Abstract
Purpose
Cone rod dystrophy (CRD) is a group of inherited retinopathies characterized by the loss of cone and rod photoreceptor cells, which results in poor vision. This study aims to clinically and genetically characterize the segregating CRD phenotype in two large, consanguineous Pakistani families.
Methods
Funduscopy, optical coherence tomography (OCT), electroretinography (ERG), color vision, and visual acuity assessments were performed to evaluate the retinal structure and function of the affected individuals. Exome sequencing was performed to identify the genetic cause of CRD. Furthermore, the mutation's effect was evaluated using purified, bacterially expressed ADP-ribosylation factor-like protein 3 (ARL3) and mammalian cells.
Results
Fundus photography and OCT imaging demonstrated features that were consistent with CRD, including bull's eye macular lesions, macular atrophy, and central photoreceptor thinning. ERG analysis demonstrated moderate to severe reduction primarily of photopic responses in all affected individuals, and scotopic responses show reduction in two affected individuals. The exome sequencing revealed a novel homozygous variant (c.296G>T) in ARL3, which is predicted to substitute an evolutionarily conserved arginine with isoleucine within the encoded protein GTP-binding domain (R99I). The functional studies on the bacterial and heterologous mammalian cells revealed that the arginine at position 99 is essential for the stability of ARL3.
Conclusions
Our study uncovers an additional CRD gene and assigns the CRD phenotype to a variant of ARL3. The results imply that cargo transportation in photoreceptors as mediated by the ARL3 pathway is essential for cone and rod cell survival and vision in humans.
Keywords: ARL3, cone rod dystrophy, retinitis pigmentosa, autosomal recessive
Retinal dystrophies constitute a group of clinically and genetically heterogeneous eye disorders and can cause permanent blindness.1 These disorders include photoreceptor dystrophies (including cone and cone rod dystrophies), macular dystrophies, choroidal dystrophies, and systemic disorders with associated retinal degeneration.2 Cone rod dystrophies (CRD), which result in a loss of cone and rod photoreceptor cells, constitute a very important but rare class of retinal dystrophies. Worldwide prevalence of CRD ranges from 1 in every 30,000 to 40,000 individuals. CRD first clinically manifests during the first two decades of life as poor visual acuity. Severe or advanced cases of CRD also present with nystagmus, photoaversion, and central scotomas. CRD, reportedly, has multiple modes of inheritance, including autosomal recessive, autosomal dominant, and X-linked recessive.3,4 The high variability of clinical symptoms combined with genetic heterogeneity complicates the prognosis. Thus, accurate diagnosis, along with precise recognition of the underlying genetic defect, is crucial for efficient management of this group of retinal disorders.
Here we report findings of two consanguineous families cosegregating an autosomal recessive form of CRD and a homozygous point mutation (c.296G>T; p.Arg99Ile) in ADP-ribosylation factor-like protein 3 (ARL3), a small G protein belonging to ADP-ribosylation factor (Arf) family. In the mammalian retina, ARL3 is mainly localized on the connecting cilium myoid region of inner segments of cone photoreceptors and acts as an allosteric factor for the release of lipidated proteins bound to PDE6δ (δ subunit of phosphodiesterase) and UNC119a/b in a GTP-dependent manner.5 The release function of ARL3 is regulated by its interaction with retinitis pigmentosa 2 (RP2) and ARL13b.6,7 Recently, two homozygous missense variants (p.Arg149Cys and p.Arg149His) of ARL3 were reported to cause Joubert syndrome (Online Mendelian Inheritance in Man [OMIM] #213300), which is characterized by hypoplasia of the cerebellar vermis, developmental delays, renal anomalies, and rod-cone dystrophy in human families.8 Furthermore, two independent studies reported a heterozygous variant p.(Tyr90Cys) of ARL3 associated with dominantly inherited retinitis pigmentosa in two European-descent pedigrees.9,10 Similarly, impaired development and degeneration of photoreceptors phenotype was observed in Arl3 knockout mice.11–13 In contrast to these previous reports, affected individuals of our two families that are homozygous for p.(Arg99Ile) have a nonsyndromic cone-dominant retinal dystrophy.
Materials and Methods
Subjects
This observational study was conducted under institutional review board–approved protocols in accordance with the Declaration of Helsinki for the release of clinical information, family history, and blood extraction for genetic testing. Informed consent was obtained after the study risks and benefits were thoroughly explained. We evaluated the medical history via interviews to identify other possible clinical features beyond vision impairment and rule out potential environmental causes. Funduscopy (TRC-50EX; Topcon, Corp., Tokyo, Japan), optical coherence tomography (3D Optical Coherence Tomography, DRI OCT Triton; Topcon, Corp.), color vision using the Ishihara Chart, visual acuity, and full field electroretinography (ERG) using skin sensor strips and a handheld portable device (Test Protocol# ISCEV 5 Step Light First cd, RETeval portable ERG; Konan Medical, Irvine, CA, USA) were performed by an ophthalmologist to characterize the vision disorder segregating in both families. The ERG data were then compared with the normative values of 50 normal controls.
Whole Exome Sequencing and Sanger Confirmation
Whole genomic DNA was extracted from the whole blood using standard methods. Whole exome sequencing using the genomic DNA sample of one affected member from each family was performed as previously described.14 Briefly, genomic libraries were recovered for exome enrichment using the NimbleGen EZ Exome V2 kit (Roche Diagnostics, San Francisco, CA, USA). The libraries were sequenced on an Illumina HiSeq4000 (Illumina, San Diego, CA, USA), and generated approximately 30 million paired end reads that were each 100 bases long. Data analysis was performed using the Broad Institute's Genome Analysis Toolkit (GATK) (Broad Institute, Cambridge, MA, USA) and the reads were aligned with the Illumina Chastity Filter with the Burrows Wheeler Aligner (BWA) (Broad Institute).15,16 The variant sites were called using the GATK. Single nucleotide variant (SNV) calls were filtered using the variant quality score recalibration method.15 Sanger sequencing was performed using standard methods to determine the cosegregation of the ARL3 variant with vision impairment in both families. Primers for Sanger sequencing were designed using Primer3. PCR amplification and DNA sequencing were performed as previously described.17 Pathogenic variant nomenclature was assigned in accordance with GenBank Accession number NM_004311, with nucleotide position 1 corresponding to the A of the ATG initiation codon.
Bioinformatics Analysis
To evaluate the protein evolutionary sequence conservation, we used clustal omega multiple sequence alignment and the Phyre2 server was used for protein 3D structure.
Bacterial Expression
Human WT ARL3 or the p.Arg99Ile mutant proteins were expressed in BL21(DE3) bacteria that had used the pET3C vector were grown overnight in ZY auto-induction medium.18 The cells were harvested and lysed with two passes through a French press and were then pelleted by centrifugation at 32,000 rpm (100,000g). Equal protein (20 μg) of whole cell lysate (pre-spin), supernatant, and resuspended pellet were run on SDS-PAGE and stained with Coomassie blue.19
Heterologous Cell Expression
Empty vector (pcDNA3.1), human ARL3 in pcDNA3.1, or ARL3 with p.Arg99Ile in pcDNA3.1 were transiently expressed in COS7, or HeLa, using Lipofectamine 2000. The cells were harvested 24 hours after transfection, lysed in buffer containing 1% CHAPS, 100 mM NaCl, and 25 mM HEPES, pH 7.4, and then clarified through centrifugation at 14,000 rpm. Lysates (20 μg) were run on a 15% SDS-PAGE gel. The membrane was blocked with 5% milk and then incubated with our ARL3 antibody and immunoblots developed with horseradish peroxidase–conjugated secondary (sheep anti-rabbit) antibody.20
Results
Clinical Description of CRD Phenotype Segregating in Two Large Pakistani Families
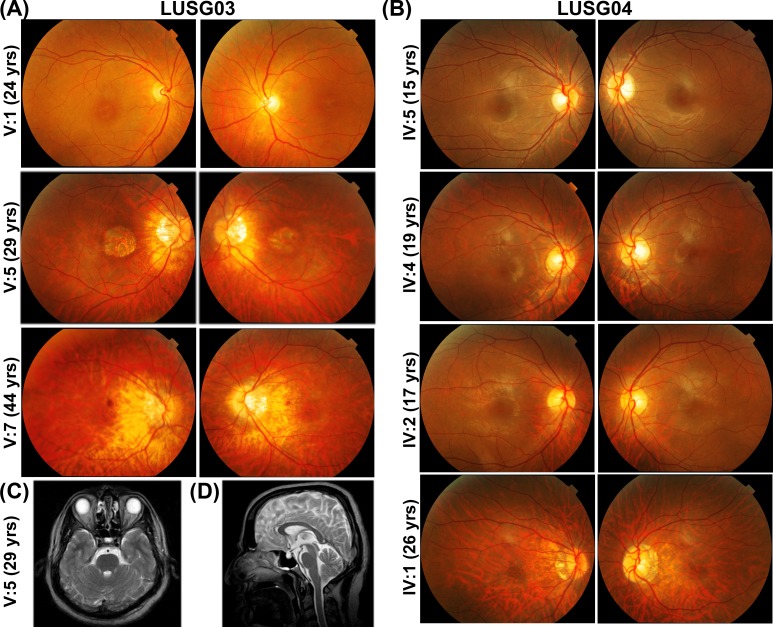
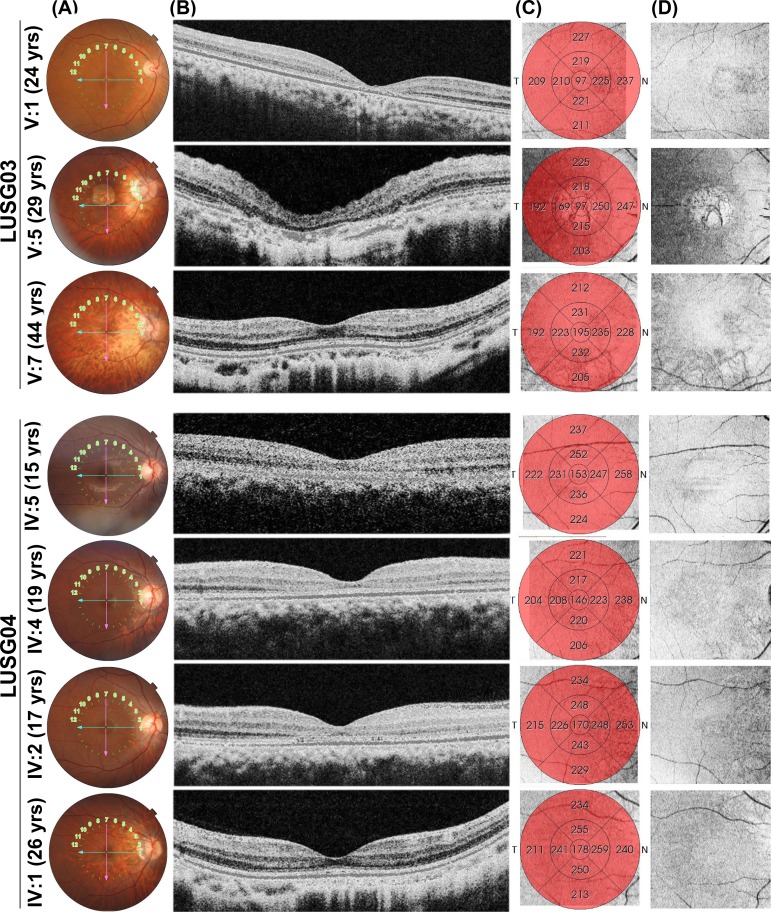
Two large, consanguineous families, LUSG03 and LUSG04, both affected with vision impairment were enrolled from the Sindh province of Pakistan (Fig. 1A). The neonatal clinical records of the affected individuals were not available at the time of enrollment. According to family medical histories, all eight affected individuals had bilateral vision impairment. We evaluated all LUSG03 and LUSG04 participating members to identify and characterize signs and symptoms of their vision impairment. No cornea opacity, color vision, or night blindness problems were noted among the affected individuals. Visual field assessment revealed normal peripheral vision, but central field vision was compromised among all the affected individuals. Affected individual V:1 (24 years old) of family LUSG03 had both moderate myopia and bilateral maculopathy without RPE atrophy, whereas individual V:5 (29 years old) had RPE atrophy. A third affected individual V:7 (44 years old) only had myopic fundus changes and foveal and outer nuclear layer thinning in the parafoveal region (Figs. 2A, 3). The fundus examination of the youngest affected individual (IV:5; 15 years old) of family LUSG04 was essentially normal, and the OCT imaging revealed foveal thinning with otherwise normal retinal architecture in both eyes (Figs. 2B, 3). Both affected individuals IV:4 (19 years old) and IV:2 (17 years old) of family LUSG04 had bull's eye maculopathy with no flecks in either eye (Figs. 2B, 4). Last, the oldest affected individual IV:1 (26 years old) of family LUSG04 demonstrated myopic fundus changes without frank myopic degeneration (Fig. 2B); moreover, his OCT imaging further confirmed myopic changes, including excess posterior curvature and retinal and choroidal thinning (Fig. 3).
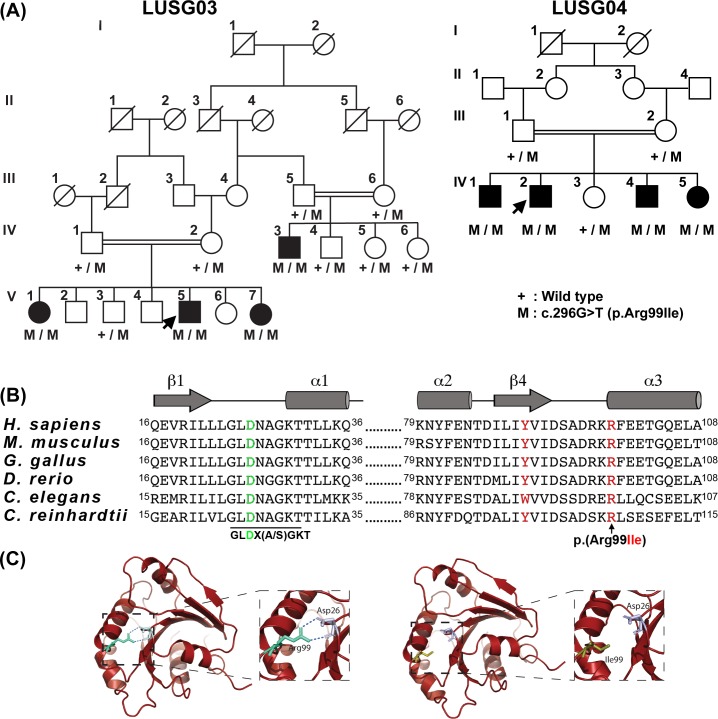
Figure 1.
A missense variant of ARL3 is associated with cone rod dystrophy in two large consanguineous Pakistani families. (A) Segregation of ARL3 allele in families LUSG03 and LUSG04. Filled and empty symbols represent affected and unaffected individuals, respectively. Double lines indicate consanguineous marriages. The genotypes (wild type, heterozygous, and homozygous mutant) of the ARL3 mutant allele are also shown for each of the participating families members. Arrows indicate probands. (B) Human ARL3 has six exons. The c.296G>T variant is located in exon 5. The ARL3 protein has a GTP-binding domain, which harbors the p.(Arg99Ile) variant. ClustalW multiple amino acid sequence alignment of ARL3 orthologs shows that p.Arg99 (red) is highly conserved across species. Shown also is the p.Tyr90 variant (red), known to be mutated in two families with dominantly inherited retinitis pigmentosa. The positively charged guanidino side chain of Arg99 is required to form ionic bonds to stabilize the orientation of the negatively charged side chain of Asp26 (green), which is central to the canonical G-1 motif (aka P-loop) found in GTP-binding proteins, conserved in the ARF family in the sequence GLD26X(A/S)GKT. (C) A homology model of the GTP-binding domain of ARL3. Replacement of arginine with isoleucine at residue 99 variant may alter/reduce the affinity of the protein for guanine nucleotides, resulting in an apoprotein that are much less stable in solution.
Figure 2.
Color fundus photography of the right and left macula, respectively, of individuals with cone dystrophy from families LUSG03 (A) and LUSG04 (B). Myopic fundus changes were variably present in LUSG03 V:5, V:7 (A) and LUSG04 IV:1, IV:4, and IV:5 (B), including tilted optic disc, alpha zone atrophy, mild temporal disc pallor, and parapapillary RPE thinning consistent with early staphyloma formation. Macular atrophy was observed in LUSG03 V:5 and bull's eye lesions are visible in LUSG03 V:1, V:7 and LUSG04 IV:1, IV:2, and IV:4. Brain magnetic resonance imaging axial T2 image (C) and sagittal T2 image (D) of LUSG03 individual IV:5, demonstrating no increased interpeduncular fossa, normal cerebellar peduncles, and normal vermis.
Figure 3.
Spectral domain OCT (Topcon) horizontal raster scans (B) with color fundus photography reference images (A), ETDRS retinal thickness maps (C), and en face infrared reference “shadowgram” imaging (D) of right eyes for individuals from families LUSG03 and LUSG04, demonstrating anatomic changes consistent with cone dystrophy. Reference image from LUSG03 V:5 shows frank geographic atrophy with overlying photoreceptor degeneration. The cone outer-segment tip (COST) line is variably diminished, from subtle (LUSG03 V:1, V:7 and LUSG04 IV:1, IV:5) to bull's eye parafoveal loss with outer nuclear layer thinning (LUSG04 IV:2, IV:4). Retinal thickness maps demonstrate diffuse retinal thinning in all patients, particularly in those with bull's eye lesions or macular atrophy (LUSG03 V:5). Increased posterior curvature from myopia is seen among LUSG03 V:5, V:7 and LUSG04 IV:1, IV:5. En face photoreceptor layer infrared shadowgram shows hypointense round lesions in LUSG04 IV:2, IV:4, isointense fovea in LUSG03 V:7 and LUSG04 IV:1, and central hyperintensity surrounded by a ring of hypointensity in LUSG03 V:1, V:5 and LUSG04 IV:5.
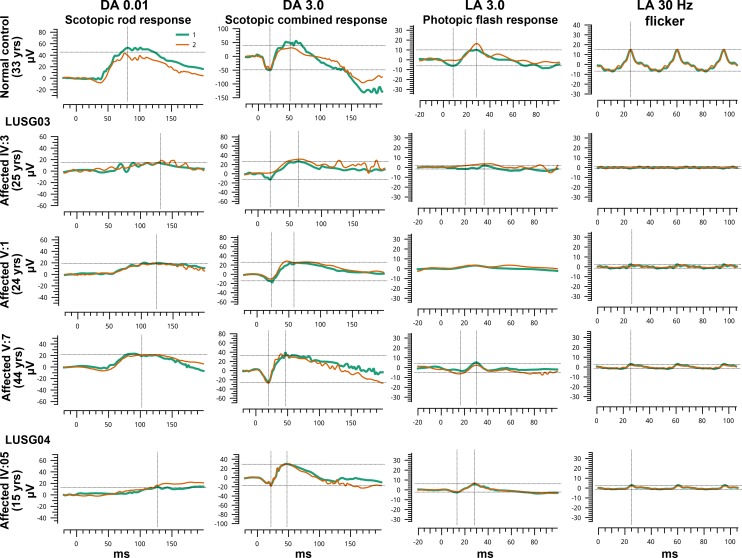
Figure 4.
Full-field ERG responses for one normal and three affected individuals homozygous for ARL3 p.(Arg99Ile) variant. Two responses (green and orange) have been superimposed for each ERG condition to show reproducibility. ERG responses of affected individuals of both families revealed moderately to severe reduction in photopic amplitudes OU, but normal scotopic, except IV:05 of family LUSG04 who had poor scotopic rod responses but normal scotopic combined responses OU (possibly insufficient dark adaptation time) and IV:03 of family LUSG03.
ERG analysis of individuals within the family LUSG03 demonstrated moderate to severe reduction in photopic responses with preserved scotopic responses (Fig. 4; Table), with the exception of individuals IV:3 and V:5, in whom scotopic responses were mildly reduced (Supplementary Table S1). ERGs from individual IV:05 of family LUSG04 had moderately reduced photopic amplitudes OU and poor scotopic rod responses but normal scotopic combined responses OU (Fig. 4; Table; Supplementary Table S1). However, the length of dark adaptation was not optimal in this case. Color vision deficits were observed in affected individuals in both families (Table). Taking into consideration fundus photography (Fig. 2) and spectral domain OCT imaging findings (Fig. 3) including bull's eye macular lesions, macular atrophy, parafoveal and central photoreceptor thinning, absence of lipofuscin flecks, and normal vision at birth, no corneal haze, and primarily attenuated phototopic responses, individuals demonstrated features that were most consistent with CRD. The findings were typically symmetrical between both eyes.
Table.
Clinical Details of the Families Segregating the ARL3 Variant
|
ID |
Disease Status |
ARL3
Genotype |
Age (y) |
Age of Onset of Symptoms |
Best-Corrected Visual Acuity: OS;OD |
Refraction |
Color Vision |
Photophobia: OS/OD |
ERG Status |
| LUSG-03 | |||||||||
| IV:4 | Normal | WT/M | 8 | NA | 6/6;6/6 | 0.00 | Normal | Neg | Normal rod and cone response |
| V:3 | Normal | WT/M | 32 | NA | 6/6;6/6 | 0.00 | Normal | Neg | Normal rod and cone response |
| IV:3 | Affected | M/M | 25 | First decade | 6/36;6/36 | −5.5/−5.5 | Normal | ++/++ | Mildly reduced scotopic amplitudes with normal implicit time, severely reduced photopic amplitudes and very delayed implicit times OU |
| V:1 | Affected | M/M | 24 | First decade | 6/36;6/36 | −5.25/−5.50 | Impaired | ++/++ | Normal scotopic, reduced photopic amplitudes (severe OD, moderate OS), delayed implicit times OU |
| V:5 | Affected | M/M | 29 | First decade | 6/60;6/60 | −5.75/−5.50 | Normal | +/+ | Normal scotopic, severely reduced photopic amplitudes OU |
| V:7 | Affected | M/M | 44 | First decade | 6/24;6/24 | −6.50/−6.0 | Impaired | +/+ | Normal scotopic, severely reduced photopic amplitudes OU |
| LUSG-04 | |||||||||
| IV:1 | Affected | M/M | 30 | First decade | 6/36;6/36 | −5.0/−5.0 | Normal | +/+ | Reduced amplitudes OU |
| IV:2 | Affected | M/M | 25 | First decade | 6/24;6/24 | −4.0/−4.75 | Normal | +/+ | Reduced amplitudes OU |
| IV:5 | Affected | M/M | 11 | First decade | 6/9;6/9 | −3.25/−2.50 | Impaired | +/+ | Moderately reduced photopic amplitudes OU, poor scotopic rod responses but normal scotopic combined responses OU (possibly insufficient dark adaptation time) |
M, mutant; NA, not applicable; Neg, negative; WT, wild type.
ARL3 Missense Variant Is Associated With CRD Phenotypes in Both Families
To determine the genetic cause of this CRD phenotype, whole exome sequencing was performed for individuals V:1 and IV:5 of families LUSG03 and LUSG04, respectively. Autosomal recessive inheritance, both homozygous and compound heterozygous, was assumed during the filtering stages (Supplementary Table S2). A novel transition variant (c.296G>T) was found in the ARL3 gene for each individual (Fig. 1A). Sanger sequencing of ARL3 revealed segregation of the c.296G>T variant with the cone dystrophy phenotype in participating individuals from both families (Fig. 1A). In humans, ARL3 is composed of six exons, encoding a 182-residue GTP-binding protein (Fig. 1B). The predicted p.(Arg99Ile) missense change, due to c.296G>T variant, was present in the GTP-binding domain of ARL3 (Fig. 1B). This variant was present in an evolutionarily conserved region (Fig. 1B) and absent in the NHLBI 6500 exome variants and the ExAC and gnomAD databases, as well as in 270 ethnically matched chromosomes, and is predicted to be pathogenic by various in silico algorithms (Supplementary Table S3).
There was no apparent relationship found between the LUSG03 and LUSG04 families. However, as both families had the same pathogenic variant, segregating with the CRD phenotype, we performed a haplotype analysis using 35 single nucleotide polymorphisms surrounding the ARL3 variant to determine whether Founder's effect played a role in this reoccurring variant. The results revealed common ancestry for the c.296G>T allele in both Pakistani families (Supplementary Table S4).
ARL3 Missense Variant Affects the Stability of the Encoded Protein
To determine the effect of p.(Arg99Ile) variant on the encoded ARL3, we performed molecular modeling using the Phyre2 software and numerous structures available on the Protein Database (e.g., 1FZQ). The positively charged guanidino side chain of Arg99 is essential for the formations of ionic bonds that stabilize the orientation of the negatively charged side chain of Asp26, which is central to the canonical G-1 motif (aka P-loop) found in GTP-binding proteins, conserved in the ARF family in the sequence GLD26X(A/S)GKT, and also involved in direct binding of the β-phosphate of GXP (GTP or GDP) and magnesium in the nucleotide binding site. We noticed that both Arg99 and Asp26 were conserved in 27 of the 30 members of the human ARF family, with differences only occurring in currently uncharacterized proteins. Molecular modeling suggested that the p.(Arg99Ile) variant may alter/reduce the affinity of the protein for guanine nucleotides, resulting in an apoprotein that is much less stable in solutions (Fig. 1C).
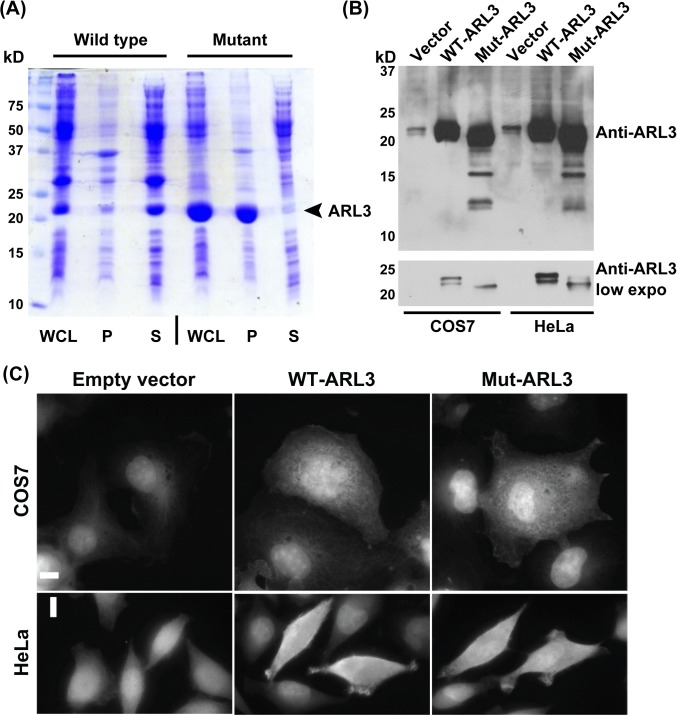
Next, to functionally evaluate the consequences of the p.Arg99Ile variant on the encoded protein, we expressed, as previously described,20,21 both wild-type and mutant ARL3 proteins in bacteria and mammalian cells (Figs. 5A, 5C). When expressed in bacterial cells, the overwhelming majority (estimated by eye from multiple preparations at >90%) of wild-type ARL3 remains in the supernatant (S100) after centrifugation at 100,000g and is soluble, whereas almost all of the ARL3[R99I] was found in the pellet (Fig. 5A), which indicates insolubility of the protein. This dramatic redistribution of ARL3 due to the insoluble fraction had not been seen previously from the mutation of several other residues of the closest paralogs, ARL3 and ARL2,22 or of other members of the ARF family, with the exception of ARL13B, which has a relatively low affinity for guanine nucleotides and is insoluble when expressed in bacteria.23 Exogenous expression of ARL3 or p.Arg99Ile in COS7 or HeLa cells revealed that the mutant migrated somewhat faster than either the endogenous or overexpressed wild-type proteins (Fig. 5B). Faster-migrating immunoreactive bands were also evident and consistent with ARL3[R99I] degradation (Fig. 5B). These characteristics are consistent with a less stable protein or one more accessible to proteases. However, no apparent difference in the cytosolic localization of ARL3[R99I] was observed in transiently transfected COS7 or HeLa cells compared with ARL3 (Fig. 5C). Thus, these studies suggest that the p.Arg99Ile variant associated with cone rod dystrophy in LUSG03 and LUSG04 families affects the stability of the encoded ARL3 and likely consequently negatively impacts its ability to bind guanine nucleotides with high affinity to allow it to remain folded in a solution.
Figure 5.
The p.Arg99Ile variant affects the stability of the encoded protein. (A) SDS-PAGE stained with Coomassie blue revealed the vast majority of ARL3 to be soluble, as evident by its presence in the 100,000g supernatant (S100). In contrast, virtually all of the R99I mutant is in the pellet (P100), indicating an insoluble protein. (B) Western blot analysis revealed increased evidence of degradation of ARL3 harboring p.Arg99Ile variant when expressed in mammalian (COS7, and HeLa) cells. Immunoblotting with anti-ARL3 antibody revealed endogenous ARL3 (vector lane), whereas strong expression was observed when COS7 and HeLa were transiently transfected with ARL3 cDNA constructs encoding either wild-type or p.Arg99Ile mutant protein. The p.Arg99Ile mutant expresses to comparable levels as the wild-type protein in both cell types; however, it is all found to migrate faster than either of the endogenous or overexpressed wild-type bands. In addition, smaller immunoreactive bands, consistent with ARL3-p.Arg99Ile degradation, can be seen in the prolonged exposure (upper) image. These results are consistent with a less stable protein or a protein more accessible to proteases. (C) Expression of wild-type and mutated forms of human GFP-tagged ARL3 in COS7 and HeLa cells revealed no apparent difference in the distribution of encoded proteins. Scale bars apply to all panels and denote 10 μm.
Discussion
This study of two families reports a novel missense variant p.(Arg99Ile) in ARL3 affecting seven individuals with CRD. In both families, in which the p.(Arg99Ile) variant segregated, the earliest signs of CRD (foveal thinning; LUSG04 IV:5) were observed during the second decade of life (15 years old). Additionally, the oldest individuals in both families demonstrated retinal thinning. Previous studies have demonstrated the role of ARL3 in the transport of cargo to photoreceptors.11,24,25 ARL3 acts as a GTPase, switching between GTP-bound (on) and GDP-bound (off) states and is involved with the traffic of proteins and lipids from their site of synthesis within inner segments to their site of action in outer segments.25
Recently, missense variants in ARL3 were reported to cause Joubert syndrome, characterized by hypoplasia of the cerebellar vermis, developmental delay, renal anomalies, and rod-cone dystrophy in human families.8 Significantly reduced levels of INPP5E and NPHP3 were observed in the cilia of the fibroblast cells derived from the ARL3-deficient patients.8 In collaboration, we further assessed the visual impairment of an affected individual with Joubert syndrome secondary to a ARL3 homozygous p.(Arg149His) variant. Full-field ERGs showed evidence of severe loss of rod and cone function in both eyes, making it impossible to confirm definitively if this was a primary cone-rod or rod-cone dystrophy. Clinical data of our two large, consanguineous human families were consistent with a nonsyndromic CRD phenotype cosegregating with a missense variant p.(Arg99Ile) of ARL3. We re-evaluated affected individuals of our families for additional ciliopathy phenotypes. Detailed clinical evaluation and kidney ultrasound revealed no features of Joubert syndrome. Furthermore, brain magnetic resonance imaging of one subject was normal, and evidence of molar teeth was not present (Figs. 2C, 2D). Brainstem, ventricle, cerebellum, and cerebellar peduncle morphology were essentially normal.
Our bioinformatic analysis and in vitro studies suggest that the p.Arg99Ile variant results in a less stable protein (based on insolubility of the mammalian protein when expressed in bacteria, altered electrophoretic migration, and increased degradation after expression in cultured mammalian, all compared with wild-type ARL3; Fig. 5) and loss of side chain interactions in the folded protein (Fig. 1C), which is predicted to compromise the binding of guanine nucleotides and might also indirectly affect the interaction with RP2 and ARL13b in vivo, leading to defects in the transportation of myristoylated and prenylated proteins in the photoreceptors.26
It is unclear why the ARL3 p.Arg99Ile mutation specifically led to deterioration of the cone and rod photoreceptors rather than a multisystem ciliopathy phenotype such as Joubert syndrome in these families. In contrast to p.Arg99, the p.Arg149Cys/His variants are predicted to have no direct effect on nucleotide binding but rather, based on structural studies, they compromise the specific interaction with ARL13b,8 and any downstream pathways that depend on activation of ARL3 by the guanine nucleotide exchange factor (GEF) activity of ARL13b. Moreover, loss of ARL13b is known to cause Joubert syndrome.27 The ARF family consists of six ARFs, 22 ARLs, and two SARs.28 Although 16 GEFs have been identified to act on the six ARFs, only one GEF has been identified for the 22 GEFs (ARL13b acting as such on ARL3).23,29 Thus, we speculate that we will find several other proteins capable of activating ARL3, leading to the regulation of currently unknown pathways; however, making predictions that link specific mutations to specific phenotypes is risky. It is plausible that impaired interaction with ARL13b due to p.Arg149Cys/His variants could yield a widespread loss of the ability to activate ARL3 and thus cause Joubert syndrome.
On the other hand, due to the limitations of in vitro studies, including protein folding differences between prokaryotes and eukaryotes, chaperone activity, and cell type–specific interactors, the in vivo impact of the p.Arg99Ile variant cannot be inferred with confidence at this time. Due to restricted phenotypic outcome, it is plausible that in vivo p.Arg99Ile might have a milder impact, perhaps a weakened affinity for nucleotides or a greater tendency toward degradation/protease action, but retention of most functions, and thus may be a hypomorphic variant of ARL3. However, further studies using animal knock-in model will be needed to address the specific roles of ARL3 in the development of retinal sensory epithelium, cytoskeletal organization, and ARL3-mediated signaling in photoreceptor cells and other cell types. In conclusion, our clinical and molecular analysis of ARL3 allele may prove useful for future genetic diagnosis and counseling, as well as for molecular epidemiology studies of CRDs in humans.
Supplementary Material
Acknowledgments
The authors thank the participating patients and their families, as well as the health care professionals involved in the patient care; Sairah Yousaf for technical assistance; Haris Rapjut for assistance with the ERGs; and Mariya Ahmed and Dimitria Gomes for constructive comments on the manuscript.
Supported by Higher Education Commission of Pakistan NRPU Grant 2835 to AMW. This study was also supported by National Institutes of Health: National Institute on Deafness and Other Communication Disorders Grant R01DC016295 (to ZMA) and National Institute of General Medical Sciences Grant R35GM122568 (to RAK and CS). JAS is funded by Kidney Research UK and the Northern Counties Kidney Research Fund.
Disclosure: S.A. Sheikh, None; R.A. Sisk, None; C.R. Schiavon, None; Y.M. Waryah, None; M.A. Usmani, None; D.H. Steel, None; J.A. Sayer, None; A.K. Narsani, None; R.B. Hufnagel, None; S. Riazuddin, None; R.A. Kahn, None; A.M. Waryah, None; Z.M. Ahmed, None
References
- 1.Bessette AP, DeBenedictis MJ, Traboulsi EI. Clinical characteristics of recessive retinal degeneration due to mutations in the CDHR1 gene and a review of the literature. Ophthalmic Genet. 2018;39:51–55. doi: 10.1080/13816810.2017.1363244. [DOI] [PubMed] [Google Scholar]
- 2.Bryant L, Lozynska O, Maguire AM, Aleman TS, Bennett J. Prescreening whole exome sequencing results from patients with retinal degeneration for variants in genes associated with retinal degeneration. Clin Ophthalmol. 2018;12:49–63. doi: 10.2147/OPTH.S147684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulanger-Scemama E, El Shamieh S, Demontant V, et al. Next-generation sequencing applied to a large French cone and cone-rod dystrophy cohort: mutation spectrum and new genotype-phenotype correlation. Orphanet J Rare Dis. 2015;10:85. doi: 10.1186/s13023-015-0300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grayson C, Bartolini F, Chapple JP, et al. Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum Mol Genet. 2002;11:3065–3074. doi: 10.1093/hmg/11.24.3065. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz N, Lane A, Jovanovic K, et al. Arl3 and RP2 regulate the trafficking of ciliary tip kinesins. Hum Mol Genet. 2017;26:2480–2492. doi: 10.1093/hmg/ddx143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephen LA, Elmaghloob Y, Ismail S. Maintaining protein composition in cilia. Biol Chem. 2017;399:1–11. doi: 10.1515/hsz-2017-0168. [DOI] [PubMed] [Google Scholar]
- 8.Alkanderi S, Molinari E, Shaheen R, et al. ARL3 mutations cause Joubert syndrome by disrupting ciliary protein composition. Am J Hum Genet. 2018;103:612–620. doi: 10.1016/j.ajhg.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtan JP, Teigen K, Aukrust I, Bragadottir R, Houge G. Dominant ARL3-related retinitis pigmentosa. Ophthalmic Genet. 2019;40:124–128. doi: 10.1080/13816810.2019.1586965. [DOI] [PubMed] [Google Scholar]
- 10.Strom SP, Clark MJ, Martinez A, et al. De novo occurrence of a variant in ARL3 and apparent autosomal dominant transmission of retinitis pigmentosa. PLoS One. 2016;11:e0150944. doi: 10.1371/journal.pone.0150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanke-Gogokhia C, Wu Z, Gerstner CD, Frederick JM, Zhang H, Baehr W. Arf-like protein 3 (ARL3) regulates protein trafficking and ciliogenesis in mouse photoreceptors. J Biol Chem. 2016;291:7142–7155. doi: 10.1074/jbc.M115.710954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanke-Gogokhia C, Wu Z, Sharif A, Yazigi H, Frederick JM, Baehr W. The guanine nucleotide exchange factor Arf-like protein 13b is essential for assembly of the mouse photoreceptor transition zone and outer segment. J Biol Chem. 2017;292:21442–21456. doi: 10.1074/jbc.RA117.000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am J Pathol. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riazuddin S, Hussain M, Razzaq A, et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaworek TJ, Kausar T, Bell SM, et al. Molecular genetic studies and delineation of the oculocutaneous albinism phenotype in the Pakistani population. Orphanet J Rare Dis. 2012;7:44. doi: 10.1186/1750-1172-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Bowzard JB, Sharer JD, Kahn RA. Assays used in the analysis of Arl2 and its binding partners. Methods Enzymol. 2005;404:453–467. doi: 10.1016/S0076-6879(05)04040-1. [DOI] [PubMed] [Google Scholar]
- 20.Cavenagh MM, Breiner M, Schurmann A, et al. ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J Biol Chem. 1994;269:18937–18942. [PubMed] [Google Scholar]
- 21.Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell. 2006;17:2476–2487. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francis JW, Newman LE, Cunningham LA, Kahn RA. A trimer consisting of the tubulin-specific chaperone D (TBCD), regulatory GTPase ARL2, and beta-tubulin is required for maintaining the microtubule network. J Biol Chem. 2017;292:4336–4349. doi: 10.1074/jbc.M116.770909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanova AA, Caspary T, Seyfried NT, et al. Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF) J Biol Chem. 2017;292:11091–11108. doi: 10.1074/jbc.M117.784025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright KJ, Baye LM, Olivier-Mason A, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright ZC, Singh RK, Alpino R, Goldberg AF, Sokolov M, Ramamurthy V. ARL3 regulates trafficking of prenylated phototransduction proteins to the rod outer segment. Hum Mol Genet. 2016;25:2031–2044. doi: 10.1093/hmg/ddw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Qin Y, Yu S, et al. Pathogenic mutations in retinitis pigmentosa 2 predominantly result in loss of RP2 protein stability in humans and zebrafish. J Biol Chem. 2017;292:6225–6239. doi: 10.1074/jbc.M116.760314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantagrel V, Silhavy JL, Bielas SL, et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sztul E, Chen PW, Casanova JE, et al. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol Biol Cell. 2019;30:1249–1271. doi: 10.1091/mbc.E18-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotthardt K, Lokaj M, Koerner C, Falk N, Giessl A, Wittinghofer A. A. G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. Elife. 2015;4(e11859) doi: 10.7554/eLife.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.