Abstract
Sulfide:quinone oxidoreductase (SQR) is a monotopic membrane flavoprotein present in all domains of life, with multiple roles including sulfide detoxification, homeostasis and energy generation by providing electrons to respiratory or photosynthetic electron transport chains. A type III SQR from the hyperthermophilic archeon Caldivirga maquilingensis has been previously characterized, and its C-terminal amphipathic helices were demonstrated to be responsible for membrane binding. Here, the oligomeric state of this protein was experimentally evaluated by size exclusion chromatography, native gels and crosslinking, and found to be a monomer-dimer-trimer equilibrium. Remarkably, mutant and truncated variants unable to bind to the membrane are able to maintain their oligomeric association. Thus, unlike other related monotopic membrane proteins, the region involved in membrane binding does not influence oligomerization. Furthermore, by studying heterodimers between the WT and mutants, it was concluded that membrane binding requires an oligomer with at least two copies of the protein with intact C-terminal amphipathic helices. A structural homology model of the C. maquilingensis SQR was used to define the flavin- and quinone-binding sites. CmGly12, CmGly16, CmAla77 and CmPro44 were determined to be important for flavin binding. Unexpectedly, CmGly299 is only important for quinone reduction despite its proximity to bound FAD. CmPhe337 and CmPhe362 are also important for quinone binding apparently by direct interaction with the quinone ring, whereas CmLys359, postulated to hydrogen bond to the quinone, seems to have a more structural role. The results presented differentiate the Type III CmSQR from some of its counterparts classified as Type I, II and III.
Keywords: sulfide:quinone, oxidoreductase, quinone reductase, oligomeric state, flavoprotein, Caldivirga maquilingensis
CLASSIFICATION: Bioenergetics, Biochemistry, Biological Sciences
1. Introduction
Sulfide (S2-, HS− and H2S) is found in marine and soil environments and is endogenously produced by eukaryotic and prokaryotic cells as a product of cysteine catabolism. A key enzyme in the maintenance of sulfide homeostasis and bioenergetics is sulfide:quinone oxidoreductase (SQR), which is present in many bacteria, archaea and in the mitochondria of eukaryotic cells [1]. SQR catalyzes the two-electron oxidation of sulfide to elemental sulfur and reduces quinone in the membrane. The sulfur is released either as a highly insoluble octameric ring, S8, or as short chains of polysulfide (HS-(Sn)-SH), which result from the reaction of elemental sulfur with H2S. The resulting sulfur is stored in cytoplasmic or periplasmic globules [2, 3].
SQRs belong to the “two dinucleotide binding domains flavoprotein” (tDBDF) superfamily, characterized by the presence of two Rossmann fold domains known to stabilize the adenosine moieties of dinucleotides (e.g. FAD and NADH) [4]. SQRs are typically oligomeric flavoproteins with multiple copies of a single subunit of molecular mass of about 50 kDa. SQRs are associated with the prokaryotic cytoplasmic or periplasmic membrane or the inner mitochondrial membrane, and have been characterized from Bacteria [5–10], Archaea [11, 12] and Eukarya [13–16]. Dimeric and trimeric forms of SQRs have been observed in X-ray structures, postulating the C-terminal amphipathic helices as the domain responsible for oligomerization [11, 17–19], similar to other monotopic tDBDFs such as type 2 NADH dehydrogenases (NDH-2s) [20–22].
SQRs are classified into six types based on sequence and structural alignments [23]. The amino acid residues required for the binding of the FAD cofactor and substrates are distinguished for each group as part of the classification criteria. In types I and V, for which crystal structures are available, a conserved cysteine is involved in FAD binding [11, 24, 25]. Type III SQRs, like Caldivirga maquilingensis SQR (CmSQR), lack the flavin-binding cysteine, but have a conserved tyrosine or tryptophan in its place, and are structurally more related to flavocytochrome c sulfide dehydrogenases (FCSDs) than to other SQRs [23].
Previous studies demonstrated that the C-terminal domain of CmSQR is important for membrane binding [12]. Replacing four hydrophobic amino acids in the last amphipathic helix is enough to eliminate membrane binding of the protein. This soluble quadruple-mutant is inactive due to the change in a single leucine residue, CmLeu379, that affects quinone reduction. Recently, several amino acids near the quinone polar head have been shown to be important for quinone binding and catalysis in the homologous Acidithiobacillus ferrooxidans SQR (AfSQR) [26], for which there is an X-ray structure [25]. No structure is available for CmSQR, but CmLeu379 aligns with AfPhe410 which is at the quinone binding site.
In the present work, we investigated whether the C-terminal amphipathic helices in CmSQR are critical for oligomerization and whether protein oligomerization is necessary for membrane binding. Additionally, the roles of several conserved residues in the redox reactions were investigated to better define the flavin- and ubiquinone-binding sites.
2. Materials and Methods
2.1. Sequence-structure analysis.
Genes encoding SQRs were retrieved from the National Center for Biotechnology Information (NCBI) and Department of Energy Joint Genome Initiative (JGI) databases. The sequences were aligned using MUSCLE [27] and conserved residues were identified with Jalview [28]. To predict the 3D homology model of C. maquilingensis SQR, the online server for protein fold recognition Phyre2 (Protein Homology/Analogy Recognition Engine) was used [29]. The location of residues in the structure was analyzed using VMD [30] and the software Ligplot+ [31] was used for helping visualization and representation of interactions between ligands and surrounding residues.
2.2. Construction of expression plasmids and site-directed mutagenesis.
The SQR from Caldivirga maquilingensis IC-167 [32] strain was previously cloned into pET22b (Apr, Novagen) [12]. A Quik Change site-directed mutagenesis kit (Stratagene) was used to construct the different point mutations, and the final plasmids were transformed into E. coli C43 (DE3) strain (Avidis, France) containing the pRARE plasmid (Cmr, Novagen).
2.3. Cell growth, enzyme expression and purification.
Cell growth and protein purification were performed as previously described [12]. E. coli C43 cells containing the expression plasmids were grown in LB medium with 100 μg/ml ampicillin and 20 μg/ml chloramphenicol at 37 °C, and expression was induced by addition of 1 mM IPTG (isopropyl-D-thiogalactoside) when cells reached an OD600 ~ 0.7. Purification was carried out at 4 °C. Cells were harvested and resuspended in buffer A (50 mM sodium phosphate, pH 7.5, 300 mM NaCl) with 5 mM MgSO4, DNase I and a protease inhibitor cocktail (Sigma). Then, cells were disrupted by passing three times through a microfluidizer at a pressure of 80,000 psi. The cell extract was centrifuged at 14,000 ×g for 10 min to remove unbroken cells. The supernatants were collected and directly added to 5 ml Ni-NTA resin (Qiagen) pre-equilibrated with buffer A plus 10 mM imidazole. The protein bound to the resin was washed with buffer A plus 50 mM imidazole and then eluted with buffer A with and 200 mM imidazole. Fractions were concentrated by filtration, and the imidazole was removed by dialysis against buffer A. Alternatively, membranes were obtained after centrifugation of disrupted cells at 230,000 ×g for 4 h. Pellets were resuspended in buffer A plus the protease inhibitor cocktail, and then solubilized by the addition of a stock solution of 20% DDM (dodecyl-β-D-maltoside) dropwise to a final concentration of 1%. The suspension was incubated at 4 °C for 2 h with mild agitation and then cleared by centrifugation at 230,000 ×g for 1 h. The supernatant was added to Ni-NTA resin (Qiagen) and membrane bound proteins were purified following a similar protocol, with 0.05% DDM added to all buffers. The purified protein samples were stored frozen at −80 °C after the addition of glycerol to a final concentration of 10%.
2.4. Native PAGE.
Non-denaturing electrophoresis was performed in 4–20% precast Tris-Glycine gels (Nusep) in the presence of Tris-Glycine native running buffer supplemented with 0.01% DDM. For Blue-Native PAGE, 0.02% of Coomassie Brilliant Blue G was added to the cathode buffer until samples ran for 1 cm into the gel, following this the coomassie-stained buffer was removed and replaced with clear Tris-Glycine native running buffer. The chamber was placed in a mix of ice-water at 4°C, to avoid heating of the samples, and gels were run for 4 h at 150 mV [33].
2.5. Crosslinking assays.
For crosslinking assays, 20 μM of purified protein were incubated at room temperature for 5 minutes with 1.25 mM of glutaraldehyde (Sigma) in 10 mM HEPES buffer, pH 8.2, containing 0.05% DDM. The reaction was quenched with 100 mM Tris-HCl buffer, pH 7.4, and the crosslinked protein samples were analyzed by SDS-PAGE. Crosslinking experiments with 3,3′-dithiobis(sulfosuccinimidyl propionate) (DTSSP), were carried out according to manufacturer’s directions at room temperature (Pierce, Thermo Fisher Scientific). A 50-fold molar excess of DTSSP (prepared in DMSO) was added to the protein in buffer A. Alternatively, 4 μM of protein were incubated with 1 ml of 10 mg/ml 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane vesicles (see preparation below), in the presence or absence of β-mercaptoethanol and/or the crosslinker.
2.6. Membrane vesicles preparation.
Membrane vesicles were prepared by resuspending a dried film of POPC in 50 mM sodium phosphate pH 7.5, to a final concentration of 10 mg/ml POPC in buffer A. Resuspension was followed by sonication and a series of freeze-thaw cycles, as established previously [34]. No extrusion steps were performed since no specific vesicle size was required.
2.7. Western blot analysis.
After SDS-PAGE, protein bands were transferred to a PVDF membrane and the WesternBreeze Chromogenic Western Blot Immunodetection Kit (Life Technologies) was used for detection. His-tagged proteins were detected using a monoclonal anti-polyhistidine–alkaline phosphatase antibody produced in mouse (Sigma). FLAG-tagged proteins were detected by chemoluminescence using a monoclonal ANTI-FLAG M2-peroxidase (HRP) antibody produced in mouse (Sigma), followed by development with a mix of 1.25 mM luminol, 22.5 pM coumaric acid and 2.6 mM of H2O2 [35].
2.8. Gel-filtration chromatography.
Gel-filtration analysis was carried out using a Superdex 200 10/300 GL column (GE Healthcare) previously equilibrated at 4 °C with buffer A plus 5 % glycerol and 0.05% DDM. The column was calibrated using BioRad molecular standards and elution was monitored by following absorption at 280 nm and 450 nm (FAD peak) on an ÄKTA FPLC system (GE healthcare).
2.9. Enzyme activity assays.
SQR activity was measured at 60° C. The 200 μL reaction mixture contained buffer A with 0.05% DDM, 100 μM decylubiquinone (Sigma), and 5 μg (0.55 μM) of the purified enzyme. The reaction was started with the addition of 250 μM sodium sulfide, prepared freshly with N2-flushed buffer A. The reaction progress was monitored for 3 min by the decrease in absorption of decylubiquinone at 275 nm [36]. An extinction coefficient of 12.4 cm−1 mM−1 was used to determine the extent of reduction of the quinone [37].
2.10. Determination of kinetic constants.
Kinetic parameters were determined using nonlinear least square analysis (Origin8.0) of the data fitted to the Michaelis–Menten rate equation (v = Vmax (S) / (Km + S), where v is the velocity, Vmax is the maximum velocity, S is the substrate concentration and Km is the Michaelis-Menten. The enzyme rates are expressed as a turnover number (kcat) based on μmol quinone reduced s–1 μmol FAD–1.
2.11. Other analytical methods.
Protein concentration was determined using the BCA protein assay (Thermo Scientific, Pierce Protein Research Products). The flavin content of the isolated SQR protein was determined in a dual-wavelength spectrophotometer (Agilent Technologies spectrophotometer 8453), using an extinction coefficient of 11.3 cm–1 mM–1 for the oxidized flavin [38] after extraction from the protein by treatment of the sample with 5 % trichloroacetic acid [39]. The redox state of the flavin in the intact protein was also monitored by the fluorescence excitation spectrum using a fluorescence spectrophotometer (Cary Eclipse, Agilent Technologies) (emission wavelength of 520 nm), which was recorded aerobically at room temperature before and after the addition of different concentrations of sulfide (reductant) and quinone (oxidant). It is worth noticing that C. maquilingensis SQR oxidation by oxygen is undetectable at room temperature.
3. Results
3.1. The homology model of CmSQR resembles FCSD, NDH-2 and other SQRs.
A CmSQR homology model was constructed using the web-based bioinformatics server Phyre2. This allows the prediction of protein structure based on identification of homologous sequences, generation of a multiple-sequence alignment and secondary structure predictions, which are then combined into a query hidden Markov model (HMM) that is scanned against a fold library of HMMs of known structure. The highest scoring alignments are used to generate three-dimensional backbone models, and the final three-dimensional structural model is generated after modeling of loops and fitting of side-chains. 97% of CmSQR was modelled at >90% confidence. Six templates were finally selected to model the protein based on heuristics to maximize confidence, percentage identity and alignment coverage. These were the flavin binding subunit of FCSDs from Thioalkalivibrio paradoxus (PDB ID: 5NLT) and A. vinosum (PDB ID: 1FCD), the SQRs from A. ferrooxidans (PDB ID: 3T31) and A. aeolicus (PDB ID: 3HYW), and Saccharomyces cerevisiae (PDB ID: 4G73) and Plasmodium falciparum (PDB ID: 5JWA) NDH-2s.
3.2. Oligomerization of CmSQR does not involve the membrane binding domain.
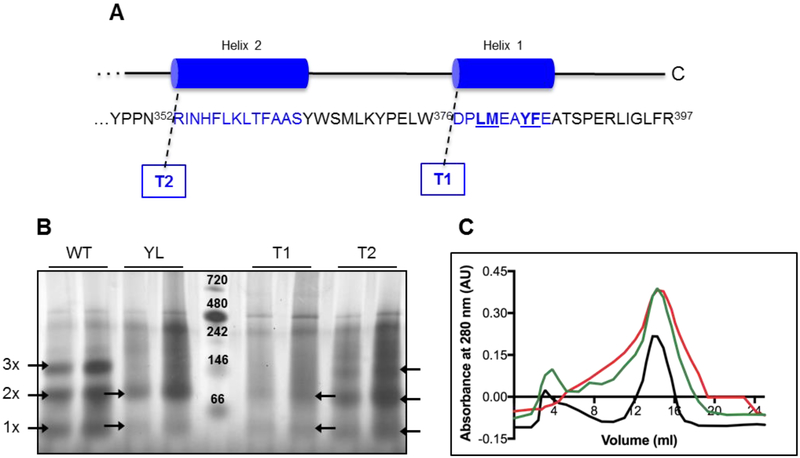
In order to determine the oligomeric state of CmSQR, and the importance of the C-terminal region for oligomerization, a previously developed set of mutants was used [12]. These are soluble variants, unable to bind to the membrane, lacking either four hydrophobic residues from the last C-terminal helix (quadruple-mutant or YL), the complete last C-terminal helix (truncated 1 or T1) or the last two C-terminal helices (truncated 2 or T2) (Figure 1A).
Figure 1. Oligomeric state of WT and soluble mutants by Blue Native-PAGE.
(A) Soluble mutants were previously constructed by truncation of one (T1) or both (T2) C-terminal helices, and by point mutations of the four amino acids underlined in the last helix (YL) [12]. (B) 5 (left) and 10 μg (right) of each protein sample were loaded in native buffer plus Coomassie Brilliant Blue G. Arrows indicate bands corresponding to the monomer (1x), dimer (2x) and trimer (3x) complexes (about 46, 88 and 139 kDa, respectively). The bands of the protein ladder (center lane) represent: Apoferritin band 1 (720 kDa), Apoferritin band 2 (480 kDa), β-phycoerythrin (242 kDa), lactate dehydrogenase (146 kDa), bovine serum albumin (66 kDa); from top to bottom. (C) Chromatograms of the purified WT (black), T1 (green) and T2 (red) SQRs showing the profile of elution monitored at 280 nm.
The oligomeric state of the WT and soluble mutants were evaluated in blue native polyacrylamide gels (BN-PAGE) [40]. The wild-type (WT) and Truncated 2 (T2) proteins, showed bands corresponding to monomer, dimer and trimer states. The Truncated 1 (T1) and quadruple mutant (YL) showed bands corresponding to monomer and dimer (Figure 1A and 1B). Additional bands between 242 and 480 kDa are observed in all the protein samples, which suggest that all the SQR variants are present in multiple oligomeric states. The WT and soluble mutants were also examined by size exclusion chromatography (SEC). In all cases, the major peak corresponds to a dimer (83–89 kDa) (Figure 1C).
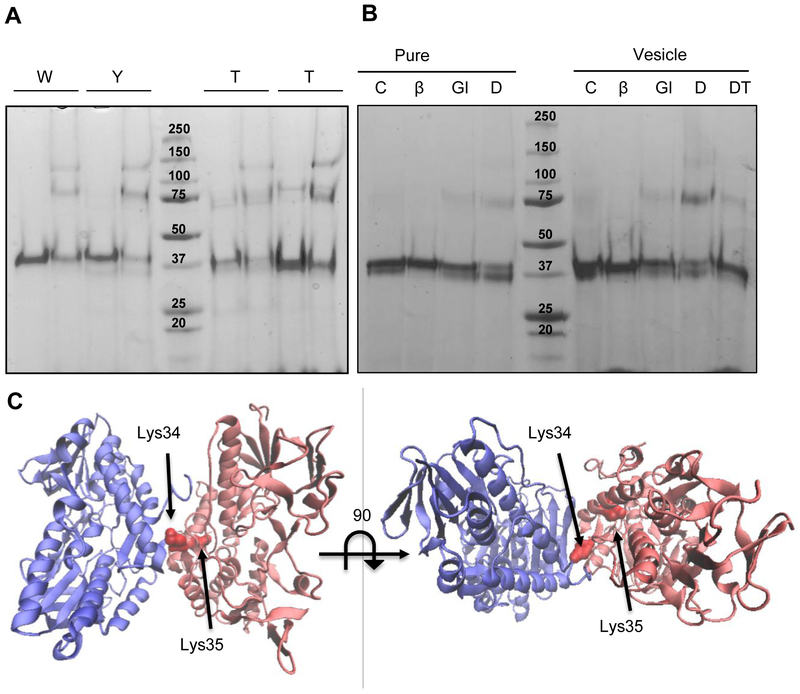
In addition, the oligomeric states of the WT and soluble variants were examined by chemical crosslinking using glutaraldehyde and DTSSP followed by SDS-PAGE. Incubation with glutaraldehyde resulted in WT, YL and T2 variants showed bands corresponding to monomer, dimer and trimer (Figure 2A). Crosslinking with DTSSP, which forms crosslinks between primary amines located up to 12 Å apart (Figure 2C) showed similar band patterns (Figure 2B). Crosslinking was also examined in the presence of liposomes to determine if hydrophobic regions involved in membrane binding also contribute to protein-protein interactions in the oligomers as suggested for the A. ferrooxidans SQR [25]. A band corresponding to the CmSQR dimer is clearly observed after crosslinking in the presence of liposomes (Figure 2B), suggesting that the interfaces for oligomerization and for membrane binding are independent and distinct.
Figure 2. Oligomeric state of WT and soluble mutants by crosslinking assay.
(A) 4 μM (0.2 mg/ml) of WT and mutated protein was run in an SDS-PAGE after a 5 min incubation in the absence (−) or presence (+) of 1.25 mM glutaraldehyde. (B) 4 μM (0.2 mg/ml) of WT protein were incubated in the absence (C: control) or presence of different crosslinkers (Glu: glutaraldehyde; DT: DTSSP) and β-mercaptoethanol (DTβ: DTSSP+ β-mercaptoethanol), without or with lipid vesicles as indicated. Controls were run without and with β-mercaptoethanol (βm). (C) C. maquilingensis SQR model (red) overlapped with the structure of the A. ferrooxidans SQR dimer (blue) indicating the putative lysines involved in DTSSP crosslinking.
Taken together, BN-PAGE, SEC and crosslinking results show that CmSQR exists in solution as an equilibrium primarily consisting of monomers, dimers and trimers, and that the domains involved in forming the oligomers are distinct from the C-terminal membrane-binding domain.
3.3. CmSQR oligomerization is necessary for membrane binding.
It has been proposed that variations between the C-terminal domain of monotopic proteins result in differences in the way these proteins bind to the membrane. For example, for A. aeolicus SQR, it has been suggested that membrane insertion stabilizes oligomerization of the trimer and vice versa [19]. The yeast NDH-2 (Ndi1), a tDBD flavoprotein, forms a larger hydrophobic patch when it dimerizes which facilitates membrane binding [20], while the bacterial version shows two separate membrane binding regions that do not merge to a single hydrophobic patch in the dimer [22].
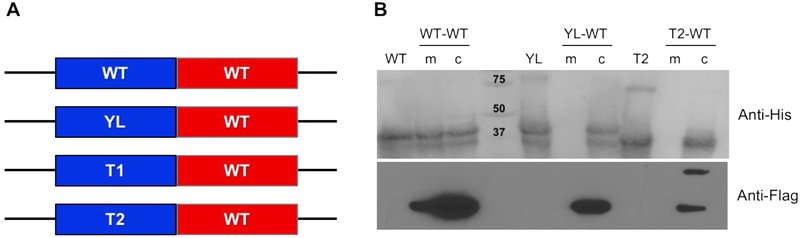
The relationship between the state of oligomerization of the CmSQR and membrane binding was investigated by examining the behavior of hetero-oligomers of differentially tagged CmSQR variants. One gene copy encoding a His-tagged WT protein was co-expressed with a second gene copy encoding a FLAG-tagged WT or FLAG-tagged soluble mutant variant, unable to bind to the membrane (Figure 3A). Following expression, cells were isolated and the membrane and cytoplasmic fractions were separated, and affinity chromatography was used to isolate the CmSQR in each fraction. All samples were processed using a Ni-NTA column, isolating any oligomer containing the His-tagged WT protein. The purified protein was analyzed by SDS-PAGE and Western blots using anti-His antibodies or anti-FLAG antibodies (Figure 3B). Preparations were also made of pure His-tagged WT, His-tagged YL variant and His-tagged T2 variant.
Figure 3. Oligomeric state of WT and soluble mutants by double-tag protein approach.
(A) Constructs containing His-tagged (blue) and Flag-tagged (red) SQR homo- and heterodimers. (B) Ni-NTA purified samples were run in SDS-PAGE, transferred to PVDF membranes and developed by anti-His-tag and anti-Flag-tag antibodies. WT, YL and T2 lanes correspond to His-tagged homodimers (synthesized using single-copy constructs). WT-WT, YL-WT and T2-WT lanes correspond to His-tagged and FLAG-tagged homo- and heterodimers, purified from membrane (m) or cytosolic (c) fractions.
Previously, it has been shown that the WT CmSQR can be found in both the cytoplasmic and membrane fraction, whereas the YL and T2 CmSQR variants are only found as soluble proteins in the cytoplasmic fraction. These results were confirmed (Figure 3B; WT, YL and T2 lanes). Co-expression of WT His-CmSQR and YL FLAG-CmSQR should result in a mixture of oligomeric forms of CmSQR. After Ni-NTA affinity chromatography each isolated oligomer must contain at least one copy of His-CmSQR. This will include the homodimer WT-WT and the heterodimer WT-YL. Western blotting (Figure 3B) shows both WT His-CmSQR and YL FLAG-CmSQR in the cytoplasmic fraction. However, the membrane fraction only contains WT His-CmSQR (Figure 3B). This shows that the WT-YL heterodimer is not present in the membrane. Similar results were obtained T2 CmSQR variant (Figure 3B), showing that the WT-T2 heterodimer does not bind to the membrane. This shows that WT-YL heterodimer and YL-YL homodimer do not bind to the membrane. Since the WT-WT homodimer does bind to the membrane, it is concluded that at least two copies of the WT CmSQR must be present in an oligomer to facilitate membrane binding of CmSQR. Only the oligomeric states of CmSQR are capable of binding to the membrane.
3.4. Identification of residues of CmSQR involved in FAD or quinone binding.
Since there is no crystal structure for a type III SQR, mutational studies were performed based on sequence and structural alignments made using the homology model for CmSQR.
3.4.1. FAD binding site characterization.
Type III SQRs lack the characteristic Cys residue seen for FAD binding in the better-known type I and type V proteins, and a Tyr or Trp residue has been described as taking the place of this Cys [135]. However, in the homology model of CmSQR, the tyrosine thought to correspond to the covalent Cys, CmTyr126, is not close to the flavin cofactor.
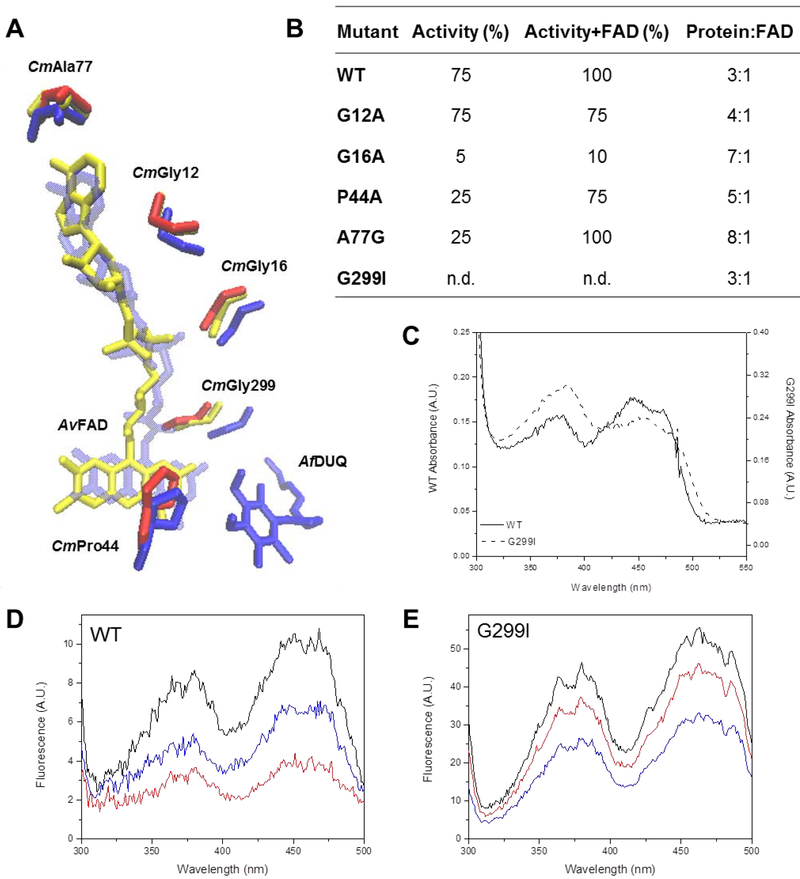
To investigate the FAD binding site in CmSQR, amino acids interacting with the cofactor in aligned structures were first observed using the Ligplot+ software, which allows a simplified view of all different interactions between a protein and its cofactors [31]. Residues that interact with the FAD cofactor and are conserved in A. vinosum FCSD and C. maquilingensis, A. ferrooxidans, A. aeolicus and A. ambivalens SQRs, were identified and mutated in CmSQR. In all five analyzed structures, Gly12, Gly16, Ala77 and Gly299 (C. maquilingensis numbering) can be seen at hydrogen bond distance to the adenine ring, pyrophosphate moiety and isoalloxazine ring, respectively (Figure 4A).
Figure 4. Residues involved in the putative FAD binding site.
(A) Overlapped residues from structures from A. vinosum FCSD (yellow), A. ferrooxidans (blue) are compared to those from the C. maquilingensis SQR model (red). DUQ: decylubiquinone. (B) WT and mutants were assayed for activity in the absence and presence of additional FAD as indicated. The FAD content of each protein was quantified after extraction by TCA treatment. 100% activity corresponds to 0.76 ± 0.03 μM of reduced DUQ per μM protein−1 per sec−1. Data are expressed as average ± SD of at least three independent experiments. Standard deviations for all the activity values were lower than 10 %. (C) FAD UV-Visible spectrum of the G299I mutant compared to WT. (D) WT and (E) G299I fluorescence excitation spectra before (black) and after addition of 200 μM of Na2S (red) and 200 μM of DUQ (blue).
CmGly12 and CmGly16 are close to the phosphate groups connecting the adenosine and the isoalloxazine ring. These were initially mutated to Ala, adding the smallest side chain possible. Gly12Ala only showed a slight drop in activity and FAD content, however addition of external flavin did not restore activity or bound flavin. Gly16Ala resulted in a loss of FAD content to less than half of the WT and a 20-fold drop in activity that could not be recovered by addition of external flavin (Figure 4B). This residue was also substituted for Ile but resulted in a severe reduction in yield, suggesting an even stronger effect in protein stability and folding due to the larger side chain.
CmAla77, which is adjacent to the adenine portion of FAD was mutated to Gly, removing the −CH3 side chain. Ala77Gly resulted in the loss of over half the flavin content and a 4-fold reduction in activity compared to the WT protein. FAD content and activity however, were almost fully restored after addition of external flavin (Figure 4B), in accordance with a role in flavin binding.
CmGly299 is predicted to be about 3 Å from the isoalloxazine ring (Figure 4A). Replacing this residue by Ile resulted in complete loss of activity, although the FAD content was unchanged. However, the flavin spectrum for this mutant is red shifted and shows a change in the relative intensity of the beta and gamma FAD absorption peaks (Figure 4C), indicating a perturbation in the cofactor environment [41]. Mutating the equivalent Gly residue in A. ferrooxidans SQR (AfGly322) affects both the rates of FAD and quinone reduction [26]. The redox half-reactions, (a) reduction of FAD by sulfide, and (b) reoxidation of FADH2 by quinone, were examined with the Gly200Ile CmSQR mutant. The FAD cofactor in this mutant was reduced by sulfide, but no reoxidation was observed after quinone addition (Figures 4D and E), indicating the presence of this larger side chain drastically affects the quinone reduction step.
Previous studies identified the equivalent of CmSQR Pro44 in S. cerevisiae Ndi1 and A. ferrooxidans SQR, as being important for FAD and quinone binding, respectively (Figure 4A) [26, 42]. This residue is less than 5 Å from the isoalloxazine and quinone rings in A. ferrooxidans SQR and S. cerevisiae Ndi1. Mutation to Ala (CmPro44Ala) led to 50% loss of the flavin content of CmSQR compared to WT, and 75% lower specific activity, and these were substantially restored after addition of external FAD. CmPro44 was additionally changed to Phe but no recombinant protein was observed upon expression.
In sum, three residues in CmSQR have been shown to be important for FAD binding, CmGly16, CmAla77 and CmPro44, based on loss of bound flavin due to the mutations. Rescue of enzyme activity was observed in the presence of added flavin for the CmAla77Gly and CmPro44Ala mutants, but not for CmGly16Ala.
3.4.2. Quinone binding site characterization..
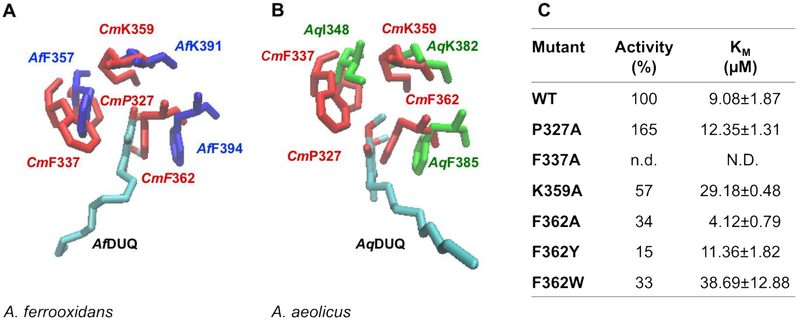
There are no identifiable quinone binding motifs described for these monotopic membrane binding proteins. However two available X-ray structures for type I SQRs contain bound quinone, from A. aeolicus and A. ferrooxidans (PDB IDs: 3HYW and 3T31) [19, 25] and these were used as a guide. In the crystal structures for A. ferrooxidans [25, 26] and A. aeolicus [19], the quinone ring is located between two hydrophobic residues: AfPhe357 and AfPhe394 for A. ferrooxidans SQR, and AqIle348 and AqPhe385 for the A. aeolicus enzyme. The residue corresponding to AfPhe357/AqIle348 position in CmSQR are CmPro327 or CmPhe337, based on sequence or structural alignments, respectively. Mutating CmPro327 to Ala, resulted in a slight increase in Km for the quinone but a surprising improvement in enzyme activity (Figure 5C). On the other hand, when CmPhe337 was mutated to Ala, activity was undetectable.
Figure 5. Residues involved in the putative quinone binding site.
C. maquilingensis SQR structural model (red) overlapped with (A) A. ferrooxidans SQR (Af, blue) and (B) A. aeolicus (Aq, green) SQR crystal structures, shows conserved residues surrounding the quinone substrate. (C) Activity and Km for DUQ of WT and mutants. 100% activity corresponds to 0.76 ± 0.03 μM of reduced DUQ per μM protein−1 per sec−1; n.d.: not detected; N.D.: not determined. Data are expressed as average ± SD of at least three independent experiments. Standard deviations for all the activity values were lower than 10 %.
Based on sequence and structural alignments, CmPhe362 corresponds to AqPhe385 and AfPhe394. In each structure, the aromatic ring is stacked with the quinone ring. When CmPhe362 was replaced by Ala, Tyr or Trp, activity dropped by up to 85% compared to WT and the Km for quinone increased, presumably due to the increase in size of the amino acid placed at this position.
A third conserved residue, CmLys359, also appears to be important for quinone binding, corresponding to AqLys382 and AfLys391. This lysine residue has been postulated to hydrogen bond to the quinone through its backbone amide, and to the FAD by its side chain amino group, yielding a completely inactive enzyme when replaced by Ala in the AfSQR [26]. However, replacement of CmLys359 by Ala, only caused about 50% loss of activity, and a 3-fold increase in Km for the quinone.
In sum, the residues selected in the CmSQR homology model as being involved in quinone binding are in agreement with what has been observed with both the A. ferrooxidans and A. aeolicus SQR structures [24, 25]. This involves the presence of two hydrophobic residues sandwiching the quinone ring (CmPhe362 and CmPhe337) and a nearby conserved Lys (CmLys359).
4. Discussion
4.1. Oligomeric state and membrane attachment in CmSQR.
SQRs have been described previously as dimers or trimers, or as equilibrium mixtures containing monomers and dimers [25] or monomers, dimers and trimers [19]. Dimeric and trimeric X-ray structures are available for type I SQRs [19, 25, 43]. A dimer has been described for a type V SQR from A. ambivalens, but the crystallized form lacks a large portion of its C-terminal [11]. The C-terminal domain is observed at the oligomer interface in A. aeolicus and A. ferrooxidans SQR structures [19, 25]. In the case of the A. aeolicus enzyme, it has been proposed that membrane insertion stabilizes the trimeric form and that the trimer creates an adequate surface for membrane binding. The membrane attachment assures that SQR exclusively reduces hydrophobic quinones and not other soluble cytoplasmic electron acceptors [19]. A. ferrooxidans SQR however, has been suggested to be a monomer in its membrane-bound form, attaching through the same region needed for oligomerization [25]. The recently reported structure of the human SQR has been used to support a proposal that this type II SQR is monomeric with a coplanar membrane-binding surface. [16].
The current work shows that the isolated type III CmSQR is oligomeric in vitro in solution and exists in a monomer-dimer-trimer equilibrium. However, only the oligomeric forms of CmSQR is able to bind to the membrane, since the membrane-binding domain of a single monomeric CmSQR is not sufficient for membrane attachment.
It is useful to compare the SQR structures to other related monotopic flavoproteins from tDBDF superfamily [4] such as NDH-2s [20–22]. The crystal structure and experimental data on S. cerevisiae NDH-2 (Ndi1) show that enzyme dimerization brings together the C-terminal domains into one large membrane-anchoring structure [20, 21]. In contrast, the bacterial NDH-2 dimer has two spatially separated membrane-anchoring regions that are not individually altered by dimerization [22]. In CmSQR oligomerization is not affected by mutations that eliminate membrane binding, clearly demonstrating that the oligomeric protein-protein interface is independent of and distinct from the membrane binding domain. However, formation of at least a dimer is required for membrane attachment since the membrane binding domains from at least two monomers is necessary for attachment to the membrane. Thus, CmSQR is similar to bacterial NDH-2 in that there are separate protein domains for oligomerization and membrane binding, but resembles Ndi1 insofar as the membrane binding regions from both monomers within a dimer combine into a larger hydrophobic patch.
4.2. FAD binding site in CmSQR.
Four amino acids at or near the putative FAD binding site were selected based on conservation between FCSDs and SQRs: CmGly12, CmGly16, CmAla77 and CmGly299. Mutating CmGly16, has a large effect on FAD content. CmAla77 is also important for keeping the cofactor bound to the enzyme and full recovery of activity is observed upon addition of external flavin (Figures 4A and B). Mutating CmGly299, on the other hand, did not alter the flavin content but prevented quinone reduction by the reduced flavin. Surprisingly, the equivalent residue in A. ferrooxidans (AfGly322) has been described to also affect FAD reduction [26]. CmPro44, absent in FCSDs but conserved in SQRs and NDH-2s, also influenced FAD content, reflecting additional similarities between these two families.
4.3. Quinone binding site in CmSQR.
As expected from the structures of the A. aeolicus and A. ferrooxidans SQRs, in CmSQR two hydrophobic residues (Phe337 and Phe362) and a Lys (359) residue appear to be important in determining the steady state kinetics of quinone reductase activity (Figure 5). CmPhe337 is essential for catalysis, whereas mutations in CmPhe362 diminish but do not eliminate activity. This differs to what has been observed for A. ferrooxidans, where both Phe residues sandwiching the quinone are equally important for activity [26].
As seen for the A. ferrooxidans SQR, mutations in CmPhe362 and CmLys359 affected the Km for quinone, consistent with the suggestion that altering these residue changes the dimensions of the quinone binding pocket [26] (Figure 5). Surprisingly, CmLys359Ala remains highly active, unlike mutations to the equivalent AfLys391, which completely inactivate the enzyme possibly due to its proposed strong interactions with FAD [26].
CmLeu379 has been previously shown to be crucial for CmSQR activity and postulated to interact with the quinone ring [12]. As suggested by the homology model, it is more likely this residue be located close to the hydrophobic tail of the quinone.
Summarizing, three residues are suggested to interact with the quinone substrate in CmSQR. CmPhe337 and CmPhe362 likely stabilize the quinone by stacking with the quinone ring, and CmLys359 may contribute to delimiting the quinone binding pocket.
Highlights.
Caldivirga maquilingensis Type III Sulfide:Quinone Oxidoreductase (CmSQR) exists in solution as an equilibrium primarily consisting of monomers, dimers and trimers.
The domains involved in forming the oligomers in CmSQR are distinct from the C-terminal membrane-binding domain.
Membrane binding requires a CmSQR oligomer with at least two copies of the protein with intact C-terminal amphipathic helices.
Acknowledgements
We thank members of the Gennis laboratory for their help and useful discussions. This work was supported by grants from the US National Institutes of Health, GM095600 and HL16101.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shahak Y. a. H., G. (2008) Sulfide Oxidation from Cyanobacteria to Humans: Sulfide–Quinone Oxidoreductase (SQR) in Advances in Photosynthesis and Respiration (Hell R, Dahl S, Knaff D, Leustek T, ed) pp. 319–335, Springer, Heidelberg, Germany. [Google Scholar]
- 2.Guiral M, Tron P, Aubert C, Gloter A, Iobbi-Nivol C & Giudici-Orticoni MT (2005) A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus, J Biol Chem. 280, 42004–15. [DOI] [PubMed] [Google Scholar]
- 3.Griesbeck C, Hauska G and Schütz M (2000) Biological Sulfide Oxidation: Sulfide-Quinone Reductase (SQR), the Primary Reaction in Recent Research Developments in Microbiology (Pandalai SG, ed) pp. 179–203, Research Signpost, Trivandrum, India. [Google Scholar]
- 4.Ojha S, Meng EC & Babbitt PC (2007) Evolution of function in the “two dinucleotide binding domains” flavoproteins, PLoS Comput Biol. 3, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schutz M, Shahak Y, Padan E & Hauska G (1997) Sulfide-quinone reductase from Rhodobacter capsulatus. Purification, cloning, and expression, J Biol Chem. 272, 9890–4. [DOI] [PubMed] [Google Scholar]
- 6.Arieli B, Shahak Y, Taglicht D, Hauska G & Padan E (1994) Purification and characterization of sulfide-quinone reductase, a novel enzyme driving anoxygenic photosynthesis in Oscillatoria limnetica, J Biol Chem. 269, 5705–11. [PubMed] [Google Scholar]
- 7.Shibata H & Kobayashi S (2006) Characterization of a HMT2-like enzyme for sulfide oxidation from Pseudomonas putida, Can J Microbiol. 52, 724–30. [DOI] [PubMed] [Google Scholar]
- 8.Shibata H, Suzuki K & Kobayashi S (2007) Menaquinone reduction by an HMT2-like sulfide dehydrogenase from Bacillus stearothermophilus, Can J Microbiol. 53, 1091–100. [DOI] [PubMed] [Google Scholar]
- 9.Shen J, Peng H, Zhang Y, Trinidad JC & Giedroc DP (2016) Staphylococcus aureus sqr Encodes a Type II Sulfide:Quinone Oxidoreductase and Impacts Reactive Sulfur Speciation in Cells, Biochemistry. 55, 6524–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duzs A, Toth A, Nemeth B, Balogh T, Kos PB & Rakhely G (2018) A novel enzyme of type VI sulfide:quinone oxidoreductases in purple sulfur photosynthetic bacteria, Appl Microbiol Biotechnol. 102, 5133–5147. [DOI] [PubMed] [Google Scholar]
- 11.Brito JA, Sousa FL, Stelter M, Bandeiras TM, Vonrhein C, Teixeira M, Pereira MM & Archer M (2009) Structural and functional insights into sulfide:quinone oxidoreductase, Biochemistry. 48, 5613–22. [DOI] [PubMed] [Google Scholar]
- 12.Lencina AM, Ding Z, Schurig-Briccio LA & Gennis RB (2013) Characterization of the Type III sulfide:quinone oxidoreductase from Caldivirga maquilingensis and its membrane binding, Biochim Biophys Acta. 1827, 266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson MR, Melideo SL & Jorns MS (2012) Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite, Biochemistry. 51, 6804–15. [DOI] [PubMed] [Google Scholar]
- 14.Vande Weghe JG & Ow DW (1999) A fission yeast gene for mitochondrial sulfide oxidation, J Biol Chem. 274, 13250–7. [DOI] [PubMed] [Google Scholar]
- 15.Theissen U & Martin W (2008) Sulfide: quinone oxidoreductase (SQR) from the lugworm Arenicola marina shows cyanide- and thioredoxin-dependent activity, FEBS J. 275, 1131–9. [DOI] [PubMed] [Google Scholar]
- 16.Jackson MR, Loll PJ & Jorns MS (2019) X-Ray Structure of Human Sulfide:Quinone Oxidoreductase: Insights into the Mechanism of Mitochondrial Hydrogen Sulfide Oxidation, Structure. 27, 794–805 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcia M, Ermler U, Peng G & Michel H (2010) A new structure-based classification of sulfide:quinone oxidoreductases, Proteins: Structure, Function, and Bioinformatics. 78, 1073–1083. [DOI] [PubMed] [Google Scholar]
- 18.Marcia M, Langer JD, Parcej D, Vogel V, Peng G & Michel H (2010) Characterizing a monotopic membrane enzyme. Biochemical, enzymatic and crystallization studies on Aquifex aeolicus sulfide:quinone oxidoreductase, Biochimica et Biophysica Acta (BBA) - Biomembranes. 1798, 2114–2123. [DOI] [PubMed] [Google Scholar]
- 19.Marcia M, Ermler U, Peng G & Michel H (2009) The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration, Proc Natl Acad Sci U S A. 106, 9625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Li W, Li J, Wang J, Ge J, Xu D, Liu Y, Wu K, Zeng Q, Wu JW, Tian C, Zhou B & Yang M (2012) Structural insight into the type-II mitochondrial NADH dehydrogenases, Nature. 491, 478–82. [DOI] [PubMed] [Google Scholar]
- 21.Iwata M, Lee Y, Yamashita T, Yagi T, Iwata S, Cameron AD & Maher MJ (2012) The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates, Proc Natl Acad Sci U S A. 109, 15247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heikal A, Nakatani Y, Dunn E, Weimar MR, Day CL, Baker EN, Lott JS, Sazanov LA & Cook GM (2014) Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation, Mol Microbiol. 91, 950–64. [DOI] [PubMed] [Google Scholar]
- 23.Marcia M, Ermler U, Peng G & Michel H (2010) A new structure-based classification of sulfide:quinone oxidoreductases, Proteins. 78, 1073–83. [DOI] [PubMed] [Google Scholar]
- 24.Marcia M, Langer JD, Parcej D, Vogel V, Peng G & Michel H (2010) Characterizing a monotopic membrane enzyme. Biochemical, enzymatic and crystallization studies on Aquifex aeolicus sulfide:quinone oxidoreductase, Biochim Biophys Acta. 1798, 2114–23. [DOI] [PubMed] [Google Scholar]
- 25.Cherney MM, Zhang Y, Solomonson M, Weiner JH & James MN (2010) Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification, J Mol Biol. 398, 292–305. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Qadri A & Weiner JH (2016) The quinone-binding site of Acidithiobacillus ferrooxidans sulfide: quinone oxidoreductase controls both sulfide oxidation and quinone reduction, Biochem Cell Biol. 94, 159–66. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res. 32, 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterhouse AM, Procter JB, Martin DM, Clamp M & Barton GJ (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench, Bioinformatics. 25, 1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis, Nat Protoc. 10, 845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey W, Dalke A & Schulten K (1996) VMD: visual molecular dynamics, J Mol Graph. 14, 33–8, 27–8. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA & Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery, J Chem Inf Model. 51, 2778–86. [DOI] [PubMed] [Google Scholar]
- 32.Itoh T, Suzuki K, Sanchez PC & Nakase T (1999) Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines, International journal of systematic bacteriology. 49 Pt 3, 1157–63. [DOI] [PubMed] [Google Scholar]
- 33.Ma J & Xia D (2008) The use of blue native PAGE in the evaluation of membrane protein aggregation states for crystallization, J Appl Crystallogr. 41, 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingolfsson HI, Sanford RL, Kapoor R & Andersen OS (2010) Gramicidin-based fluorescence assay; for determining small molecules potential for modifying lipid bilayer properties, J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stott RA (1998) Enhanced chemiluminescence immunoassay, Methods Mol Biol. 80, 197–205. [DOI] [PubMed] [Google Scholar]
- 36.Shahak Y, Klughammer C, Schreiber U, Padan E, Herrman I & Hauska G (1994) Sulfide-quinone and sulfide-cytochrome reduction in Rhodobacter capsulatus, Photosynth Res. 39, 175–81. [DOI] [PubMed] [Google Scholar]
- 37.Morton RA (1965) Quinones as a Biological Catalysts, Endeavour. 24, 81–6. [DOI] [PubMed] [Google Scholar]
- 38.Weyler W & Salach JI (1985) Purification and properties of mitochondrial monoamine oxidase type A from human placenta, J Biol Chem. 260, 13199–207. [PubMed] [Google Scholar]
- 39.Susin S, Abian J, Sanchez-Baeza F, Peleato ML, Abadia A, Gelpi E & Abadia J (1993) Riboflavin 3′- and 5′-sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris), J Biol Chem. 268, 20958–65. [PubMed] [Google Scholar]
- 40.Wittig I, Braun HP & Schagger H (2006) Blue native PAGE, Nat Protoc. 1, 418–28. [DOI] [PubMed] [Google Scholar]
- 41.Ghisla S & Massey V (1986) New flavins for old: artificial flavins as active site probes of flavoproteins, Biochem J. 239, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Yamashita T, Nakamaru-Ogiso E, Hashimoto T, Murai M, Igarashi J, Miyoshi H, Mori N, Matsuno-Yagi A, Yagi T & Kosaka H (2011) Reaction mechanism of single subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae: evidence for a ternary complex mechanism, J Biol Chem. 286, 9287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherney MM, Zhang Y, James MN & Weiner JH (2012) Structure-activity characterization of sulfide:quinone oxidoreductase variants, J Struct Biol. 178, 319–28. [DOI] [PubMed] [Google Scholar]