Abstract
Fish are known for the outstanding variety of their sex determination mechanisms and sex chromosome systems. The western (Gambusia affinis) and eastern mosquitofish (G. holbrooki) are sister species for which different sex determination mechanisms have been described: ZZ/ZW for G. affinis and XX/XY for G. holbrooki. Here, we carried out restriction-site associated DNA (RAD-) and pool sequencing (Pool-seq) to characterize the sex chromosomes of both species. We found that the ZW chromosomes of G. affinis females and the XY chromosomes of G. holbrooki males correspond to different linkage groups, and thus evolved independently from separate autosomes. In interspecific hybrids, the Y chromosome is dominant over the W chromosome, and X is dominant over Z. In G. holbrooki, we identified a candidate region for the Y-linked melanic pigmentation locus, a rare male phenotype that constitutes a potentially sexually antagonistic trait and is associated with other such characteristics, e.g., large body size and aggressive behavior. We developed a SNP-based marker in the Y-linked allele of GIPC PDZ domain containing family member 1 (gipc1), which was linked to melanism in all tested G. holbrooki populations. This locus represents an example for a color locus that is located in close proximity to a putative sex determiner, and most likely substantially contributed to the evolution of the Y.
Keywords: Mosquitofish, sex determination, pool sequencing, gipc1, sox9b
TELEOST sex determination is perplexingly variable, ranging from genetic XX/XY, ZZ/ZW, and polygenic to environmentally regulated systems and hermaphroditism (Devlin and Nagahama 2002; Mank et al. 2006; Kikuchi and Hamaguchi 2013). The high diversity of teleosts, with over 26,000 species (Nelson et al. 2016) by far the largest clade of vertebrates, has been extensively studied at many levels of their biological organization, but surprisingly little is known about the genes on the sex chromosomes that determine sex or fulfill sex-linked functions. In only a few teleosts, extensive gene expression analyses, positional cloning, and functional analyses led to the identification of the sex chromosome and the sex-determination locus. Master sex-determining genes of fish were found to have evolved either by allelic diversification on proto-sex chromosomes or by neofunctionalization of autosomal gene duplicates that moved to the prospective sex chromosomal regions (Herpin and Schartl 2015). So far, most identified fish sex determination genes fall into only two groups, which are implicated in the conserved genetic network regulating gonadal development. These genes are either members of the transforming growth factor-β signaling pathway, for instance, anti-Müllerian hormone, its type II receptor, and gonadal soma derived factor, or transcription factors, namely dmrt1 and sox3 (Matsuda et al. 2002; Nanda et al. 2002; Smith et al. 2009; Sutton et al. 2011; Kamiya et al. 2012; Myosho et al. 2012; Eshel et al. 2014; Takehana et al. 2014; Li et al. 2015). As an exception, the sex determination gene of salmonids is a duplicate of an immune regulatory gene that has no known function in sexual development (Yano et al. 2012, 2013). The region linked to sex determination can vary greatly in length and differentiation, ranging from no differentiation at all in fugu (Takifugu rubripes), where a single SNP in amhr2 determines sex, to a 258-kb region in medaka (Oryzias latipes), and a region estimated to several megabases in the guppy Poecilia wingei and sticklebacks (Gasterosteus aculeatus and Pungitius pungitius) (Peichel et al. 2004; Kondo et al. 2006; Shikano et al. 2011; Kamiya et al. 2012; Nanda et al. 2014; White et al. 2015; Dixon et al. 2018; Natri et al. 2019). In most cases, information on the molecular differentiation of the sex chromosomes, the structure and function of sex-linked genes, and sex chromosome evolution is scarce. Only a few studies have compared the sex chromosomes of closely related teleosts, e.g., of different species within the genus Oryzias and Gasterosteus, which highlight that, even within the same genus, different master sex-determining genes or sex-linked chromosomes were frequently recruited (Matsuda et al. 2002; Nanda et al. 2002; Kitano et al. 2009; Ross et al. 2009; Shikano et al. 2011; Myosho et al. 2012; Takehana et al. 2014).
Livebearing teleosts of the family Poeciliidae provide some well-studied, classical examples of genetic sex determination, which were already investigated in the 1920s (Schmidt 1920; Winge 1922, 1927). In this group, sex determination is often tightly linked to sex-specific coloration, as many color loci have been mapped in close proximity to the putative sex-determining loci (reviewed in Kottler and Schartl 2018). Many of these pigmentation loci are thought to be sexually antagonistic, or at least to be advantageous for only one of the sexes (reviewed in Kottler and Schartl 2018). Despite many efforts, neither a poecilid master sex determiner nor a sexually antagonistic pigmentation gene has yet been identified on the molecular level.
The eastern (Gambusia holbrooki) and western (Gambusia affinis) mosquitofish are poeciliids native to the southeastern part of the USA and the northeastern part of Mexico. They were introduced for mosquito control in many freshwater habitats worldwide, where they threaten the native fauna (Hamer et al. 2002; Pyke 2008). Phylogenetic analyses demonstrated that G. holbrooki and G. affinis are sister species, with an estimated divergence time of ∼2–7 MY (Lydeard et al. 1995; Lamatsch et al. 2015; Helmstetter et al. 2016). The natural distribution of the two species overlaps in an area extending from the Southeast of Mississippi (Mobile Bay) to the Northeast of Georgia, but hybridization between the two species is rare, suggesting the occurrence of reproductive barriers (Hubbs 1955; Black and Howell 1979; Wooten et al. 1988; Wooten and Lydeard 1990; Scribner and Avise 1993; Wilk and Horth 2016).
Within G. holbrooki populations, on average 1% of males present a spectacular melanic coloration phenotype that consists of a conspicuous jet-black spotting pattern covering the entire body and fins (Figure 1A) (Horth and Travis 2002; Horth 2003, 2004). This pattern is generated by large, irregularly distributed macromelanocytes (Figure 1B) (nomenclature according to Schartl et al. 2016a; Regan 1961; Angus 1989; Horth 2006). Studies suggest that the melanic color pattern provides an advantage to the males carrying it, as they grow faster, have a larger gonopodium, and show more aggressive behavior toward females and competitors (Martin 1977; Horth 2003; Horth et al. 2010). Although melanic males are very conspicuous, they seem to be less prone to predation than the wildtype, silvery males (Figure 1C) (Horth 2004). Female preference is polymorphic, as female G. holbrooki that are derived from populations with melanic males prefer such males and vice versa (Bisazza and Pilastro 2000). The low numbers of melanic males seem to be stabilized by negative frequency-dependent selection (Horth and Travis 2002; Horth 2003). Melanism in G. holbrooki is a candidate for a sexually antagonistic phenotype (Martin 1977; Horth 2003; Horth et al. 2010), but further studies are required to determine whether the melanic coloration is neutral or would indeed be disadvantageous for females in the sense of a truly sexually antagonistic gene. As a potentially sexually antagonistic color locus, the melanic locus might have contributed to the evolution of the G. holbrooki Y chromosome and its identification is therefore of prime interest (Kottler and Schartl 2018).
Figure 1.
Phenotype of G. holbrooki and G. affinis. (A) Melanic G. holbrooki male (top) and wildtype female (bottom) (WLC2514). (B) Detail of the caudal peduncle of a melanic G. holbrooki male (WLC2514). Large dendritic- (white arrow) and corolla- (white arrowhead) shaped macromelanocytes and smaller melanocytes of the background pattern (yellow arrow) are visible. (C) Wildtype-pigmented G. holbrooki WLC6439 male. (D) G. affinis female (top) and male (bottom) (WLC1313). (E) Detail of the caudal peduncle of a wildtype-pigmented G. holbrooki male (WLC6439); the yellow arrows point to the melanocytes of the background pattern. (F) G. holbrooki male (WLC1475) with pronounced xanthophore-derived pigmentation on the head. All pictures taken under incident light conditions. Bar, (B), (E) 500 μm. Length of males of both species is ∼2–3 and of females 4–6 cm.
Cytogenetic studies revealed that G. holbrooki lacks heteromorphic sex chromosomes (Black and Howell 1979). However, as the melanic macromelanocyte pattern is transmitted from fathers to sons, it has been generalized that all G. holbrooki most likely have an XX/XY sex determination system, in which the male-advantageous melanic locus became linked to the sex-determining locus on the Y chromosome (Angus 1989; Horth 2006). In stark contrast, G. affinis has conspicuous, morphologically differentiated sex chromosomes (Black and Howell 1979). Female G. affinis have a very large W chromosome, suggesting that the sister species of G. holbrooki evolved a ZZ/ZW sex determination mechanism (Black and Howell 1979). G. holbrooki is so far the only mosquitofish in which an XX/XY sex determination system has been postulated, while a distinct W chromosome was found not only in G. affinis, but also in G. nobilis, G. gaigei, and G. hurtadoi females (Campos and Hubbs 1971; Black and Howell 1979; Lydeard et al. 1995). The presence of different sex chromosomal systems makes G. holbrooki and G. affinis excellent model organisms for studying the evolution of sex chromosomes, Y-linked pigmentation, and other sex-linked traits.
We performed restriction-site associated DNA (RAD) and pool sequencing approaches to identify the sex linkage groups of G. holbrooki and G. affinis, revealing independent origins of the XY and ZW systems. We identified a unique allele of GIPC PDZ domain containing family member 1 (gipc1) on the G. holbrooki Y chromosome (gipc1Y), which is linked to the potentially sexually antagonistic melanic phenotype. We also present a detailed description of the melanic phenotype, which is of utmost importance in order to identify reasonable candidate genes that might underlie this phenotype in a gene set. With our newly developed SNP-based marker for the Y chromosome, we demonstrate that the Y chromosome of G. holbrooki is dominant over the W chromosome of G. affinis and the X is dominant over the Z. Our data give insights into the genetics and genomics of the complex evolution of different sex chromosome systems in closely related species.
Materials and Methods
Experimental animals/fish strains
Fish of the genus Gambusia were maintained under standard conditions as described for the related genus Xiphophorus by (Kallman (1975)). Nine G. holbrooki strains (six of which produced melanic individuals) and one G. affinis strain were included in our analyses: G. affinis (WLC1313), geographic origin: Pena blanca Lake, Arizona 31°24’01’’N 111°05’20’’W. G. holbrooki (WLC1475), geographic origin: Tamiami trail, Everglades, Florida. G. holbrooki (WLC1253), geographic origin: Fort Lauderdale, Florida. G. holbrooki (WLC2105), geographic origin: Everglades City, Florida. G. holbrooki (WLC2514), geographic origin: Florida City, Florida 25°23’54.2’’N 80°34’20.2’’W. G. holbrooki (WLC6439), geographic origin: Astouin, Camargue, France. G. holbrooki (WLC5589), substrain established from WLC2514. G. holbrooki (WLC6475), substrain established from WLC2105. G. holbrooki (WLC4598), substrain established from WLC1475. G. holbrooki (WLC6636), established from WLC4598. The observed sex ratio in our strains was 1:1.
Ethanol fixed individuals of wild caught G. holbrooki from Florida were kindly supplied by Ingo Schlupp, University of Oklahoma (Norman, OK). from his collection: ISO8042 (geographic origin: Kissimmee), ISO8043 (geographic origin: Satellite Beach), ISO8046 (geographic origin: Lake Eustis), and ISO8048 (geographic origin: Bulow Creek).
Males of G. holbrooki strains WLC1475, 1253, 2105, and 2514 are melanic, but occasionally nonmelanic males occur. In WLC1475, only a single nonmelanic male besides melanic males and nonmelanic females was recorded since its establishment in 1992. In the other strains, a higher frequency was noted. Nonmelanic males were removed promptly to keep the melanic strains monomorphic. Males of WLC6439 are nonmelanic (wildtype-pigmented). In WLC4598 and WLC6636, melanic pigmentation and sex determination are uncoupled, as both males and females are melanic, although nonmelanic fish occur sometimes. WLC5589 and WLC6475 are F1 fish derived from a cross between a wildtype, nonmelanic female and a nonmelanic male of strain WLC2514 and WLC2105, respectively, resulting in exclusively nonmelanic offspring. The melanic pigmentation of G. holbrooki has been reported to be either sensitive or insensitive to temperature (Angus 1989; Horth 2006). We transferred one WLC2514 and three WLC2105 nonmelanic males to a tank with a water temperature of 21°, a temperature sufficiently low to trigger melanic pigment development in temperature-sensitive strains (Angus 1989). None of them developed any melanic pigmentation after 3 months (data not shown), indicating that our melanic strains are not sensitive to temperature.
All animals were kept and sampled in accordance with the applicable EU and national German legislation governing animal experimentation. In particular, all experimental protocols were approved through an authorization (568/300-1870/13) of the Veterinary Office of the District Government of Lower Franconia, Germany, in accordance with the German Animal Protection Law (TierSchG).
Crosses
All crosses were performed with virgin females by unassisted matings. Pregnant females were kept isolated in 1 liter aquaria with high density plastic wool and controlled daily after the onset of illumination to avoid cannibalism. Offspring were removed and raised separately. Phenotypic sex was determined after puberty. No increased mortality compared to the parental strains was observed.
Imaging of fish and pigment cells
Pictures of whole fish were taken with a Nikon S5 Pro camera with an AF-S DX NIKKOR 18-105 mm 1:3.5-5.6G ED VR lens or with a VH-Z20W zoom lens connected to a VHX-S90BE and VHX-2000D with the OP-87429 polarization illumination attachment. Details of pigment cells were imaged with the VHX-2000D. To document the pigment cell development of fish after birth, the fish were immobilized and photographed with a Leica DFC450 C camera connected to a Nikon SMZ1000 dissecting microscope. The contrast, brightness, and background of some images were adjusted with GIMP 2.10.6.
Transmission electron microscopy
For transmission electron microscopy imaging, small pieces of the caudal peduncles of one melanic and one wildtype WLC2514 male were excised and fixed in a buffered solution containing 2.5% glutaraldehyde (w/v), 0.05 M KCl, and 2.5 mM MgCl2 in 0.05 M cacodylate buffer (pH 7.2) at 4° overnight. The pieces were washed five times with 50 mM cacodylate buffer pH 7.2 for 10 min at 4° and then fixed with 2% (w/v) osmium tetroxide for 3 hr on ice. Afterward, the samples were washed five times with water at room temperature and stained in 0.5% (w/v) aqueous uranyl acetate solution at 4° overnight. The samples were then washed five times with water at room temperature and dehydrated (50%, 70%, 90% ethanol for 30 min at 4°, then three times 100% ethanol for 20 min at room temperature). They were washed two times with propylene oxide for 30 min and embedded in Epon/propylene oxide (1:1) overnight at room temperature. The final embedding was carried out with Epon and polymerized at 60° for at least 48 hr. Ultrathin sections (60–100 nm) along the longitudinal axis were made with a Leica EM UC7 microtome. The samples were inspected with a JEOL JEM-2100 transmission electron microscope at 200 kV; images were taken with TVIPS TemCAM F416 4k × 4k. The skin layers were named according to Kottler et al. (2014).
Adrenaline treatment
To investigate whether the melanocytes of melanic fish react to adrenaline by aggregating their melanosomes, fish were anesthetized and killed by decapitation. Subsequently, the trunk was kept in tank water containing 2.4 mg/ml epinephrine and moved gently. After 10 min, the pigment cells were photographed with the VHX-2000D. WLC2514 and WLC6439 fish were used.
Cytogenetics
Mitotic chromosome preparations of WLC2514 and WLC1313 fish were made directly from pooled spleen, gills, and cephalic kidney cells after exposing the fish for several hours to a 0.02% (w/v) solution of colchicine followed by hypotonic treatment [0.46% (w/v) KCl] and fixation with a mixture of methanol and acetic acid (3:1). Slides were stained with 5% Giemsa solution. C-banding using the standard BSG (barium hydroxide/saline/Giemsa) technique (Sumner 1972) was used to visualize the constitutive heterochromatin. DAPI and mithramycin staining of metaphase chromosomes were performed according to Schmid et al. (2010). To analyze meiotic chromosomes of hybrids, seminiferous tubules were exposed to hypotonic treatment followed by fixation in methanol-acetic acid (3:1) mixture. The meiotic cells from the fixed pieces of seminiferous tubules were mechanically released and allowed to spread on a hot plate in 50% acetic acid. After evaporation of acetic acid, slides were Giemsa stained and subjected to microscopic analysis. WLC2514, WLC1313, and WLC1475 fish were used.
DNA extraction
For RAD and pool sequencing, DNA from fish organs was extracted with phenol/chloroform. Tissue was lysed in 1 ml extraction buffer (0.1 M EDTA pH 8, 0.2% SDS, 0.2 M NaCl) containing 200 μg/ml proteinase K for 3 hr at 80°. Then, 500 μl phenol was added to each sample. After mixing and incubation at room temperature for 10 min, 500 μl chloroform:isoamyl alcohol (24:1) was added. The samples were incubated with occasional inversion at room temperature for 10 min and then centrifuged for 10 min at 5° (5000 × g). The upper layer was transferred to a new tube and one volume chloroform:isoamyl alcohol (24:1) was added. With occasional inverting, the samples were kept at room temperature for 10 min, and subsequently centrifuged for 10 min at 5° (5000 × g). The supernatant was transferred to a glass vial on ice and 2.5 times the sample volume of cold 100% ethanol was carefully added. The DNA was spooled with a glass rod and dissolved in TE buffer, pH 8. The DNA concentration was measured with a Qubit fluorometer (dsDNA BR Assay Kit, Invitrogen, Carlsbad, CA).
For PCR, DNA was extracted from trunk tissue of ethanol-fixed fish according to Reid et al. (2008). Lysis was carried out at 56° overnight.
RAD sequencing
For G. affinis RAD sequencing, DNA from 31 females and 30 males (WLC1313) was used. From G. holbrooki, 26 females and 30 melanic males of WLC2514 were included.
RAD-tag libraries were built from extracted DNA using Sbf1 as a single restriction enzyme, following the standard protocol described in Amores et al. (2011). All resulting libraries were sequenced on one lane of Illumina HiSeq 2500 in single end 150 bp mode. Reads were demultiplexed and quality-controlled using the process_radtags.pl script from the Stacks software version 1.44 (Catchen et al. 2011), with the following parameters: discard reads with low quality scores (-q), remove any reads with an uncalled base (-c), and rescue barcodes and RAD-tags (-r).
Demultiplexed reads were analyzed with the RADSex pipeline (version 0.2.0, Zenodo, doi: 10.5281/zenodo.2616396). The underlying principle of RADSex is to split the demultiplexed reads into groups based on exact sequence match. Therefore, markers created with RADSex are nonpolymorphic, and reads that would be grouped into the same polymorphic marker using standard analyses software are split into multiple markers with RADSex. As a consequence, RADSex markers are only influenced by a single parameter (i.e., minimum depth to consider a marker present in an individual), and markers can be compared between individuals using a straightforward presence/absence analysis. These properties enable easier cross-species comparison than with heavily parameter-driven software like Stacks. To analyze the RAD-Seq reads with RADSex, a table containing the depth of each marker in each individual from the dataset was first generated using the process command with default parameters. Then, the distribution of markers between males and females was computed using the distrib command with the minimum depth to consider a marker set to 10 (–min-cov 10), and the resulting distribution was plotted with the R package RADSex-vis. Sequences of markers significantly associated with sex were obtained with the distrib command also using a minimum depth of 10 (–min-cov 10). Markers were aligned to the poeciliid reference genome of the platyfish (Xiphophorus maculatus) [NCBI accession number: GCA_002775205.2; improved version of the genome described in Schartl et al. (2013)] with the map command using a minimum depth of 10 (–min-cov 10) and manhattan plots were generated with RADSex-vis. The manhattan plots showed -log10(passociation with sex) from a Chi-squared test on the number of males and number of females in which a marker is present for G. affinis/G. holbrooki markers aligned to the genome of X. maculatus. The Xiphophorus maculatus genome was used, as Xiphophorus is the closest relative of Gambusia, for which chromosome information is available. Xiphophorus and Gambusia have the same number of chromosomes.
Pool sequencing
For Pool-seq library preparation, individual gDNAs were pooled in equimolar ratios by sex (Supplemental Material, Table S1). Pool-seq libraries were prepared following Illumina’s protocols using the Illumina TruSeq Nano DNA HT Library Prep Kit (Illumina, San Diego). Briefly, 200 ng of each gDNA pool (pools of males and females) was fragmented to 550 bp by sonication with a M220 Focused-ultrasonicator (Covaris, Woburn, MA). Size selection was performed using SPB beads and the 3′ ends of blunt fragments were adenylated. Then, adaptors and indexes were ligated and the construction was amplified with Illumina-specific primers. Library quality was assessed using a Fragment Analyzer (Advanced Analytical Technologies, Ames) and libraries were quantified by qPCR using the Kapa Library Quantification Kit (Roche, Basel, Switzerland). Sequencing was performed on a NovaSeq S4 Lane (Illumina, San Diego, CA) using a paired-end read length of 2 × 150 nt with Illumina NovaSeq Reagent Kits. Sequencing produced 121 and 136 million paired reads for the male and female pooled libraries in G. holbrooki, respectively, and 94.7 and 94.2 million paired reads for the male and female pooled libraries in G. affinis, respectively.
For each species, reads from the male and female pools were aligned to the genomes of Xiphophorus maculatus [NCBI accession number: GCA_002775205.2; improved version of the genome described in Schartl et al. (2013)] and G. affinis (NCBI accession number: NHOQ01000000; Hoffberg et al. 2018) using BWA mem with default parameters (version 0.7.17; Li 2013). The resulting BAM files were sorted with Samtools sort (version 1.8; Li et al. 2009) with default parameters and PCR duplicates were removed with Picard tools MarkDuplicates (version 2.18.2) with the parameter REMOVE_DUPLICATES = true. A pileup file combining male and female pools was generated from the resulting BAM files using Samtools mpileup with minimum base quality set to 0 (-Q 0), and then converted to a sync file using popoolation2 (version 1201; Kofler et al. 2011) with minimum mapping quality set to 20 (–min-qual 20). This sync file was analyzed with PSASS (version 2.0.0, Zenodo, doi: 10.5281/zenodo.2615936, available at https://github.com/RomainFeron/PSASS), with parameters –range-het 0.1, –range-hom 0.01, –window-size 100000, and –output-resolution 100,000, to compute three metrics in 100-kb nonoverlapping windows over the genome: (1) FST between males and females over the window, (2) number of sex-specific SNPs, defined as SNPs with a frequency between 0.4 and 0.6 in one sex and a frequency higher than 0.99 in the other sex, and (3) reads depth for each sex. We filtered for a minimum depth of 10. We also carried out the same analysis with a 10 kb window size. Results were plotted with the R package PSASS-vis (version 2.0.0, available at https://github.com/RomainFeron/PSASS-vis).
Genes within the regions of interest were determined using the platyfish and G. affinis reference genomes. In platyfish, the genes in the regions of interest were determined with the NCBI Genome Data Viewer (https://www.ncbi.nlm.nih.gov/genome/gdv/). In G. affinis, the regions of interest were extracted from the reference genome and aligned against the NCBI nr/nt databank using the discontiguous megablast algorithm (Altschul et al. 1990) to identify potential candidate genes.
Genome assembly of a melanic G. holbrooki male
A melanic WLC2514 G. holbrooki male genome was assembled from 250 bp long Illumina paired-end reads using DiscovarDeNovo (reference https://software.broadinstitute.org/software/discovar/blog/) with standard parameters.
Sequence alignments
DNA and protein alignments of candidate genes were carried out with Clustal Omega (1.2.4) (Sievers et al. 2011). GenBank accession numbers: XP_007572354.1 (Amazon molly (Poecilia formosa) Gipc1); XP_007560319.1 (Amazon molly Sox9a); XP_007556425.2 (Amazon molly Sox9b); XP_025001585.1 (chicken (Gallus gallus) Gipc1); BAA25296.1 (chicken Sox9); XP_008436174.1 (guppy (Poecilia reticulata) Sox9a); NP_001284373.1 (guppy Sox9b); NP_005707.1 (human (Homo sapiens) Gipc1); CAA86598.1 (human Sox9); XP_023813268.1 (medaka (Oryzias latipes) Gipc1); AAX62152.1 (medaka Sox9a); AAX62151.1 (medaka Sox9b); XP_014866717.1 (Atlantic molly (Poecilia mexicana) Gipc1); NP_061241.1 (mouse (Mus musculus) Gipc1); NP_035578.3 (mouse Sox9); XP_003442153.1 (Nile tilapia (Oreochromis niloticus) Gipc1); XP_005448042.1 (Nile tilapia Sox9a); XP_003450167.1 (Nile tilapia Sox9b); XP_005801543.1 (platyfish (Xiphophorus maculatus) Gipc1); XP_005807407.1 (platyfish Sox9a); XP_005801494.1 (platyfish Sox9b). Accession numbers of G. affinis sequences: sox9b ORF, NHOQ01002125.1:61568-61995,62336-62583,62831-63594; sox9b protein sequence, PWA19450.1; gipc1 ORF, NHOQ01002357.1: 1316372-1316665,1316740-1316925,1317062- 1317242,1318142-1318254,1318339-1318420, 1318497-1318640,1318740-1318819,1319648- 1319671 [shortened because probably not correctly assembled (does not end with stop codon)]; gipc1 protein sequence, PWA17858.1 (shortened in accordance with ORF sequence).
Phylogenetic analysis
The phylogenetic tree was built with MEGA7.0.26 (Kumar et al. 2018) according to Hall (2013). The sox9a and sox9b ORF sequences were aligned with MUSCLE using the default settings. The Maximum Likelihood tree was calculated using the Tamura-Nei model with the partial deletion option (Tamura and Nei 1993). Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.3546)]. Fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position; 1000 replicates of bootstrapping were carried out. The tree was rooted on mouse sox9. GenBank accession numbers: AY870394.1 (medaka sox9a), AY870393.1 (medaka sox9b), XM_005807350.2 (platyfish sox9a), XM_005801437.2 (platyfish sox9b), XM_008437952.2 (guppy sox9a), NM_001297444.1 (guppy sox9b), XM_007560257.2 (Amazon molly sox9a), XM_007556363.2 (Amazon molly sox9b), XM_005447985.2 (Nile tilapia sox9a), XM_003450119.4 (Nile tilapia sox9b), NM_011448.4 (mouse sox9).
Preparation of cDNA
To prepare cDNA of different Gambusia strains, total RNA was extracted from skin tissues using TRIzol (Invitrogen) according to the manufacturer’s instructions. After DNase I (Thermo Fisher Scientific, Waltham, MA) treatment, cDNA was prepared with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. First strand cDNA was directly used for PCR.
Gipc1 and sox9b PCRs and sequencing
To amplify gipc1 and sox9b from cDNA of different Gambusia strains, 5′ and 3′ UTR primers were used (gipc1: forward 5′-CTTCCTAGAGCGGCGTCCAC-3′, reverse 5′-AGTGTTAACACCCGCACAGG-3′; sox9b: forward 5′-TTCAGTGCGAGCAGATAGTC-3′, reverse 5′-AGTAGTCTTGGGTGGATGTGTTG-3′). Q5 High-Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA) was used for amplification, which was carried out according to the manufacturer’s instructions with an annealing temperature of 62° and an elongation time of 1 min. The PCR products were visualized by gel electrophoresis, purified with the GenElute PCR Clean-Up Kit (Sigma-Aldrich, St. Louis, MO), and sequenced by a sequencing provider (Eurofins, Ebersberg, Germany). For cloning, a TA-overhang was added to the PCR products using a Taq DNA polymerase. Subsequently, the PCR products were cloned into pGEM-T Easy vector (Promega, Madison, WI) following the manufacturer’s instructions. Plasmid DNA was extracted with the GenElute HP Plasmid Miniprep Kit (Sigma-Aldrich) and sequenced by Eurofins. Sequences were inspected with SnapGene Viewer Version 4.2.11.
For genotyping the genomic DNA of 71 fish from different strains (Table S2), SNP Pol (high discrimination of single nucleotides) DNA polymerase (Genaxxon, Ulm, Germany) was used with primers specific for gipc1Y (forward 5′-GCTCCAGGTCGGGCTCAGC-3′, reverse 5′-AGCTCGTCCACGGTGGCGA-3′) and sox9bY (forward 5′-ACCACCCTTGGTGGAGCAGC-3′, reverse 5′-GAAGAGGGGCTGTAGTGCTGTAA-3′). The PCRs were carried out according to the manufacturer’s instructions with an annealing temperature of 62° and an elongation time of 30 s. PCR products were inspected by gel electrophoresis.
SNP Pol DNA polymerase and gel electrophoresis were used as well to distinguish between G. holbrooki sox9bX (primers: forward 5′-GCAGCGAAGTCATCTCCAACATC-3′, reverse 5′-GCTCTCCTGCTCCACCAAGGT-3′), sox9bY (primers: forward 5′-GCTGCGAAGTCATCTCCAACATC-3′, reverse 5′-GCTCTGCTGCTCCACCAAGGG-3′), and G. affinis sox9b (primers: forward 5′-AGCAGCGAAGTCATCTCCAACATT-3′, reverse 5′-GCTCTCCTGCTCCACCAAGGC-3′; based on a resequencing of sox9b of G. affinis strain WLC1313).
PCR using DNA of F1 hybrids to determine sex chromosome genotypes was carried out using the sox9bY primers [forward 5′-GCTGCGAAGTCATCTCCAACATC-3′, reverse 5′-GCTCTGCTGCTCCACCAAGGG-3′; SNP Pol DNA polymerase PCR protocol (Genaxxon, Ulm, Germany)] and the amtW primers [Gaf88 (Lamatsch et al. 2015); standard Taq DNA polymerase PCR protocol].
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Figure S1 presents the results of the RAD-sequencing analysis. Figure S2 shows the analysis of G. affinis and G. holbrooki pooled-sequencing reads aligned to the genome of X. maculatus using 10 kb windows. Figure S3 represents a molecular phylogeny of fish sox9. Figure S4 are alignments of male G. holbrooki and G. affinis gipc1 and sox9b alleles and protein sequences. Figure S5 are multiple species alignments of Gipc1 and Sox9 protein sequences. Figure S6 contains PCR results demonstrating that sox9bY was generated by allelic diversification. Figure S7 contains pictures of melanic and nonmelanic G. holbrooki. Table S1 gives information on the fish used for pool sequencing. Table S2 presents the results of the allele-specific PCRs. Table S3 contains the annotated loci within the 100 kb windows with the highest density of sex-specific SNPs. Table S4 contains the sex chromosome genotypes of F1 fish (pedigree WLC20074) from a cross between a G. affinis female (WLC1313) and a G. holbrooki male (WLC2514) determined by PCR. Table S5 gives information on the number of offspring of nonmelanic females and nonmelanic males derived from melanic strains.
RAD-seq and pool sequencing data are available at Sequence Read Archive, grouped under the same Bioproject (accession number: PRJNA563257). The genome assembly of a melanic G. holbrooki male is available at NCBI, accession number VOPI00000000. Gipc1 and sox9b cDNA sequences are available at GenBank (accession numbers: MK721001-MK721004). Supplemental material available at figshare: https://doi.org/10.25386/genetics.7957376.
Results
Sex chromosomes are not heterochromatic in G. affinis
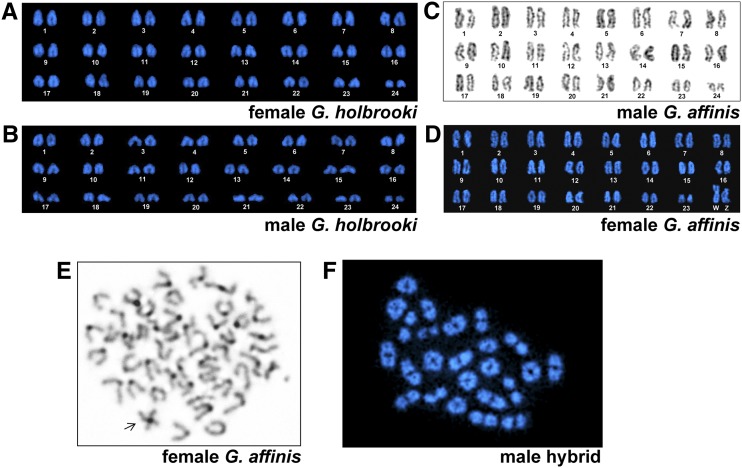
A previous study described a large W chromosome in the karyotype of female G. affinis, but found no heteromorphic sex chromosomes in G. holbrooki (Black and Howell 1979). To confirm this for our study material, we carried out cytogenetic analyses of female and male G. affinis and G. holbrooki. Both species have 2n = 48 chromosomes (Figure 2) (Black and Howell 1979). In accordance with this earlier study, no heteromorphic sex chromosomes were discernible in G. holbrooki male and female chromosome spreads, while female G. affinis had a large W chromosome (Figure 2, A–D). The W chromosome is the largest and only biarmed chromosome of the karyotype (Figure 2D). Despite the considerable morphological differentiation in size, C-banding of female metaphases revealed that the W is not heterochromatic except for its centromeric region (Figure 2E). This indicates that evolution of the W did not develop through canonical heterochromatinization.
Figure 2.
Cytogenetic analyses in the eastern (Gambusia holbrooki) and western (G. affinis) mosquitofish. (A) DAPI-stained female karyotype of G. holbrooki (WLC2514). (B) DAPI-stained male karyotype of G. holbrooki (WLC2514). No distinct heteromorphic sex chromosomes can be identified in the karyotype. (C) Giemsa-stained karyotype of male G. affinis (WLC1313). (D) DAPI-stained karyotype of female G. affinis (WLC1313) displaying distinguishable heteromorphic sex chromosomes (ZW). (E) C-banding of female metaphase of G. affinis (WLC1313) showing a large W chromosome (arrow). (F) DAPI stained metaphase I from a male F1 hybrid of a G. affinis female (WLC1313) and a melanic G. holbrooki male (WLC1475) revealing the expected number of paired bivalents. The chromosomes in (A–D) are arranged based on their size.
The Y and W chromosome correspond to different linkage groups
The chromosomes of F1 hybrids between G. affinis females and G. holbrooki males form 24 bivalents during meiosis, which suggests that the genomes of the two species are highly homologous (Figure 2F). To identify the sex linkage groups of G. holbrooki and G. affinis, we first analyzed the RAD sequencing data from each species using RADSex.
In G. affinis, 433 markers were significantly associated with phenotypic sex (P < 0.05, Chi-squared test with Bonferroni correction), 397 of which (92%) were present in all females and absent from all males, confirming that this species has a ZZ/ZW sex determination system (Figure S1A). In G. holbrooki, 309 markers were significantly associated with phenotypic sex (P < 0.05, Chi-squared test with Bonferroni correction), 195 of which (63%) were present in all males and absent from all females, confirming that this species has an XX/XY sex determination system (Figure S1B). When aligning the significant RADSex markers from G. affinis to the genome of the platyfish, which is closely related to Gambusia and has the same number of chromosomes (Walter et al. 2004; Hrbek et al. 2007), 337 markers (78%) were aligned to LG1 (NC_036443.1) (Figure S1C), suggesting that this linkage group is homologous to the sex chromosome in the genome of G. affinis. The platyfish genome was used, as the current G. affinis genome does not contain chromosome information. The same analysis for G. holbrooki revealed 249 markers (81%) significantly associated with sex were aligned to LG16 of X. maculatus (NC_036458.1) (Figure S1D), suggesting that LG16 is homologous to the sex chromosome in the genome of G. holbrooki. Overall, these results show that, despite being sister species, G. affinis and G. holbrooki have different sex determining systems and different sex chromosomes.
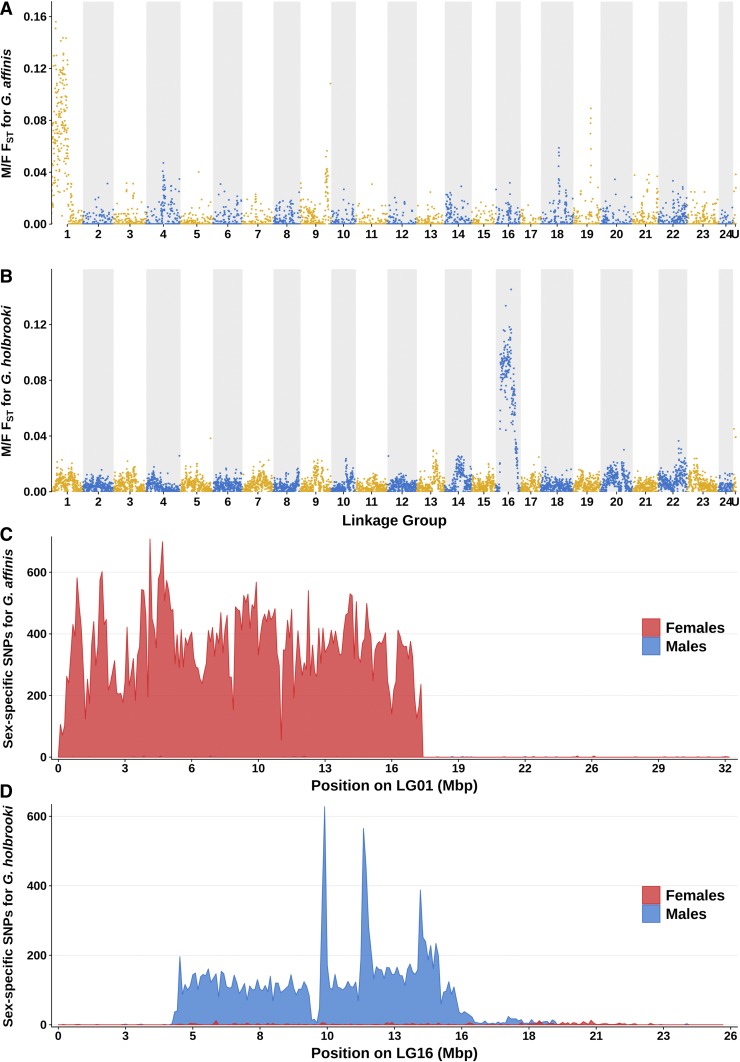
To confirm the results from the RAD sequencing, and to identify differences between the sex chromosomes at a higher resolution, we generated pool sequencing data for a pool of at least 30 males and a pool of at least 30 females of each species (Table S1) and aligned the reads to the genome of the platyfish and the genome of G. affinis. The resulting BAM files were analyzed with PSASS to output male/female FST, number of sex-specific SNPs, and reads depth for each sex in 100 kb nonoverlapping windows over the genomes. In the following, we focus on the mapping results against the X. maculatus genome, as no chromosome information was available for the G. affinis genome.
Male/female FST was highest on platyfish LG1 for G. affinis (Figure 3A) and highest on platyfish LG16 for G. holbrooki (Figure 3B), confirming the results from the RAD sequencing analysis. The FST was computed for the respective entire 100 kb window. Using a 10 kb window size did yield similar results (Figure S2). In G. affinis, we identified 64,290 female-specific SNPs (FSS) and 827 male-specific SNPs (MSS). Female-specific SNPs are only found in females and male-specific SNPs only in males. Among the FSS, 63,323 (98%) were aligned to LG1 of X. maculatus in a region spanning from 0 to 17.7 Mb (Figure 3C), suggesting that this region is homologous to a highly differentiated region in the sex chromosome pair of G. affinis. In G. holbrooki, we identified 1581 FSS and 17,231 MSS, among which 15,956 (93%) were aligned to LG16 of X. maculatus in a region spanning from 4.31 to 16.1 Mb (Figure 3D), suggesting that this region is homologous to a highly differentiated region in the sex chromosome pair of G. holbrooki. While the distribution of FSS in G. affinis was homogeneous in the region on LG1 (Figure 3C), there were three peaks of MSS in G. holbrooki located within the region of LG16 ∼10.2, 11.9, and 14 Mb, suggesting that the regions homologous to these locations have a higher male/female differentiation on the sex chromosomes of G. holbrooki (Figure 3D). The FSS and MSS that did not align to LG1 and LG16, respectively, were not clustered on any other linkage group.
Figure 3.
Analysis of G. affinis and G. holbrooki pooled-sequencing reads aligned to the genome of X. maculatus. Male/female FST was computed in nonoverlapping 100 kb windows from G. affinis (A) and G. holbrooki (B) pooled-sequencing reads aligned to X. maculatus genome. Odd linkage groups (LGs) are colored in yellow with light background, and even LGs are colored blue with dark background. For G. affinis, a large region at the first half of LG1 shows a high male/female FST compared to the rest of the genome, while for G. holbrooki, a large region in the center of LG16 has the highest male/female FST. These results indicate that around half of LG1 of X. maculatus is homologous to the sex chromosome of G. affinis, while half of LG16 of X. maculatus is homologous to the sex chromosome of G. holbrooki. The number of female-specific SNPs (FSS, in red) and male-specific SNPs (MSS, in blue) was similarly computed, and results are shown for LG1 for G. affinis (C) and LG16 for G. holbrooki (D). For G. affinis, a region spanning from 0 to 17.7 Mb on LG1 was strongly and homogeneously enriched in FSS compared to the rest of the genome, while for G. holbrooki, the region enriched in MSS was located between 4.31 and 16.1 Mb on LG16, and showed three peaks of MSS density ∼10.2, 11.9, and 14 Mb.
Taken together, these results show a ZZ/ZW sex-determining system with a 17.7 Mb sex-differentiated region homologous to the first half of LG1 of X. maculatus in G. affinis, and an XX/XY sex-determining system with an 11.8 Mb region homologous to the middle part of LG16 of X. maculatus in G. holbrooki.
A Y-specific allele of GIPC PDZ domain containing family member 1 (gipc1) is linked to G. holbrooki melanic pigmentation
To identify candidate genes for Gambusia sex determination and pigmentation based on published literature on gene function, we investigated the annotated loci within the platyfish and G. affinis genomic areas with the highest density of female- or male-specific SNPs from our pool sequencing analysis. In G. affinis, the 100 kb window with the peak density of female-specific SNPs (708 SNPs when aligned to the platyfish genome and 1058 SNPs when aligned to the G. affinis reference genome assembly) contains two annotated miRNAs, a lncRNA, and piggyBac transposable element-derived protein 3-like (pgbd3-like) in platyfish and a lncRNA and gata5-like in G. affinis (Table S3). The distance between pgbd3- and gata5-like is ∼87 kb in the platyfish genome. In the G. affinis genome, pgbd3- and gata5-like are not located on the same scaffold. In a previous study, a female-specific G. affinis marker in the 3′UTR of aminomethyl transferase (amt) was discovered (Lamatsch et al. 2015). Platyfish amt is located ∼4.5 Mb downstream of pgbd3-like on chromosome 1. In the G. affinis genome, amt and pgbd3-like are located on separate scaffolds. Based on the available information, it is not possible to further narrow down a region that may harbor a female sex-determining gene.
In G. holbrooki, three distinct male-specific SNP peaks are clearly visible on LG16 (Figure 3D). The window containing the highest peak (628 SNPs when aligned to the platyfish and 683 SNPs when aligned to the G. affinis reference genome) spans 10 loci in the platyfish and 8 in the G. affinis genome, among them gipc1 (also named synectin, neuropilin-1-interacting protein, or GAIP interacting protein, C terminus) (Table S3). Gipc1 interacts with other proteins by its peptide-binding PDZ domain and is involved in vesicular trafficking, thereby affecting, for instance, blood vessel and nervous system formation (De Vries et al. 1998; Cai and Reed 1999; Chittenden et al. 2006; Naccache et al. 2006; Varsano et al. 2006; Valdembri et al. 2009; Sanchez et al. 2011; Katoh 2013). Gipc1 might also play a role in the sorting of tyrosine-related protein 1 (Tyrp1) to the melanosomes, as it binds the cytoplasmic tail of this protein (Liu et al. 2001). Tyrp1 is required for pigment synthesis and melanosome formation in fish (Braasch et al. 2009; Krauss et al. 2014).
The second highest peak (565 and 546 SNPs, respectively) contains only one locus, the transcription factor SRY-related HMG box9-like (sox9-like), in both platyfish and G. affinis (Table S3). Phylogenetic analysis revealed that this gene corresponds to the teleost paralog sox9b (Figure S3). Sox9, the main target of the mammalian sex-determining gene SRY, is crucial for testis formation, making this gene an excellent candidate for G. holbrooki sex determination (Wagner et al. 1994; Sekido and Lovell-Badge 2008; Capel 2017; Gonen et al. 2018). Sox9 is also expressed in neural crest cells and required for the development of neural crest cell derivates like bone and cartilage cells (Wagner et al. 1994; Bi et al. 1999; Yan et al. 2005). Moreover, it has been shown that Sox9 plays a role in melanocyte differentiation and melanoma formation (Passeron et al. 2007; Shakhova et al. 2015). Within the third peak, three loci were found, none of which, to our knowledge, has been implicated in sex determination or pigmentation so far (Table S3).
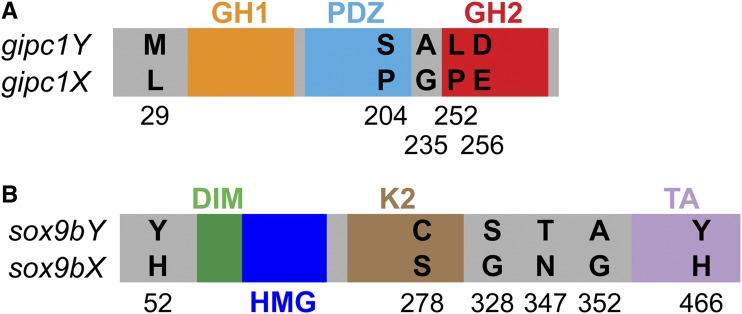
As gipc1 and sox9b are potential candidate genes for pigmentation and sex determination, we further investigated both genes, first utilizing our preliminary genome assembly of a melanic G. holbrooki WLC2514 male. In the genome assembly, two alleles of both genes were present. The two gipc1 alleles differ in six positions within the open reading frame (ORF), translating into five potential amino acid changes, one of which is located in the PDZ and two in the GH2 domain (Figure 4A and Figure S4, A and B). The sox9b alleles have 10 variable positions in the ORF, leading to six nonsynonymous amino acid changes (Figure 4B and Figure S4, C and D). The transactivation domains K2 and TA each contain one nonconservative change (Figure 4B and Figure S4D). Multiple species alignments revealed which of the alleles are most likely Y-linked, as the putative Y-linked proteins contain rare or unique amino acid changes (Figure S5). The amino acids at position 204, 235, 252, and 256 are fully conserved in all Gipc1 protein sequences from fish to human except for Gipc1Y (Figure S5A). In Sox9bY, the H > Y at position 52 and 466 are unusual, as the histidine at position 52 is conserved in all Sox9b sequences and the one at position 466 is present in all of the investigated fish Sox9a/b sequences, as well as in human, mouse, and chicken Sox9 (Figure S5B). Moreover, position 278 is a serine in almost all of the sequences except for Nile tilapia Sox9b and G. holbrooki Sox9bY (Figure S5B). A glycine is conserved in all fish Sox9a and Sox9b sequences at position 352, but not in Sox9bY (Figure S5B).
Figure 4.
Differences between the X- and Y-encoded Gipc1 and Sox9b proteins found in a melanic WLC2514 male. (A) Amino acid differences between Gipc1Y and Gipc1X; domains according to (Katoh 2013). (B) Amino acid differences between Sox9bY and Sox9bX; domains according to Jo et al. (2014). A PQA (proline-, glutamine- and alanine-rich) domain is not present in mosquitofish Sox9b. DIM, dimerization domain; GH1, GIPC homology one domain; GH2, GIPC homology two domain; HMG, high mobility group domain; K2, transactivation domain; PDZ, postsynaptic density-95/Discs large/zona occludens-1 domain; TA, transactivation domain. Protein and domains are not drawn to scale.
To confirm these results, we designed primers in the 5′ and 3′ untranslated regions of both genes based on our genome assembly. We then amplified gipc1 and sox9b from cDNA prepared from skin tissue of six individuals from different G. holbrooki strains and directly sequenced the PCR products. This allowed us to investigate whether the fish were heterozygous at the sites predicted in our genome assembly.
We found that the predicted six heterozygous sites between gipc1X and gipc1Y were present in all melanic fish, suggesting that gipc1Y might be closely linked to the melanic locus (Table 1). They were not linked to being male, as the heterozygous sites could not be found in the wildtype WLC6439 male sequence (Table 1).
Table 1. SNPs at predicted heterozygous sites detected by direct sequencing in gipc1 G. holbrooki.
| Gipc1 | SNPs at predicted heterozygous sitesa | |||||||
|---|---|---|---|---|---|---|---|---|
| 85 | 610 | 702 | 704 | 755 | 768 | |||
| gipc1X (genome prediction) | T | C | G | G | C | G | ||
| gipc1Y (genome prediction) | A | T | A | C | T | C | ||
| predicted amino acid change X > Y | L > M | P > S | silent | G > A | P > L | E > D | ||
| Strain | Sex | Phenotype | ||||||
| WLC2514 | male | melanic | T/A | C/T | G/A | G/C | C/T | G/C |
| WLC2105 | male | melanic | T/A | C/T | G/A | G/C | C/T | G/C |
| WLC1475 | male | melanic | T/A | C/T | G/A | G/C | C/T | G/C |
| WLC4598 | female | melanic | T/A | C/T | G/A | G/C | C/T | G/C |
| WLC6439 | male | wildtype | T | C | G | G | C | G |
| WLC2514 | female | wildtype | T | C | G | G | C | G |
Positions according to G. holbrooki gipc1 ORF sequences (Figure S4A); SNPs predicted by genome assembly (see Figure S4)
In sox9b, no heterozygous sites fully associated with sex or melanic pigmentation were found (Table 2). The melanic WLC2514, WLC2105, and WLC1475 males shared the heterozygous sites found in the preliminary genome assembly of the melanic WLC2514 male, except for the silent SNP at position 513 (Table 2). The melanic WLC4598 female, the wildtype female from the Florida population (WLC2514), and the wildtype-pigmented male from the European population (WLC6439) did not have these heterozygous sites (Table 2). The sequences of the gipc1 and sox9b alleles of the melanic WLC2514 male were confirmed by cloning the PCR products and sequencing (GenBank accession numbers: MK721001-MK721004).
Table 2. SNPs at predicted heterozygous sites detected by direct sequencing in sox9b G. holbrooki ORFs.
| Sox9b | SNPs at predicted heterozygous sitesa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 154 | 364 | 513 | 675 | 832 | 982 | 1040 | 1055 | 1174 | 1396 | |||
| sox9bX (genome prediction) | C | C | C | G | A | G | A | G | C | C | ||
| sox9bY (genome prediction) | T | T | T | T | T | A | C | C | T | T | ||
| predicted amino acid change X > Y | H > Y | silent | silent | silent | S > C | G > S | N > T | G > A | silent | H > Y | ||
| Strain | Sex | Phenotype | ||||||||||
| WLC2514 | male | melanic | C/T | C/T | C/T | G/T | A/T | G/A | A/C | G/C | C/T | C/T |
| WLC2105 | male | melanic | C/T | C/T | T | G/T | A/T | G/A | A/C | G/C | C/T | C/T |
| WLC1475 | male | melanic | C/T | C/T | T | G/T | A/T | G/A | A/C | G/C | C/T | C/T |
| WLC4598 | female | melanic | C | C | T | G | A | G | A | G | C | C |
| WLC6439 | male | wildtype | C | C | T | G | A | G | A | G | C | C |
| WLC2514 | female | wildtype | C | C | C | G | A | G | A | G | C | C |
Positions according to G. holbrooki sox9b ORF sequences (Figure S4C); SNPs predicted by genome assembly (see Figure S4).
Next, we designed primers specific for gipc1Y and sox9bY and carried out allele-specific PCRs using DNA from 71 fish from different strains as template (Table S2). Gipc1Y was identified in all melanic fish, while sox9bY was found in 25 of 28 investigated melanic individuals (Table S2). The three melanic fish lacking sox9bY were melanic females, a rare incidence in G. holbrooki (Angus 1989). Gipc1Y and sox9bY were not detected in any of the wildtype-pigmented fish (Table S2).
To find out if sox9bY is a Y-specific duplicate of sox9bX, we designed primers that distinguish between G. holbrooki sox9bX, sox9bY, and G. affinis sox9b. PCRs using DNA from F1 individuals of a cross between a G. affinis female and a melanic G. holbrooki male as template revealed that melanic F1 males have the G. affinis sox9b and G. holbrooki sox9bY, but not sox9bX (Figure S6). This suggests that sox9bY was generated by allelic diversification, not duplication, of sox9b (Figure S6). Such an analysis was not possible for gipc1, as no suitable primers could be designed.
The Y-linked macromelanocyte pattern of G. holbrooki is composed of a divergent pigment cell lineage
G. affinis females and males are grayish silvery, as are most G. holbrooki fish (Figure 1, A, C, and D). The uniform pigmentation is caused by melanocytes (Figure 1E), iridophores, and xanthophores, which are distributed over the whole body. In melanic G. holbrooki, macromelanocytes are superimposed on this background pattern (Figure 1B). Some macromelanocytes have a compact shape, while others are highly dendritic (Figure 1B). Compared with the background melanocytes, they are considerably darker, and, with an average diameter of 167 μm (up to 300 μm), almost twice as large. Transmission electron microscopy imaging revealed that the macromelanocytes are located in the dermis and hypodermis, where they are frequently associated with iridophores and xanthophores (Figure S7A). Their melanosomes show the same structure as those of the normal melanocytes (Figure S7, A–D). The first macromelanocytes are usually already discernible 1 or 2 days after birth, and quickly increase in size (Figure S7, E–H). At older age, some melanic males develop pronounced yellow areas, which are usually located on the head and belly (Figure 1F).
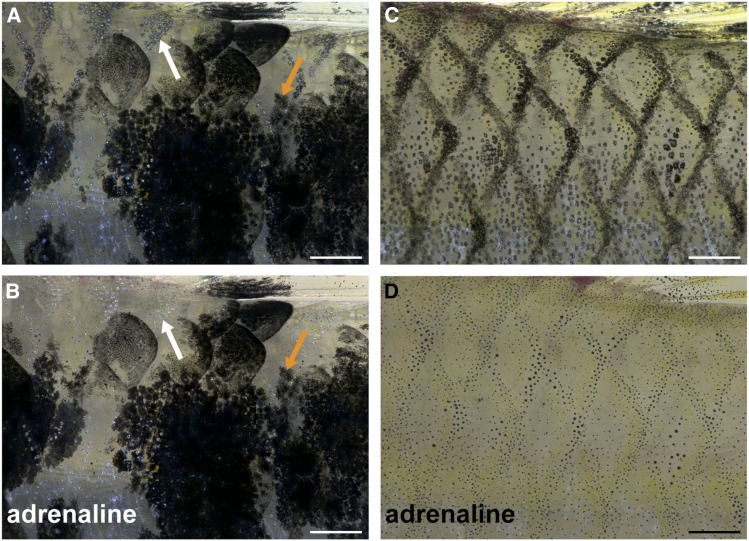
To investigate whether macromelanocytes can aggregate their melanosomes like normal melanocytes, we treated melanic and nonmelanic (wildtype) fish with epinephrine (adrenaline). While the background melanocytes of wildtype and melanic males aggregated their melanosomes upon exposure to epinephrine, the macromelanocytes did not change their appearance (Figure 5). As epinephrine is a potent physiological substance to elicit melanosome contraction, we conclude that the macromelanocytes in contrast to the background melanocytes are compromised with respect to this function. This information is crucial in order to identify potential candidate loci underlying this phenotype, e.g., gipc1. Gipc1 affects a protein involved in melanosome development (Liu et al. 2001).
Figure 5.
Effect of adrenaline on melanocytes and macromelanocytes of G. holbrooki. Skin from a melanic male (WLC2514) before (A) and after (B) treatment with adrenaline. The melanosomes of the background melanocytes (white arrows) aggregate, while those of the macromelanocytes do not (orange arrows). Wildtype, nonmelanic male (WLC6439) skin before (C) and after (D) treatment with adrenaline. The melanocytes aggregate their melanosomes. Pictures were taken under incident light conditions below the dorsal fin. Bar, 500 μm.
G. holbrooki Y is dominant over G. affinis W and X is dominant over Z
To determine dominance relationships among sex chromosomes, we set up reciprocal crosses between G. affinis and G. holbrooki. All crosses resulted in viable and fertile offspring. The brood size of the interspecific crosses and backcross matings was comparable to the brood size of the purebred parental species.
In platyfish, the W chromosome is dominant over the Y chromosome, as WY fish develop as females (Kallman 1973; Volff and Schartl 2002). If, like in platyfish, the G. affinis W chromosome would be dominant over the G. holbrooki Y chromosome, we would expect 25% WY melanic females, 25% WX nonmelanic (wildtype) females, 25% ZY melanic males, and 25% ZX nonmelanic (wildtype) males in crosses between ZW G. affinis females and XY melanic G. holbrooki males (note that the W and Y do not correspond to the same linkage group). Deviating from this expectation, only one melanic F1 female and 280 melanic F1 males were found in a total of 568 F1 fish (Table 3). This suggests that almost all WY F1 fish become males. Support for this comes from the cytogenetic analysis, which revealed that melanic F1 males had two different karyotypes: some with a W chromosome and others without a morphologically recognizable sex chromosome (data not shown).
Table 3. Segregation of offspring pigmentation and sex from crosses between G. affinis females and melanic G. holbrooki males.
| Parental ♀ | Parental ♂ | Pedigree # | F1♀ | F1 ♀ | F1♂ | F1 ♂ |
|---|---|---|---|---|---|---|
| melanic | wt | melanic | wt | |||
| G. aff. WLC1313 | G. hol. WLC2105 | 2353 | 1 | 22 | 51 | 21 |
| G. aff. WLC1313 | G. hol. WLC1475 | 5964/I | 0 | 74 | 118 | 30 |
| G. aff. WLC1313 | G. hol. WLC1475 | 5964/II | 0 | 51 | 44 | 0 |
| G. aff. WLC1313 | G. hol. WLC1475 | 6244 | 0 | 56 | 43 | 0 |
| G. aff. WLC1313 | G. hol. WLC1475 | 6245 | 0 | 33 | 24 | 0 |
| Σ | 1 | 236 | 280 | 51 | ||
| Σ | 568 | |||||
wt, nonmelanic wildtype-pigmented fish.
The nonmelanic F1 of our crosses consisted of 236 females and 51 males (Table 3). In some of the crosses, no nonmelanic F1 males were obtained at all (Table 3). This indicated that, at variable frequencies depending on the cross, many or most ZX fish become females. We further investigated this by crossing XX G. holbrooki females with ZZ G. affinis males. Their offspring consisted of 83% females despite the uniform ZX genotype and several broods consisted exclusively of females (Table 4).
Table 4. Sex ratio of offspring between G. holbrooki females and G. affinis males.
| Parental ♀ | Parental ♂ | Pedigree # | Brood | F1 ♀ | F1 ♂ |
|---|---|---|---|---|---|
| G. hol. WLC1475 | G. aff. WLC1313 | 5964r/II | 1 | 22 | 0 |
| 2 | 25 | 0 | |||
| 3 | 11 | 5 | |||
| 4 | 28 | 6 | |||
| 5 | 13 | 6 | |||
| 6 | 8 | 12 | |||
| 7 | 34 | 6 | |||
| G. hol. WLC1475 | G. aff. WLC1313 | 5964r/I | 1 | 33 | 0 |
| Σ | 174 | 35 | |||
| Σ | 209 | ||||
To confirm that WY fish usually develop as males and ZX fish as females, we genotyped F1 nonmelanic females and melanic males from one cross of G. affinis x G. holbrooki for the G. affinis W chromosome and the G. holbrooki Y chromosome using molecular markers. In this cross, all WY F1 fish were males and all ZX F1 fish developed as female (Table S4).
Crosses suggest that female-to-male sex reversal occurs in G. holbrooki
All of our melanic G. holbrooki strains occasionally produce nonmelanic male offspring. Nonmelanic males derived from melanic fathers can be the result of a recombination or deletion event leading to the loss of the melanic locus from the Y or by a female-to-male sex reversal in XX fish. To test this, we carried out crosses between such exceptional males and nonmelanic G. holbrooki females (Table S5). Variation in these crosses was high, as they resulted in 0–87.7% male offspring (average 29.2%). We genotyped 15 of these males as being negative for gipc1Y and sox9bY (Table S2). The generally lower percentage of male offspring and the consistency with which such nonmelanic males occur in our melanic strains suggests that at least some of these males are XX.
Discussion
Gambusia affinis and G. holbrooki provide the unique opportunity to study different sex chromosome systems in two sister species. The sex determination mechanism of the last common ancestor of G. holbrooki and G. affinis is not known. Both systems might have evolved in parallel or a transition might have occurred in the common ancestor, separating the lineages from each other. Higher differentiation of the W than the Y chromosome, at least with respect to size, does not indicate that the W is older than the Y. Large W chromosomes are also found in other nonamniotes with young sex chromosomes (Schartl et al. 2016b).
The finding from our interspecific crosses that WY hybrids usually become males demonstrates that the sex determination mechanism of G. holbrooki is dominant over the one of G. affinis. This indicates that the Y harbors a strong, dominant male determiner. Whether sex-determination in G. affinis is mediated by a dosage-sensitive locus on the Z similar to the situation in chicken (Smith et al. 2009), or a dominant female-sex determiner on the W, still needs to be determined.
Sex chromosome turnover driven by sexually antagonistic selection requires that the new sex determiner is genetically dominant over the old one, meaning that a new Y chromosome can only invade a ZW sex determination system if the Y is dominant over the W (van Doorn and Kirkpatrick 2007). Thus, we predict that the Y from G. holbrooki may invade G. affinis in the hybrid zone between the two species and the W will be eventually lost. This is supported by a recent study using microsatellite markers, which provided evidence for nonlinear gene flow within the hybrid zone of the two species (Wilk and Horth 2016). The distortion of the sex ratio caused by interspecific crosses might lead to a decreased fitness of hybrid populations and multiple introgression events could even lead to WW offspring that might not be viable. A previous study using microsatellite markers found that the heterozygosity of hybrid mosquitofish in the Mobile Bay area is low, and that the G. holbrooki phenotype is more abundant (Wilk and Horth 2016). This motivates future studies to further sample the region where the two species are in contact, to determine which sex chromosomes are present in the hybrid zone, and to carry out crosses to find out whether WW fish are viable. Hybrid speciation and introgression are prevalent in fish evolution and have been documented in particular in poeciliids (Meyer et al. 2006; Roberts et al. 2009; Cui et al. 2013; Kang et al. 2013).
Our study provides an important first step toward identifying the sex determination locus in G. holbrooki and G. affinis and the Y-linked pigmentation locus in G. holbrooki by pinpointing regions on the sex chromosomes that are especially highly differentiated. These regions were identified by pool sequencing, which defines such areas in the genome more precisely than RAD-seq and is therefore a promising method to investigate sex determination systems in nonmodel organisms. G. affinis and G. holbrooki confirm that highly differentiated sex chromosomes can be either heteromorphic (ZW) or homomorphic (XY). Like in G. holbrooki, the sex chromosome pair of medaka appears homomorphic, but, on the molecular level, the medaka Y is highly differentiated over a region of at least 3 Mb (Takeda 2008; Wilson et al. 2014). Even between populations, the accumulation of repetitive elements and pseudogenes on the sex chromosomes can vary greatly (Nanda et al. 2014).
As a potentially sexually antagonistic Y-linked locus, the melanic locus might have played a major role in the evolution of the G. holbrooki Y chromosome (Kottler and Schartl 2018). Studies suggest that the low percentage of melanic males in natural populations is maintained by negative frequency-dependent selection (Horth and Travis 2002; Horth 2003). Large numbers of melanic males seem to trigger increased male and female mortality rates (Horth and Travis 2002; Horth 2003). One possible explanation for this would be the aggressive behavior of melanic males, leading to, ultimately detrimental, stress (Horth and Travis 2002; Horth 2003). Our genotyping results suggest that gipc1Y either contributes to the formation of the melanic color pattern or is closely linked to the melanic locus. In zebrafish, a dominant mutation in tyrp1a leads to defects in eumelanin production and melanosome morphology, ultimately resulting in melanocyte death (Krauss et al. 2014). A similar phenotype is observed in tyrp1a and tyrp1b knockdown zebrafish (Braasch et al. 2009). Gipc1 is involved in the intracellular transport of newly synthetized Tyrp1 protein, but Tyrp1 is probably only one of many Gipc1 interaction partners within the melanocytes and the effect of the detected mutations, or of an overexpression of Gipc1, is difficult to predict (Liu et al. 2001).
Because of the repressed recombination on the Y, genotyping of more populations with melanic males is required to confirm the linkage between the melanic phenotype and gipc1Y. In general, the detection of an array of SNPs differing between the X and Y suggests that a large part of the Y is differentiated from the X, as it is the case in the G. affinis W and Z. This implies that the expression and/or coding sequences of many genes on the sex chromosomes are most likely altered. It is therefore possible that the melanic phenotype is caused by several additive Y-linked mutations that became fixed on the Y due to the repressed recombination. For instance, cAMP-dependent protein kinase catalytic subunit alpha, which encodes a subunit of protein kinase A (PKA), is located close to gipc1. PKA regulates melanosome movement via cAMP regulation, and mutations at this locus leading to permanently high internal levels of cAMP could explain the insensitivity of the macromelanocytes to adrenaline (Fujii 2000; Rodionov et al. 2003). The temperature-sensitivity of the melanic phenotype in some G. holbrooki populations indicates that the expression of the causative locus, or loci, is affected and that melanocytes are transformed into macromelanocytes at lower temperatures (Angus 1989; Horth 2006). However, further studies are necessary to confirm that the temperature-sensitive melanic phenotype is caused by the same locus. The melanic locus, or a locus closely linked to it, affects xanthophores as well, as older G. holbrooki males frequently develop yellow areas on the head and trunk. In our strains, macromelanocytes were first visible shortly after birth, well before males could be distinguished from females by their secondary sexual characteristics. This demonstrates that, in contrast to, for instance, the famous male pigment patterns of the guppy, the melanic pattern is not under the control of high androgen levels that are only established after reaching sexual maturity (Kottler and Schartl 2018).
Occasionally, our melanic G. holbrooki strains produce nonmelanic males. While some of these males might have lost the melanic locus on the Y, the sex bias to female in our crosses suggests that many of these males are XX. As we currently only have a marker for the melanic locus (gipc1Y) but not for the Y chromosome in general, we could not investigate yet whether most of these males indeed lack a Y chromosome. Male development in the absence of the Y can be induced by autosomal modifiers that occur with varying frequency and penetrance within a population. This has been reported from other poeciliid fish of the related genus Xiphophorus (Kallman 1984; Schartl et al. 2011). Recurrence of XX males in crosses from female-to-male sex-reversed XX fishes due to autosomal modifiers was also observed in medaka (Nanda et al. 2003). Another possibility would be that the environment affects sex determination in Gambusia.
Sox9bY appeared as a reasonable sex determination candidate gene in our study, but was not linked to male sex in all of our fish, as wildtype-pigmented males lacked sox9bY. However, we currently do not know whether melanic and wildtype males share the same sex chromosome. If melanic males have a distinct Y chromosome, sox9bY would be an excellent candidate for their sex-determining locus.
Taken together, understanding how different sex determination mechanisms in closely related species evolved will shed light on one of the most fundamental and fascinating aspects of vertebrate genome evolution.
Acknowledgments
We thank Ingo Schlupp (Norman, OK) for generously supplying specimen; Georg Schneider, Joachim Schürger, and Petra Weber, Wuerzburg, for breeding and maintenance of Gambusia, and Daniela Bunsen, Claudia Gehrig, and Christian Stigloher, Imaging Core Facility, Biocenter Wuerzburg, for technical support. This project was supported by funds to Y.G. and M.S. from the “Agence Nationale de la Recherche” and the “Deutsche Forschungsgemeinschaft” (ANR/DFG, PhyloSex project, 2014-2016). V.A.K. was supported through a SCIENTIA fellowship of the Free State of Bavaria. The GeT, Toulouse (C.L.-R., J.L.), and the Montpellier GenomiX (MGX) France (L.J., H.P.) core facilities were supported by France Génomique National infrastructure, funded as part of “Investissement d’avenir” program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.7957376.
Communicating editor: C. Peichel
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amores A., Catchen J., Ferrara A., Fontenot Q., and Postlethwait J. H., 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188: 799–808. 10.1534/genetics.111.127324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus R. A., 1989. Inheritance of melanistic pigmentation in the eastern mosquitofish. J. Hered. 80: 387–392. 10.1093/oxfordjournals.jhered.a110880 [DOI] [Google Scholar]
- Bi W., Deng J. M., Zhang Z., Behringer R. R., and de Crombrugghe B., 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85–89. 10.1038/8792 [DOI] [PubMed] [Google Scholar]
- Bisazza A., and Pilastro A., 2000. Variation of female preference for male coloration in the eastern mosquitofish Gambusia holbrooki. Behav. Genet. 30: 207–212. 10.1023/A:1001914208075 [DOI] [PubMed] [Google Scholar]
- Black D. A., and Howell W. M., 1979. The North American mosquitofish, Gambusia affinis: a unique case in sex chromosome evolution. Copeia 1979: 509–513. 10.2307/1443231 [DOI] [Google Scholar]
- Braasch I., Liedtke D., Volff J. N., and Schartl M., 2009. Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell Melanoma Res. 22: 839–850. 10.1111/j.1755-148X.2009.00614.x [DOI] [PubMed] [Google Scholar]
- Cai H., and Reed R. R., 1999. Cloning and characterization of neuropilin-1-interacting protein: a PSD-95/Dlg/ZO-1 domain-containing protein that interacts with the cytoplasmic domain of neuropilin-1. J. Neurosci. 19: 6519–6527. 10.1523/JNEUROSCI.19-15-06519.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos H. H., and Hubbs C., 1971. Cytomorphology of six species of gambusiine fishes. Copeia 1971: 566–569. 10.2307/1442462 [DOI] [Google Scholar]
- Capel B., 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 18: 675–689. 10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- Catchen J. M., Amores A., Hohenlohe P., Cresko W., and Postlethwait J. H., 2011. Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda) 1: 171–182. 10.1534/g3.111.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden T. W., Claes F., Lanahan A. A., Autiero M., Palac R. T. et al. , 2006. Selective regulation of arterial branching morphogenesis by synectin. Dev. Cell 10: 783–795. 10.1016/j.devcel.2006.03.012 [DOI] [PubMed] [Google Scholar]
- Cui R., Schumer M., Kruesi K., Walter R., Andolfatto P. et al. , 2013. Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution 67: 2166–2179. 10.1111/evo.12099 [DOI] [PubMed] [Google Scholar]
- De Vries L., Lou X., Zhao G., Zheng B., and Farquhar M. G., 1998. GIPC, a PDZ domain containing protein, interacts specifically with the C terminus of RGS-GAIP. Proc. Natl. Acad. Sci. USA 95: 12340–12345. 10.1073/pnas.95.21.12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin R. H., and Nagahama Y., 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364. 10.1016/S0044-8486(02)00057-1 [DOI] [Google Scholar]
- Dixon G., Kitano J., and Kirkpatrick M., 2018. The origin of a new sex chromosome by introgression between two stickleback fishes. Mol. Biol. Evol. 36: 28–38. 10.1093/molbev/msy181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel O., Shirak A., Dor L., Band M., Zak T. et al. , 2014. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15: 774 10.1186/1471-2164-15-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R., 2000. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 13: 300–319. 10.1034/j.1600-0749.2000.130502.x [DOI] [PubMed] [Google Scholar]
- Gonen N., Futtner C. R., Wood S., Garcia-Moreno S. A., Salamone I. M. et al. , 2018. Sex reversal following deletion of a single distal enhancer of Sox9. Science 360: 1469–1473. 10.1126/science.aas9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., 2013. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30: 1229–1235. 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
- Hamer A., Lane S., and Mahony M., 2002. The role of introduced mosquitofish (Gambusia holbrooki) in excluding the native green and golden bell frog (Litoria aurea) from original habitats in south-eastern Australia. Oecologia 132: 445–452. 10.1007/s00442-002-0968-7 [DOI] [PubMed] [Google Scholar]
- Helmstetter A. J., Papadopulos A. S., Igea J., Van Dooren T. J., Leroi A. M. et al. , 2016. Viviparity stimulates diversification in an order of fish. Nat. Commun. 7: 11271 10.1038/ncomms11271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A., and Schartl M., 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16: 1260–1274. 10.15252/embr.201540667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffberg S. L., Troendle N. J., Glenn T. C., Mahmud O., Louha S. et al. , 2018. A high-quality reference genome for the invasive mosquitofish Gambusia affinis using a Chicago library. G3 (Bethesda) 8: 1855–1861. 10.1534/g3.118.200101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horth L., 2003. Melanic body colour and aggressive mating behaviour are correlated traits in male mosquitofish (Gambusia holbrooki). Proc. Biol. Sci. 270: 1033–1040. 10.1098/rspb.2003.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horth L., 2004. Predation and the persistence of melanic male mosquitofish (Gambusia holbrooki). J. Evol. Biol. 17: 672–679. 10.1111/j.1420-9101.2004.00710.x [DOI] [PubMed] [Google Scholar]
- Horth L., 2006. A sex-linked allele, autosomal modifiers and temperature-dependence appear to regulate melanism in male mosquitofish (Gambusia holbrooki). J. Exp. Biol. 209: 4938–4945. 10.1242/jeb.02599 [DOI] [PubMed] [Google Scholar]
- Horth L., and Travis J., 2002. Frequency-dependent numerical dynamics in mosquitofish. Proc. Biol. Sci. 269: 2239–2247. 10.1098/rspb.2002.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horth L., Binckley C., Wilk R., Reddy P., and Reddy A., 2010. Color, body size, and genitalia size are correlated traits in eastern mosquitofish (Gambusia holbrooki). Copeia 2010: 196–202. 10.1643/CG-09-044 [DOI] [Google Scholar]
- Hrbek T., Seckinger J., and Meyer A., 2007. A phylogenetic and biogeographic perspective on the evolution of poeciliid fishes. Mol. Phylogenet. Evol. 43: 986–998. 10.1016/j.ympev.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Hubbs C. L., 1955. Hybridization between fish species in nature. Syst. Zool. 4: 1–20. 10.2307/2411933 [DOI] [Google Scholar]
- Jo A., Denduluri S., Zhang B., Wang Z., Yin L. et al. , 2014. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 1: 149–161. 10.1016/j.gendis.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallman K. D., 1973. The sex-determining mechanism of the platyfish, Xiphophorus maculatus, pp. 19–28 in Genetics and Mutagenesis of Fish, Springer-Verlag, New York. [Google Scholar]
- Kallman K. D., 1975. The platyfish, Xiphophorus maculatus, pp. 81–132 in Handbook of Genetics Springer, Berlin, Germany. [Google Scholar]
- Kallman K. D., 1984. A new look at sex determination in poeciliid fishes, pp. 95–171 in Evolutionary Genetics of Fishes Springer, Berlin, Germany. [Google Scholar]
- Kamiya T., Kai W., Tasumi S., Oka A., Matsunaga T. et al. , 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8: e1002798 10.1371/journal.pgen.1002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. H., Schartl M., Walter R. B., and Meyer A., 2013. Comprehensive phylogenetic analysis of all species of swordtails and platies (Pisces: genus Xiphophorus) uncovers a hybrid origin of a swordtail fish, Xiphophorus monticolus, and demonstrates that the sexually selected sword originated in the ancestral lineage of the genus, but was lost again secondarily. BMC Evol. Biol. 13: 25 10.1186/1471-2148-13-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M., 2013. Functional proteomics, human genetics and cancer biology of GIPC family members. Exp. Mol. Med. 45: e26 10.1038/emm.2013.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., and Hamaguchi S., 2013. Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dyn. 242: 339–353. 10.1002/dvdy.23927 [DOI] [PubMed] [Google Scholar]
- Kitano J., Ross J. A., Mori S., Kume M., Jones F. C. et al. , 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461: 1079–1083. 10.1038/nature08441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Pandey R. V., and Schlotterer C., 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27: 3435–3436. 10.1093/bioinformatics/btr589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Hornung U., Nanda I., Imai S., Sasaki T. et al. , 2006. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 16: 815–826. 10.1101/gr.5016106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler V. A., and Schartl M., 2018. The colorful sex chromosomes of teleost fish. Genes (Basel) 9 pii: E233. 10.3390/genes9050233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler V. A., Koch I., Flötenmeyer M., Hashimoto H., Weigel D. et al. , 2014. Multiple pigment cell types contribute to the black, blue, and orange ornaments of male guppies (Poecilia reticulata). PLoS One 9: e85647 10.1371/journal.pone.0085647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss J., Geiger-Rudolph S., Koch I., Nusslein-Volhard C., and Irion U., 2014. A dominant mutation in tyrp1A leads to melanophore death in zebrafish. Pigment Cell Melanoma Res. 27: 827–830. 10.1111/pcmr.12272 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., and Tamura K., 2018. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamatsch D. K., Adolfsson S., Senior A. M., Christiansen G., Pichler M. et al. , 2015. A transcriptome derived female-specific marker from the invasive Western mosquitofish (Gambusia affinis). PLoS One 10: e0118214 10.1371/journal.pone.0118214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv :1303.3997v2.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Sun Y., Zhao J., Shi H., Zeng S. et al. , 2015. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in nile Tilapia, Oreochromis niloticus. PLoS Genet. 11: e1005678 10.1371/journal.pgen.1005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. F., Kandala G., and Setaluri V., 2001. PDZ domain protein GIPC interacts with the cytoplasmic tail of melanosomal membrane protein gp75 (tyrosinase-related protein-1). J. Biol. Chem. 276: 35768–35777. 10.1074/jbc.M103585200 [DOI] [PubMed] [Google Scholar]
- Lydeard C., Wooten M. C., and Meyer A., 1995. Molecules, morphology, and area cladograms - a cladistic and biogeographic analysis of Gambusia (teleostei, Poeciliidae). Syst. Biol. 44: 221–236. 10.2307/2413708 [DOI] [Google Scholar]
- Mank J. E., Promislow D. E., and Avise J. C., 2006. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol. J. Linn. Soc. Lond. 87: 83–93. 10.1111/j.1095-8312.2006.00558.x [DOI] [Google Scholar]
- Martin R. G., 1977. Density dependent aggressive advantage in melanistic male mosquitofish Gambusia affinis holbrooki (Girard). Fla. Sci. 40: 393–400. [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C. et al. , 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. 10.1038/nature751 [DOI] [PubMed] [Google Scholar]
- Meyer A., Salzburger W., and Schartl M., 2006. Hybrid origin of a swordtail species (Teleostei: Xiphophorus clemenciae) driven by sexual selection. Mol. Ecol. 15: 721–730. 10.1111/j.1365-294X.2006.02810.x [DOI] [PubMed] [Google Scholar]
- Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y. et al. , 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. 10.1534/genetics.111.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache S. N., Hasson T., and Horowitz A., 2006. Binding of internalized receptors to the PDZ domain of GIPC/synectin recruits myosin VI to endocytic vesicles. Proc. Natl. Acad. Sci. USA 103: 12735–12740 (erratum: Natl. Acad. Sci. USA 103: 15272). 10.1073/pnas.0605317103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C. et al. , 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99: 11778–11783. 10.1073/pnas.182314699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Hornung U., Kondo M., Schmid M., and Schartl M., 2003. Common spontaneous sex-reversed XX males of the medaka Oryzias latipes. Genetics 163: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Schories S., Tripathi N., Dreyer C., Haaf T. et al. , 2014. Sex chromosome polymorphism in guppies. Chromosoma 123: 373–383. 10.1007/s00412-014-0455-z [DOI] [PubMed] [Google Scholar]
- Natri H. M., Merilä J., and Shikano T., 2019. The evolution of sex determination associated with a chromosomal inversion. Nat. Commun. 10: 145 10.1038/s41467-018-08014-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. S., Grande T. C., and Wilson M. V., 2016. Fishes of the World, John Wiley & Sons, Hoboken, NJ: 10.1002/9781119174844 [DOI] [Google Scholar]
- Passeron T., Valencia J. C., Bertolotto C., Hoashi T., Le Pape E. et al. , 2007. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc. Natl. Acad. Sci. USA 104: 13984–13989. 10.1073/pnas.0705117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Ross J. A., Matson C. K., Dickson M., Grimwood J. et al. , 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. 10.1016/j.cub.2004.08.030 [DOI] [PubMed] [Google Scholar]
- Pyke G. H., 2008. Plague minnow or mosquito fish? A review of the biology and impacts of introduced Gambusia species. Annu. Rev. Ecol. Evol. Syst. 39: 171–191. 10.1146/annurev.ecolsys.39.110707.173451 [DOI] [Google Scholar]
- Regan J. D., 1961. Melanodimorphism in the poeciliid fish, Gambusia affinis (Baird and Girard). Am. Midl. Nat. 65: 139–143. 10.2307/2423009 [DOI] [Google Scholar]
- Reid S. M., Wilson C. C., Mandrak N. E., and Carl L. M., 2008. Population structure and genetic diversity of black redhorse (Moxostoma duquesnei) in a highly fragmented watershed. Conserv. Genet. 9: 531–546. 10.1007/s10592-007-9367-2 [DOI] [Google Scholar]
- Roberts R. B., Ser J. R., and Kocher T. D., 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326: 998–1001. 10.1126/science.1174705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov V., Yi J., Kashina A., Oladipo A., and Gross S. P., 2003. Switching between microtubule- and actin-based transport systems in melanophores is controlled by cAMP levels. Curr. Biol. 13: 1837–1847. 10.1016/j.cub.2003.10.027 [DOI] [PubMed] [Google Scholar]
- Ross J. A., Urton J. R., Boland J., Shapiro M. D., and Peichel C. L., 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5: e1000391 10.1371/journal.pgen.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez N. S., Hill C. R., Love J. D., Soslow J. H., Craig E. et al. , 2011. The cytoplasmic domain of TGFbetaR3 through its interaction with the scaffolding protein, GIPC, directs epicardial cell behavior. Dev. Biol. 358: 331–343. 10.1016/j.ydbio.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl, M., D. Galiana-Arnoux, C. Schultheis, A. Böhne, and J.-N. Volff, 2011 A primer on sex determination, in Ecology and Evolution of Poecliid Fishes, edited by A. Pilastro, J. Evans, I. Schlupp, University of Chicago Press, Chicago, IL. [Google Scholar]
- Schartl M., Walter R. B., Shen Y., Garcia T., Catchen J. et al. , 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat. Genet. 45: 567–572. 10.1038/ng.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M., Larue L., Goda M., Bosenberg M. W., Hashimoto H. et al. , 2016a What is a vertebrate pigment cell? Pigment Cell Melanoma Res. 29: 8–14. 10.1111/pcmr.12409 [DOI] [PubMed] [Google Scholar]
- Schartl M., Schmid M., and Nanda I., 2016b Dynamics of vertebrate sex chromosome evolution: from equal size to giants and dwarfs. Chromosoma 125: 553–571. 10.1007/s00412-015-0569-y [DOI] [PubMed] [Google Scholar]
- Schmidt J., 1920. Racial investigations. IV. The genetic behavior of a secondary sexual character. C. R. Trav. Lab. Carlsberg 14: 1–8. [Google Scholar]
- Schmid M., Steinlein C., Bogart J., Feichtinger W., León P. et al. , 2010. The chromosomes of Terraranan frogs. Insights into vertebrate cytogenetics. Cytogenet. Genome Res. 568: 130–131. 10.1159/000301339 [DOI] [PubMed] [Google Scholar]
- Scribner K. T., and Avise J. C., 1993. Cytonuclear genetic architecture in mosquitofish populations and the possible roles of introgressive hybridization. Mol. Ecol. 2: 139–149. 10.1111/j.1365-294X.1993.tb00103.x [DOI] [Google Scholar]
- Sekido R., and Lovell-Badge R., 2008. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453: 930–934. 10.1038/nature06944 [DOI] [PubMed] [Google Scholar]
- Shakhova O., Cheng P., Mishra P. J., Zingg D., Schaefer S. M. et al. , 2015. Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma. PLoS Genet. 11: e1004877 10.1371/journal.pgen.1004877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T., Natri H. M., Shimada Y., and Merila J., 2011. High degree of sex chromosome differentiation in stickleback fishes. BMC Genomics 12: 474 10.1186/1471-2164-12-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K. et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G. et al. , 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461: 267–271. 10.1038/nature08298 [DOI] [PubMed] [Google Scholar]
- Sumner A. T., 1972. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75: 304–306. 10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Sutton E., Hughes J., White S., Sekido R., Tan J. et al. , 2011. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J. Clin. Invest. 121: 328–341. 10.1172/JCI42580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H., 2008. Draft genome of the medaka fish: a comprehensive resource for medaka developmental genetics and vertebrate evolutionary biology. Dev. Growth Differ. 50: S157–S166. 10.1111/j.1440-169X.2008.00992.x [DOI] [PubMed] [Google Scholar]
- Takehana Y., Matsuda M., Myosho T., Suster M. L., Kawakami K. et al. , 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5: 4157 10.1038/ncomms5157 [DOI] [PubMed] [Google Scholar]
- Tamura K., and Nei M., 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- Valdembri D., Caswell P. T., Anderson K. I., Schwarz J. P., Konig I. et al. , 2009. Neuropilin-1/GIPC1 signaling regulates alpha5beta1 integrin traffic and function in endothelial cells. PLoS Biol. 7: e25 10.1371/journal.pbio.1000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn G. S., and Kirkpatrick M., 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912. 10.1038/nature06178 [DOI] [PubMed] [Google Scholar]
- Varsano T., Dong M. Q., Niesman I., Gacula H., Lou X. et al. , 2006. GIPC is recruited by APPL to peripheral TrkA endosomes and regulates TrkA trafficking and signaling. Mol. Cell. Biol. 26: 8942–8952. 10.1128/MCB.00305-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff J. N., and Schartl M., 2002. Sex determination and sex chromosome evolution in the medaka, Oryzias latipes, and the platyfish, Xiphophorus maculatus. Cytogenet. Genome Res. 99: 170–177. 10.1159/000071590 [DOI] [PubMed] [Google Scholar]
- Wagner T., Wirth J., Meyer J., Zabel B., Held M. et al. , 1994. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111–1120. 10.1016/0092-8674(94)90041-8 [DOI] [PubMed] [Google Scholar]
- Walter R. B., Rains J. D., Russell J. E., Guerra T. M., Daniels C. et al. , 2004. A microsatellite genetic linkage map for Xiphophorus. Genetics 168: 363–372. 10.1534/genetics.103.019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Kitano J., and Peichel C. L., 2015. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol. Biol. Evol. 32: 1981–1995. 10.1093/molbev/msv078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R. J., and Horth L., 2016. A genetically distinct hybrid zone occurs for two globally invasive mosquito fish species with striking phenotypic resemblance. Ecol. Evol. 6: 8375–8388. 10.1002/ece3.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. A., High S. K., McCluskey B. M., Amores A., Yan Y. L. et al. , 2014. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198: 1291–1308. 10.1534/genetics.114.169284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge Ö., 1922. One-sided masculine and sex-linked inheritance in Lebistes reticulatus. J. Genet. 12: 145–162. 10.1007/BF02983078 [DOI] [Google Scholar]
- Winge Ö., 1927. The location of eighteen genes in Lebistes reticulatus. J. Genet. 18: 1–43. 10.1007/BF03052599 [DOI] [Google Scholar]
- Wooten M. C., and Lydeard C., 1990. Allozyme variation in a natural contact zone between Gambusia affinis and Gambusia holbrooki. Biochem. Syst. Ecol. 18: 169–173. 10.1016/0305-1978(90)90054-J [DOI] [Google Scholar]
- Wooten M. C., Scribner K. T., and Smith M. H., 1988. Genetic-variability and systematics of Gambusia in the southeastern united-states. Copeia 1988: 283–289. 10.2307/1445867 [DOI] [Google Scholar]
- Yan Y. L., Willoughby J., Liu D., Crump J. G., Wilson C. et al. , 2005. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132: 1069–1083. 10.1242/dev.01674 [DOI] [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E. et al. , 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. 10.1016/j.cub.2012.05.045 [DOI] [PubMed] [Google Scholar]
- Yano A., Nicol B., Jouanno E., Quillet E., Fostier A. et al. , 2013. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 6: 486–496. 10.1111/eva.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Figure S1 presents the results of the RAD-sequencing analysis. Figure S2 shows the analysis of G. affinis and G. holbrooki pooled-sequencing reads aligned to the genome of X. maculatus using 10 kb windows. Figure S3 represents a molecular phylogeny of fish sox9. Figure S4 are alignments of male G. holbrooki and G. affinis gipc1 and sox9b alleles and protein sequences. Figure S5 are multiple species alignments of Gipc1 and Sox9 protein sequences. Figure S6 contains PCR results demonstrating that sox9bY was generated by allelic diversification. Figure S7 contains pictures of melanic and nonmelanic G. holbrooki. Table S1 gives information on the fish used for pool sequencing. Table S2 presents the results of the allele-specific PCRs. Table S3 contains the annotated loci within the 100 kb windows with the highest density of sex-specific SNPs. Table S4 contains the sex chromosome genotypes of F1 fish (pedigree WLC20074) from a cross between a G. affinis female (WLC1313) and a G. holbrooki male (WLC2514) determined by PCR. Table S5 gives information on the number of offspring of nonmelanic females and nonmelanic males derived from melanic strains.
RAD-seq and pool sequencing data are available at Sequence Read Archive, grouped under the same Bioproject (accession number: PRJNA563257). The genome assembly of a melanic G. holbrooki male is available at NCBI, accession number VOPI00000000. Gipc1 and sox9b cDNA sequences are available at GenBank (accession numbers: MK721001-MK721004). Supplemental material available at figshare: https://doi.org/10.25386/genetics.7957376.