (not quantified), respectively.
Describing Glycans in the Data Reporting Template
Naming Conventions
Currently, there is no standard way of naming N-glycans. Common names using the G0F and Oxford naming conventions were used. In cases where, to the authors' knowledge, there is no existing name for a glycan, every attempt was made to derive it from naming conventions, summarized below.
Common Name
All N-glycans have two core GlcNAcs;
High mannose glycans are named ManX where X is the number of mannoses after the two core GlcNAcs;
F is fucose, number after F indicates number of fucoses. No number indicates the presence of core fucose only;
Gx is galactose and x is the number of terminal Gal connected to two GlcNAcs, G0 is a biantennary complex glycan with two terminal GlcNAcs, G1 is a biantennary complex glycan with two GlcNAcs and one terminal Gal, G2 is a biantennary complex glycan with two GlcNAcs and two terminal Gal;
Gx-yN means y GlcNAc is missing, e.g. G1-N is biantennary complex glycan with one terminal GlcNAc and one terminal Gal;
S (NeuAc) or (NeuGc) is sialic acid. (NeuAc) or (NeuGc) indicates type of sialic acid;
Number in parenthesis indicates linkage: F(6) or S(6) means a α1–6-linked core fucose or α2,6-linked sialic acid, respectively;
Number in square brackets is the location of residue, e.g. (3) or (6) indicates that the residue is in the α1,3 or α1,6 mannose arm, respectively; and
xaGal is α-1,3-linked galactose, x is the number of residues;
Oxford Name
All N-glycans have two core GlcNAcs;
F at the start of the abbreviation indicates a core fucose, (6) after the F indicates that the fucose is α1–6 linked to the inner GlcNAc;
Mx, number (x) of mannose on core GlcNAcs;
Ax, number of antenna (GlcNAc) on trimannosyl core; A2, biantennary with GlcNAcs as β1–2 linked; A3, triantennary with a GlcNAc linked β1–2 to both mannose and the third GlcNAc linked β1–4 to the α1–3 linked mannose; A3′, triantennary with a GlcNAc linked β1–2 to both mannose and the third GlcNAc linked β1–6 to the α1–6 linked mannose; A4, GlcNAcs linked as A3 with additional GlcNAc β1–6 linked to α1–6 mannose;
B, bisecting GlcNAc linked β1–4 to α1–3 mannose;
Gx, number (x) of linked galactose on antenna, (4) or (3) after the G indicates that the Gal is β1–4 or β1–3 linked; [3]G1 and [6]G1 indicates that the galactose is on the antenna of the α1–3 or α1–6 mannose;
Gax, number (x) of linked alpha galactose on antenna; and
Sx, number (x) of sialic acids linked to galactose; the numbers 3 or 6 in parentheses after S indicate whether the sialic acid is in an α2–3 or α2–6 linkage.
Monosaccharide Composition
The monosaccharide composition is inside a square bracket. Small letters were used to avoid confusion with elements (hydrogen, nitrogen, fluorine, etc.): h = hexose, n = N-acetylhexosamine, f = deoxyhexose (e.g. fucose), a = NeuAc, g = NeuGc. Number after the letter denotes the number of residues. For example, [h6n4f1a1] has 6 hexoses, 4 N-acetylhexosamine, 1 fucose, 1 NeuAc. For sulfonated glycans, S = sulfur.
Structure Conventions
Two structure notations were used: the Consortium for Functional Glycomics (CFG) (25) and the Oxford Glycobiology Institute (UOXF) notations. Glycoworkbench 2.1 (Eurocarb) (26) was used to draw the structures. The differences are in the monosaccharide residue representations (for example, NeuAc is purple diamond in CFG but purple star in UOXF notations) and linkage notations (angle represents linkage in UOXF). A revised CFG format has been introduced but it was not used in this reporting template.
Calculations for Derived Attributes of NISTmAb
Calculations for the glycan attributes were estimated from the median results and based on an earlier reported method (27):
- Galactosylation = Sum of (% abundance)×(galactosylation factor) for all glycans with terminal galactose where the factor is the fraction of antennae that are galactosylated. For example, the galactosylation factor of G0F is 0, G1F is 0.5, G2F is 1.
- α Galactosylation = As above but for α galactosylation only
- Sialylation = Sum of (% abundance) × (sialylation factor) for all glycans with NeuAc or NeuGc sialic acid where the factor is the fraction of antanaee that are sialylated.
- NeuAc sialylation = As above, but for NeuAc only
- NeuGc sialylation = As above, but for NeuGc only
- Fucosylation = Sum of (% abundance) for all glycans with fucose residues.
- Core fucosylation only = Sum of (% abundance) for all glycans with 1 fucose. This calculation assumes that the first fucose is always a core fucose.
- Difucosylation = Sum of (% abundance) for all glycans with 2 fucose residues.
Bisecting GlcNAc = Sum of (% abundance) for all glycans with bisecting GlcNAc.
High mannose level = Sum of (% abundance) for all high-mannose glycans.
Sialic Acid/Galactose ratio = Sialylation/Galactosylation.
RESULTS
Overview
Because the participating labs selected their own methods of analysis and these methods can differ in many ways, merits and drawbacks of the methods can only be discussed in general terms. There are two principal factors distinguishing output: the number of different glycans reported and how the structures of those glycans were determined. High mass accuracy mass spectrometry is currently capable of determining compositions of glycans over a wide range of abundance and therefore can yield the largest number of different glycans, it is limited in isomer identification because spectra of different isomers are often indistinguishable and do not generally contain sufficient information for full structure determination. For example, glycopeptide fragmentation appears incapable of yielding complete glycan structural information. On the other hand, defined structures may be determined using chromatographic methods, coupled with standard materials and labeling or use of enzymes (exoglycosidases) capable to removing selected outer glycans. However, only a limited number of standard glycans are available and enzymatic methods are of limited use in complex glycan mixtures. In the absence of direct structural information, structures are generally represented based on biological inference, which are, in effect, informed guesses.
Sample preparation methods, ranging from enzymatic glycan release and labeling to protein digestion constitute a major source of variation. The effectiveness of these methods depends as much on laboratory skill than specific method and are, in effect, a hidden source of variation in these studies.
Differences in the objectives of the labs is a major contributor to the diversity of methods and results. Probably the most critical measurements are made by biopharmaceutical companies who rely on glycan determinations for both product quality and government approval. Consequently, they generally use well-established, conservative methods that often involve several targeted, derivatized glycans with precisely defined established chromatographic methods. Further, they may limit their number of analytes to only the major glycans considered. Other groups, such as instrument companies and some academic institutions seek to maximize the number of glycans identified, which generally sacrifices structural information. Other labs made these measurements for various educational and internal quality control purposes whereas others wish to develop or demonstrate new methods of analysis.
Demographics of Laboratories
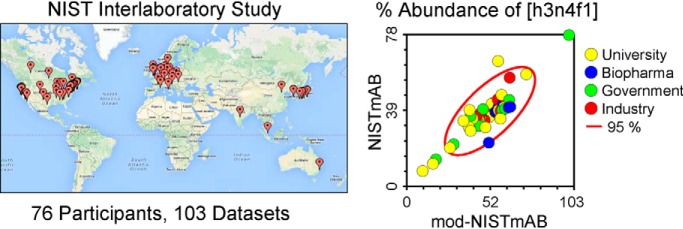
One hundred eighteen laboratories responded to the call for participation. However, because of challenges with timing, personnel, shipping (bad weather, customs delay), legal, technical (instrument, freezer malfunction), and other issues, several laboratories had to drop out of the study. Samples were sent to 90 laboratories; 76 laboratories submitted 103 reports. Supplemental Fig. S1A shows a map of the participating laboratories from Europe (42%) and North America (38%), Asia (18%), and Australia (2%). Laboratories were primarily from the industry sector, with almost half of these laboratories from biopharmaceutical companies, as shown in supplemental Fig. S1B.
Glycosylation Analysis Methods Used by Participating Laboratories
Table I summarizes the glycosylation analysis methods used by laboratories in this study. Out of 103 reports, 74% analyzed released glycans, 20% used glycopeptides, and 6% used intact protein and protein fragments. Fluorescently-labeled glycans were commonly analyzed by LC-FD or LC-FD-MS methods except for APTS-labeled glycans, which were analyzed by CE-LIF exclusively. Reduced and permethylated glycans were analyzed solely by MS (using MALDI, direct infusion- (DI) and LC-electrospray ionization techniques). Hydrophilic interaction liquid chromatography (HILIC) is the commonly used chromatographic method for glycans labeled with 2-AB, glycosylamine, and procainamide fluorophores whereas porous graphitized carbon (PGC) was used for reduced glycans.
In Table I, glycopeptides were typically analyzed without derivatization (18 out of 21 reports) by reversed-phase LC-MS (16 out of 21 reports). MS techniques (MS mass, MS/MS fragmentation data, or a combination) were frequently used for identification whereas MS peak area, MS intensity, and summation of isotope peaks were used for quantification. Supplemental Fig. S2 shows an example of fragmentation data of glycopeptides using LC-MS/MS analysis. HCD spectra at 40% normalized collision energy (supplemental Fig. S2A) shows the peptide backbone (supplemental Fig. S2B), oxonium ions (supplemental Figs. S2C and S2J), and glycan fragmentation (supplemental Fig. S2D–S2I). Oxonium ions have been used to screen for the presence of glycopeptides and glycan motifs (28,29). The laboratory used oxonium ions from a HexNAc (m/z 168.07, m/z 186.08, and m/z 204.09) to screen for the presence of glycopeptides (supplemental Fig. S2C). Presence of m/z 512.20 oxonium ion is specific for antenna fucosylation (30) (supplemental Fig. S2E); m/z 290.09 and m/z 308.10 oxonium ions are specific for NeuGc residues (supplemental Fig. S2F); and m/z 528.19 is indicative of a trisaccharide having 2 hexoses and 1 N-acetylhexosamine., e.g. Gal-Gal-GlcNAc (supplemental Fig. S2G and S2H). For high mannose glycans, the absence of GlcNAc antenna could result in hexose oxonium ions m/z 127.04, m/z 145.05, and m/z 163.06 (supplemental Fig. S2J). The laboratory screened glycopeptides for high-mannose glycans using m/z 163.06.
Intact proteins and protein fragments were analyzed mostly using LC-MS, as shown in Table I. Separation was performed by CE, RP, SEC, or PGC; identification was performed by MS mass or MS/MS fragmentation with exoglycosidases; and quantification by MS intensity.
Supplemental Table S1 lists the analytical approaches by laboratory sector. Here, a disparity could be observed in the choice of methods. Biopharmaceutical company laboratories preferred the well-established method involving fluorophore-labeled glycans for their analysis (19 out of 21 laboratories); whereas two used glycopeptide analyses. University laboratories, however, primarily used either more generic mass spectrometry-based methods of glycopeptide analysis (14 out of 32 laboratories) or non-fluorescent glycan (12 out of 32 laboratories) approaches. Protein fragment (n = 3) and intact protein (n = 2) techniques were used to demonstrate the technology - they are not listed in supplemental Table S1 to protect laboratory anonymity.
Glycan Identification
The data reporting template (24) included 54 glycan compositions for 68 glycan structures, however an additional 62 compositions and 71 other structures were reported by the participating laboratories. A total of 116 compositions and 139 structures were identified. Supplemental Table S2 lists the glycan compositions and isomers identified by laboratories in this study (24). Supplemental Table S3 lists all the quantified and derived values for NISTmAb and mod-NISTmAb for glycan compositions and glycan structures (24). Independent of this study, glycosylation of NISTmAb was previously characterized by three laboratories using HILIC with fluorescence detection of 2-AB-labeled N-glycans and collectively found 24 glycan peaks (28, 31). Another work using 1D- and 2D-LC-MS/MS for the analysis of glycopeptides found 60 glycan masses on NISTmAb (16).
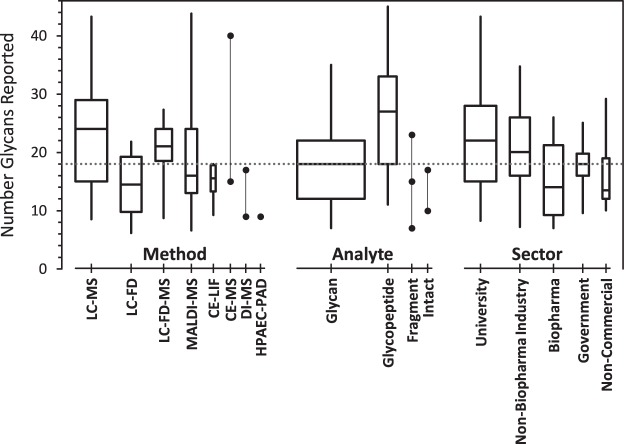
The number of glycan compositions reported by each laboratory ranged from 4 to 48. Most reports listed about the same number of glycan compositions for each of the two samples. Fig. 1 summarizes the number of unique glycan compositions reported for NISTmAb and/or mod-NISTmAb samples as a function of the laboratory's analytical method, analyte, and organizational type. On average, more compositions were reported by laboratories (1 using MS-based methods, (2 analyzing glycopeptides, and (3 that were university-based. However, the wide range in the number of compositions reported within most of the groups suggests that the technology is not the major determinant.
Fig. 1.
Number of unique glycan compositions reported, grouped by method, analyte, and sector. The boxes span the central 50% of reported values, 25% to 75%; the whiskers span the central 90%, 5% to 95%; the central line marks the median, 50%. Box widths are proportional to the number of reports. Groups within each category are presented in order of decreasing number of reports. Solid circles represent individual results within categories of fewer than six reports. The dotted line marks the median number of compositions reported in the 103 reports provided by 76 laboratories.
Fig. 2 summarizes the proportion of compositions that were reported with isomeric information as a function of identification method. On the average, laboratories that used exoglycosidases reported the greatest number of isomeric information, followed by retention times, then by FD, then by MS/MS. Surprisingly, about one-fourth of the data sets that nominally used accurate mass (MS) identification identified some isomers.
Fig. 2.
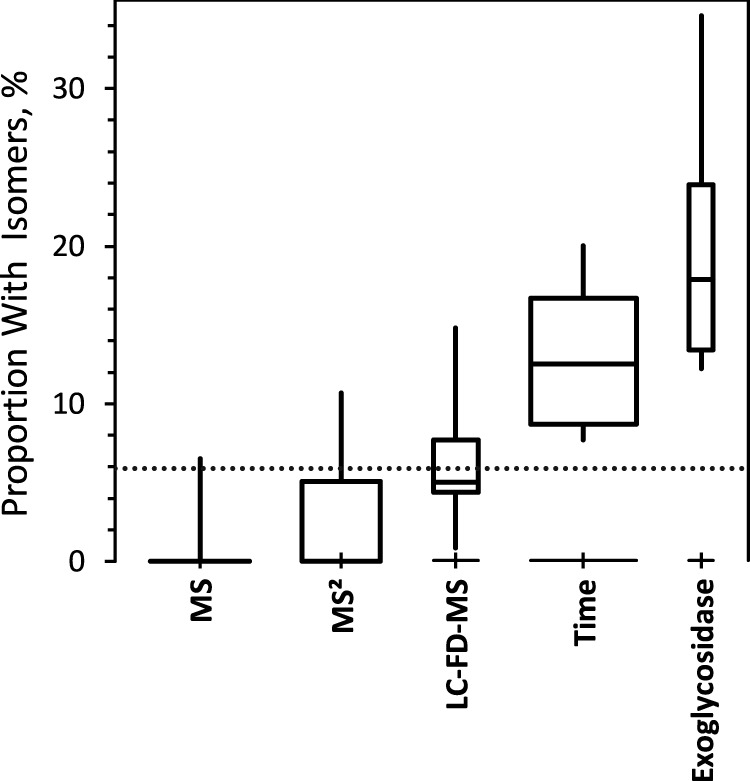
Proportion of glycan compositions reported as isomers. The boxes span the central 50% of reported values, 25% to 75%; the whiskers span the central 90%, 5% to 95%; the central line marks the median, 50%. Box widths are proportional to the number of reports. Categories are presented in order of increasing median proportion. The dotted line marks the median proportion of compositions reported as isomers.
Glycan Quantification
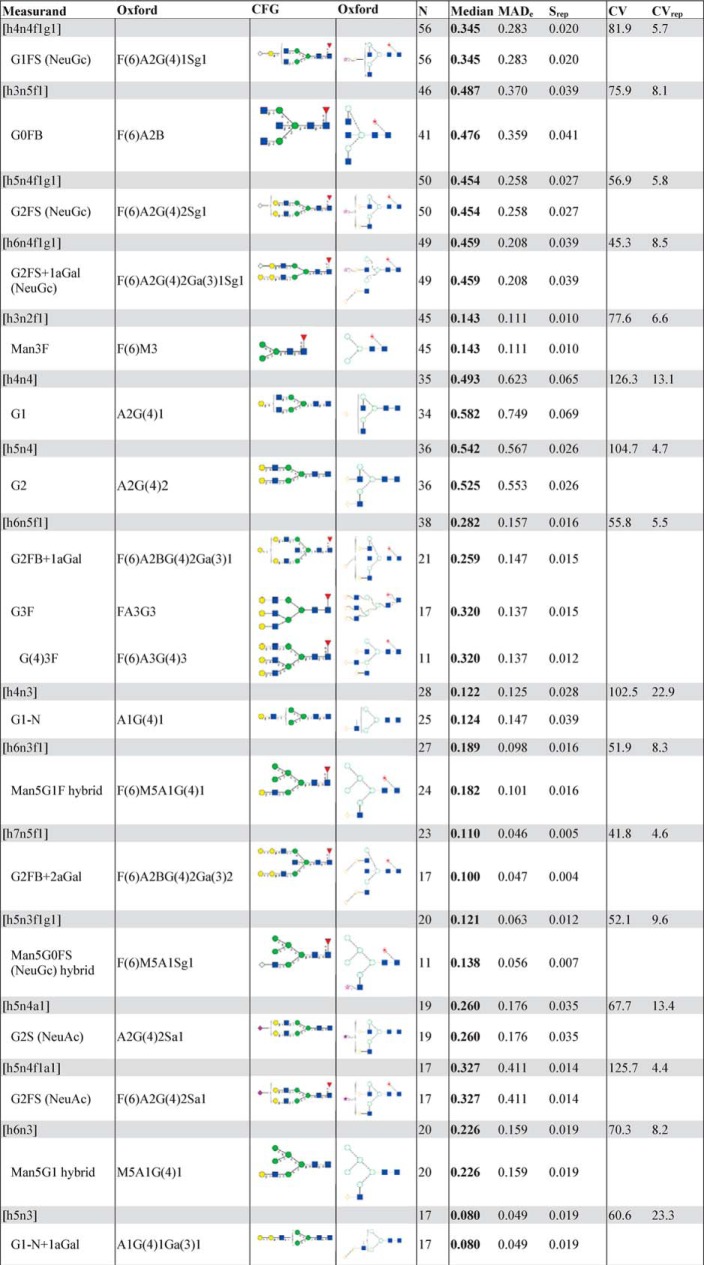
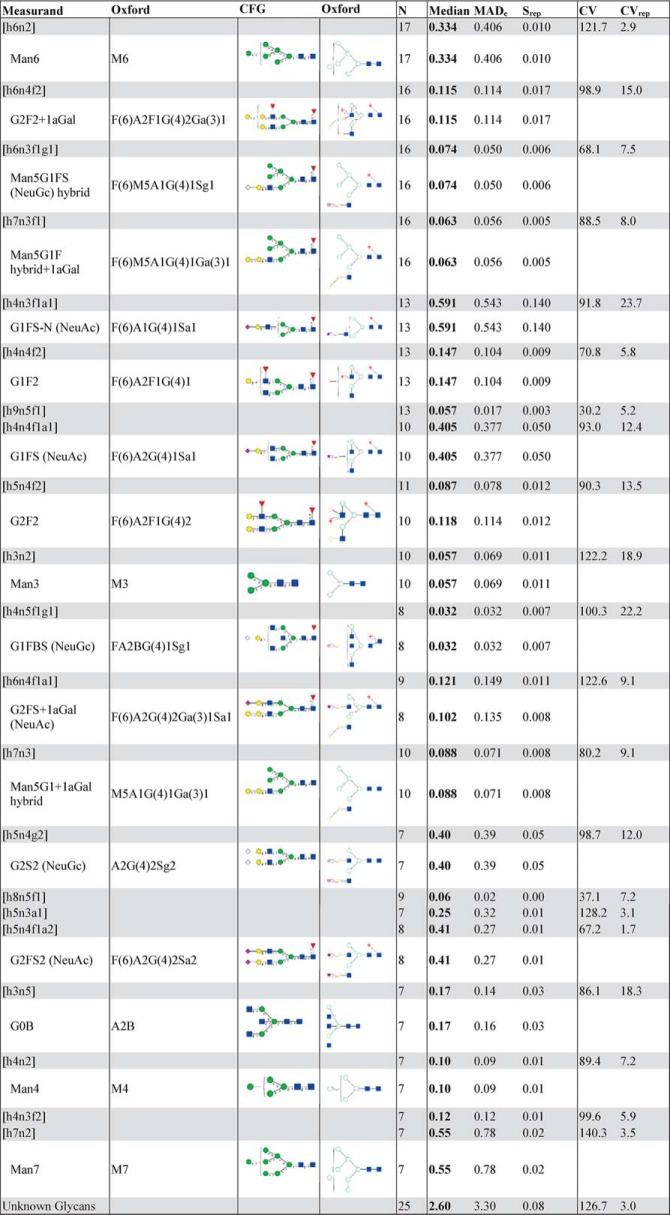
Laboratories were asked to report quantitative values for each glycoform as proportions relative to the sum of all glycoforms detected. Table II lists the consensus median abundances of glycan compositions and glycan structures on NISTmAb that were reported at least six times. The three compositions [h3n4f1], [h4n4f1] and [h5n4f1] are the most commonly reported and the most abundant compositions; together, they account for more than 85% of the total signal intensity. Although the normalization factors (the sum of signals) for the different data sets are not based on the same compositions nor the same number of compositions, the dominance of [h3n4f1] and [h4n4f1] ensures that the reported proportions are comparable across data sets. However, the differences among the normalization factors are a source of variability. Other approaches such as normalizing to the most abundant few glycoforms could be pursued in future studies.
Table II. Community's consensus abundances of glycans in NISTmAb PS 8670 reported by laboratories at least six times. Glycan compositions are arranged by decreasing number of values (N). Supplementary Table S2 lists all glycan structures and names. Supplementary Table S3 lists all the community's consensus values.
n = number of values; consensus median = consensus median (50th percentile or 2nd quartile, expressed as percent of total composition) of the distribution of the reported results; MADE = median absolute deviation; Srep = a robust estimate of the expected repeatability, the median SD for glycan compositions with at least six results; CV = robust coefficient of variation (MADE/median); CVrep = (Srep/median).
Laboratories were asked to report their results as percent abundances normalized such that they summed to 100% per sample. Supplemental Fig. S3 shows a histogram of the sum of the unique glycan composition values for NISTmAb and mod-NISTmAb as reported by the laboratories (n = 206, two samples per laboratory). Although results in most data sets summed to 100%, the sums ranged from 88% to 122%. Some laboratories assigned percent abundances to unidentified glycans; the sum of these values was reported as one entry called “Unknown Glycans” and added to the abundance sum for that sample.
The number of replicate values per reported composition or structure per sample ranged from one to nine. The nature of these values ranged from purely technical replicates (multiple measurements of the same preparation) to process replicates (single measurements of multiple independent preparations). In all cases where two or more replicate values were reported, the values were summarized as their mean and standard deviation (SD).
Some data sets reported replicate values equal to zero, not detected (ND), and not quantified (NQ). These values can be ignored when there are no quantitative values in a set of replicates but cause numerical instability when there is at least one quantitative value in the set. Various options were explored for handling these situations in a uniform manner including: treating non-numerical results as zero, replacing zeros and non-numerical values with the data set's minimum reported value (MRV, the smallest reported numerical value of a data set), or replacing them with the data set's limit of reporting (LoR, the extrapolated smallest value of a data set). Replacement with the LoR provided slightly smaller SDs than replacement by the MRV. Supplemental Fig. S4 shows the LoR for one set of results. Gray lines are traces of the unique non-zero values reported in each set of results, where the numbers are ordered by decreasing value. If the true amounts of the minor glycans are randomly distributed and all results reflect the same level of analytical effort, a best-fit line to the right-tail of the trace estimates the LoR for that set. As shown in supplemental Fig. S5, most of the LoRs agree well for MRVs above about 0.05%. Below this value, many of the sets contain a few values many-fold smaller than their LoR. This may reflect special interest in selected glycan components rather than reporting issues with the less-abundant glycans. The LoR values in the data set may be more representative of the analytical sensitivity of a measurement system than is the MRV.
Derived Attribute Quantities in NISTmAb
Table III shows the degree of galactosylation, sialylation, fucosylation; levels of bisecting GlcNAc and high-mannose; and the sialic acid/galactose ratio in NISTmAb. These values are estimated from the consensus median values of the glycan compositions. Calculations are based on previous works by Wuhrer (27) and are designed to reflect biosynthetic pathways (32) and, to some extent, enzyme activity. In addition, these glycosylation traits relate to differences in effector functions of monoclonal antibodies and circulation half-time for other therapeutic glycoproteins.
Table III. Derived attribute quantities for NISTmAb PS 8670, estimated from the consensus median values of the glycan compositions.
| Features | Number of Labs | 25% | Median | 75% |
|---|---|---|---|---|
| Galactosylation | 32 | 31.78 | 36.21 | 43.30 |
| alpha-Galactosylation | 13 | 3.00 | 3.77 | 4.97 |
| Sialylation | 18 | 2.26 | 3.48 | 6.98 |
| NeuAc sialylation | 6 | 0.71 | 1.25 | 2.83 |
| NeuGc sialylation | 12 | 1.55 | 2.23 | 4.16 |
| Core fucosylation only | 37 | 92.36 | 103.95 | 118.23 |
| Antenna fucosylation | 3 | 0.25 | 0.38 | 0.73 |
| Bisecting GlcNAc | 7 | 1.49 | 2.17 | 3.83 |
| High mannose | 6 | 1.04 | 1.92 | 3.42 |
| Sialic Acid/Galactose Ratio | 0.07 | 0.10 | 0.16 |
All antennae were assumed to be available for galactosylation by most galactosyltransferases. Antennae galactosylation may be a reasonable proxy for enzyme activity and may therefore reflect regulation of galactosylation process in a biological system. Thus, galactosylation levels were expressed by calculating the number of galactosylated (occupied) antennae divided by the total number of antennae of the specific glycan. Only glycans identified with galactose residues were included in the calculations. For biantennary glycans, the galactosylation levels are 0.0, 0.5, and 1.0 for 0, 1, and 2 galactoses, respectively. For triantennary glycans, the galactosylation levels are 0.0, 0.33, 0.67, and 1.0, reflecting the presence of 0, 1, 2, or 3 galactosylated antennae. For NISTmAb, the median degree of galactosylation is 36.2% with alpha-galactosylation at 3.8%, as shown in Table III.
For sialylation levels, the same principle is applied, i.e. the sialylation per antenna was calculated. For NISTmAb, NeuAc and NeuGc sialylation were observed at a medium value of 1.3% and 2.2%, respectively. The ratio of sialic acid per galactose was calculated as 0.1%. This value reflects the sialylation activity, i.e. whether available acceptor positions have been sialylated.
Monofucosylation was interpreted as core fucosylation. NISTmAb had very high levels of core fucosylation (median of 104%; values exceeding 100% are artifacts of the variable normalization factors). The antenna fucosylation (manifested as difucosylation) at 0.38% was calculated separately because the interaction between core and antenna fucosylation is assumed to be minimal.
Issues with Glycosylation Analysis Methods
Laboratories reported challenges in identifying and quantifying glycans. Some laboratories reported their analysis at the composition level only and did not differentiate isomeric species; some laboratories analyzed at the glycan isomer level and had challenges in identifying co eluting or same mass species. These issues are usually method dependent, as shown in supplemental Table 4. Some glycan structures were supported by MS/MS and other structures were inferred from similar structures, e.g. triantennary structures. Consequently, some abundance values were assigned to triantennary structures instead of bisecting glycans. The same laboratory observed a discrepancy for glycan G1FS N (NeuGc). Compared with quantitative data from subunit analysis, the glycopeptide abundance was higher than subunit abundance. Overall, the laboratory observed that glycopeptide abundances were in good accordance with the subunit data with slightly lower values for G0F and G1F in the glycopeptide analysis.
One laboratory analyzed protein fragments by LC MS that had masses up to 25 kDa. One or two nominal mass differences were challenging to distinguish using their technique.
Another laboratory analyzed 2-AB glycans using HILIC LC FD with comparison to retention time of standards. It was difficult to distinguish glycans that coelute, e.g. G2 and Man6. Moreover, the laboratory was unable to identify glycan peaks present in the samples but absent in their lab-designated standard sample.
Supplemental Table S4 lists the advantages of certain methods as described by laboratories. Sialic acid specific derivatization of 2-AB or ethyl esterified glycans analyzed by LC FD or MALDI MS could confirm presence of terminal α2 6 linked NeuGc in glycans.
Intact protein analysis could give the G0F/G1F, G1F/G2F, G0/G0F, G0F N/G0F, G2F+1aGal/G2F glycoforms present in the monoclonal antibodies. Due to the cleavage of glycopeptides or glycans from the protein, analysis using these two analytes could not provide this specific information.
2-AB glycans analyzed by LC FD using glucose units and APTS labeled glycans analyzed by multiplexed capillary gel electrophoresis (xCGE) using migration time of standards and exoglycosidases could distinguish between isomers.
Additional Information on NISTmAb
Some laboratories performed unique analyses, resulting in additional information on the glycosylation of NISTmAb
Absolute Glycan Amounts
One laboratory determined absolute glycan amounts in the samples by employing isotopic dilution methods, using 13C-labeled N-glycans as internal standards followed by MALDI-TOF MS analysis. For example, the absolute amounts of three glycans in NISTmAb were reported to be:
G0Fa: (626.7 ± 7.5) pmol per 100 μg NISTmAb
G2F: (110.8 ± 5.9) pmol per 100 μg NISTmAb
G2: (19.0 ± 3.8) pmol per 100 μg NISTmAb
Glycoforms in Intact Samples
One laboratory analyzed intact mAb samples using LC-MS and identified glycoforms on the two Fc portions that were analyzed. Example glycoforms are G0/G0F, G0F/G1F, G1F/G2F, G0F-N/G0F, and G2F+1aGal/G2F. Abundance values for these glycoforms are shown in supplemental Table S3, bottom rows.
Unknown Modifications
One laboratory found an unknown delta mass of + 1856 Da at 0.40% abundance in NISTmAb by LC-MS. Another laboratory detected + 54 Da unidentified protein modification using 1H-NMR and MS. The latter found a glycan present in <3% with no branching at the central β-Mannose, i.e. there is only one arm present with a terminal NeuGc and a proximal Fuc.
Unglycosylated Forms
One laboratory used protein fragment analysis by C4-LC-MS and observed the unglycosylated form of the samples at 0.60% abundance in NISTmAb. Another participant used glycopeptide analysis by C18-LC-MS and detected the unglycosylated form at 0.91% abundance in NISTmAb, as confirmed by high mass accuracy MS (< 3 parts per million (ppmb) mass deviation).
Glycan Motifs
One laboratory employed lectin microarrays on intact proteins (33). The participant found that NISTmAb has more core fucosylation whereas mod-NISTmAb has slightly more terminal fucosylation. Both samples hint at the presence of α2,6 sialylation and show similar patterns for hybrid/lower order mannose N-glycans, indicating no strong presence of complex glycans.
DISCUSSION
Sample Ratios Demonstrate Comparability
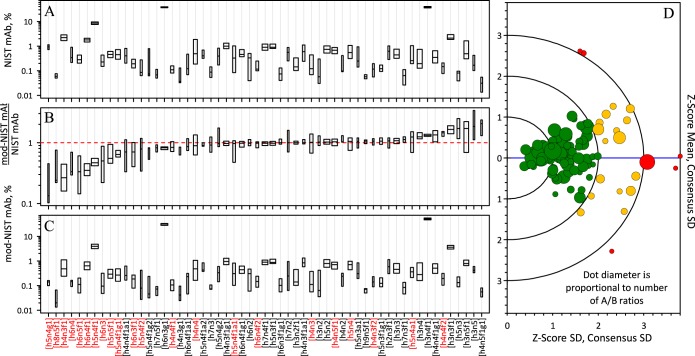
To examine the between-data set differences in the measurements of the two samples, mod-NISTmAb/NISTmAb ratios were calculated for the 57 glycan compositions that were reported at least six times for either NISTmAb or mod-NISTmAb. Since the number and identity of the reported glycan compositions in the two samples were nearly the same within each data set, these ratios are insensitive to the normalization factors. Figs. 3A and 3C display the NISTmAb and mod-NISTmAb measurement distributions of these compositions as boxplots. Fig. 3B displays the mod-NISTmAb/NISTmAb ratios. Supplemental Table S3 lists the values of these ratios.
Fig. 3.
Summary results for the 57 most frequently reported unique glycan compositions. Box plots for A) mod-NISTmAb, B) mod-NISTmAb/NISTmAb ratio, and C) NISTmAb PS 8670. Glycan compositions in red have terminal β1,4-gal as their dominant structure. Each box represents the distribution of the central 50% of the mean of the reported replicate values for one glycan. The horizontal middle line in each box represents the consensus median. The width of each box is proportional to the square root of the number of values defining the distribution. The dashed red line in the display of the Fig. 3B denotes the expected ratio, 1.0, when a glycan result is the same in mod-NISTmAb as it is in NISTmAb. Glycans are sorted in order of increasing mod-NISTmAb/NISTmAb ratio. D) Targetplot summary of mod-NISTmAb/NISTmAb ratios relative to the consensus medians. Each dot represents one set of results. Dot diameter is proportional to number of mod-NISTmAb/NISTmAb ratios reported. The dots are color-coded by distance from the (0, 0) origin: dots within two comparability units are colored green, between two and three units are colored yellow, and greater than three units are colored red. The “Z-score Mean” axis displays the average bias estimated as the mean of the “Z-score” values of the ratios. The “Z-Score SD.” axis displays the variability of individual bias estimates, estimated as the standard deviation of the Z-scores.
Every box in Fig. 3 spans the central 50% of the reported or calculated values, with the horizontal middle line denoting the consensus median. The compositions are sorted in order of increasing mod-NISTmAb/NISTmAb median. The width of each box is proportional to the square root of the number of values defining the distribution, so the wider the box, the more laboratories reported that glycan. For example, 102 of the 103 data sets identified [h3n4f1] and [h4n4f1], which have the widest boxes. The dashed red line in Fig. 3B denotes the expected ratio, 1.0, when a glycan result is the same in mod-NISTmAb as it is in NISTmAb. Ideally, glycan structures with terminal β1,4-gal should fall below this red line because β1,4-galactosidase, an enzyme that specifically cleaves terminal β1,4-gal, was added to a portion of the mod-NISTmAb. Glycan compositions colored red in the x axis have terminal β1,4-gal as their dominant structure. As expected, most of these glycans fall below the red line, i.e. they have lower abundance in mod-NISTmAb than in NISTmAb.
Fig. 3D displays the average variability and bias of the mod-NISTmAb/NISTmAb ratios relative to the consensus medians in a form sometimes called a “targetplot” (34). Each dot marks the summary score for the unique glycan compositions in one set of results. The vertical axis displays the mean bias or “concordance” of the ratios: zi = (∑j(xij − x̄j)/sj)/nj, where xij is the ratio for the jth composition reported in the ith data set, x̄j is the consensus location of the jth composition, sj is the consensus dispersion for that composition, and nj is the number of data sets that report values for that composition in both samples. Because the distributions of the ratios for most compositions are heavily-tailed, the consensus location and dispersion are estimated using robust estimators: the median for location and the scale-adjusted median absolute deviation from the median (MADE) for dispersion. The horizontal axis displays the variability of bias estimates, estimated as the SD. or “apparent precision” of the biases:
The semicircles mark one, two, and three “comparability” distances from the ideal (zi, s(zi)) value of (0, 0):
The targetplot dots are color-coded by distance from the (0, 0) origin: dots within two comparability units are colored green, between two and three units are colored yellow, and greater than three units are colored red. These codes roughly indicate “Good”, “Moderate”, and “Questionable” agreement with the consensus mod-NISTmAb/NISTmAb ratio estimates.
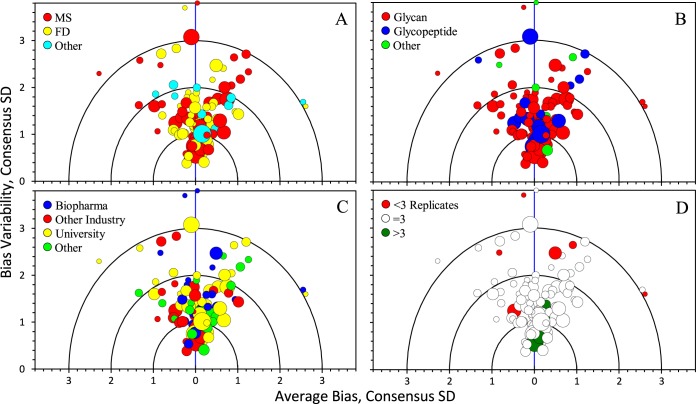
Fig. 4 displays the same targetplot colored by analyte, analytical technique, organizational type, and number of replicates. No systematic trend was apparent in any of the parameters.
Fig. 4.
Targetplot summary of mod-NISTmAb/NISTmAb ratios relative to the consensus medians. Each dot represents one set of results. Dot diameter is proportional to number of ratios reported. The “Average Bias” axis displays values estimated as the mean of the “Z-scores” of the ratios. The “Bias Variability axis displays values estimated as the standard deviation of the Z-scores. The subplots are colored by: A) analytical technique, B) analyte, C) laboratory type, and D) number of replicates.
Youden Two-Sample Plots of Glycan Compositions
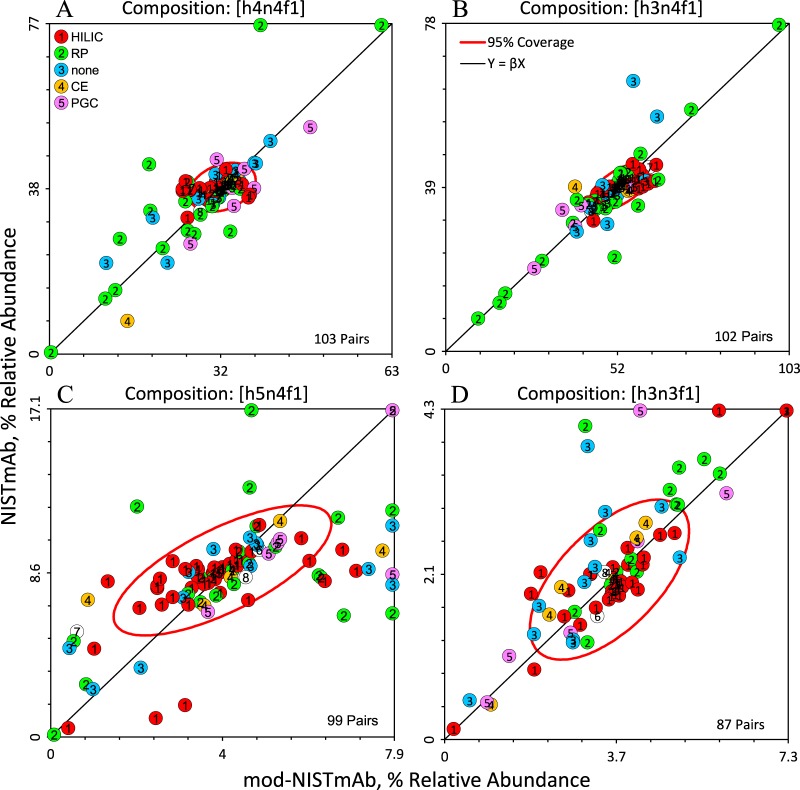
Fig. 5 shows Youden two-sample plots for the four most abundant glycan compositions, [h3n4f1], [h4n4f1], [h5n4f1] and [h3n3f1]. Supplemental Fig. S6 presents similar plots for all compositions with quantitative results for both samples in at least six data sets. Each dot in these panels represents one (mod-NISTmAb, NISTmAb) pair from one data set. The median of each sample is used as the univariate estimate of distribution location because it is not as vulnerable to extreme values as is the mean. The center of each panel (for [h3n4f1], the values (52, 39)) represents the consensus location of the resulting bivariate distribution. The ellipse in each panel is constructed to enclose data pairs that are consistent with the consensus medians at an approximate 68% (one sigma) level of confidence. These ellipses are defined by (1 the univariate medians, (2 the MADE robust estimates of the univariate SDs, (3 the bivariate correlation between the two distributions, and (4 a factor that provides a stated coverage probability.
Fig. 5.
Youden two-sample plots for the four most abundant glycan compositions in NISTmAb. Each panel displays the bivariate distribution for one composition. A) [h4n4f1], B) [h3n4f1], C) [h5n4f1], and D) [h3n3f1] (see Materials and Methods for key). The panels are centered on the univariate medians and scaled to display all values from 0 to twice the median. Values that are greater than twice the median are assigned a value of twice the median. The ellipse includes about 68% of the pairs (≈ 1 SD). The diagonal line represents the expected relationship when measurement systems have the same bias for both samples. Each symbol represents the (mod-NISTmAb, NISTmAb) pair for one data set. Symbols are coded and labeled by separation technique.
When the two samples present similar measurement challenges, Youden two-sample plots graphically separate random within-data set measurement imprecision from systematic between-data set bias (35, 36). Pairs of measurements that reflect the same proportional bias will fall along the 45° line. This often indicates a calibration issue. Pairs of measurements that lie well away from the line indicate sample-specific interferences or measurement systems that are not in adequate statistical control.
The dots in Fig. 5 are colored by separation method. Essentially all laboratories that used HILIC separation are within the ellipse for [h3n4f1] and [h4n4f1], are within the ellipse or lie along the diagonal for [h3n3f1] but show significant scatter for [h5n4f1]. The pattern of off-diagonal results for [h5n4f1] suggests that for this composition the two samples presented several measurement systems with different measurement challenges.
Measurement Repeatability Better for More Abundant Glycans
Metrological repeatability is defined as the variation in measurements taken by a single person or instrument on the same sample, under the same conditions, and in a short period of time. Fig. 6 shows a scatterplot of the relationship between measurement repeatability, estimated as the coefficient of variation expressed as percentage (CV), and glycan amount, estimated as the mean of the replicates, for one exemplar laboratory. The black line represents a simple consensus power-law: CV = 5.0 × Mean−0.35 (or, expressed as SD = 0.050 × Mean0.65). Note that CV is not constant for all glycan amounts but rather generally increases with decreasing amount. This trend is closely related to Horwitz's observation that the interlaboratory study CV generally increases with decreasing analyte concentration regardless of the analytical method or number of laboratories (37). It has been speculated that this empirical trend arises more from cost-benefit considerations than intrinsic analytical limitations (38).
Fig. 6.
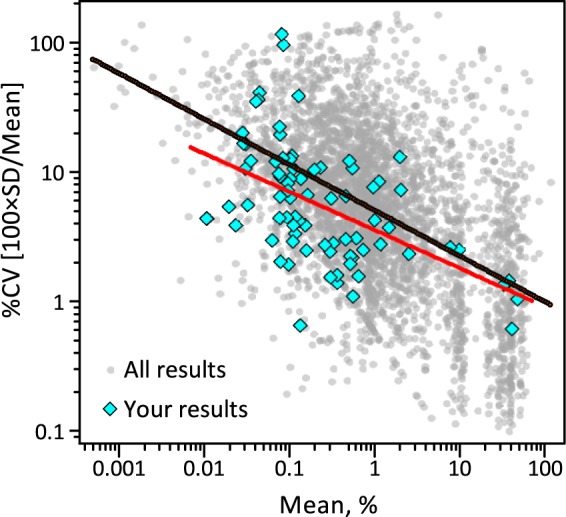
Scatterplot of the relationship between measurement repeatability, estimated as the CV, and glycan amount, estimated as the mean of the replicates. The black line represents a consensus power curve fit to all available (mean of replicates, relative standard deviation) pairs, denoted by the light gray dots: CV = 5.0 × Mean−0.35 (or SD = 0.050 × Mean0.65). The red line is the power curve fit to the pairs, denoted by the blue diamonds, reported in one data set. The measurement repeatability or this data set is somewhat better than average.
Repeatable Measurements are Closer to Community's Consensus Values
The extent of agreement between a given laboratory's reported values and the study's consensus median values is a strong function of the laboratory's measurement repeatability. Thus, establishing within-laboratory repeatability is critical to the harmonization of glycosylation analysis methods between-laboratories. Fig. 7 shows a scatterplot of the closeness to consensus of the reported medians as a function of measurement repeatability. “Closeness” is estimated as the relative absolute difference between a given result mean and the median of the means provided in all 103 reports: 100× Mean-Consensus Median /Mean. The symbols are coded by the user-stated nature of the reported replicates. Because of the great variability in the results for the various glycans, the over-all repeatability for each laboratory is estimated as the median of the repeatabilities of the reported unique glycan compositions.
Fig. 7.
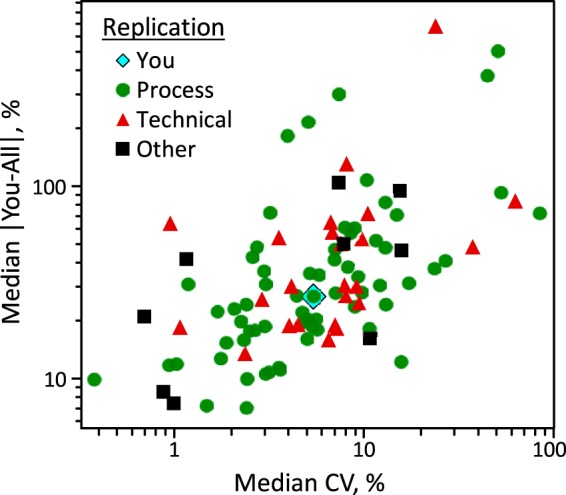
Scatterplot of the closeness to consensus of the reported values as a function of measurement repeatability estimated as CV. The symbols are coded by the user-stated nature of the reported replicates. The plot shows the data point, in blue diamond, of one data set.
Trueness of the Consensus Estimates
Metrological trueness is the closeness of a result to the best available approximation to its (unknowable) true value. Area-based estimates for 27 resolved NISTmAb peaks of defined composition have been published (22). Unique correspondences can be established between glycan compositions and most of the peak assignments, Fig. 8 demonstrates that the study's consensus medians agree well with published values for composition levels of 1% or more. While agreement diverges with decreasing abundance, only three of the published values are not contained within the study's central 50% distribution.
Fig. 8.
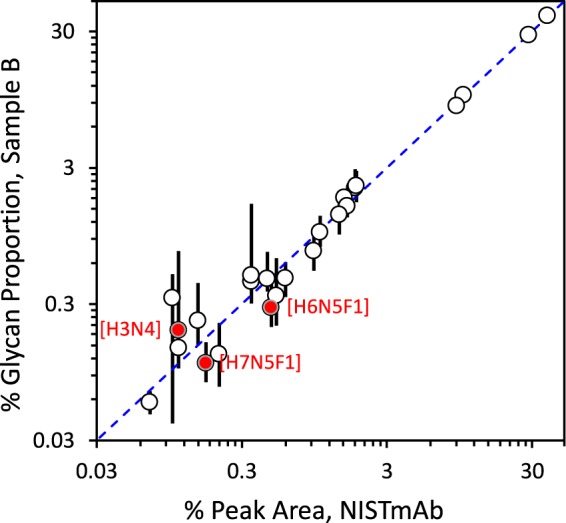
Comparison Consensus Medians to Published Peak Areas22. Each symbol represents this interlaboratory study's consensus median % proportion as a function of the published peak areas for one composition or defined group of compositions. The bars span the central 50% of the distribution of reported values. The solid red circles denote compositions where the central 50% of the values does not include the published peak area. The dashed line represents equality between the two estimates.
In broad terms, a large proportion of methods used chromatography for separation followed by identification either by mass spectrometry or chromatographic retention times. Some laboratories combined both for identification. Laboratories that used MS only reported more distinct glycan compositions. Laboratories that used MS with exoglycosidases, RT, FD, and/or MS/MS reported isomers. However, the range in the number of reported compositions within each category is quite large.
CONCLUSION
Community consensus medians for 57 glycan compositions in NISTmAb were obtained from 103 reports of 76 laboratories. Levels of sialylation (NeuAc and NeuGc), galactosylation (including alpha-galactosylation), and fucosylation (core and antenna) were calculated from consensus medians. These values could be useful in comparing analytical methods for determining glycosylation of a publicly-available material. A unique advantage of using NISTmAb is that because it was produced in NS0 cells, a rich array of glycan compositions with low-abundant variants including NeuGc and alpha-Gal-containing glycans is observed.
More importantly, this study provides an overview of the current state of glycosylation determination in the glycomics community:
Most biopharmaceutical company laboratories analyzed glycosylation using fluorescently-labeled glycans whereas most academic laboratories prefer glycopeptide analysis and non-fluorescently-labeled glycan analysis. On average, biopharmaceutical company laboratories reported lower number of glycan compositions than university laboratories.
Glycan compositions identified using different methods for determining glycosylation ranged from 4 to 48 compositions. Laboratories that used glycopeptides as analyte, on the average, reported the greatest number of glycan compositions.
Agreement to the community's consensus medians did not depend on a specific method or laboratory type but on the measurement repeatability. The better a laboratory's measurement precision the more likely that the laboratory's mean values will agree with the community consensus. Thus, establishing within-laboratory repeatability is critical to the harmonization of glycosylation analysis methods between-laboratories.
Most methods used in the different laboratories could be corrected by calibration methods when and if a standard becomes available.
Measurement repeatability was better for more abundant glycans. The CVs increased with decreasing glycan abundances. This work is the first attempt to analyze data from the interlaboratory study. Further data mining studies on this large data set could be valuable to the glycomics communty to uncover underlying systematic trends. Examples of potential studies include comparisons of identification methods, quantification methods, or normalization methods. Other areas to explore include assigning degrees of confidence in identification methods (e.g. identification by MS1 alone versus identification by MS1 with one, two, or three orthogonal values).
The study shows a clear need for harmonization of glycosylation analysis methods. Further understanding of causes of deviations would be useful in developing a harmonized method for glycosylation analyses of mAbs.
Finally, we note that the generally unsatisfactory state of confident identification of less abundant glycan structures. While the increasing sensitivity of mass spectrometry-based methods have revealed an increasingly large number of glycan compositions, the ability to elucidate their structures have not kept pace. In many cases, glycan structures are routinely inferred through biological reasoning. The use of exoglycosidases is helpful in excluding candidates, but of limited value for minor glycans or complex mixtures. Perhaps the emerging field of ion mobility mass spectrometry can aid the distinction of isomers.
DATA AVAILABILITY
Summary results as provided by the participants, expressed as percentage total glycan, are provided in the Supplemental Information.
Supplementary Material
Acknowledgments
Thanks to NISTers Mike Tarlov, Dean Ripple, Karen Phinney, John Schiel, John Bowden, David Bunk, Jeff Hudgens, and Paul Rudnick for help in study design; Luke Arbogast, Ioannis Karageorgos, Yuxue Liang, Eric Yan, Qian Dong, and Yamil Simon for their help in preliminary experiments; Jeanice Brown Thomas, Randy Isenburg, Dawn Bruchey, Paul Thompson, and Jeff Hudgens for their help in logistics; and Honeyeh Zube and J'aime Maynard for help in legal matters. We thank the following for their help in the study and study design: Chuck Hague and Cara Wong (BioMarin); Dongbo Bu (Chinese Academy of Sciences); Joon Ho Eom (Korea National Institute of Food and Drug Safety); Joe Zaia and Nancy Leymarie (Boston U); Manfred Wuhrer and David Falck (Leiden U); Louise Royle (Ludger Ltd.); CT Yuen (NIBSC); Carlito Lebrilla (U California Davis); Pauline Rudd (NIBRT); Ron Orlando and Parastoo Azadi (U Georgia); John Cipollo (FDA); Yehia Mechref (Texas Tech U); Albert Heck (Utrecht U); Paul Rudnick (Spectragen Informatics); Eoin Cosgrave (Seattle Genetics); Doug Sheeley (NIH); Nuno Bandeira (U San Diego); and Thuy Tran and Vern Reinhold (U New Hampshire). Thanks to Lai-Xi Wang (U Maryland); Chris Barton (Shire); Ellen Guthrie, Paula Magnelli and Beth Mcleod (NEB); Sam Tep, Justin Hyche, and Ted Haxo (Prozyme); and Charles Nwosu (Takeda) for their help in modifying NISTmAb. We thank Manfred Wuhrer, Pauline Rudd, Mustafa Varoglu (SAGE Therapeutics), and Ron Orlando for their help in publicity.
Footnotes
* This work was supported by: 1. Science Foundation Ireland Starting Investigator Research under Grant 13/SIRG/2164 to R.S.; 2. Swedish Research Council under Grant 8266 to G.L. and J.N.; 3. Alberta Glycomics Centre to C.C.; 4. National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) under Grant K01-DK101632 to J.W.F.; 5. Australian Research Council Centre of Excellence for Nanoscale BioPhotonics (CNBP) under Grant CE1401000003 to N.H.P. and A.E.-D.; 6. European Union (EU) FP7 program HighGlycan under Grant 278535 to R.O.F. and P.M.R.; 7. National Institutes of Health (NIH) Research Resource for Biomedical Glycomics under Grant P41GM10349010 and Grant 1S10OD018530 to P.A.; 8. Austrian Research Promotion Agency Laura Bassi Center of Expertise under Grant 822757 to C.G.-G.; 9. National Natural Science Foundation of China under Grant 31600650, 31671369, and 31770775D to D.B., S.S., C.H. and Y.L.; 10. Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) “The concert of dolichol-based glycosylation: from molecules to disease models” under Grant FOR2509 to E.R.; 11. European Union (EC) “IMforFuture” under Grant 721815 to E.R.; 12. German Federal Ministry of Education and Research (BMBF) “Die Golgi Glykan Fabrik 2.0” under Grant 031A557 to S.C. and E.R.; 13. Health Canada, Government of Canada to T.D.C. and M.C.; 14. European Union FP7 GastricGlycoExplorer ITN under Grant 316929 and the Swedish Research Council under Grant 621–2013-5895 to N.G.K.; 15. National Research Council of Science and Technology Creative Allied Project (CAP) under Grant NTM2371511 to J.S.Y.; 16. Hungarian Government project under Grant NKFIH-K 116263 and BIONANO GINOP-2.3.2- 15–2016-00017 to A.G.; and 17. National Research Foundation of Korea under Grant NRF-2013M3A9B6075933 to H.J.A.
 This article contains supplemental Figures, Tables, and Discussion.
This article contains supplemental Figures, Tables, and Discussion.
DISCLAIMER: The authors declare that they have no conflicts of interest with the contents of this article. Any mention of commercial products is for information only; it does not imply recommendation or endorsement by any of the participating institutions. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the participating institutions.
Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
a Definition of naming convention of glycans is in the Experimental Procedures section.
b ppm is a unit of mass measurement error (Δm/z)/(m/z) * 1000000, expressed as unified atomic mass units (Daltons) divided by charge.
1 The abbreviations used are:
- MS
- generic mass spectrometry or first stage mass spectrometry
- AA
- aminobenzoic acid
- AB
- aminobenzamide
- APTS
- 9-aminopyrene-1,4–6-trisulfonate
- C4
- C4 (butyl) desalting column
- C8
- C8 (octyl) desalting column
- CE
- capillary electrophoresis
- CFG
- Consortium for Functional Glycomics
- DI
- direct infusion
- Exo
- exoglycosidase
- Fab
- antigen-binding fragment of a monoclonal antibody
- Fc
- crystallizable fragment of a monoclonal antibody
- FD
- fluorescence detection
- GU
- glucose units
- HILIC
- hydrophilic interaction liquid chromatography
- HPAEC-PAD
- high performance anion exchange chromatography with pulsed amperometric detection
- IC
- ion chromatography
- IdeS
- immunoglobulin G-degrading enzyme
- LC
- liquid chromatography
- LIF
- laser-induced fluorescence detection
- LoR
- limit of reporting
- mAb
- monoclonal antibody
- MALDI
- matrix-assisted laser desorption/ionization
- MRV
- minimum reported value
- MS/MS
- tandem mass spectrometry
- MSn
- nth stage MS
- MT
- migration time
- ND
- not detected
- NISTIR
- NIST internal report
- NMR
- nuclear magnetic resonance
- NQ
- not quantified
- PA
- peak area/integration
- PGC
- porous graphitized carbon
- PH
- peak height
- PNGase F
- Peptide-N-Glycosidase F
- PS
- primary sample
- RP
- reversed-phase
- RT
- retention time
- SEC
- size-exclusion chromatography
- UOXF
- Oxford Glycobiology Institute
- xCGE
- multiplexed capillary gel electrophoresis.
REFERENCES
- 1. Yang S., Li Y., Shah P., and Zhang H. (2013) Glycomic analysis using glycoprotein immobilization for glycan extraction. Anal. Chem. 85, 5555–5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vreeker G. C. M., and Wuhrer M. (2017) Reversed-phase separation methods for glycan analysis. Anal. Bioanal. Chem. 409, 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruhaak L. R., Zauner G., Huhn C., Bruggink C., Deelder A. M., and Wuhrer M. (2010) Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem. 397, 3457–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dotz V., Haselberg R., Shubhakar A., Kozak R. P., Falck D., Rombouts Y., Reusch D., Somsen G. W., Fernandes D. L., and Wuhrer M. (2015) Mass spectrometry for glycosylation analysis of biopharmaceuticals. Trac-Trend Anal. Chem. 73, 1–9 [Google Scholar]
- 5. O'Flaherty R., Muniyappa M., Walsh I., Stockmann H., Hutson R., Saldova R., and Rudd P. M. (2016) High-throughput sequential glycoprofiling of six abundant glycoproteins IgG, IgA, IgM, transferrin, haptoglobin and alpha-1-antitrypsin in ovarian cancer. Glycobiology 26, 1430–1431 [Google Scholar]
- 6. Beck A., Wagner-Rousset E., Ayoub D., Van Dorsselaer A., and Sanglier-Cianferani S. (2013) Characterization of therapeutic antibodies and related products. Anal. Chem. 85, 715–736 [DOI] [PubMed] [Google Scholar]
- 7. Beck A., Wagner-Rousset E., Bussat M. C., Lokteff M., Klinguer-Hamour C., Haeuw J. F., Goetsch L., Wurch T., Van Dorsselaer A., and Corvaia N. (2008) Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr. Pharm. Biotechno. 9, 482–501 [DOI] [PubMed] [Google Scholar]
- 8. Hecht E. S., McCord J. P., and Muddiman D. C. (2015) Definitive screening design optimization of mass spectrometry parameters for sensitive comparison of filter and solid phase extraction purified, INLIGHT plasma N-glycans. Anal. Chem. 87, 7305–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker S. H., Taylor A. D., and Muddiman D. C. (2013) Individuality normalization when labeling with isotopic glycan hydrazide tags (INLIGHT): a novel glycan-relative quantification strategy. J. Am. Soc. Mass Spectr. 24, 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Y. L., Shihab T., Zhou S. Y., Wooding K., and Mechref Y. (2016) LC-MS/MS of permethylated N-glycans derived from model and human blood serum glycoproteins. Electrophoresis 37, 1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mechref Y., and Muddiman D. C. (2017) Recent advances in glycomics, glycoproteomics and allied topics. Anal. Bioanal. Chem. 409, 355–357 [DOI] [PubMed] [Google Scholar]
- 12. Zhou S. Y., Hu Y. L., Veillon L., Snovida S. I., Rogers J. C., Saba J., and Mechref Y. (2016) Quantitative LC-MS/MS glycomic analysis of biological samples using aminoxyTMT. Anal. Chem. 88, 7515–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang N., Goonatilleke E., Park D., Song T., Fan G. R., and Lebrilla C. B. (2016) Quantitation of site-specific glycosylation in manufactured recombinant monoclonal antibody drugs. Anal. Chem. 88, 7091–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hong Q. T., Ruhaak L. R., Stroble C., Parker E., Huang J. C., Maverakis E., and Lebrilla C. B. (2015) A method for comprehensive glycosite-mapping and direct quantitation of serum glycoproteins. J. Proteome Res. 14, 5179–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giorgetti J., D'Atri V., Canonge J., Lechner A., Guillarme D., Colas O., Wagner-Rousset E., Beck A., Leize-Wagner E., and Francois Y. N. (2018) Monoclonal antibody N-glycosylation profiling using capillary electrophoresis - Mass spectrometry: Assessment and method validation. Talanta 178, 530–537 [DOI] [PubMed] [Google Scholar]
- 16. Dong Q., Yan X., Liang Y., and Stein S. E. (2016) In-depth characterization and spectral library building of glycopeptides in the tryptic digest of a monoclonal antibody using 1D and 2D LC-MS/MS. J. Proteome Res. 15, 1472–1486 [DOI] [PubMed] [Google Scholar]
- 17. Wada Y., Azadi P., Costello C. E., Dell A., Dwek R. A., Geyer H., Geyer R., Kakehi K., Karlsson N. G., Kato K., Kawasaki N., Khoo K. H., Kim S., Kondo A., Lattova E., Mechref Y., Miyoshi E., Nakamura K., Narimatsu H., Novotny M. V., Packer N. H., Perreault H., Peter-Katalinic J., Pohlentz G., Reinhold V. N., Rudd P. M., Suzuki A., and Taniguchi N. (2007) Comparison of the methods for profiling glycoprotein glycans–HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 17, 411–422 [DOI] [PubMed] [Google Scholar]
- 18. Wada Y., Dell A., Haslam S. M., Tissot B., Canis K., Azadi P., Backstrom M., Costello C. E., Hansson G. C., Hiki Y., Ishihara M., Ito H., Kakehi K., Karlsson N., Hayes C. E., Kato K., Kawasaki N., Khoo K. H., Kobayashi K., Kolarich D., Kondo A., Lebrilla C., Nakano M., Narimatsu H., Novak J., Novotny M. V., Ohno E., Packer N. H., Palaima E., Renfrow M. B., Tajiri M., Thomsson K. A., Yagi H., Yu S. Y., and Taniguchi N. (2010) Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1. Mol. Cell. Proteomics 9, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thobhani S., Yuen C. T., Bailey M. J., and Jones C. (2009) Identification and quantification of N-linked oligosaccharides released from glycoproteins: an inter-laboratory study. Glycobiology 19, 201–211 [DOI] [PubMed] [Google Scholar]
- 20. Leymarie N., Griffin P. J., Jonscher K., Kolarich D., Orlando R., McComb M., Zaia J., Aguilan J., Alley W. R., Altmann F., Ball L. E., Basumallick L., Bazemore-Walker C. R., Behnken H., Blank M. A., Brown K. J., Bunz S. C., Cairo C. W., Cipollo J. F., Daneshfar R., Desaire H., Drake R. R., Go E. P., Goldman R., Gruber C., Halim A., Hathout Y., Hensbergen P. J., Horn D. M., Hurum D., Jabs W., Larson G., Ly M., Mann B. F., Marx K., Mechref Y., Meyer B., Möginger U., Neusüâ C., Nilsson J., Novotny M. V., Nyalwidhe J. O., Packer N. H., Pompach P., Reiz B., Resemann A., Rohrer J. S., Ruthenbeck A., Sanda M., Schulz J. M., Schweiger-Hufnagel U., Sihlbom C., Song E., Staples G. O., Suckau D., Tang H., Thaysen-Andersen M., Viner R. I., An Y., Valmu L., Wada Y., Watson M., Windwarder M., Whittal R., Wuhrer M., Zhu Y., and Zou C. (2013) Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: The ABRF Glycoprotein Research Multi-Institutional Study 2012. Mol. Cell. Proteomics 12, 2935–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reusch D., Haberger M., Maier B., Maier M., Kloseck R., Zimmermann B., Hook M., Szabo Z., Tep S., Wegstein J., Alt N., Bulau P., and Wuhrer M. (2015) Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 1: Separation-based methods. Mabs 7, 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Formolo T., Ly M., Levy M., Kilpatrick L., Lute S., Phinney K., Marzilli L., Brorson K., Boyne M., Davis D., and Schiel J. (2015) Determination of the NISTmAb Primary Structure. In State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2. Biopharmaceutical Characterization: The NISTmAb Case Study (Schiel John E., Davis Darryl L., Borisov Oleg V., eds.) Chapter 1, pp. 1–62, American Chemical Society [Google Scholar]
- 23. Schiel J. E., and Turner A. (2018) The NISTmAb Reference Material 8671 lifecycle management and quality plan. Anal. Bioanal. Chem. 410, 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Leoz M. L. A., Duewer D. L., and Stein S. E. (2017) NIST Interlaboratory Study on the Glycosylation of NISTmAb, a Monoclonal Antibody Reference Material. In NIST Internal Report (NISTIR), 8186 [Google Scholar]
- 25. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Marth J. D., Bertozzi C. R., Hart G. W., and Etzler M. E. (2009) Symbol nomenclature for glycan representation. Proteomics 9, 5398–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 27. de Jong S. E., Selman M. H., Adegnika A. A., Amoah A. S., van Riet E., Kruize Y. C., Raynes J. G., Rodriguez A., Boakye D., von Mutius E., Knulst A. C., Genuneit J., Cooper P. J., Hokke C. H., Wuhrer M., and Yazdanbakhsh M. (2016) IgG1 Fc N-glycan galactosylation as a biomarker for immune activation. Sci. Rep. 6, 28207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halim A., Westerlind U., Pett C., Schorlemer M., Ruetschi U., Brinkmalm G., Sihlbom C., Lengqvist J., Larson G., and Nilsson J. (2014) Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 [DOI] [PubMed] [Google Scholar]
- 29. Yu J., Schorlemer M., Gomez Toledo A., Pett C., Sihlbom C., Larson G., Westerlind U., Nilsson J., and Distinctive M. S. (2016) /MS Fragmentation Pathways of Glycopeptide-Generated Oxonium Ions Provide Evidence of the Glycan Structure. Chemistry 22, 1114–1124 [DOI] [PubMed] [Google Scholar]
- 30. Pompach P., Ashline D. J., Brnakova Z., Benicky J., Sanda M., and Goldman R. (2014) Protein and Site Specificity of Fucosylation in Liver-Secreted Glycoproteins. J. Proteome Res. 13, 5561–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prien J. M., Stöckmann H., Albrecht S., Martin S. M., Varatta M., Furtado M., Hosselet S., Wang M., Formolo T., Rudd P. M., and Schiel J. E. (2015) Orthogonal Technologies for NISTmAb N-Glycan Structure Elucidation and Quantitation. In State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2. Biopharmaceutical Characterization: The NISTmAb Case Study. Schiel John E., Davis Darryl L., Borisov Oleg V., eds.) Chapter 4, pp. 185–235, American Chemical Society [Google Scholar]
- 32. Benedetti E., Pucic-Bakovic M., Keser T., Wahl A., Hassinen A., Yang J. Y., Liu L., Trbojevic-Akmacic I., Razdorov G., Stambuk J., Klariæ L., Ugrina I., Selman M. H. J., Wuhrer M., Rudan I., Polasek O., Hayward C., Grallert H., Strauch K., Peters A., Meitinger T., Gieger C., Vilaj M., Boons G. J., Moremen K. W., Ovchinnikova T., Bovin N., Kellokumpu S., Theis F. J., Lauc G., and Krumsiek J. (2017) Network inference from glycoproteomics data reveals new reactions in the IgG glycosylation pathway. Nat. Commun. 8, 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pilobello K. T., Krishnamoorthy L., Slawek D., and Mahal L. K. (2005) Development of a lectin microarray for the rapid analysis of protein glycopatterns. ChemBioChem 6, 985–989 [DOI] [PubMed] [Google Scholar]
- 34. Duewer D. L., Kline M. C., Sharpless K. E., Thomas J. B., Gary K. T., and Sowell A. L. (1999) Micronutrients measurement quality assurance program: helping participants use interlaboratory comparison exercise results to improve their long-term measurement performance. Anal. Chem. 71, 1870–1878 [DOI] [PubMed] [Google Scholar]
- 35. Youden W. J. (1959) Graphical diagnosis of interlaboratory test results. Industrial Quality Control XV, 29–33 [Google Scholar]
- 36. Shirono K., Iwase K., Okazaki H., Yamazawa M., Shikakume K., Fukumoto N., Murakami M., Yanagisawa M., and Tsugoshi T. (2013) A study on the utilization of the Youden plot to evaluate proficiency test results. Accredit. Qual. Assur. 18, 161–174 [Google Scholar]
- 37. Horwitz W. (1982) Evaluation of analytical methods used for regulation of foods and drugs. Anal. Chem. 54, 67–76 [Google Scholar]
- 38. Thompson M., and Lowthian P. J. (1997) The Horwitz function revisited. J. AOAC. Int. 80, 676–679 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary results as provided by the participants, expressed as percentage total glycan, are provided in the Supplemental Information.