Abstract
Because of variation in linkage phase and heterozygosity among individuals, some individuals produce genetically more variable gametes than others. With the availability of genomic EBVs (GEBVs) or estimates of SNP-effects together with phased genotypes, differences in gametic variability can be quantified by simulating a set of virtual gametes of each selection candidate. Previous results in dairy cattle show that gametic variance can be large. Here, we show that breeders can increase the probability of breeding a top-ranking genotype and response to recurrent selection by selecting parents that produce more variable gametes, using the index where is the standardized normal truncation point belonging to selected proportion p, and SDgGEBV is the SD of the GEBV of an individual’s gametes. Benefits of the index were considerably larger in an ongoing selection program with equilibrium genetic parameters than in an initially unselected population. Superiority of the index over selection on GEBV increased strongly with the magnitude of the indicating that benefits of the index may vary considerably among populations. Compared to selection on ordinary GEBV, the probability of breeding a top-ranking individual can be increased by ∼36%, and response to selection by ∼3.6% when selection is strong (P = 0.001) based on values for the Holstein-Friesian dairy cattle population. Two-stage selection, with a preselection on GEBV and a final selection on the index, considerably reduced computational requirements with little loss of benefits. Response to multiple generations of selection and inheritance of the SDgEBV require further study.
Keywords: Mendelian sampling, GEBV, genomic selection, response to selection, virtual gametes, gametic breeding value, within-family variation, usefulness criterion
GENETIC improvement in livestock and crop populations relies on recurrent selection of parents of the next generation in outbred populations, or on the identification of elite parents to produce a new commercial variety for clonal reproduction. In outbred populations, recurrent selection of parents based on estimated breeding values (EBV) is widely used to maximize response to selection in the short term. This is because the EBV of a parent predicts the mean phenotype of its offspring. The focus on EBV, however, partly obscures the mechanism of genetic improvement and the central role of Mendelian-sampling therein. An alternative perspective is that genetic improvement of populations requires the offspring generation to be better, on average, than the parent generation. Genetic improvement, therefore, ultimately relies on selection for Mendelian sampling deviations, i.e., on selection for deviations of the offspring breeding value from the parent-average breeding value (Wray and Thompson 1990; Woolliams et al. 1999). The central role of Mendelian sampling deviations is also illustrated by the fact that any breeding value can be decomposed fully into Mendelian sampling deviations of ancestors (Thompson 1977).
In the classical infinitesimal model, the Mendelian sampling variance on the gametes produced by noninbred individuals in outbred panmictic mating populations equals one-quarter of the additive genetic variance. Hence, apart from limiting inbreeding, there are no opportunities to accelerate response to selection in outbred populations by increasing the Mendelian sampling variance in the infinitesimal model.
The actual Mendelian sampling variance, however, differs among individuals (beyond the effect of inbreeding). Because of linkage and finite genome size, the effective number of segregating segments in the meiosis is limited (Stam 1980; Hill and Weir 2011). Together with variation among individuals in heterozygosity and linkage-phase, this leads to variation among individuals in the Mendelian sampling variance on their gametes, both in panmictic outbred populations (Segelke et al. 2014; Bonk et al. 2016) and in crosses between inbred lines (Schnell and Utz 1976; Bernardo 2014). In other words, relative to the breeding value of the parent, the mean breeding value of gametes is zero, but some parents produce more variable gametes (and thus offspring) than others.
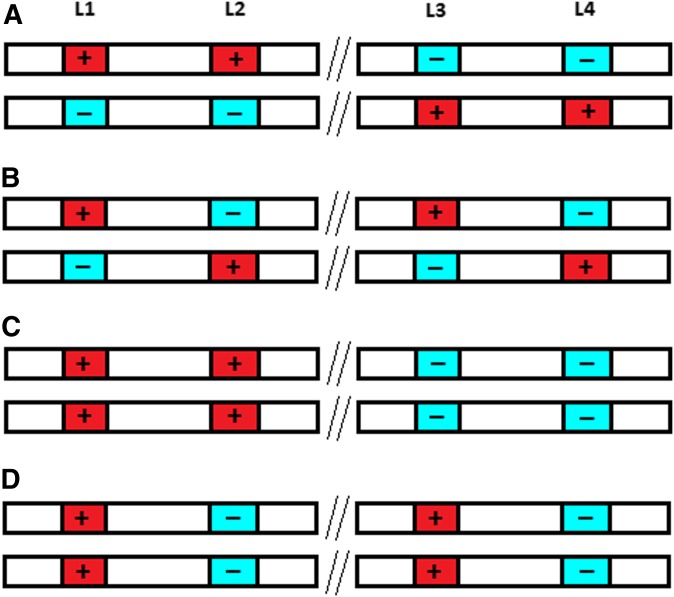
Figure 1 illustrates this phenomenon, showing that heterozygotes in coupling phase for closely linked loci have the highest standard deviation (SD) in the genetic merit of their gametes. While all four individuals in Figure 1 have the same genetic merit, only the heterozygous coupling-phase individual produces gametes carrying all four favorable alleles.
Figure 1.
Four diploid individuals (A–D), with two pairs of loci (L1–L4). Locus 1 and 2 are closely linked, and so are locus 3 and 4, while locus 1 and 2 are unlinked to locus 3 and 4. The unlinked loci are separated by //. The favorable allele is indicated by “+.” All individuals have the same genetic merit (“4+” and “4−“). Individual A is both in coupling phase and heterozygous, and produces 25% “4+” gametes, 50% “2+,2−” gametes and 25% “4−” gametes. All other individuals produce 100% “2+,2− gametes.” Hence, individual A produces gametes with most variation in genetic merit, and is also the only individual producing the most favorable combination of alleles (“4+”).
After selection in the offspring generation, offspring of parents with greater Mendelian sampling variance will show a greater within-family selection differential. This suggests that variation in the Mendelian sampling variance among potential parents can be used to accelerate response to recurrent selection, or to increase the probability of breeding a top-ranking individual or commercial variety (Schnell and Utz 1976; Bernardo 2014; Segelke et al. 2014). In other words, the response to selection, and the probability of breeding a top-ranking individual, can be increased by selecting parents not only on EBV, but also on the Mendelian sampling variance on their gametes.
In the context of development of inbred lines, such as in maize breeding, plant breeders have long realized the relevance of selecting crosses that produce variable offspring. About 40 years ago, Schnell and Utz (1976) proposed the usefulness criterion (UC), which represents the expected genotypic mean of the selected inbred offspring of biparental crosses among inbred lines. The UC increases with the genetic variance among the offspring of a cross. More recent work has investigated the benefit of selecting parents for the development of inbred lines, combining the UC with genomic information (Zhong and Jannink 2007; Bernardo 2014; Lehermeier et al. 2017; Allier et al. 2019a,b; Beckett et al. 2019). These studies show that the genetic level of double haploids and recombinant inbred lines can be improved by selecting crosses that yield more genetically variable offspring. (See Discussion for other strategies to increase long-term response in plant breeding).
For recurrent selection in outbred populations, such as in livestock or tree breeding, selection of parents with greater Mendelian sampling variance has received little attention, probably because it is difficult to use in the absence of genomic information. Before the genomic era, Van Raden et al. (1984) and Woolliams and Meuwissen (1993) investigated the benefit of selecting parents with a lower accuracy of their EBV. Given their EBV, such parents also produce more variable offspring. Benefits of this strategy, however, and application in breeding, were limited.
The availability of genomic information allows us to estimate the Mendelian sampling variance on the gametes of an individual, either by simulation or by theoretical prediction (Bernardo 2014; Segelke et al. 2014; Lian et al. 2015; Bonk et al. 2016; Lehermeier et al. 2017). With simulation, for example, one can create a virtual sample of the gametes of a selection candidate and estimate the variance in the genomic EBVs (GEBV) of these gametes. Thus, for breeding schemes with an existing genomic reference population, a known linkage map, and the availability of phased genotypes, it only requires computing time to obtain the SD of the gametic GEBV for all selection candidates.
The central “parameter” to assess the benefit of recurrent selection of parents that produce more variable gametes is the distribution of the gametic variability of individuals. At present, however, empirical knowledge of this distribution is very limited for panmictic outbred populations, and available only for the Holstein Friesian dairy cattle population. Segelke et al. (2014) used simulation to find the distribution of the SD of gametic GEBV in this population. Though they do not explicitly present the SD of among individuals, results in their Figures 2 and 4 indicate that the coefficient of variation (CV) of ranges from ∼0.10 through ∼0.14 for the traits protein yield, fat yield, somatic cell score, and still birth. These values are similar to results of Bonk et al. (2016), who find a CV of the Mendelian sampling variance of ∼0.20 in the same population. (Note that the CV of the variance is twice the CV of the SD, so that results of both studies are in agreement). With an approximate normal distribution of , and an average of 0.5, these results indicate that the SD of gametic GEBV of individuals ranges from ∼0.32 to ∼0.68 (μ ± 3 SD, using ). This range suggests considerable variation among Holstein-Friesian individuals in the SD of the GEBV of their gametes, where extreme individuals may differ by a factor of two. Moreover, empirical results in Segelke et al. (2014) suggest that the variability of the gametes of an individual is independent of the GEBV of the individual, in contrast to findings in biparental crosses in maize (Bernardo 2014). The variability in is unknown for other traits and populations, but may be larger for species with smaller genomes, and for traits determined by fewer genes.
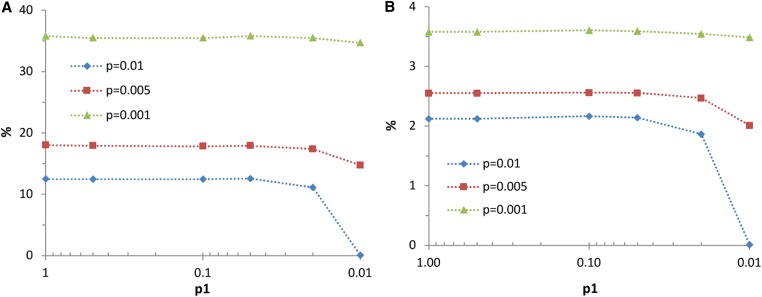
Figure 2.
Results of two-stage selection, for the selected population scenario, and for three overall selected proportions (p). Selection in the first stage is on GEBV (I1), while selection in the second stage is on Index 5. Lines show the superiority (%) of two-stage selection, relative to selection on GEBV, as a function of the selected proportion in the first stage (p1). Moving from left to right on the x-axis represents increasing preselection. With p1 = 1, there is no preselection, so that selection is entirely in the second stage on Index 5 and needs to be computed for all candidates. With P = 0.01, there is no second stage selection when p1 is also 0.01, so that selection is entirely in the first stage on GEBV. (A) % increase in the probability that offspring are in the top p fraction. (B) % increase in response to selection. For = 0.1. Note that the x-axis is on a logarithmic scale. Results are averages of 20,000 replicates.
Segelke et al. (2014) considered the use of variation in in mating schemes, and illustrated this phenomenon for the mating of two sires and three dams. However, they did not quantify the benefits of systematic selection of parents with high for response to recurrent selection in an outbred population, or for the probability of breeding a top-ranking individual.
Here we investigate the potential to increase response to recurrent selection and the probability of breeding a top-ranking genotype by selecting parents with higher Mendelian sampling variance in an outbred population. Building on the concept underlying the UC, we propose a number of indices for the selection of parents of the next generation of an outbred population, and identify the optimum index of an individual’s GEBV and the variability of its gametes using simulation. We quantify the benefits of selection on the optimum index for response to recurrent selection and for the probability of breeding a top-ranking genotype, for a range of selection intensities and CVs of . Because the magnitude of the Mendelian sampling variance, expressed relative to the full genetic variance, increases in selected populations (Bulmer 1971), we consider both initially unselected and selected populations (Bijma et al. 2018). Finally, we consider the prospects of selection in two stages to reduce the computation requirements for selection on Mendelian sampling variance.
Materials and Methods
We consider an outbred population with an existing genomic reference population, where GEBV for a polygenic additive trait are available on all selection candidates. We assume that the GEBV is an unbiased predictor of the true breeding value, , which is a property of a Best Linear Unbiased Predictor (BLUP; Henderson 1975). With this assumption, the response to selection in true breeding value is equal to the change in mean GEBV. We directly model GEBVs and SD of gametic GEBV of selection candidates based on the empirical distribution found by Segelke et al. (2014) in dairy cattle, without simulating actual genomes. Hence, we do not simulate the QTL, SNPs, linkage disequilibrium, chromosome structure, and recombination underlying the GEBV. We take this approach because results of Segelke et al. (2014) represent the best current knowledge of the distribution of gametic GEBV for an outbred population, and because response in true breeding value is equal to response in GEBV with Best Linear Unbiased Prediction (Henderson 1975; see Discussion for a more detailed motivation of this choice.) Because response to selection follows directly from the change in mean GEBV, we do not model the true breeding values, but simply calculate response to selection as the change in mean GEBV due to selection. Moreover, given the coefficient of variation of , the relative benefit of selecting for higher gametic variability is independent of the heritability of the trait (see section Scenarios and simulations below). For this reason, we do not consider heritability values.
Furthermore, we assume that the SD of the GEBV of the gametes produced by each selection candidate is known. We make this assumption, because an accurate estimate of the SD in gametic GEBV can be obtained by simulating a sufficiently large sample of virtual gametes for each selection candidate, assuming that accurately phased genotypes are available for all selection candidates (see Discussion). Thus all selection candidates have a known value for their GEBV and for the variability of the GEBV of their gametes. We will use the symbol for the GEBV of an individual, for the GEBV of a gamete, and for the SD in GEBV of the gametes of an individual.
Model for variation in gametic variability
Figures 5 and 6 in Segelke et al. (2014) show that the distribution of is close to log-normal; is restricted to positive values and shows slight positive skewness (after removal of the well-known effect of the DGAT1 gene on fat%). Moreover, Figure 2 in Segelke et al. (2014) shows that the GEBV and of individuals are independent in dairy cattle. For this reason, we simulated from a log-normal distribution, and independent of GEBV.
Without loss of generality, we assume that the variance of GEBV in the unselected base population is equal to 1, . Thus the mean Mendelian sampling variance of GEBV equals , the mean variance of gametic GEBV equals , and the mean SD of gametic GEBV equals ∼, ≅ . (The mean is not precisely equal to , because when varies among individuals). The GEBV of selection candidates were drawn from . The variance in the GEBV of the gametes produced by each selection candidate was drawn from a log-normal distribution, with mean and a variance, , depending on the scenario of interest (see below); . The log-normal distribution avoids negative values of gametic variances, and results in slightly positive skewness and excess kurtosis of the SD of gametic GEBV. For example, for a coefficient of variation of of 20% (Bonk et al. 2016), skewness of equals 0.30 and excess kurtosis equals 0.16. By visual inspection, the resulting distribution is very similar to that for fat yield (corrected for DGAT1) presented in panel 2 of Figure 6 of Segelke et al. (2014).
Selection indices
We investigated the performance of several selection indices for two criteria: (1) The probability that offspring in the next generation exceed a predefined GEBV threshold. (2) The mean GEBV of the selected offspring in the next generation. The first criterion measures the probability of breeding a top-ranking genotype, while the second criterion measures response to recurrent selection at the population level. Note that the second criterion is an analogy of the usefulness criterion (UC) used for the selection of biparental crosses in plant breeding. However, our selection indices will differ from the UC because we considered the selection of single parents of the next generation of an outbred population from a set of available selection candidates, rather than selection of biparental crosses to be made for inbred development (see also Discussion).
The default index was the ordinary GEBV,
In addition, we considered three types of selection indices of an individual’s GEBV and the variability of its gametes. First, we used empirical linear indices of and of a candidate, where the latter was weighted by an empirically obtained regression coefficient. Second, we used theoretically motivated linear indices of and . Finally we used theoretically motivated nonlinear indices based on probabilities derived from the normal distribution.
Empirical indices:
Indices 2 through 4 used empirical index weights estimated from simulated data. Hence, we used separate simulations to find the weights on in indices 2 through 4, which were independent of the simulations used later to evaluate the indices. To find the empirical index weights, we regressed the “success” of an individual’s offspring on its and ,
where b1 and b2 are regression coefficients (see Appendix A for details). The relative weight on in indices 2 through 4 was the ratio of the estimated regression coefficients;
For Index 2, offspring “success” (yoff) was defined as the fraction of offspring of the candidate that rank in the top p fraction of GEBVs in the next generation,
For Index 3, offspring “success” was defined as the mean GEBV of the selected offspring of a candidate,
For Index 4, offspring “success” was defined as the contribution (c) of the candidate to the mean GEBV of all selected offspring in the next generation,
The contribution of the candidate to the mean GEBV of all selected offspring was calculated as the product of the probability that an offspring of the candidate ranks in the top p fraction of GEBVs in the next generation and the mean GEBV of those top offspring. A table of empirical regression coefficients is given in Appendix A.
Theoretical linear indices:
Indices 5 through 8 were theoretically motivated linear indices. (Derivations are in Appendix B). Index 5 aimed to maximize the probability of breeding a top-ranking genotype, and was proportional to the linearly predicted probability that an offspring of the candidate ranked in the top p fraction of GEBVs in the next generation,
where xp is the truncation point of a standard normal distribution belonging to the upper-tail proportion p. Index 6 aimed to maximize response to recurrent selection, and was the predicted mean GEBV of the selected offspring of a candidate,
where ip is the selection intensity belonging to selected proportion p in a recurrent testing program (Falconer and Mackay 1996). Indices 5 and 6 are based on a linear approximation of the within-family SD in GEBV (; See Appendix B). To investigate the impact of this approximation, we also considered indices 7 and 8, which are analogous to indices 5 and 6, but use directly rather than linearizing it in ,
where . Hence, indices 7 and 8 use the expected GEBV SD among the offspring of the selection candidate when it is mated to a randomly chosen other parent (who has an expected of 0.25). Note that index 8 is analogous to the UC (see Discussion for a mathematical expression of the UC), but it uses a gametic variance equal to 0.25 for the unknown mate of the selection candidate, and it differs by a factor of two (which does not affect the selection of parents).
Theoretical nonlinear indices:
The theoretically motivated nonlinear indices 9 and 10 used the probability that offspring of a candidate were selected (or ranked in the top p fraction), calculated from the normal distribution. Index 9 aimed to maximize the probability of breeding a top-ranking individual, and was the probability that an offspring of the candidate ranked in the top p fraction of GEBVs in the next generation,
where is the cumulative normal distribution function, being the expected mean GEBV of the offspring of the candidate, and is the absolute truncation point in the offspring generation belonging to the top p fraction. Index 10 was the predicted contribution of the candidate to the mean GEBV of all selected individuals in the offspring generation,
where the first term is the probability that an offspring is selected, and the second term is the predicted mean of those selected offspring. In the second term, is the selection intensity specifically for the offspring of this candidate, which follows from and the mean GEBV and of those offspring. Note that Index 10 is a theoretical analogy of Index 4.
Scenarios and simulations
We used simulation to quantify the benefits of selecting on indices I2 through I10 vs. selection on GEBV (I1), and to identify the best index. The relative benefit of selecting on will depend on the coefficient of variation of among individuals, , and on the intensity of selection. The importance of relative to the GEBV increases with its CV. Since breeders are usually interested in the relative benefits of innovations (e.g., percentage increase in response) rather than the absolute benefit, we measured the differences among individuals in variability of their gametes by the , rather than the SD of . (The CV is also independent of the unit of measurement). Likewise, with increasing selection intensity the benefit of producing extreme offspring increases. The accuracy of GEBV is irrelevant here, since response to selection was modeled entirely in terms of GEBV. Based on values found by Segelke et al. (2014; see Introduction), we considered CVs of of 0.05, 0.10, 0.15, and 0.20. These values correspond to a range of of 0.425 to 0.575 for a CV of 0.05, to 0.20 to 0.80 for a CV of 0.2 (using μ ± 3 SD). To vary the intensity of selection, we considered selected proportions (p) of 0.5, 0.2, 0.1, 0.05, 0.01, 0.005, and 0.001 for both sexes.
Each index was used to select the best 100 individuals (50 males and 50 females) as parents from 100/p selection candidates. Selected parents were mated at random, and a total of 100/p offspring were simulated using normally distributed gametic GEBV with the appropriate . The threshold used to define the top p fraction in the offspring generation was based on the offspring of parents selected on ordinary GEBV (I1). Then, for parents selected on indexes 2 through 10, we estimated (i) the probability of breeding a top-ranking genotype as the fraction of offspring exceeding this selection threshold, and (ii) response to recurrent selection as the mean GEBV of the best 100 offspring. Hence, in the results on the probability of breeding a top-ranking genotype, p refers to the definition of “top,” whereas in the results on response to recurrent selection, p refers to the proportion of the candidates that is selected to become parent of the next generation. Results were expressed relative to selection on ordinary GEBV (I1), and were based on 10,000 replicates.
With respect to the selection history of the population, we considered two scenarios: a scenario where selection candidates come from an unselected population (“unselected population”), and a scenario where parents come from an selected population (“selected population”) in which recombination balances the gametic phase disequilibrium generated by selection, leading to equilibrium genetic parameters (known as the “Bulmer equilibrium,” after Bulmer 1971). Directional selection reduces the between-family variance compared to the within-family segregation variance (Mendelian sampling variance). Under the infinitesimal model, selection does not affect the within-family segregation variance, while the between-family and full genetic variance reach an equilibrium in about three generations (Bulmer 1971; see also Discussion). Simulations for the selected population scenario, therefore, consisted of three generations of truncation selection on GEBV to obtain the equilibrium variance of GEBVs, and a fourth generation where parents were selected on one of the indices. Hence, the indices were judged based on response to selection in generation 4. Because Segelke et al. (2014) found no correlation between the GEBV and the of individuals, we assumed that selection on GEBV in generations 1 through 3 does not affect the in generation 4.
Empirical indices I2-4 were omitted for the selected population scenario for two reasons: (i) those indices had been estimated from data on initially unselected populations, so they were probably suboptimal for a selected population. (ii) Results from initially unselected population indicated that theoretically motivated indices were superior to empirical indices (See Results section below).
Two-stage selection
Practical implementation of the above indices requires the estimation of for each selection candidate. While this is essentially straightforward, and can be done either by simulating virtual gametes (Bernardo 2014; Segelke et al. 2014) or by deterministic prediction (Bonk et al. 2016), it may be computer intensive when the number of candidates is large. With simulation, for example, a large number of gametes has to be simulated for each candidate to accurately estimate the variance in GEBVs among those gametes. To investigate opportunities to reduce computational effort without losing the benefit of selecting for , we considered a two-stage selection scenario. Selection in the first stage was for GEBV, while selection in the second stage was for Index 5. (This choice was motivated by the results, see below). Such preselection may greatly reduce computational requirements, because needs to be estimated only for the individuals that enter the second stage.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study. The R-codes used for simulation are available in Supplemental Material, Files S1 through S3. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11106125.
Results
Differences between the indices were small, both for the probability that offspring rank within the top p fraction and for response to recurrent selection. Moreover, empirical indices were not systematically superior over theoretically motivated indices (Tables S1 and S2). We, therefore, show results for Index 5 only, which was among the best in all scenarios, both for the probability to breed a top-ranking genotype and for response to recurrent selection.
Initially unselected population
Table 1 shows the relative increase in the probability of breeding a top-ranking genotype for the unselected population scenario. Compared to selection on ordinary GEBV, this probability increased when a smaller fraction was defined as “top” (smaller p), and when the CV of was larger. For a CV of 0.1 (as found in dairy cattle; Segelke et al. 2014), the probability that an offspring ranked within the top 0.1% (P = 0.001) increased by 19% when parents were selected on Index 5 instead of ordinary GEBV. The probability of breeding a top-ranking offspring increased very strongly with the CV of . Hence, the benefits of selecting for may depend strongly on the species and on the population history.
Table 1. Percentage increase in the probability to breed a top-ranking genotype, for the unselected population scenario.
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| 0.5 | 0 | 0 | 0 | 0 |
| 0.2 | 0 | 0 | 1 | 2 |
| 0.1 | 0 | 1 | 3 | 6 |
| 0.05 | 1 | 3 | 6 | 10 |
| 0.01 | 2 | 8 | 17 | 31 |
| 0.005 | 3 | 10 | 25 | 45 |
| 0.001 | 4 | 19 | 48 | 94 |
Values are the relative increase (%) in the number of individuals in the offspring generation that are in the top p fraction for GEBV when parents are selected on Index 5, compared to selection on ordinary GEBV (I1). The top p-fraction was defined based on the offspring of parents selected on ordinary GEBV.
This is the value found in dairy cattle (Segelke et al. 2014).
Table 2 shows the relative increase in response to recurrent selection for the unselected population scenario. Compared to selection on ordinary GEBV, selection of parents on Index 5 increased the mean GEBV of the selected offspring. The relative increase was larger when selection was stronger (smaller p), and when the CV of was larger. The increases in response to selection were smaller than those in the probability of breeding a top-ranking individual (Table 2 vs. Table 1). For example, for and P = 0.001, the mean GEBV of selected offspring increased by only 2%, whereas the probability of breeding a top-ranking individual increased by 19% when parents were selected on Index 5 instead of GEBV.
Table 2. Percentage increase in response to recurrent selection, for the unselected population scenario.
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| 0.5 | 0.3 | 0.2 | 0.2 | −0.2 |
| 0.2 | 0.1 | 0.3 | 0.9 | 1.6 |
| 0.1 | 0.2 | 0.6 | 1.4 | 2.4 |
| 0.05 | 0.2 | 0.8 | 1.7 | 3.2 |
| 0.01 | 0.3 | 1.3 | 3.1 | 5.8 |
| 0.005 | 0.3 | 1.4 | 3.6 | 7.1 |
| 0.001 | 0.5 | 2.0 | 5.2 | 10.8 |
Values are the relative increase (%) in the mean GEBV of selected offspring when parents are selected on Index 5, relative to selection of parents on ordinary GEBV; .
This is the value found in dairy cattle (Segelke et al. 2014).
Selected population
Table 3 shows the probabilities of breeding a top-ranking individual for the selected population scenario. The benefit of selecting on Index 5 instead of ordinary GEBV was larger than for an initially unselected population (Table 3 vs. Table 1). For a CV of 0.1, for example, the probability of breeding an offspring that ranked within the top 0.1% (P = 0.001) increased by 36% when selecting on Index 5 instead of the GEBV. The corresponding value for an initially unselected population was 19% (Table 1).
Table 3. Percentage increase in the probability to breed a top-ranking genotype, for the selected population scenario.
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| 0.5 | 0 | 0 | 0 | 0 |
| 0.2 | 0 | 1 | 2 | 3 |
| 0.1 | 1 | 2 | 4 | 8 |
| 0.05 | 1 | 4 | 10 | 16 |
| 0.01 | 3 | 13 | 29 | 53 |
| 0.005 | 4 | 18 | 42 | 78 |
| 0.001 | 8 | 36 | 89 | 175 |
Values are the relative increase (%) in the number of individuals in the offspring generation that are in the top p fraction for GEBV when parents are selected on Index 5, compared to selection on ordinary GEBV. The top p-fraction was defined based on the offspring of parents selected on ordinary GEBV.
This is the value found in dairy cattle (Segelke et al. 2014).
Table 4 shows the responses to selection for the selected population scenario. Benefits of selecting on Index 5 instead of GEBV were ∼70% greater than for an initially unselected population (Table 4 vs. Table 2). For example, for a CV of 0.1 and strong selection (P = 0.001), the mean GEBV of selected offspring increased by 3.6% when parents were selected on Index 5 instead of ordinary GEBV. The corresponding value for an initially unselected population was 2% (Table 2).
Table 4. Percentage increase in response to recurrent selection, for the selected population scenario.
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| 0.5 | −0.2 | −0.2 | 0.0 | 0.0 |
| 0.2 | 0.3 | 0.6 | 1.3 | 2.1 |
| 0.1 | 0.3 | 0.9 | 1.9 | 3.6 |
| 0.05 | 0.3 | 1.3 | 2.9 | 5.2 |
| 0.01 | 0.6 | 2.2 | 5.1 | 9.9 |
| 0.005 | 0.6 | 2.5 | 6.2 | 12.1 |
| 0.001 | 0.8 | 3.6 | 9.1 | 18.2 |
Values are the relative increase (%) in the mean GEBV of selected offspring when parents are selected on Index 5, relative to selection of parents on ordinary GEBV; .
This is the value found in dairy cattle (Segelke et al. 2014).
Two-stage selection
Figure 2A shows the relative increase in the probability of breeding a top-ranking genotype as a function of the degree of preselection on GEBV, for Index 5 and the selected population scenario. Results show that very strong preselection can be applied with little loss in the probability of breeding a top-ranking genotype. For example, for a total selected proportion of P = 0.001, preselection with p1 = 0.01 (so that p2 = 0.1) showed no significant reduction in the probability of breeding a top-ranking genotype. Hence, for this scheme, preselection allowed a 100-fold reduction in the computation effort for estimating , without meaningful loss of benefit. Figure 2B shows similar results for response to selection.
Discussion
We have investigated the opportunities to increase the probability of breeding a top-ranking genotype, and the response to recurrent selection, by selecting parents with greater Mendelian sampling variance on their gametes, for outbred panmictic populations. Ten selection indices were compared, most of which gave similar results. A simple index with a relative weight of on the SD of gametic GEBV of the selection candidates was near optimal in all scenarios (Index 5; is the truncation point of a standard-normal distribution belonging to an upper-tail fraction p). Benefits of selection for increased with the intensity of selection, and were larger for populations with a history of selection than for initially unselected populations. For input values based on results found in dairy cattle (Segelke et al. 2014), selection for considerably increased the probability of breeding a top-ranking genotype, while benefits for response to selection were limited. For practical implementation, preselection on ordinary GEBV can be used to substantially reduce computational requirements.
Selection indices
We compared three types of selection indices of the GEBV of an individual and the Mendelian sampling variance on its gametes: (i) empirical indices estimated from simulated data (I2-4), (ii) theoretically motivated linear indices (I5-8), and (iii) theoretically motivated nonlinear indices (I9,10). For each type, we considered both indices that aimed to maximize the probability of breeding a top-ranking genotype (I2,5,7,9), and indices that aimed to maximize response to selection (I3,4,6,8,10). All indices are approximations, because the distribution of GEBV in the offspring generation is a complex mixture of distributions. For the probability of breeding a top-ranking genotype, empirical indices yielded similar results as theoretically motivated indices, while empirical indices were slightly worse for response to recurrent selection (Tables S1 and S2). Nonlinear indices were more complex than linear indices, but showed similar results.
For the theoretically motivated linear indices, results were very similar for indices targeting a top-ranking individual (I5,7) and indices targeting response to recurrent selection (I6,8). For the first category, the index weight depends on the standardized truncation point (x), while, for the second category, the index weight depends on selection intensity (i). However, meaningful benefit of selection for higher gametic variability was observed only when selection was reasonably strong (say ). With strong selection, x and i have a similar value, so that indices 5 and 7 are similar to indices 6 and 8.
Results were also very similar for indices that were a linear function of the SD of the gametic GEBV of the candidate (; I5,6), and indices that were a linear function of the Mendelian SD of the GEBV of its offspring (; I7,8). The first class involves an extra approximation, because they approximate the by a linear function of (see Appendix B). The similarity of results, however, indicates that this additional approximation had little impact.
Santos et al. (2019) recently proposed the selection index (labeled RTPA). They state that this index refers to the genetic level of the selected offspring of a candidate in the next generation, but do not provide a proof thereof. This index, however, ignores that the GEBV SD in the offspring of a mating follows from the gametic variances of the parents, , not from the SD, . For this reason, our Index 6 includes a factor √2 (See Appendix B for the derivation thereof).
Selected vs. unselected populations
The benefit of selecting for higher Mendelian sampling variance was considerably larger for populations undergoing recurrent genomic selection than for initially unselected populations (Table 3 and Table 4 vs. Table 1 and Table 2). This occurs because selection reduces the between-family variance relative to the Mendelian sampling variance (the “Bulmer effect,” after Bulmer 1971). This effect is particularly strong for the variance of GEBV, because genomic selection yields a very strong reduction in the variance of GEBV when the update of the reference population in each generation is relatively small. For a trait determined by many loci of small effect and equal selection intensity in both sexes, the variance of GEBV may be modeled as
where k is the relative reduction in the variance of the selection criterion (i.e., the GEBV here) due to selection, and is the full variance of the GEBV in the unselected base population. With truncation selection on Gaussian GEBV, k = i(i-x), with i the selection intensity, and x the standardized truncation point (Cochran 1951). Values of k are typically between ∼0.6 and ∼0.93. In around three generations, the asymptotes to an equilibrium value of
For a selected proportion of 5%, for example, k = 0.86, the equilibrium variance is only 54% of the full variance, and 0.50/0.54 = 92% of the variance in GEBVs is due to Mendelian sampling. Hence, Mendelian sampling is the dominant source of variance in GEBV with recurrent genomic selection. This explains the greater relevance of variation among individuals in the Mendelian sampling variance of their gametes for populations undergoing recurrent genomic selection. Allier et al. (2019b) investigated the relevance of the Bulmer effect in on the context of hybrid development in plant breeding.
Simulation method
We directly simulated the of the selection candidates from the distribution found by Segelke et al. (2014) in dairy cattle, without simulating the underlying genomes of the selection candidates. Hence, we did not simulate the QTL, SNPs, linkage disequilibrium, chromosome structure, and recombination underlying the GEBV. We chose this approach based on the following reasoning: our simulations should agree as well as possible with our current knowledge of reality. The central “parameter” here is the bivariate distribution of GEBV and gametic variability of individuals. At present, our knowledge of this distribution is very limited for outbred populations; the distribution has been quantified only for Holstein Friesian dairy cattle (Segelke et al. 2014; Bonk et al. 2016). For this reason, we have mimicked that distribution as closely as possible.
We chose to mimic this distribution by directly simulating the gametic variance from a log-normal distribution. Alternatively, we could have simulated a detailed genomic architecture, which would have required many detailed assumptions, such as the number of QTL and their positions, LD, linkage, and the distribution of QTL-effects. Since we have very limited knowledge of these details, we would need to compare the resulting distribution of with the available empirical distribution, to judge whether the detailed simulations are realistic. If not, we would have needed to tune the simulations until the distribution of agrees with the empirical knowledge. Hence, if done properly, detailed simulations would have yielded the same bivariate distribution of GEBV and that we chose to simulate directly, and, therefore, also the same results, albeit at greater computational cost.
Furthermore, we assumed that the is known for each selection candidate. Hence, we assumed that selection candidates have accurately phased genotypes, so that the can be estimated accurately by simulating a sufficiently large sample of virtual gametes for each candidate. Phasing errors will reduce the benefit of selection on . In livestock populations, phased genotypes are typically produced as a byproduct of the imputation of missing marker data. The most important livestock breeds have large genomic reference populations, strong LD, known pedigree, and genotypic records on the parents of the selection candidates. Hence, for such populations phasing is accurate, as illustrated by the small error rate of imputation (Druet and Georges 2015). Phasing accuracy may be considerably lower for highly polymorphic populations with little LD (Bukowicki et al. 2016) and for polyploid species (He et al. 2018).
The magnitude of and its relationship with the GEBV
In our simulations, we assumed a CV of of ∼10% and independence of from the GEBV, as found by Segelke et al. (2014) in a dairy cattle population. The variability of the gametic GEBV of a parent and its relationship to the level of the GEBV of the parent will depend on the population structure. Bernardo (2014) analyzed the variance in GEBV among double haploids resulting from 45 virtual F1-crosses of 10 selected inbred lines in maize. He found a triangular relationship between the mean and the variance; double haploids of F1-crosses with an intermediate GEBV level showed the largest variance, whereas F1-crosses with a high or low GEBV showed very little variance among their double haploid offspring. Hence, this contrasts the findings of Segelke et al. (2014), who found independence of the mean and the variance in GEBV of offspring. This difference probably originates at least in part from the population structure, which consisted of a single panmictic outbred population in Segelke et al. (2014) and of 10 selected inbred lines in Bernardo (2014). Selection among inbred lines may create substantial differences in allele frequency between inbreds of high vs. low genetic merit. The F1s originating from crosses between opposite extremes will then show the highest heterozygosity, and thus greater variability in their double haploid offspring (Bernardo 2014). In contrast, recurrent selection for a highly polygenic trait in an outbred population generates only small changes in allele frequency within a generation, so that heterozygosity in parents with extreme GEBV may differ hardly from the average heterozygosity. The latter would lead to approximate independence of GEBV and, as found in dairy cattle.
The variability of the gametic GEBV of a parent will probably also depend on the method to estimate GEBV. Segelke et al. (2014) estimated marker effects using SNP-BLUP, which is equivalent to GBLUP with back solving of SNP-effects (Hayes et al. 2009). With GBLUP, the will depend on the genomic relationship matrix (GRM) used to back-solve the SNP effects. GRM that (implicitly) assume greater variance of SNP-effects at loci with lower MAF, such as Van Raden (2008) method 2, predict large SNP-effects for rare alleles (Bouwman et al. 2017), and may therefore yield higher than GRM assuming independence of SNP-effect variance and allele frequency, such as Van Raden (2008) method 1. Moreover, with GBLUP, the effect of a QTL is distributed over multiple markers, resulting in many markers of small effect spread all over the genome. Variable selection methods in contrast, such as BayesB or Bayesian Lasso (reviewed in Gianola et al. 2009), attempt to find a limited number of marker loci of large effect to explain the full genetic variance. One would expect larger for variable selection methods than for GBLUP, because the number of relevant loci is smaller. However, when GBLUP distributes the effect of a QTL over a limited number of closely linked marker loci, recombination among these loci will be rare, and they may contribute equally to the as a single locus identified by a variable selection method. Hence, the benefit of variable selection methods for the detection of individuals with a high requires further research.
Variation in the gametic variability among individuals originates from variation in heterozygosity and linkage (Bernardo 2014; Segelke et al. 2014; Bonk et al. 2016; Figure 1). Segelke et al. (2014) indeed found that was smaller in individuals with higher inbreeding coefficients, but the pedigree and genomic inbreeding coefficients explained only very little of the variance in (0.25–5.3%). Hence, parents with a high cannot be identified accurately based on their genome-wide inbreeding coefficients. The ability to predict may be increased by using a weighted average of locus specific heterozygosity, the weights being the variance in GEBV due to the locus. Nevertheless, variation in linkage-phase among individuals may still contribute the majority of the variation in among individuals (Bernardo 2014; Bonk et al. 2016; e.g., individual A vs. B in Figure 1).
Variation in the recombination rate among individuals (e.g., Kong et al. 2004) may be an additional source of variance in gametic variability among individuals. Battagin et al. (2016) performed simulations to explore the potential of manipulating recombination rates to increase response to recurrent selection. Figure 3 in Battagin et al. (2016) suggests that higher recombination rates result in somewhat higher genetic variance, but do not change the genic variance in the short term. Hence, whether or not parents with higher recombination rates also have greater cannot be concluded based on results in Battagin et al. (2016). Our distribution of is based on findings of Segelke et al. (2014), who simulated gametes based on the recombination map of dairy cattle. Hence, in these simulations, recombination rates were the same for all individuals, meaning that our distribution of does not include a potential contribution of variation in the recombination rate among individuals. If higher recombination rates indeed go together with greater , then there is a synergy between selection for greater Mendelian sampling variance and the building of optimal genotypes over generations (see also below).
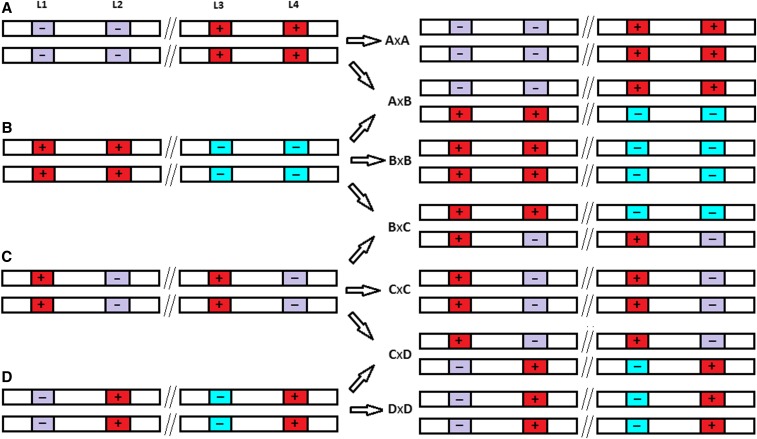
Figure 3.
The use of mating to create offspring with greater Mendelian sampling variance on their gametic GEBV. Locus 1 and 2 are closely linked, and so are locus 3 and 4, while locus 1 and 2 are unlinked to locus 3 and 4. The unlinked loci are separated by //. The favorable allele is indicated by “+.” A through D indicate four types of parents. Mating within type yields homozygous offspring. Both the mating AxB and CxD create fully heterozygous offspring. However, only the AxB mating creates an offspring with a large Mendelian sampling variance on its gametic GEBV (see individual A in Figure 1). Mating BxC yields offspring with lower heterozygosity than mating CxD, but those offspring nevertheless have higher Mendelian sampling variance on their gametic GEBV.
Hybrid development in plants and the UC:
While selection of parents with variable gametes has received little attention in the improvement of outbred populations, such as in animal and tree breeding, it has a long history in the development of hybrid crosses in plant breeding (e.g., Schnell and Utz 1976; Bernardo 2014; Lehermeier et al. 2017; Allier et al. 2019a,b; Becket et al. 2019). Schnell and Utz (1976) proposed the UC, which is a measure of the short-term improvement that can be achieved when developing inbreds out of an F1. The UC measures the mean of the selected inbreds of an F1 parent. With selection on estimated marker effects (e.g., GEBV), the UC depends on the mean estimated breeding value of the parents of the F1, , on the intensity of selection (i), and on the SD in estimated genetic values among the inbreds developed from the F1 (; Zhong and Jannink 2007),
Hence, the UC is similar to the indices proposed here (particularly I6 and I8), but considers inbred line development rather than response to recurrent selection of single parents in an outbred population. The UC cannot be used for the selection of single parents in outbred populations, because the is a property of the offspring, which depends also on the prospective mate of the selection candidate (the mate is typically unknown at the time of selection). For this reason, we developed indices that either ignored the mate (I5,6) or assumed an average mate (I7,8). Both indices yielded very similar results.
Lehermeier et al. (2017) showed that selection on the UC is superior over selection on GEBV for the development of inbreds from biparental crosses. They compared two types of UC: one based on the SD of estimated genomic values (GEBV) of the offspring of an F1 (as in the above expression for the UC), and one based on the SD of the true genetic values among the offspring of the F1 (i.e., replacing by ; note that > ). Their results show that selection for a UC based on the true genetic values is superior. This result arises because selection among the offspring was based on their true genotypic values in the simulations in Lehermeier et al. (2017; Christina Lehermeier, personal communication), which requires either known QTL-effects or field testing of genotypes. In outbred populations, true genetic values are typically unknown, and selection for an index based on the SD of true genetic effects would overestimate the value of Mendelian sampling variance for the selection of parents. For this reason, we proposed indices based on the SD of the estimated (genomic) breeding values.
Inheritance of gametic variability
Here, we have assessed the benefit of selection for based on response to a single generation of selection. If the on the gametes of an individual is created de novo each generation, rather than inherited from the parents, then the benefits of selection for in a single generation probably also reflect the benefits over multiple generations. However, when is partly inherited, response to selection may change over generations. Because the of an individual depends on its heterozygosity and linkage phase, inheritance of will depend on the transfer of heterozygosity and linkage phase from parents to offspring.
It may seem that heterozygosity is not inherited when mating is random, because a parent transmits only a single allele (or haplotype) to its offspring while the other allele originates from the mate. However, while the inbreeding coefficient is indeed not inherited, heterozygosity is partly “heritable.” With random mating, 50% of the offspring of a heterozygous parent are also heterozygous themselves, irrespective of the allele frequency in the population. This occurs because . Hence, whenever population allele frequency deviates from 0.5 and mating is random, heterozygous parents produce offspring with above-average heterozygosity, indicating “inheritance” of heterozygosity. With random mating, the “heritability” of heterozygosity may be derived by linking heterozygosity in a single parent and its offspring, similar to parent-offspring regression for ordinary quantitative traits; . Substituting , and and solving for yields
(Note that this result is identical to the heritability of a trait determined entirely by dominance, e.g., Equations 8.3b, 8.4 and 10.1 with a = 0, d = 1 and VE = 0 in Falconer and Mackay 1996). The is a V-shaped function of allele frequency, where when P = 0.5, and when p approaches 0 or 1. For example, for P = 0.1, it follows that and , so that heterozygosity in the offspring of a heterozygote equals , as also shown above. Hence, offspring of a parent heterozygous for a rare allele “inherit” almost 50% of the heterozygosity of their parent. This result suggests that offspring of parents heterozygous for rare alleles may also inherit part of the of their parent, which would increase the benefits of selecting on Index 5 over multiple generations compared to the values presented here.
Effects of inheritance of the linkage phase are more complex. For closely linked loci, parents will transmit their linkage phase to their offspring. However, this does not necessarily imply inheritance of . Individual A in Figure 3, for example, is in coupling phase and produces an offspring with high when mated to individual B. However, individual A itself has a of zero. Stochastic simulation will probably be required to quantify the inheritance of due to the combination of linkage and heterozygosity.
While high gametic variability is desired in the selection of parents of the next generation of breeding individuals, producers in agriculture typically prefer a uniform population. Hence, parents of production individuals should ideally have a low gametic variability, and outliers on the bottom end of the scale are particularly undesirable. In most livestock populations, selection of breeding individuals is separated from the selection of parents of production animals. In dairy cattle, for example, selection of breeding bulls is done by breeding companies, whereas selection of bulls of ordinary cows is done by farmers. Hence, breeding companies may select bulls with high gametic variability, while farmers may select the opposite bulls. Nevertheless, because gametic variability is partly inherited, selection of breeding individuals with high gametic variability may increase variability in production herds in the next generation. When production individuals are hybrids descending from inbred lines, such as in maize, there seems to be little room for a conflict between selection of parents with variable gametes and the uniformity of the hybrids.
Genotype building and long-term response
Compared to selection on GEBV, selection of parents for higher Mendelian sampling variance looks one additional generation ahead, since response is realized only after selection in the offspring generation. Selection methods that aim to build superior genotypes over multiple generations have a long history in inbred development from crosses in plant breeding (Dudley 1984a,b; Bernardo 2014; Daetwyler et al. 2015; Goiffon et al. 2017; Müller et al. 2018), and have also received some attention in animal breeding (Cole and Van Raden 2011; Kemper et al. 2012).
Selection methods that consider a single additional generation, such as I5 and the UC, may seem very different from those aiming to build an optimum genotype over many generations. However, selection for higher Mendelian sampling variance may also be interpreted as a way to select for combinations of favorable alleles within gametes, i.e., as an analogy of genotype building strategies. Figure 1 illustrates that selection for higher Mendelian sampling variance favors heterozygous individuals that are in coupling phase for closely linked loci, but that may be in repulsion phase for unlinked loci. While all four individuals in Figure 1 have the same GEBV, only the heterozygous coupling-phase individual produces gametes that carry all four favorable alleles. This individual also has the highest SD in the GEBV of its gametes. Hence, selection methods that consider a single additional generation, such as the indices proposed here, may also accelerate the process of bringing the favorable alleles together.
Genotype building strategies aim to create an optimal genotype over multiple generations, either from single individuals (Daetwyler et al. 2015; Müller et al. 2018) or from multiple individuals (Cole and Van Raden 2011; Kemper et al. 2012; Goiffon et al. 2017). These studies assumed marker or QTL-effects to remain constant over generations, which implies additivity of QTL. Compared to selection on GEBV, genotype building strategies typically show increased long-term gain, reduced short term gain, and less loss of genetic diversity. The utility of genotype building strategies for the improvement of polygenic traits by recurrent selection in outbred populations seems to be limited for two reasons. First, the (apparent) SNP-effects change over generations due to erosion of LD and relationship information (Habier et al. 2007) and due to nonadditive genetic effects. For example, the presence of directional dominance (d > 0) at a QTL leads to an apparent negative epistatic interaction between the favorable alleles of two SNP-loci that are in incomplete LD with each other and with the QTL (de los Campos et al. 2019). Such spurious epistasis leads to diminishing return of selection for the favorable SNP alleles, so that long-term response falls short of its prediction based on SNP-effects in the initial generation. Such diminishing return of selection on SNP-effects is likely to be systematic, because the wide-spread observation of inbreeding depression and hybrid vigor suggest that directional dominance is common. Contrary to the expectation of Daetwyler et al. (2015), genotype building strategies probably suffer more from overprediction than schemes with selection on GEBV, I5 or the UC, because they consider several generations, whereas selection on GEBV, I5 or the UC considers only one or two generations. Second, a reduction in short-term gain may be acceptable in the context of hybrid development in plants, but probably not for outbred populations such as in trees or livestock, where the goal is to create continued genetic progress at the population level and where the breeding and production populations often overlap considerably, particularly in ruminants (Daetwyler et al. 2015).
Selection of parents with higher Mendelian sampling variance may be a practical strategy intermediate of selection of GEBV and genotype building, particularly when combined with a restriction on the increase of average coancestry in the population (see also Kemper et al. 2012). The impact of selection of parents with more variable gametes on the increase in coancestry (and thus on inbreeding) will depend on the correlation of between relatives. While relatives have similar GEBV, it is unknown at present whether they also have similar . However, since relatives share haplotypes and full siblings share genotypes, they may also show similar . Nevertheless, compared to selection for GEBV, selection for will tend to increase the rate of inbreeding only when the correlation between of relatives exceeds that of the GEBV, which seems unlikely.
Shaping the selection candidates
Table 3 and Table 4 show that the benefit of selecting for greater Mendelian sampling variance increases strongly when differences in among individuals become larger. This raises the question whether breeders can create selection candidates with a high , by using a specific mating strategy. Together with selection for, e.g., Index 5, this would speed up the process of combining good alleles within individuals, which would accelerate response over generations. The use of a mating strategy that maximizes heterozygosity in the offspring, where marker-based heterozygosities are weighted by the (apparent) effect of the marker, will increase heterozygosity at SNPs that explain the GEBV, and will probably increase the in the offspring. Such a mating strategy may be interpreted as a generalization of the classes of loci method (Dudley 1984a,b; see above). However, as illustrated in Figure 3, the in a prospective offspring depends not only on heterozygosity in the offspring, but also on linkage and the linkage phase in the parents (Bonk et al. 2016). In principle, optimum matings could be identified by simulation, where virtual offspring are simulated for all potential matings to estimate their . However, this is computationally demanding because the number of potential matings is large. An analytical approach, similar to Bonk et al. (2016), would also be possible, but may be equally computationally demanding. A simple approximate indicator for promising matings would be valuable here.
Genotypic value vs. breeding value
We have considered an additive model. This makes sense when the objective is to increase response to recurrent selection, because the average effect of nonadditive interactions is included in the additive genetic effect, while the remaining dominance and epistatic deviations do not contribute to response to recurrent selection in the short term (Falconer and Mackay 1996). Also when interest is in breeding a top-ranking individual to be used as parent in an outbred population, such as sire selection in dairy cattle, the use of an additive model makes sense because the relevant criterion is the breeding value of the sire, not its genotypic value. However, when the objective is to breed a top-ranking genotype for clonal reproduction, such as in maize, interest is in the full genotypic value, including effects due to dominance and epistasis. Estimation of the Mendelian sampling variance on the genotypic value of prospective offspring, rather than on the gametic GEBV of the candidate, requires the simulation of offspring genotypes of potential mating pairs rather than gametes of single selection candidates (Bonk et al. 2016). This substantially increases computational requirements because the number of potential mating pairs is much larger than the number of selection candidates. When nonadditive genetic effects are small to moderate in size, we expect that computational requirements can be reduced substantially by selecting in three stages: a first selection on GEBV, a second selection on Index 5, and a final selection on an adjusted version of Index 7, in which the additive Mendelian SD on GEBV is replaced by the corresponding SD of the full genotypic value of the prospective offspring.
Conclusion
Breeders can increase the probability of breeding a top-ranking genotype and response to recurrent selection by selecting parents on an index of the GEBV and the gametic variability of selection candidates. Benefits depend strongly on the variation in gametic variability, and may thus differ considerably among populations. Response to multiple generations of selection, and the inheritance of the gametic variability, need further study.
Acknowledgments
This study was supported financially by the Dutch Ministry of Economic Affairs (TKI Agri & Food) and the Breed4Food partners Cobb Europe, CRV, Hendrix Genetics and Topigs Norsvin. The authors declare that they have no competing interests.
Author contributions: P.B. conceived the study, derived the selection indices, performed the simulations and drafted the manuscript. M.P.L.C. and Y.C.J.W. assisted in the design of the study and contributed to the structuring and writing of the manuscript. All authors read and approved the final manuscript.
Appendix A
Empirical Indices
To find empirical linear selection indices of and , we used regression of the success of offspring on the information sources in the index,
where is a measure of the success of the offspring of a sire, the GEBV of the sire, the SD among the GEBVs of the gametes of the sire, and b1 and b2 the corresponding regression coefficients. For simplicity, we simulated selection in males only. As measures of success of the offspring of a sire, we used: (i) the fraction of offspring of the sire that ranked in the top p proportion of GEBVs in the offspring generation , (ii) the mean GEBV of the selected offspring of the sire , and (iii) the contribution of a sire to the mean of all selected offspring in the next generation . The was motivated by the idea that a poor sire might have very few selected offspring, but those offspring must nevertheless have had reasonably high GEBV otherwise they would not have been selected. Use of might result in selection of such sires, while would punish such sires.
Regression coefficients and were estimated from simulated data. Each of a total of 105 sires was mated to 100 dams, each dam producing a single offspring. GEBV of sires and dams were drawn from a normal distribution, and gametic variances from a log-normal distribution. Subsequently, gametic GEBV of parents were drawn from . GEBV of the offspring were the sum of the gametic GEBV of the sire and dam. In the offspring generation, individuals were selected on GEBV with selected proportion p. Then, , and were calculated for each sire, and the corresponding regression coefficients and were estimated by least squares (Table A1, Table A2, Table A3). Because regression coefficients may depend both on the and on the selected proportion, they were estimated separately for each scenario. Finally, the standardized weight on was calculated as
Table A1. Empirical regression coefficients for the fraction of offspring of the sire that ranked in the top p proportion of GEBVs in the offspring generation .
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10 | 0.15 | 0.20 |
| 0.5 | 0.00 | 0.00 | 0.00 | 0.00 |
| 0.2 | 0.84 | 0.84 | 0.84 | 0.83 |
| 0.1 | 1.25 | 1.28 | 1.30 | 1.32 |
| 0.05 | 1.73 | 1.67 | 1.72 | 1.69 |
| 0.01 | 2.56 | 2.29 | 2.50 | 2.57 |
| 0.005 | 3.05 | 2.51 | 2.71 | 2.81 |
| 0.001 | 3.43 | 3.11 | 3.21 | 3.55 |
Table A2. Empirical regression coefficients for the mean GEBV of the selected offspring of the sire .
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| 0.5 | 2.43 | 2.31 | 2.38 | 2.35 |
| 0.2 | 3.36 | 3.46 | 3.46 | 3.41 |
| 0.1 | 3.63 | 3.05 | 2.96 | 2.90 |
| 0.05 | 2.86 | 2.87 | 2.80 | 2.76 |
| 0.01 | 3.20 | 3.10 | 3.06 | 3.04 |
| 0.005 | 3.37 | 3.06 | 3.23 | 3.21 |
| 0.001 | 3.58 | 3.58 | 3.39 | 3.78 |
This is the value found in dairy cattle Segelke et al. (2014).
Table A3. Empirical regression coefficients for the contribution of a sire to the mean of all selected offspring in the next generation .
| CV of the SD of gametic GEBV | ||||
|---|---|---|---|---|
| p | 0.05 | 0.10a | 0.15 | 0.20 |
| 0.5 | 0.86 | 0.79 | 0.82 | 0.81 |
| 0.2 | 1.11 | 1.12 | 1.11 | 1.11 |
| 0.1 | 1.41 | 1.42 | 1.45 | 1.50 |
| 0.05 | 1.83 | 1.77 | 1.82 | 1.81 |
| 0.01 | 2.62 | 2.34 | 2.55 | 2.65 |
| 0.005 | 3.08 | 2.54 | 2.75 | 2.87 |
| 0.001 | 3.43 | 3.17 | 3.24 | 3.58 |
This is the value found in dairy cattle Segelke et al. (2014).
Appendix B
Theoretical Linear Indices
To facilitate presentation of the derivations of the indices, the order of indices here differs from the main text.
Index 7 is a linear combination of the GEBV of the candidate and the within-family SD in GEBV of its offspring,
and serves to predict the probability that an offspring of the candidate ranks in the top p fraction of GEBV in the offspring generation. The goal is to find and so that I is proportional to this probability. Assuming an approximate normal distribution of the GEBV of the offspring of a candidate, the probability that an offspring of a certain candidate is selected follows from
where τp is the threshold value for the top p fraction of GEBV in the offspring generation, is the mean GEBV of the offspring of this candidate, and σ is the SD in the GEBV of the offspring of this candidate. To find b1 and b2, we linearized this probability in and using partial derivatives. To obtain a single value for b1 and b2, rather than a value specific for each candidate, those partial derivatives are calculated using population averages (i.e., P is substituted by p). This yields
where , and
Using , which is the standard normal density at xp, and yields
Using , and yields
When the candidate has multiple mates, the use of ignores the contribution of variation in GEBV among those mates to the within-family variance. However, as soon as selection is reasonably strong (say p < 0.1), variation in GEBV among the mates will be small (see Discussion), so that the error will be small.
Dividing and by yields
where is the standardized truncation point in the offspring of the candidate. Substituting this term by corresponding population parameter, i.e., using yields Index 7,
Index 5 is obtained by expressing in Equation 7 as a linear function of . Linearizing using a first-order Taylor-series yields . Substitution into Index 7 and dropping constants yields Index 5,
Index 8 predicts the mean GEBV of the selected offspring of a candidate, which is the sum of the mean offspring GEBV before selection and the within-family selection differential,
Dropping the constant and multiplying by two yields Index 8,
Index 6 is obtained by substituting the Taylor-series approximation used for Index 5 into Index 8, and dropping constants,
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11106125.
Communicating editor: W. Valdar
Literature Cited
- Allier A., Lehermeier C., Charcosset A., Moreau L., and Teyssèdre S., 2019a Improving short- and long-term genetic gain by accounting for within-family variance in optimal cross-selection. Front. Genet. 10: 1006. 10.1186/s12711-016-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allier A., Moreau L., Charcosset A., Teyssèdre S., and Lehermeier C., 2019b Usefulness criterion and post-selection parental contributions in multi-parental crosses: application to polygenic trait introgression. G3 (Bethesda). 9: 1469–1479. 10.1186/s12711-016-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battagin M., Gorjanc G., Faux A. M., Johnston S. E., and Hickey J. M., 2016. Effect of manipulating recombination rates on response to selection in livestock breeding programs. Genet. Sel. Evol. 48: 44 10.1186/s12711-016-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett T.J., Rocheford T. R., and Mohammadi M.. 2019. Reimagining maize inbred potential: identifying breeding crosses using genetic variance of simulated progeny. Crop Sci. 59: 1457–1468. 10.2135/cropsci2014.01.0088 [DOI] [Google Scholar]
- Bernardo R., 2014. Genomewide selection of parental inbreds: classes of loci and virtual biparental populations. Crop Sci. 54: 2586–2595. 10.2135/cropsci2014.01.0088 [DOI] [Google Scholar]
- Bijma P., Wientjes Y. C. J., and Calus M. P. L., 2018. Increasing genetic gain by selecting for higher Mendelian sampling variance. Proceeding From World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, pp. 11–47. [Google Scholar]
- Bonk S., Reichelt M., Teuscher F., Segelke D., and Reinsch N., 2016. Mendelian sampling covariability of marker effects and genetic values. Genet. Sel. Evol. 48: 36 10.1186/s12711-016-0214-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman A. C., Hayes B. J., and Calus M. P., 2017. Estimated allele substitution effects underlying genomic evaluation models depend on the scaling of allele counts. Genet. Sel. Evol. 49: 79. 10.1111/1755-0998.12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowicki M., Franssen S. U., and Schlötterer C., 2016. High rates of phasing errors in highly polymorphic species with low levels of linkage disequilibrium. Mol. Ecol. Resour. 16: 874–882. 10.1111/1755-0998.12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M., 1971. The effect of selection on genetic variability. Am. Nat. 105: 201–211. 10.1111/1755-0998.12516 [DOI] [Google Scholar]
- Cochran W. G., 1951. Improvement by Means of Selection. John Hopkins University, Baltimore. [Google Scholar]
- Cole J. B., and Van Raden P. M., 2011. Use of haplotypes to estimate Mendelian sampling effects and selection limits. J. Anim. Breed. Genet. 128: 446–455. 10.1111/j.1439-0388.2011.00922.x [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Hayden M. J., Spangenberg G. C., and Hayes B. J., 2015. Selection on optimal haploid value increases genetic gain and preserves more genetic diversity relative to genomic selection. Genetics 200: 1341–1348. 10.1534/genetics.115.178038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Sorensen D. A., and Toro M. A., 2019. Imperfect linkage disequilibrium generates phantom epistasis (& perils of big data). G3 (Bethesda) 9: 1429–1436. 10.1534/g3.119.400101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druet T., and Georges M., 2015. LINKPHASE3: an improved pedigree-based phasing algorithm robust to genotyping and map errors. Bioinformatics 31: 1677–1679. 10.1093/bioinformatics/btu859 [DOI] [PubMed] [Google Scholar]
- Dudley J. W., 1984a A method of identifying lines for use in improving parents of a single cross 1. Crop Sci. 24: 355–357. 10.2135/cropsci1984.0011183X002400020034x [DOI] [Google Scholar]
- Dudley J. W., 1984b A method for identifying populations containing favorable alleles not present in elite germplasm 1. Crop Sci. 24: 1053–1054. 10.2135/cropsci1984.0011183X002400060011x [DOI] [Google Scholar]
- Falconer D. S., and Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Longman Group, Essex, UK. [Google Scholar]
- Gianola D., de los Campos G., Hill W. G., Manfredi E., and Fernando R., 2009. Additive genetic variability and the Bayesian alphabet. Genetics 183: 347–363. 10.1534/genetics.109.103952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goiffon M., Kusmec A., Wang L., Hu G., and Schnable P., 2017. Improving response in genomic selection with a population-based selection strategy: optimal population value selection. Genetics 206: 1675–1682. 10.1534/genetics.116.197103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., and Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. 10.1534/genetics.107.081190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Visscher P. M., and Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. 10.1017/S0016672308009981 [DOI] [PubMed] [Google Scholar]
- He D., Saha S., Finkers R., and Parida L., 2018. Efficient algorithms for polyploid haplotype phasing. BMC Genomics 19: 110 10.1186/s12864-018-4464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. R., 1975. Best linear unbiased estimation and prediction under a selection model. Biometrics 31: 423–447. 10.2307/2529430 [DOI] [PubMed] [Google Scholar]
- Hill W. G., and Weir B. S., 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 93: 47–64. 10.1017/S0016672310000480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K. E., Bowman P. J., Pryce J. E., Hayes B. J., and Goddard M. E., 2012. Long-term selection strategies for complex traits using high-density genetic markers. J. Dairy Sci. 95: 4646–4656. 10.3168/jds.2011-5289 [DOI] [PubMed] [Google Scholar]
- Kong A., Barnard J., Gudbjartsson D. F., Thorliefsson G., Jonsdottir G. et al. , 2004. Recombination rate and reproductive success in humans. Nat. Gen. 36: 1203. 10.3168/jds.2011-5289 [DOI] [PubMed] [Google Scholar]
- Lehermeier C., Teyssèdre S., and Schön C. C., 2017. Genetic gain increases by applying the usefulness criterion with improved variance prediction in selection of crosses. Genetics 207: 1651–1661. 10.1534/g3.118.200091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian L., Jacobson A., Zhong S., and Bernardo R., 2015. Prediction of genetic variance in biparental maize populations: genomewide marker effects vs. mean genetic variance in prior populations. Crop Sci. 55: 1181–1188. 10.2135/cropsci2014.10.0729 [DOI] [Google Scholar]
- Müller D., Schopp P., and Melchinger A. E., 2018. Selection on expected maximum haploid breeding values can increase genetic gain in recurrent genomic selection. G3 (Bethesda) 8: 1173–1181. 10.1534/g3.118.200091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos D. J. A., Cole J. B., Lawlor T. J. Jr., Van Raden P. M., Tonhati H. et al. , 2019. Variance of gametic diversity and its application in selection programs. J. Dairy Sci. 102: 5279–5294. 10.3168/jds.2018-15971 [DOI] [PubMed] [Google Scholar]
- Schnell F. W., and Utz H. F., 1976. F1 Leistung und Elternwahl in der Zuchtung von Selbstbefruchtern, p. 243–248 in Ber Arbeitstag Arbeitsgem Saatzuchtleiter. BAL Gumpenstein, Gumpenstein, Austria. [Google Scholar]
- Segelke D., Reinhardt F., Liu Z., and Thaller G., 2014. Prediction of expected genetic variation within groups of offspring for innovative mating schemes. Genet. Sel. Evol. 46: 42 10.1186/1297-9686-46-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P., 1980. The distribution of the fraction of the genome identical by descent in finite random mating populations. Genet. Res. 35: 131–155. 10.1017/S0016672300014002 [DOI] [Google Scholar]
- Thompson R., 1977. The estimation of heritability with unbalanced data: ii. data available on more than two generations. Biometrics 33: 497–504. 10.2307/2529364 [DOI] [Google Scholar]
- Van Raden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Van Raden P. M., Freeman A. E., and Rothschild M. F., 1984. Maximizing genetic gain under multiple-stage selection 1. J. Dairy Sci. 67: 1761–1766. 10.3168/jds.S0022-0302(84)81502-7 [DOI] [Google Scholar]
- Woolliams J. A., and Meuwissen T. H. E., 1993. Decision rules and variance of response in breeding schemes. Anim. Sci. 56: 179–186. 10.1017/S0003356100021231 [DOI] [Google Scholar]
- Woolliams J. A., Bijma P., and Villanueva B., 1999. Expected genetic contributions and their impact on gene flow and genetic gain. Genetics 153: 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N. R., and Thompson R., 1990. Prediction of rates of inbreeding in selected populations. Genet. Res. 55: 41–54. 10.1017/S0016672300025180 [DOI] [PubMed] [Google Scholar]
- Zhong S., and Jannink J. L., 2007. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 177: 567–576. 10.1017/S0016672300025180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study. The R-codes used for simulation are available in Supplemental Material, Files S1 through S3. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11106125.