Abstract
Harnessing the process of natural selection to obtain and understand new microbial phenotypes has become increasingly possible due to advances in culturing techniques, DNA sequencing, bioinformatics, and genetic engineering. Accordingly, Adaptive Laboratory Evolution (ALE) experiments represent a powerful approach to both investigate the evolutionary forces influencing strain phenotypes, performance, and stability, and to acquire production strains that contain beneficial mutations. In this review, we summarize and categorize the applications of ALE to various aspects of microbial physiology pertinent to industrial bioproduction by collecting case studies that highlight the multitude of ways in which evolution can facilitate the strain construction process. Further, we discuss principles that inform experimental design, complementary approaches such as computational modeling that help maximize utility, and the future of ALE as an efficient strain design and build tool driven by growing adoption and improvements in automation.
1. Introduction
Adaptive Laboratory Evolution, or ALE, experiments are an increasingly popular technique for both improving microbial phenotypes and investigating biological phenomena. ALE experiments can be traced back to reports of controlled evolution studies in the earliest parts of the last century (Atwood et al., 1951; Bennett and Hughes, 2009; Novick and Szilard, 1950) and are methodologically straightforward. In their simplest form, experiments consist of prolonged culturing of cells in a chosen environment to naturally select for those which acquire beneficial mutations. Given the speed with which beneficial mutations can arise and fix, it is safe to say that many biologists have ‘adaptively evolved’ their lab microbe of choice simply via the unavoidable cycles of growth/plating/freezing involved in cell culturing. ALE works robustly in microbes due to the ease with which large populations (108 - 1010 cells) of rapidly dividing (20 min - <10 hour generation time) cells can be maintained; typical mutation rates and genome sizes ensure extensive sampling of the adaptive space, providing ample genetic diversity from which beneficial mutants will be naturally enriched (Gresham and Dunham, 2014). Furthermore, the rising use of ALE has been driven by the increasing availability of low cost, high throughput DNA sequencing (Shendure et al., 2017), which, when paired with appropriate bioinformatics tools, facilitates the increasingly critical mutation-identification step in the ALE process. Similarly, genome engineering is a key complementary technology (R. Liu et al., 2015), enabling the introduction of desired mutations into strains of interest for causal determination or phenotypic alteration.
Evolution’s reliance on natural selection to enrich for mutants with increased fitness allows strain optimization to be performed without requiring a priori knowledge of the genetic alterations necessary to effect such changes (Fig. 1). With perfect information on an organism’s genotype-to-phenotype mapping, desired traits could simply be engineered in rather than evolved, but the incredible complexity of biological systems renders our current knowledgebase insufficient to inform such rational design. ALE thus serves as a powerful technique for strain construction, complementing or even replacing rational design approaches which frequently induce stressed cellular states that negatively impact phenotype. It is important to note that, despite some interchangeable use of the terms, ALE is distinct from the field of ‘directed evolution’ (Packer and Liu, 2015; Arnold, 2018) – ALE finds whatever genome-wide mutations aid in the fitness of actively growing cultures, while directed evolution typically targets a particular gene for mutagenesis and then screens resultant variants for a phenotype of interest, often independent of fitness effects. However, both approaches fall under the category of ‘evolutionary engineering’ (Sauer, 2001; Shepelin et al., 2018) given that they generate new phenotypes via random mutation followed by selection or screening.
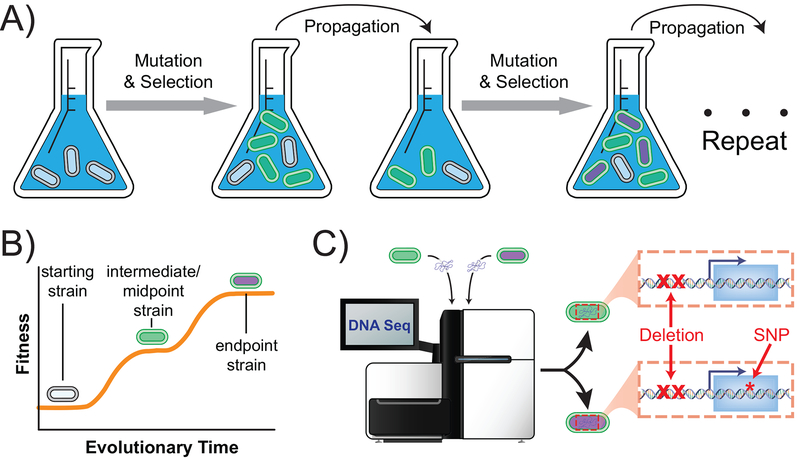
Figure 1. Adaptive Laboratory Evolution.
A) Microbes are cultured in a desired growth environment for an extended period of time, allowing natural selection to enrich for mutant strains (altered coloration) with improved fitness. This example depicts ALE via serial propagation of batch cultures. B) Evolved strains are characterized for phenotypic improvements relative to the ancestral strain, using whatever “fitness” metric is appropriate given the evolutionary environment. C) Evolved strains have their DNA sequenced to reveal the adaptive mutations enabling phenotypic improvement. This example case depicts the fixation of two successive mutations targeting the same genetic region.
Key to the success of ALE experiments is the fitness advantage bestowed by adaptive mutations, allowing the mutant strains to outcompete their ancestor and dominate the population. However, it is important to note that “fitness” is not a well-defined term, but rather depends on the growth environment employed in the ALE experiment. For example, if batch cultures are kept propagating in constant exponential-phase and with excess nutrients, then fitness is essentially equivalent to growth rate (Sandberg et al., 2014). If, instead, exponential-phase growth is not constantly maintained, then fitness expands to include factors other than just growth rate, such as survival in stationary phase or decreased lag phase (Wiser and Lenski, 2015). This sensitive environmental dependence of fitness thus allows different traits to be selected for based on the specifics of the ALE procedure. In addition to batch culturing, the most popular alternative growth method is continuous culture (e.g, chemostats or turbidostats) (Gresham and Dunham, 2014). Other ALE culturing methods have also been implemented for specific purposes, but have yet to see widespread use (See Section 4).
Once a desired growth environment has been carefully selected, an ALE experiment enables the generation of a whole lineage of mutant strains with improved fitness relative to the ancestral (i.e., starting) strain (Fig. 1B) and sequencing enables causal mutation determination (Fig. 1C). Selective sweeps often occur as the result of single beneficial mutations overtaking the population, but cultures contain significant amounts of genetic diversity from which intrapopulation competition can lead to clonal interference (Maddamsetti et al., 2015). It is common practice to save frozen stocks of cells periodically over the course of an ALE experiment, allowing for characterization of both endpoint strains and evolutionary intermediates. Although midpoints are perforce less fit than endpoints (and, in turn, might have less of a tradeoff for alternate phenotypic properties), including them in characterizations can greatly aid in understanding the adaptive steps taken by the evolving ancestor to eventually arrive at an endpoint. Furthermore, by virtue of their smaller number of genetic alterations, midpoints can allow for easier discrimination of the minimum mutational set necessary to enable a bioprocess-relevant beneficial phenotype. In concert with phenotypic characterization (or screening), which establishes the fitness improvements realized over the course of the ALE, genetic characterization allows determination of the causal mutations that enable improved fitness in the ALE environment. To aid in causal mutation identification, independent replicates starting from the same ancestral strain are often evolved simultaneously – genes or genetic regions found to mutate multiple times across the independent replicates are almost surely adaptive (Bailey et al., 2015). Such causal mutations can thus be distinguished from neutral or even deleterious hitchhiker mutations that might otherwise require categorization via site-directed mutagenesis and growth profiling of the resultant mutants. Causal establishment of ALE mutations via reintroduction to a starting strain was possible and demonstrated in the mid 2000s (Herring et al., 2006), and this process is now more efficient with high-throughput genome engineering (R. Liu et al., 2015).
This review will focus on the use of ALE in industrial biotechnology and related fields by compiling a comprehensive collection of relevant studies, analyzing their collective contributions, and highlighting several case studies of importance. An analysis of the most common methods used for ALE and the strains utilized will be included. The review will also touch on the emergence of automation and bioinformatics in the field and the impact this has had. Previous reviews relating to ALE have addressed the underlying genetic and metabolic basis for adaptive evolution experiments (Conrad et al., 2011; Gresham and Dunham, 2014; Long et al., 2015; Long and Antoniewicz, 2018; Remigi et al., 2019), the role of systems biology and in silico evolution (Hansen et al., 2017; Hindré et al., 2012; Papp et al., 2011), overall strategies to improve metabolic performance (Rabbers et al., 2015; Shepelin et al., 2018; Williams et al., 2016) and tolerance (Peabody et al., 2014), as well as other industrially relevant strain properties (Bachmann et al., 2015; Dragosits and Mattanovich, 2013; Portnoy et al., 2011/8; Stella et al., 2019; Winkler and Kao, 2014; Fernández-Cabezón et al., 2019). This work provides an updated review and categorization of studies useful for metabolic engineering, as well as other selected foundational studies using laboratory evolution that are relevant to industrial biotechnology. As such, studies focused on the use of ALE in understanding antimicrobial resistance and other fundamental principles of evolutionary biology will not be covered here, as they have been reviewed elsewhere (Hughes and Andersson, 2015; Kawecki et al., 2012; Palmer and Kishony, 2013; Remigi et al., 2019).
2. Applications of ALE
ALE has become a valuable tool in metabolic engineering for strain development and optimization by reliably facilitating microbial fitness improvements, via both predictable and non-intuitive mechanisms. However, specific mutations themselves are rarely predictable given current knowledge, and the adaptive strategies employed are often strain-specific. Supplemental Table 1 outlines a collection of ALE-based manuscripts that will serve as case studies and a source of content for this review. The different uses of ALE can be roughly categorized into five general application areas (Figure 2): 1) growth rate optimization, 2) increasing tolerance, 3) substrate utilization, 4) increasing product yield/titer, and 5) general discovery. Although these categories and applications are not mutually exclusive and ALE experiments often improve multiple strain properties simultaneously, it serves as a useful heuristic.
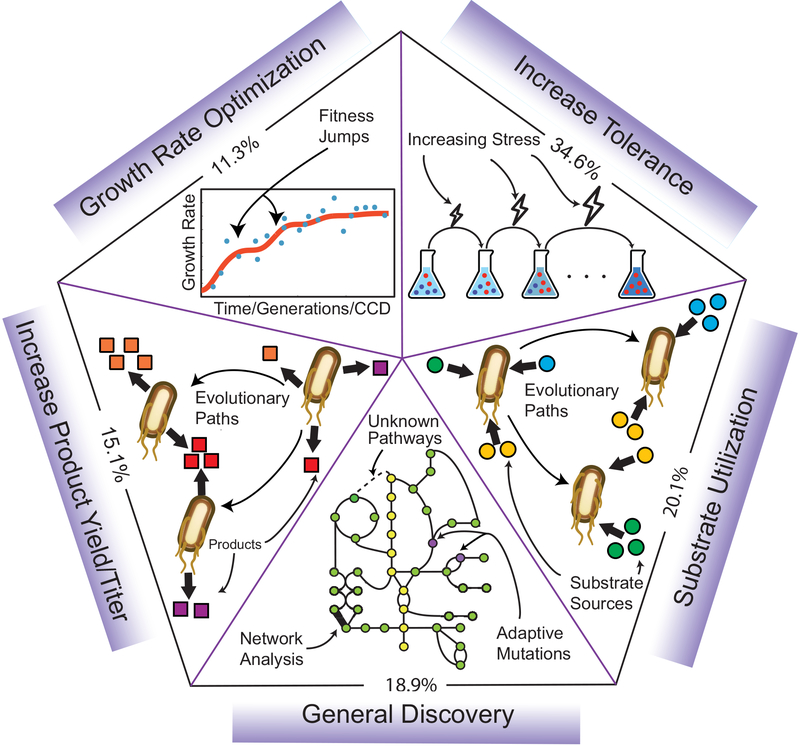
Figure 2: A Categorization of ALE Studies.
A diagram of different categories of use for laboratory evolution experiments, detailing the percent makeup of studies examined (159 total). ‘Growth rate optimization’ illustrates fitness development over the course of an ALE, with noticeable fitness jumps. ‘Increase tolerance’ illustrates an experimental schematic of an initially mixed population acquiring beneficial mutations (red cells) that promote cell survival in a constantly increasing external stress environment (e.g., pH, antibiotics, temperature, etc). ‘Substrate utilization’ and ‘Increase Product Yield/titer’ illustrate evolutionary pathways enabled via ALE that enhance the organism’s ability to make use of alternative nutrient sources (colored circles), and to increase production of metabolites of interest (colored squares), respectively. Lastly, ‘General Discovery’ encompasses studies that examined ALEs at a genetic or systems level in greater detail.
Several features are revealed through a categorization of ALE studies relevant to industrial biotechnology. First, the predominant use of ALE in this collection involves increasing an organism’s ability to tolerate stresses (55 out of 159 included studies, 35%). Tolerance evolutionary engineering has significant industrial implications due to the toxicity that many biorenewables have on microorganisms’ overall production capacity and viability (Lee and Kim, 2015; Qi et al., 2019; S. Wang et al., 2018). Most starting strains and production chassis have never had to deal with high amounts of desired product chemicals in their native environments, thus improved tolerance mechanisms are almost always beneficial for a successful strain design. Second, there is significant focus on metabolite production as well as nutrient uptake, with 15% and 20% of the studies, respectively. In metabolic engineering the two concepts are frequently linked, given that sufficient nutrient uptake is a prerequisite for any metabolite biosynthesis, but there are distinct cases where microorganisms are designed and/or evolved to either utilize poorly accessible substrates or produce non-native metabolites. Nutrient uptake is particularly relevant in industrial settings where poor uptake of desired feedstocks can lead to inefficient conversion and inhibition of metabolic pathways (Görke and Stülke, 2008; Ingram and Doran, 1995). Lastly, the general discovery category contains studies covering an array of topics that are relevant to industrial biotechnology and can be investigated with ALE, including systems biology, evolutionary modeling, and genome dynamics (Barrick and Lenski, 2013). This general category could be significantly expanded via inclusion of purely computational or theoretical rather than empirical evolution studies, but that is beyond the scope of this review.
Each category will be individually addressed in the following sections to outline a general summary of their applications, highlight case studies useful for demonstration, and summarize the lessons learned from each collection of studies.
2.1. Growth Rate Optimization
While “fitness” is often a complex trait with many contributing factors, growth rate (ln(2)/doubling time) is generally the most important fitness determinant in ALE experiments (Vasi et al., 1994). Indeed, although the other ALE use categories are more specialized, the enrichment for adaptive mutants would not be possible without a growth advantage relative to the starting strain, regardless of the specifics of the environment. A key reason for the use of microbes in biotech and research is their short generation time, and ALE allows for rapid improvement of this desirable trait. ALE has been applied to optimize the growth rate of various industrially relevant microbial species (Hong et al., 2011; LaCroix et al., 2014; Pfeifer et al., 2017; Yu et al., 2013), to ameliorate growth defects in engineered strains (Aguilar et al., 2018; Carroll and Marx, 2013; Radek et al., 2017; Thyer et al., 2018; Zelle et al., 2011), or to study how and why growth rate changes over the course of evolution (Barrick et al., 2009; Ferea et al., 1999; Lenski et al., 2015, 1998, 1991; Lenski and Travisano, 1994; Weikert et al., 1997; Wong and Liao, 2009).
ALE can significantly increase a strain’s growth rate, even in frequently used culturing environments for which one might expect growth to already be close to optimal. For example, a simple ~1 month ALE experiment in glucose minimal medium resulted in growth rate improvements of up to 60% for the commonly used lab strain and engineering chassis E. coli K-12 MG1655 (LaCroix et al., 2014). This rapid growth phenotype was achieved predominantly through mutations to only three genetic regions, suggesting their easy use as allelic knock-ins to improve growth on glucose of production strains derived from MG1655, as has been demonstrated in the case of an RNA polymerase mutation (Rugbjerg et al., 2018). Such causal mutations can even have cross-phylum fitness implications – evolution on glucose minimal media led to mutations in pyruvate kinase and the CorA magnesium transporter in both C. glutamicum (Pfeifer et al., 2017) and MG1655. Moreover, the metabolic flux state of the MG1655 evolved strains was found to be remarkably unperturbed - rather than changing relative pathway usage, faster growth was enabled by proportional increases to all intracellular rates of central carbon metabolism (Long et al., 2017). ALE has been used to successfully improve growth capabilities of a variety of non-bacterial species (Supplemental Figure 1), most notably yeast, where a similar volume of ALE work has been performed (Mans et al., 2018). As with E. coli, improved growth rates in different organisms frequently come along with other beneficial traits, such as evolved yeast strains outperforming rationally designed ones in several phenotypic metrics (Hong et al., 2011), and evolved microalgae achieving improved lipid productivity (Yu et al., 2013).
In addition to optimizing industrially or scientifically relevant wild-type strains, ALE has been successfully applied to fix fitness defects in strains rationally designed for a specific metabolic phenotype. An increased growth rate, in most cases, also makes a strain more robust to perturbations resulting from genetic alterations, as mutations can restore microbes back to their wild-type physiology and activate flux through alternate energy generation pathways (McCloskey et al., 2018). In one study, an engineered M. extorquens strain with a foreign metabolic pathway (resulting in abnormal morphology and significantly decreased growth rate) was subjected to laboratory evolution (Carroll and Marx, 2013). ALE resulted in the isolation of evolved strains with up to 150% growth rate improvements, elimination of morphological abnormalities, and insight into the metabolic consequences resulting from introduction of the foreign pathway. In another study, ALE was used in concert with strain engineering to impart novel metabolic capabilities onto yeast (Zelle et al., 2011). The native malic enzyme in S. cerevisiae is unable to function anaplerotically via pyruvate carboxylation, nor is heterologous expression of the E. coli malic enzyme sufficient to activate this ability, but ALE successfully selected for a mutant enzyme that enabled this trait. ALE can also reliably ameliorate fitness defects resulting from ambitious, large-scale strain alterations - a genomically recoded E. coli with all 300+ amber stop codons removed was evolved to achieve strains with both significantly faster (>40%) growth rates and improved incorporation of nonstandard amino acids, opening the door to novel protein production capabilities (Wannier et al., 2018). Similarly, a genome-reduced E. coli with more than 1 million basepairs removed from its chromosome grew 3-fold slower than the wild-type in glucose minimal media, and yet a <1,000 generation ALE experiment almost completely abolished this severe fitness deficit (Choe et al., 2019).
ALE for improvement in growth rate thus serves not only to increase the ease and speed of microbial culturing, but also as a technique for targeted selection on specific rate-limiting system components. This is especially important when a strain has been engineered and rate-limiting constraints now significantly impact fitness, which will become an increasingly frequent occurrence as genome engineering techniques continue to improve. With proven efficacy across a number of species and the frequent occurrence of concomitant desirable traits such as increased productivity/yield, growth optimization stands as a key application for ALE experiments. Although the following sections delve into more specialized uses, it is important to note that growth rate improvements underlie these ALE outcomes as well.
2.2. Increase Tolerance
When exposed to stress, cells respond by activating stress responses, which require a reallocation of cellular resources to enact and thus inhibit growth potential. Cellular stress can be caused by a complex growth environment that induces some form of metabolic misregulation, or by exposure to specific inhibitory chemicals or biomolecules. Common stressors of both basic and applied interest introduce a diverse range of challenges for growing cells to overcome, and ALE has been established as an efficient tool to surmount such challenges. ALE studies have investigated the adaptive responses to many of these stressful environments: different levels of pH (Fletcher et al., 2017; Kildegaard et al., 2014; Zorraquino-Salvo et al., 2014), osmotic pressure (Dhar et al., 2011; Dragosits et al., 2013; Stoebel et al., 2009; Tilloy et al., 2014; Winkler et al., 2014), temperature (Blaby et al., 2012; Caspeta et al., 2014; Deatherage et al., 2017; Riehle et al., 2003; Rudolph et al., 2010; Sandberg et al., 2014; Sleight and Lenski, 2007; Tenaillon et al., 2012), UV irradiation (Alcántara-Díaz et al., 2004; González-Ramos et al., 2016), inhibitors (most frequently, metabolic byproducts produced during biomass pretreatment) (Adamo et al., 2012; Almario et al., 2013; Atsumi et al., 2010; Avrahami-Moyal et al., 2012; Cakar et al., 2005; Goodarzi et al., 2010; Haft et al., 2014; Henson et al., 2018; Horinouchi et al., 2010; Jiang et al., 2012; Koppram et al., 2012; Linville et al., 2013; Luan et al., 2013; Minty et al., 2011; McCarthy et al., 2017; Qin et al., 2016; Reyes et al., 2012; Reyes et al., 2013; Royce et al., 2015; Sehnem et al., 2013; Shao et al., 2011; Wallace-Salinas and Gorwa-Grauslund, 2013; Wang et al., 2018; Xu et al., 2018; Zhu et al., 2015); and nutritional stressors (Bachmann et al., 2012; Brennan et al., 2015; Cakar et al., 2009; Dhar et al., 2013; Hawkins and Doran-Peterson, 2011; Jansen et al., 2005; Jung et al., 2017; Lee et al., 2013; Mundhada et al., 2017; Pereira et al., 2015; Summers et al., 2012). Utilization of ALE to understand stress responses and overcome growth inhibition is one of the most popular and historically relevant applications (Figure 2), frequently increasing the utility and robustness of industrially valuable strains. Moreover, stress tolerance ALEs are particularly efficacious due to the complex, global physiological response stresses often induce – tuning a large number of gene expression levels to restore robust growth is currently beyond the realm of rational design, but ALE can reliably uncover point mutations in global regulators which achieve just that (Sandberg et al., 2014).
Compounds with potential biotechnological value can often not be efficiently metabolized or produced in living systems due to toxicity. For example, many carboxylic acids are promising biorenewables but induce membrane damage, and attempts to rationally engineer greater membrane integrity have failed to improve carboxylic acid production. Royce et al. addressed this with an ALE experiment of E. coli onto octanoic acid, wherein strains were obtained that had improved tolerance not just to octanoic acid but also several other carboxylic acids and butanol isomers, with concomitant 5 fold higher production titer (Royce et al., 2015). This highlights how stress mitigation can simultaneously improve production properties, and how certain classes of stressors trigger similar physiological responses that cause particular mutations to be beneficial across multiple environments. In another study, knowledge of S. cerevisiae metabolism led to an ALE experiment designed for osmotic stress tolerance - this pushed the yeast metabolic state more towards glycerol production and away from ethanol, resulting in evolved strains with improved properties for lower-alcohol wine production (Tilloy et al., 2014).
Causal mutations in tolerance-evolved strains are straightforward to identify with DNA sequencing, but interpreting the molecular causality underlying a mutation’s phenotypic impact is often more challenging. A study by Caspeta et al. stands as a successful example of ALE results elucidating the molecular mechanisms of adaptation – yeast was evolved for improved thermotolerance, yielding strains that grew >50% faster at 40°C (Caspeta et al., 2014). Mutational convergence in independently evolved replicates revealed that a gene inactivation changed the sterol composition of the cells from ergosterol to fecosterol, optimizing membrane fluidity at the elevated growth temperatures. ALE can also be used to increase tolerance to multiple stressors at once, as demonstrated by a study in which an industrial yeast strain was evolved for improved tolerance to both biomass hydrolysate inhibitors and high temperature, creating a robust strain for ethanol production via simultaneous saccharification and fermentation (Wallace-Salinas and Gorwa-Grauslund, 2013). However, it should be noted that tolerance to one stress often comes with tradeoffs in a separate growth environment, depending on the particular adaptive mutations acquired – while strains evolved to high-temperature typically lose fitness at low-temperature, one ALE-identified mutation managed to avoid such a tradeoff (Rodriguez-Verdugo et al., 2014).
A significant effort to overcome the toxicity of a nutrient stressor, specifically L-serine in high concentration, was seen in the studies performed by Mundhada et al. L-serine is a promising target for commercial bioproduction but induces inhibition of various cellular processes such as peptidoglycan synthesis and cell division, and the reactive byproducts such as acrylates can hinder cell growth (Zhang and Newman, 2008; Zhang et al., 2010; de Lorenzo et al., 2015). An initial study succeeded in achieving L-serine tolerance in E. coli by employing random mutagenesis and selection, but it was not able to meet the demand for extensive serine production (Mundhada et al., 2016). By implementing ALE, E. coli lacking L-serine degradation pathways (which makes cells much more susceptible to toxicity) were evolved to increasing concentrations of L-serine that reached 100 g/L, resulting in a titer much higher than the mutagenized strain with the same pathway (Mundhada et al., 2017).
Evolutionary engineering for improved stress tolerance stands as a dominant ALE application for metabolic engineering and industrial biotechnology. As previously stated, wild-type cells are rarely well-poised to deal with the burdens caused by growth environments with high concentrations of an atypical but desired bioproduction chemical. Where rational engineering approaches can fall short due to a lack of requisite knowledge, ALE enables mechanistically naïve but reliable improvements to industrially valuable strain properties. Mutational results from ALE experiments can then be used to inform further strain design attempts, while also elucidating basic tenants of biological systems. Further, attempts to rationally design strains for a specific phenotype often induce fitness defects which can be countervailed by evolution. Even if specific tolerance mechanisms can be rationally engineered into a strain, necessary gene expression levels can be hard to infer. In contrast, in a single experiment, ALE can find both the mechanisms underlying fitness defects and the gene expression levels necessary to enable a functional and robust phenotype. ALE also enables acclimation to growth environments with complex combinations of stressors that rational design cannot currently address given our existing biological knowledge base. Importantly, when stress on strains is reduced via ALE, the result is typically not only growth rate increases, but also phenotypic improvements in industrially relevant properties such as production or degradation capabilities.
2.3. Substrate Utilization
Efficient substrate uptake is necessary to enable satisfactory production of metabolites and additional products of interest in most, it not all, industrial applications. There exists a very direct link between organism fitness and the ability to efficiently uptake and metabolize rate-limiting growth nutrients, thus ALE experiments with a properly designed selective environment readily uncover mutations conducive to robust growth on substrates of interest. Primary motivations for improving substrate utilization of an organism are tied to current market surpluses/prices (Ji et al., 2011; Lin and Tanaka, 2006) and harnessing the energy of inaccessible or toxic byproducts to improve yield of commodity chemicals (Plácido and Capareda, 2016). The most ubiquitous organism studied to improve uptake is the yeast Saccharomyces cerevisiae (Cadière et al., 2011; Garcia Sanchez et al., 2010; Guimarães et al., 2008; Kim et al., 2012; Smith et al., 2014; Sonderegger and Sauer, 2003; Zha et al., 2014; Zhang et al., 2018; Zhou et al., 2012), with the CEN.PK strain and its derivatives the most common (de Kok et al., 2012; Ho et al., 2017; Jansen et al., 2004; Klimacek et al., 2014; Kuyper et al., 2005, 2004; Marques et al., 2017; Merico et al., 2011; Novy et al., 2014; Ochoa-Estopier et al., 2011; Papapetridis et al., 2018; Scalcinati et al., 2012; Strucko et al., 2018; van Rossum et al., 2016), mainly due to its widespread use in industry, robust fermentative capability, and inherent ethanol tolerance. There has been considerable interest in generating microbial strains that can utilize the fermentable sugars in lignocellulosic biomass (Clark et al., 2012; Sanderson, 2011); S. cerevisiae has again been the driving force in discovery to this end (Klimacek et al., 2014; Marques et al., 2017; Zha et al., 2014), with some work done in E. coli strains (Lee and Palsson, 2010; Rajaraman et al., 2016; Sandberg et al., 2017, 2016; Utrilla et al., 2012) and other various bacteria and yeast (Cordova et al., 2016; Latif et al., 2015; Moser et al., 2017).
Saccharomyces cerevisiae, due to its anaerobic fermentative capabilities and tolerance to low pH and phage infection, has been the species of interest for industrial production of specialty metabolites (Moysés et al., 2016). Although S. cerevisiae does not natively catabolize xylose or other pentose sugars readily available in plant biomass, there has been considerable work to engineer strains capable of co-utilizing these compounds (Chu and Lee, 2007; Van Vleet and Jeffries, 2009). Adaptive laboratory evolution has been used in tandem with other metabolic engineering techniques to further drive substrate utilization. In (Klimacek et al., 2014), previously constructed S. cerevisiae strains with enhanced expression of heterologous genes involved in xylose metabolism were evolved via a two phase process. ALE selection for anaerobic xylose fermentation led to a strain with faster growth but undesirable metabolic byproducts, and a subsequent ALE on this strain under batch culture conditions ameliorated these phenotypic defects. The final strain grew more than 500% faster than the original progenitor despite fewer than 100 total generations of evolution, highlighting the impressive speed with which ALE can realize significant fitness improvements.
ALE has been used to tackle challenges in the utilization of plant biomass as a nutrient source, such as the presence of toxic or inhibitory byproducts like furfural and acetate (Bellissimi et al., 2009; Heer and Sauer, 2008). A study by Rajaraman et al. addressed this with a chemostat ALE on several E. coli strains with acetate as the sole carbon source (Rajaraman et al., 2016). Evolved strains had a specific growth rate increase of ~25% and significantly altered expression for a number of genes, enabled by a single amino acid substitution in the RNA polymerase complex subunit RpoA; unlike with rational engineering of specific genes for altered expression, ALE can find point mutations changing global expression patterns in a net beneficial way not predictable with current protein modeling techniques. In addition to improving growth rate on non-optimal nutrients, a properly designed ALE experiment can also enable growth on completely non-permissive substrates. With a passage protocol in which glycerol concentration was steadily lowered while L-1,2-propanediol was increased, E. coli strains were obtained capable of growth solely on the latter substrate, despite the ancestral strain’s inability to uptake or metabolize this nonnative carbon source (Lee and Palsson, 2010). A study that significantly expanded this approach involved automation of the process, weaning cultures off of a helper substrate (which generated diversity), and selecting for strains that could utilize a number of non-native carbon sources by directly targeting promiscuity properties of enzymes (Guzmán et al., 2019; Notebaart et al., 2018). Impressively, growth on these new substrates was enabled by the acquisition of only one or two point mutations – completely ‘novel’ phenotypes for a particular strain can readily be acquired via evolution if the growth environment provides the necessary fitness benefit for such an occurrence.
Although ALE has proven efficacy for enabling growth on non-native substrates, access to a new nutrient niche can require large jumps across the fitness landscape that render a phenotype infeasible to select for on reasonable timeframes – famously, it took 15 years for one of twelve Long Term Evolution Experiment populations to acquire a citrate-consuming phenotype, and the other eleven populations are still unable to consume citrate after 30 total years (Leon et al., 2018). Rational design with heterologous pathways can facilitate such fitness landscape leaps, but clever use of ALE alone can sometimes suffice. Szapponos et al. utilized metabolic modeling to identify non-native growth substrates with similar catabolic pathways, then performed a multi-step ALE to select for a complex metabolic innovation (Szappanos et al., 2016). Specifically, E. coli cannot natively grow on propylene glycol (PG) or ethylene glycol (EG), but culturing of a hypermutable variant resulted in isolation of PG+, but not EG+, strains. However, selecting for EG growth using PG+ mutant strains did successfully yield the EG+ phenotype, indicating an increase in adaptation rate of more than two orders of magnitude. Innovative niche expansions can thus be achieved via stepwise evolution, with complimentary technologies such as metabolic modeling providing the information necessary to choose suitable substrate targets.
ALE has seen significant use as a tool for improving substrate uptake and metabolism, often countervailing the growth-inhibitory effects of toxic byproducts or fixing metabolic network misregulation in non-optimal growth environments. In this way, the utility of ALE has been established in use cases ranging from improving wild-type strain growth on a sub-optimal substrate, to facilitating proper metabolic assimilation of heterologous pathways in engineered strains, to imparting novel growth phenotypes even in unengineered strains - in each case improving substrate uptake as a coupled property. The mutational mechanisms range from single-nucleotide polymorphisms and small indels (insertions or deletions) in specific enzymes that enable pure innovation (e.g., utilizing a novel substrate), to altered transporter activity that leads to more efficient import and routing of substrates into central metabolism, to regulatory mutations which globally rebalance the metabolic network, shutting down cellular content unnecessary for bioreactor growth and freeing up resources for growth-coupled processes like substrate uptake. ALE can thus greatly complement traditional engineering techniques for altering microbial response to various feedstocks, creating more efficient organisms with improved chemical conversion to industrially desirable products.
2.4. Product Titer/Yield Optimization
Maximizing the bioproduction of metabolites of interest is a difficult task, and rational approaches to strain design are frequently insufficient to achieve desired productivity (Guimarães et al., 2008; Kuyper et al., 2005, 2004; Shepelin et al., 2018). A main source of difficulty lies in the conflict between fast, robust strain growth and the fitness-counterproductive repurposing of cellular resources to produce large amounts of a desirable compound. Nevertheless, ALE studies with proper experimental design can be used to overcome this conflict and optimize metabolite production and increase titer (Basso et al., 2011; Charusanti et al., 2012; Fong et al., 2005a; Fu et al., 2013; Grabar et al., 2006; Jiang et al., 2013; Lee et al., 2016, 2014; Lu et al., 2012; Luo et al., 2019; Mahr et al., 2015; Otero et al., 2013; Pontrelli et al., 2018; Reyes et al., 2014; Royce et al., 2015; Shen et al., 2012; Smith and Liao, 2011; Vilela et al., 2015; Wang et al., 2012; Wisselink et al., 2007; Zhang et al., 2007; Zhao et al., 2013; Zhou et al., 2005, 2003). Given that an ALE experiment’s efficacy is contingent on the fitness advantage bestowed by adaptive mutations, it is essential to tie metabolite production to fitness in some way. The most straightforward way to do this is to genetically alter strains to couple production to overall energy and biomass generation rates in a given environment, and computational studies suggest a multitude of possible growth-coupled products that go far beyond simple and intuitive strain designs (Burgard et al., 2003; Klamt and Mahadevan, 2015; Pharkya et al., 2004; von Kamp and Klamt, 2017). However, given the limited scope of molecules for which this has been executed and validated (i.e., coupling production to overall energy generation), innovative methods (e.g., using a selective environment where production of the molecule of interest provides some protective or fitness-beneficial function) and advanced selection systems (e.g., fluorescent reporters + FACS sorting) are often needed to improve production for a wide range of molecules. Success cases for multiple forms of production optimization via growth coupling will now be discussed.
Genetic alterations that modify the metabolic network of a cell can force growth-sustaining flux through desired pathways, thereby coupling cell viability to the production of a compound of interest. Significant computational work has been performed to enable prediction of specific gene knockouts or heterologous pathway insertions that result in growth-coupling (Burgard et al., 2003; Feist et al., 2010; Jensen et al., 2019; Klamt and Mahadevan, 2015; Pharkya et al., 2004; von Kamp and Klamt, 2017), and such approaches have been experimentally validated (Alper et al., 2005; Fong et al., 2005a; Yim et al., 2011) and are growing in utility due to advances in genome-scale metabolic modeling techniques (King et al., 2017). In a study by Jantama et al., this modeling-informed rational design was combined with ALE to optimize an E. coli strain with gene knockouts that forced NAD+ regeneration to occur via malate- and succinate-producing pathways under anaerobic growth conditions (Jantama et al., 2008). By thus coupling production to NADH oxidation, necessary for the growth-essential maintenance of ATP generation, ALE successfully selected for strains with robust minimal medium growth and improved malate and succinate titer. Further, physiological characterization of evolved strains pointed to additional knockouts to decrease byproduct formation, ultimately yielding strains with even greater chemical yields. Properly designed growth-coupling strategies can even see utility across diverse organisms and enzyme types (Jensen et al., 2019), as exemplified by recent work coupling SAM-dependent methylation to growth (Luo et al., 2019). By tying production of the essential amino acid cysteine to the activity of methyltransferases, Luo et al. were able to use ALE to select for both E. coli and S. cerevisiae strains with mutations providing 2-fold increases in heterologous methyltransferase activity. Ensuring that the methyltransferase activity was rate-limiting for growth forced adaptive mutations to preferentially target this enzymatic bottleneck; in vivo enzyme engineering was thus undertaken by the evolving cells, with demonstrated success for improving activity of both N- and O-type methyltransferases.
Innovative selection strategies and systems have also been demonstrated, which expand the range of compounds whose production can be optimized using ALE beyond those that can be directly growth-coupled. One such selective approach involves utilizing interspecies competition or cooperation (i.e., syntrophy). Charusanti et al. serially propagated MRSA alongside an antibiotic-producing microorganism, harnessing competition to achieve an S. clavuligerus strain with constitutive holomycin expression (Charusanti et al., 2012). The production of this molecule, though energetically costly, was nevertheless selectable with ALE due to the competitive co-culturing growth environment utilized. Similarly, through the use of syntrophic coupling and co-cultures, Lloyd et al. designed and evolved mutually reliant strains to optimize the sharing and production of non-trivial compounds (Lloyd et al., 2019). Environmental manipulation in monocultures has also been successfully employed, as in a study by Reyes et al. wherein exposure to hydrogen peroxide selected for more than 3-fold increases in carotenoid production due to these compounds’ antioxidant properties (Reyes et al., 2014). Such approaches change the meaning of “fitness” to now include production of the molecule of interest as a factor, but other selective techniques are possible – a noteworthy study in this category used FACS to apply multi-dimensional selection to a strain engineered with a biosensor for valine-induced fluorescence (Mahr et al., 2015). Biosensors provide a powerful way to link otherwise-obscured intracellular states to a screenable output (D. Liu et al., 2015), and by screening out and serially propagating only the most highly fluorescent cells, Mahr et al. increased valine titer by ~25% alongside growth rate improvements and decreased byproduct formation, despite maximum possible growth rate being higher if the cells had acquired mutations to eliminate their energetically wasteful expression of fluorescent proteins.
When paired with properly selected experimental methodology and biological design, ALE’s ability to increase bioproduction of desired molecules has been established. Though more difficult to select for than simple growth rate improvements, fitness can still be tied to titer increases in a variety of circumstances: when the produced molecule can enable better growth or survivability in spite of production costs; when evolution can countervail phenotypic defects resulting from metabolic engineering for a novel production phenotype; or when growth rate maximization is not the sole driving force behind selection. An important factor to keep in mind is the stability of production strains – though production phenotypes can be selectable under carefully designed ALE conditions, industrial-scale bioreactor growth can open the door for population-takeover by mutant strains which genetically purge the production machinery (Rugbjerg and Sommer, 2019). Engineering techniques can improve strain stability by removing mobile genetic elements or error-prone polymerases and thus reducing mutation rate (Csorgo et al., 2012), but, fortunately, this is not a strict necessity – a 1,4-BDO bioproduction strain (Yim et al., 2011) which was further improved using ALE has been used at industrial scales by Genomatica, and found to remain genetically stable over the course of a bioprocess cycle (personal communication, John D. Trawick). As our knowledge of metabolic and regulatory networks grows and techniques for engineering and strain design continue to improve, we are poised to see an increased adoption of ALE as a complementary technology for optimizing microbial production strains.
2.5. General Discovery
By enabling observation of evolutionary outcomes in a controlled laboratory setting, ALE facilitates research into basic bioprocesses in addition to its more applied uses as an engineering and strain design/optimization tool. ALE studies have yielded insight into important evolutionary phenomena such as clonal interference and regulatory rewiring (Kao and Sherlock, 2008; Oud et al., 2013; Sniegowski and Gerrish, 2010; Maddamsetti et al., 2015), rate and mechanism of mutation development (Araya et al., 2010; Chang et al., 2013; Dunham et al., 2002; Notley-McRobb and Ferenci, 1999; Yona et al., 2012; Lind et al., 2017; Ene et al., 2018; Lauer et al., 2018), and response to genetic perturbation or sub-optimal growth environment (Charusanti et al., 2010; Conrad et al., 2010; Fong et al., 2005b; Wang et al., 2010; Giusy M. Adamo et al., 2012; Deng and Fong, 2011; Szamecz et al., 2014; Wenger et al., 2011; Wright et al., 2011). ALE has also been used to study fundamental aspects of biology such as regulatory gene network response to external stressors or a new environment (Quan et al., 2012; Zhu et al., 2015), as well as the impact of genome shuffling and recombination techniques on the construction of industrially pertinent strains (Peabody et al., 2016; Reyes et al., 2012; Winkler and Kao, 2012). When paired with in silico modeling and algorithmic approaches frequently used in systems-level analyses, ALE additionally serves as an empirical hypothesis tester (Dekel and Alon, 2005; Ibarra et al., 2002).
Several studies have been dedicated to uncovering the molecular framework of an organism’s response to a genetic perturbation, with obvious implications for strain engineering. Gene essentiality and the impact of gene knockouts has been analyzed using ALE (Charusanti et al., 2010; Tokuyama et al., 2018), wherein strain physiology immediately post knockout can be compared against the physiological state following the strain’s adaptive evolution. For example, Charusanti et al. investigated the adaptive response of multiple E. coli lineages following knockout of the important phosphoglucose isomerase (pgi) gene (Charusanti et al., 2010), a frequent target for disruption in engineered strains (Chin and Cirino, 2011; Papapetridis et al., 2018; Shiue et al., 2015). Distinct phenotypic outcomes were reached across the different evolved endpoints, and examination of the flux state via carbon-13 metabolic flux analysis determined that the dominant mechanism of adaptation was to overcome key bottlenecks in cofactor metabolism and substrate uptake that were induced by the knockout (Long et al., 2018). Studies such as these examining evolutionary response to genetic perturbation are increasingly yielding insight into the regulatory architecture of metabolic networks (McCloskey et al., 2018).
Modifications to ALE experiment methodology have been explored for the purposes of speeding adaptation, increasing mutation rate, and maintaining beneficial mutations. Bacterial asexual reproduction leads to clonal interference, preventing many beneficial mutations from reaching fixation within the evolving populations. Chu et al. investigated induced horizontal gene transfer as a method to speed adaptation and circumvent clonal interference, and found that genetic exchange between distinct co-cultured E. coli strains could significantly aid in adaptation to growth on novel carbon sources, contingent on the specifics of donor vs. host strain similarity and evolutionary environment (Chu et al., 2018). Contrasting with horizontal gene transfer, another technique to increase the accessibility of adaptive mutations is simply to increase the mutation rate, which can be induced with chemical mutagens (Lee et al., 2011) or naturally result from a DNA repair gene deactivation (LaCroix et al., 2014; Lenski et al., 2015); in all cases, mutation rate positively correlated with the speed of adaptation and magnitude of fitness gains. However, Couce et al. found that, even under conditions of strong selection and with improving population fitness, hypermutability leads to steady decay of the genome and can cause ‘genomic draft,’ whereby deleterious mutations hitchhike and cannot be purged from the population (Couce et al., 2017). Care should thus be taken when evolving hypermutable strains – though hypermutability can facilitate the speed and magnitude of ALE fitness increases, and even make leaps across the fitness landscape accessible by providing a large mutational pool with complex epistatic interactions, there are downsides in regards to genome stability and even adaptive potential, if the mutation rate is inordinately high (Sprouffske et al., 2018).
The novel or improved microbial phenotypes achievable with ALE makes it a powerful engineering tool, and studying the genetic and metabolic mechanisms underlying these phenotypic changes can yield valuable biological discoveries. Moreover, modifications to the selection protocol and evolutionary environment are easily explorable with ALE, allowing investigation into methods by which adaptation can be facilitated. Insights gained through the use of ALE can then be employed in other design strategies for novel solutions in industrial settings.
3. Strain Types and Generations
As long as an organism is reproducibly culturable in laboratory conditions it can be subjected to ALE, though the majority of studies analyzed in this review utilize either bacteria or yeast, specifically E. coli and S. cerevisiae (Supplemental Figure 1A). This is due to their fast growth, genetic tractability, and status as model organisms, providing the necessary knowledge base with which to interpret evolutionary outcomes. Using different organisms such as algae can allow for evolved phenotypes not achievable with bacteria or yeast, but ALE studies focusing on less standard microbial species are comparatively scant. Although much less relevant to metabolic engineering, higher organisms like fruit flies or even mice have been selectively competed and bred under controlled conditions (Barrett et al., 2019; Folk and Bradley, 2005), though these non-microbial ALE studies are beyond the scope of this review. ALE for mammalian production systems is limited given their slow growth rates and more involved culturing requirements, but some work has been done examining the genetic changes which accumulate in CHO cell lines under various culturing conditions (Feichtinger et al., 2016). Earlier examples without genetic analysis also exist, such as adapting hybridoma cells to lower serum concentrations (Lee et al., 1991). However, larger scale DNA sequencing is needed to reliably identify the potential causal mutations from such mammalian selection experiments, and epigenetic factors can further complicate the interpretation of results.
A critical parameter in an ALE experiment is the length of time for which populations of strains are evolved. Generations is the most commonly used metric for evolutionary time, though alternatives exist that include more data on the selective protocol employed, such as Cumulative Cell Divisions, which factors in population bottlenecks resulting from serial passage (Lee et al., 2011; Sandberg et al., 2014). Most ALE studies have a duration of 100–500 generations of growth (Supplemental Figure 1B), though significantly reduced (<50 generations) and increased (>60,000 generations) timescales have also seen use. The decision to stop an ALE experiment after a certain number of generations is always somewhat arbitrary - though evolved strains get labelled ‘endpoints,’ this does not mean further fitness gains are not achievable with increased experiment duration. The Long Term Evolution Experiment (LTEE) provides the best example of this; E. coli has been kept adapting to glucose minimal media for over 30 years, and competition assays on strains spanning 60,000 generations of evolution revealed that fitness gains best fit a power-law model, indicating a decreasing rate of improvement over time but no asymptotic limit (Lenski et al., 2015). Despite fitness gains being achievable indefinitely, for practical applications it is important to strike an appropriate balance between acquiring a fit endpoint and not wasting resources drawing out an experiment excessively. For example, Hua et al. evolved E. coli onto lactate growth and found ~60% fitness improvements with less than 250 generations of ALE, while increasing experiment duration to 900 generations only resulted in a further fitness gain of less than 20% (Hua et al., 2007).
When an ALE experiment is performed in a way that facilitates easy fitness tracking over time, it is typically sufficient to set an ‘endpoint’ based on subjective valuation of when fitness starts to plateau. If fitness-tracking is not possible, the studies collected herein establish ALEs of a several hundred generation timescale as suitable for most purposes. Further, modeling approaches have been developed to optimize ALE experimentation and best decide passage protocols and when to end an experiment (Bittihn et al., 2017; LaCroix et al., 2017). Such approaches will better enable automation and optimization of ALE use in the strain design process.
4. ALE Protocols and Experimental Setup
The evolutionary environment employed in an ALE experiment determines the selective pressure guiding adaptation, and is thus a critical choice when a particular phenotypic outcome is sought. Batch culturing is both the simplest and most popular ALE method (Supplemental Figure 1C), though there are significant differences between various batch culturing methods. Often it is desirable for fitness and growth rate to be equivalent, and serial batch culture propagation is a way to enforce this if cultures are passed while still in exponential phase and with excess nutrients. Without limited resources or shifts in growth phase between lag/exponential/stationary, improvements to maximum growth rate determine which strains come to dominate the population. Unfortunately, as growth rate increases over the course of the ALE, changes must be made either to the passage frequency or passage volume used for propagation if stationary phase is to be avoided (Charusanti et al., 2010; Fong and Palsson, 2004). Increasing passage frequency can be prohibitively difficult without automation, while decreasing passage volume leads to tighter population bottlenecks and a potential loss in adaptive mutations. Given these issues, many researchers opt for batch culture propagation of fixed volumes at fixed intervals (generally once per day), as in the LTEE (Lenski et al., 1991). This is a more easily maintainable experimental setup, but by virtue of going through cycles of resource excess followed by depletion selection occurs for more than just growth rate: decreases in lag phase and survival in stationary phase also significantly impact fitness. With a dynamic environment such as this, selection can also lead to co-existing specialist strains that thrive in different phases of the daily cycle (Rozen et al., 2009).
Non-batch ALE culturing setups are dominated by chemostat use, but other methods also exist and are practiced. Chemostats are the second most popular culturing technique for ALEs, and this method differs from batch culturing in ways that alter the selective forces at play. Chemostats culture cells in a bioreactor, enabling tight control over environmental parameters like pH and oxygenation, and maintain continuous growth via a constant influx of media at a set dilution rate (Gresham and Dunham, 2014). Although this can facilitate undesirable adaptive events, such as bacterial persistence due to adhesive wall growth (Rao and Rao, 2004), and can be more difficult to maintain multiple replicates in parallel, it can also be used to select for particular phenotypes. For example, Koppram et al. evolved yeast for improved biomass inhibitor tolerance via both batch and chemostat culture, and found that batch culture selected more for improvements in growth rate, glucose consumption, and ethanol productivity, while chemostat evolution favored improved inhibitor conversion rates (Koppram et al., 2012). Other culturing methods have seen occasional use in ALE experiments for specific purposes, such as long term culturing in a single flask to select for a growth advantage in stationary phase (Westphal et al., 2018), or growth on oversized petri dish with antibiotic gradients to allow spatiotemporal tracking competing lineages (Baym et al., 2016). Serial propagation of colonies streaked onto plates, by virtue of the clonal bottlenecks at each step, is not a form of ‘adaptive’ evolution without some secondary screening measure influencing the transfer process, but can be used for mutation accumulation studies (Shibai et al., 2017).
5. Automation and ALE
Although ALE experiments can be performed manually without significant difficulty and require little setup or specialized equipment, automating the process can facilitate greatly increased experimental capabilities, throughput, and clarity in outcome. In terms of expanding capabilities while running an ALE experiment, automation allows for complex culturing procedures which cannot be reasonably performed manually (e.g. propagating cultures many times a day, or splitting many independent cultures different thresholds) as well as for dynamic alterations to passage protocols (e.g., alternating substrates or temperature from flask to flask) (Sandberg et al., 2017; Wong et al., 2018). Automation also allows for improved monitoring of physiological properties of evolving cultures, enabling researchers to better understand the evolutionary dynamics at play and to make informed decisions on when and how many times to sequence for adaptive mutations. Furthermore, eliminating repetitive manual ALE passage work saves time and effort and reduces researcher fatigue, which can lead to mistakes and premature stoppage of an experiment where a mutational jump in fitness is still possible. Perhaps the most important aspect of automated ALE is the ability to efficiently execute multiple independent replicates of the same experiment at one time. With this ability to “play the same game” repeatedly, evolved genotypes can be more easily interpreted by leveraging cross-replicate mutational comparisons (e.g., identifying genetic hitchhikers which lack statistically significant independent recurrence), and multiple equivalent fitness-optimized clones are obtainable from which a strain with desirable properties can be selected, spanning the range of intracellular states enabling improved growth (e.g., a high uptake rate vs. a high yield clone).
Automation can speed up the time it takes to obtain a desirable strain by virtue of less stringent population bottlenecks and sustained exponential-phase growth, translating to faster mutant fixation. The tight environmental control of automated systems can also obviate an important issue intrinsic to manually performed batch culture propagation (Gresham and Dunham, 2014), namely the difficulty in maintaining a strict selection pressure that evolves cells under essentially invariant conditions. For example, automation has been used to select for growth rate improvement in a 2,000 generation ALE in which the cells never experienced nutrient limitation or left exponential growth phase, all while maintaining large passage volumes (LaCroix et al., 2014). When contrasted with the manually-performed Long Term Evolution Experiment (Lenski et al., 1991), this automated ALE saw equivalent fitness gains on a real-world timescale nearly 10 times shorter.
A number of automated ALE devices have been developed that span the range of culturing methods: large volume batch culture (Sandberg et al., 2014; LaCroix et al., 2014; Wong et al., 2018), microtiter plates (Horinouchi et al., 2014; Radek et al., 2017), chemostats/turbidostats (Blaby et al., 2012; Toprak et al., 2013), or microfluidics (Rotem et al., 2016). Such demonstrations establish the broad scope of technologies which can be part of automated strain construction workflows (Chao et al., 2017). Putting some of the current capabilities of such automated systems in context, gains in throughput enabled by automation allowed for more than 30 independent replicates to be evolved simultaneously on the same robotic system, or for large culture volumes to be propagated multiple times per day, minimizing the risk of missing out on adaptive mutations due to population bottlenecks (LaCroix et al., 2017). With real-time tracking of growth response to environmental particulars, automation allows a ‘sweet spot’ of chemical concentration to be maintained, such as keeping cultures in a constantly growth-inhibiting but non-lethal antibiotic environment to study mechanisms of resistance evolution (Toprak et al., 2013). This ability to modify an ALE environment on the fly opens up a number of experimental possibilities - tolerance to certain chemicals can be incrementally built up, resulting in optimized bioproduction strains (Mohamed et al., 2017), or the growth environment can be rapidly alternated to yield strains with desirable diauxic phenotypes (Sandberg et al., 2017). The information needed to develop inexpensive, automated culturing systems is also becoming publicly available (Wong et al., 2018), contrasting with systems using industrial-grade hardware (LaCroix et al., 2014), and will doubtlessly lead to more widespread and innovative adoption of ALE as a powerful technique for both biological discovery and metabolic engineering.
6. Conclusions and Future Directions
Adaptive laboratory evolution stands as a powerful tool available to strain engineers and biologists, yielding strains with phenotypic improvements that stem from random mutation and natural selection rather than rational engineering. This renders ALE a multi-functional technique: 1) it enables laboratory investigation of evolutionary processes, expanding the biological knowledge base that informs successful strain engineering through identification of beneficial mutations associated to a specific selection pressure, or those which counteract production goals; 2) it complements rational design approaches by improving engineered strain properties, countervailing the fitness defects frequently introduced by genetic manipulation; and 3) it can replace rational design approaches when a desired phenotype is selectable due to a fitness benefit in a particular environment. Furthermore, with a sizable and well-annotated collection of causal mutations, phenotypes of interest could be designed by directly engineering ALE-identified alleles into a strain to induce a particular cellular state. ALE can thus be of great utility in the Design-Build-Test-Learn cycle employed in strain construction (R. Liu et al., 2015), augmenting or even replacing the Design and Build steps (Figure 3).
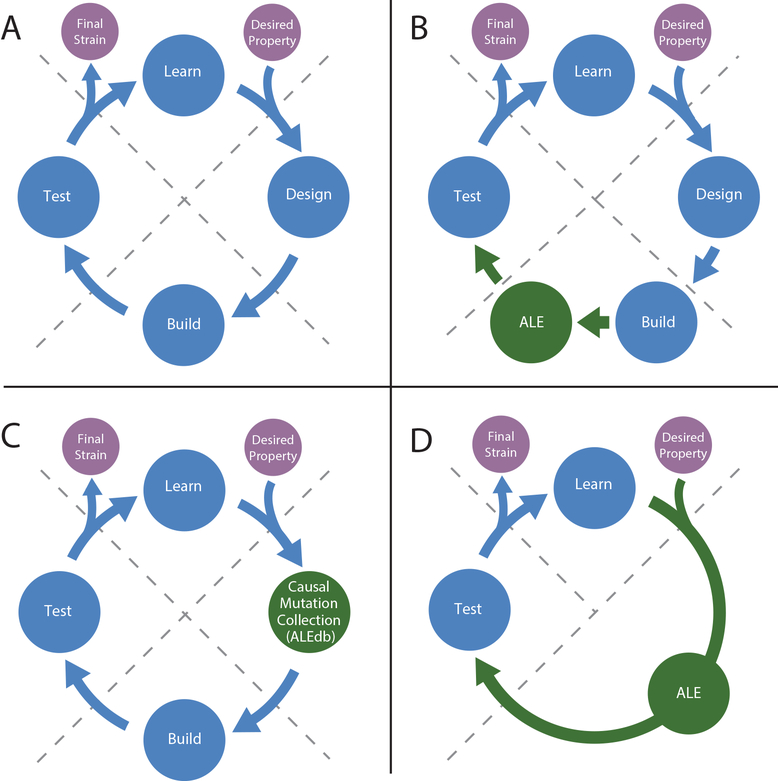
Figure 3: ALE for use in the Design, Build, Test, Learn cycle.
A) The typical Design, Build, Test, Learn cycle used in metabolic engineering to generate a strain with a desired property. B) Augmentation of the cycle where ALE is included in the Build step to rescue a strain that has decreased fitness due to a perturbation, or to optimize a strain after removal or addition of genetic content. C) Augmentation of the cycle where a collection of mutations (e.g., ALEdb (Phaneuf et al., 2018)) associated to a particular phenotype is leveraged for the Design step D) Augmentation of the cycle where ALE can be used to completely replace the Design and Build steps and a desirable strain is acquired directly from ALE when a phenotype can be tied to selection without engineering.
ALE can serve as a powerful tool, but several principles constrain such experiments. First and foremost, ALE only has efficacy with achieving evolutionarily selectable phenotypes. Growth rate improvements are inherently selectable, making tolerance and substrate-utilization ALEs straightforward approaches for phenotypic improvement, but desired phenotypes can be fitness-counterproductive for cells (e.g., overproduction of some metabolite), preventing evolution from selecting for such traits. In these cases, approaches must be taken to alter the environment and/or genome of the starting strain in a way that links the property of interest to a growth advantage, e.g. via metabolic growth-coupling, or by using a growth environment where the otherwise-wasteful metabolite production now confers a benefit. Additionally, the choice of organism is key; model organisms are genetically tractable and have the knowledgebase with which to interpret mutational results, but more rarely-used species can have atypical physiological properties that make them ideal for a particular phenotype. The overall goals of the study are paramount to consider when selecting ALE experimental parameters, such as replicate number, evolutionary duration, serial passage bottleneck size, etc. – e.g., if a particular strain phenotype is all that’s desired then replicates are less important, but if one wants to decipher molecular mechanisms then replicates greatly facilitate identifying regions of convergent evolution. Fortunately, tools exist to aid in the selection of parameters for optimizing results from an ALE experiment without wasting time or resources (LaCroix et al., 2017).
Advances in genetic engineering techniques are facilitating ambitious and novel strain designs, which can benefit from ALE as a method by which to understand limitations in these highly perturbed systems and uncover fitness-restoring mutations. Some prominent examples of studies enabled by advanced genetic engineering include synthetic minimal cells (Hutchison et al., 2016) and genome-wide codon repurposing (Wannier et al., 2018). ALE can aid in these efforts by mitigating the fitness defects of such highly altered strains, as demonstrated in the case of an E. coli strain with nearly 25% of its genome removed (Choe et al., 2019). Additionally, improvements in strain engineering techniques allow for particular phenotypes to be linked to other properties, such as biosensors that cause cellular fluorescence upon production of a desired compound. Biosensors hold great promise as a complementary ALE technology, but advancements are needed in their dynamic ranges and saturation points, compatibility across organisms, and evolutionary robustness to ‘cheaters’ that purge the sensing components, all of which currently limit efficacy in host strains (Williams et al., 2016). Nevertheless, biosensors enable enrichment for traits other than simple growth rate when the selective environment is properly designed (Mahr et al., 2015). Modifications to the ALE environment can also select for complex phenotypes, such as the evolution of multicellularity in a unicellular yeast as a result of predator-induced selection against small or individual cells (Ratcliff et al., 2015). The inherent dependence of ‘fitness’ on the particulars of the evolutionary environment makes ALE capable of selecting for diverse and novel traits.
The growing body of ALE work and the increased throughput of studies driven by automation necessitate bioinformatics pipelines and databases to fully realize its potential as an efficient tool in the strain engineering process. The ability to generate hundreds of relevant clones and populations in any given experiment (which can contain thousands of unique mutations), and the ease and cost-efficiency with which they can be sequenced, makes mutational analysis and data handling non-trivial. In addition, new technologies are enabling increased clarity on genetic alterations, such as long-read sequencing that reveals genomic rearrangements and expansions difficult to discover with shotgun sequencing alone (Pollard et al., 2018). There is a need to coalesce the growing body of studies (~102), relevant strains (~104), and mutations (~105–6) derived from ALE to learn the overarching principles and mutational types that drive phenotypic changes. Work towards meeting this challenge has appeared in the form of a database which collates ALE-derived mutations across a variety of species, growth environments, and selection schemes (Phaneuf et al., 2018). Such mutation collections essentially represent “parts lists” for cell engineers (Fig. 3C), and are ripe for computational probing with Big Data techniques that can extract salient evolutionary features. Inclusion of mutations identified in environmental and clinical isolates can also expand this rich source of allelic parts for strain engineering. The grand promise of such collections is that ALE wet lab experiments may not be required to engineer a desirable trait - mutations could be mined directly from a structured database with query tools to isolate alleles which convey desired physiological features.
In conclusion, ALE has facilitated a number of strain engineering efforts, and the rapid improvements in complementary technologies leave it poised for accelerated adoption, improved utility, and establishment as an essential method in the metabolic engineer’s experimental toolbox. Causal mutations identified in evolved strains serve both as a way to interpret the biochemical method by which fitness and any associated production or growth-coupled property was increased, and as genetic parts that can be introduced into related strains to induce a desired phenotype; importantly, these mutations are also patentable and can form the basis for intellectual property generation.
Supplementary Material
Supplemental Figure 1 - Categorization of species, timescales of evolutions studies, and experimental culturing setups used in ALE studies.
Supplemental Table 1 - A collection and categorization of ALE studies relevant to industrial biotechnology. Excel file .xlsx
Acknowledgements
We would like to thank Zachary A. King, Nathan E. Lewis, and John D. Trawick for informative discussions and insights related to the review. Funding for this work was provided by the Novo Nordisk Foundation through the Center for Biosustainability at the Technical University of Denmark under Grant NNF10CC1016517. Funding was also provided by the NIH National Institute of Allergy and Infectious Diseases under Grant no. U01AI124316. TES was supported in part through the National Science Foundation Graduate Research Fellowship (grant DGE1144086).
References
- Adamo GM, Brocca S, Passolunghi S, Salvato B, Lotti M, 2012. Laboratory evolution of copper tolerant yeast strains. Microb. Cell Fact 11, 1 10.1186/1475-2859-11-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo GM, Lotti M, Tamás MJ, Brocca S, 2012. Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 158, 2325–2335. 10.1099/mic.0.058024-0 [DOI] [PubMed] [Google Scholar]
- Aguilar C, Martínez-Batallar G, Flores N, Moreno-Avitia F, Encarnación S, Escalante A, Bolívar F, 2018. Analysis of differentially upregulated proteins in ptsHIcrr- and rppH-mutants in Escherichia coli during an adaptive laboratory evolution experiment. Appl. Microbiol. Biotechnol 102, 10193–10208. 10.1007/s00253-018-9397-3 [DOI] [PubMed] [Google Scholar]
- Alcántara-Díaz D, Breña-Valle M, Serment-Guerrero J, 2004. Divergent adaptation of Escherichia coli to cyclic ultraviolet light exposures. Mutagenesis 19, 349–354. 10.1093/mutage/geh039 [DOI] [PubMed] [Google Scholar]
- Almario MP, Reyes LH, Kao KC, 2013. Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol. Bioeng 110, 2616–2623. 10.1002/bit.24938 [DOI] [PubMed] [Google Scholar]
- Alper H, Jin Y-S, Moxley JF, Stephanopoulos G, 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng 7, 155–164. 10.1016/j.ymben.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Araya CL, Payen C, Dunham MJ, Fields S, 2010. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11, 88 10.1186/1471-2164-11-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold FH, 2018. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed Engl 57, 4143–4148. 10.1002/anie.201708408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Wu T-Y, Machado IMP, Huang W-C, Chen P-Y, Pellegrini M, Liao JC, 2010. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol 6, 449 10.1038/msb.2010.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood KC, Schneider LK, Ryan FJ, 1951. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 37, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami-Moyal L, Engelberg D, Wenger JW, Sherlock G, Braun S, 2012. Turbidostat culture of Saccharomyces cerevisiae W303–1A under selective pressure elicited by ethanol selects for mutations in SSD1 and UTH1. FEMS Yeast Res. 12, 521–533. 10.1111/j.1567-1364.2012.00803.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann H, Pronk JT, Kleerebezem M, Teusink B, 2015. Evolutionary engineering to enhance starter culture performance in food fermentations. Curr. Opin. Biotechnol 32, 1–7. 10.1016/j.copbio.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Bachmann H, Starrenburg MJC, Molenaar D, Kleerebezem M, van Hylckama Vlieg JET, 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22, 115–124. 10.1101/gr.121285.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF, Rodrigue N, Kassen R, 2015. The effect of selection environment on the probability of parallel evolution. Mol. Biol. Evol 32, 1436–1448. 10.1093/molbev/msv033 [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Laurent S, Mallarino R, Pfeifer SP, Xu CCY, Foll M, Wakamatsu K, Duke-Cohan JS, Jensen JD, Hoekstra HE, 2019. Linking a mutation to survival in wild mice. Science 363, 499–504. 10.1126/science.aav3824 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE, 2013. Genome dynamics during experimental evolution. Nat. Rev. Genet 14, 827–839. 10.1038/nrg3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF, 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247. 10.1038/nature08480 [DOI] [PubMed] [Google Scholar]
- Basso TO, de Kok S, Dario M, do Espirito-Santo JCA, Müller G, Schlölg PS, Silva CP, Tonso A, Daran J-M, Gombert AK, van Maris AJA, Pronk JT, Stambuk BU, 2011. Engineering topology and kinetics of sucrose metabolism in Saccharomyces cerevisiae for improved ethanol yield. Metab. Eng 13, 694–703. 10.1016/j.ymben.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, Kishony R, 2016. Spatiotemporal microbial evolution on antibiotic landscapes. Science 353, 1147–1151. 10.1126/science.aag0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissimi E, van Dijken JP, Pronk JT, van Maris AJA, 2009. Effects of acetic acid on the kinetics of xylose fermentation by an engineered, xylose-isomerase-based Saccharomyces cerevisiae strain. FEMS Yeast Res. 9, 358–364. 10.1111/j.1567-1364.2009.00487.x [DOI] [PubMed] [Google Scholar]
- Bennett AF, Hughes BS, 2009. Microbial experimental evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol 297, R17–25. 10.1152/ajpregu.90562.2008 [DOI] [PubMed] [Google Scholar]
- Bittihn P, Hasty J, Tsimring LS, 2017. Suppression of Beneficial Mutations in Dynamic Microbial Populations. Phys. Rev. Lett 118, 028102 10.1103/PhysRevLett.118.028102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Lyons BJ, Wroclawska-Hughes E, Phillips GCF, Pyle TP, Chamberlin SG, Benner SA, Lyons TJ, Crécy-Lagard V. de, Crécy E. de, 2012. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl. Environ. Microbiol 78, 144–155. 10.1128/AEM.05773-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan TCR, Williams TC, Schulz BL, Palfreyman RW, Krömer JO, Nielsen LK, 2015. Evolutionary Engineering Improves Tolerance for Replacement Jet Fuels in Saccharomyces cerevisiae. Appl. Environ. Microbiol 81, 3316–3325. 10.1128/AEM.04144-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard AP, Pharkya P, Maranas CD, 2003. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng 84, 647–657. 10.1002/bit.10803 [DOI] [PubMed] [Google Scholar]
- Cadière A, Ortiz-Julien A, Camarasa C, Dequin S, 2011. Evolutionary engineered Saccharomyces cerevisiae wine yeast strains with increased in vivo flux through the pentose phosphate pathway. Metab. Eng 13, 263–271. 10.1016/j.ymben.2011.01.008 [DOI] [PubMed] [Google Scholar]
- Cakar ZP, Alkim C, Turanli B, Tokman N, Akman S, Sarikaya M, Tamerler C, Benbadis L, François JM, 2009. Isolation of cobalt hyper-resistant mutants of Saccharomyces cerevisiae by in vivo evolutionary engineering approach. J. Biotechnol 143, 130–138. 10.1016/j.jbiotec.2009.06.024 [DOI] [PubMed] [Google Scholar]
- Cakar ZP, Seker UOS, Tamerler C, Sonderegger M, Sauer U, 2005. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res. 5, 569–578. 10.1016/j.femsyr.2004.10.010 [DOI] [PubMed] [Google Scholar]
- Carroll SM, Marx CJ, 2013. Evolution after introduction of a novel metabolic pathway consistently leads to restoration of wild-type physiology. PLoS Genet. 9, e1003427 10.1371/journal.pgen.1003427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Petranovic D, Nielsen J, 2014. Biofuels. Altered sterol composition renders yeast thermotolerant. Science 346, 75–78. 10.1126/science.1258137 [DOI] [PubMed] [Google Scholar]
- Chang S-L, Lai H-Y, Tung S-Y, Leu J-Y, 2013. Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS Genet. 9, e1003232 10.1371/journal.pgen.1003232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao R, Mishra S, Si T, Zhao H, 2017. Engineering biological systems using automated biofoundries. Metab. Eng 42, 98–108. 10.1016/j.ymben.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BØ, 2010. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 6, e1001186 10.1371/journal.pgen.1001186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charusanti P, Fong NL, Nagarajan H, Pereira AR, Li HJ, Abate EA, Su Y, Gerwick WH, Palsson BO, 2012. Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. PLoS One 7, e33727 10.1371/journal.pone.0033727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Cirino PC, 2011. Improved NADPH supply for xylitol production by engineered Escherichia coli with glycolytic mutations. Biotechnol. Prog 27, 333–341. 10.1002/btpr.559 [DOI] [PubMed] [Google Scholar]
- Choe D, Lee JH, Yoo M, Hwang S, Sung BH, Cho S, Palsson B, Kim SC, Cho B-K, 2019. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun 10, 935 10.1038/s41467-019-08888-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu BCH, Lee H, 2007. Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol. Adv 25, 425–441. 10.1016/j.biotechadv.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Chu HY, Sprouffske K, Wagner A, 2018. Assessing the benefits of horizontal gene transfer by laboratory evolution and genome sequencing. BMC Evol. Biol 18, 54 10.1186/s12862-018-1164-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JH, Luque R, Matharu AS, 2012. Green chemistry, biofuels, and biorefinery. Annu. Rev. Chem. Biomol. Eng 3, 183–207. 10.1146/annurev-chembioeng-062011-081014 [DOI] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, Cho B-K, Knight EM, Lewis NE, Landick R, Palsson BØ, 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. U. S. A 107, 20500–20505. 10.1073/pnas.0911253107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Lewis NE, Palsson BØ, 2011. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol 7, 509 10.1038/msb.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova LT, Lu J, Cipolla RM, Sandoval NR, Long CP, Antoniewicz MR, 2016. Co-utilization of glucose and xylose by evolved Thermus thermophilus LC113 strain elucidated by (13)C metabolic flux analysis and whole genome sequencing. Metab. Eng 37, 63–71. 10.1016/j.ymben.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couce A, Caudwell LV, Feinauer C, Hindre T, Feugeas JP, Weigt M, Lenski RE, Schneider D, Tenaillon O, 2017. Mutator genomes decay, despite sustained fitness gains, in a long-term experiment with bacteria. Proc Natl Acad Sci U S A. 114, E9026–E9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorgo B, Feher T, Timar E, Blattner FR, Posfai G, 2012. Low-mutation-rate, reduced-genome Escherichia coli: an improved host for faithful maintenance of engineered genetic constructs. Microb Cell Fact. 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, Kepner JL, Bennett AF, Lenski RE, Barrick JE, 2017. Specificity of genome evolution in experimental populations ofEscherichia colievolved at different temperatures. Proc. Natl. Acad. Sci. U. S. A 114, E1904–E1912. 10.1073/pnas.1616132114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U, 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592. 10.1038/nature03842 [DOI] [PubMed] [Google Scholar]
- de Kok S, Nijkamp JF, Oud B, Roque FC, de Ridder D, Daran J-M, Pronk JT, van Maris AJA, 2012. Laboratory evolution of new lactate transporter genes in a jen1Δ mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res. 10.1111/j.1567-1364.2012.00787.x [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Sekowska A, Danchin A, 2015. Chemical reactivity drives spatiotemporal organisation of bacterial metabolism. FEMS Microbiol. Rev 39, 96–119. 10.1111/1574-6976.12089 [DOI] [PubMed] [Google Scholar]
- Deng Y, Fong SS, 2011. Laboratory evolution and multi-platform genome re-sequencing of the cellulolytic actinobacterium Thermobifida fusca. J. Biol. Chem 286, 39958–39966. 10.1074/jbc.M111.239616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R, Sägesser R, Weikert C, Wagner A, 2013. Yeast adapts to a changing stressful environment by evolving cross-protection and anticipatory gene regulation. Mol. Biol. Evol 30, 573–588. 10.1093/molbev/mss253 [DOI] [PubMed] [Google Scholar]
- Dhar R, Sägesser R, Weikert C, Yuan J, Wagner A, 2011. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J. Evol. Biol 24, 1135–1153. 10.1111/j.1420-9101.2011.02249.x [DOI] [PubMed] [Google Scholar]
- Dragosits M, Mattanovich D, 2013. Adaptive laboratory evolution -- principles and applications for biotechnology. Microb. Cell Fact 12, 64 10.1186/1475-2859-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I, 2013. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol. Syst. Biol 9, 643 10.1038/msb.2012.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D, 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A 99, 16144–16149. 10.1073/pnas.242624799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Farrer RA, Hirakawa MP, Agwamba K, Cuomo CA Bennett RJ, 2018. Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. Proc. Natl. Acad. Sci. U. S. A 115, E8688–E8697. 10.1073/pnas.1806002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feichtinger J, Hernandez I, Fischer C, Hanscho M, Auer N, Hackl M, Jadhav V, Baumann M, Krempl PM, Schmidl C, Farlik M, Schuster M, Merkel A, Sommer A, Heath S, Rico D, Bock C, Thallinger GG, Borth N, 2016. Comprehensive genome and epigenome characterization of CHO cells in response to evolutionary pressures and over time. Biotechnol Bioeng. 113, 2241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, Palsson BØ, 2010. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng 12, 173–186. 10.1016/j.ymben.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO, Rosenzweig RF, 1999. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. U. S. A 96, 9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cabezón L, Cros A, Nikel PI, 2019. Evolutionary Approaches for Engineering Industrially Relevant Phenotypes in Bacterial Cell Factories. Biotechnol. J, 10.1002/biot.201800439 [DOI] [PubMed] [Google Scholar]
- Fletcher E, Feizi A, Bisschops MMM, Hallström BM, Khoomrung S, Siewers V, Nielsen J, 2017. Evolutionary engineering reveals divergent paths when yeast is adapted to different acidic environments. Metab. Eng 39, 19–28. 10.1016/j.ymben.2016.10.010 [DOI] [PubMed] [Google Scholar]
- Folk DG, Bradley TJ, 2005. Adaptive evolution in the lab: unique phenotypes in fruit flies comprise a fertile field of study. Integr. Comp. Biol 45, 492–499. 10.1093/icb/45.3.492 [DOI] [PubMed] [Google Scholar]
- Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BO, 2005a. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng 91, 643–648. 10.1002/bit.20542 [DOI] [PubMed] [Google Scholar]
- Fong SS, Joyce AR, Palsson BØ, 2005b. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 15, 1365–1372. 10.1101/gr.3832305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SS, Palsson BØ, 2004. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat. Genet 36, 1056–1058. 10.1038/ng1432 [DOI] [PubMed] [Google Scholar]
- Fu W, Guðmundsson O, Paglia G, Herjólfsson G, Andrésson OS, Palsson BO, Brynjólfsson S, 2013. Enhancement of carotenoid biosynthesis in the green microalga Dunaliella salina with light-emitting diodes and adaptive laboratory evolution. Appl. Microbiol. Biotechnol 97, 2395–2403. 10.1007/s00253-012-4502-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Sanchez R, Karhumaa K, Fonseca C, Sànchez Nogué V, Almeida JR, Larsson CU, Bengtsson O, Bettiga M, Hahn-Hägerdal B, Gorwa-Grauslund MF, 2010. Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol. Biofuels 3, 13 10.1186/1754-6834-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ramos D, Gorter de Vries AR, Grijseels SS, van Berkum MC, Swinnen S, van den Broek M, Nevoigt E, Daran J-MG, Pronk JT, van Maris AJA, 2016. A new laboratory evolution approach to select for constitutive acetic acid tolerance in Saccharomyces cerevisiae and identification of causal mutations. Biotechnol. Biofuels 9, 173 10.1186/s13068-016-0583-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S, 2010. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol. Syst. Biol 6, 378 10.1038/msb.2010.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Stülke J, 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol 6, 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- Grabar TB, Zhou S, Shanmugam KT, Yomano LP, Ingram LO, 2006. Methylglyoxal bypass identified as source of chiral contamination in l(+) and d(−)-lactate fermentations by recombinant Escherichia coli. Biotechnol. Lett 28, 1527–1535. 10.1007/s10529-006-9122-7 [DOI] [PubMed] [Google Scholar]
- Gresham D, Dunham MJ, 2014. The enduring utility of continuous culturing in experimental evolution. Genomics 104, 399–405. 10.1016/j.ygeno.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães PMR, François J, Parrou JL, Teixeira JA, Domingues L, 2008. Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl. Environ. Microbiol 74, 1748–1756. 10.1128/AEM.00186-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán GI, Sandberg TE, LaCroix RA, Nyerges Á, Papp H, de Raad M, King ZA, Hefner Y, Northen TR, Notebaart RA, Pál C, Palsson BO, Papp B, Feist AM, 2019. Enzyme promiscuity shapes adaptation to novel growth substrates. Mol. Syst. Biol 15, e8462 10.15252/msb.20188462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft RJF, Keating DH, Schwaegler T, Schwalbach MS, Vinokur J, Tremaine M, Peters JM, Kotlajich MV, Pohlmann EL, Ong IM, Grass JA, Kiley PJ, Landick R, 2014. Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc. Natl. Acad. Sci. U. S. A 111, E2576–85. 10.1073/pnas.1401853111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen ASL, Lennen RM, Sonnenschein N, Herrgård MJ, 2017. Systems biology solutions for biochemical production challenges. Curr. Opin. Biotechnol 45, 85–91. 10.1016/j.copbio.2016.11.018 [DOI] [PubMed] [Google Scholar]
- Hawkins GM, Doran-Peterson J, 2011. A strain of Saccharomyces cerevisiae evolved for fermentation of lignocellulosic biomass displays improved growth and fermentative ability in high solids concentrations and in the presence of inhibitory compounds. Biotechnol. Biofuels 4, 49 10.1186/1754-6834-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heer D, Sauer U, 2008. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb. Biotechnol 1, 497–506. 10.1111/j.1751-7915.2008.00050.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson WR, Campbell T, DeLorenzo DM, Gao Y, Berla B, Kim SJ, Foston M, Moon TS, Dantas G, 2018. Multi-omic elucidation of aromatic catabolism in adaptively evolved Rhodococcus opacus. Metab. Eng 49, 69–83. 10.1016/j.ymben.2018.06.009 [DOI] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BØ, 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet 38, 1406–1412. 10.1038/ng1906 [DOI] [PubMed] [Google Scholar]
- Hindré T, Knibbe C, Beslon G, Schneider D, 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol 10, 352–365. 10.1038/nrmicro2750 [DOI] [PubMed] [Google Scholar]
- Hong K-K, Vongsangnak W, Vemuri GN, Nielsen J, 2011. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc. Natl. Acad. Sci. U. S. A 108, 12179–12184. 10.1073/pnas.1103219108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P-W, Swinnen S, Duitama J, Nevoigt E, 2017. The sole introduction of two single-point mutations establishes glycerol utilization inSaccharomyces cerevisiaeCEN.PK derivatives. Biotechnol. Biofuels 10, 10 10.1186/s13068-016-0696-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T, Minamoto T, Suzuki S, Shimizu H, Furusawa C, 2014. Development of an automated culture system for laboratory evolution. J. Lab. Autom 19, 478–482. 10.1177/2211068214521417 [DOI] [PubMed] [Google Scholar]
- Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, Hirasawa T, Yomo T, Shimizu H, 2010. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 11, 579 10.1186/1471-2164-11-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q, Joyce AR, Palsson BØ, Fong SS, 2007. Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Appl. Environ. Microbiol 73, 4639–4647. 10.1128/AEM.00527-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, Andersson DI, 2015. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat. Rev. Genet 16, 459–471. 10.1038/nrg3922 [DOI] [PubMed] [Google Scholar]
- Hutchison CA 3rd, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi Z-Q, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC, 2016. Design and synthesis of a minimal bacterial genome. Science 351, aad6253. 10.1126/science.aad6253 [DOI] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO, 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189. 10.1038/nature01149 [DOI] [PubMed] [Google Scholar]
- Ingram LO, Doran JB, 1995. Conversion of cellulosic materials to ethanol. FEMS Microbiol. Rev 16, 235–241. 10.1016/0168-6445(94)00083-B [DOI] [Google Scholar]
- Jansen MLA, Daran-Lapujade P, de Winde JH, Piper MDW, Pronk JT, 2004. Prolonged maltose-limited cultivation of Saccharomyces cerevisiae selects for cells with improved maltose affinity and hypersensitivity. Appl. Environ. Microbiol 70, 1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen MLA, Diderich JA, Mashego M, Hassane A, de Winde JH, Daran-Lapujade P, Pronk JT, 2005. Prolonged selection in aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity. Microbiology 151, 1657–1669. 10.1099/mic.0.27577-0 [DOI] [PubMed] [Google Scholar]
- Jantama K, Haupt MJ, Svoronos SA, Zhang X, Moore JC, Shanmugam KT, Ingram LO, 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng 99, 1140–1153. 10.1002/bit.21694 [DOI] [PubMed] [Google Scholar]
- Jensen K, Broeken V, Lærke Hansen AS, Sonnenschein N, Herrgård MJ, 2019. OptCouple: Joint simulation of gene knockouts, insertions and medium modifications for prediction of growth-coupled strain designs. Metabolic Engineering Communications e00087 10.1016/j.mec.2019.e00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Li S, Hu Y, Xu Q, Huang H, 2012. Adaptive evolution for fast growth on glucose and the effects on the regulation of glucose transport system in Clostridium tyrobutyricum. Biotechnol. Bioeng 109, 708–718. 10.1002/bit.23346 [DOI] [PubMed] [Google Scholar]
- Jiang L-Y, Chen S-G, Zhang Y-Y, Liu J-Z, 2013. Metabolic evolution of Corynebacterium glutamicum for increased production of L-ornithine. BMC Biotechnol. 13, 47 10.1186/1472-6750-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X-J, Huang H, Ouyang P-K, 2011. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv 29, 351–364. 10.1016/j.biotechadv.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Jung YH, Kim S, Yang J, Seo J-H, Kim KH, 2017. Intracellular metabolite profiling of Saccharomyces cerevisiae evolved under furfural. Microb. Biotechnol 10, 395–404. 10.1111/1751-7915.12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KC, Sherlock G, 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet 40, 1499–1504. 10.1038/ng.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC, 2012. Experimental evolution. Trends Ecol. Evol 27, 547–560. 10.1016/j.tree.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Kildegaard KR, Hallström BM, Blicher TH, Sonnenschein N, Jensen NB, Sherstyk S, Harrison SJ, Maury J, Herrgård MJ, Juncker AS, Forster J, Nielsen J, Borodina I, 2014. Evolution reveals a glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance. Metab. Eng 26, 57–66. 10.1016/j.ymben.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Kim SR, Ha S-J, Kong II, Jin Y-S, 2012. High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab. Eng 14, 336–343. 10.1016/j.ymben.2012.04.001 [DOI] [PubMed] [Google Scholar]
- King ZA, O’Brien EJ, Feist AM, Palsson BO, 2017. Literature mining supports a next-generation modeling approach to predict cellular byproduct secretion. Metab. Eng 39, 220–227. 10.1016/j.ymben.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Klamt S, Mahadevan R, 2015. On the feasibility of growth-coupled product synthesis in microbial strains. Metab. Eng 30, 166–178. 10.1016/j.ymben.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Klimacek M, Kirl E, Krahulec S, Longus K, Novy V, Nidetzky B, 2014. Stepwise metabolic adaption from pure metabolization to balanced anaerobic growth on xylose explored for recombinant Saccharomyces cerevisiae. Microb. Cell Fact 13, 37 10.1186/1475-2859-13-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppram R, Albers E, Olsson L, 2012. Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol. Biofuels 5, 32 10.1186/1754-6834-5-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT, 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5, 925–934. 10.1016/j.femsyr.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Kuyper M, Winkler AA, van Dijken JP, Pronk JT, 2004. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 4, 655–664. 10.1016/j.femsyr.2004.01.003 [DOI] [PubMed] [Google Scholar]
- LaCroix RA, Palsson BO, Feist AM, 2017. A Model for Designing Adaptive Laboratory Evolution Experiments. Appl. Environ. Microbiol 83 10.1128/AEM.03115-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM, 2014. Discovery of key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal media using adaptive laboratory evolution. Appl. Environ. Microbiol 10.1128/AEM.02246-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif H, Sahin M, Tarasova J, Tarasova Y, Portnoy VA, Nogales J, Zengler K, 2015. Adaptive Evolution of Thermotoga maritima Reveals Plasticity of the ABC Transporter Network. Appl. Environ. Microbiol 81, 5477–5485. 10.1128/AEM.01365-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S, Avecilla G, Spealman P, Sethia G, Brandt N Levy SF, Gresham D, 2018. Single-cell copy number variant detection reveals the dynamics and diversity of adaptation. PloS Biol. 16, e3000069 10.1371/journal.pbio.3000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-H, Feist AM, Barrett CL, Palsson BØ, 2011. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PLoS One 6, e26172 10.1371/journal.pone.0026172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-H, Palsson BØ, 2010. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol. Appl. Environ. Microbiol 76, 4158–4168. 10.1128/AEM.00373-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Seo J, Kim E-S, Lee H-S, Kim P, 2013. Adaptive evolution of Corynebacterium glutamicum resistant to oxidative stress and its global gene expression profiling. Biotechnol. Lett 35, 709–717. 10.1007/s10529-012-1135-9 [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim M-S, Lee J-H, Kim TW, Bae SS, Lee S-M, Jung HC, Yang T-J, Choi AR, Cho Y-J, Lee J-H, Kwon KK, Lee HS, Kang SG, 2016. Adaptive engineering of a hyperthermophilic archaeon on CO and discovering the underlying mechanism by multi-omics analysis. Sci. Rep 6, 22896 10.1038/srep22896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-M, Jellison T, Alper HS, 2014. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol. Biofuels 7, 122 10.1186/s13068-014-0122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kim HU, 2015. Systems strategies for developing industrial microbial strains. Nat. Biotechnol 33, 1061 10.1038/nbt.3365 [DOI] [PubMed] [Google Scholar]
- Lenski RE, Mongold JA, Sniegowski PD, Travisano M, Vasi F, Gerrish PJ, Schmidt TM, 1998. Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie Van Leeuwenhoek 73, 35–47. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC, 1991. Long-Term Experimental Evolution in Escherichia coli. I. Adaptation and Divergence During 2,000 Generations. Am. Nat 138, 1315–1341. 10.1086/285289 [DOI] [Google Scholar]
- Lenski RE, Travisano M, 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. U. S. A 91, 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Wiser MJ, Ribeck N, Blount ZD, Nahum JR, Morris JJ, Zaman L, Turner CB, Wade BD, Maddamsetti R, Burmeister AR, Baird EJ, Bundy J, Grant NA, Card KJ, Rowles M, Weatherspoon K, Papoulis SE, Sullivan R, Clark C, Mulka JS, Hajela N, 2015. Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with Escherichia coli. Proc. Biol. Sci 282, 20152292 10.1098/rspb.2015.2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon D, D’Alton S, Quandt EM, Barrick JE, 2018. Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides. PLoS Genet. 14, e1007348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linville JL, Rodriguez M Jr, Land M, Syed MH, Engle NL, Tschaplinski TJ, Mielenz JR, Cox CD, 2013. Industrial robustness: understanding the mechanism of tolerance for the Populus hydrolysate-tolerant mutant strain of Clostridium thermocellum. PLoS One 8, e78829 10.1371/journal.pone.0078829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Tanaka S, 2006. Ethanol fermentation from biomass resources: current state and prospects. Appl. Microbiol. Biotechnol 69, 627–642. 10.1007/s00253-005-0229-x [DOI] [PubMed] [Google Scholar]
- Lind PA, Farr AD, Rainey PB, 2017. Evolutionary convergence in experimental Pseudomonas populations. ISME J. 11, 589–600. 10.1038/ismej.2016.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Evans T, Zhang F, 2015. Applications and advances of metabolite biosensors for metabolic engineering. Metab. Eng 31, 35–43. 10.1016/j.ymben.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Liu R, Bassalo MC, Zeitoun RI, Gill RT, 2015. Genome scale engineering techniques for metabolic engineering. Metab. Eng 32, 143–154. 10.1016/j.ymben.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Lloyd CJ, King ZA, Sandberg TE, Hefner Y, Olson CA, Phaneuf PV, O’Brien EJ, Sanders JG, Salido RA, Sanders K, Brennan C, Humphrey G, Knight R, Feist AM, 2019. The genetic basis for adaptation of model-designed syntrophic co-cultures. PLoS Comput. Biol 15, e1006213 10.1371/journal.pcbi.1006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A, Liti G, Luptak A, Tenaillon O, 2015. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet 16, 567–582. 10.1038/nrg3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR, 2018. How adaptive evolution reshapes metabolism to improve fitness: recent advances and future outlook. Curr. Opin. Chem. Eng 22, 209–215. 10.1016/j.coche.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR, 2018. Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 115, 222–227. 10.1073/pnas.1716056115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR, 2017. Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab. Eng 44, 100–107. 10.1016/j.ymben.2017.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan G, Cai Z, Li Y, Ma Y, 2013. Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production. Biotechnol. Biofuels 6, 137 10.1186/1754-6834-6-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Wei L, Zhu K, Wei D, Hua Q, 2012. Combining metabolic engineering and adaptive evolution to enhance the production of dihydroxyacetone from glycerol by Gluconobacter oxydans in a low-cost way. Bioresour. Technol 117, 317–324. 10.1016/j.biortech.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Luo H, Hansen ASL, Yang L, Schneider K, Kristensen M, Christensen U, Christensen HB, Du B, Özdemir E, Feist AM, Keasling JD, Jensen MK, Herrgård MJ, Palsson BO, 2019. Coupling S-adenosylmethionine-dependent methylation to growth: Design and uses. PLoS Biol. 17, e2007050 10.1371/journal.pbio.2007050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddamsetti R, Lenski RE, Barrick JE, 2015. Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with Escherichia coli. Genetics 200, 619–631. 10.1534/genetics.115.176677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr R, Gätgens C, Gätgens J, Polen T, Kalinowski J, Frunzke J, 2015. Biosensor-driven adaptive laboratory evolution of l-valine production in Corynebacterium glutamicum. Metab. Eng 32, 184–194. 10.1016/j.ymben.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Mans R, Daran J-MG, Pronk JT, 2018. Under pressure: evolutionary engineering of yeast strains for improved performance in fuels and chemicals production. Curr. Opin. Biotechnol 50, 47–56. 10.1016/j.copbio.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Marques WL, Mans R, Marella ER, Cordeiro RL, van den Broek M, Daran J-MG, Pronk JT, Gombert AK, van Maris AJA, 2017. Elimination of sucrose transport and hydrolysis in Saccharomyces cerevisiae: a platform strain for engineering sucrose metabolism. FEMS Yeast Res. 17 10.1093/femsyr/fox006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Ai C, Blum P, 2018. Enhancement of Metallosphaera sedula Bioleaching by Targeted Recombination and Adaptive Laboratory Evolution. Adv. Appl. Microbiol 104, 135–165. 10.1016/bs.aambs.2018.03.002 [DOI] [PubMed] [Google Scholar]
- McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO, 2018. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun 9, 3796 10.1038/s41467-018-06219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico A, Ragni E, Galafassi S, Popolo L, Compagno C, 2011. Generation of an evolved Saccharomyces cerevisiae strain with a high freeze tolerance and an improved ability to grow on glycerol. J. Ind. Microbiol. Biotechnol 38, 1037–1044. 10.1007/s10295-010-0878-3 [DOI] [PubMed] [Google Scholar]
- Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, Xie B, McConnell CA, Ward RJ, Schwartz DR, Rouillard J-M, Gao Y, Gulari E, Lin XN, 2011. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb. Cell Fact 10, 18 10.1186/1475-2859-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed ET, Wang S, Lennen RM, Herrgård MJ, Simmons BA, Singer SW, Feist AM, 2017. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb. Cell Fact 16, 204 10.1186/s12934-017-0819-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser JW, Prielhofer R, Gerner SM, Graf AB, Wilson IBH, Mattanovich D, Dragosits M, 2017. Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris. Microb. Cell Fact 16, 49 10.1186/s12934-017-0661-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moysés DN, Reis VCB, de Almeida JRM, de Moraes LMP, Torres FAG, 2016. Xylose Fermentation by Saccharomyces cerevisiae: Challenges and Prospects. Int. J. Mol. Sci 17, 207 10.3390/ijms17030207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundhada H, Schneider K, Christensen HB, Nielsen AT, 2016. Engineering of high yield production of L-serine in Escherichia coli. Biotechnol. Bioeng 113, 807–816. [DOI] [PubMed] [Google Scholar]
- Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT, 2017. Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution. Metab. Eng 39, 141–150. 10.1016/j.ymben.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Notebaart RA, Kintses B, Feist AM, Papp B, 2018. Underground metabolism: network-level perspective and biotechnological potential. Curr. Opin. Biotechnol 49, 108–114. 10.1016/j.copbio.2017.07.015 [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, Ferenci T, 1999. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol 1, 45–52. [DOI] [PubMed] [Google Scholar]
- Novick A, Szilard L, 1950. Experiments with the Chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. U. S. A 36, 708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novy V, Krahulec S, Wegleiter M, Müller G, Longus K, Klimacek M, Nidetzky B, 2014. Process intensification through microbial strain evolution: mixed glucose-xylose fermentation in wheat straw hydrolyzates by three generations of recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 7, 49 10.1186/1754-68347-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Estopier A, Lesage J, Gorret N, Guillouet SE, 2011. Kinetic analysis of a Saccharomyces cerevisiae strain adapted for improved growth on glycerol: Implications for the development of yeast bioprocesses on glycerol. Bioresour. Technol 102, 1521–1527. 10.1016/j.biortech.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Otero JM, Cimini D, Patil KR, Poulsen SG, Olsson L, Nielsen J, 2013. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS One 8, e54144 10.1371/journal.pone.0054144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJA, Daran J-M, 2013. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A 110, E4223–31. 10.1073/pnas.1305949110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer MS, Liu DR, 2015. Methods for the directed evolution of proteins. Nat. Rev. Genet 16, 379–94. 10.1038/nrg3927 [DOI] [PubMed] [Google Scholar]
- Palmer AC, Kishony R, 2013. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet 14, 243–248. 10.1038/nrg3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetridis I, Verhoeven MD, Wiersma SJ, Goudriaan M, van Maris AJA, Pronk JT, 2018. Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res. 18 10.1093/femsyr/foy056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Notebaart RA, Pál C, 2011. Systems-biology approaches for predicting genomic evolution. Nat. Rev. Genet 12, 591–602. 10.1038/nrg3033 [DOI] [PubMed] [Google Scholar]
- Peabody GL, Winkler J, Kao KC, 2014. Tools for developing tolerance to toxic chemicals in microbial systems and perspectives on moving the field forward and into the industrial setting. Curr. Opin. Chem. Eng 6, 9–17. 10.1016/j.coche.2014.08.001 [DOI] [Google Scholar]
- Peabody G, Winkler J, Fountain W, Castro DA, Leiva-Aravena E, Kao KC, 2016. Benefits of a Recombination-Proficient Escherichia coli System for Adaptive Laboratory Evolution. Appl. Environ. Microbiol 82, 6736–6747. 10.1128/AEM.01850-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SR, Sànchez I Nogué V, Frazão CJR, Serafim LS, Gorwa-Grauslund MF, Xavier AMRB, 2015. Adaptation of Scheffersomyces stipitis to hardwood spent sulfite liquor by evolutionary engineering. Biotechnol. Biofuels 8, 50 10.1186/s13068-015-0234-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer E, Gätgens C, Polen T, Frunzke J, 2017. Adaptive laboratory evolution of Corynebacterium glutamicum towards higher growth rates on glucose minimal medium. Sci. Rep 7, 16780 10.1038/s41598-017-17014-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phaneuf PV, Gosting D, Palsson B, Feist A, 2018. ALEdb 1.0: A Database of Mutations from Adaptive Laboratory Evolution. bioRxiv. 10.1101/320747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharkya P, Burgard AP, Maranas CD, 2004. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 14, 2367–2376. 10.1101/gr.2872004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plácido J, Capareda S, 2016. Conversion of residues and by-products from the biodiesel industry into value-added products. Bioresources and Bioprocessing 3, 23 10.1186/s40643-016-0100-1 [DOI] [Google Scholar]
- Pollard MO, Gurdasani D, Mentzer AJ, Porter T, Sandhu MS, 2018. Long reads: their purpose and place. Hum. Mol. Genet 27, R234–R241. 10.1093/hmg/ddy177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontrelli S, Fricke RCB, Sakurai SSM, Putri SP, Fitz-Gibbon S, Chung M, Wu H-Y, Chen Y-J, Pellegrini M, Fukusaki E, Liao JC, 2018. Directed strain evolution restructures metabolism for 1-butanol production in minimal media. Metab. Eng 49, 153–163. 10.1016/j.ymben.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Portnoy VA, Bezdan D, Zengler K, 2011/8. Adaptive laboratory evolution — harnessing the power of biology for metabolic engineering. Curr. Opin. Biotechnol 22, 590–594. 10.1016/j.copbio.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Qin D, Hu Y, Cheng J, Wang N, Li S, Wang D, 2016. An auto-inducible Escherichia coli strain obtained by adaptive laboratory evolution for fatty acid synthesis from ionic liquid-treated bamboo hydrolysate. Bioresour. Technol 221, 375–384. 10.1016/j.biortech.2016.09.024 [DOI] [PubMed] [Google Scholar]
- Qi Y, Liu H, Chen X, Liu L, 2019. Engineering microbial membranes to increase stress tolerance of industrial strains. Metab. Eng 53, 24–34. 10.1016/j.ymben.2018.12.010 [DOI] [PubMed] [Google Scholar]
- Quan S, Ray JCJ, Kwota Z, Duong T, Balázsi G, Cooper TF, Monds RD, 2012. Adaptive evolution of the lactose utilization network in experimentally evolved populations of Escherichia coli. PLoS Genet. 8, e1002444 10.1371/journal.pgen.1002444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbers I, van Heerden JH, Nordholt N, Bachmann H, Teusink B, Bruggeman FJ, 2015. Metabolism at evolutionary optimal States. Metabolites 5, 311–343. 10.3390/metabo5020311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radek A, Tenhaef N, Müller MF, Brüsseler C, Wiechert W, Marienhagen J, Polen T, Noack S, 2017. Miniaturized and automated adaptive laboratory evolution: Evolving Corynebacterium glutamicum towards an improved d-xylose utilization. Bioresour. Technol 245, 1377–1385. 10.1016/j.biortech.2017.05.055 [DOI] [PubMed] [Google Scholar]
- Rajaraman E, Agarwal A, Crigler J, Seipelt-Thiemann R, Altman E, Eiteman MA, 2016. Transcriptional analysis and adaptive evolution of Escherichia coli strains growing on acetate. Appl. Microbiol. Biotechnol 100, 7777–7785. 10.1007/s00253-016-7724-0 [DOI] [PubMed] [Google Scholar]
- Rao VSH, Rao PRS, 2004. Global stability in chemostat models involving time delays and wall growth. Nonlinear Anal. Real World Appl 5, 141–158. 10.1016/S1468-1218(03)00022-1 [DOI] [Google Scholar]
- Ratcliff WC, Fankhauser JD, Rogers DW, Greig D, Travisano M, 2015. Origins of multicellular evolvability in snowflake yeast. Nat. Commun 6, 6102 10.1038/ncomms7102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi P, Masson-Boivin C, Rocha EPC, 2019. Experimental Evolution as a Tool to Investigate Natural Processes and Molecular Functions. Trends Microbiol. 10.1016/j.tim.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Reyes LH, Abdelaal AS, Kao KC, 2013. Genetic determinants for n-butanol tolerance in evolved Escherichia coli mutants: cross adaptation and antagonistic pleiotropy between n-butanol and other stressors. Appl. Environ. Microbiol 79, 5313–5320. 10.1128/AEM.01703-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC, 2012. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab. Eng 14, 579–590. 10.1016/j.ymben.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Reyes LH, Gomez JM, Kao KC, 2014. Improving carotenoids production in yeast via adaptive laboratory evolution. Metab. Eng 21, 26–33. 10.1016/j.ymben.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Riehle MM, Bennett AF, Lenski RE, Long AD, 2003. Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol. Genomics 14, 47–58. 10.1152/physiolgenomics.00034.2002 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Verdugo A, Carrillo-Cisneros D, Gonzalez-Gonzalez A, Gaut BS, Bennett AF, 2014. Different tradeoffs result from alternate genetic adaptations to a common environment. Proc. Natl. Acad. Sci. U. S. A 111, 12121–12126. 10.1073/pnas.1406886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Serohijos A, Chang C, Wolfe J, Fischer A, Mehoke T, Zhang H, Tao Y, Ung L, Choi J-M, Kolawole A, Koehler S, Wu S, Thielen P, Cui N, Demirev P, Giacobbi N, Julian T, Schwab K, Lin J, Smith T, Pipas J, Wobus C, Feldman A, Weitz D, Shakhnovich E, 2016. Tuning the course of evolution on the biophysical fitness landscape of an RNA virus. bioRxiv. 10.1101/090258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce LA, Yoon JM, Chen Y, Rickenbach E, Shanks JV, Jarboe LR, 2015. Evolution for exogenous octanoic acid tolerance improves carboxylic acid production and membrane integrity. Metab. Eng 29, 180–188. 10.1016/j.ymben.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Rozen DE, Philippe N, Arjan de Visser J, Lenski RE, Schneider D, 2009. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol. Lett 12, 34–44. 10.1111/j.1461-0248.2008.01257.x [DOI] [PubMed] [Google Scholar]
- Rudolph B, Gebendorfer KM, Buchner J, Winter J, 2010. Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem 285, 19029–19034. 10.1074/jbc.M110.103374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugbjerg P, Feist AM, Sommer MOA, 2018. Enhanced Metabolite Productivity of Escherichia coli Adapted to Glucose M9 Minimal Medium. Front Bioeng Biotechnol 6, 166 10.3389/fbioe.2018.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugbjerg P, Sommer MOA, 2019. Overcoming genetic heterogeneity in industrial fermentations. Nature biotechnology. 10.1038/s41587-019-0171-6 [DOI] [PubMed] [Google Scholar]
- Sandberg TE, Lloyd CJ, Palsson BO, Feist AM, 2017. Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies. Appl. Environ. Microbiol 83 10.1128/AEM.00410-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg TE, Long CP, Gonzalez JE, Feist AM, Antoniewicz MR, Palsson BO, 2016. Evolution of E. coli on [U-13C]Glucose Reveals a Negligible Isotopic Influence on Metabolism and Physiology. PLoS One 11, e0151130 10.1371/journal.pone.0151130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM, 2014. Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol 31, 2647–2662. 10.1093/molbev/msu209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K, 2011. Lignocellulose: A chewy problem. Nature 474, S12–4. 10.1038/474S012a [DOI] [PubMed] [Google Scholar]
- Sauer U, 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol 73, 129–169. [DOI] [PubMed] [Google Scholar]
- Scalcinati G, Otero JM, Van Vleet JRH, Jeffries TW, Olsson L, Nielsen J, 2012. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res. 12, 582–597. 10.1111/j.1567-1364.2012.00808.x [DOI] [PubMed] [Google Scholar]
- Sehnem NT, Machado A. da S., Leite FCB, Pita W. de B., de Morais MA Jr, Ayub MAZ, 2013. 5-Hydroxymethylfurfural induces ADH7 and ARI1 expression in tolerant industrial Saccharomyces cerevisiae strain P6H9 during bioethanol production. Bioresour. Technol 133, 190–196. 10.1016/j.biortech.2013.01.063 [DOI] [PubMed] [Google Scholar]
- Shao X, Raman B, Zhu M, Mielenz JR, Brown SD, Guss AM, Lynd LR, 2011. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl. Microbiol. Biotechnol 92, 641–652. 10.1007/s00253-011-3492-z [DOI] [PubMed] [Google Scholar]
- Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, Waterston RH, 2017. DNA sequencing at 40: past, present and future. Nature 550, 345–353. 10.1038/nature24286 [DOI] [PubMed] [Google Scholar]
- Shen Y, Chen X, Peng B, Chen L, Hou J, Bao X, 2012. An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl. Microbiol. Biotechnol 96, 1079–1091. 10.1007/s00253-012-4418-0 [DOI] [PubMed] [Google Scholar]
- Shepelin D, Hansen ASL, Lennen R, Luo H, Herrgård MJ, 2018. Selecting the Best: Evolutionary Engineering of Chemical Production in Microbes. Genes 9 10.3390/genes9050249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibai A, Takahashi Y, Ishizawa Y, Motooka D, Nakamura S, Ying B-W, Tsuru S, 2017. Mutation accumulation under UV radiation in Escherichia coli. Sci. Rep 7, 14531 10.1038/s41598-017-15008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiue E, Brockman IM, Prather KLJ, 2015. Improving product yields on D-glucose in Escherichia coli via knockout of pgi and zwf and feeding of supplemental carbon sources. Biotechnol. Bioeng 112, 579–587. 10.1002/bit.25470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleight SC, Lenski RE, 2007. Evolutionary adaptation to freeze-thaw-growth cycles in Escherichia coli. Physiol. Biochem. Zool 80, 370–385. 10.1086/518013 [DOI] [PubMed] [Google Scholar]
- Smith J, van Rensburg E, Görgens JF, 2014. Simultaneously improving xylose fermentation and tolerance to lignocellulosic inhibitors through evolutionary engineering of recombinant Saccharomyces cerevisiae harbouring xylose isomerase. BMC Biotechnol. 14, 41 10.1186/1472-6750-14-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Liao JC, 2011. An evolutionary strategy for isobutanol production strain development in Escherichia coli. Metab. Eng 13, 674–681. 10.1016/j.ymben.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, 2010. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos. Trans. R. Soc. Lond. B Biol. Sci 365, 1255–1263. 10.1098/rstb.2009.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderegger M, Sauer U, 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol 69, 1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske K, Aguilar-Rodriguez J, Sniegowski P, Wagner A, 2018. High mutation rates limit evolutionary adaptation in Escherichia coli. PloS Genet. 14, e1007324 10.1371/journal.pgen.1007324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella RG, Wiechert J, Noack S, Frunzke J, 2019. Evolutionary engineering of Corynebacterium glutamicum. Biotechnol. J e1800444 10.1002/biot.201800444 [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Hokamp K, Last MS, Dorman CJ, 2009. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 5, e1000671 10.1371/journal.pgen.1000671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucko T, Zirngibl K, Pereira F, Kafkia E, Mohamed ET, Rettel M, Stein F, Feist AM, Jouhten P, Patil KR, Forster J, 2018. Laboratory evolution reveals regulatory and metabolic trade-offs of glycerol utilization in Saccharomyces cerevisiae. Metab. Eng 47, 73–82. 10.1016/j.ymben.2018.03.006 [DOI] [PubMed] [Google Scholar]
- Summers ZM, Ueki T, Ismail W, Haveman SA, Lovley DR, 2012. Laboratory evolution of Geobacter sulfurreducens for enhanced growth on lactate via a single-base-pair substitution in a transcriptional regulator. ISME J. 6, 975–983. 10.1038/ismej.2011.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Boross G, Kalapis D, Kovács K, Fekete G, Farkas Z, Lázár V, Hrtyan M, Kemmeren P, Groot Koerkamp MJA, Rutkai E, Holstege FCP, Papp B, Pál C, 2014. The genomic landscape of compensatory evolution. PLoS Biol. 12, e1001935 10.1371/journal.pbio.1001935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos B, Fritzemeier J, Csorgo B, Lazar V, Lu X, Fekete G, Balint B, Herczeg R, Nagy I, Notebaart RA, Lercher MJ, Pal C, Papp B, 2016. Adaptive evolution of complex innovations through stepwise metabolic niche expansion. Nat. Commun 7, 11607 10.1038/ncomms11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS, 2012. The molecular diversity of adaptive convergence. Science 335, 457–461. 10.1126/science.1212986 [DOI] [PubMed] [Google Scholar]
- Thyer R, Shroff R, Klein DR, d’Oelsnitz S, Cotham VC, Byrom M, Brodbelt JS, Ellington AD, 2018. Custom selenoprotein production enabled by laboratory evolution of recoded bacterial strains. Nat. Biotechnol 36, 624–631. 10.1038/nbt.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilloy V, Ortiz-Julien A, Dequin S, 2014. Reduction of ethanol yield and improvement of glycerol formation by adaptive evolution of the wine yeast Saccharomyces cerevisiae under hyperosmotic conditions. Appl. Environ. Microbiol 80, 2623–2632. 10.1128/AEM.03710-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama K, Toya Y, Horinouchi T, Furusawa C, Matsuda F, Shimizu H, 2018. Application of adaptive laboratory evolution to overcome a flux limitation in an Escherichia coli production strain. Biotechnol. Bioeng 10.1002/bit.26568 [DOI] [PubMed] [Google Scholar]
- Toprak E, Veres A, Yildiz S, Pedraza JM, Chait R, Paulsson J, Kishony R, 2013. Building a morbidostat: an automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nat. Protoc 8, 555–567. 10.1038/nprot.nprot.2013.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrilla J, Licona-Cassani C, Marcellin E, Gosset G, Nielsen LK, Martinez A, 2012. Engineering and adaptive evolution of Escherichia coli for D-lactate fermentation reveals GatC as a xylose transporter. Metab. Eng 14, 469–476. 10.1016/j.ymben.2012.07.007 [DOI] [PubMed] [Google Scholar]
- van Rossum HM, Kozak BU, Niemeijer MS, Dykstra JC, Luttik MAH, Daran J-MG, van Maris AJA, Pronk JT, 2016. Requirements for Carnitine Shuttle-Mediated Translocation of Mitochondrial Acetyl Moieties to the Yeast Cytosol. MBio 7 10.1128/mBio.00520-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vleet JH, Jeffries TW, 2009. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotechnol 20, 300–306. 10.1016/j.copbio.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Vasi F, Travisano M, Lenski RE, 1994. Long-Term Experimental Evolution in Escherichia coli. II. Changes in Life-History Traits During Adaptation to a Seasonal Environment. Am. Nat 144, 432–456. 10.1086/285685 [DOI] [Google Scholar]
- Vilela L. de F., de Araujo VPG, Paredes R. de S., Bon E.P. da S., Torres FAG, Neves BC, Eleutherio ECA, 2015. Enhanced xylose fermentation and ethanol production by engineered Saccharomyces cerevisiae strain. AMB Express 5, 16 10.1186/s13568-015-0102-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kamp A, Klamt S, 2017. Growth-coupled overproduction is feasible for almost all metabolites in five major production organisms. Nat. Commun 8, 15956 10.1038/ncomms15956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace-Salinas V, Gorwa-Grauslund MF, 2013. Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol. Biofuels 6, 151 10.1186/1754-6834-6-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Spira B, Zhou Z, Feng L, Maharjan RP, Li X, Li F, McKenzie C, Reeves PR, Ferenci T, 2010. Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol. Evol 2, 478–487. 10.1093/gbe/evq035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun X, Yuan Q, 2018. Strategies for enhancing microbial tolerance to inhibitors for biofuel production: A review. Bioresour. Technol 258, 302–309. 10.1016/j.biortech.2018.03.064 [DOI] [PubMed] [Google Scholar]
- Wang X, Khushk I, Xiao Y, Gao Q, Bao J, 2018. Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl. Microbiol. Biotechnol 102, 377–388. 10.1007/s00253-017-8627-4 [DOI] [PubMed] [Google Scholar]
- Wang Y, Tian T, Zhao J, Wang J, Yan T, Xu L, Liu Z, Garza E, Iverson A, Manow R, Finan C, Zhou S, 2012. Homofermentative production of D-lactic acid from sucrose by a metabolically engineered Escherichia coli. Biotechnol. Lett 34, 2069–2075. 10.1007/s10529-012-1003-7 [DOI] [PubMed] [Google Scholar]
- Wannier TM, Kunjapur AM, Rice DP, McDonald MJ, Desai MM, Church GM, 2018. Adaptive evolution of genomically recoded Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 115, 3090–3095. 10.1073/pnas.1715530115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikert C, Sauer U, Bailey JE, 1997. Use of a glycerol-limited, long-term chemostat for isolation of Escherichia coli mutants with improved physiological properties. Microbiology 143 (Pt 5), 1567–1574. 10.1099/00221287-143-5-1567 [DOI] [PubMed] [Google Scholar]
- Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F, 2011. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 7, e1002202 10.1371/journal.pgen.1002202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal LL, Lau J, Negro Z, Moreno IJ, Ismail Mohammed W, Lee H, Tang H, Finkel SE, Kram KE, 2018. Adaptation of Escherichia coli to long-term batch culture in various rich media. Res. Microbiol 169, 145–156. 10.1016/j.resmic.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Williams TC, Pretorius IS, Paulsen IT, 2016. Synthetic Evolution of Metabolic Productivity Using Biosensors. Trends Biotechnol. 34, 371–381. 10.1016/j.tibtech.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Winkler JD, Garcia C, Olson M, Callaway E, Kao KC, 2014. Evolved osmotolerant Escherichia coli mutants frequently exhibit defective N-acetylglucosamine catabolism and point mutations in cell shape-regulating protein MreB. Appl. Environ. Microbiol 80, 3729–3740. 10.1128/AEM.00499-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler JD, Kao KC, 2014. Recent advances in the evolutionary engineering of industrial biocatalysts. Genomics 104, 406–411. 10.1016/j.ygeno.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Winkler J, Kao KC, 2012. Harnessing recombination to speed adaptive evolution in Escherichia coli. Metab. Eng 14, 487–495. 10.1016/j.ymben.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Wiser MJ, Lenski RE, 2015. A Comparison of Methods to Measure Fitness in Escherichia coli. PLoS One 10, e0126210 10.1371/journal.pone.0126210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink HW, Toirkens MJ, del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJA, 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Appl. Environ. Microbiol 73, 4881–4891. 10.1128/AEM.00177-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong BG, Mancuso CP, Kiriakov S, Bashor CJ, 2018. A generalizable experimental framework for automated cell growth and laboratory evolution. BioRxiv. [Google Scholar]
- Wong WW, Liao JC, 2009. Microbial maximal specific growth rate as a square-root function of biomass yield and two kinetic parameters. Metab. Eng 11, 409–414. 10.1016/j.ymben.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Wright J, Bellissimi E, de Hulster E, Wagner A, Pronk JT, van Maris AJA, 2011. Batch and continuous culture-based selection strategies for acetic acid tolerance in xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res. 11, 299–306. 10.1111/j.1567-1364.2011.00719.x [DOI] [PubMed] [Google Scholar]
- Xu C, Sun T, Li S, Chen L, Zhang W, 2018. Adaptive laboratory evolution of cadmium tolerance in Synechocystis sp. PCC 6803. Biotechnol. Biofuels 11, 205 10.1186/s13068-018-1205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S, 2011. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol 7, 445–452. 10.1038/nchembio.580 [DOI] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O, 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. U. S. A 109, 21010–21015. 10.1073/pnas.1211150109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhao Q, Miao X, Shi J, 2013. Enhancement of lipid production in low-starch mutants Chlamydomonas reinhardtii by adaptive laboratory evolution. Bioresour. Technol 147, 499–507. 10.1016/j.biortech.2013.08.069 [DOI] [PubMed] [Google Scholar]
- Zelle RM, Harrison JC, Pronk JT, van Maris AJA, 2011. Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains. Appl. Environ. Microbiol 77, 732–738. 10.1128/AEM.02132-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J, Shen M, Hu M, Song H, Yuan Y, 2014. Enhanced expression of genes involved in initial xylose metabolism and the oxidative pentose phosphate pathway in the improved xylose-utilizing Saccharomyces cerevisiae through evolutionary engineering. J. Ind. Microbiol. Biotechnol 41, 27–39. 10.1007/s10295-013-1350-y [DOI] [PubMed] [Google Scholar]
- Zhang W, Cheng Y, Li Y, Du G, Xie G, Zou H, Zhou J, Chen J, 2018. Adaptive Evolution Relieves Nitrogen Catabolite Repression and Decreases Urea Accumulation in Cultures of the Chinese Rice Wine Yeast Strain Saccharomyces cerevisiae XZ-11. J. Agric. Food Chem 66, 9061–9069. 10.1021/acs.jafc.8b01313 [DOI] [PubMed] [Google Scholar]
- Zhang X, El-Hajj ZW, Newman E, 2010. Deficiency in L-serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J. Bacteriol 192, 5515–5525. 10.1128/JB.00748-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jantama K, Moore JC, Shanmugam KT, Ingram LO, 2007. Production of L - alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol 77, 355–366. 10.1007/s00253-007-1170-y [DOI] [PubMed] [Google Scholar]
- Zhang X, Newman E, 2008. Deficiency in l-serine deaminase results in abnormal growth and cell division of Escherichia coli K-12. Mol. Microbiol 69, 870–881. 10.1111/j.1365-2958.2008.06315.x [DOI] [PubMed] [Google Scholar]
- Zhao J, Xu L, Wang Y, Zhao X, Wang J, Garza E, Manow R, Zhou S, 2013. Homofermentative production of optically pure L-lactic acid from xylose by genetically engineered Escherichia coli B. Microb. Cell Fact 12, 57 10.1186/1475-2859-12-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cheng J-S, Wang BL, Fink GR, Stephanopoulos G, 2012. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng 14, 611–622. 10.1016/j.ymben.2012.07.011 [DOI] [PubMed] [Google Scholar]
- Zhou S, Shanmugam KT, Ingram LO, 2003. Functional replacement of the Escherichia coli D-(−)-lactate dehydrogenase gene (ldhA) with the L-(+)-lactate dehydrogenase gene (ldhL) from Pediococcus acidilactici. Appl. Environ. Microbiol 69, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yomano LP, Shanmugam KT, Ingram LO, 2005. Fermentation of 10% (w/v) sugar to D: (−)-lactate by engineered Escherichia coli B. Biotechnol. Lett 27, 1891–1896. 10.1007/s10529-005-3899-7 [DOI] [PubMed] [Google Scholar]
- Zhu L, Li Y, Cai Z, 2015. Development of a stress-induced mutagenesis module for autonomous adaptive evolution of Escherichia coli to improve its stress tolerance. Biotechnol. Biofuels 8, 93 10.1186/s13068-015-0276-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorraquino-Salvo V, Quinones-Soto S, Kim M, Rai N, Tagkopoulos I, 2014. Deciphering the genetic and transcriptional basis of cross-stress responses in Escherichia coli under complex evolutionary scenarios. bioRxiv. 10.1101/010595 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 - Categorization of species, timescales of evolutions studies, and experimental culturing setups used in ALE studies.
Supplemental Table 1 - A collection and categorization of ALE studies relevant to industrial biotechnology. Excel file .xlsx