Abstract
Although early growth response-1 (EGR-1) has been shown as a key transcription factor in controlling cell growth, proliferation, differentiation, and angiogenesis, its role in the development of esophageal cancer is poorly understood despite the high frequency of this disease in many parts of the world. Here, immunohistochemistry showed that EGR-1 is overexpressed in 80% of esophageal tumor tissues examined. Furthermore, EGR-1 is constitutively expressed in all esophageal cancer cell lines analyzed. Esophageal squamous carcinoma WHCO1 cells stably transfected with EGR-1 short hairpin RNA displayed a 55% reduction in EGR-1 protein levels, 50% reduction in cell proliferation, a 50% reduction in cyclin-dependent kinase 4 levels, and a 2-fold induction in p27Kip1 levels associated with a G2-M cell cycle arrest. EGR-1 knockdown also caused a marked induction in IκBα expression, an effect also observed in GROβ RNA interference-expressing WHCO1 cells, because EGR-1 lies downstream of GRO/CXCR2 signaling. Furthermore, p65 mRNA levels were also reduced in cells treated with either short hairpin RNA EGR-1 or small interfering RNA EGR-1. Immunohistochemical analysis indicated that p65 is elevated in 78% (n = 61) of esophageal tumor sections analyzed. Moreover, nuclear factor-κB inhibition with either sodium salicylate or p65 RNA interference led to a significant reduction in GROα and GROβ expression. These results indicate that EGR-1 and nuclear factor-κB mediate GRO/CXCR2 proliferative signaling in esophageal cancer and may represent potential target molecules for therapeutic intervention.
Introduction
Chemokines, a superfamily of small cytokine-like proteins, selectively regulate the recruitment and trafficking of leukocyte subsets to inflammation sites through chemoattraction (1). There is sufficient evidence to show that chemokines also play a significant role in cancer in addition to its role in development and inflammatory responses. Blocking GROα or its receptor CXCR2 results in reduction in melanoma cell proliferation and inhibition of tumor formation in a severe combined immunodeficient mouse model (2, 3). Conversely, constitutive overexpression of GROα, GROβ, or GROγ in various melanoma cell lines enhances their colony-forming activity and tumorigenicity in nude mice (2, 4, 5). In a previous study, we showed the presence of a functional autocrine chemokine loop in esophageal cancer cells involving GROα/GROβ and their cognate receptor, CXCR2 (6). Signaling along this pathway was associated with extracellular signal-regulated kinase 1/2 phosphorylation and elevated expression of EGR-1. Blockade of the GRO/CXCR2 signaling loop using either an antagonist of CXCR2 (SB225002) or knockdown of GROβ significantly reduced the proliferation of esophageal cancer cells in culture. This earlier study highlighted the central role of GRO/CXCR2 signaling in maintaining the continued proliferation of esophageal cancer cells. Although we have shown the importance of GRO/CXCR2 signaling in esophageal cancer cells, many questions around this pathway still remain. The role of EGR-1 in the GRO/CXCR2 signaling loop was not characterized, and the factors driving the expression of GROα and GROβ in these cells have yet to be elucidated. Moreover, the factors responsible for the decreased proliferation of esophageal cancer cells in response to blockade of GRO/CXCR2 signaling remain unknown.
To begin to address these questions further for esophageal cancer, it is clear that the role of EGR-1 in GRO/CXCR2 signaling needs to be explored in more detail. Early growth response-1 (EGR-1; also known as NGFI-A, Zif268, Krox24, and Tis8) is a serum-inducible zinc finger transcription factor that plays a critical role in controlling cell growth, proliferation, differentiation, and apoptosis (7–10). As an immediate-early gene that is rapidly induced in response to a wide range of extracellular stimuli, EGR-1 is thought to transduce extracellular signals into a rapid transcriptional response (11). Evidence also suggests that EGR-1 is involved in the development of human cancers such as prostate carcinomas that express high levels of EGR-1 (12–14), and there are tantalizing reports implicating EGR-1 in colon cancer (15). This study sought to examine the role of EGR-1 in the GRO/CXCR2 signaling loop in esophageal cancer.
Results
Overexpression of EGR-1 in Esophageal Squamous Cell Carcinoma
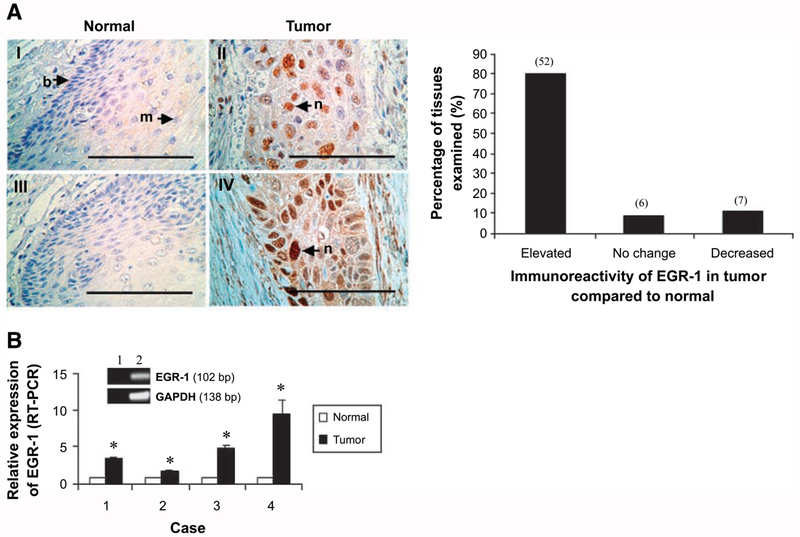
Because EGR-1 was previously implicated in GRO/CXCR2 signaling and cellular proliferation in esophageal cancer (6), we determined the expression of EGR-1 in this cancer by immunohistochemistry. EGR-1 levels were elevated in 80% (n = 65) of tumor sections analyzed compared with the matched normal tissue (Fig. 1A). Furthermore, EGR-1 was primarily located in the nuclei of tumor cells, with cytoplasmic staining observed in some tissue sections. To test whether EGR-1 mRNA levels were correspondingly elevated, real-time reverse transcription-PCR (RT-PCR) was done using specific primers designed to quantitate EGR-1 mRNA levels in samples of normal and tumor esophageal tissues. The 102 bp product generated by real-time RT-PCR (Fig. 1B) was sequenced to verify its identity. Real-time quantitative RT-PCR using total RNA isolated from esophageal tumor and adjacent normal tissues indicated that EGR-1 mRNA levels were elevated (between 1.8-fold and 9.6-fold; P < 0.01) in four esophageal tumor samples analyzed (Fig. 1B). These findings show that esophageal squamous cell carcinomas overexpress EGR-1, which is primarily located in nuclei, suggesting a role of this zinc finger transcription factor in the development and/or maintenance of this poorly understood cancer.
FIGURE 1.
Expression of EGR-1 in normal and tumor esophageal tissues. A. Formalin-fixed, paraffin-embedded esophageal tumor tissues (top, II and IV) and adjacentnormal tissues (I and III) were subjected to immunohistochemical staining using polyclonal antibodies to human EGR-1 as described in Materials and Methods. EGR-1 was primarily located in the nucleus (see arrows in II and IV). Bar, 10 μm. b, basal cells of esophageal epithelium; m, mature epithelial cells; n, nucleus. An analysis of the stained sections shows that 80% (52 of 65) of the tumor tissues express EGR-1 at higher levels than observed in the corresponding normal tissues (right). B. Total RNA extracted from biopsies (normal and tumor tissue) obtained from patients diagnosed with esophageal squamous cell carcinoma was subjected to real-time RT-PCR, and the PCR products were analyzed by agarose gel electrophoresis; real-time RT-PCR was carried out with EGR-1-specific primers as described in Materials and Methods. Data represent the standardized expression of EGR-1 mRNA in esophageal squamous cell carcinoma tumor samples and adjacent uninvolved normal esophageal samples. Bars, SD for each sample analyzed in triplicate. *, P < 0.05, Student’s t test relative to normal control. Inset, real-time RT-PCR products separated on the agarose gel for EGR-1 and glyceraldehyde-3-phosphate dehydrogenase (lane 2) and the water blank control (lane 1).
EGR-1 Promotes Proliferation of Cultured Esophageal Cancer Cells
To investigate the functional role of EGR-1 in esophageal tumorigenesis, we then examined the expression of EGR-1 in esophageal cancer cell lines. Real-time quantitative RT-PCR of total RNA isolated from cultured esophageal cancer cell lines (WHCO1, WHCO5, WHCO6, and KYSE 450) revealed that EGR-1 was constitutively expressed in all four cell lines (data not shown). WHCO1 was selected as a model system to further explore the role of EGR-1 in esophageal cancer because we have previously generated stable GROα and GROβ knockdown clones of this cell line (6).
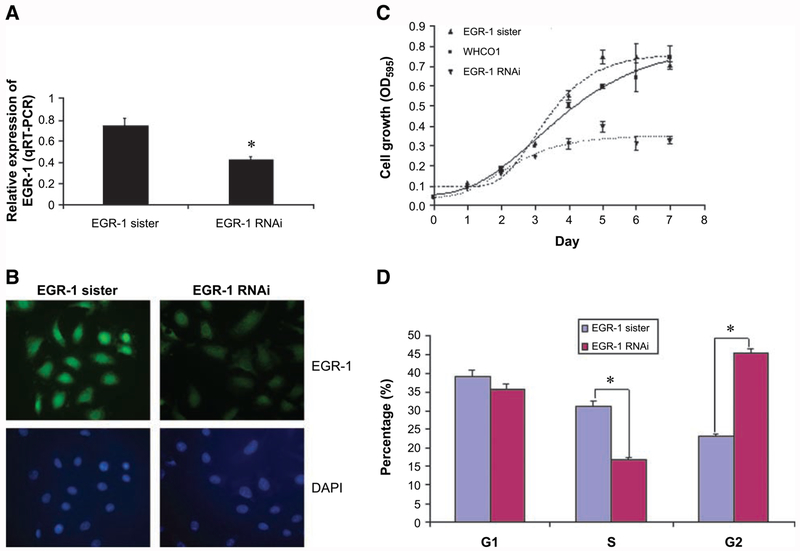
Using short hairpin RNA (shRNA)-specific targeting EGR-1, expression plasmids were constructed to develop stable clones of WHCO1 in which EGR-1 had been knocked down. Using real-time quantitative RT-PCR analysis, the WHCO1 EGR-1 RNA interference (RNAi) clone was shown to display a 50% reduction (P < 0.05) in EGR-1 mRNA levels relative to the sister control containing a random nucleotide sequence of similar composition to the EGR-1 RNAi (Fig. 2A). Furthermore, fluorescent immunocytochemistry indicated that the EGR-1 RNAi-expressing clone displayed lower levels of nuclear EGR-1 than the cells transfected with its sister control (Fig. 2B), indicating that EGR-1 expression levels had been successfully suppressed in this clone. Furthermore, a 55% reduction in EGR-1 protein expression was detected by Western blot analysis (Fig. 3A). As we expected, WHCO1 cells con-tabling EGR-1 RNAi showed a 50% reduction (P < 0.05) in cell growth (Fig. 2C), suggesting that the transcription factor, EGR-1, is essential for the sustained proliferation of WHCO1 cells. These results are consistent with previous reports in which a DNAzyme targeting the 5′-untranslated region of EGR-1 mRNA significantly inhibits human beast carcinoma growth and angiogenesis in nude mice (16). Moreover, cell cycle analysis revealed that EGR-1 knockdown cells undergo G2-M arrest in the cell cycle with a decreased S-phase fraction (Fig. 2D). Our next objective was to identify some of the EGR-1 target genes that may contribute to the regulation of the cell cycle in esophageal cancer cells and potentially affect cellular proliferation.
FIGURE 2.
Inhibitory effect of EGR-1 RNAi on cellular proliferation of esophageal cancer. A. To explore the potential role of EGR-1 in cellular proliferation of esophageal cancer, WHCO1 cells were stably transfected with either EGR-1 RNAi or EGR-1 sister control. Total cellular RNA was isolated from clones transfected with either EGR-1 RNAi or its sister control and subjected to real-time RT-PCR using primers to EGR-1. Data represent the standardized expression of EGR-1 mRNA in the indicated cell lines. Bars, SD for each sample analyzed in triplicate. *, P < 0.05, Student’s t test relative to normal control. B. Immunofluorescence staining was done using an EGR-1-specific antibody to determine the EGR-1 protein level and its subcellular localization as described in Materials and Methods. C. To detect the effect of silencing EGR-1 on the proliferation of WHCO1 cells, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was done as described in Materials and Methods and the absorbance was measured at 595 nm using a microtiter plate reader. Each point represents four analyses with the experiment repeated. D. To determine the effect of silencing EGR-1 on the progress of cell cycle, cell cycle analysis was carried out using propidium iodide staining as described in Materials and Methods and each sample was done in triplicate. *, P < 0.05.
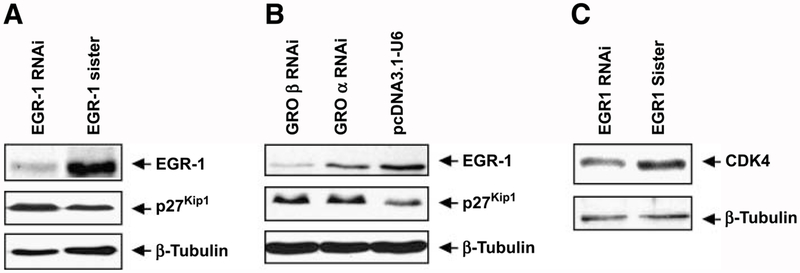
FIGURE 3.
Interference with EGR-1 signaling affects expression of key cell cycle regulators. Whole-cell lysates prepared from WHCO1 cells stably transfected with either EGR-1 RNAi, EGR-1 sister, GROα RNAi, GROβ RNAi, or vector control were separated on 7% to 15% gradient SDS-PAGE. Western blot analysis was carried out using antibodies specific to either EGR-1 or CDK4 or p27Kip1 as described in Materials and Methods.
EGR-1 Alters Expression Level of Key Cell Cycle Regulators
Because sequence analysis showed a potential EGR-1 binding site in the promoter region of genes encoding cyclin-dependent kinase (CDK) 4 and p27Kip1, key regulators of cell cycle, we hence determined the potential role of EGR-1 in the transcriptional regulation of CDK4 and p27Kip1 in esophageal cancer cells. Western blot analysis using whole-cell lysates from the EGR-1 RNAi-expressing clone and its sister control showed a 55% reduction in EGR-1 protein levels and a 2-fold induction in p27Kip1 levels in the EGR-1 knockdown clone relative to the sister control (Fig. 3A). Because our previous studies showed that depletion of GROβ in the medium of WHCO1 cells resulted in a significant reduction of EGR-1 mRNA levels (6), the levels of EGR-1 and p27Kip1 in GROβ RNAi-expressing and GROα RNAi-expressing WHCO1 clones (generated in previous study) were also determined. Western blot analysis confirmed that EGR-1 protein levels were reduced by 85% and 40% in GROβ RNAi-expressing and GROα RNAi-expressing WHCO1 clones, respectively, relative to that in vector control WHCO1 cells (Fig. 3B). Consistent with the results obtained in EGR-1 RNAi clone, the p27Kip1 levels were also strongly elevated in GROβ RNAi-expressing and GROα RNAi-expressing WHCO1 cells in which a 50% reduction in proliferation was observed in the GROβ knockdown clone (observed previously in ref. 6; Fig. 3B). Furthermore, knockdown of EGR-1 in the WHCO1 cells was associated with a 50% reduction in CDK4 levels compared with the EGR-1 sister control cells, consistent with the reduced proliferation observed in these cells (Fig. 3C). Using commercially available small interfering RNA (siRNA), we were able to transiently knock down EGR-1 (by 60%) in another esophageal cancer cell line, KYSE 450 (Supplementary Fig. S1A). This was associated with a 60% increase in p27Kip1 levels and a 30% decrease in CDK4 levels (Supplementary Fig. S1A) as was observed in WHCO1 cells (Fig. 3). Although poly (ADP-ribose) polymerase was largely intact, a small increase (1.8-fold) in poly(ADP-ribose) polymerase cleavage was observed in both KYSE 450 and WHCO1 cells, in which EGR-1 had been knocked down, relative to sister control RNAi (Supplementary Fig. S1), implicating the induction of low levels of apoptosis. Taken together, these results clearly indicated that EGR-1 contributes to the proliferation of esophageal cancer cells via the transcriptional activation of CDK4 and inhibition of p27Kip1 under conditions where the GRO/CXCR2/EGR-1 loop is engaged.
EGR-1 Contributes to Activation of Nuclear Factor-κB
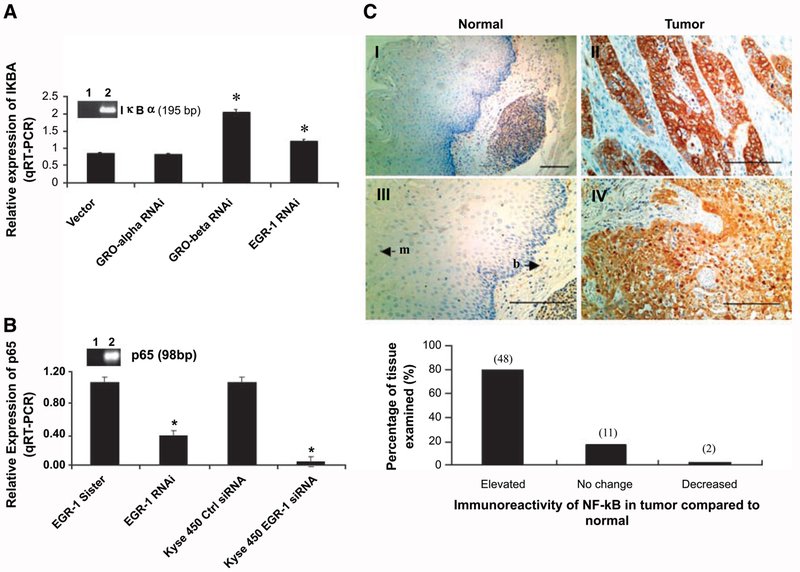
To gain a better understanding of the GRO/CXCR2/EGR-1 signaling loop, we examined other reported targets of EGR-1 that could affect this pathway. IκBα, a specific inhibitor of nuclear factor-κB (NF-κB), which plays a key role in retaining NF-κB in the cytoplasm of unstimulated cells (17, 18), has been identified as a potential target gene of EGR-1 (14, 19). We investigated the role of EGR-1 in IκBα transcription. Specific primers were designed to quantitate IκBα mRNA levels in vector control WHCO1 and WHCO1 clones containing EGR-1 RNAi, GROα RNAi, and GROβ RNAi by real-time RT-PCR. The PCR products were sequenced to verify their identity. Real-time quantitative RT-PCR indicated that IκBα mRNA levels increased by 2-fold in the GROβ RNAi-containing clone and by 50% in the EGR-1 RNAi-containing clone (P < 0.05; Fig. 4A), suggesting that EGR-1 may be involved in the transcriptional inhibition of IκBα. Furthermore, p65 levels (a subunit of the major NF-κB heterodimer complex in most cells) in WHCO1 cells stably transfected with shRNA EGR-1 or KYSE 450 cells transiently transfected with siRNA EGR-1 was decreased by 60% or >95%, respectively, as measured by quantitative real-time PCR (Fig. 4B). This suggested that EGR-1 contributed to NF-κB activation by increasing the expression level of p65 and facilitated its activation by repressing the transcription of IκBα. Because EGR-1 has been found to be overexpressed in 80% of esophageal tumor sections analyzed by immunohistochemistry (Fig. 1A), we determined the expression of p65 in esophageal cancer by immunohistochemistry. As we expected, the immunohistochemical analysis using specific antibody to p65 showed that NF-κB/p65 was elevated in 78% (n = 61) of esophageal tumor sections examined (Fig. 4C). Interestingly, 83% of sections analyzed displayed elevated levels of both EGR-1 and p65. These results implicate EGR-1 in the activation of the NF-κB pathway in esophageal cancer via transcriptional inhibition of IκBα and increased transcription of p65.
FIGURE 4.
EGR-1 contributes to constitutive activation of NF-κB in esophageal cancer. A. Total RNA extracted from WHCO1 and WHCO1 clones containing GROα RNAi, GROβ RNAi, EGR-1 RNAi was subjected to real-time RT-PCR carried out with IκBα-specific primers as described in Materials and Methods, and the PCR products were analyzed by agarose gel electrophoresis (lane 1, water blank; lane 2, IκBα). Data represent the standardized expression of IκBα mRNA in WHCO1 transfected with vector and WHCO1 clones containing GROα RNAi, GROβ RNAi, and EGR-1 RNAi. Bars, SD for each sample measured in triplicate. *, P < 0.05, Student’s t test relative to vector control WHCO1. B. Total RNA extracted from EGR-1 stable clones and KYSE 450 transiently transfected with EGR-1 siRNA was subjected to real-time RT-PCR using p65-specific primers as described in Materials and Methods. Data represent the standardized expression of p65 mRNA in the indicated cell lines. Bars, SD for each sample measured in triplicate. C. Formalin-fixed, paraffin-embedded esophageal tumor tissues (II and IV) and adjacentnormal tissues (I and III) were subjected to immunohistochemical staining using monoclonal antibody to human p65 as described in Materials and Methods; p65 was located in both cytoplasm and nucleus. Bar, 10 μm. An analysis of the stained sections shows that 78% (48 of 61) of the tumor tissues express p65 at higher levels than observed in the corresponding normal tissues (bottom).
Activated NF-κB Plays a Crucial Role in Maintaining GRO/CXCR2 Autocrine Loop
Although our previous studies implicate GROα and GROβ in cellular proliferation of esophageal cancer (6), the molecular mechanism involved in the transcriptional regulation of these chemokines in this disease is less clear. Because EGR-1 contributes to constitutive activation of NF-κB via transcriptional inhibition of IκBα and transcriptional activation of p65, and GROα and GROβ have been identified to be downstream targets of the NF-κB pathway (20), we determined the effect of EGR-1 and NF-κB on the expression of GROα and GROβ in esophageal cancer cells. Using real-time quantitative RT-PCR analysis, a 30% reduction in GROα mRNA levels and a 40% reduction in GROβ mRNA levels in the EGR-1 RNAi-expressing clone were observed (P < 0.05; Supplementary Fig. S2), suggesting that EGR-1 is indeed involved in the expression of GROα and GROβ in esophageal cancer. The regulatory control of EGR-1 on GROα and GROβ levels is probably mediated by the NF-κB pathway, because analysis of the promoter regions of both GROα and GROβ genes failed to identify an EGR-1-binding site.
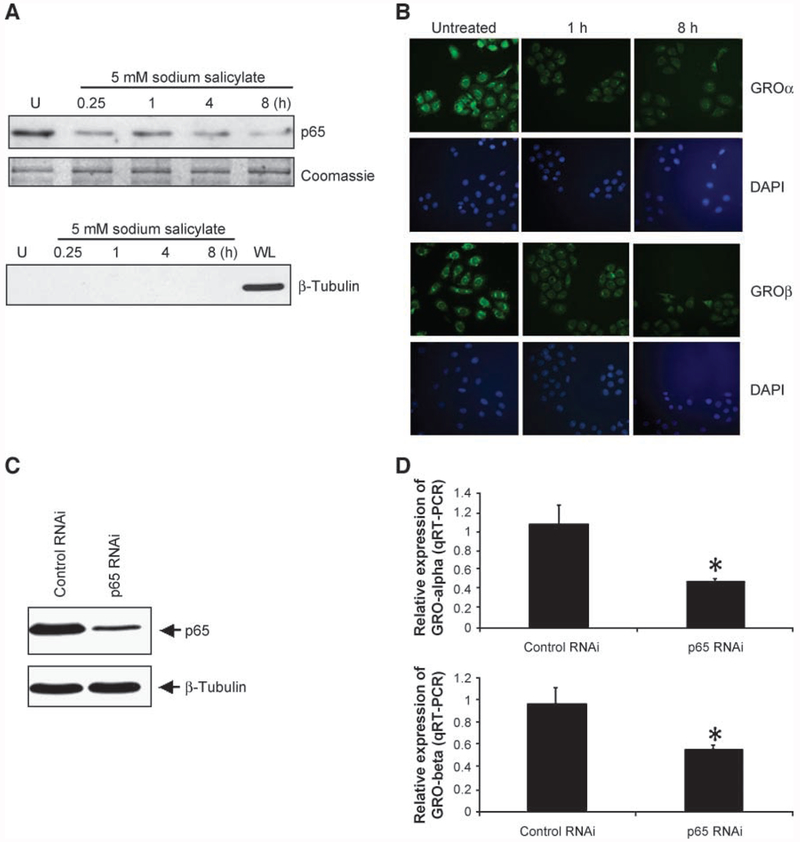
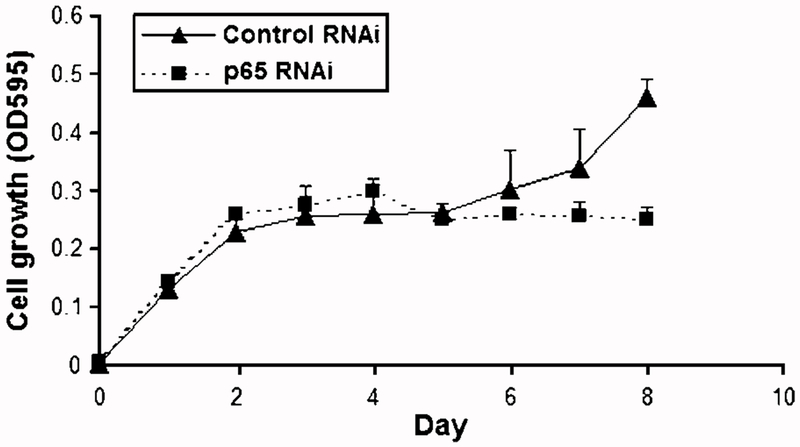
It is known that sodium salicylate functions as an inhibitor of IKK-β, which in turn phosphorylates IκB, leading to its degradation and subsequent translocation of NF-κB to the nucleus (21). To explore the role of NF-κB in up-regulation of GROα and GROβ, WHCO1 cells were treated with 5 mmol/L sodium salicylate, and the NF-κB/p65 and GROα/β levels were determined by Western blot and fluorescent immunocytochemistry, respectively. Nuclear p65 levels were significantly reduced by sodium salicylate within 15 min post-treatment and this effect was sustained for 8 h (Fig. 5A, top). Moreover, immunofluorescence staining using antibodies specific to either GROα or GROβ showed that the levels of both GROα and GROβ were correspondingly decreased in response to sodium salicylate treatment (Fig. 5B, top and bottom, respectively). Although these results strongly showed that the expression of GROα and GROβ correlates with the level of nuclear p65, we cannot exclude nonspecific effects of sodium salicylate on the other signaling pathways. To address this issue, we used a RNAi lentiviral system to specifically inhibit the expression of p65 in WHCO1 cells and isolated stably transfected cells using fluorescence-activated cell sorting. Using Western blot analysis, we showed that stably transfected WHCO1 cells containing shRNA targeting p65 showed a 70% decrease in p65 levels compared with cells transfected with control RNAi only (Fig. 5C). These cells displayed a 60% reduction in the levels of GROα mRNA and a 40% reduction in the levels of GROβ mRNA relative to control cells (Fig. 5D). Furthermore, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay revealed a 40% reduction (P < 0.05) in proliferation of the p65 knockdown cells only after day 7 (Fig. 6). Taken together, these results indicated that NF-κB plays a key role in the transcriptional activation of GROα and GROβ and maintenance of this proliferative autocrine loop in esophageal cancer.
FIGURE 5.
Activation of NF-κB is essential for GROα and GROβ expression. A. Nuclear extracts prepared from WHCO1 treated with 5 mmol/L sodium salicylate for indicated times were subjected to 10% SDS-PAGE, and Western blot analysis was done using monoclonal antibody to p65 (top). Western blot analysis of β-tubulin was carried out using nuclear extracts and whole-cell lysate to determine cytoplasmic contamination of nuclear extracts as described in Materials and Methods. WL, whole-cell lysate. B. WHCO1 cells were grown on coverslips in the same dish for preparation of nuclear extracts and exposed to 5 mmol/L sodium salicylate for the indicated times, and immunofluorescence staining was done to detect GROα (top) and GROβ (bottom) protein levels using polyclonal antibodies to either GROα or GROβ. C. Whole-cell lysates prepared from WHCO1 cells stably transfected with either p65 RNAi or control RNAi were separated on 10% gradient SDS-PAGE. Western blot analysis was carried out using monoclonal antibody specific to p65 as described in Materials and Methods. D. Total RNA isolated from p65 RNAi-expressing WHCO1 cells and control RNAi WHCO1 cells was subjected to real-time RT-PCR using GROα- and GROβ-specific primers as described in Materials and Methods. Data represent the standardized expression of GROα and GROβ mRNA in the indicated cells. Bars, SD for each sample measured in triplicate. *, P < 0.05, Student’s t test relative to normal control.
FIGURE 6.
NF-κB contributes significantly to proliferation in esophageal cancer cells. To determine the effect of p65 knockdown on the proliferation of WHCO1 cells, cells were maintained in serum-free DMEM, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was done as described in Materials and Methods, and absorbance was measured at 595 nm using a microtiter plate reader. Each point represents four analyses with the experiment done in duplicate.
Discussion
This study extends our understanding of the GRO/CXCR2 chemokine autocrine loop described previously in esophageal cancer cells (6). Considering that engagement of the GRO/CXCR2 autocrine signaling pathway triggers a complex network of signaling events, we were able to define a series of steps that constitute a self-sustaining loop for GRO/CXCR2 in esophageal cancer cells, comprising GRO/CXCR2/EGR-1 /NF-κB/GRO. EGR-1 and NF-κB constitute critical components of this autocrine pathway, with blockade of either GRO (shown previously in ref. 6) or EGR-1 resulting in reduced cell proliferation.
EGR-1 is considered a convergence point for many signaling cascades, including the Ras-Raf-MEK-extracellular signal-regulated kinase signaling pathway (22), which plays a central role in receiving and transducing signals transmitted by ligand-activated growth factor receptors and integrins. The results in this study show that esophageal squamous cell carcinomas frequently overexpress this zinc finger transcription factor facilitating the proliferation of esophageal cancer cells in culture. Although the data presented here are the first describing the contribution of EGR-1 overexpression to esophageal tumorigenesis, other studies have implicated EGR-1 in the progression of breast, colon, and prostate cancers (13–15, 23, 24).
Our results presented here show that elevated levels of EGR-1 in esophageal cancer cells is associated with activation of NF-κB via the repression of IκBα as well as elevated expression of p65, because reducing EGR-1 in WFICOl cells with either EGR-1 shRNAi or EGR-1 siRNAi substantially increased IκBα mRNA and substantially decreased p65 mRNA levels in these cells. This is consistent with previous evidence indicating that IκBα and p65 may be potential targets of EGR-1 (19, 25). The modest induction (50%) in IκBα expression in an EGR-1 knockdown clone and lack of IκBα induction in the GROα knockdown clone may reflect the incomplete knockdown of the EGR-1 and GROα mRNA levels in these clones. The reduction in GROα and GROβ expression in esophageal cancer cells treated with either sodium salicylate or p65 RNAi suggests a key role for NF-κB in maintaining the GRO/CXCR2 autocrine loop that facilitates cellular proliferation in esophageal tumorigenesis. The data presented in this study also suggest that EGR-1 may contribute to the activation of NF-κB in the majority of primary esophageal squamous cell carcinomas, because our immunohistochemistry results show that 83% esophageal tumor samples overexpress both EGR-1 and NF-κB (p65) compared with normal esophageal epithelial tissue.
G1-S and G2-M transitions are key checkpoints of the cell cycle and are normally regulated by cyclins, CDKs, and inhibitors of CDKs. The cyclin D1/cdk4 complex regulates mid to late G1 by phosphorylating retinoblastoma, which allows E2F to activate S-phase genes (26). Data presented here clearly indicate a 50% reduction in cell proliferation in EGR-1 knockdown cells and a decreased S-phase fraction in cell cycle with G2-M arrest in these cells. This correlates significantly with CDK4 levels. This is the first evidence that suggests that the GRO/CXCR2 autocrine loop in esophageal cancer cells affects CDK4 levels in these cells and perhaps other cancer cells with a functional GRO/CXCR2 autocrine loop. This direct effect of a signaling cascade on CDK4 expression is similar to the observation by Lee and Kay (27) who reported increased expression of CDK4 in corneal endothelial cells treated with fibroblast growth factor-2. Furthermore, blockade of the GRO/CXCR2 loop in this study was also associated with elevated expression of p27Kip1. Taken together, our results suggest that the GRO/CXCR2 signaling loop contributes to cellular proliferation by directly affecting CDK4 expression and repressing p27 levels. The small increase in apoptosis observed in cells in which EGR-1 levels had been knocked down with either shRNAi or siRNAi (to EGR-1) could also contribute to the reduced proliferation observed. Our findings may therefore suggest a model system in which EGR-1 and NF-κB may play a key role in transducing GRO/CXCR2 signals into a survival response and facilitating sustained proliferation.
NF-κB and EGR-1 may be convergent therapeutic targets that should be considered in the future treatment of esophageal cancer. For example, curcumin inhibits migration of human colon cancer cells (28) and suppresses cellular proliferation in mantle cell lymphoma (29) by suppressing the NF-κB pathway. An approach targeting the zinc finger transcription factor EGR-1 in breast cancer has already shown considerable promise. DNAzymes (catalytic single-stranded DNA) targeting the EGR-1 mRNA inhibit EGR-1 and fibroblast growth factor-2 expression, block endothelial cell growth, and consequently suppress neovascularization and tumor angiogenesis in several animal models (16). Although the results of the present study have highlighted the feasibility of inhibiting the transcriptional activity of EGR-1 as a therapeutic strategy in esophageal squamous cell carcinoma, the functional pleiotropy of EGR-1 should be considered in such a strategy.
Materials and Methods
Tissue Samples
All biopsies were obtained from patients who had undergone esophagectomies at Groote Schuur Hospital with histologically confirmed esophageal squamous cell carcinoma. Ethics approval was granted by the University Human Ethics Committee.
Real-time Quantitative RT-PCR
Total RNA was prepared from four paired normal and esophageal squamous cell carcinoma biopsies and from esophageal carcinoma cell lines using TRIzol (Invitrogen) according to the manufacturer’s instructions and quantified by UV absorbance at 260 nm in a DU650 spectrophotometer (Beckman Instrument). cDNA was synthesized using 5 μg total RNA and 2.5 μmol/L oligo(dT)20 primer using the SuperScript III RT-PCR system (Invitrogen). The target primers for amplifying EGR-1, IκBα, and p65 were EGR-1 forward primer 5′-AGCAGCACCTTCAACCCTCA-3′ and reverse primer 5′-CAGCACCTTCTCGTTGTTCAGA-3′ (30), IκBα forward primer 5′-CGGAAACCAGCCTCTCAAT-3′ and reverse primer 5′-TTCAGCCCACACTTTACGC-3′, and p65 forward primer 5′-CCCCACGAGCTTGTAGGAAAG-3′ and reverse primer 5′-CCAGGTTCTGGAAACTGTGGAT-3′. Each 20 μL of reaction mixture for real-time RT-PCR contained 1 mmol/L MgCl2 (for amplifying EGR-1) or 2 mmol/L MgCl2 (for amplifying IκBα), 0.5 μmol/L of each primer, and 1 μL LightCycler FastStart DNA Master SYBR Green I (Roche Diagnostics South Africa). Primers and conditions used for amplifying GROα and GROβ by real-time RT-PCR were described previously (6). Real-time quantitative RT-PCR analysis was carried out with LightCycler II (Roche Diagnostics). The glyceraldehyde-3-phosphate dehydrogenase gene was used as internal control to standardize and to test the RNA integrity with sequence for forward primer 5′-GAAGGCTGGGGCTCATTT-3′ and reverse primer 5′-CAGGAGGCATTGCTGATGAT-3′ (31). All experiments for real-time RT-PCR were done in triplicate, data were analyzed using the comparative Ct method (32), and results are shown as fold induction of mRNA.
Immunohistochemical Analysis
Samples for immunohistochemical analysis were sectioned from archived, paraffin-embedded tissues obtained from esophageal cancer patients who had undergone esophagectomies. The slides were dewaxed in xylol and incubated with 1:20 dilution of normal goat serum (DAKOCytomation Denmark) for 30 min and then incubated with either 1:1,000 dilution of anti-EGR-1 polyclonal antibody or 1:100 dilution of anti-p65 monoclonal antibody (Santa Cruz Biotechnology) at 4°C overnight. For EGR-1 staining, after rinsing in PBS, slides were incubated with 1:400 dilution of DAKO EnVision+ peroxidase (DAKOCytomation Denmark) at room temperature for 30 min; for p65 staining, slides were first incubated with biotinylated link antibody (DAKOCytomation Denmark) at room temperature for 30 min, washed in TBS for 2 × 5 min, and then incubated with streptavidin peroxidase (DAKOCytomation Denmark) for 30 min and washed in TBS for 2 × 5 min. The color was developed using 3,3′-diaminobenzidine (DAKOCytomation Denmark). Counter-staining was done with hematoxylin. The stained tissue sections were analyzed independently by two pathologists.
The criteria used for quantitating immunohistochemical staining included the staining intensity and percentage of cells stained. A range of 0 to 3 was used for classifying the intensity of staining: 0, absence of staining; 1, weak staining; 2, moderate staining; and 3, intense staining. The numbers of cells stained were recorded according to the following classification: a, <25% of cells stained; b, 25% to 50% of cells stained; c, 51% to 75% of cells stained; and d, >75% of cells stained.
Cell Culture
Four esophageal cancer cell lines, WHCO1, WHCO5, WHCO6 (33), and KYSE 450 (obtained commercially from Deutsche Sammlung von Mikroorganismen und Zellkulturen), originally established from surgical specimens of primary esophageal squamous cell carcinomas and WHCO1 cells expressing EGR-1 RNAi, EGR-1 sister control, p65 RNAi, and control RNAi were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air in DMEM containing 10% FCS. 293FT cells (Invitrogen) were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air in DMEM containing 10% FCS, 0.1 mmol/L MEM nonessential amino acids, 2 mmol/L ≻1-glutamine, 1% penicillin-streptomycin, and 500 μg/mL geneticin. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays were carried out using the Cell Proliferation Kit I (Roche Diagnostics South Africa) as described by the manufacturer using 1.5 × 103 cells plated in 96-well plates. The spectrophotometric absorbance of samples was measured at 595 nm using a microtiter plate reader.
RNA Interference
To create the shRNA plasmid constructs, complementary strands of oligonucleotides specifically targeting EGR-1 and its sister control were synthesized. For EGR-1 shRNA, oligo1 5′-ACAGCGCTCCAGTACCCGCTTCAAGAGAGCGGGTACTGGAGCGCTGTTTTTTT-3′ and oligo2 5′-AATTAAAAAAACAGCGCTCCAGTACCCGCTCTCTTGAAGCGGGTACTGGAGCGCTGTGGCC-3′; for EGR-1 sister control, oligo1 5′-ACACCGCTCCAGTACGCGCTTCAAGAGAGCGCGTACTGGAGCGGTGTTTTTTT-3′ and oligo2 5′-AATTAAAAAAACACCGCTCCAGTACGCGCTCTCTTGAAGCGCGTACTGGAGCGGTGTGGCC-3′. After annealing, double-stranded oligonucleotides were cloned into ApaI and EcoRI linearized pSilencer 1.0-U6 vector (Ambion). The insert together with U6 promoter was released by digestion with NotI and KpnI. The CMV promoter in pcDNA3.1(+) vector was released by digestion with NotI and MulI and replaced by the U6 promoter/oligonucleotide insert to obtain pcDNA3.1-U6/EGR-1RNAi and pcDNA3.1-U6/EGR-1 sister. The inserts were confirmed by DNA sequence analysis. The U6 promoter-driven shRNAs express the sense and antisense strands of EGR-1 shRNA that have a termination signal consisting of six thymidines.
To generate the shRNA lentiviral constructs, complementary strands of oligonucleotides specifically targeting p65 and control RNAi were synthesized. For p65 siRNA, oligo1 5′-/5Phos/CGCGTCCCCGAAGAGTCCTTTCAGCGGATTCAAGAGATCCGCTGAAAGGACTCTTCTTTTTGGAAAT-3′ and oligo2 5′-/5Phos/CGATTTCCAAAAAGAAGAGTCCTTTCAGCGGATCTCTTGAATCCGCTGAAAGGACTCTTCGGGGA-3′; for control RNAi, oligo1 5′-/5Phos/CGCGTCCCCTACAAGTCCTCGACTATCGTTCAAGAGACGATAGTCGAGGACTTGTATTTTTGGAAAT-3′ and oligo2 5′-/5Phos/CGATTTCCAAAAATACAAGTCCTCGACTATCGTCTCTTGAACGATAGTCGAGGACTTGTAGGGGA-3′. After annealing, double-stranded oligonucleotides were cloned into MluI and ClaI linearized pLVTHM transfer vector (Addgene). Sequences of inserts were confirmed by DNA sequence analysis.
Transfection and Immunofluorescence
Stable transfection of WHCO1 cells with either pcDNA3.1-U6/EGR-1RNAi or pcDNA3.1-U6/EGR-1 sister using FuGENE 6 Transfection Reagent (Roche Diagnostics South Africa) was carried out as recommended by the manufacturer.
To detect the expression level of EGR-1 in positive clones selected with G418, cells were cultured on coverslips, fixed and permeabilized with methanol and 4% paraformaldehyde, and incubated with 1:100 dilution of rabbit anti-EGR-1 polyclonal antibody (Santa Cruz Biotechnology) in blocking buffer at 4°C overnight. After incubation with primary antibody, cells were washed five times in PBS (pH 7.4) and incubated with 1:100 dilution of FITC-conjugated goat anti-rabbit antibody (Zymed) for 2 h at room temperature. After five washes in PBS, nuclei were stained with 4′,6-diamidino-2-phenylindole (Sigma). Fluorescence was observed by using a ×40 objective lens on an inverted microscope (Zeiss Axiovert 200M). Fluorescent immunocytochemical analysis of GROα and GROβ in WHCO1 cells treated with 5 mmol/L sodium salicylate was done as described previously (6).
KYSE 450 Transient Transfection
Cells were plated at a density of 150,000 per 35 mm dish and left overnight to settle. The following day, medium was removed from the cells and a cocktail of transfectin (Bio-Rad) containing either EGR-1 or control siRNA (Santa Cruz Biotechnology) was added according to the manufacturer’s specifications. Following a 6 h incubation with siRNA, medium was changed and cells were cultured for a further 48 h. Proteins were then harvested as described previously.
Production of Lentiviral Particles, Infection, and Cell Sorting
Transfer vector pLVTHM, packaging plasmid psPAX2, and envelop plasmid pMD2.G (Addgene) were all purified using Endo-free Maxiprep Kit (Qiagen) as described by the manufacturer’ s instruction. Lentiviral particles were generated using 293FT cells (Invitrogen) as described by manufacturer’s instruction (Addgene).5 Day 1: Plate 6 × 106 293FT cells in a T75 flask with 10 mL medium and incubate at 37°C in a humidified atmosphere of 5% CO2 and 95% air overnight. Day 2: Prepare the following plasmid mixture in a polypylene microcentrifuge tube, 7.5 μg pLVTHM RNAi vector, 3.75 μg psPAX2 packaging plasmid, and 1.25 μg pMD2.G envelop plasmid, and make up to 100 μL with serum-free Opti-MEM; add 30 μL FuGENE HD to 370 μL Opti-MEM, mix well, add the FuGENE HD dilution directly to the plasmid mixture, mix by flicking the tube, incubate for 30 min at room temperature, gently add DNA:FuGENE mixture drop-wise to cells (50-80% confluency), swirl the flask to disperse mixture evenly, and incubate cells for 24 h at 37°C in a humidified atmosphere of 5% CO2 and 95% air. Day 3: Replace with 8 mL fresh complete medium to remove the transfection reagent and continue to incubate for 24 h at 37°C in a humidified atmosphere of 5% CO2 and 95% air. Day 4: Harvest medium containing lentiviral particles, spin at 1,250 rpm for 5 min at room temperature, filter supernatant with 0.45 μm filter (Millex-HV, polyvinylidene difluoride; Millipore), and store at −80°C. Lentiviral titer was determined as described previously (34). WHCO1 cells were infected with lentiviral particles as described by the manufacturer’s instruction (Addgene). Day 1: Plate 2 × 106 WHCO1 cells per 100 mm dish and incubate at 37°C in a humidified atmosphere of 5% CO2 and 95% air overnight. Day 2: Replace medium with fresh medium containing lentiviral particles (ratio between lentiviral particles and target cells used in this study is ~2:1), add polybrene to a final concentration of 2.5 μg/mL, and incubate at 37°C in a humidified atmosphere of 5% CO2 and 95% air overnight. Day 3: Change medium with fresh complete medium and continue to incubate for another 24 h. Day 4: Harvest cells and sort the cells with higher levels of GFP by FACScan.
Western Blot Analysis
All the cells used in this study were rinsed three times with ice-cold PBS and scraped off the plate in radioimmunoprecipitation assay buffer [150 mmol/L NaCl, 1% Triton X-100, 0.1% SDS, 25 mmol/L Tris-HCl (pH 7.5), 1% sodium deoxycholate, 1 mmol/L Na3VO4, 20 μg/mL pepstatin, 5 μg/mL aprotinin, and 1 mmol/L phenylmethylsulfonyl fluoride], sonicated for 5 s with a probe sonicator (Heat System-Ultrasonics), and centrifuged for 10 min at 13,000 × g in a microcentrifuge. Nuclear lysates were prepared by harvesting cells by trypsinization followed by centrifugation. The resulting pellet was rinsed twice with 1× PBS and treated with lysis buffer with NP-40. Following centrifugation, 20 volumes of lysis buffer without NP-40 was added and the lysate was centrifuged again. The pellet was then resuspended in radioimmunoprecipitation assay buffer. The protein concentration of the lysates was determined using the BCA Protein Assay Kit (Pierce). Protein (20-50 μg/sample) was electrophoresed on 7% to 15% SDS-PAGE and electrophoretically transferred to nitrocellulose membrane (Hybond-ECL; Amersham Pharmacia Biotech UK) at 4°C for 1.5 h. Blots were incubated for 1 h with 5% nonfat dry milk to block nonspecific binding sites and then incubated with either 1:500 dilution of rabbit polyclonal antibody against EGR-1 (Santa Cruz Biotechnology), 1:1,000 dilution of rabbit polyclonal antibody to p27Kip1 (Santa Cruz Biotechnology), 1:500 dilution of mouse monoclonal antibody to CDK4 (Santa Cruz Biotechnology), 1:500 dilution of mouse monoclonal antibody to p65 (Santa Cruz Biotechnology), 1:1,000 dilution of rabbit polyclonal antibody to poly(ADP-ribose) polymerase (Santa Cruz Biotechnology) at 4°C overnight. The immunoreactivity was detected using peroxidase-conjugated antibody and visualized by enhanced chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Pierce). The blots were stripped before reprobing with antibody to β-tubulin (Santa Cruz Biotechnology).
Nuclear extracts were prepared from WHCO1 cells treated with 5 mmol/L sodium salicylate from the indicated times as described previously (35).
Analysis of Cell Cycle
Cells (1 × 106) were plated in 100 mm culture dish in DMEM supplemented with 10% FCS and incubated in a 37°C incubator in a humidified atmosphere of 5% CO2 and 95% air for 48 h. Cells were harvested and washed once with 10 mL cold PBS, resuspended at 2 × 106 cells in 1 mL ice-cold PBS, and fixed by adding 9 mL ice-cold 70% ethanol. Cells were spun down at 1,000 rpm for 5 min, washed twice with 1 mL cold PBS, resuspended in 200 μL PBS containing 50 μg/mL DNase-free RNase A, incubated for 30 min at room temperature, and then analyzed by flow cytometry 20 min after incubation with 10 μg/mL propidium iodide. Data analyzed using Flow Jo software (version 7.1.3).
Statistical Analysis
The Student’s t test was used for statistical significance of differences in the expression of EGR-1, IκBα, GROα, and GROβ between groups. P < 0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Prof. Nor Hayati Othman (Universiti Sains Malaysia) and Dr. Ryan Soldin (University of Cape Town) for pathologic evaluation of all the slides, Dr. Gabi Walther (Thoracic Surgery, Groote Schuur Hospital) for providing tissue biopsy specimens, Heather McLeod and Nafiesa Allie for expert technical assistance in immunohistochemistry, Drs. Virna D. Leaner and Howard Donninger for many fruitful discussions and proofreading, Dr. Sharon Prince for reagents, Dr. Dirk Lang and Liz van der Merwe for assistance with immunofluorescence analysis, Wei Wei for assistance with cell cycle analysis, and Dr. Veale (University of Witwatersrand) for providing the WHCO cell lines.
Grant support: Medical Research Council (South Africa) and University of Cape Town, NRF Joint China/South Africa Science and Technology Agreement GUN 2075479, and NIH grant 5RO1 CA 112020.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Ebnet K, Vestweber D. Molecular mechanisms that control leukocyte extravasion: the selections and the chemokines. Histochem Cell Biol 1999;112:1–23. [DOI] [PubMed] [Google Scholar]
- 2.Luan J, Shattuck-Brandt R, Haghnegahdar H, et al. Mechanism and biological significance of constitutive expression of MGSA/GRO chemokines in malignant melanoma tumor progression. J Leukoc Biol 1997;62:588–97. [DOI] [PubMed] [Google Scholar]
- 3.Norgauer J, Metzner B, Schraufstatter I. Expression and growth-promoting function of the IL-8 receptor β in human melanoma cells. J Immunol 1996; 156:1132–7. [PubMed] [Google Scholar]
- 4.Balentien E, Mufson BE, Shattuck RL, Derynck R, Richmond A. Effects of MGSA/GROα on melanocyte transformation. Oncogene 1991;6:1115–24. [PubMed] [Google Scholar]
- 5.Owen JD, Strieter R, Burdick M, et al. Enhanced tumor-forming capacity for immortalized melanocytes expressing melanoma growth stimulatory activity/growth-regulated cytokine β and γ proteins. Int J Cancer 1997;73:94–103. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Hendricks DT, Wamunyokoli F, Parker MI. A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer Res 2006;66:3071–7. [DOI] [PubMed] [Google Scholar]
- 7.Muthukkumar S, Nair P, Sells SF, Maddiwar NG, Jacob RJ, Rangnekar VM. Role of EGR-1 in thapsigargin-inducible apoptosis in the melanoma cell line A375–6. Mol Cell Biol 1995;15:6262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen HQ, Hoffman-Liebermann B, Liebermann DA. The zinc finger transcription factor EGR-1 is essential for and restricts differentiation along the macrophage lineage. Cell 1993;72:197–209. [DOI] [PubMed] [Google Scholar]
- 9.Santiago FS, Atkins DG, Khachigian LM. Vascular smooth muscle cell proliferation and regrowth after mechanical injury in vitro are EGR-1/NGFI-A-dependent. Am J Pathol 1999;155:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukhatme VP, Cao X, Chang LC, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 1988;53:37–43. [DOI] [PubMed] [Google Scholar]
- 11.Khachigian LM, Collins T. Inducible expression of Egr-1-dependent genes: a paradigm of transcriptional activation in vascular endothelium. Circ Res 1997;81: 457–61. [DOI] [PubMed] [Google Scholar]
- 12.Thigpen AE, Cala KM, Guileyardo JM, Molberg KH, McConnell JD, Russell DW. Increased expression of early growth response-1 messenger ribonucleic acid in prostatic adenocarcinoma. J Urol 1996;155:975–81. [PubMed] [Google Scholar]
- 13.Eid MA, Kumar MV, Iczkowski KA, Bostwick DG, Thindall DJ. Expression of early growth response genes in human prostate cancer. Cancer Res 1998;58: 2461–8. [PubMed] [Google Scholar]
- 14.Svaren J, Ehrig T, Abdulkadir SA, Ehrengruber MU, Watson MA, Milbrandt J. EGR-1 target genes in prostate carcinoma cells identified by microarray analysis. J Biol Chem 2000;275:38524–31. [DOI] [PubMed] [Google Scholar]
- 15.Chen A, Xu J, Johnson AC. Curcumin inhibits human colon cancer cell growth by suppressing gene expression of epidermal growth factor receptor through reducing the activity of the transcription factor EGR-1. Oncogene 2006;25:278–87. [DOI] [PubMed] [Google Scholar]
- 16.Fahmy RG, Dass CR, Sun LQ, Chesterman CN, Khachigian LM. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat Med 2003;9:1026–32. [DOI] [PubMed] [Google Scholar]
- 17.Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS Jr. IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev 1992;6:1899–913. [DOI] [PubMed] [Google Scholar]
- 18.Ganchi PA, Sun SC, Greene WC, Ballard DW. IκB/MAD-3 masks the nuclear localization signal of NF-κB p65 and requires the transactivation domain to inhibit NF-κB p65 DNA binding. Mol Biol Cell 1992;3:1339–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virolle T, Krones-Herzig A, Baron V, Gregorio GD, Adamson ED, Mercola D. EGR-1 promotes growth and survival of prostate cancer cells. J Biol Chem 2003;278:11802–10. [DOI] [PubMed] [Google Scholar]
- 20.Joshi-Barve SS, Rangnekar VV, Sells SF, Rangnekar MV. Interleukin-1-inducible expression of gro-β via NF-κB activation is dependent upon tyrosine kinase signaling. J Biol Chem 1993;268:18018–29. [PubMed] [Google Scholar]
- 21.Yin MJ, Yamamoto Y, Gaynor RB. The anti-inflammatory agents aspirin and salicylate inhibit the activity of IκB kinase-β. Nature 1998;396:77–80. [DOI] [PubMed] [Google Scholar]
- 22.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol 2002;193:287–92. [DOI] [PubMed] [Google Scholar]
- 23.Baron V, Duss S, Rhim J, Mercola D. Antisense to the early growth response-1 gene (EGR-1) inhibits prostate tumor development in TRAMP mice. Ann N Y Acad Sci 2003;1002:197–216. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell A, Dass CR, Sun LQ, Khachigian LM. Inhibition of human breast carcinoma proliferation, migration, chemoinvasion and solid tumour growth by DNAzymes targeting the zinc finger transcription factor EGR-1. Nucleic Acids Res 2004;32:3065–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cogswell PC, Mayo MW, Baldwin AS Jr. Involvement of Egr-1/Re1A synergy in distinguishing T cell activation from tumor necrosis factor α-induced NFκB1 transcription. J Exp Med 1997;185:491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Beer DG. Molecular biology of upper gastrointestinal malignancies. Semin Oncol 2004;31:476–86. [DOI] [PubMed] [Google Scholar]
- 27.Lee HT, Kay EP. Regulatory role of PI3-kinase on expression of Cdk4 and p27, nuclear localization of Cdk4, and phosphorylation of p27 in corneal endothelial cells. Invest Ophthalmol Vis Sci 2003;44:1521–8. [DOI] [PubMed] [Google Scholar]
- 28.Su CC, Chen GW, Lin JG, Wu LT, Chung JG. Curcumin inhibits cell migration of human colon cancer Colo 205 cells through the inhibition of nuclear factor κB/p65 and down-regulates cyclooxygenase-2 and matrix metalloproteinase-2 expression. Anticancer Res 2006;26:1281–8. [PubMed] [Google Scholar]
- 29.Shishodia S, Amin HM, Lai R, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive NF-κB activation, induces G1/S arrest, suppresses proliferation, and induces apoptosis in mantle cell lymphoma. Biochem Pharmacol 2005; 70:700–13. [DOI] [PubMed] [Google Scholar]
- 30.Ning W, Li CJ, Kaminski N, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A 2004;101:14895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carraro G, Albertin G, Forneris M, Nussdorfer GG. Similar sequence-free amplification of human glyceraldehyde-3-phoshpate dehydrogenase for real time RT-PCR application. Mol Cell Probes 2005;19:181–6. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 33.Veale RB, Thornley AL. Atypical cytokeratins synthesized by human oesophageal carcinoma cells in culture. S Afr J Sci 1984;80:260–7. [Google Scholar]
- 34.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc 2006;1:241–5. [DOI] [PubMed] [Google Scholar]
- 35.Lee KA, Green MR. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Ezymol 1990;181:20–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.