Abstract
Life-history traits or “fitness components”—such as age and size at maturity, fecundity and fertility, age-specific rates of survival, and life span—are the major phenotypic determinants of Darwinian fitness. Analyzing the evolution and genetics of these phenotypic targets of selection is central to our understanding of adaptation. Due to its simple and rapid life cycle, cosmopolitan distribution, ease of maintenance in the laboratory, well-understood evolutionary genetics, and its versatile genetic toolbox, the “vinegar fly” Drosophila melanogaster is one of the most powerful, experimentally tractable model systems for studying “life-history evolution.” Here, I review what has been learned about the evolution and genetics of life-history variation in D. melanogaster by drawing on numerous sources spanning population and quantitative genetics, genomics, experimental evolution, evolutionary ecology, and physiology. This body of work has contributed greatly to our knowledge of several fundamental problems in evolutionary biology, including the amount and maintenance of genetic variation, the evolution of body size, clines and climate adaptation, the evolution of senescence, phenotypic plasticity, the nature of life-history trade-offs, and so forth. While major progress has been made, important facets of these and other questions remain open, and the D. melanogaster system will undoubtedly continue to deliver key insights into central issues of life-history evolution and the genetics of adaptation.
Keywords: FlyBook, life-history evolution, fitness components, fitness, variation, selection, adaptation, trade-offs, plasticity
ADAPTATION by natural selection is based on heritable variation in Darwinian fitness, i.e ., genetic and phenotypic variation in net fitness (Lewontin 1974; Roff 1992, 2002; Stearns 1992; Charlesworth 1994, 2013). Net fitness can be approximated by the expected lifetime reproductive success of a genotype (Charlesworth 1994), but this requires information on, for example, lifetime fecundity and fertility, and can thus be difficult to estimate in many organisms (Clutton-Brock 1988; Fowler et al. 1997; Charlesworth and Hughes 2000). Studies of the evolution and genetics of Darwinian fitness have therefore mostly relied on analyzing life-history traits or fitness components, i.e., phenotypic characters that affect an organism’s survival and reproduction, a field called life-history evolution (Cole 1954; Knight and Robertson 1957; Lewontin 1965; Stearns 1976, 1978, 1992, 2000; Charlesworth 1980, 1994, 2003, 2013; Clutton-Brock 1988; Partridge and Harvey 1988; Rose 1991; Roff 1992, 2002; Charlesworth and Hughes 2000; Houle 2001; Flatt and Heyland 2011; Fabian and Flatt 2012). The analysis of the causes and consequences of genetic and phenotypic variation in life-history traits is central to our understanding of natural selection, and adaptation (Stearns 1976, 1992; Charlesworth 1994).
A life-history trait or fitness component can be technically defined as a phenotypic character for which an increased trait value causes an increase in net fitness when all other traits are being held constant (Charlesworth and Hughes 2000); together, these fitness components determine the multivariate phenotype called fitness (Knight and Robertson 1957; Lande 1982; Lande and Arnold 1983; Charlesworth 1993a). Major life-history traits include, e.g., size at birth, developmental rate, age and size at reproductive maturity, number and size of offspring, age- or size-specific schedules of fecundity and fertility, age- or size-specific schedules of survival, and life span (Knight and Robertson 1957; Stearns 1976, 1992; Roff 1992, 2002). These traits are connected to each other through phenotypic, physiological, and/or genetic correlations, especially so-called trade-offs, i.e., negative correlations between fitness components (Stearns 1989a, 1992; Charlesworth 1990; Roff 1992, 2002; Houle 2001; Zera and Harshman 2001; Flatt 2011; Flatt and Heyland 2011).
While evolutionary ecologists have analyzed life-history evolution predominantly from a phenotypic point of view (Stearns 1976, 1978, 1992, 2000; Roff 1992, 2002), there is also a rich tradition of investigating the quantitative and population genetics of fitness components (Lewontin 1974; Charlesworth 1980, 1994, 2003, 2015; Roff 1992; Stearns 1992; Charlesworth and Hughes 2000). At the theoretical level, this work has led to the development of mathematical models for selection in age-structured populations with overlapping generations (Charlesworth 1980, 1994, 2003), whereas experiments—most of them carried out in Drosophila melanogaster—have produced fundamental insights into the origin, amount, and maintenance of genetic variation for fitness-related traits and the genetics of life-history adaptations (Mukai 1964; Prout 1971a,b; Lewontin 1974; Simmons and Crow 1977; Hedrick and Murray 1983; Mackay 1985; Sved 1989; Houle et al. 1994b; Latter and Sved 1994; Charlesworth and Hughes 2000; Charlesworth 2015). Moreover, laboratory studies of large-effect mutants and transgenes have examined the genetic basis of growth, size, reproduction, and life span in several model organisms including Drosophila (Tatar 1999, 2000; Clancy et al. 2001; Stearns and Partridge 2001; Tatar et al. 2001a, 2003; Partridge and Gems 2002; Oldham and Hafen 2003; Flatt et al. 2005; Partridge et al. 2005a; Edgar 2006; Mirth and Riddiford 2007; Flatt and Schmidt 2009; Flatt and Heyland 2011).
Here, I review what has been learned about the genetics and evolution of life-history traits in the vinegar fly D. melanogaster (Figure 1). Due to its rapid generation time, small size, high fertility, and short life; cosmopolitan distribution; ease of sampling, laboratory breeding, and manipulation; well-understood development and physiology; and its versatile genetic toolbox [e.g., balancer chromosomes, classical mutants, transgenes, RNA interference (RNAi), and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9], this species has proved to be one of the most powerful models for investigating the genetics and other aspects of life-history evolution and adaptation.
Figure 1.
The vinegar fly (D. melanogaster), here depicted sitting on a ripe banana in a kitchen, is a human commensal (Lachaise et al. 1988; Keller 2007; Markow 2015; Mansourian et al. 2018) and represents the probably most intensely studied model organism, having first been bred in the laboratory in the early 1900s (Kohler 1994; Mohr 2018). As reviewed here, this holometabolous insect has been widely used in studies of life-history evolution, genetics of fitness components, correlated responses to selection and trade-offs, and the evolution of aging. Figure credit: Chloé Schmidt (University of Manitoba).
Consequently, the D. melanogaster system has been widely used to address many fundamental questions in evolutionary biology, including for example:
What is the mutation rate for fitness-related traits and for total fitness?
How much genetic variation is there for fitness components?
What evolutionary processes maintain variation in fitness components?
Which loci and polymorphisms underpin variation in fitness components?
What are the causes of trade-offs between fitness components?
Given trade-offs between growth, survival, and reproduction, how does selection optimize overall life history (the so-called “general life-history” or “reproductive effort” problem)?
How does aging (senescence) evolve?
What are the patterns of life-history plasticity and genotype-by-environment interactions for fitness-related traits?
The long history of the D. melanogaster model, and its sophisticated experimental techniques, mean that these and other major questions have been examined in more detail than in most other organisms. Consequently, a vast amount of information on the genetics and evolution of fitness components in D. melanogaster is available; my aim here is to provide a summary and point of entry into this large body of literature.
Overview of D. melanogaster Life History
For natural selection to occur two conditions—one phenotypic and one genetic—must be fulfilled (Stearns 1992): the phenotypic condition is that individuals must vary in reproductive success (i.e., fitness, as determined by the phenotypic components of fitness = life-history traits); the genetic condition is that there must be heritable variation for the trait under selection and that the trait is correlated with reproductive success (Robertson 1966). Before discussing the genetics of fitness components, I summarize some general aspects of the life cycle and phenotypic life history of D. melanogaster.
The following description is mainly based on Ashburner et al. (2005); additional references are given where appropriate. For more details see Ashburner et al. (2005) on the general biology, life cycle, and development of D. melanogaster; Parsons (1975), Roff (1992), Prasad and Joshi (2003), and David et al. (2004) on life history; Parsons (1975), Powell (1997), David et al. (2004), Markow and O’Grady (2008), Markow (2015), and Mansourian et al. (2018) on aspects of the natural history and ecology; and Lachaise et al. (1988) and Keller (2007) on the evolutionary history and biogeography of D. melanogaster.
The egg-to-adult life cycle
The vinegar fly D. melanogaster is a human commensal (Figure 1) of eastern sub-Saharan African origin (Lachaise et al. 1988; Keller 2007); it migrated out of Africa ∼12,000–19,000 years ago and subsequently became cosmopolitan (Li and Stephan 2006; Laurent et al. 2011; Duchen et al. 2013). It is a holometabolous insect that undergoes a complete metamorphosis from its larval form to its adult (imago) state (Figure 2). Adults breed on and larvae develop in rotten, fermenting fruit (Lachaise et al. 1988; Keller 2007), with yeasts that grow on the fruit being nutritionally critical for proper larval development (Sang 1978; Begon 1982). D. melanogaster is commonly referred to as a (or the) “fruit fly”; however, this is not entirely accurate since it does not directly feed on fruits (unlike flies of the family Tephritidae), but rather on microbes (yeasts and bacteria) associated with them. The name vinegar fly is derived from the fact that D. melanogaster is strongly attracted to acetic acid, the compound that gives vinegar its pungent smell, and which accumulates in fermenting fruits (Jouandet and Gallio 2015). Interestingly, recent work suggests that marula fruit, an important human staple food in the ancestral African range of D. melanogaster, might be the ancestral host and might have driven the close commensalism between flies and humans (Mansourian et al. 2018; also cf. Lachaise et al. 1988).
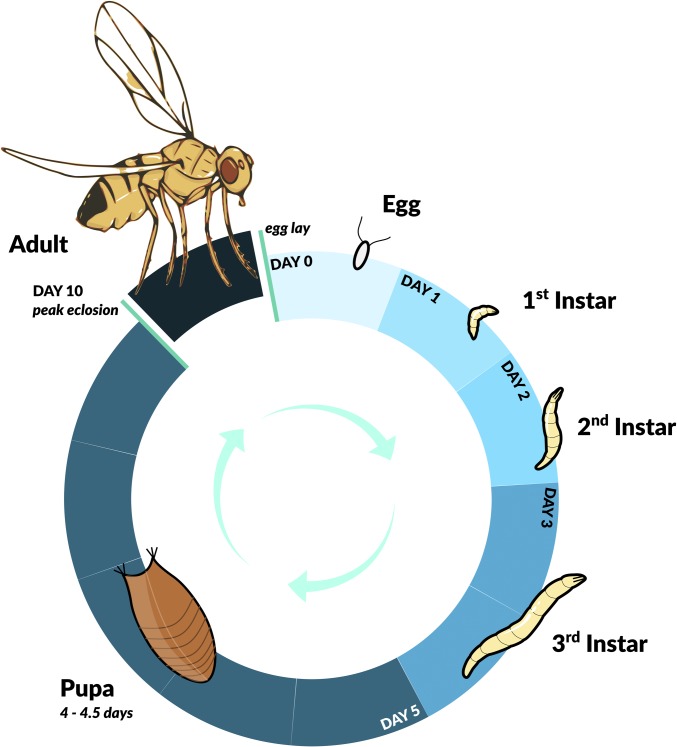
Figure 2.
The preadult life cycle of D. melanogaster. At 25° the developmental cycle, from the fertilized egg to the adult fly (imago), proceeds through three larval instar stages and one pupal stage, and takes ∼10 days. For a depiction of the adult part of the life cycle see Figure 3. See main text for further details. Figure credit: Chloé Schmidt (University of Manitoba).
Development from the egg to the adult takes ∼9–10 days under optimal standard conditions in the laboratory (∼25°, 60% humidity, nutritious food, and no overcrowding) (Figure 2). There can be considerable variation in egg-to-adult development time within large cohorts of flies or among wild-type strains, in the range of ∼8–16 days after egg laying (AEL). The adult eclosion peak typically occurs ∼10–11 days AEL, and by 15–16 days AEL > 99% of the viable flies have eclosed (Welbergen and Sokolowski 1994; Flatt 2004a; Flatt and Kawecki 2007). As mentioned above, females lay eggs on decaying fruit in the wild or on food medium in the laboratory; larvae hatch ∼22–24 hr AEL (Markow et al. 2009). The proportion of eggs that produce larvae (hatchability) is ∼90% at the beginning of life but then decreases with age (David et al. 1974, 1975; Klepsatel et al. 2013a). The larvae go through three larval stages (instars; L1–L3) in ∼4 days (L1 and L2: 24 hr, and L3: 48 hr) (Bakker 1959) (Figure 2). At the L1 stage larvae feed on the surface, whereas upon molting to the L2 stage the larvae burrow into the food. Approximately 5 days AEL, the larvae stop feeding, leave the food, and begin to wander around in search of an optimal site for pupariation. The pupal period, i.e., the time from pupariation to adult eclosion, lasts ∼4–4.5 days. The proportion of egg-to-adult survival (“viability”) is often ∼70–80% (Welbergen and Sokolowski 1994; Zwaan et al. 1995a; Gasser et al. 2000; Flatt 2004a; Flatt and Kawecki 2007).
For reviews of the physiology of growth and development, the attainment of “critical size,” and the genetics of size control, which are not discussed here, see Oldham and Hafen (2003), Prasad and Joshi (2003), Edgar (2006), Mirth and Riddiford (2007), Mirth and Shingleton (2012), and Ghosh et al. (2013), and references therein.
The adult life history
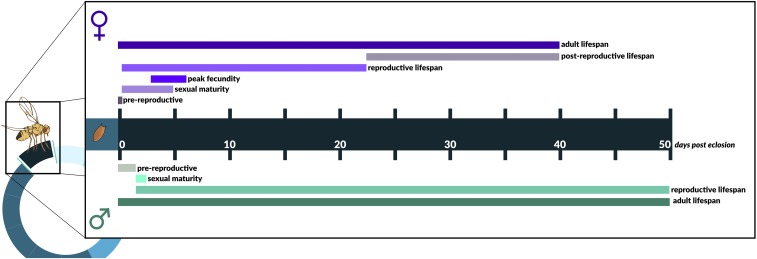
Upon eclosion, females are sexually unreceptive for ∼8–12 hr; they reach sexual maturity within 1–4 (typically 3–4) days after eclosion, while males become mature ∼2 days posteclosion (Pitnick et al. 1995; Klepsatel et al. 2013a) (Figure 3). Female and male size at maturity (using thorax length as a size proxy) is ∼0.9–1.15 and ∼0.85–0.95 mm, respectively (the total body length is ∼2–3 mm) (Robertson and Reeve 1952; David et al. 1977; Roff 1981; Telonis-Scott et al. 2005; Klepsatel et al. 2014); body size depends positively on development time (Alpatov 1929; Robertson 1960a).
Figure 3.
The adult life history of D. melanogaster. The figure gives (very approximate) timelines for the major life-history events and stages, including reproductive maturation, reproductive activity, and the overall life span of female and male flies. The durations of the different events and phases are mainly based on values obtained under optimal, protected laboratory conditions; however, estimates can vary widely among studies (i.e., depending on laboratory conditions, populations and strains assayed, etc.) and might therefore not be representative of the situation in the wild. See main text for further details; see Figure 2 for a depiction of the preadult life cycle. Figure credit: Chloé Schmidt (University of Manitoba).
Parentage analyses show that D. melanogaster females in natural populations typically mate with several males, even though the last male has the largest share of paternity (“last male sperm precedence”) (e.g., Giardina et al. 2017; Laturney et al. 2018; and references therein). Giardina et al. (2017) analyzed mating rates in a wild population and inferred that female flies mate a bit less than once per day, and that mating takes place in the early morning or late afternoon. For reviews of courtship and mating behavior see Spieth (1974) and Greenspan and Ferveur (2000).
Females can lay up to ∼100 eggs per day during peak fecundity, which is typically reached between ∼3 and 5 days after eclosion, and might produce ∼1000–3000 eggs in a lifetime (Shapiro 1932; Gowen and Johnson 1946; McMillan et al. 1970a,b; David et al. 1974; Klepsatel et al. 2013a). Age-specific and lifetime fecundity can be highly variable among individual females, laboratory strains, or wild populations measured in the laboratory (Bergland et al. 2012; Klepsatel et al. 2013a,b; Durham et al. 2014; Fabian et al. 2015). Under optimal, sheltered laboratory conditions lifetime reproductive success (total number of progeny surviving to adulthood) can be ∼500–1500 offspring (Partridge et al. 1986; Partridge 1988; Klepsatel et al. 2013a; Nguyen and Moehring 2015); in females, peak fecundity is often highly correlated with lifetime fecundity (Gowen and Johnson 1946; but cf. Klepsatel et al. 2013a). For example, the average number of offspring produced by females has been found to be in the order of ∼600 (mean: 615; range 0–1455) and 1700 (mean: 1699; range: 426–3198) for offspring produced by males (cf. Partridge et al. 1986; Partridge 1988). These numbers likely represent upper bounds and are probably markedly different from the situation in the field, e.g., because of environmental fluctuations (e.g., in food availability), environmentally imposed (“extrinsic”) mortality, and the presumably short lifetime of flies in the wild (Partridge 1988).
The maximum daily rate of egg production scales positively with the number of ovarioles, structures that represent “production lines” for making eggs within the paired ovaries (David 1970; King 1970; Boulétreau-Merle et al. 1982); females have ∼15–20 ovarioles per ovary, so ∼30–40 in total (King 1970; Wayne et al. 1997). Female fecundity increases with body size and, as mentioned above, size increases with development time (Alpatov 1929; Robertson 1960a; David and Bocquet 1974; Roff 1981). For a review of lifetime reproductive success in D. melanogaster see Partridge (1988), for behavioral aspects of reproduction see Markow and O’Grady (2005), and for reproductive ecology see Markow and O’Grady (2008).
In the laboratory, the adult life span of vinegar flies is on average ∼30–40 days for females and typically ∼5–10 days longer for males, but there can be tremendous variation among individuals, lines, populations, and environmental conditions, in the range of ∼3–90 days (Pearl and Parker 1924; Rose 1984b; Chippindale et al. 1993; Partridge et al. 1999a; Klepsatel et al. 2013a; Durham et al. 2014; Fabian et al. 2015; Ivanov et al. 2015); some wild-caught strains have a mean life span of > 80 days when measured in the laboratory (Linnen et al. 2001) (Figure 3). Unmated females and males live ∼10–20 days longer than mated flies (“survival cost of mating”; Partridge et al. 1986, 1987; Fowler and Partridge 1989; Partridge and Fowler 1992).
Under protected laboratory conditions flies can also exhibit postreproductive life span (PRLS) (Rogina et al. 2007; Mueller et al. 2009; Khazaeli and Curtsinger 2010; Klepsatel et al. 2013a). For example, Klepsatel et al. (2013a) tracked the life histories of individual female flies from three wild-caught populations and found that females have a reproductive life span of ∼20–22 days, during which they were fertile and produced viable offspring, followed by a postreproductive period of ∼14–15 days, during which fertility dropped to zero and which made up ∼40% of total life span. Because the duration of this period was not correlated with other fitness components, the authors concluded that PRLS observed under optimal, protected laboratory conditions likely represents a nonadaptive, random “add-on” at the end of reproductive life rather than a correlate of selection on reproductive fitness (Klepsatel et al. 2013a; cf. Reznick et al. 2005).
In the wild, the adult survival of flies is likely dramatically shorter than under optimal laboratory conditions. In a mark–recapture study, Rosewell and Shorrocks (1987) estimated average adult survival rates per day (φ) of 0.66 for females and 0.72 for males, giving a mean life expectancy (−1/ln[φ]) of ∼2.4–3 days. However, sample sizes in this study were small and the robustness of these estimates remains somewhat unclear [also see Boesiger (1968) and Boulétreau (1978)]. Notably, under some environmental conditions adult flies can persist for 6–9 months in the laboratory or under winter conditions in outdoor enclosures (see discussion of “reproductive dormancy” below). These observations indicate that vinegar flies possess the somatic ability to live much longer than usually thought, possibly also in the wild.
Quantitative Variation in Life-History Traits
I have already mentioned the large variation of fitness components in D. melanogaster; we now discuss the patterns, causes, and maintenance of this variability. As is true for phenotypic characters in general, phenotypic variation (VP) for life-history traits is due to genetic differences [genetic variation (VG)] and/or nongenetic differences [environmental variation (VE)] among individuals (Fisher 1918; Stearns 1992; Falconer and Mackay 1996; Roff 1997; Lynch and Walsh 1998; Charlesworth and Charlesworth 2010; Walsh and Lynch 2018). Because evolution by natural selection requires heritable variation in fitness-related traits among individuals in a population (Robertson 1966; Charlesworth and Edwards 2018; Walsh and Lynch 2018), the genetic component of phenotypic variation is the major driver of life-history evolution (Roff 1992; Stearns 1992). Life-history traits represent so-called quantitative traits, i.e., characters such as body height in humans for which variation is not discrete but continuous (or at least approximately so). This continuous distribution has two causes: the summing over of (i) the effects of many loci of typically small effect (polygeny) and (ii) environmental effects that influence the trait (East 1910; Fisher 1918; Stearns 1992; Falconer and Mackay 1996; Charlesworth and Charlesworth 2010; Charlesworth and Edwards 2018).
Below, I first discuss variation and covariation in fitness components with an emphasis on phenotypes (phenotypic variation within and among populations, focusing mainly on clines; correlations and trade-offs between fitness components; and patterns of life-history plasticity and genotype-by-environment interactions); then, I summarize what we know about the amount and maintenance of genetic variability in life-history traits. For background on quantitative genetics see Falconer and Mackay (1996), Roff (1997), Lynch and Walsh (1998), and Walsh and Lynch (2018).
Phenotypic patterns of life-history variation and covariation
Numerous studies, too many to discuss in detail, have measured phenotypic variation in fitness components by assaying these traits under standard laboratory conditions or, to examine plasticity, across different environmental conditions (e.g., Pearl and Parker 1921, 1922, 1924; Pearl 1932; Gowen and Johnson 1946; Buzzati-Traverso 1955; Knight and Robertson 1957; Robertson 1957b, 1960a; Kenyon 1967; McMillan et al. 1970b; Lewontin 1974; Parsons 1975; David and Capy 1988; Tanaka and Yamazaki 1990; Gebhardt and Stearns 1992, 1993a,b; Draye et al. 1994; Draye and Lints 1995, 1996; James et al. 1997; Prasad and Joshi 2003; David et al. 2004; Gibert et al. 2004; Mackay 2004; Schmidt et al. 2005a,b; Schmidt and Paaby 2008; Klepsatel et al. 2013a,b, 2014; Behrman et al. 2015; Fabian et al. 2015; Hangartner et al. 2015; Mackay and Huang 2018; and Lewontin et al. 2003, which provide a compilation of Dobzhansky’s classical work in D. pseudoobscura).
Such studies have measured fitness components, depending on their aims, using a variety of approaches, including measurements performed on individuals or groups (e.g., cohorts) of flies from laboratory mass culture (e.g., wild-derived flies in population cages), on strains derived from mass culture (e.g., laboratory wild-type strains such as Oregon R, Canton-S, or Samarkand), isofemale lines (established as full-sib families from a single inseminated wild-caught female), inbred lines (yielding homozygous estimates of fitness components), wild chromosome extraction lines (maintained over balancer chromosomes), recombinant inbred lines, mutation accumulation (MA) lines, and so forth (also see discussion below; e.g., Muller 1928; L’Héritier and Teissier 1933; Mukai 1964; Parsons and Hosgood 1967; Mukai et al. 1972; Lewontin 1974; Parsons 1975; Mackay 1985, 2001a,b, 2004; Tanaka and Yamazaki 1990; Charlesworth and Hughes 2000; Gayon and Veuille 2001; David et al. 2005; Mackay and Huang 2018).
Together, this vast body of work has revealed that there generally exist large amounts of phenotypic variation for components of fitness, both within and among populations, including traits such as development time, larval competitive ability, viability, size at eclosion, age-specific fecundity and fertility, lifetime reproductive success, age-specific mortality, life span, various stress resistance traits, reproductive dormancy, and so forth. Much of this variation is genetically based and therefore can, at least potentially, respond to selection (Roff and Mousseau 1987; Houle 1992, 1998; Charlesworth and Hughes 2000; Charlesworth 2015), as we shall discuss below.
The large extent of within-population life-history variability is well exemplified by considering the Drosophila Genetic Reference Panel (DGRP), a set of 205 inbred lines, derived from a population collected at a farmers’ market in Raleigh (NC). Dozens of genome-wide association studies (GWAS) have used these lines to map the genetic basis of phenotypic variance for a multitude of traits including several major fitness components; many of these mapping efforts have been aided by the fact that the DGRP lines often differ markedly, sometimes even extremely, from each other for various fitness-related traits (Mackay and Huang 2018). For example, Durham et al. (2014) used the DGRP panel to measure mated life span, age-specific fecundity (at weeks 1, 3, 5, and 7), and lifetime fecundity and to estimate the amounts of phenotypic, genetic, and environmental variation: the authors observed massive amounts of variation at all levels, with phenotypic coefficients of variation (CVP = ratio of phenotypic SD divided by the mean) for these traits ranging between ∼39 and 264%. However, estimates of phenotypic and genetic variances (and covariances) for fitness components are expected to be quite different in panels of inbred lines as compared to outbred populations, due to homozygosity and inbreeding depression in the former [e.g., see Charlesworth and Charlesworth (2010)].
Major patterns of differentiation for life-history traits have also been observed among natural populations of D. melanogaster when measured in the laboratory (e.g., Lemeunier et al. 1986; Coyne and Beecham 1987; David and Capy 1988; James and Partridge 1995; James et al. 1995, 1997; de Jong and Bochdanovits 2003; David et al. 2004; Gibert et al. 2004; Schmidt et al. 2005a,b; Hoffmann and Weeks 2007; Schmidt and Paaby 2008; Klepsatel et al. 2013a,b, 2014; Adrion et al. 2015; Fabian et al. 2015; Hangartner et al. 2015). So-called clines provide a particularly compelling example of among-population variation of fitness components, as we discuss next.
Among-population variation and life-history clines:
Clines are defined as systematic changes in the frequency of phenotypes or genotypes among populations that are spread along continuous environmental gradients through space, e.g., across latitude (Huxley 1938; Charlesworth and Charlesworth 2010). They are often thought to be driven by spatially varying selection, especially when similar clines are found repeatedly in different species or across distinct geographic areas within species (Haldane 1948; Levene 1953; Mayr 1963; Dobzhansky 1970; Slatkin 1975; Felsenstein 1976; Endler 1977, 1986; Hedrick 1986; Charlesworth and Charlesworth 2010; Adrion et al. 2015; Yeaman 2015). However, they can also arise from nonadaptive factors such as population structure and demography, for example admixture (Endler 1977; Bergland et al. 2016; Flatt 2016).
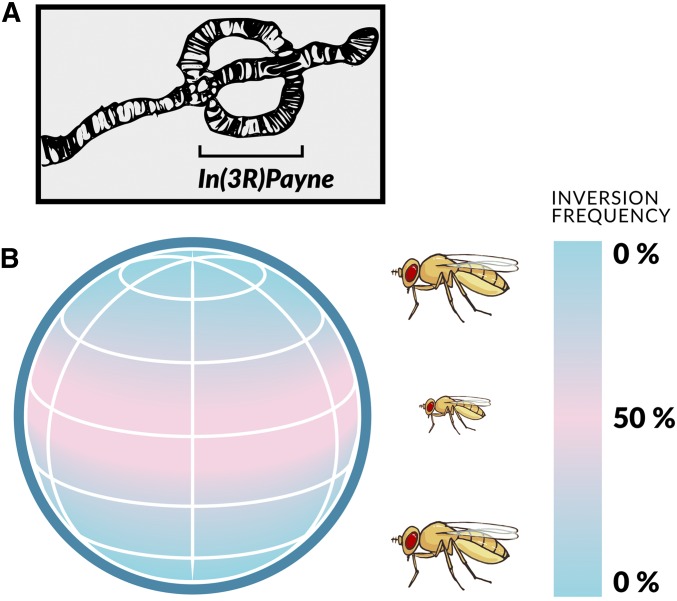
While clines have been investigated in many organisms, they have been particularly well studied in D. melanogaster (Figure 4; e.g., Lemeunier et al. 1986; David and Capy 1988; van Delden and Kamping 1997; de Jong and Bochdanovits 2003; David et al. 2004; Gibert et al. 2004; Hoffmann and Weeks 2007; Schmidt and Paaby 2008; Fabian et al. 2012, 2015; Klepsatel et al. 2014; Adrion et al. 2015; Hangartner et al. 2015; Kapun et al. 2016a,b; Durmaz et al. 2018, 2019).
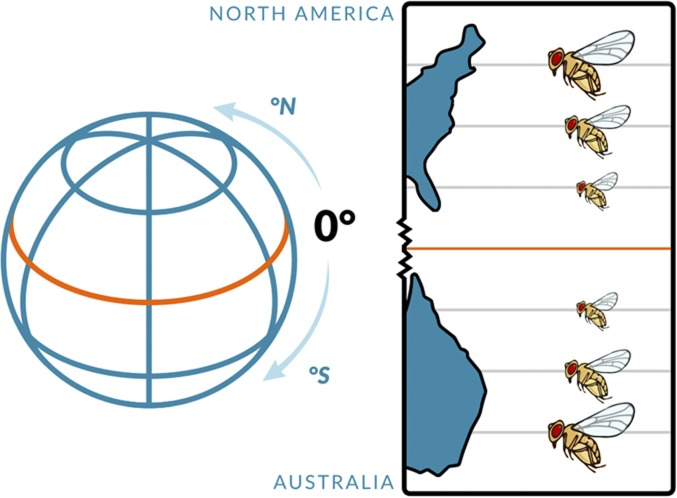
Figure 4.
Latitudinal life-history clines in D. melanogaster. On multiple continents and subcontinents, spanning temperate to subtropical/tropical regions, fly populations exhibit major differences in fitness components across latitudes. For example, in the northern hemisphere there exists a well-established latitudinal cline for body size along the North American east coast, with flies being larger in temperate populations (e.g., Maine) but smaller in subtropical/tropical areas (e.g., Florida). This pattern is matched, in an upside-down manner, in the southern hemisphere, for example along the Australian east coast. Such parallel clines exist for several fitness-related traits and imply that these clines are (at least partly) shaped by spatially varying selection. For example, high-latitude flies are typically not only larger but also less fecund, more stress-resistant, and longer-lived than flies from subtropical/tropical locales. See main text for further details; also see Figure 6. Figure credit: Chloé Schmidt (University of Manitoba).
Many studies have observed various fitness components to vary latitudinally (and sometimes also altitudinally) among natural populations of D. melanogaster, including clines in developmental rate (James and Partridge 1995; Van’t Land et al. 1999); larval growth efficiency (Robinson and Partridge 2001); viability (Folguera et al. 2008); body size and size-related traits (David and Bocquet 1975; Coyne and Beecham 1987; Capy et al. 1993; Imasheva et al. 1994; James et al. 1995, 1997; Van’t Land et al. 1999; Klepsatel et al. 2014; Fabian et al. 2015; Kapun et al. 2016b; Durmaz et al. 2019); ovariole number (David and Bocquet 1975; Capy et al. 1993); fecundity (Schmidt et al. 2005a; Schmidt and Paaby 2008; Fabian et al. 2015); egg size (Azevedo et al. 1996); starvation, desiccation, and cold- and heat-stress resistance (Da Lage et al. 1990; Karan et al. 1998; Hoffmann et al. 2002, 2005; Frydenberg et al. 2003; Durmaz et al. 2018; Rajpurohit et al. 2018); the propensity to undergo reproductive dormancy under cold conditions (Schmidt et al. 2005a,b; Schmidt and Paaby 2008); and life span (Schmidt and Paaby 2008; Fabian et al. 2015; Durmaz et al. 2018; also see Mitrovski and Hoffmann 2001; Sgrò et al. 2013).
For several of the above-mentioned traits, patterns of clinal differentiation have been observed in a parallel manner on multiple continents and subcontinents that span across both temperate and subtropical/tropical areas; thus it seems likely that in many cases these clines are shaped, at least in part, by spatially varying selection (Mayr 1963; David and Capy 1988; de Jong and Bochdanovits 2003; Rako et al. 2006; Hoffmann and Weeks 2007; Fabian et al. 2012, 2015; Klepsatel et al. 2014; Adrion et al. 2015; Kapun et al. 2016a,b). For example, flies from temperate regions often tend to be phenotypically and genetically larger, less fecund, more stress-resistant, and longer-lived than flies from subtropical/tropical climates (Figure 4).
This combination of fitness-related traits suggests a hypothetical selection regime whereby at high latitudes seasonal stresses (overwinter survival and ephemeral food resources) impose strong selection for increased stress resistance, metabolic reserves, and somatic maintenance at the cost of reduced fecundity, whereas warm climates with ample feeding and breeding opportunities favor increased larval competitive ability and high fecundity, at the expense of reduced size and decreased maintenance (Paaby and Schmidt 2009; cf. Sevenster and Van Alphen 1993; James and Partridge 1995; de Jong and Bochdanovits 2003). These opposite sets of trait values might be viewed as representing “pro maintenance and survival” vs. “pro reproduction” life-history “modes” or “strategies” (Flatt et al. 2013), which reflect local adaptation and life-history trade-offs across geography (Paaby et al. 2014; Fabian et al. 2015; Kapun et al. 2016b; Durmaz et al. 2018, 2019).
The direct causes of spatially varying selection underlying life-history clines remain poorly understood in most cases (Charlesworth and Charlesworth 2010; Kapun et al. 2016a,b; Durmaz et al. 2018, 2019). A possible exception is body size, a major fitness proxy: around the world, flies from high-latitude populations are larger than those from low-latitude populations [Figure 4; Misra and Reeve 1964 (in D. subobscura); David and Bocquet 1975; Coyne and Beecham 1987; David and Capy 1988; James et al. 1995, 1997; Klepsatel et al. 2014; Fabian et al. 2015; Kapun et al. 2016b; Durmaz et al. 2019], as is also the case for many other animals (“Bergmann’s rule”; Mayr 1963). The principal determinant underlying this pattern in flies appears to be temperature. Consistent with temperature being the causative factor, flies bred at lower temperature in experimental evolution studies in the laboratory evolve genetically larger size [Anderson 1973 (D. pseudoobscura); Cavicchi 1978, 1989, 1991; Huey et al. 1991; Partridge et al. 1994a; Santos et al. 1994; Bochdanovits and de Jong 2003a; de Jong and Bochdanovits 2003; Prasad and Joshi 2003] and show evidence for local adaptation to the cold (Partridge et al. 1994a; Bochdanovits and de Jong 2003b; cf. Nunney and Cheung 1997). Similarly, high-altitude flies are typically significantly larger than those from low elevations, with the parallelism between latitude and altitude being most parsimoniously explained by temperature (Pitchers et al. 2013; Klepsatel et al. 2014; Fabian et al. 2015; Lack et al. 2016).
What drives this pattern (Atkinson and Sibly 1997)? Ectotherms such as insects commonly exhibit an inverse relationship between developmental temperature and size [“temperature–size rule” (TSR); Atkinson 1994]. Several competing hypotheses have been put forward to account for the TSR phenomenon, including inevitable biophysical constraints on growth vs. various adaptive mechanisms (Ghosh et al. 2013). For the case of larval development in D. melanogaster, Ghosh and colleagues (2013) found that, in contrast to the hawkmoth Manduca sexta, the signal to terminate growth is initiated at a smaller (so-called) critical size at higher temperatures. Together with the evidence above, this suggests that the TSR represents an adaptive response to temperature, yet at the proximate level it is apparently achieved via distinct mechanisms in different species (Ghosh et al. 2013).
In contrast, some thermal experimental evolution studies in D. subobscura and D. melanogaster have failed to faithfully replicate the clinal patterns observed in natural populations, suggesting that temperature might not be the sole factor driving clinality (Santos et al. 2005; Kellermann et al. 2015). While these findings do not necessarily rule out that temperature is the causal factor in the wild (Huey and Rosenzweig 2009), they suggest that the natural pattern of clinal selection is much more complex than the one imposed in experimental evolution (cf. Kapun et al. 2016a,b).
Clines in D. melanogaster can thus serve as an insightful test bed for probing the causes and consequences of life-history adaptations in wild populations of flies. Beyond their phenotypic effects, clines have also been extensively studied in terms of population genomics (Turner et al. 2008, Kolaczkowski et al. 2011, Fabian et al. 2012, Bergland et al. 2014; Kapun et al. 2014, 2016a; Reinhardt et al. 2014; Adrion et al. 2015; Machado et al. 2016).
Correlations between life-history traits and trade-offs:
To function properly, and to survive and reproduce, organisms must work as well-integrated entities. Although it can be conceptually or practically convenient to define and measure single traits, organisms are obviously not collections of well separable, independent characters (Stearns 1984, 1989b; 1992; Wagner 2001; Pigliucci and Preston 2004; Flatt et al. 2005; Flatt and Heyland 2011; Martin et al. 2015). This also applies to fitness components, which are commonly tightly integrated through various developmental, physiological, and genetic mechanisms that result in positive or negative correlations (covariances) between life-history traits (Bell and Koufopanou 1986; Stearns 1989a, 1992; Zera and Harshman 2001; Prasad and Joshi 2003; Flatt et al. 2005, 2013; Roff 2007; Roff and Fairbairn 2007; Flatt 2011; Flatt and Heyland 2011; Hughes and Leips 2017). Life-history traits should thus be viewed from a multivariate perspective as a set of interrelated traits that jointly determine reproductive success or fitness (Lande 1982; Lande and Arnold 1983; Charlesworth 1993a; Roff 2007).
Correlations between traits arise from genetic and/or nongenetic (environmental) sources of covariance; to the extent that they are genetically determined, they imply that the traits cannot evolve independently (Stearns 1989a, 1992; Falconer and Mackay 1996; Roff 1997, 2002, 2007). While phenotypic correlations do not necessarily reflect underlying genetic correlations and can arise from environmental sources, they can be reasonably good predictors of genetic correlations, especially for morphological traits (“Cheverud’s conjecture”; Roff and Mousseau 1987; Cheverud 1988; Roff 1995).
The best-known type of life-history correlation are “trade-offs,” defined as negative correlations between components of fitness (Stearns 1989a, 1992; Roff 1992, 2007, 2002; Prasad and Joshi 2003; Hughes and Leips 2017). For example, levels of reproductive effort might trade off with growth or survival (“costs of reproduction”; reproductive effort model; Fisher 1930; Williams 1966). To the extent that phenotypic trade-offs are rooted in negative genetic correlations, they might represent evolutionary (genetic) constraints: selection for an increased value of one fitness-related trait causes a correlated decrease of the value of the other fitness component and might thus constrain their independent evolution.
Phenotypic and genetic correlations between fitness components in D. melanogaster are pervasive, and have commonly been observed in quantitative genetic studies and analyses of correlated responses to selection in the laboratory [see below and Prasad and Joshi (2003)]. For example, as mentioned above, high- and low-latitude flies typically differ with respect to the values of several life-history traits, suggesting multivariate correlations and trade-offs within and, due to local adaptation along the cline, among populations (Paaby and Schmidt 2009; Flatt et al. 2013; Paaby et al. 2014; Fabian et al. 2015; Durmaz et al. 2019). Although there are many exceptions and patterns can be extremely variable across studies (Rose and Charlesworth 1981a; Giesel et al. 1982; Roff and Mousseau 1987; Gromko 1995), the following correlations tend often to be observed [see Stearns and Partridge (2001), Prasad and Joshi (2003), and section on correlated responses below]: usually positive correlations between development time, size at eclosion, ovariole number, and fecundity; commonly a negative correlation between life span and early fecundity; sometimes a trade-off between early and late-life fecundity; often a positive correlation between total (lifetime) fecundity and life span; and typically no consistent correlations between development time and/or size with life span (Alpatov 1929; Robertson 1957a,b, 1960; David and Bocquet 1974, 1975; Rose and Charlesworth 1981a,b; Giesel et al. 1982; Luckinbill et al. 1984; Rose 1984b, 1991; Bell and Koufopanou 1986; Roff and Mousseau 1987; Tucić et al. 1988; Tanaka and Yamazaki 1990; Roff 1992; Stearns 1992; Zwaan et al. 1995a,b; Lefranc and Bundgaard 2000; Stearns et al. 2000; Stearns and Partridge 2001; Harshman 2003; Prasad and Joshi 2003; Rose et al. 2004; Schmidt et al. 2005a,b; Bergland et al. 2008; Schmidt and Paaby 2008; Flatt and Schmidt 2009; Paaby and Schmidt 2009; Flatt 2011; Fabian et al. 2015).
Fitness components in flies are also frequently correlated with traits that confer the ability to resist and survive various stresses, and environmental insults [reviewed in Prasad and Joshi (2003); cf. section on correlated responses]. Although stress resistance traits are not usually defined as life-history traits sensu stricto, they represent important fitness components because they contribute to somatic maintenance and thus to survival. Such stress resistance traits include, for example, resistance to starvation, desiccation, oxidative stress, and cold and heat, with resistance being measured as survival after exposure (Service 1987; Rose et al. 1992, 2004; Lin et al. 1998; Hoffmann and Harshman 1999; Harshman et al. 1999a; Harshman and Haberer 2000; Hoffmann et al. 2001, 2005; Salmon et al. 2001; Wang et al. 2001, 2006; Prasad and Joshi 2003; Arking 2006; Rion and Kawecki 2007; Goenaga et al. 2010; Tower 2011; Kellermann et al. 2012a,b; Hansen et al. 2013; Kalra and Parkash 2014; Wit et al. 2015). Correlations between classical fitness components and stress resistance traits are often seen in long-lived genotypes (long-lived mutants; flies selected for increased life span), where increased life span and reduced early life fecundity go together with increased resistance to one or multiple stressors (Hoffmann and Harshman 1999; Rion and Kawecki 2007; Flatt and Schmidt 2009; Flatt 2011; Hansen et al. 2013).
What is the genetic basis of life-history correlations and trade-offs? Genetic correlations between fitness components measure the degree to which two life-history traits are affected by one or several loci as the result of pleiotropy and/or linkage disequilibrium (LD) among the loci affecting the trait (Stearns 1992; Falconer and Mackay 1996; Roff 1997, 2007; Conner and Hartl 2004; Hughes and Leips 2017). Given that recombination can break down LD readily, pleiotropy is likely the predominant factor in causing stable (nontransient) genetic correlations (Roff 1997, 2007). Pleiotropic effects on the values of two traits can either be positive (+, +), negative (−, −), or antagonistic (+, − or −, +); positive and negative pleiotropy cause positive genetic correlations, while antagonistic pleiotropic effects cause negative genetic correlations. A major but underappreciated caveat is that a lack of genetic correlation does not always imply a lack of pleiotropy: if alleles or loci vary in the signs and magnitudes of their pleiotropic effects, the effects might cancel each other out, resulting in a net correlation of zero (“variable pleiotropy”; Gromko et al. 1991; Gromko 1995; Falconer and Mackay 1996; Lyman and Mackay 1998; Flatt and Kawecki 2004). Only “consistent” pleiotropic effects will, on average, lead to significant correlations between fitness components.
Experimental work in D. melanogaster and other species has revealed that mutational correlations between fitness components arising from deleterious de novo mutations are typically positive: this is because most deleterious mutations affect two or more fitness components negatively in the same direction, with different mutations doing so to a different extent (Houle et al. 1994b; Keightley and Ohnishi 1998; Pletcher et al. 1998; also see below). Yet, in terms of standing genetic variance in equilibrium populations, we expect to find negative genetic correlations (trade-offs) for at least some pairs of fitness components (Charlesworth 1990, 1993b, 1994). Because selection exhausts VA for net fitness, alleles with unconditionally beneficial effects on two or more fitness components should become fixed, whereas alleles with deleterious pleiotropic effects on multiple fitness-related traits should be eliminated (Hazel 1943). Hence, the remaining standing variance for fitness components might represent segregating alleles that exhibit antagonistic pleiotropy (AP) and that cause negative genetic covariances between life-history traits (Hazel 1943; Dickerson 1955; Robertson 1955; Charlesworth 1980, 1990, 1993b, 1994; Rose 1982, 1985; Houle 1991; Curtsinger et al. 1994; Roff 1997; Lynch and Walsh 1998; Charlesworth and Hughes 2000).
Many studies in D. melanogaster and other organisms support the existence of negative genetic correlations between fitness components consistent with trade-offs and AP (also cf. section on correlated responses). For instance, the genetically based trade-off between early fecundity and life span, observed in many selection experiments, lends strong support to a central explanation for the evolution of senescence (aging): under the AP hypothesis of the evolution of aging, due to Medawar (1946, 1952) and Williams (1957), senescence evolves because, when the force of selection declines with age, selection favors alleles with beneficial effects on early life fitness components even when they have deleterious effects late in life (Charlesworth 1980; Rose and Charlesworth 1981a,b; Rose 1991; Charlesworth 1993b, 1994; Partridge and Barton 1993; Rose and Bradley 1998; Stearns and Partridge 2001; Flatt and Promislow 2007; Flatt and Schmidt 2009; Charlesworth and Charlesworth 2010; Flatt 2011; Gaillard and Lemaître 2017; Austad and Hoffman 2018; Flatt and Partridge 2018). Overall, the existence of AP alleles in D. melanogaster (and other model organisms such as Caenorhabditis elegans) is well supported by laboratory analyses of large-effect mutants and transgenic constructs [see Tatar et al. (2001a) and reviews in Tatar et al. (2003), Kenyon (2005), Partridge et al. (2005a), Flatt and Schmidt (2009), Paaby and Schmidt (2009), Flatt (2011), Flatt et al. (2013), Hughes and Leips (2017), and Austad and Hoffman (2018)], and a growing body of evidence suggests that segregating polymorphisms in natural populations has pleiotropic effects on fitness components consistent with genetic trade-offs (Paaby and Schmidt 2009; Mackay 2010; Paaby et al. 2014; Durmaz et al. 2019).
Although it seems clear that at the genetic-level trade-offs are caused by genetic correlations due to AP (and/or LD), little is known about their underlying proximate causes, especially their physiological underpinnings (Scheiner et al. 1989; Rose and Bradley 1998; Leroi 2001; Zera and Harshman 2001; Barnes and Partridge 2003; Flatt et al. 2005, 2008; Flatt and Kawecki 2007; Harshman and Zera 2007; Roff 2007; Flatt and Heyland 2011; Flatt 2011; Metcalf 2016; Hughes and Leips 2017). This is a major unresolved problem that limits our understanding of the functional constraints that act on the evolution of life histories (Box 1).
Box 1. What is the functional nature of life-history trade-offs?
“It would be instructive to know not only by what physiological mechanisms a just apportionment is made between the nutriment devoted to the gonads and that devoted to the rest of the parental organism, but also what circumstances in the life-history and environment would render profitable the diversion of a greater or lesser share of the available resources towards reproduction.” R. A. Fisher (1930, p. 43–44).
Genetic correlations are merely statistical descriptions (Stearns 1989a, 1992; Falconer and Mackay 1996; Roff 2007): many experiments have found correlations consistent with trade-offs, but they are not informative about their causes (Charlesworth 1990; Houle 1991; Partridge and Barton 1993; Gromko 1995; Hughes and Leips 2017). Notably, theory shows that the relationship between genetic covariances (or the genetic variance–covariance matrix, G) and underlying functional constraints might be complex and indirect, so that only little about the former can be learned from the latter (Pease and Bull 1988; Charlesworth 1990; Houle 1991). In equilibrium populations, positive genetic correlations for trait pairs embedded in a higher-dimensional system of fitness components do not rule out that these traits are involved in negative genetic correlations and thus subject to constraint; therefore, finding positive genetic correlations does not exclude trade-offs (Pease and Bull 1988; Charlesworth 1990, 1993b). On the other hand, negative genetic correlations indicate that constraints might be at play, yet there are always some positive genetic correlations between particular trait pairs expected to be present at equilibrium (Charlesworth 1990). Hence, analyses of genetic correlations (or G) shed little light on functional constraints (Pease and Bull 1988; Charlesworth 1990). This is echoed by Houle (1991), who modeled genetically based resource allocation trade-offs between life-history traits: “The form of G does not necessarily reveal the constraint on resource acquisition inherent in the system, and therefore studies estimating G do not test for the existence of life-history tradeoffs. Characters may evolve in patterns that are unpredictable from G.” Similarly, using simulations, Gromko (1995) found that correlated responses to selection can be strictly constrained even when the genetic correlation is zero.
The rather crude insights into trade-offs gained from analyzing correlations thus call for detailed analyses of “functional architecture,” i.e., the pathways that connect genotypes to phenotypes (Houle 1991, 2001; Chippindale et al. 1993, 1997; Finch and Rose 1995; Rose and Bradley 1998; Leroi 2001; Barnes and Partridge 2003; Flatt et al. 2005; Roff 2007; Flatt and Heyland 2011; Hughes and Leips 2017; Ng’oma et al. 2017; Flatt and Partridge 2018).
For instance, it is usually assumed that at the physiological level, genetically based trade-offs might manifest themselves as allocation trade-offs between processes that compete for energetic resources such as growth, reproduction, survival, and somatic maintenance (Fisher 1930; Williams 1966; Sibly and Calow 1985; van Noordwijk and de Jong 1986; Scheiner et al. 1989; Houle 1991; de Jong and van Noordwijk 1992; Perrin and Sibly 1993; Rose and Bradley 1998; Houle 2001; Metcalf 2016; Nestel et al. 2016; Ng’oma et al. 2017); this is commonly called the “Y model” (de Jong and van Noordwijk 1992). However, while allocation trade-offs are commonly invoked, they are rarely firmly established; and while AP-effect loci underlying genetic trade-offs might be involved in resource acquisition and/or allocation, they could have effects that are independent of resource allocation, i.e., involving other types of physiological and/or structural constraints (Scheiner et al. 1989; Tatar and Carey 1995; Leroi 2001; Barnes and Partridge 2003; Flatt 2009, 2011; Tatar 2011; Metcalf 2016; Hughes and Leips 2017). Indeed, despite some evidence in favor of resource allocation trade-offs [reviewed in Boggs (2009), Nestel et al. (2016), and Ng’oma et al. (2017)], studies that have examined the physiological basis of life-history trade-offs in flies, i.e., by measuring details of resource acquisition vs. allocation and/or energy metabolism, have found little or no evidence for the classical resource allocation model (Djawdan et al. 1996; Simmons and Bradley 1997; Min et al. 2006; O’Brien et al. 2008; Grandison et al. 2009a; also cf. Flatt 2011; Tatar 2011; Ng’oma et al. 2017). Moreover, several studies have found that trade-offs can be “uncoupled,” e.g., by genetic or dietary manipulation [reviewed in Flatt (2011) and Flatt and Partridge (2018)]. In sum, phenotypic and genetic correlations are only remotely connected to constraints on life-history evolution (Charlesworth 1990; Houle 1991; also cf. Roff and Fairbairn 2007; Conner 2012; Metcalf 2016), and the expression of trade-offs can be highly dynamic and contingent (Stearns 1989a, 1992; Flatt 2011; Ng’oma et al. 2017; Hughes and Leips 2017).
Environmental variation and life-history plasticity:
Another fundamental aspect of the quantitative genetics of life-history traits is phenotypic plasticity, i.e., the ability of a single genotype to produce multiple phenotypes across environments (Stearns 1989c, 1992; Scheiner 1993; Via et al. 1995; Roff 1997; Flatt 2005). Plasticity, which is mechanistically due to differences in gene expression across environments, represents the environmental (nongenetic) variance in the phenotype (VE), i.e., all phenotypic variation that is due to environmental effects (Falconer and Mackay 1996; Flatt 2005). Although it is commonly assumed that most plasticity is adaptive, environmentally induced variation can be neutral or even maladaptive (Steiner and Tuljapurkar 2012; Acasuso-Rivero et al. 2019); in fact, plasticity might often be physiologically inevitable and/or caused by random environmental changes, including “microenvironmental” variation (“developmental noise”; Flatt 2005). For introductions regarding plasticity see chapter 3 in Stearns (1992) and chapter 6 in Roff (1997); for book-length treatments see Schlichting and Pigliucci (1998), Pigliucci (2001), West-Eberhard (2003), DeWitt and Scheiner (2004), and Whitman and Ananthakrishnan (2009).
There are several methods for conceptualizing and analyzing plasticity (Stearns 1992; Scheiner 1993; Via et al. 1995; Roff 1997): for quantitative traits in continuous environments, a convenient way is to measure a genotype’s phenotypic value across several values of the environmental parameter (e.g., temperature): the resulting curve (function) maps the phenotype to the environment and is called the genotype’s “reaction norm.” The steeper the slope of the reaction norm, the higher the degree of plasticity (Stearns 1992; Roff 1992, 1997; Flatt 2005): most life-history traits are typically highly sensitive to changes in the environment (Price and Schluter 1991; Houle 1992; Travis 1994; Nylin and Gotthard 1998; Flatt et al. 2013). Genetic variation for plasticity is present when the genotypes in a population differ in the slope and/or curvature of their reaction norms across environments: this is called “genotype–environment interaction” (GxE), and the model for partitioning the total phenotypic variance of a trait can then be written as VP = VG + VE + VGxE (Stearns 1992; VE + VGxE = VPL = plastic variance).
Life-history plasticity can be evolutionarily important for at least four reasons. First, if there are significant amounts of GxE for fitness components, and if there exist recurring, predictable environmental changes (i.e., reliable environmental cues), selection might favor genotypes with “optimal” (fitness maximizing) phenotypic responses to changes in the environment (“optimal reaction norm”: Stearns and Koella 1986; cf. Via et al. 1995; Rueffler et al. 2006). Selection for adaptive plastic life-history responses is expected to cause an erosion of genetic variation in reaction norms at the population level, leading to a genetically “canalized” bundle of reaction norms (Flatt 2005; Acasuso-Rivero et al. 2019). However, even though life-history traits are expected to be highly sensitive to environment on theoretical grounds (Price and Schluter 1991; Houle 1992), a recent meta-analysis has found that they might not necessarily be more or less plastic than other (morphological or behavioral) traits (Acasuso-Rivero et al. 2019). Importantly, this analysis suggests that plasticity of life-history traits, despite the proximity of these traits to fitness, might often be neutral or maladaptive (Acasuso-Rivero et al. 2019; cf. Steiner and Tuljapurkar 2012); convincing cases of adaptive life-history plasticity thus require robust empirical evidence (Travis 1994; Flatt et al. 2013; see below). Second, plasticity changes the genetic response of the trait(s) to selection across environments (Stearns et al. 1991; Stearns 1992): it does so by modulating (i) how genetic variation for fitness components is expressed across environments (depending on the environment, phenotypic differences between genotypes can be blurred, amplified, or their phenotypic ranking reversed) and (ii) the phenotypic expression (the magnitude and/or sign) of genetic correlations between different environments. Importantly, plasticity and GxE can change, for example, a negative correlation in one environment into a positive correlation in another (Stearns 1989c, 1992), which explains why the expression of trade-offs is often dynamic and contingent. Third, a related point is that under changing environments, GxE interactions for fitness components and net fitness can maintain genetic variation (Mukai 1988; Gillespie 1991; Stearns 1992; Mackay 2010). Fourth, plasticity can allow for compensation among fitness components, so that a fitness reduction through a plastic change in one trait might be balanced by increased fitness through a plastic response in another trait (Stearns 1992; Flatt 2005).
In D. melanogaster, life-history plasticity and reaction norms have been particularly well investigated with regard to the effects of temperature, nutrition, and crowding [Imai 1933; Parsons 1961; David et al. 1983; Gebhardt and Stearns 1988, 1993a,b (D. mercatorum); Zwaan et al. 1992; Chippindale et al. 1993; Delpuech et al. 1995; Huey et al. 1995; Zamudio et al. 1995; Crill et al. 1996; De Moed et al. 1997; James et al. 1997; Nunney and Cheung 1997; Prasad and Joshi 2003; Gibert et al. 2004; Rose et al. 2004; Trotta et al. 2006; Tatar 2007, 2011; de Jong and Van der Have 2009; Flatt and Schmidt 2009; Whitman and Ananthakrishnan 2009; Schmidt 2011; Flatt et al. 2013; Klepsatel et al. 2013b; Flatt 2014; Clemson et al. 2016; Mathur and Schmidt 2017; van Heerwaarden and Sgrò 2017], and also with respect to adult reproductive dormancy (Saunders et al. 1989; Tatar and Yin 2001; Tatar et al. 2001b; Schmidt et al. 2005a,b; Schmidt and Paaby 2008; Flatt et al. 2013).
In terms of thermal plasticity, a multitude of studies has established that flies raised at cool temperatures (e.g., ≤ 18°) develop more slowly; show reduced viability; eclose as adults at a larger size (e.g., larger thoraces and wings, and increased dry weight); exhibit reduced sexual dimorphism for size; have reduced ovariole number, decreased early fecundity, and lower “thermal fecundity performance”; produce eggs that are larger; are more cold- and starvation-resistant; and live longer than flies raised at warmer temperatures (e.g., ≥ 25°) [Alpatov and Pearl 1929; Alpatov 1930; Maynard Smith 1958 (in D. subobscura); Zwaan et al. 1992; David et al. 1994, 1997, 2011; Partridge et al. 1994a,b; Delpuech et al. 1995; James and Partridge 1995; Azevedo et al. 1996; Crill et al. 1996; De Moed et al. 1997; James et al. 1997; Nunney and Cheung 1997; French et al. 1998; Trotta et al. 2006; Folguera et al. 2008; Klepsatel et al. 2013b, 2019; Fallis et al. 2014; Mathur and Schmidt 2017; Ørsted et al. 2019]. With respect to fecundity performance and reproductive fitness, a temperature of ∼25° seems to be invariably optimal (Klepsatel et al. 2013b, 2019).
Thermal plasticity of gene expression (“transcriptional plasticity”) has also been studied, for example in the context of population-level plasticity, thermal reaction norms and GxE (Zhou et al. 2012; Carreira et al. 2013; Chen et al. 2015), fluctuating temperatures (Sørensen et al. 2016), and latitudinal clines [Levine et al. 2011; Chen et al. 2012; Zhao et al. 2016; Porcelli et al. 2016 (D. subobscura); Clemson et al. 2016). Such studies can be informative about the mechanisms underlying thermal life-history plasticity.
Notably, Nunney and Cheung (1997) found that size changes in response to rearing temperature are accompanied by changes in early fecundity and longevity, which support the hypothesis that the thermal reaction norm for size represents an adaptive plastic response. It is noteworthy in this context that the plastic response of size to temperature seems to parallel the genetic response of size to thermal laboratory selection and presumably, in the case of latitudinal clines (see above), to natural selection (Nunney and Cheung 1997).
The idea that thermal reaction norms are shaped by selection, and that this selection erodes genetic variation for thermal plasticity, is consistent with the observation that population-level reaction norms for six populations (from Africa and Europe, spanning tropical and temperate areas) assayed for size-related traits, ovariole number, and fecundity performance across seven fluctuating temperature regimes (ranging on average from 14 to 30°) were remarkably parallel, with little evidence for GxE (Klepsatel et al. 2013b). Similarly, an analysis of 19 populations across the eastern Australian cline did not find any latitudinal differentiation in plasticity for developmental time, thorax length, and wing size (James et al. 1997). Yet, many studies have observed significant amounts of variation for plasticity of fitness components in response to thermal change, both within and among populations [Parsons 1977 (D. simulans), 1978; Murphy et al. 1983 (D. simulans); Scheiner et al. 1989; Gebhardt and Stearns 1993a; Van’t Land et al. 1999; Vieira et al. 2000; Lazzaro et al. 2008; Klepsatel et al. 2013b; Fallis et al. 2014; Mathur and Schmidt 2017; Lafuente et al. 2018; Ørsted et al. 2019], including latitudinal differentiation in plasticity. For example, Mathur and Schmidt (2017) found that, in contrast to low‐latitude North American populations, high‐latitude populations are more responsive to cold exposure and exhibit more rapid recovery from chill coma in response to cold temperatures in the field, suggesting differential patterns of local adaptation for adaptive plasticity along the cline. In terms of uncovering the genetic basis of thermal plasticity for body size, important recent progress has been made by Lafuente et al. (2018) who applied a GWAS approach to the DGRP lines.
Another, and perhaps the most fundamental, environmental factor in the life of the vinegar fly is nutrition, which is obviously critical for development, growth, survival, and reproduction; it is also central to the notion of resource allocation trade-offs discussed above (cf. Nestel et al. 2016). Effects of diet quality and quantity on many aspects of fly development, growth, physiology, and life history have been studied in great detail, beginning > 100 years ago and using a variety of different natural fruit, and various laboratory-made and chemically defined (“holidic”), food media (Delcourt and Guyenot 1910; Northrop 1917; Baumberger 1919; Sturtevant 1921; Beadle et al. 1938; Tatum 1939; Sang 1956, 1978; Robertson 1960a; Sang and King 1961; Begon 1982; Ashburner et al. 2005; Bass et al. 2007; Tatar 2007, 2011; Lee and Micchelli 2013; Piper et al. 2014). Most of the early work [reviewed in Sang (1978)] focused on the dietary requirements for proper larval development, growth, and the attainment of adult size, a research tradition that has become increasingly molecular, and has led to modern studies of growth control and metabolism (Britton and Edgar 1998; Britton et al. 2002; reviewed in Edgar 2006; Géminard et al. 2006; Baker and Thummel 2007; Leopold and Perrimon 2007; Tennessen and Thummel 2011; Hansen et al. 2013). Research on how nutrition impacts Drosophila life history has focused on numerous aspects, including the effects of malnutrition, overfeeding, dietary restriction (DR), calories, the ratio (“balance”) of diet components, and of specific nutrients such as essential amino acids.
Not surprisingly, food shortage during development (e.g., reduction or deprivation of yeast, or whole-food dilution) increases developmental time, and decreases body size and size-related traits (Beadle et al. 1938; Bubliy et al. 2001; Tu and Tatar 2003; Layalle et al. 2008; Vijendravarma et al. 2011; Klepsatel et al. 2018). Beadle et al. (1938) found that larvae stopped growing and died after a few days if starved before 70–72 hr AEL but, when starved after this time, larvae would grow into very small adults which, as shown by Tu and Tatar (2003), have 50% fewer ovarioles, greatly reduced fecundity, and shortened life span. Conversely, larval overfeeding, e.g., on a high-sugar diet, markedly prolongs development, reduces size, and increases fat storage (Palanker Musselman et al. 2011; Reis 2016); similarly, overfeeding in the adult stage leads to weight gain, increased fat storage, and can shorten life span (Skorupa et al. 2008; Morris et al. 2012).
Given the importance of fecundity and adult survival in determining fitness, the effects of diet on these adult traits are especially interesting. Generally, adult food deprivation or very restricted diets reduce fecundity and life span, whereas high food levels increase fecundity but decrease life span (Hollingsworth and Burcombe 1970; Chapman and Partridge 1996; Good and Tatar 2001; Tatar 2011). Interestingly, within a range of relatively low-to-intermediate food concentrations (usually in the adult stage; but see below), one can observe so-called DR effects (specifically defined as reduced food intake without malnutrition); the hallmarks of such DR effects are that they extend life span but reduce fecundity. This phenomenon, first discovered in rats in 1935, has been examined by many studies in Drosophila, with a strong focus on understanding the molecular mechanisms underlying DR-induced longevity [for the first demonstration of DR in Drosophila see Chippindale et al. (1993); also see Chapman and Partridge (1996), Mair et al. (2003, 2005), Magwere et al. (2004), Bross et al. (2005), Burger et al. (2007), Min et al. (2007), Ja et al. (2009), and Burger et al. (2010); reviewed in Partridge et al. (2005b,c), Piper et al. (2005, 2011), Pletcher et al. (2005), Tatar (2007, 2011), Mair and Dillin (2008), Flatt (2014), Tatar et al. (2014), Hoedjes et al. (2017), and Kapahi et al. (2017)]. While DR is usually implemented at the adult stage, several studies have found that a (not too strongly) restricted juvenile diet can also extend adult life span (Economos and Lints 1984; May et al. 2015; Stefana et al. 2017); remarkably, depending on the adult diet, larval yeast DR can double median life span, a carry-over effect caused by the larval diet-induced suppression of toxic, life span-shortening lipids produced by the adults (Stefana et al. 2017).
Initially it was thought that the effects of adult DR might be caused by reduced intake of calories (“caloric restriction”), but it was later found that they are in fact independent of calories (Mair et al. 2005; also cf. Min et al. 2007; Tatar 2011); careful studies that manipulated yeast and sugar in different combinations discovered that DR extends life span because of the reduced amount of dietary yeast (relative to sugar), the major source of protein in the fly diet, suggesting that the main determinant is the ratio of protein to carbohydrate (Lee et al. 2008; Skorupa et al. 2008; Bruce et al. 2013; Lee 2015; Tatar et al. 2014). These insights were aided by the advent of the “nutritional geometry” framework, providing a quantitative method for examining multidimensional dietary responses (Lee et al. 2008; Simpson and Raubenheimer 2012; Flatt 2014, Tatar et al. 2014). The role of yeast is also underscored by observations showing that the quality and species of dietary yeast fungi has profound effects on fly life history (Begon 1982; Bass et al. 2007; Anagnostou et al. 2010; Grangeteau et al. 2018). In contrast, sugar has overall rather little effect on life span but increasing amounts reduce fecundity (Bass et al. 2007; Min et al. 2007). Subsequently, research in this area has led to the realization that dietary proteins (and essential amino acids) from yeast are crucial for egg production, but that they have major life span-shortening effects (Min and Tatar 2006; Grandison et al. 2009a; Lee et al. 2014; Piper et al. 2017; also cf. Flatt 2009; Tatar et al. 2014; Hoedjes et al. 2017).
From an evolutionary genetic perspective, several studies have reported substantial amounts of genetic variance for dietary plasticity within and among populations (or strains) of D. melanogaster for various fitness-related and metabolic traits, and for the DR response itself, e.g., using quantitative genetics estimation or experimental evolution approaches [Gebhardt and Stearns 1988 (D. mercatorum); Hillesheim and Stearns 1991; Gebhardt and Stearns 1993a; Bergland et al. 2008; Grandison et al. 2009b; Reed et al. 2010; Dick et al. 2011; Metaxakis and Partridge 2013; Zajitschek et al. 2016, 2019; Ng’oma et al. 2019]. Thus, fly populations are expected to be able to rapidly adapt to specific diets or changing dietary conditions.
In terms of molecular mechanisms, insights into the physiological underpinnings of DR and dietary plasticity have come from analyses of gene expression changes (Pletcher et al. 2002; Carsten et al. 2005; Gershman et al. 2007; Ding et al. 2014; Whitaker et al. 2014; Stanley et al. 2017; Zandveld et al. 2017; Hemphill et al. 2018), and many studies have sought to identify genes that underlie DR-induced longevity using mutants and transgenes, with growing (but still ambiguous) evidence for an involvement of genes in the insulin/insulin-like growth factor signaling (IIS)/target of rapamycin (TOR) pathways [reviewed in Tatar et al. (2014); also cf. discussion in Flatt (2009), Hoedjes et al. (2017), and Flatt and Partridge (2018)]. However, there is still little evidence for any specific gene to be functionally required for the life span increase under DR (Tatar 2007, 2011; Flatt 2014; Tatar et al. 2014).
Evolutionary biologists have been particularly interested in DR because it might represent an example of adaptive plasticity and underpin dynamic resource allocation trade-offs. Specifically, it has been hypothesized that DR is an adaptive response to temporary food shortage or starvation, whereby the organism withdraws energetic resources away from costly reproductive functions and reallocates them into somatic maintenance and survival functions until nutritional conditions that are more favorable for reproduction have returned (Holliday 1989; Masoro and Austad 1996; Kirkwood and Shanley 2005; also cf. Flatt 2011, 2014), a prediction consistent with the “disposable soma” theory for the evolution of aging and life histories (Kirkwood 1977), and supported by a theoretical model (Shanley and Kirkwood 2000). However, there is increasing evidence that this interpretation in terms of adaptive resource reallocation might not be correct: (i) flies that have been genetically sterilized, which should bring many (but perhaps not all) physiological activities geared toward reproduction to a halt, exhibit full life span expansion when exposed to DR (Mair et al. 2004); (ii) females flies that have evolved on and adapted to a DR diet evolve reduced life span without showing a concomitant evolutionary increase in fecundity [Zajitschek et al. 2019; males in contrast evolve higher reproductive success without reduced survival (Zajitschek et al. 2016)]; and (iii) flies kept under DR conditions and then switched back to a full diet perform worse in terms of survival and fecundity than flies continuously kept on a rich diet (McCracken et al. 2019). Thus, while in many species DR increases life span (Nakagawa et al. 2012) and usually reduces fecundity (Moatt et al. 2016), the intuitively appealing idea that this response has evolved as a flexible and adaptive resource reallocation strategy might be wrong. Clearly, from an evolutionary point of view, much more work is required to determine the potential fitness costs and benefits of DR, and to elucidate the adaptive or nonadaptive nature of this kind of dietary plasticity.
Plastic responses to larval crowding in laboratory cultures, which can have major effects on fitness-related traits, have also been investigated [reviewed in Prasad and Joshi (2003)]. Strong crowding (hundreds of larvae vs. 50–100 larvae per vial) leads to a major food shortage and hence intense competition, and increases the levels of noxious metabolic waste (ammonia) produced by the larvae (Shiotsugu et al. 1997; Borash et al. 1998). The phenotypic consequences of increased larval crowding include increased larval and pupal mortality, markedly prolonged development time, increased pupation height, often dramatically reduced adult size at eclosion, reduced fecundity, decreased lipid content, and reduced starvation resistance [reviewed in Prasad and Joshi (2003); cf. Joshi (1997), Shiotsugu et al. (1997), Mueller and Joshi (2000), and Borash and Ho (2001)]. The effects of larval crowding on life span likely depend on a balance between exposure to toxic metabolic waste vs. food limitation: the former can decrease life span (Shiotsugu et al. 1997), while the latter (if not too severe) increases life span (see above; Chippindale et al. 1993) so that larval crowding is sometimes found to extend adult life span (Miller and Thomas 1958; Lints and Lints 1969; Zwaan et al. 1991; Klepsatel et al. 2018). This life span-extending effect of larval crowding has recently been explained as being due to the reduced availability of dietary yeast caused by increased larval competition (Klepsatel et al. 2018; see above). Effects of adult crowding on life history are less well understood but can include reduced fecundity and life span (cf. Prasad and Joshi 2003).
One of the most interesting cases of life-history plasticity in the fly is reproductive dormancy (Figure 5); dormancy refers to a state of environmentally induced arrest of growth, development, and activity accompanied by decreased metabolic function. and which promotes somatic persistence [reviewed in Tatar and Yin (2001), Emerson et al. (2009a), Schmidt (2011), and Flatt et al. (2013)].
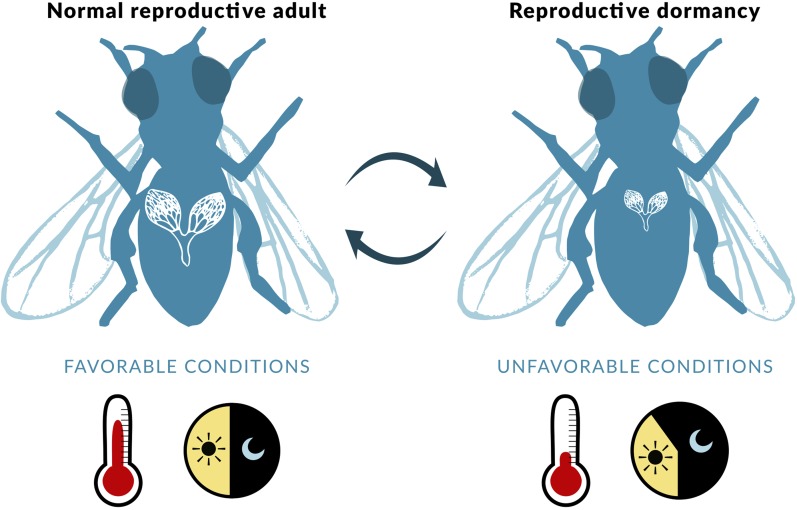
Figure 5.
Adult reproductive dormancy in D. melanogaster. In response to cool temperatures and short day lengths, some populations of vinegar flies can undergo a plastic, reversible state of adult reproductive dormancy (often referred to as reproductive diapause). This syndrome is associated with ovarian arrest in females (causing small, nonvitellogenic ovaries, as illustrated in the figure) or arrested spermatogenesis in males, increased levels of stress resistance, and greatly improved adult survival. See main text for further details. Figure credit: Chloé Schmidt (University of Manitoba).
D. melanogaster enters such a state of dormancy in response to low temperature (≤ 12–13°) and reduced (short-day) photoperiod (≤ 12 hr light); under such conditions, dormant flies are characterized by ovarian arrest (halted vitellogenesis) in females and arrested spermatogenesis in males, improved resistance to oxidative and heat stress (as well as other stressors), negligible rates of senescence, and increased adult life span (Saunders et al. 1989, 1990; Saunders and Gilbert 1990; Tatar and Yin 2001; Tatar et al. 2001b; Schmidt et al. 2005a,b; Schmidt and Paaby 2008; Emerson et al. 2009a,b; Schmidt 2011; Kubrak et al. 2016; Lirakis et al. 2018). The effects of low temperature on dormancy induction tend to be stronger than the effects of photoperiod (Emerson et al. 2009b).
Because in the vinegar fly—in contrast to some other insects including other Drosophila species—this state is rapidly induced, rather “shallow,” and can easily be “broken” by increasing temperature and lengthening the photoperiod (Saunders et al. 1989; Saunders and Gilbert 1990; Saunders and Bertossa 2011), it might not present a proper “diapause” but rather a state of “quiescence”; on the other hand, because it is under neuroendocrine control and seems to be adaptive (see below), it does exhibit some major hallmarks of proper diapause (cf. Tatar et al. 2001b; Tatar and Yin 2001; Flatt et al. 2005, 2013).
Interestingly, there is geographic (clinal) and genetic variation for dormancy expression in D. melanogaster: while genotypes from some populations can enter dormancy readily in response to low temperatures and short-day photoperiod, others have low or zero dormancy propensity (Williams and Sokolowski 1993; Schmidt et al. 2005a,b, 2008; Schmidt and Paaby 2008; Emerson et al. 2009b; Fabian et al. 2015). This pattern is most clearly seen along the North American east coast where the propensity of dormancy expression follows a latitudinal cline: flies from temperate, seasonal high-latitude populations (e.g., from Maine) show much greater dormancy inducibility than flies from subtropical/tropical low-latitude populations (e.g., from Florida) (Williams and Sokolowski 1993; Schmidt et al. 2005a,b, 2008; Schmidt and Paaby 2008; Emerson et al. 2009b).
In Europe, the propensity to undergo dormancy exhibits a similar but considerably shallower north–south latitudinal cline as compared to North America (Pegoraro et al. 2017). Along the Australian east coast, dormancy expression is also clinal but the pattern is nonlinear, with dormancy incidence being lowest in subtropical Australia, and then increasing toward both temperate and tropical Australia (Lee et al. 2011); however, the pattern is similar to the cline along the North American east coast when considering a similar latitudinal range as in North America.
When experimentally isolated in the laboratory and measured under nondormancy-inducing conditions, Schmidt et al. (2005b) found that “high-dormancy” genotypes (i.e., strains that always undergo dormancy under dormancy-inducing conditions) have lower early fecundity, improved resistance to starvation and cold stress, reduced age-specific mortality, and longer life span as compared to “low-dormancy” genotypes, suggesting that the ability to undergo dormancy forms part of a pleiotropic, polymorphic life-history syndrome (cf. Flatt et al. 2013).
Several lines of evidence suggest that D. melanogaster can overwinter in temperate areas on several continents, despite this species being an ancestrally tropical insect (Izquierdo 1991; Mitrovski and Hoffmann 2001; Boulétreau-Merle and Fouillet 2002; Boulétreau-Merle et al. 2003; Hoffmann et al. 2003); this is consistent with population genetic studies that have observed temporally persistent population structure, implying that flies might overwinter locally [Ives (1945, 1970) for North America; also cf. Izquierdo (1991) and references therein). This has led to the hypothesis that dormancy in D. melanogaster represents an overwintering strategy (Williams and Sokolowski 1993; Schmidt et al. 2005a,b), similar to the winter diapause observed in northern Drosophila species (Lumme 1978; also cf. Tatar and Yin 2001; Tatar et al. 2001b; Flatt et al. 2013). This notion is supported by population cage experiments by Schmidt and Conde (2006) who observed that under stressful conditions (alternating bouts of starvation and cold stress), the frequency of genotypes able to express dormancy increased over time relative to the frequency of nondormant genotypes, whereas under favorable control conditions the opposite pattern was found. In favor of the overwintering hypothesis, Paul Schmidt (personal communication) has observed that flies can live for up to 6 months under standard dormancy conditions, and Marko Brankatschk (personal communication) has found that adult flies can live up to 9 months when kept under temperatures fluctuating ∼8° and on a plant diet, which, in contrast to a yeast-based diet, confers increased cold tolerance (also cf. Brankatschk et al. 2018).
Perhaps consistent with the idea that dormancy is an adaptation of temperate populations of D. melanogaster to cold climates, Fabian et al. (2015) found that dormancy inducibility was < 2% among 119 lines from across 10 sub-Saharan African populations; this low diapause propensity was independent of whether the lines came from lowland or from high-altitude populations (also cf. Schmidt 2011). However, Zonato et al. (2017) and Lirakis et al. (2018) have recently reported positive dormancy induction in African strains, even though non-African admixture was not ruled out as an explanation; according to Zonato et al. (2017), dormancy might represent an ancestral adaptation to unfavorable seasonal changes (cf. Fabian et al. 2015). Whichever the case may be—overwintering adaptation or a stress response to other unfavorable conditions—the evidence to date suggests that dormancy represents a case of adaptive life-history plasticity.
The physiological mechanisms underlying dormancy have been probed using gene expression analyses (Kubrak et al. 2014; Zhao et al. 2016). These, as well as several genetic studies, have identified a major role of neuroendocrine pathways in affecting dormancy, especially the IIS pathway and secondary downstream hormones such as juvenile hormone and ecdysone [reviewed Tatar and Yin (2001), Flatt et al. (2005), Emerson et al. (2009b), Schmidt (2011), and Flatt et al. (2013)]. For North American populations, Schmidt et al. (2008) have mapped diapause propensity to the couch potato locus, a gene involved in neural development and possibly endocrine regulation; and a study by Williams et al. (2006) has implicated the IIS signaling gene PI3 kinase in explaining natural variation in dormancy inducibility. Notably, loss-of-function laboratory mutations in the IIS pathway “phenocopy” important aspects of dormancy [Tatar et al. (2001a); reviewed in Tatar and Yin (2001), Flatt et al. (2005, 2013), Emerson et al. (2009b), and Schmidt (2011)]. Interestingly, and beyond dormancy, growing evidence suggests that variation in IIS makes a major contribution to life-history clines (Paaby et al. 2010, 2014; Fabian et al. 2012; Flatt et al. 2013; Durmaz et al. 2019). In European populations, dormancy has been linked to a polymorphism at the timeless locus, a gene involved in the circadian clock (Sandrelli et al. 2007; Tauber et al. 2007; Zonato et al. 2018).
Together, the literature above suggests that many fitness components in D. melanogaster are highly plastic. Yet, we still know little about the potential inevitability, costs, and benefits of life-history plasticity and the evolutionary forces that shape it.
The amount and maintenance of genetic variation for fitness components
Populations of D. melanogaster harbor extensive amounts of genetic variation in fitness components. This is evidenced by a large number of quantitative genetic studies (and laboratory evolution; see below). For reviews see Lewontin (1974), Simmons and Crow (1977), Crow and Simmons (1983), Hedrick and Murray (1983), Charlesworth (1987), Roff and Mousseau (1987), and Mukai (1988); the treatment here mainly follows the syntheses of Charlesworth and Hughes (2000) and Charlesworth (2015).
To examine the effects of de novo mutations or of standing genetic variation on the variability of life-history traits, quantitative genetic studies of Drosophila have analyzed net fitness using genotypic (sometimes also interspecific) competition assays or, the majority of them, fitness components under laboratory conditions (Reed and Reed 1948, 1950; Knight and Robertson 1957; Mukai 1964, 1984, 1988; Prout 1971a,b; Sved 1971, 1989; Bundgaard and Christiansen 1972; Lewontin 1974; Mukai et al. 1974; Simmons and Crow 1977; Jungen and Hartl 1979; Haymer and Hartl 1982, 1983; Crow and Simmons 1983; Hedrick and Murray 1983; Kusakabe and Yamazaki 1984; Yamazaki and Hirose 1984; Mackay 1985; Charlesworth 1987, 2015; Tanaka and Yamazaki 1990; Crow 1993; Houle et al. 1994b, 1997; Latter and Sved 1994; Fowler et al. 1997; Charlesworth and Hughes 2000).
Many of these studies have taken advantage of balancer chromosomes, using them to isolate wild-type chromosomes and study their effects on fitness components (Muller 1928; cf. Lewontin 1974; Simmons and Crow 1977; Charlesworth and Hughes 2000; Charlesworth 2015). Balancer chromosomes carry multiple inversions that suppress crossing over along a particular chromosome when in heterozygous state with a homologous wild-type chromosome; moreover, most balancer stocks carry recessive lethals (thus preventing the homozygosity of the balancer chromosome) and dominant phenotypic marker mutations (Muller 1928; Ashburner et al. 2005). This powerful technique can be used to study the effects of de novo mutations on variation in fitness components (the mutational component of the genetic variance, VM; Simmons and Crow 1977): fully or partially recessive deleterious (including lethal) mutations that occur spontaneously on the wild-type chromosome can be completely sheltered from selection by being kept continuously in a nonrecombining, heterozygous state over the balancer, by backcrossing male heterozygotes to females from the balancer strain (Muller 1928; Lewontin 1974), in MA experiments (Mukai 1964, 1969; Mukai et al. 1972; Ohnishi 1977; Houle et al. 1994b, 1997; Charlesworth et al. 2004; Ávila et al. 2006; Keightley et al. 2009; Schrider et al. 2013; also cf. Charlesworth and Charlesworth 2010). Another application is to use balancers to extract a set of independent wild-type chromosomes from natural populations and measure trait values of fitness components of flies homozygous for such chromosomes. Moreover, by performing intercrosses between such sets of chromosomes (“diallel” crosses), one can measure the effects on fitness components of one of the major chromosomes (either the X, second, or third) independent of the other chromosomes. This information can be used to estimate the additive and dominance components of genetic variance, VA and VD (Mukai et al. 1974; Mukai and Nagano 1983; Kusakabe and Mukai 1984; Mukai 1988; Hughes 1995; Charlesworth and Hughes 2000; Charlesworth 2015). A third method is to compete nonlethal wild-type chromosomes against a balancer in population cages to estimate the fitness effects of the wild homozygous chromosomes relative to the heterozygous chromosomes (Sved 1971, 1975; Wilton and Sved 1979; Latter and Sved 1994; cf. Fowler et al. 1997 for an extension of this method).
What have we learned from > 50 years of quantitative genetic studies of Drosophila life history? One emerging conclusion is that newly arising deleterious mutations make a major contribution to standing genetic variation in net fitness and genetic load; this is revealed by the unmasking of concealed variability, which is not detectable in outbred populations, among homozygotes upon inbreeding (Charlesworth and Charlesworth 2010). For example, if a set of wild-type second or third chromosomes in Drosophila is made homozygous, often ∼30% of the chromosomal homozygotes are lethal (Simmons and Crow 1977; Crow 1993; Charlesworth and Charlesworth 2010). However, some estimates can be considerably higher: for instance, Mukai and Nagano (1983) found a frequency of lethal-carrying chromosomes of 0.55 for a population from Florida [also cf. Simmons and Crow (1977) for a review]. In contrast to the second and third chromosome, only a few lethals are found on the X chromosome because X-linked lethal alleles are exposed to selection in (hemizygous) males and thus are rapidly eliminated from the population (Crow 1993; Charlesworth and Charlesworth 2010). Crosses between distinct lines, each with different lethal chromosomes, produce viable heterozygous offspring in close to 100% of cases. This implies that most recessive lethal variants affect distinct genes and that the high frequency of chromosomes with lethal mutations is caused by individually rare mutations distributed over many loci (Crow 1993; Charlesworth and Charlesworth 2010). Based on a literature analysis, Charlesworth and Hughes (2000) conclude that deleterious mutations typically lead to reductions of net fitness when in homozygous or heterozygous states of at least ∼1% and ∼5%, respectively [see Simmons and Crow (1977), Crow and Simmons (1983), Crow (1993), García-Dorado and Caballero (2000), and Charlesworth and Charlesworth (2010)].
A related inference is that most deleterious mutations affecting fitness are likely to be partially recessive, not fully recessive or additive (Charlesworth and Hughes 2000; Charlesworth and Charlesworth 2010). For example, even fully recessive lethal mutations (with a dominance coefficient h = 0) have average heterozygous fitness effects of ∼2–3% (Crow 1993; García-Dorado and Caballero 2000), and mutations with small homozygous effects on fitness have h-values that are considerably larger than zero. For example, estimates from MA experiments suggest that such mildly deleterious mutations have dominance coefficients in the range of h = 0.1–0.4, with an average of ∼0.20–0.25 (Crow and Simmons 1983; Crow 1993; Charlesworth and Hughes 2000; García-Dorado and Caballero 2000; Charlesworth and Charlesworth 2010; Charlesworth 2015). The literature summarized above thus suggests that most standing variation in net (total) fitness in populations of D. melanogaster is fueled by the input of mildly deleterious, partially recessive mutations (Simmons and Crow 1977; Crow and Simmons 1983; Charlesworth and Hughes 2000; Charlesworth 2015).
The accumulation of deleterious mutations is also fundamentally important for the evolution of aging or senescence. According to the MA theory of aging (Medawar 1952), mutations with detrimental effects confined to old age classes are effectively neutral and attain higher equilibrium allele frequencies under mutation–selection balance than early acting deleterious alleles [reviewed in Rose (1991), Charlesworth (1993b, 2000, 2001), Partridge and Barton (1993), Hughes and Reynolds (2005), Partridge and Gems (2002), Flatt and Schmidt (2009), Paaby and Schmidt (2009), Charlesworth and Charlesworth (2010), and Flatt and Partridge (2018)]. Analyses of the age-specific properties of quantitative genetic parameters in D. melanogaster (e.g., VA, VD, and inbreeding depression) provide support for this theory (Hughes and Charlesworth 1994; Charlesworth and Hughes 1996; Hughes et al. 2002; Reynolds et al. 2007; Hughes 2010; Felix et al. 2012; Durham et al. 2014). However, alternative models predict similar patterns of age-specific genetic variance under both MA and AP (Moorad and Promislow (2009); yet, there is currently little evidence for the kind of age-specific changes in allelic effects and dominance variance assumed by these models (Hughes 2010). Importantly, by using the DGRP lines to examine the age-specific fitness effects of individual single-nucleotide polymorphisms (SNPs), Durham et al. (2014) found a major increase in the number of SNPs affecting fecundity with increasing age, with minimal overlap among SNPs across ages. Thus, there is overall quite strong empirical evidence for MA in maintaining genetic variance in senescence.
Interestingly, the selective effects of new, mildly detrimental mutations in Drosophila, as inferred from quantitative genetic studies, are markedly underestimated by analyses of DNA sequence variability: this implies the existence of mutations with relatively large fitness effects that make a major but underappreciated contribution to mutational input and standing variation for fitness (Charlesworth 2015). This could be because genomic analyses might miss contributions of transposable element insertions or of other large insertion/deletions (Charlesworth 2015). A contribution from inversion polymorphisms seems more unlikely because most quantitative genetic studies have been performed with inversion-free lines (Charlesworth 2015) but can, generally speaking, not be ruled out as an important source of fitness variation in natural populations (Kapun and Flatt 2019).
For detrimental mutations, mutation rates (U) have also been estimated from MA experiments, either using assays of fitness components (mainly viability) or sequencing analyses [Charlesworth et al. (2004), Ávila et al. (2006), and Haag-Liautard et al. (2007); for earlier estimates see Houle et al. (1992, 1994a)], with UD being defined as the total mutation rate per diploid genome to deleterious alleles for autosomal sites. MA experiments have produced an extremely large range of estimates for UD, from 0.02 to 1.2 (Charlesworth et al. 2004; Nishant et al. 2009; Charlesworth and Charlesworth 2010). The most current estimate, based on combining estimates for U from DNA sequence comparisons across species with direct estimates of the mutation rate from DNA sequences in MA lines, is a genome-wide deleterious mutation rate of ∼1.2 for the diploid genome (Haag-Liautard et al. 2007), but this estimate must be regarded as provisional as it is subject to large uncertainty (Charlesworth 2015).
Newly arisen deleterious mutations typically impact multiple fitness components simultaneously (Charlesworth and Hughes 2000; Charlesworth 2015). This inference is supported by several facts. First, many MA experiments have consistently found positive mutational correlations between multiple fitness components (Houle et al. 1994b; Martorell et al. 1998; Pletcher et al. 1998; Keightley and Ohnishi 1998). Second, as a consequence, the effects of deleterious mutations on individual fitness components are typically only a fraction of their effects on net (total) fitness, with an average effect of a mutation on a trait of ∼25–40% of its overall effect on fitness (Charlesworth and Hughes 2000; Charlesworth 2015). Third, the fitness reduction caused by making wild chromosomes from natural populations homozygous is typically a multiple of the homozygous reduction seen for viability (Crow and Simmons 1983; Latter and Sved 1994). This means that most deleterious mutations have pleiotropic effects on several fitness components that go in the same direction, thus causing positive correlations.
While standing variation for net fitness seems to be dominated by an input of partially recessive mildly deleterious mutations, most studies have found that genetic variance for fitness components is dominated by additive variance, with a relatively minor contribution of nonadditive variance (e.g., VD) (Kusakabe and Mukai 1984; Charlesworth 1987; Roff and Mousseau 1987; Mukai 1988; Houle 1992; Crnokrak and Roff 1995; Charlesworth and Hughes 2000; Hill et al. 2008). Consistent with additivity, most life-history traits seem to be affected by many loci of small effect; however, in some cases traits exhibit rather large values of VD (relative to VA and inbreeding depression) that are compatible with the existence of a few genes with relatively large effects (Charlesworth and Hughes 2000; cf. Hughes 1995, 1997).
Importantly, while narrow-sense heritabilities (h2 = VA/VP) for life-history traits are typically lower (average: ∼10–12% and range: ∼0–60%) than those for morphological or physiological traits (Roff and Mousseau 1987), Houle (1992) has found that fitness components harbor significantly more additive variance than morphological traits when VA is divided by the square of the trait mean (VA/m2 = mean-standardized additive variance = square of the coefficient of additive variation, CVA2) instead of being divided by VP [for estimates of CVA and CVD see, e.g., Charlesworth and Hughes (2000)]. Because heritability estimates can suffer from inherent positive correlations between the numerator (VA) and components of VP in the denominator, the mean-scaled additive variance is a better predictor of “evolvability,” i.e., the potential to respond to selection, than heritability (Hansen et al. 2011).
This leads to an interesting conundrum (Houle 1992; Charlesworth and Hughes 2000; Charlesworth 2015). Fisher’s Fundamental Theorem of Natural Selection implies that selection should exhaust additive genetic variance for fitness, so that there should be no additive genetic variance for fitness in an equilibrium population under selection alone (Fisher 1930; cf. Charlesworth 1987). This leads to the prediction that traits closely connected to fitness should exhibit very little or no additive genetic variance (Robertson 1955; Mousseau and Roff 1987; Roff and Mousseau 1987; Price and Schluter 1991; Houle 1992). How can this discrepancy be explained?
Several explanations have been put forward to account for the low heritability of fitness components and their excess of additive genetic variance (Charlesworth 1987, 2015; Mukai 1988; Price and Schluter 1991; Houle 1992; Charlesworth and Hughes 2000). The fact that for fitness components, as compared to other traits, estimates of h2 are much lower than estimates of CVA2 might be explained by higher amounts of residual variation VR (VP = VA + VR, where VR is the sum of nonadditive genetic variance components and environmental variance) (Houle 1992). Because VD is expected to contribute relatively little to VR (but see below), this implies that VE makes a major contribution to the variability of fitness components (also see section on plasticity). A large amount of VE could result from fitness traits typically integrating (environmental) variability over the entire lifetime of an organism, maybe much more so than other traits (Price and Schluter 1991; Houle 1992). This argument is supported by theoretical work suggesting that much phenotypic variation in fitness components might be due to neutral stochastic (environmental or demographic) “noise” (Steiner and Tuljapurkar 2012). Yet, a meta-analysis of published data on trait plasticity has found that life-history traits are not necessarily more plastic than other traits (Acasuso-Rivero et al. 2019).
A second more fundamental explanation for the high values of VA for fitness components (as revealed by estimates of CVA2) is that life-history traits are influenced by many more loci than other traits (Houle 1992, 2001; the “target size” hypothesis). In support of this, Houle (1998) found high correlations between mutational target sizes and mutational variances, suggesting that traits with a complex highly polygenic architecture, such as fitness components, are larger targets for mutational input than more simple traits. The mutational target size hypothesis is clearly an appealing explanation for the high levels of VA for fitness components, but the large amounts of VA appear to contradict the expectation that there should be no additive variance for fitness in an equilibrium population.
Generally, the available quantitative genetic estimates suggest that there is too much additive genetic variance for fitness components and more inbreeding depression than can be explained by mutational input and mutation-selection balance alone (Kusakabe and Mukai 1984; Charlesworth 1987, 2015; Mukai 1988; Charlesworth and Hughes 2000). For example, the value of VA for viability—estimated for second chromosomes subject to MA and inbreeding depression—is expected to be much greater than 0.001, while estimates of VA from natural populations are at least twice as large, suggesting that > 50% of the additive variance for fitness components is not accounted for by mutation (Mukai 1988; Charlesworth et al. 2004, Charlesworth and Charlesworth 2010).
What then explains the high amount of (additive) variance in fitness components? This issue bears directly on one of the two central questions of population genetics (Lewontin 1974; Mackay 2010; Charlesworth and Charlesworth 2010): (i) how much variation is there in natural populations and (ii) what evolutionary forces maintain this variation? While the first problem can be considered solved, especially due to the advent of DNA sequencing, the maintenance of variation in quantitative polygenic traits remains incompletely understood (Johnson and Barton 2005) (Box 2).
Box 2. How is genetic variation for fitness maintained?
Two broad types of explanation have been proposed to explain the maintenance of variation in natural populations (Dobzhansky 1955; Lewontin 1974). The “classical hypothesis” posits that most novel variation in populations is caused by the input of rare deleterious mutations (Muller 1950); it essentially represents a model of purifying selection and mutation–selection balance. The “balanced hypothesis,” on the other hand, suggests that most loci are polymorphic and that balancing selection maintains several alleles in the population (Dobzhansky 1955).
As argued above, the data at hand rule out the idea that the observed variability in fitness components in Drosophila is solely due to mutation–selection balance (Charlesworth 1987; Mukai 1988; Charlesworth and Hughes 2000; Charlesworth 2015). Thus, it is likely that some types of balancing selection make a major contribution to the maintenance of variation in fitness components, as Dobzhansky (1955) had envisaged.
Yet, classical models of balancing selection, involving overdominance (heterozygote advantage), seem in most cases to be incompatible with the data (Charlesworth and Hughes 2000): quantitative genetic studies often find that there is too much additive relative to dominance variance, and too much variance of chromosomal homozygotes as compared to chromosomal heterozygotes for this model to explain the maintenance of variation for net fitness (Kusakabe and Mukai 1984; Mukai 1988; Houle 1992).
However, an interesting possibility compatible with the data is AP (see Roff 1997; Charlesworth and Hughes 2000; Mackay 2010): if two alleles at a locus affect two traits, with the directionality of their homozygous effects on a given trait being opposite, and if the allele that is beneficial with respect to a given trait is dominant over the deleterious allele (“dominance reversal”), heterozygote advantage for net fitness can result (Rose 1982, 1985; Curtsinger et al. 1994; Charlesworth and Hughes 2000; also cf. Hazel 1943). Thus, in such a situation, there can be additive genetic variance for fitness components among loci even if there is no VA for net fitness. Theory suggests that this mechanism might lead to the maintenance of stable polymorphisms (Rose 1982, 1985; Curtsinger et al. 1994; also cf. Charlesworth and Hughes 1996; Roff 1997). This hypothesis is consistent with empirical estimates of the ratio of dominance to additive genetic variance for life-history traits (Roff 1997; Charlesworth and Hughes 2000) and empirical observations of pleiotropic effects of natural polymorphisms on various life-history traits (Paaby et al. 2014; Durmaz et al. 2019; also see section on correlated responses).
A similar uncoupling between the effects of a locus on a fitness component and fitness can occur under spatially and/or temporally varying selection, GxE, or frequency-dependent selection (Mukai et al. 1974; Mukai 1988; Gillespie 1991; Stearns 1992; Charlesworth and Hughes 2000). For example, Wittmann et al. (2017) have recently developed a model of seasonally fluctuating selection, based on AP with dominance reversal, which can explain the maintenance of multilocus polymorphisms; this might account for seasonal fluctuations of allele frequencies and fitness components in temperate populations of D. melanogaster (Bergland et al. 2014; Behrman et al. 2015). This model suggests that the conditions under which temporally varying selection can maintain variation are less restrictive than previously thought. Similarly, as discussed in the context of clines above, spatially varying selection provides an attractive mechanism for explaining the maintenance of balanced polymorphisms (Levene 1953; Slatkin 1975; Felsenstein 1976; Endler 1977; Hedrick 1986; Adrion et al. 2015; Yeaman 2015; Kapun and Flatt 2019), and trade-offs due to AP might explain the maintenance of spatial variation in fitness components (e.g., Paaby and Schmidt 2009; Paaby et al. 2014; Durmaz et al. 2018, 2019).
Moreover, both theory and experiments show that frequency- or density-dependent selection—due to interactions among individuals—as well as gene flow (migration) can also generally have a major impact on the maintenance of genetic variation [e.g., discussed in Lewontin (1955), Barton and Turelli (1989), Sokolowski et al. (1997), Bürger (2000), Mappes et al. (2008), Saltz et al. (2012), and references therein].
Finally, what is the role of epistasis in maintaining variation (e.g., Mackay 2010, 2014)? Despite evidence for pervasive epistasis among individual loci in Drosophila, also at the level of fitness (e.g., Yamamoto et al. 2009; Huang et al. 2012; Corbett-Detig et al. 2013), most genetic variation is additive (Hill et al. 2008). These notions are not incompatible as epistasis can generate substantial additive variance, especially when alleles are at extreme frequencies (Hill et al. 2008; Mackay 2014). Nonetheless, epistasis might have important implications for the maintenance of variation. For example, with suppressing epistasis it is possible that the amount of stabilizing selection is overestimated and mutational variance is underestimated (Yamamoto et al. 2009; Mackay 2014). If this is true, the inference that mutation–selection balance is insufficient to explain the maintenance of variation might need to be revisited. Moreover, in finite populations subject to inbreeding and drift, epistatic variance can become converted into additive variance, which can contribute to the response to selection (e.g., Mackay 2014; Hill 2017).
Life-History Evolution in the Laboratory
The fact that populations of D. melanogaster often harbor large amounts of variation available for selection to act upon is most powerfully illustrated by selection experiments in the laboratory [reviewed in Lewontin (1974), Wright (1984), and Powell (1997)], a framework called laboratory evolution, laboratory selection, or experimental evolution (sensu lato). The forerunners of this approach were the early population cage experiments of L’Héritier and Teissier (L’Héritier and Teissier 1933; also cf. Gayon and Veuille 2001), and Wright and Dobzhansky (1946).
Essentially three types of laboratory evolution experiments can be distinguished (e.g., Bennett and Lenski 1999): (i) laboratory natural selection (= experimental evolution sensu stricto; sometimes called “quasi-natural” selection), where the researcher surveys evolutionary changes of replicated populations in response to different experimentally imposed conditions (e.g., environmental, demographic, genetic, social, etc.); (ii) artificial selection (or “selective breeding”), where the researcher directly selects breeding individuals based on their phenotype or genotype for further breeding (the approach classically used in animal and plant breeding); and (iii) laboratory culling, where the experimenter imposes an extreme environmental condition (e.g., usually causing high mortality), representing a stringent selective screen, with only a small proportion of the individuals contributing to the next generation [this design has elements of both (i) and (ii)]. For general reviews of these methods see Hill and Caballero (1992), Rose et al. (1996), Bennett and Lenski (1999), Scheiner (2002), Fuller et al. (2005), Kawecki et al. (2012), and the edited volume by Garland and Rose (2009).
As reviewed in detail by Prasad and Joshi (2003) and as briefly summarized below, laboratory evolution experiments have contributed greatly to our understanding of many aspects of life-history evolution and evolutionary genetics in general (e.g., Lewontin 1974; Stearns 1992; Roff 1992, 1997; Charlesworth 1994; Powell 1997; Harshman and Hoffmann 2000; Stearns and Partridge 2001; Harshman 2003; Rose et al. 2004; Burke and Rose 2009; Garland and Rose 2009).
Laboratory selection on fitness components
Many studies have imposed laboratory selection on fitness components in flies, either by direct artificial selection or indirectly in response to specific environmental conditions. Fitness components that have been successfully subjected to artificial selection include developmental time (Clarke et al. 1961; Prout 1962; Bakker 1969; Zwaan et al. 1995a), age at sexual maturity [Hudak and Gromko 1989; Promislow and Bugbee 2000 (D. simulans)], copulation duration (Gromko et al. 1991), size at eclosion [= size at maturity; Robertson and Reeve 1952; Robertson 1959, 1960b; Druger 1962 (D. pseudoobscura); Hillesheim and Stearns 1991, 1992; Partridge and Fowler 1993; Partridge et al. 1999b; Turner et al. 2011], ovariole number (Robertson 1957a; Engstrom 1971), (early) fecundity (Bell et al. 1955; Rose and Charlesworth 1981b; Rose 1984a; Reeve and Fairbairn 1999; Charlesworth et al. 2007), egg size (Bell et al. 1955; Parsons 1964; Schwarzkopf et al. 1999), late-life fertility and life span (Rose and Charlesworth 1981b; Luckinbill et al. 1984; Rose 1984b; Partridge and Fowler 1992; Partridge et al. 1999a; Remolina et al. 2012; May et al. 2019), direct family selection on life span without selection for late-life fertility (Zwaan et al. 1995b), and various stress resistance traits (Service et al. 1985; Hoffmann and Parsons 1993; Hoffmann and Harshman 1999; Harshman et al. 1999a; Rose et al. 2004; Rion and Kawecki 2007).
Life-history evolution in Drosophila has also been investigated in response to several environmental conditions using laboratory natural selection (experimental evolution sensu stricto), including adaptation to laboratory culture conditions [evolutionary “domestication”; Matos et al. 2000 (in D. subobscura); Sgrò and Partridge 2000, 2001; Houle and Rowe 2003; Simões et al. 2007 (in D. subobscura); cf. Promislow and Tatar 1998), temperature [Anderson 1973 (in D. pseudoobscura); Cavicchi 1978; Cavicchi et al. 1989, 1991; Huey et al. 1991; Partridge et al. 1994a,b; James and Partridge 1995; Azevedo et al. 1996; Bochdanovits and de Jong 2003a; Tobler et al. 2015], nutrition (including selection for starvation resistance, adaptation to different larval and adult diets, DR, etc.; Robertson 1960b,c; Hillesheim and Stearns 1991, 1992; Chapman et al. 1994; Hoffmann and Harshman 1999; Nusbaum and Rose 1999; Bochdanovits and de Jong 2003a; Rion and Kawecki 2007; Kolss et al. 2009; Kristensen et al. 2011; Vijendravarma et al. 2011, 2012; Zajitschek et al. 2016, 2019; Hoedjes et al. 2019; May et al. 2019), humidity (Kennington et al. 2003), levels of extrinsic (environmental) mortality at the adult stage (Gasser et al. 2000; Stearns et al. 2000), reverse life-history evolution back to the ancestral state (Teotónio and Rose 2000; Teotónio et al. 2009), and so forth.
A growing number of so-called “Evolve and Resequence” (E&R) studies have examined the genomic basis of adaptive responses in laboratory evolution experiments in Drosophila using next-generation resequencing technology [reviewed in Schlötterer et al. (2014), (2015)], including investigations of the evolution of development time and correlated traits (Burke et al. 2010), size (Turner et al. 2011), life span and late-life fertility (Remolina et al. 2012; Carnes et al. 2015; Fabian et al. 2018; Hoedjes et al. 2019), egg size (Jha et al. 2015), starvation (Hardy et al. 2017) and desiccation resistance (Kang et al. 2016), and thermal adaptation (both in D. melanogaster and D. simulans; Orozco-terWengel et al. 2012; Tobler et al. 2013; Kapun et al. 2014; Barghi et al. 2019). These studies have yielded major insights into the dynamics of allele frequency changes, linked selection due to hitchhiking (“genetic drift”), and the identity of genomic regions and loci that might underpin adaptation.
What general conclusions can be drawn from the work on laboratory evolution in the fly? The studies above imply that life-history adaptation in Drosophila is often very rapid and, at least initially, not limited by mutational input: it appears to proceed mainly from a large store of standing variation (often involving alleles at intermediate frequencies, consistent with balancing selection), likely via polygenic responses at many loci and maybe also soft sweeps [see data and discussion in Charlesworth et al. (2007), Teotónio et al. (2009), Burke et al. (2010), Pritchard and Di Rienzo (2010), Pritchard et al. (2010), Orozco-terWengel et al. (2012), Messer and Petrov (2013), Charlesworth (2015), Garud et al. (2015), Barghi et al. (2019), Höllinger et al. (2019), and Kelly and Hughes (2019)]. This conjecture is not new but is now supported by much stronger empirical evidence: Lewontin (1974) stressed that “… the results of artificial selection experiments remain the strongest evidence we have of widespread genetic variation for genes that are relevant to characters of adaptive significance” (p. 93) and “Certainly the most extreme form of the classical hypothesis, which allows only a handful of rare mutations to be heterozygous in each individual, is contradicted by the selection results. Some substantial number of loci contributing to adaptive morphological and physiological characters must be segregating at intermediate allelic frequencies” (p. 94).
Correlated responses to selection and trade-offs
Laboratory evolution in Drosophila has also illuminated our understanding of correlations between fitness components and trade-offs (Harshman 2003; Prasad and Joshi 2003). A powerful way to establish genetic trade-offs is to apply selection on a focal life-history trait and to identify which other traits exhibit correlated responses to selection (Roff 1997; but cf. Gromko et al. 1991; Gromko 1995). Table 1 summarizes the results of 30 studies of direct and correlated life-history responses in D. melanogaster across 12 laboratories [Stearns and Partridge (2001) and Stearns and Medzhitov (2016); also cf. reviews in Bell and Koufopanou (1986), Stearns (1992), Harshman (2003), Prasad and Joshi (2003), and Rose et al. (2004)]. Overall, the results from these experiments suggest that:
Table 1. Summary of selection experiments on D. melanogaster life history [updated based on a figure in the Ph.D. thesis of Zwaan (1993) and first published in modified form by Stearns and Partridge (2001)].
| Correlated response to selection | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Larval competition | Development time | Body size | Longevity | Early fecundity | Late fecundity | Starvation resistance/fat content | Desiccation resistance | ||
| Selected trait | Larval competition | ||||||||
| Development time | + | + | 0 | + | − | ||||
| Body size | − | + | +/− | + | − | ||||
| Longevity | 0 | 0 | 0/− | − | 0/− | + | |||
| Early fecundity | + | + | 0/− | 0/− | − | 0 | |||
| Late fecundity | 0/+ | 0/+ | +/− | − | 0/+ | 0/+ | |||
| Starvation resistance/fat content | 0/+ | 0/+ | + | ||||||
| Desiccation resistance | 0/+ | 0 | + | − | + | ||||
The table represents a correlation matrix where the rows represent the focal life-history traits on which selection was applied and the columns represent the traits in which a correlated evolutionary response to selection was measured. Included are also two physiological traits (starvation and desiccation resistance) that also represent components of fitness contributing to somatic maintenance (survival); the expression of these traits can depend on nutritional conditions. The signs (0, +, and −) indicate the sign/direction of the correlated response; sometimes responses were not consistent (or even opposite) across different laboratories (e.g., 0/−, 0/+, or +/−) (also cf. Ackermann et al. 2001). The data in the matrix are based on results from (in alphabetical order) Buck et al. (2000), Chippindale et al. (1994, 1996, 1998), Force et al. (1995), Gasser et al. (2000), Graves et al. (1992), Harshman et al. (1999a,b), Hillesheim and Stearns (1991, 1992), Hoffmann and Parsons (1989, 1993), Luckinbill et al. (1984, 1988), Mueller (1987), Partridge and Fowler (1992, 1993), Partridge et al. (1999a,b), Reeve and Fairbairn (1999), Remolina et al. (2012), Rose (1984b), Rose and Charlesworth (1981b), Rose et al. (1992), Service et al. (1985, 1988), Stearns et al. (2000), and Zwaan et al. (1995a,b). Note that the matrix above does not represent a comprehensive or complete summary of the vast literature on correlated life-history responses in D. melanogaster. For reviews and discussion also see Stearns and Partridge (2001), Prasad and Joshi (2003), Harshman (2003), and Stearns and Medzhitov (2016).
Flies selected for prolonged development have larger size at eclosion; conversely, flies selected for larger size exhibit increased development time.
Females selected for increased early fecundity take longer to develop and are larger; conversely, flies selected for prolonged development or larger size exhibit increased early fecundity.
Flies selected for increased longevity have decreased early fecundity.
Flies selected for late-life fertility—and thus increased life span—exhibit improved stress (starvation or desiccation) resistance; conversely, selection for increased stress resistance is typically correlated with longer life span.
Importantly, several of these responses (or their absence) have been consistently observed across independent studies and laboratories: e.g., selection for longer life span (either by postponing reproduction or via direct selection for life span) typically leads to the correlated evolution of reduced early fecundity (Luckinbill et al. 1984; Rose 1984b; Zwaan et al. 1995b; Partridge et al. 1999a; Remolina et al. 2012). Fewer data exist about the correlated response of life span to selection for increased early fecundity (or early fitness more generally): while Reeve and Fairbairn (1999) observed no consistent effect on life span of selection for increased early fecundity, Stearns et al. (2000) found that after 50 generations of experimental evolution under high adult mortality, thus imposing strong selection on early fitness, flies had evolved faster development, earlier age at maturity, smaller size at eclosion, higher early fecundity, and shorter life span. The notion that selection for increased early fecundity might lead to the correlated evolution of shorter life span is also supported by two selection experiments in Tribolium flour beetles (Sokal 1970; Mertz 1975). Thus, it seems likely that the trade-off between early fecundity and life span might be symmetrical [but see Reeve and Fairbairn (1999)].
Another consistent pattern across most experiments is that selection for increased late-life fecundity/increased life span does not affect development time and/or body size, or vice versa, suggesting that development does not have a major impact on longevity (Luckinbill et al. 1984, 1988; Chippindale et al. 1994; Zwaan et al. 1995a,b; Partridge et al. 1999a; Buck et al. 2000; cf. discussion in McCulloch and Gems 2003).
These experiments thus provide robust evidence that fitness components are intimately connected through genetic (and the resulting developmental, physiological, and phenotypic) correlations (Stearns 1989a, 1992; Charlesworth 1990). The observed trade-offs are consistent with (but not proof of) AP alleles underlying genetic correlations; in particular, the commonly observed trade-off between early fecundity and longevity is consistent with the AP hypothesis of the evolution of aging (Medawar 1946, 1952; Williams 1957; Rose and Charlesworth 1981a,b; Rose 1991; Charlesworth 1993b, 1994; Partridge and Barton 1993; Stearns and Partridge 2001; Rose et al. 2004; Hughes and Reynolds 2005; Flatt and Promislow 2007; Flatt and Schmidt 2009; Charlesworth and Charlesworth 2010; Gaillard and Lemaître 2017; Austad and Hoffman 2018; Flatt and Partridge 2018).
Inadvertent selection, GxE, and interpretational caveats
Although we have learned a great deal from the data above, the interpretation of life-history selection experiments is not always straightforward (Partridge and Barton 1993; Harshman and Hoffmann 2000; Prasad and Joshi 2003).
As indicated in Table 1, some correlated responses are inconsistent across studies and laboratories. One explanation for this might be variable pleiotropy: high variability in the signs and magnitudes of pleiotropic effects across selected loci can cause inconsistent correlated responses to selection, thus rendering the correlated responses difficult to predict; a related issue is that correlated responses to selection in the laboratory are not always well predicted by knowledge of genetic correlations (Gromko et al. 1991; Gromko 1995; cf. Flatt and Kawecki 2004).
Inadvertent selection or unintended differences between selection regimes can also cause interpretational problems. As discussed by Promislow and Tatar (1998), some selection experiments for postponed senescence might have been subject to inadvertent selection for early life performance (rapid development and high early fecundity) and to strongly relaxed selection on the later part of adult life, due to the typical, discrete 2-week culture regime of the base stocks used in many of these experiments: under such regimes alleles for adult fitness traits that are expressed after 4 days of adult age are sheltered from selection, possibly leading to the accumulation of late-acting mutations (cf. Harshman and Hoffmann 2000; Sgrò and Partridge 2000, 2001). In such a situation, correlations between early and late fitness components might not be due AP, but could instead arise from LD between the newly accumulated mutations and alleles affecting early or late-age traits under direct selection (Promislow and Tatar 1998; also cf. Clark 1987).
GxE interactions can also obscure correlated responses and might arise from unintended differences between selection regimes, for example differences in nutrition, and from selection lines being measured under assay conditions that differ from those in the selection regime (Leroi et al. 1994a,b; Rose et al. 1996; Harshman and Hoffmann 2000; Ackermann et al. 2001). Similarly, correlations might be masked by GxE interactions when populations are exposed to novel conditions, for instance when wild-caught flies are brought into the laboratory and they are adapting to the laboratory conditions (Service and Rose 1985; Harshman and Hoffmann 2000; Matos et al. 2000; Sgrò and Partridge 2000; Simões et al. 2007; Klepsatel et al. 2013a).
Some of the inconsistent (or occasionally even opposite) life-history responses among different laboratories in Table 1 might thus—at least in part—be due to differences between selection and assay environments within the same laboratory, or to differences between assay environments among laboratories, i.e., a type of genotype- (or selection regime) by-environment interaction (Harshman and Hoffmann 2000; Ackermann et al. 2001). In support of this notion, when life-history selection lines from laboratories in Basel, Groningen, Irvine, and London were all measured in each of these laboratories, Ackermann et al. (2001) found clear interactions between origin and assay environment for early fecundity, a highly environmentally sensitive (plastic) trait (also cf. Charlesworth et al. 2007; May et al. 2019). In contrast, for longevity, development time and body size interactions between selection regime and assay environment were not significant, yet in several cases measurements in different environments would have led to opposite conclusions for these traits with respect to the effects (or direction) of selection.
A selection experiment by Partridge et al. (1999a) set out to avoid some of these difficulties by using a long-standing (> 28 years), laboratory-adapted, outbred base population maintained in population cages with overlapping generations as the starting material for a selection experiment on age at reproduction and by measuring selection under conditions identical to those during selection. Importantly, similar to earlier experiments, breeding from older adult flies led to an evolutionary increase in life span (albeit not increased late-life fertility) at the expense of reduced early life fertility, thus strongly suggesting that the trade-off between early fecundity and longevity is driven by AP. However, in contrast to some earlier studies (see Table 1), no correlated responses were found for developmental traits such as development time, larval competitive ability, and size at eclosion (Partridge et al. 1999a).
The Loci Underlying Variation in Fitness Components
Understanding how variation in fitness components, or ultimately in fitness itself, maps into genetic variation at underlying causative loci is a central and long-standing problem of evolutionary genetics (Lewontin 1974; Charlesworth and Hughes 2000; Houle 2001; Mackay 2010; Barrett and Hoekstra 2011; Rockman 2012). Genes and alleles that affect fitness components in D. melanogaster have been identified and studied by both molecular and evolutionary biologists, using two quite distinct genetic research traditions and approaches (Rose et al. 2011; cf. discussion in Stearns et al. 1993; Stern 2000; de Jong and Bochdanovits 2003; Flatt 2004b).
The first approach, which might be called “Mendelian genetics” (or “transmission genetics”, “molecular genetics,” or “functional genetics”), has mainly employed large-effect, laboratory-generated, complete (amorphic) or partial (hypomorphic) loss-of-function mutants (e.g., isolated from mutant screens) or transgenic constructs (overexpression, RNAi, etc.) to examine genetic effects upon fitness components, in particular on life span (Stearns et al. 1993; Stearns and Kaiser 1993, 1996; Orr and Sohal 1994; Kaiser et al. 1997; Lin et al. 1998; Tatar 1999, 2000; Rogina et al. 2000; Silbermann and Tatar 2000; Tower 2000; Clancy et al. 2001; Stearns and Partridge 2001; Tatar et al. 2001a, 2003; Partridge and Gems 2002; Harshman 2003; Oldham and Hafen 2003; Flatt and Kawecki 2004; Hwangbo et al. 2004; Kenyon 2005; Partridge et al. 2005a; Ford and Tower 2006; Flatt and Schmidt 2009; Paaby and Schmidt 2009; Magwire et al. 2010; Tower 2011).
This work has contributed significantly to our understanding of the developmental, physiological, and molecular underpinnings of traits such as growth, size, and life span and their regulation. For example, it has led to the important realization that molecular signaling pathways such as the IIS pathway can have evolutionarily conserved effects on life span [reviewed in Flatt (2004b) and Partridge (2018)]. However, while these analyses have illuminated major aspects of the “functional architecture” of fitness-related traits, the genes identified in such experiments do not necessarily harbor segregating alleles that contribute to standing genetic variation for traits in natural, evolving populations (Flatt 2004; Flatt and Schmidt 2009; Birney 2016; Vonesch et al. 2016; Fabian et al. 2018; Durmaz et al. 2019).
The second approach, taken by evolutionary and quantitative geneticists, has thus focused on the effects of genetic variation in outbred populations of D. melanogaster, and might be called “experimental evolutionary genetics” or “experimental population genetics” (including “quantitative genetics”) (Lewontin 1974; Falconer and Mackay 1996; Roff 1997; Lynch and Walsh 1998; Charlesworth and Hughes 2000; Mackay 2001a,b, 2004, 2010; Harshman 2003; Flatt 2004b; Flatt and Kawecki 2004; Geiger-Thornsberry and Mackay 2004; Mackay et al. 2006; Flatt and Schmidt 2009; Paaby and Schmidt 2009; Barrett and Hoekstra 2011; Rose et al. 2011; Walsh and Lynch 2018). For instance, numerous studies have used quantitative trait locus (QTL) mapping to identify chromosomal regions and individual genes that affect fitness components such as ovariole number, fecundity, and life span (Leips and Mackay 2000, 2002; Vieira et al. 2000; Mackay 2001a,b, 2004, 2010; Gockel et al. 2002; Calboli et al. 2003; Geiger-Thornsberry and Mackay 2004; Leips et al. 2006; Mackay et al. 2006, 2009; Wilson et al. 2006; Bergland et al. 2008; Flatt and Schmidt 2009; Paaby and Schmidt 2009). Figure 6 illustrates a specific example of a natural, balanced life-history polymorphism that underpins clinal adaptation, and Table 2 lists a selection of examples of naturally occurring life-history variants that were identified using the second, “evolutionary” approach [for more comprehensive reviews see Flatt and Schmidt (2009) and Paaby and Schmidt (2009)]. Generally, there are still relatively few cases of naturally segregating life-history loci and polymorphisms that have been identified and studied in detail.
Figure 6.
An example of a naturally segregating life-history polymorphism in D. melanogaster. (A) A polytene third chromosome of a fly heterozygous for the In(3R)Payne inversion polymorphism, i.e., the fly carries one normal noninverted third chromosome (standard arrangement) and one homologous chromosome with the inverted arrangement. In the region spanned by the inversion, the inverted and standard arrangements have paired by forming a loop structure (inversion loop). (B) Around the world, the In(3R)Payne inversion polymorphism is typically at intermediate frequency in warm climates but decreases in frequency toward temperate regions. Experiments show that the inversion confers small body size, decreased stress resistance, and shorter life span, while flies carrying the standard arrangement are characterized by the opposite phenotypes. This balanced polymorphism makes a major contribution to the well-known clines for these fitness components. Because the inversion affects several quantitative fitness-related traits that are likely to be affected by multiple loci inside the inversion, this polymorphism has been hypothesized to represent a life-history “supergene.” Also see Figure 4. Figure credit: Chloé Schmidt (University of Manitoba).
Table 2. Examples of natural variants or polymorphisms affecting D. melanogaster life history.
| Gene | Variant type | Origin of variant | Affected trait(s) | Analysis method | Reference |
|---|---|---|---|---|---|
| bellwether (blw) | AG/GT polymorphism in promoter region | Identified in a selection experiment for longevity | Life span | Transgenic constructs | Garcia et al. (2017) |
| catecholamines up (catsup) | QTL alleles; naturally segregating SNPs and indel polymorphisms | QTL mapping between inbred strains and alleles from a single population | Life span, locomotory behavior, sensory bristle number; (alleles have nonpleiotropic effects on these traits) | QTL mapping; association mapping; DCT; mutant alleles | Carbone et al. (2006), Mackay et al. (2006) |
| CG9509 | SNPs in cis-regulatory (enhancer) region | Several natural populations | Larval growth rate, body weight, wing size, insecticide and female cold tolerance | Hypomorphic allele; transgenic RNAi; association study | Glaser-Schmitt and Parsch (2018) |
| couch potato (cpo) | SNPs | Latitudinal cline | Reproductive dormancy | QTL mapping; DCT | Schmidt et al. (2008) |
| dopa decarboxylase (Ddc) | QTL alleles; naturally segregating SNPs | Single natural population | Life span | QTL mapping; DCT | Pasyukova et al. (2000), Leips and Mackay (2002), De Luca et al. (2003) |
| Drip (aquaporin) | QTL alleles; molecular nature unknown | Single natural population | Fecundity | QTL mapping; DCT; transgenic RNAi | Bergland et al. (2012) |
| foxo | Clinally varying two-SNP variant; GG vs. AT haplotype, identified from population genomic analyses of the cline | Latitudinal cline | Viability, body size, size-related traits, starvation resistance | Recombinant outbred populations that differ in allelic state using DGRP lines | Durmaz et al. (2019) |
| I am not deady yet (Indy) | Hoppel transposon insertion variant | Several natural populations | Life span, fecundity | Insertion homo- and heterozygotes from natural lines | Zhu et al. (2014) |
| Insulin-like receptor (InR) | Indel polymorphism | Latitudinal cline | Body size, fecundity, life span | Lines that differ in allelic state, isolated from natural populations with balancers | Paaby et al. (2010, 2014) |
| In(3R)Payne (see Figure 6) | Inversion polymorphism | Latitudinal cline | Body size, size-related traits, cold-shock mortality, starvation resistance, life span | Association study; inversion and noninverted homokaryotype lines isolated from natural populations | Weeks et al. (2002); Rako et al. (2006) Kapun et al. (2016b), Durmaz et al. (2018) |
| Menin1 (Mnn1) | SNP/amino acid polymorphism | Latitudinal cline | Hatch rate, UV sensitivity, chill coma recovery, starvation resistance | Transgenic lines differing in allelic state | Svetec et al. (2019) |
| methuselah (mth) | Wild-derived alleles | Several natural populations; latitudinal cline | Life span, fecundity, oxidative stress resistance | QCT | Paaby and Schmidt (2008); also cf. Schmidt et al. (2000) |
| neurofibromin 1 (Nf1) | Indel polymorphism | Latitudinal cline | Wing size, developmental time | Association study | Lee et al. (2013) |
| Phosphatidylinositol 3- kinase (Pi3-kinase, PI3K) (Dp110) | Autosomal recessive vs. dominant natural variants; molecular nature unknown | Natural populations | Reproductive dormancy | Segregation analysis; DCT; QCT; transgenic constructs | Williams et al. (2006) |
| shuttle craft (stc) | QTL alleles | QTL mapping between inbred strains | Life span | DCT; QCT | Pasyukova et al. (2004) |
| S6 kinase (S6K) | QTL alleles | QTL mapping between inbred strains | Total protein levels, glycogen storage, life span, immune response | QCT | Cho et al. (2010) |
| tailup (tup) | QTL alleles | QTL mapping between inbred strains | Life span | DCT; QCT | Mackay et al. (2006) |
The table provides examples of naturally segregating alleles and polymorphisms that affect fitness components in D. melanogaster. In several of these cases there is evidence suggesting that these polymorphisms have pervasive pleiotropic effects upon multiple fitness-related traits and that they are maintained by some sort of balancing selection, for instance due to spatially varying (clinal) selection and/or antagonistic pleiotropy/trade-offs. For more detailed reviews see Paaby and Schmidt (2009) and Flatt and Schmidt (2009). For the example of the inversion polymorphism In(3R)Payne, mentioned in the table, see Figure 6. DCT, deficiency complementation testing/mapping; Indel, insertion/deletion; DGRP, Drosophila Genetic Reference Panel; QCT, quantitative complementation testing; RNAi, RNA interference.
The “molecular” vs. “evolutionary” approaches have become increasingly unified through modern genomic and genetic methods (Rose et al. 2011), including fine-scale mapping from QTL to quantitative trait nucleotides, GWAS, scans for signatures of selection down to SNPs, and E&R experiments (Mackay et al. 2009, 2012; Barrett and Hoekstra 2011; Rose et al. 2011; Rockman 2012; Schlötterer et al. 2014, 2015; Adrion et al. 2015; Stephan 2016; Mackay and Huang 2018). For example, GWAS have identified many candidate loci (typically SNPs) for fitness components such as development time, body size and size-related traits, ovariole number, age-specific and lifetime fecundity, life span, oxidative stress resistance, and chill coma recovery (Jordan et al. 2012; Mackay et al. 2012; Weber et al. 2012; Burke et al. 2014; Durham et al. 2014; Ivanov et al. 2015; Highfill et al. 2016; Vonesch et al. 2016; Lobell et al. 2017; Lafuente et al. 2018; Mackay and Huang 2018).
Functional analysis and validation of the phenotypic effects of natural alleles can now be achieved with a variety of methods (see e.g., Table 2), including—at the level of nucleotides—with CRISPR/Cas9 gene editing, e.g., coupled with quantitative complementation or reciprocal hemizygosity tests (Stern 2014; Turner 2014; cf. Ding et al. 2016 for an example); yet, functional genetic analysis and validation can be technically challenging, and are still relatively rarely done by evolutionary geneticists.
Several major conclusions have emerged from the work reviewed above. First, as stated by Mackay et al. (2009): “QTL alleles with large effects are rare… the bulk of genetic variation for quantitative traits is due to many loci with [small] effects.” [cf. discussion in Rockman (2012); but see below]. Second, fitness components and other quantitative traits have highly complex genetic architectures, i.e., they are affected by many loci whose effects can be highly contingent, e.g., sex-, environment-, or genetic background-specific (cf. Mackay 2010). Third, many alleles with effects on fitness components have pleiotropic effects, which are often AP effects; this is consistent with the notion that variance for fitness might be maintained by AP/trade-offs; it also supports the AP theory of the evolution of aging (cf. Mackay 2010). Fourth, genome-wide analyses (GWAS and E&R) have found relatively little overlap between “canonical” candidate loci, as identified by the first approach, and loci that harbor segregating alleles in natural or laboratory populations (Remolina et al. 2012; Vonesch et al. 2016; Fabian et al. 2018).
There may be several reasons for this. One reason could be that the analyses using the first approach are biased toward detecting large effects. A second reason might be that, because fitness components are so polygenic, mutant screens might be far away from “saturation.” A third idea is that canonical large-effect candidate genes are functionally so important that they are under strong selective constraints in natural populations and therefore do not harbor any standing variation (Remolina et al. 2012; Fabian et al. 2018; Flatt and Partridge 2018). Fourth, as pointed out by Charlesworth and Hughes (2000) and Charlesworth (2015), some fitness components seem to be affected by relatively few loci of major effect, especially when they harbor dominance variance, implying that the search for large-effect life-history polymorphisms might be fruitful. Indeed, several examples suggest that segregating life-history polymorphisms can have relatively large effects (Schmidt et al. 2008; Bergland et al. 2012; Paaby et al. 2014; Durmaz et al. 2019). However, with pervasive suppressing epistasis or linked antagonistic-effect loci, the effects of isolated variants might exceed their net effects in their native genomic background and context (Bernstein et al. 2019; also cf. Lewontin 1974).
Conclusions
Here, I have summarized a large body of work concerned with the genetics of fitness components and the evolution of life-history traits in D. melanogaster. To conclude, I would like to highlight a few general key points:
Outbred populations of D. melanogaster typically harbor substantial amounts of variation for life-history traits and other fitness components. Although selection is expected to rapidly and relentlessly erode additive genetic variance for fitness, fitness components exhibit a lot of variability, in fact substantially more additive genetic variance relative to the trait mean than morphological or other traits. Fitness components thus have a high degree of evolvability, i.e., a large potential to respond to selection.
A fundamental and still not satisfactorily resolved question is how this large pool of variability in fitness components is maintained, despite strong selection. One major idea is that fitness components, due to their highly polygenic architecture, receive more mutational input than other, less complex traits. However, while mutation–selection balance certainly plays an important role in maintaining life-history variation, it cannot fully account for it; some sort of balancing selection must be invoked. Yet, classical balancing selection via overdominance is in most cases inconsistent with the available data. It is therefore probable that other types of balancing selection—e.g., AP with dominance reversal, causing fitness overdominance; spatially and/or temporally varying selection; frequency-dependent selection; and/or GxE interactions—contribute significantly to maintaining variation in fitness components. Resolving the details of this issue is a central problem of evolutionary genetics and at the heart of the long-standing debate between the classical hypothesis, due to Muller, and the “balanced” hypothesis, due to Dobzhansky (Lewontin 1974).
Consistent with large stores of standing variation for fitness components maintained by balancing selection in populations of D. melanogaster, laboratory evolution experiments do not appear to be limited (at least not initially) by mutational input and often produce rapid, strong responses to selection, with evidence that alleles at intermediate frequencies play a major role in the response. At least in the short-run, life-history adaptation in D. melanogaster thus appears to proceed mainly via polygenic responses at many loci and possibly also through soft sweeps.
Trade-offs between fitness components, observed as negative genetic correlations or as correlated responses to selection, are consistent with AP; studies of candidate genes using large-effect mutants or transgenes, or of naturally occurring, segregating life-history polymorphisms, suggest that AP is common. Such AP might be a major source of balancing selection (see above). Yet, still little is known about the mechanistic basis of life-history trade-offs, for example to what extent they impinge on resource allocation, a concept that is commonly invoked but rarely tested.
Studies of latitudinal clines also suggest that balancing (spatially varying) selection and trade-offs across geography (local adaptation) play a major role in the maintenance of life-history polymorphisms.
Both quantitative genetic analyses and laboratory evolution experiments have provided qualitative support for the MA and AP theories of the evolution of senescence.
Relatively few alleles and polymorphisms underpinning life-history evolution have been identified and examined in detail. Most fitness components seem to be influenced by many genes with small, often pleiotropic effects; a few traits may be influenced by a small number of polymorphic genes with relatively large effects. Knowing about the transmission and quantitative genetic properties of life-history loci is critical for our ability to test population and quantitative genetic models of selection, adaptation, the evolution of senescence, genetic architecture, and so forth.
As the major progress reviewed above illustrates, studies of D. melanogaster have greatly illuminated many important problems in life-history evolution and evolutionary genetics. Yet, despite many decades of work, several fundamental issues—for example the nature of balancing selection that maintains standing variance for fitness components—remain fascinating open questions in need of more future work.
Acknowledgments
I wish to thank Steve Stearns, Tad Kawecki, and Marc Tatar for having stimulated my interest in life-history evolution as a student; Teri Markow and Trudy Mackay for their kind invitation to contribute this paper; two reviewers for their valuable comments; Chloé Schmidt for providing the illustrations; and the members of my laboratory for stimulating discussions. Research in my group has been generously supported by grants and fellowships from the Swiss National Science Foundation (PP00P3_165836, 310030E-164207, and 31003A_182262), the European Molecular Biology Organization, the Marie Skłodowska-Curie Actions (H2020-MSCA), and the Novartis Foundation for Medical-Biological Research. Parts of this paper were written while I was an EvoPAD (Evolutionary Processes in Adaptation and Disease) Visiting Professor at the Institute for Evolution and Biodiversity, University of Münster, supported by a Mercator Fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft).
Footnotes
Communicating editor: T. Mackay
Literature Cited
- Acasuso-Rivero C., Murren C. J., Schlichting C. D., and Steiner U. K., 2019. Adaptive phenotypic plasticity for life-history and less fitness-related traits. Proc. Biol. Sci. 286: 20190653 10.1098/rspb.2019.0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M., Bijlsma R., James A. C., Partridge L., Zwaan B. J. et al. , 2001. Effects of assay conditions in life history experiments with Drosophila melanogaster. J. Evol. Biol. 14: 199–209. 10.1046/j.1420-9101.2001.00281.x [DOI] [Google Scholar]
- Adrion J. R., Hahn M. W., and Cooper B. S., 2015. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 31: 434–444. 10.1016/j.tig.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpatov W. W., 1929. Growth and variation of the larvae of Drosophila melanogaster. J. Exp. Zool. 52: 407–437. 10.1002/jez.1400520303 [DOI] [Google Scholar]
- Alpatov W. W., 1930. Phenotypical variation in body and cell size of Drosophila melanogaster. Biol. Bull. 58: 85–103. 10.2307/1537121 [DOI] [Google Scholar]
- Alpatov W. W., and Pearl R., 1929. Experimental studies on the duration of life. XII. Influence of temperature during the larval period and adult life on the duration of the life of the imago of Drosophila melanogaster. Am. Nat. 63: 37–67. 10.1086/280236 [DOI] [Google Scholar]
- Anagnostou C., Dorsch M., and Rohlfs M., 2010. Influence of dietary yeasts on Drosophila melanogaster life-history traits. Entomol. Exp. Appl. 136: 1–11. 10.1111/j.1570-7458.2010.00997.x [DOI] [Google Scholar]
- Anderson W. W., 1973. Genetic divergence in body size among experimental populations of Drosophila pseudoobscura kept at different temperatures. Evolution 27: 278–284. 10.1111/j.1558-5646.1973.tb00673.x [DOI] [PubMed] [Google Scholar]
- Arking R., 2006. The Biology of Aging - Observations and Principles. Oxford University Press, Oxford. [Google Scholar]
- Ashburner M., Golic K. C., and Hawley R. S., 2005. Drosophila – A Laboratory Handbook, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Atkinson D., 1994. Temperature and organism size – a biological law for ectotherms? pp. 1–58 in Advances in Ecological Research, edited by Begon M. and Fitter A. H.. Academic Press, New York. [Google Scholar]
- Atkinson D., and Sibly R. M., 1997. Why are organisms usually bigger in colder environments? Making sense of a life history puzzle. Trends Ecol. Evol. 12: 235–239. 10.1016/S0169-5347(97)01058-6 [DOI] [PubMed] [Google Scholar]
- Austad S. N., and Hoffman J. M., 2018. Is antagonistic pleiotropy ubiquitous in aging biology? Evol. Med. Public Health 2018: 287–294. 10.1093/emph/eoy033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávila V., Chavarrías D., Sánchez E., Manrique A., López-Fanjul C. et al. , 2006. Increase of the spontaneous mutation rate in a long-term experiment with Drosophila melanogaster. Genetics 173: 267–277. 10.1534/genetics.106.056200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo R. B. R., French V., and Partridge L., 1996. Thermal evolution of egg size in Drosophila melanogaster. Evolution 50: 2338–2345. 10.1111/j.1558-5646.1996.tb03621.x [DOI] [PubMed] [Google Scholar]
- Baker K. D., and Thummel C. S., 2007. Diabetic larvae and obese flies - emerging studies of metabolism in Drosophila. Cell Metab. 6: 257–266. 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker K., 1959. Feeding period, growth, and pupation in larvae of Drosophila melanogaster. Entomol. Exp. Appl. 2: 171–186. 10.1111/j.1570-7458.1959.tb00432.x [DOI] [Google Scholar]
- Bakker K., 1969. Selection for rate of growth and its influence on competitive ability of larvae of Drosophila melanogaster. Neth. J. Zool. 19: 541–595. 10.1163/002829669X00035 [DOI] [Google Scholar]
- Barghi N., Tobler R., Nolte V., Jakšić A. M., Mallard F. et al. , 2019. Genetic redundancy fuels polygenic adaptation in Drosophila. PLoS Biol. 17: e3000128 10.1371/journal.pbio.3000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes A. I., and Partridge L., 2003. Costing reproduction. Anim. Behav. 66: 199–204. 10.1006/anbe.2003.2122 [DOI] [Google Scholar]
- Barrett R. D., and Hoekstra H. E., 2011. Molecular spandrels: tests of adaptation at the genetic level. Nat. Rev. Genet. 12: 767–780 [corrigenda: Nat. Rev. Genet. 13: 70 (2012)]. 10.1038/nrg3015 [DOI] [PubMed] [Google Scholar]
- Barton N. H., and Turelli M., 1989. Evolutionary quantitative genetics: how little do we know? Annu. Rev. Genet. 23: 337–370. 10.1146/annurev.ge.23.120189.002005 [DOI] [PubMed] [Google Scholar]
- Bass T. M., Grandison R. C., Wong R., Martinez P., Partridge L. et al. , 2007. Optimization of dietary restriction protocols in Drosophila. J. Gerontol. A 62: 1071–1081. 10.1093/gerona/62.10.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger J. P., 1919. A nutritional study of insects, with special reference to microörganisms and their substrata. J. Exp. Zool. 28: 1–81. 10.1002/jez.1400280102 [DOI] [Google Scholar]
- Beadle G. W., Tatum E. L., and Clancy C. W., 1938. Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol. Bull. 75: 447–462. 10.2307/1537573 [DOI] [Google Scholar]
- Begon M., 1982. Yeasts and Drosophila, pp. 345–384 in The Genetics and Biology of Drosophila, Vol. 3B, edited by Ashburner M., Carson H. L., and Thompson J. N. Jr. Academic Press, New York. [Google Scholar]
- Behrman E. L., Watson S. S., O’Brien K. R., Heschel M. S., and Schmidt P. S., 2015. Seasonal variation in life history traits in two Drosophila species. J. Evol. Biol. 28: 1691–1704. 10.1111/jeb.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. E., Moore C. H., and Warren D. C., 1955. The evaluation of new methods for the improvement of quantitative characteristics. Cold Spring Harb. Symp. Quant. Biol. 20: 197–212. 10.1101/SQB.1955.020.01.019 [DOI] [PubMed] [Google Scholar]
- Bell G., and Koufopanou V., 1986. The cost of reproduction. Oxf. Surv. Evol. Biol. 3: 83–131. [Google Scholar]
- Bennett A. F., and Lenski R. E., 1999. Experimental evolution and its role in evolutionary physiology. Am. Zool. 39: 346–362. 10.1093/icb/39.2.346 [DOI] [Google Scholar]
- Bergland A. O., Genissel A., Nuzhdin S. V., and Tatar M., 2008. Quantitative trait loci affecting phenotypic plasticity and the allometric relationship of ovariole number and thorax length in Drosophila melanogaster. Genetics 180: 567–582. 10.1534/genetics.108.088906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Chae H., Kim Y.-J., and Tatar M., 2012. Fine-scale mapping of natural variation in fly fecundity identifies neuronal domain of expression and function of an aquaporin. PLoS Genet. 8: e1002631 10.1371/journal.pgen.1002631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., and Petrov D. A., 2014. Genomic evidence of rapid and stable Adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775 10.1371/journal.pgen.1004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Tobler R., González J., Schmidt P., and Petrov D., 2016. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol. Ecol. 25: 1157–1174. 10.1111/mec.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein M. R., Zdraljevic S., Andersen E. C., and Rockman M. V., 2019. Tightly linked antagonistic-effect loci underlie polygenic phenotypic variation in C. elegans. Evol. Lett. 3: 462–473. 10.1002/evl3.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., 2016. The mighty fruit fly moves into outbred genetics. PLoS Genet. 12: e1006388 10.1371/journal.pgen.1006388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J., and Edgar B., 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125: 2149–2158. [DOI] [PubMed] [Google Scholar]
- Britton J., Lockwood W., Li L., Cohen S., and Edgar B., 2002. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2: 239–249. 10.1016/S1534-5807(02)00117-X [DOI] [PubMed] [Google Scholar]
- Bochdanovits Z., and de Jong G., 2003a Experimental evolution in Drosophila melanogaster: interaction of temperature and food quality selection regimes. Evolution 57: 1829–1836. 10.1111/j.0014-3820.2003.tb00590.x [DOI] [PubMed] [Google Scholar]
- Bochdanovits Z., and de Jong G., 2003b Temperature dependence of fitness components in geographical populations of Drosophila melanogaster: changing the association between size and fitness. Biol. J. Linn. Soc. Lond. 80: 717–725. 10.1111/j.1095-8312.2003.00271.x [DOI] [Google Scholar]
- Boesiger E., 1968. Estimation globale de l’age des femelles de Drosophila melanogaster capturées dans les populations naturelles. C. R. Seances Soc. Biol. Fil. 162: 358–361. [PubMed] [Google Scholar]
- Boggs C. L., 2009. Understanding insect life histories and senescence through a resource allocation lens. Funct. Ecol. 23: 27–37. 10.1111/j.1365-2435.2009.01527.x [DOI] [Google Scholar]
- Borash D. J., and Ho G. T., 2001. Patterns of selection: stress resistance and energy storage in density-dependent populations of Drosophila melanogaster. J. Insect Physiol. 47: 1349–1356. 10.1016/S0022-1910(01)00108-1 [DOI] [PubMed] [Google Scholar]
- Borash D. J., Gibbs A. G., Joshi A., and Mueller L. D., 1998. A genetic polymorphism maintained by natural selection in a temporally varying environment. Am. Nat. 151: 148–156. 10.1086/286108 [DOI] [PubMed] [Google Scholar]
- Boulétreau J., 1978. Ovarian activity and reproductive potential in a natural population of Drosophila melanogaster. Oecologia 35: 319–342. 10.1007/BF00345140 [DOI] [PubMed] [Google Scholar]
- Boulétreau-Merle J., and Fouillet P., 2002. How to overwinter and be a founder: egg-retention phenotypes and mating status in Drosophila melanogaster. Evol. Ecol. 16: 309–332. 10.1023/A:1020216230976 [DOI] [Google Scholar]
- Boulétreau-Merle J., Allemand R., Cohet Y., and David J. R., 1982. Reproductive strategy in Drosophila melanogaster: significance of a genetic divergence between temperate and tropical populations. Oecologia 53: 323–329. 10.1007/BF00389008 [DOI] [PubMed] [Google Scholar]
- Boulétreau-Merle J., Fouillet P., and Varaldi J., 2003. Divergent strategies in low temperature environment for the sibling species Drosophila melanogaster and D. simulans: overwintering in extension border areas of France and comparison with African populations. Evol. Ecol. 17: 523–548. 10.1023/B:EVEC.0000005632.21186.21 [DOI] [Google Scholar]
- Brankatschk M., Gutmann T., Knittelfelder O., Palladini A., Prince E. et al. , 2018. A temperature-dependent switch in feeding preference improves Drosophila development and survival in the cold. Dev. Cell 46: 781–793.e4 (erratum: Dev. Cell 47: 257–259). 10.1016/j.devcel.2018.05.028 [DOI] [PubMed] [Google Scholar]
- Bross T. G., Rogina B., and Helfand S. L., 2005. Behavioral, physical, and demographic changes in Drosophila populations through dietary restriction. Aging Cell 4: 309–317. 10.1111/j.1474-9726.2005.00181.x [DOI] [PubMed] [Google Scholar]
- Bruce K. D., Hoxha S., Carvalho G. B., Yamada R., Wang H. D. et al. , 2013. High carbohydrate-low protein consumption maximizes Drosophila lifespan. Exp. Gerontol. 48: 1129–1135. 10.1016/j.exger.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubliy O. A., Loeschcke V., and Imasheva A. G., 2001. Genetic variation of morphological traits in Drosophila melanogaster under poor nutrition: isofemale lines and offspring–parent regression. Heredity 86: 363–369. 10.1046/j.1365-2540.2001.00837.x [DOI] [PubMed] [Google Scholar]
- Buck S., Vettraino J., Force A. G., and Arking R., 2000. Extended longevity in Drosophila is consistently associated with a decrease in developmental viability. J. Gerontol. A 55: B292–B301. 10.1093/gerona/55.6.B292 [DOI] [PubMed] [Google Scholar]
- Bundgaard J., and Christiansen F. B., 1972. Dynamics of polymorphisms: I. Selection components in an experimental population of Drosophila melanogaster. Genetics 71: 439–460. [DOI] [PubMed] [Google Scholar]
- Burger J. M., Hwangbo D. S., Corby-Harris V., and Promislow D. E., 2007. The functional costs and benefits of dietary restriction in Drosophila. Aging Cell 6: 63–71. 10.1111/j.1474-9726.2006.00261.x [DOI] [PubMed] [Google Scholar]
- Burger J. M., Buechel S. D., and Kawecki T. J., 2010. Dietary restriction affects lifespan but not cognitive aging in Drosophila melanogaster. Aging Cell 9: 327–335. 10.1111/j.1474-9726.2010.00560.x [DOI] [PubMed] [Google Scholar]
- Bürger R., 2000. The Mathematical Theory of Selection, Recombination, and Mutation. John Wiley & Sons, New York. [Google Scholar]
- Burke M. K., and Rose M. R., 2009. Experimental evolution with Drosophila. Am. J. Physiol. 296: R1847–R1854. [DOI] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R. et al. , 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467: 587–590. 10.1038/nature09352 [DOI] [PubMed] [Google Scholar]
- Burke M. K., King E. G., Shahrestani P., Rose M. R., and Long A. D., 2014. Genome-wide association study of extreme longevity in Drosophila melanogaster. Genome Biol. Evol. 6: 1–11. 10.1093/gbe/evt180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzati-Traverso A. A., 1955. Evolutionary changes in components of fitness and other polygenic traits in Drosophila melanogaster populations. Heredity 9: 153–186. 10.1038/hdy.1955.18 [DOI] [Google Scholar]
- Calboli F. C., Kennington W. J., and Partridge L., 2003. QTL mapping reveals a striking coincidence in the positions of genomic regions associated with adaptive variation in body size in parallel clines of Drosophila melanogaster on different continents. Evolution 57: 2653–2658. 10.1111/j.0014-3820.2003.tb01509.x [DOI] [PubMed] [Google Scholar]
- Capy P., Pla E., and David J. R., 1993. Phenotypic and genetic variability of morphometrical traits in natural populations of Drosophila melanogaster and D simulans. I. Geographic variations. Genet. Sel. Evol. 25: 517–536. 10.1186/1297-9686-25-6-517 [DOI] [Google Scholar]
- Carbone M., Jordan K., Lyman R., Harbison S., Leips J. et al. , 2006. Phenotypic variation and natural selection at Catsup, a pleiotropic quantitative trait gene in Drosophila. Curr. Biol. 16: 912–919. 10.1016/j.cub.2006.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes M. U., Campbell T., Huang W., Butler D. G., Carbone M. A. et al. , 2015. The genomic basis of postponed senescence in Drosophila melanogaster. PLoS One 10: e0138569 10.1371/journal.pone.0138569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira V. P., Imberti M. A., Mensch J., and Fanara J. J., 2013. Gene-by-temperature interactions and candidate plasticity genes for morphological traits in Drosophila melanogaster. PLoS One 8: e70851 10.1371/journal.pone.0070851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsten L. D., Watts T., and Markow T. A., 2005. Gene expression patterns accompanying a dietary shift in Drosophila melanogaster. Mol. Ecol. 14: 3203–3208. 10.1111/j.1365-294X.2005.02654.x [DOI] [PubMed] [Google Scholar]
- Cavicchi S., 1978. Investigation on early divergence between populations of Drosophila melanogaster kept at different temperatures. Genetica 48: 81–87. 10.1007/BF00127503 [DOI] [Google Scholar]
- Cavicchi S., Guerra D., Natali V., Pezzoli C., and Giorgi G., 1989. Temperature-related divergence in experimental populations of Drosophila melanogaster. II. Correlation between fitness and body dimensions. J. Evol. Biol. 2: 235–251. 10.1046/j.1420-9101.1989.2040235.x [DOI] [Google Scholar]
- Cavicchi S., Giorgi G., Natali V., and Guerra D., 1991. Temperature related divergence in experimental populations of Drosophila melanogaster. 4. Fourier and centroid analysis of wing shape and relationship between shape variation and fitness. J. Evol. Biol. 4: 141–159. 10.1046/j.1420-9101.1991.4010141.x [DOI] [Google Scholar]
- Chapman T., and Partridge L., 1996. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc. Biol. Sci. 263: 755–759. 10.1098/rspb.1996.0113 [DOI] [PubMed] [Google Scholar]
- Chapman T., Trevitt S., and Partridge L., 1994. Remating and Male-Derived Nutrients in Drosophila melanogaster. J. Evol. Biol. 7: 51–69. 10.1046/j.1420-9101.1994.7010051.x [DOI] [Google Scholar]
- Charlesworth B., 1980. Evolution in Age-Structured Populations. Cambridge University Press, Cambridge. [Google Scholar]
- Charlesworth B., 1987. The heritability of fitness, pp. 21–40 in Sexual Selection: Testing the Alternatives, edited by Bradbury J. W. and Andersson M. John Wiley & Sons, New York. [Google Scholar]
- Charlesworth B., 1990. Optimization models, quantitative genetics, and mutation. Evolution 44: 520–538. 10.1111/j.1558-5646.1990.tb05936.x [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1993a Natural selection on multivariate traits in age-structured populations. Proc. Biol. Sci. 251: 47–52. 10.1098/rspb.1993.0007 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1993b Evolutionary mechanisms of senescence. Genetica 91: 11–19. 10.1007/BF01435984 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1994. Evolution in Age-Structured Populations. Cambridge University Press, Cambridge: 10.1017/CBO9780511525711 [DOI] [Google Scholar]
- Charlesworth B., 2000. Fisher, Medawar, Hamilton and the evolution of aging. Genetics 156: 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2001. Patterns of age-specific means and genetic variances of mortality rates predicted by the mutation accumulation theory of aging. J. Theor. Biol. 210: 47–65. 10.1006/jtbi.2001.2296 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2003. The population genetics of life-history evolution, pp. 216–232 in The Evolution of Population Biology, edited by Singh R. S. and Uyenoyama M. K.. Cambridge University Press, Cambridge. [Google Scholar]
- Charlesworth, B., 2013 Fitness and Selection. eLS (Encyclopedia of Life Sciences). Accessed November 7, 2019. Available at: 10.1002/9780470015902.a0005444.pub3 [DOI] [Google Scholar]
- Charlesworth B., 2015. Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc. Natl. Acad. Sci. USA 112: 1662–1669 (erratum: Proc. Natl. Acad. Sci. USA 112: E1049). 10.1073/pnas.1423275112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., and Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts and Company Publishers, Greenwood Village, CO. [Google Scholar]
- Charlesworth B., and Edwards A. W. F., 2018. A century of variance. Significance 15: 20–25. 10.1111/j.1740-9713.2018.01170.x [DOI] [Google Scholar]
- Charlesworth B., and Hughes K. A., 1996. Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl. Acad. Sci. USA 93: 6140–6145. 10.1073/pnas.93.12.6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., and Hughes K. A., 2000. The maintenance of genetic variation in life-history traits, pp. 369–392 in Evolutionary Genetics - From Molecules to Morphology, edited by Singh R. S. and Krimbas C. B.. Cambridge University Press, Cambridge. [Google Scholar]
- Charlesworth B., Borthwick H., Bartolomé C., and Pignatelli P., 2004. Estimates of the genomic mutation rate for detrimental alleles in Drosophila melanogaster. Genetics 167: 815–826. 10.1534/genetics.103.025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Miyo T., and Borthwick H., 2007. Selection responses of means and inbreeding depression for female fecundity in Drosophila melanogaster suggest contributions from intermediate-frequency alleles to quantitative trait variation. Genet. Res. 89: 85–91. 10.1017/S001667230700866X [DOI] [PubMed] [Google Scholar]
- Chen J., Nolte V., and Schlötterer C., 2015. Temperature-related reaction norms of gene expression: regulatory architecture and functional implications. Mol. Biol. Evol. 32: 2393–2402. 10.1093/molbev/msv120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lee S. F., Blanc E., Reuter C., Wertheim B. et al. , 2012. Genome-wide transcription analysis of clinal genetic variation in Drosophila. PLoS One 7: e34620 10.1371/journal.pone.0034620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., 1988. A comparison of genetic and phenotypic correlations. Evolution 42: 958–968. 10.1111/j.1558-5646.1988.tb02514.x [DOI] [PubMed] [Google Scholar]
- Chippindale A. K., Leroi A. M., Kim S. B., and Rose M. R., 1993. Phenotypic plasticity and selection in Drosophila life history evolution. I. Nutrition and the cost of reproduction. J. Evol. Biol. 6: 171–193. 10.1046/j.1420-9101.1993.6020171.x [DOI] [Google Scholar]
- Chippindale A. K., Hoang D. T., Service P. M., and Rose M. R., 1994. The evolution of development in Drosophila melanogaster selected for postponed senescence. Evolution 48: 1880–1899. 10.1111/j.1558-5646.1994.tb02221.x [DOI] [PubMed] [Google Scholar]
- Chippindale A. K., Chu T. J. F., and Rose M. R., 1996. Complex trade-offs and the evolution of starvation resistance in Drosophila melanogaster. Evolution 50: 753–766. 10.1111/j.1558-5646.1996.tb03885.x [DOI] [PubMed] [Google Scholar]
- Chippindale A. K., Leroi A. M., Saing H., Borash D. J., and Rose M. R., 1997. Phenotypic plasticity and selection in Drosophila life history evolution. 2. Diet, mates and the cost of reproduction. J. Evol. Biol. 10: 269–293. 10.1007/s000360050023 [DOI] [Google Scholar]
- Chippindale A. K., Gibbs A. G., Sheik M., Yee K. J., Djawdan M. et al. , 1998. Resource acquisition and the evolution of stress resistance in Drosophila melanogaster. Evolution 52: 1342–1352. 10.1111/j.1558-5646.1998.tb02016.x [DOI] [PubMed] [Google Scholar]
- Cho I., Horn L., Felix T. M., Foster L., Gregory G. et al. , 2010. Age- and diet-specific effects of variation at S6 kinase on life history, metabolic, and immune response traits in Drosophila melanogaster. DNA Cell Biol. 29: 473–485. 10.1089/dna.2009.0997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D. J., Gems D., Harshman L. G., Oldham S., Stocker H. et al. , 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106. 10.1126/science.1057991 [DOI] [PubMed] [Google Scholar]
- Clark A. G., 1987. Senescence and the genetic correlation hangup. Am. Nat. 129: 932–940. 10.1086/284686 [DOI] [Google Scholar]
- Clarke J. M., Smith J. M., and Sondhi K. C., 1961. Asymmetrical response to selection for rate of development in Drosophila subobscura. Genet. Res. 2: 70–81. 10.1017/S0016672300000550 [DOI] [Google Scholar]
- Clemson A. S., Sgrò C. M., and Telonis-Scott M., 2016. Thermal plasticity in Drosophila melanogaster populations from eastern Australia: quantitative traits to transcripts. J. Evol. Biol. 29: 2447–2463. 10.1111/jeb.12969 [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T. H. (Editor), 1988. Reproductive Success: Studies of Individual Variation in Contrasting Breeding Systems. University of Chicago Press, Chicago. [Google Scholar]
- Cole L. C., 1954. The population consequences of life history phenomena. Q. Rev. Biol. 29: 103–137. 10.1086/400074 [DOI] [PubMed] [Google Scholar]
- Conner J. K., 2012. Quantitative genetic approaches to evolutionary constraint: how useful? Evolution 66: 3313–3320. 10.1111/j.1558-5646.2012.01794.x [DOI] [PubMed] [Google Scholar]
- Conner J. K., and Hartl D. L., 2004. A Primer of Ecological Genetics. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Corbett-Detig R. B., Zhou J., Clark A. G., Hartl D. L., and Ayroles J. F., 2013. Genetic incompatibilities are widespread within species. Nature 504: 135–137. 10.1038/nature12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., and Beecham E., 1987. Heritability of two morphological characters within and among natural populations of Drosophila melanogaster. Genetics 117: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill W. D., Huey R. B., and Gilchrist G. W., 1996. Within- and between-generation effects of temperature on the morphology and physiology of Drosophila melanogaster. Evolution 50: 1205–1218. 10.1111/j.1558-5646.1996.tb02361.x [DOI] [PubMed] [Google Scholar]
- Crnokrak P., and Roff D. A., 1995. Dominance variance: associations with selection and fitness. Heredity 75: 530–540. 10.1038/hdy.1995.169 [DOI] [Google Scholar]
- Crow J. F., 1993. Mutation, mean fitness, and genetic load. Oxf. Surv. Evol. Biol. 9: 3–42. [Google Scholar]
- Crow J. F., and Simmons M. J., 1983. The mutation load in Drosophila, pp. 1–35 in The Genetics and Biology of Drosophila, Vol. 3c, edited by Ashburner M., Carson H. L., and Thompson J. N. Jr. Academic Press, New York. [Google Scholar]
- Curtsinger J. W., Service P. M., and Prout T., 1994. Antagonistic pleiotropy, reversal of dominance, and genetic polymorphism. Am. Nat. 144: 210–228. 10.1086/285671 [DOI] [Google Scholar]
- D Lage J. L., Capy P., and David J. R., 1990. Starvation and desiccation tolerance in Drosophila melanogaster: differences between European, North African and Afrotropical populations. Genet. Sel. Evol. 22: 381–391. 10.1186/1297-9686-22-4-381 [DOI] [Google Scholar]
- David J., and Bocquet C., 1974. Nouvelles particularities génétiques des souches Japonaises de Drosophila melanogaster. Arch. Zool. Exp. Gén. 115: 489–503. [Google Scholar]
- David J., Cohet Y., and Fouillet P., 1975. The variability between individuals as a measure of senescence: A study of the number of eggs laid and the percentage of hatched eggs in the case of Drosophila melanogaster. Exp. Gerontol. 10: 17–25. 10.1016/0531-5565(75)90011-X [DOI] [PubMed] [Google Scholar]
- David J., Bocquet C., and De Scheemaecker-Louis M., 1977. Genetic latitudinal adaptation of Drosophila melanogaster: new discriminative biometrical traits between European and equatorial African populations. Genet. Res. 30: 247–255. 10.1017/S0016672300017651 [DOI] [Google Scholar]
- David J., Moreteau B., Gauthier J., Pétavy G., Stockel A. et al. , 1994. Reaction norms of size characters in relation to growth temperature in Drosophila melanogaster: an isofemale lines analysis. Genet. Sel. Evol. 26: 229–251. 10.1186/1297-9686-26-3-229 [DOI] [Google Scholar]
- David J. R., 1970. Le nombre d’ovarioles chez la Drosophila: relation avec la fecondite et valeur adaptive. Arch. Zool. Exp. Gén. 111: 357–370. [Google Scholar]
- David J. R., and Bocquet C., 1975. Evolution in a cosmopolitan species: genetic latitudinal clines in Drosophila melanogaster wild populations. Experientia 31: 164–166. 10.1007/BF01990682 [DOI] [PubMed] [Google Scholar]
- David J. R., and Capy P., 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4: 106–111. 10.1016/0168-9525(88)90098-4 [DOI] [PubMed] [Google Scholar]
- David J. R., Biémont C., and Fouillet P., 1974. Sur la forme des courbes de ponte de Drosophila melanogaster et leur adjustment à des modèles mathématiques. Arch. Zool. Exp. Gén. 115: 263–277. [Google Scholar]
- David J. R., Allemand R., van Herrewege J., and Cohet Y., 1983. Ecophysiology: abiotic factors, pp. 105–170 in The Genetics and Biology of Drosophila, Vol. 3d, edited by Ashburner M., Carson H. L., and Thompson J. N.. Academic Press, London. [Google Scholar]
- David J. R., Gibert P., Gravot E., Petavy G., Morin J.-P. et al. , 1997. Phenotypic plasticity and developmental temperature in Drosophila: analysis and significance of reaction norms of morphometrical traits. J. Therm. Biol. 22: 441–451. 10.1016/S0306-4565(97)00063-6 [DOI] [Google Scholar]
- David J. R., Allemand R., Capy P., Chakir M., Gibert P. et al. , 2004. Comparative Life Histories and Ecophysiology of Drosophila melanogaster and D. simulans. Genetica 120: 151–163. 10.1023/B:GENE.0000017638.02813.5a [DOI] [PubMed] [Google Scholar]
- David J. R., Gibert P., Legout H., Pétavy G., Capy P. et al. , 2005. Isofemale lines in Drosophila: an empirical approach to quantitative trait analysis in natural populations. Heredity (Edinb) 94: 3–12. 10.1038/sj.hdy.6800562 [DOI] [PubMed] [Google Scholar]
- David J. R., Yassin A., Moreteau J. C., Legout H., and Moreteau B., 2011. Thermal phenotypic plasticity of body size in Drosophila melanogaster: sexual dimorphism and genetic correlations. J. Genet. 90: 295–302. 10.1007/s12041-011-0076-8 [DOI] [PubMed] [Google Scholar]
- de Jong G., and Bochdanovits Z., 2003. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J. Genet. 82: 207–223. 10.1007/BF02715819 [DOI] [PubMed] [Google Scholar]
- de Jong G., and Van Noordwijk A. J., 1992. Acquisition and allocation of resources – genetic (co) variances, selection, and life histories. Am. Nat. 139: 749–770. 10.1086/285356 [DOI] [Google Scholar]
- Delcourt A., and Guyenot E., 1910. De la possibilité d’étudier certains diptères en milieu défini. C. R. Hebd. Seances Acad. Sci. 151: 255–257. [Google Scholar]
- Delpuech J.-M., Moreteau B., Chiche J., Pla E., Vouidibio J. et al. , 1995. Phenotypic plasticity and reaction norms in temperate and tropical populations of Drosophila melanogaster: ovarian size and developmental temperature. Evolution 49: 670–675. [DOI] [PubMed] [Google Scholar]
- De Luca M., Roshina N. V., Geiger-Thornsberry G. L., Lyman R. F., Pasyukova E. G. et al. , 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34: 429–433. 10.1038/ng1218 [DOI] [PubMed] [Google Scholar]
- De Moed G. H., De Jong G., and Scharloo W., 1997. Environmental effects on body size variation in Drosophila melanogaster and its cellular basis. Genet. Res. 70: 35–43. 10.1017/S0016672397002930 [DOI] [PubMed] [Google Scholar]
- DeWitt T. J., and Scheiner S. M. (Editors), 2004. Phenotypic Plasticity: Functional and Conceptual Approaches. Oxford University Press, New York. [Google Scholar]
- Dick K. B., Ross C. R., and Yampolsky L. Y., 2011. Genetic variation of dietary restriction and the effects of nutrient-free water and amino acid supplements on lifespan and fecundity of Drosophila. Genet. Res. 122: 582–594. [DOI] [PubMed] [Google Scholar]
- Dickerson G. E., 1955. Genetic slippage in response to selection. Cold Spring Harb. Symp. Quant. Biol. 20: 213–224. 10.1101/SQB.1955.020.01.020 [DOI] [PubMed] [Google Scholar]
- Ding F., Gil M. P., Franklin M., Ferreira J., Tatar M. et al. , 2014. Transcriptional response to dietary restriction in Drosophila melanogaster. J. Insect Physiol. 69: 101–106. 10.1016/j.jinsphys.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Berrocal A., Morita T., Longden K. D., and Stern D. L., 2016. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 536: 329–332. 10.1038/nature19093 [DOI] [PubMed] [Google Scholar]
- Djawdan M., Sugiyama T. T., Schlaeger L. K., Bradley T. J., and Rose M. R., 1996. Metabolic aspects of the trade-off between fecundity and longevity in Drosophila melanogaster. Physiol. Zool. 69: 1176–1195. 10.1086/physzool.69.5.30164252 [DOI] [Google Scholar]
- Dobzhansky T., 1955. A review of some fundamental concepts and problems of population genetics. Cold Spring Harb. Symp. Quant. Biol. 20: 1–15. 10.1101/SQB.1955.020.01.003 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1970. Genetics of the Evolutionary Process. Columbia University Press, New York. [Google Scholar]
- Draye X., and Lints F., 1995. Geographic variations of life history strategies in Drosophila melanogaster. II. Analysis of laboratory-adapted populations. Exp. Gerontol. 30: 517–532. 10.1016/0531-5565(95)00006-3 [DOI] [PubMed] [Google Scholar]
- Draye X., and Lints F., 1996. Geographic variations of life history strategies in Drosophila melanogaster. III. New data. Exp. Gerontol. 31: 717–733. 10.1016/S0531-5565(96)00073-3 [DOI] [PubMed] [Google Scholar]
- Draye X., Bullens P., and Lints F. A., 1994. Geographic variations of life history strategies in Drosophila melanogaster. I. Analysis of wild-caught populations. Exp. Gerontol. 29: 205–222. 10.1016/0531-5565(94)90052-3 [DOI] [PubMed] [Google Scholar]
- Druger M., 1962. Selection and body size in Drosophila pseudoobscura at different temperatures. Genetics 47: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen P., Živković D., Hutter S., Stephan W., and Laurent S., 2013. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics 193: 291–301. 10.1534/genetics.112.145912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham M. F., Magwire M. M., Stone E. A., and Leips J., 2014. Genome-wide analysis in Drosophila reveals age-specific effects of SNPs on fitness traits. Nat. Commun. 5: 4338 10.1038/ncomms5338 [DOI] [PubMed] [Google Scholar]
- Durmaz E., Benson C., Kapun M., Schmidt P., and Flatt T., 2018. An inversion supergene in Drosophila underpins latitudinal clines in survival traits. J. Evol. Biol. 31: 1354–1364. 10.1111/jeb.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz E., Rajpurohit S., Betancourt N., Fabian D. K., Kapun M. et al. , 2019. A clinal polymorphism in the insulin signaling transcription factor foxo contributes to life-history adaptation in Drosophila. Evolution 73: 1774–1792. 10.1111/evo.13759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- East E. M., 1910. A Mendelian interpretation of variation that is apparently continuous. Am. Nat. 44: 65–82. 10.1086/279117 [DOI] [Google Scholar]
- Economos A. C., and Lints F. A., 1984. Growth rate and life span in Drosophila. II. A biphasic relationship between growth rate and life span. Mech. Ageing Dev. 27: 143–151. 10.1016/0047-6374(84)90039-3 [DOI] [PubMed] [Google Scholar]
- Edgar B., 2006. How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7: 907–916. 10.1038/nrg1989 [DOI] [PubMed] [Google Scholar]
- Emerson K., Bradshaw W., and Holzapfel C., 2009a Complications of complexity: integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 25: 217–225. 10.1016/j.tig.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Emerson K., Uyemura A., McDaniel K., Schmidt P., Bradshaw W. et al. , 2009b Environmental control of ovarian dormancy in natural populations of Drosophila melanogaster. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 195: 825–829. 10.1007/s00359-009-0460-5 [DOI] [PubMed] [Google Scholar]
- Endler J. A., 1977. Geographic Variation, Speciation, and Clines. Princeton University Press, Princeton. [PubMed] [Google Scholar]
- Endler J. A., 1986. Natural Selection in the Wild. Princeton University Press, Princeton. [Google Scholar]
- Engstrom, L. E., 1971 Studies of the effects of two-way selection for ovariole number in Drosophila melanogaster. Ph.D. Dissertation, University of Illinois, Urbana-Champaign, IL. [Google Scholar]
- Fabian D. K., and Flatt T., 2012. Life history evolution. Nat. Edu. Knowl. 3: 24. [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S. et al. , 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21: 4748–4769. 10.1111/j.1365-294X.2012.05731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Lack J. B., Mathur V., Schlötterer C., Schmidt P. S. et al. , 2015. Spatially varying selection shapes life history clines among populations of Drosophila melanogaster from sub-Saharan Africa. J. Evol. Biol. 28: 826–840. 10.1111/jeb.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Garschall K., Klepsatel P., Santos-Matos G., Sucena É. et al. , 2018. Evolution of longevity improves immunity in Drosophila. Evol. Lett. 2: 567–579. 10.1002/evl3.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., and Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Addison Wesley Longman, Harlow. [Google Scholar]
- Fallis L. C., Fanara J. J., and Morgan T. J., 2014. Developmental thermal plasticity among Drosophila melanogaster populations. J. Evol. Biol. 27: 557–564. 10.1111/jeb.12321 [DOI] [PubMed] [Google Scholar]
- Felix T. M., Hughes K. A., Stone E. A., Drnevich J. M., and Leips J., 2012. Age-specific variation in immune response in Drosophila melanogaster has a genetic basis. Genetics 191: 989–1002. 10.1534/genetics.112.140640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1976. The theoretical population genetics of variable selection and migration. Annu. Rev. Genet. 10: 253–280. 10.1146/annurev.ge.10.120176.001345 [DOI] [PubMed] [Google Scholar]
- Finch C. E., and Rose M. R., 1995. Hormones and the physiological architecture of life history evolution. Q. Rev. Biol. 70: 1–52. 10.1086/418864 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1919. XV.—The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52: 399–433. 10.1017/S0080456800012163 [DOI] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection, Oxford University Press, Oxford: 10.5962/bhl.title.27468 [DOI] [Google Scholar]
- Flatt, T., 2004a The effects of juvenile hormone on trait architecture in Drosophila melanogaster. Ph.D. Thesis, University of Fribourg, Switzerland. [Google Scholar]
- Flatt T., 2004b Assessing natural variation in genes affecting Drosophila lifespan. Mech. Ageing Dev. 125: 155–159. 10.1016/j.mad.2004.02.004 [DOI] [PubMed] [Google Scholar]
- Flatt T., 2005. The evolutionary genetics of canalization. Q. Rev. Biol. 80: 287–316. 10.1086/432265 [DOI] [PubMed] [Google Scholar]
- Flatt T., 2009. Ageing: diet and longevity in the balance. Nature 462: 989–990. 10.1038/462989a [DOI] [PubMed] [Google Scholar]
- Flatt T., 2011. Survival costs of reproduction in Drosophila. Exp. Gerontol. 46: 369–375. 10.1016/j.exger.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Flatt T., 2014. Plasticity of lifespan: a reaction norm perspective. Proc. Nutr. Soc. 73: 532–542. 10.1017/S0029665114001141 [DOI] [PubMed] [Google Scholar]
- Flatt T., 2016. Genomics of clinal variation in Drosophila: disentangling the interactions of selection and demography. Mol. Ecol. 25: 1023–1026. 10.1111/mec.13534 [DOI] [PubMed] [Google Scholar]
- Flatt T., and Heyland A. (Editors), 2011. Mechanisms of Life History Evolution. The Genetics and Physiology of Life History Traits and Trade-Offs. Oxford University Press, Oxford, UK. [Google Scholar]
- Flatt T., and Kawecki T. J., 2004. Pleiotropic effects of methoprene-tolerant (Met), a gene involved in juvenile hormone metabolism, on life history traits in Drosophila melanogaster. Genetica 122: 141–160. 10.1023/B:GENE.0000041000.22998.92 [DOI] [PubMed] [Google Scholar]
- Flatt T., and Kawecki T. J., 2007. Juvenile hormone as a regulator of the trade-off between reproduction and life span in Drosophila melanogaster. Evolution 61: 1980–1991. 10.1111/j.1558-5646.2007.00151.x [DOI] [PubMed] [Google Scholar]
- Flatt T., and Partridge L., 2018. Horizons in the evolution of aging. BMC Biol. 16: 93 10.1186/s12915-018-0562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., and Promislow D. E. L., 2007. Still pondering an age-old question. Science 318: 1255–1256. 10.1126/science.1147491 [DOI] [PubMed] [Google Scholar]
- Flatt T., and Schmidt P. S., 2009. Integrating evolutionary and molecular genetics of aging. Biochim. Biophys. Acta 1790: 951–962. 10.1016/j.bbagen.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., Tu M.-P., and Tatar M., 2005. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays 27: 999–1010. 10.1002/bies.20290 [DOI] [PubMed] [Google Scholar]
- Flatt T., Min K. J., D’Alterio C., Villa-Cuesta E., Cumbers J. et al. , 2008. Drosophila germ-line modulation of insulin signaling and lifespan. Proc. Natl. Acad. Sci. USA 105: 6368–6373. 10.1073/pnas.0709128105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., Amdam G. V., Kirkwood T. B. L., and Omholt S. W., 2013. Life-history evolution and the polyphenic regulation of somatic maintenance and survival. Q. Rev. Biol. 88: 185–218. 10.1086/671484 [DOI] [PubMed] [Google Scholar]
- Folguera G., Ceballos S., Spezzi L., Fanara J. J., and Hasson E., 2008. Clinal variation in developmental time and viability, and the response to thermal treatments in two species of Drosophila. Biol. J. Linn. Soc. Lond. 95: 233–245. 10.1111/j.1095-8312.2008.01053.x [DOI] [Google Scholar]
- Force A. G., Staples T., Soliman S., and Arking R., 1995. Comparative biochemical and stress analysis of genetically selected Drosophila strains with different longevities. Dev. Genet. 17: 340–351. 10.1002/dvg.1020170407 [DOI] [PubMed] [Google Scholar]
- Ford D., and Tower J., 2006. Genetic manipulation of lifespan in Drosophila melanogaster, pp. 400–414 in Handbook of the Biology of Aging, Ed. 6, edited by Masoro E. J. and Austad S. N.. Academic Press, New York. [Google Scholar]
- Fowler K., and Partridge L., 1989. A cost of mating in female fruitflies. Nature 338: 760–761. 10.1038/338760a0 [DOI] [Google Scholar]
- Fowler K., Semple C., Barton N. H., and Partridge L., 1997. Genetic variation for total fitness in Drosophila melanogaster. Proc. Biol. Sci. 264: 191–199. 10.1098/rspb.1997.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V., Feast M., and Partridge L., 1998. Body size and cell size in Drosophila: the developmental response to temperature. J. Insect Physiol. 44: 1081–1089. 10.1016/S0022-1910(98)00061-4 [DOI] [PubMed] [Google Scholar]
- Frydenberg J., Hoffmann A. A., and Loeschcke V., 2003. DNA sequence variation and latitudinal associations in hsp23, hsp26 and hsp27 from natural populations of Drosophila melanogaster. Mol. Ecol. 12: 2025–2032. 10.1046/j.1365-294X.2002.01882.x [DOI] [PubMed] [Google Scholar]
- Fuller R. C., Baer C. F., and Travis J., 2005. How and when selection experiments might actually be useful. Integr. Comp. Biol. 45: 391–404. 10.1093/icb/45.3.391 [DOI] [PubMed] [Google Scholar]
- Gaillard J.-M., and Lemaître J.-F., 2017. The Williams’ legacy: a critical reappraisal of his nine predictions about the evolution of senescence. Evolution 71: 2768–2785. 10.1111/evo.13379 [DOI] [PubMed] [Google Scholar]
- Garcia J. F., Carbone M. A., Mackay T. F. C., and Anholt R. R. H., 2017. Regulation of Drosophila lifespan by bellwether promoter alleles. Sci. Rep. 7: 4109 10.1038/s41598-017-04530-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorado A., and Caballero A., 2000. On the average coefficient of dominance of deleterious spontaneous mutations. Genetics 155: 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T., and Rose M. R. (Editors), 2009. Experimental Evolution. University of California Press, Berkeley, CA. [Google Scholar]
- Garud N. R., Messer P. W., Buzbas E. O., and Petrov D. A., 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11: e1005004 10.1371/journal.pgen.1005004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser M., Kaiser M., Berrigan D., and Stearns S. C., 2000. Life-history correlates of evolution under high and low adult mortality. Evolution 54: 1260–1272. 10.1111/j.0014-3820.2000.tb00559.x [DOI] [PubMed] [Google Scholar]
- Gayon J., and Veuille M., 2001. The genetics of experimental populations: L’Heritier and Teissier’s population cages (1932–1954), pp. 77–102 in Thinking about Evolution: Historical, Philosophical, and Political Perspectives, Vol. 2, edited by Singh R. S., Krimbas C. B., Paul D. B., and Beatty J.. Cambridge University Press, Cambridge. [Google Scholar]
- Gebhardt M. D., and Stearns S. C., 1988. Reaction norms for developmental time and weight at eclosion in Drosophila mercatorum. J. Evol. Biol. 1: 335–354. 10.1046/j.1420-9101.1988.1040335.x [DOI] [Google Scholar]
- Gebhardt M. D., and Stearns S. C., 1992. Phenotypic plasticity for life-history traits in Drosophila melanogaster. III. Effect of the environment on genetic parameters. Genet. Res. 60: 87–101. 10.1017/S0016672300030780 [DOI] [PubMed] [Google Scholar]
- Gebhardt M. D., and Stearns S. C., 1993a Phenotypic plasticity for life history traits in Drosophila melanogaster. I. Effect on phenotypic and environmental correlations. J. Evol. Biol. 6: 1–16. 10.1046/j.1420-9101.1993.6010001.x [DOI] [Google Scholar]
- Gebhardt M. D., and Stearns S. C., 1993b Phenotypic plasticity for life history traits in Drosophila melanogaster. II. Epigenetic mechanisms and the scaling of variances. J. Evol. Biol. 6: 17–29. 10.1046/j.1420-9101.1993.6010017.x [DOI] [Google Scholar]
- Geiger-Thornsberry G. L., and Mackay T. F., 2004. Quantitative trait loci affecting natural variation in Drosophila longevity. Mech. Ageing Dev. 125: 179–189. 10.1016/j.mad.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Géminard C., Arquier N., Layalle S., Bourouis M., Slaidina M. et al. , 2006. Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes 55: S5–S8. 10.2337/db06-S001 [DOI] [Google Scholar]
- Gershman B., Puig O., Hang L., Peitzsch R. M., Tatar M. et al. , 2007. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol. Genomics 29: 24–34. 10.1152/physiolgenomics.00061.2006 [DOI] [PubMed] [Google Scholar]
- Ghosh S. M., Testa N. D., and Shingleton A. W., 2013. Temperature-size rule is mediated by thermal plasticity of critical size in Drosophila melanogaster. Proc. Biol. Sci. 280: 20130174 10.1098/rspb.2013.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina T. J., Clark A. G., and Fiumera A. C., 2017. Estimating mating rates in wild Drosophila melanogaster females by decay rates of male reproductive proteins in their reproductive tracts. Mol. Ecol. Resour. 17: 1202–1209. 10.1111/1755-0998.12661 [DOI] [PubMed] [Google Scholar]
- Gibert P., Capy P., Imasheva A., Moreteau B., Morin J. P. et al. , 2004. Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity. Genetica 120: 165–179. 10.1023/B:GENE.0000017639.62427.8b [DOI] [PubMed] [Google Scholar]
- Giesel J. T., Murphy P. A., and Manlove M. N., 1982. The influence of temperature on genetic interrelationships of life-history traits in a population of Drosophila melanogaster: what tangled data sets we weave. Am. Nat. 119: 464–479. 10.1086/283926 [DOI] [Google Scholar]
- Gillespie J. H., 1991. The Causes of Molecular Evolution. Oxford University Press, Oxford. [Google Scholar]
- Glaser-Schmitt A., and Parsch J., 2018. Functional characterization of adaptive variation within a cis-regulatory element influencing Drosophila melanogaster growth. PLoS Biol. 16: e2004538 10.1371/journal.pbio.2004538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gockel J., Robinson S. J. W., Kennington W. J., Goldstein D. B., and Partridge L., 2002. Quantitative genetic analysis of natural variation in body size in Drosophila melanogaster. Heredity 89: 145–153. 10.1038/sj.hdy.6800121 [DOI] [PubMed] [Google Scholar]
- Goenaga J., Fanara J. J., and Hasson E., 2010. A quantitative genetic study of starvation resistance at different geographic scales in natural populations of Drosophila melanogaster. Genet. Res. 92: 253–259. 10.1017/S0016672310000327 [DOI] [PubMed] [Google Scholar]
- Good T. P., and Tatar M., 2001. Age-specific mortality and reproduction respond to adult dietary restriction in Drosophila melanogaster. J. Insect Physiol. 47: 1467–1473. 10.1016/S0022-1910(01)00138-X [DOI] [PubMed] [Google Scholar]
- Gowen J. W., and Johnson L. E., 1946. On the mechanism of heterosis. I. Metabolic capacity of different races of Drosophila melanogaster for egg production. Am. Nat. 80: 149–179. 10.1086/281373 [DOI] [PubMed] [Google Scholar]
- Grandison R. C., Piper M. D. W., and Partridge L., 2009a Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462: 1061–1064. 10.1038/nature08619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandison R. C., Wong R., Bass T. M., Partridge L., and Piper M. D., 2009b Effect of a standardised dietary restriction protocol on multiple laboratory strains of Drosophila melanogaster. PLoS One 4: e4067 10.1371/journal.pone.0004067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeteau C., Yahou F., Everaerts C., Dupont S., Farine J.-P. et al. , 2018. Yeast quality in juvenile diet affects Drosophila melanogaster adult life traits. Sci. Rep. 8: 13070 10.1038/s41598-018-31561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. L., Toolson E. C., Jeong C., Vu L. N., and Rose M. R., 1992. Desiccation, flight, glycogen, and postponed senescence in Drosophila melanogaster. Physiol. Zool. 65: 268–286. 10.1086/physzool.65.2.30158253 [DOI] [Google Scholar]
- Greenspan R. J., and Ferveur J.-F., 2000. Courtship in Drosophila. Annu. Rev. Genet. 34: 205–232. 10.1146/annurev.genet.34.1.205 [DOI] [PubMed] [Google Scholar]
- Gromko M. H., 1995. Unpredictability of correlated response to selection - pleiotropy and sampling interact. Evolution 49: 685–693. 10.1111/j.1558-5646.1995.tb02305.x [DOI] [PubMed] [Google Scholar]
- Gromko M. H., Briot A., Jensen S. C., and Fukui H. H., 1991. Selection on copulation duration in Drosophila melanogaster: predictability of direct response vs. unpredictability of correlated response. Evolution 45: 69–81. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L. et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85 [corrigenda: Nature 453: 128 (2008)]. 10.1038/nature05388 [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1948. The theory of a cline. J. Genet. 48: 277–284. 10.1007/BF02986626 [DOI] [PubMed] [Google Scholar]
- Hangartner S. B., Hoffmann A. A., Smith A., and Griffin P. C., 2015. A collection of Australian Drosophila datasets on climate adaptation and species distributions. Sci. Data 2: 150067 10.1038/sdata.2015.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Flatt T., and Aguilaniu H., 2013. Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 17: 10–19 [corrigenda: Cell Metab. 19: 1066 (2014)]. 10.1016/j.cmet.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T., Pélabon C., and Houle D., 2011. Heritability is not evolvability. Evol. Biol. 38: 258–277. 10.1007/s11692-011-9127-6 [DOI] [Google Scholar]
- Hardy C. M., Burke M. K., Everett L. J., Han M. V., Lantz K. M. et al. , 2017. Genome-wide analysis of starvation-selected Drosophila melanogaster - a genetic model of obesity. Mol. Biol. Evol. 35: 50–65. 10.1093/molbev/msx254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman L. G., 2003. Life span extension of Drosophila melanogaster: genetic and population studies. Popul. Dev. Rev. 29: 99–126. [Google Scholar]
- Harshman L. G., and Haberer B. A., 2000. Oxidative stress resistance: a robust correlated response to selection in extended longevity lines of Drosophila melanogaster? J. Gerontol. A. 55: B415–B417. 10.1093/gerona/55.9.B415 [DOI] [PubMed] [Google Scholar]
- Harshman L. G., and Hoffmann A. A., 2000. Laboratory selection experiments using Drosophila: what do they really tell us? Trends Ecol. Evol. 15: 32–36. 10.1016/S0169-5347(99)01756-5 [DOI] [PubMed] [Google Scholar]
- Harshman L. G., and Zera A. J., 2007. The cost of reproduction: the devil in the details. Trends Ecol. Evol. 22: 80–86. 10.1016/j.tree.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Harshman L. G., Hoffmann A. A., and Clark A. G., 1999a Selection for starvation resistance in Drosophila melanogaster : physiological correlates, enzyme activities and multiple stress responses. J. Evol. Biol. 12: 370–379. 10.1046/j.1420-9101.1999.00024.x [DOI] [Google Scholar]
- Harshman L. G., Moore K. M., Sty M. A., and Magwire M. M., 1999b Stress resistance and longevity in selected lines of Drosophila melanogaster. Neurobiol. Aging 20: 521–529. 10.1016/S0197-4580(99)00091-3 [DOI] [PubMed] [Google Scholar]
- Haymer D. S., and Hartl D. L., 1982. The experimental assessment of fitness in Drosophila. I. comparative measures of competitive reproductive success. Genetics 102: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haymer D. S., and Hartl D. L., 1983. The experimental assessment of fitness in Drosophila. II. a comparison of competitive and noncompetitive measures. Genetics 104: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel L. N., 1943. The genetic basis for constructing selection indices. Genetics 28: 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 1986. Genetic polymorphism in heterogeneous environments: a decade later. Annu. Rev. Ecol. Syst. 17: 535–566. 10.1146/annurev.es.17.110186.002535 [DOI] [Google Scholar]
- Hedrick P. W., and Murray E., 1983. Selection and measures of fitness, pp. 61–104 in The Genetics and Biology of Drosophila, Vol. 3d, edited by Ashburner M., Carson H. L., and Thompson J. N. Jr. Academic Press, New York. [Google Scholar]
- Hemphill W., Rivera O., and Talbert M., 2018. RNA-sequencing of Drosophila melanogaster head tissue on high-sugar and high-fat diets. G3 (Bethesda) 8: 279–290. 10.1534/g3.117.300397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill C. A., Reeves G. A., and Macdonald S. J., 2016. Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population. BMC Genet. 17: 113 10.1186/s12863-016-0419-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., 2017. “Conversion” of epistatic into additive genetic variance in finite populations and possible impact on long-term selection response. J. Anim. Breed. Genet. 134: 196–201. 10.1111/jbg.12270 [DOI] [PubMed] [Google Scholar]
- Hill W. G., and Caballero A., 1992. Artificial selection experiments. Annu. Rev. Ecol. Syst. 23: 287–310. 10.1146/annurev.es.23.110192.001443 [DOI] [Google Scholar]
- Hill W. G., Goddard M. E., and Visscher P. M., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4: e1000008 10.1371/journal.pgen.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillesheim E., and Stearns S. C., 1991. The responses of Drosophila melanogaster to artificial selection on body weight and its phenotypic plasticity in two larval food environments. Evolution 45: 1909–1923. 10.1111/j.1558-5646.1991.tb02696.x [DOI] [PubMed] [Google Scholar]
- Hillesheim E., and Stearns S. C., 1992. Correlated responses in life-history traits to artificial selection for body weight in Drosophila melanogaster. Evolution 46: 745–752. 10.1111/j.1558-5646.1992.tb02080.x [DOI] [PubMed] [Google Scholar]
- Hoedjes K. M., Rodrigues M. A., and Flatt T., 2017. Amino acid modulation of lifespan and reproduction in Drosophila. Curr. Opin. Insect Sci. 23: 118–122. 10.1016/j.cois.2017.07.005 [DOI] [PubMed] [Google Scholar]
- Hoedjes K. M., van den Heuvel J., Kapun M., Keller L., Flatt T. et al. , 2019. Distinct genomic signals of lifespan and life history evolution in response to postponed reproduction and larval diet in Drosophila. Evol. Lett. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., and Harshman L. G., 1999. Desiccation and starvation resistance in Drosophila: patterns of variation at the species, population and intrapopulation levels. Heredity 83: 637–643. 10.1046/j.1365-2540.1999.00649.x [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., and Parsons P. A., 1989. An integrated approach to environmental stress tolerance and life-history variation: desiccation tolerance in Drosophila. Biol. J. Linn. Soc. Lond. 37: 117–136. 10.1111/j.1095-8312.1989.tb02098.x [DOI] [Google Scholar]
- Hoffmann A. A., and Parsons P. A., 1993. Selection for adult desiccation resistance in Drosophila melanogaster: fitness components, larval resistance and stress correlations. Biol. J. Linn. Soc. Lond. 48: 43–54. 10.1111/j.1095-8312.1993.tb00875.x [DOI] [Google Scholar]
- Hoffmann A. A., and Weeks A. R., 2007. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129: 133–147. 10.1007/s10709-006-9010-z [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Hallas R., Sinclair C., and Mitrovski P., 2001. Levels of variation in stress resistance in Drosophila among strains, local populations, and geographic regions: patterns for desiccation, starvation, cold resistance, and associated traits. Evolution 55: 1621–1630. 10.1111/j.0014-3820.2001.tb00681.x [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Anderson A., and Hallas R., 2002. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol. Lett. 5: 614–618. 10.1046/j.1461-0248.2002.00367.x [DOI] [Google Scholar]
- Hoffmann A. A., Scott M., Partridge L., and Hallas R., 2003. Overwintering in Drosophila melanogaster: outdoor field cage experiments on clinal and laboratory selected populations help to elucidate traits under selection. J. Evol. Biol. 16: 614–623. 10.1046/j.1420-9101.2003.00561.x [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Shirriffs J., and Scott M., 2005. Relative importance of plastic vs. genetic factors in adaptive differentiation: geographical variation for stress resistance in Drosophila melanogaster from eastern Australia. Funct. Ecol. 19: 222–227. 10.1111/j.1365-2435.2005.00959.x [DOI] [Google Scholar]
- Holliday R., 1989. Food, reproduction and longevity: is the extended lifespan of calorie-restricted animals an evolutionary adaptation? Bioessays 10: 125–127. 10.1002/bies.950100408 [DOI] [PubMed] [Google Scholar]
- Höllinger I., Pennings P. S., and Hermisson J., 2019. Polygenic adaptation: from sweeps to subtle frequency shifts. PLoS Genet. 15: e1008035 10.1371/journal.pgen.1008035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth M. J., and Burcombe J. V., 1970. The nutritional requirements for longevity in Drosophila. J. Insect Physiol. 16: 1017–1025. 10.1016/0022-1910(70)90195-2 [DOI] [PubMed] [Google Scholar]
- Houle D., 1991. Genetic covariance of fitness correlates: what genetic correlations are made of and why it matters. Evolution 45: 630–648. 10.1111/j.1558-5646.1991.tb04334.x [DOI] [PubMed] [Google Scholar]
- Houle D., 1992. Comparing evolvability and variability of quantitative traits. Genetics 130: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., 1998. How should we explain variation in the genetic variance of traits? Genetica 102/103: 241–253. 10.1023/A:1017034925212 [DOI] [PubMed] [Google Scholar]
- Houle D., 2001. Characters as the units of evolutionary change, pp. 109–140 in The Character Concept in Evolutionary Biology, edited by Wagner G. P. Academic Press, New York: 10.1016/B978-012730055-9/50015-X [DOI] [Google Scholar]
- Houle D., and Rowe L., 2003. Natural selection in a bottle. Am. Nat. 161: 50–67. 10.1086/345480 [DOI] [PubMed] [Google Scholar]
- Houle D., Hoffmaster D. K., Assimacopoulos S., and Charlesworth B., 1992. The genomic mutation rate for fitness in Drosophila. Nature 359: 58–60. 10.1038/359058a0 [DOI] [PubMed] [Google Scholar]
- Houle D., Hoffmaster D. K., Assimacopoulos S. and Charlesworth B., 1994a The genomic mutation rate for fitness in Drosophila. Nature 371: 358. [DOI] [PubMed] [Google Scholar]
- Houle D., Hughes K. A., Hoffmaster D. K., Ihara J., Assimacopoulos S. et al. , 1994b The effects of spontaneous mutation on quantitative traits. I. Variances and covariances of life history traits. Genetics 138: 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., Hughes K. A., Assimacopolous S., and Charlesworth B., 1997. The effects of spontaneous mutation on quantitative traits. II. Dominance of mutations with effects on life-history traits. Genet. Res. 70: 27–34. 10.1017/S001667239700284X [DOI] [PubMed] [Google Scholar]
- Huang W., Richards S., Carbone M. A., Zhu D., Anholt R. R. H. et al. , 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. USA 109: 15553–15559. 10.1073/pnas.1213423109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak M. J., and Gromko M. H., 1989. Response to selection for early and late development of sexual maturity in Drosophila melanogaster. Anim. Behav. 38: 344–351. 10.1016/S0003-3472(89)80095-8 [DOI] [Google Scholar]
- Huey R. B., and Rosenzweig F., 2009. Laboratory evolution meets catch-22: balancing simplicity and realism, pp. 670–701 in Experimental Evolution, edited by Garland T. and Rose M. R.. University of California Press, Berkeley, CA. [Google Scholar]
- Huey R. B., Partridge L., and Fowler K., 1991. Thermal sensitivity of Drosophila melanogaster responds rapidly to laboratory natural selection. Evolution 45: 751–756. 10.1111/j.1558-5646.1991.tb04343.x [DOI] [PubMed] [Google Scholar]
- Huey R. B., Wakefield T., Crill W. D., and Gilchrist G. W., 1995. Within- and between-generation effects of temperature on early fecundity of Drosophila melanogaster. Heredity 74: 216–223. 10.1038/hdy.1995.30 [DOI] [PubMed] [Google Scholar]
- Hughes K. A., 1995. The evolutionary genetics of male life-history characters in Drosophila melanogaster. Evolution 49: 521–537. 10.1111/j.1558-5646.1995.tb02284.x [DOI] [PubMed] [Google Scholar]
- Hughes K. A., 1997. Quantitative genetics of sperm precedence in Drosophila melanogaster. Genetics 145: 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. A., 2010. Mutation and the evolution of ageing: from biometrics to system genetics. Philos. Trans. R. Soc. Lond., B 365: 1273–1279. 10.1098/rstb.2009.0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. A., and Charlesworth B., 1994. A genetic analysis of senescence in Drosophila. Nature 367: 64–66. 10.1038/367064a0 [DOI] [PubMed] [Google Scholar]
- Hughes K. A., and Leips J., 2017. Pleiotropy, constraint, and modularity in the evolution of life histories: insights from genomic analyses. Ann. N. Y. Acad. Sci. 1389: 76–91. 10.1111/nyas.13256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. A., and Reynolds R. M., 2005. Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 50: 421–445. 10.1146/annurev.ento.50.071803.130409 [DOI] [PubMed] [Google Scholar]
- Hughes K. A., Alipaz J. A., Drnevich J. M., and Reynolds R. M., 2002. A test of evolutionary theories of aging. Proc. Natl. Acad. Sci. USA 99: 14286–14291. 10.1073/pnas.222326199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley J., 1938. Clines: an auxiliary taxonomic principle. Nature 142: 219–220. 10.1038/142219a0 [DOI] [Google Scholar]
- Hwangbo D. S., Gersham B., Tu M. P., Palmer M., and Tatar M., 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566. 10.1038/nature02549 [DOI] [PubMed] [Google Scholar]
- Imai T., 1933. The influence of temperature on variation and inheritance of bodily dimensions in Drosophila melanogaster. Wilhelm Roux Arch. Entwickl. Mech. Org. 128: 634–660. 10.1007/BF00649866 [DOI] [PubMed] [Google Scholar]
- Imasheva A. G., Bubli O. A., and Lazebny O. E., 1994. Variation in wing length in Eurasian natural populations of Drosophila melanogaster. Heredity 72: 508–514. 10.1038/hdy.1994.68 [DOI] [PubMed] [Google Scholar]
- Ivanov D. K., Escott-Price V., Ziehm M., Magwire M. M., Mackay T. F. C. et al. , 2015. Longevity GWAS using the Drosophila genetic reference panel. J. Gerontol. A 70: 1470–1478. 10.1093/gerona/glv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives P. T., 1945. The genetic structure of American populations of Drosophila melanogaster. Genetics 30: 167–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives P. T., 1970. Further genetic studies of the South Amherst population of Drosophila melanogaster. Evolution 24: 507–518. 10.1111/j.1558-5646.1970.tb01785.x [DOI] [PubMed] [Google Scholar]
- Izquierdo J. I., 1991. How does Drosophila melanogaster overwinter? Entomol. Exp. Appl. 59: 51–58. 10.1111/j.1570-7458.1991.tb01485.x [DOI] [Google Scholar]
- Ja W. W., Carvalho G. B., Zid B. M., Mak E. M., Brummel T. et al. , 2009. Water- and nutrient-dependent effects of dietary restriction on Drosophila lifespan. Proc. Natl. Acad. Sci. USA 106: 18633–18637. 10.1073/pnas.0908016106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. C., and Partridge L., 1995. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. J. Evol. Biol. 8: 315–330. 10.1046/j.1420-9101.1995.8030315.x [DOI] [Google Scholar]
- James A. C., Azevedo R. B. R., and Partridge L., 1995. Cellular basis and developmental timing in a size cline of Drosophila melanogaster. Genetics 140: 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. C., Azevedo R. B. R., and Partridge L., 1997. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics 146: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A. R., Miles C. M., Lippert N. R., Brown C. D., White K. P. et al. , 2015. Whole genome resequencing of experimental populations reveals polygenic basis of egg size variation in Drosophila melanogaster. Mol. Biol. Evol. 32: 2616–2632. 10.1093/molbev/msv136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., and Barton N., 2005. Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1411–1425. 10.1098/lrstb.2005.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G., and Van der Have T. M., 2009. Temperature dependence of development rate, growth rate and size: from biophysics to adaptation, pp. 523–588 in Phenotypic Plasticity of Insects: Mechanisms and Consequences, edited by Whitman D. W. and Ananthakrishnan T. N.. Science Publishers, Enfield: 10.1201/b10201-13 [DOI] [Google Scholar]
- Jordan K. W., Craver K. L., Magwire M. M., Cubilla C. E., Mackay T. F. C. et al. , 2012. Genome-wide association for sensitivity to chronic oxidative stress in Drosophila melanogaster. PLoS One 7: e38722 10.1371/journal.pone.0038722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., 1997. Laboratory studies of density-dependent selection: adaptations to crowding in Drosophila melanogaster. Curr. Sci. 72: 555–562. [Google Scholar]
- Jouandet G. C., and Gallio M., 2015. Catching more flies with vinegar. Elife 4: e10535 10.7554/eLife.10535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungen H., and Hartl D. L., 1979. Average fitness of populations of Drosophila melanogaster as estimated using compound-autosome strains. Evolution 33: 359–370. 10.1111/j.1558-5646.1979.tb04689.x [DOI] [PubMed] [Google Scholar]
- Kaiser M., Gasser M., Ackermann R., and Stearns S. C., 1997. P-element inserts in transgenic flies: a cautionary tale. Heredity 78: 1–11. 10.1038/hdy.1997.1 [DOI] [PubMed] [Google Scholar]
- Kalra B., and Parkash R., 2014. Trade-off of ovarian lipids and total body lipids for fecundity and starvation resistance in tropical populations of Drosophila melanogaster. J. Evol. Biol. 27: 2371–2385. 10.1111/jeb.12480 [DOI] [PubMed] [Google Scholar]
- Kang L., Aggarwal D. D., Rashkovetsky E., Korol A. B., and Michalak P., 2016. Rapid genomic changes in Drosophila melanogaster adapting to desiccation stress in an experimental evolution system. BMC Genomics 17: 233 10.1186/s12864-016-2556-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P., Kaeberlein M., and Hansen M., 2017. Dietary restriction and lifespan: lessons from invertebrate models. Ageing Res. Rev. 39: 3–14. 10.1016/j.arr.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., and Flatt T., 2019. The adaptive significance of chromosomal inversion polymorphisms in Drosophila melanogaster. Mol. Ecol. 28: 1263–1282. 10.1111/mec.14871 [DOI] [PubMed] [Google Scholar]
- Kapun M., van Schalkwyk H., McAllister B., Flatt T., and Schlötterer C., 2014. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol. Ecol. 23: 1813–1827. 10.1111/mec.12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Fabian D. K., Goudet J., and Flatt T., 2016a Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol. Biol. Evol. 33: 1317–1336. 10.1093/molbev/msw016 [DOI] [PubMed] [Google Scholar]
- Kapun M., Schmidt C., Durmaz E., Schmidt P. S., and Flatt T., 2016b Parallel effects of the inversion In(3R)Payne on body size across the North American and Australian clines in Drosophila melanogaster. J. Evol. Biol. 29: 1059–1072. 10.1111/jeb.12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan D., Dahiya N., Munjal A. K., Gibert P., Moreteau B. et al. , 1998. Desiccation and starvation tolerance of adult Drosophila: opposite latitudinal clines in natural populations of three different species. Evolution 52: 825–831. [DOI] [PubMed] [Google Scholar]
- Kawecki T. J., Lenski R. E., Ebert D., Hollis B., Olivieri I. et al. , 2012. Experimental evolution. Trends Ecol. Evol. 27: 547–560. 10.1016/j.tree.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Keightley P. D., and Ohnishi O., 1998. EMS-induced polygenic mutation rates for nine quantitative characters in Drosophila melanogaster. Genetics 148: 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S. et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. 10.1101/gr.091231.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., 2007. Drosophila melanogaster’s history as a human commensal. Curr. Biol. 17: R77–R81. 10.1016/j.cub.2006.12.031 [DOI] [PubMed] [Google Scholar]
- Kellermann V., Loeschcke V., Hoffmann A. A., Kristensen T. N., Flojgaard C. et al. , 2012a Phylogenetic constraints in key functional traits behind species’ climate niches: patterns of desiccation and cold resistance across 95 Drosophila species. Evolution 66: 3377–3389. 10.1111/j.1558-5646.2012.01685.x [DOI] [PubMed] [Google Scholar]
- Kellermann V., Overgaard J., Hoffmann A. A., Flojgaard C., Svenning J.-C. et al. , 2012b Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc. Natl. Acad. Sci. USA 109: 16228–16233. 10.1073/pnas.1207553109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V., Hoffmann A. A., Kristensen T. N., Moghadam N. N., and Loeschcke V., 2015. Experimental evolution under fluctuating thermal conditions does not reproduce patterns of adaptive clinal differentiation in Drosophila melanogaster. Am. Nat. 186: 582–593. 10.1086/683252 [DOI] [PubMed] [Google Scholar]
- Kelly J. K., and Hughes K. A., 2019. Pervasive linked selection and intermediate-frequency alleles are implicated in an evolve-and-resequencing experiment of Drosophila simulans. Genetics 211: 943–961. 10.1534/genetics.118.301824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennington W. J., Killeen J. R., Goldstein D. B., and Partridge L., 2003. Rapid laboratory evolution of adult wing area in Drosophila melanogaster in response to humidity. Evolution 57: 932–936. 10.1111/j.0014-3820.2003.tb00304.x [DOI] [PubMed] [Google Scholar]
- Kenyon A., 1967. Comparison of frequency distributions of viabilities of second with fourth chromosomes from caged Drosophila melanogaster. Genetics 55: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., 2005. The plasticity of aging: insights from long-lived mutants. Cell 120: 449–460. 10.1016/j.cell.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Khazaeli A. A., and Curtsinger J. W., 2010. Post-reproductive survival and the Faulkner Effect in Drosophila melanogaster. Drosoph. Inf. Serv. 93: 154–155. [Google Scholar]
- King R. C., 1970. Ovarian Development in Drosophila melanogaster. Academic Press, New York. [Google Scholar]
- Kirkwood T. B. L., 1977. Evolution of aging. Nature 270: 301–304. 10.1038/270301a0 [DOI] [PubMed] [Google Scholar]
- Kirkwood T. B., and Shanley D. P., 2005. Food restriction, evolution and ageing. Mech. Ageing Dev. 126: 1011–1016. 10.1016/j.mad.2005.03.021 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Galikova M., De Maio N., Ricci S., Schlötterer C. et al. , 2013a Reproductive and post-reproductive life history of wild-caught Drosophila melanogaster under laboratory conditions. J. Evol. Biol. 26: 1508–1520. 10.1111/jeb.12155 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Gáliková M., De Maio N., Huber C. D., Schlötterer C. et al. , 2013b Variation in thermal performance and reaction norms among populations of Drosophila melanogaster. Evolution 67: 3573–3587. 10.1111/evo.12221 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Gáliková M., Huber C. D., and Flatt T., 2014. Similarities and differences in altitudinal vs. latitudinal variation for morphological traits in Drosophila melanogaster. Evolution 68: 1385–1398. 10.1111/evo.12351 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Procházka E., and Gáliková M., 2018. Crowding of Drosophila larvae affects lifespan and other life-history traits via reduced availability of dietary yeast. Exp. Gerontol. 110: 298–308. 10.1016/j.exger.2018.06.016 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Girish T. N., Dircksen H., and Gáliková M., 2019. Reproductive fitness of Drosophila is maximised by optimal developmental temperature. J. Exp. Biol. 222: jeb202184 10.1242/jeb.202184 [DOI] [PubMed] [Google Scholar]
- Knight G. R., and Robertson A., 1957. Fitness as a measurable character in Drosophila. Genetics 42: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R. E., 1994. Lords of the Fly: Drosophila Genetics and the Experimental Life. University of Chicago Press, Chicago. [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., and Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolss M., Vijendravarma R. K., Schwaller G., and Kawecki T. J., 2009. Life-history consequences of adaptation to larval nutritional stress in Drosophila. Evolution 63: 2389–2401. 10.1111/j.1558-5646.2009.00718.x [DOI] [PubMed] [Google Scholar]
- Kristensen T. N., Overgaard J., Loeschcke V., and Mayntz D., 2011. Dietary protein content affects evolution for body size, body fat and viability in Drosophila melanogaster. Biol. Lett. 7: 269–272. 10.1098/rsbl.2010.0872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubrak O. I., Kučerová L., Theopold U., and Nässel D. R., 2014. The sleeping beauty: how reproductive diapause affects hormone signaling, metabolism, immune response and somatic maintenance in Drosophila melanogaster. PLoS One 9: e113051 10.1371/journal.pone.0113051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubrak O. I., Kučerová L., Theopold U., Nylin S., and Nässel D. R., 2016. Characterization of reproductive dormancy in male Drosophila melanogaster. Front. Physiol. 7: 572 [corrigenda: Front. Physiol. 8: 314 (2017)]. 10.3389/fphys.2016.00572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe S., and Mukai T., 1984. The genetic structure of natural populations of Drosophila melanogaster. XVIII. Clinal and uniform genetic variation over populations. Genetics 108: 617–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D., Cariou M. L., David J. R., Lemeunier F., Tsacas L. et al. , 1988. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 22: 159–225. 10.1007/978-1-4613-0931-4_4 [DOI] [Google Scholar]
- Lack J. B., Yassin A., Sprengelmeyer Q. D., Johanning E. J., David J. R. et al. , 2016. Life history evolution and cellular mechanisms associated with increased size in high-altitude Drosophila. Ecol. Evol. 6: 5893–5906. 10.1002/ece3.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E., Duneau D., and Beldade P., 2018. Genetic basis of thermal plasticity variation in Drosophila melanogaster body size. PLoS Genet. 14: e1007686 10.1371/journal.pgen.1007686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1982. A quantitative genetic theory of life history evolution. Ecology 63: 607–615. 10.2307/1936778 [DOI] [Google Scholar]
- Lande R., and Arnold S. J., 1983. The measurement of selection on correlated characters. Evolution 37: 1210–1226. 10.1111/j.1558-5646.1983.tb00236.x [DOI] [PubMed] [Google Scholar]
- Latter B. D., and Sved J. A., 1994. A reevaluation of data from competitive tests show high levels of heterosis in Drosophila melanogaster. Genetics 137: 509–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laturney M., van Eijk R., and Billeter J.-C., 2018. Last male sperm precedence is modulated by female remating rate in Drosophila melanogaster. Evol. Lett. 2: 180–189. 10.1002/evl3.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent S. J. Y., Werzner A., Excoffier L., and Stephan W., 2011. Approximate Bayesian analysis of Drosophila melanogaster polymorphism data reveals a recent colonization of Southeast Asia. Mol. Biol. Evol. 28: 2041–2051. 10.1093/molbev/msr031 [DOI] [PubMed] [Google Scholar]
- Layalle S., Arquier N., and Leopold P., 2008. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell 15: 568–577. 10.1016/j.devcel.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Lazzaro B. P., Flores H. A., Lorigan J. G., and Yourth C. P., 2008. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 4: e1000025 10.1371/journal.ppat.1000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. C., Kaya A., Ma S., Kim G., Gerashchenko M. V. et al. , 2014. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat. Commun. 5: 3592 10.1038/ncomms4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., 2015. Dietary protein:carbohydrate balance is a critical modulator of lifespan and reproduction in Drosophila melanogaster: a test using a chemically defined diet. J. Insect Physiol. 75: 12–19. 10.1016/j.jinsphys.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Lee K. P., Simpson S., Clissold F., Brooks R., Ballard J. et al. , 2008. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc. Natl. Acad. Sci. USA 105: 2498–2503. 10.1073/pnas.0710787105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Sgrò C. M., Shirriffs J., Wee C. W., Rako L. E. A. et al. , 2011. Polymorphism in the couch potato gene clines in eastern Australia but is not associated with ovarian dormancy in Drosophila melanogaster. Mol. Ecol. 20: 2973–2984. 10.1111/j.1365-294X.2011.05155.x [DOI] [PubMed] [Google Scholar]
- Lee S. F., Eyre-Walker Y. C., Rane R. V., Reuter C., Vinti G. et al. , 2013. Polymorphism in the neurofibromin gene, Nf1, is associated with antagonistic selection on wing size and development time in Drosophila melanogaster. Mol. Ecol. 22: 2716–2725. 10.1111/mec.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., and Micchelli C. A., 2013. Development and characterization of a chemically defined food for Drosophila. PLoS One 8: e67308 10.1371/journal.pone.0067308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc A., and Bundgaard J., 2000. The influence of male and female body size on copulation duration and fecundity in Drosophila melanogaster. Hereditas 132: 243–247. 10.1111/j.1601-5223.2000.00243.x [DOI] [PubMed] [Google Scholar]
- Leips J., and Mackay T. F. C., 2000. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155: 1773–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips J., and Mackay T. F. C., 2002. The complex genetic architecture of Drosophila life span. Exp. Aging Res. 28: 361–390. 10.1080/03610730290080399 [DOI] [PubMed] [Google Scholar]
- Leips J., Gilligan P., and Mackay T. R. C., 2006. Quantitative trait loci with age-specific effects on fecundity in Drosophila melanogaster. Genetics 172: 1595–1605. 10.1534/genetics.105.048520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier F., David J. R., and Tsacas L., 1986. The melanogaster species group, pp. 148–256 in Genetics and Biology of Drosophila, Vol. 3e, edited by Ashburner M., Carson H. L., and Thompson J. N.. Academic Press, New York. [Google Scholar]
- Leopold P., and Perrimon N., 2007. Drosophila and the genetics of the internal milieu. Nature 450: 186–188. 10.1038/nature06286 [DOI] [PubMed] [Google Scholar]
- Leroi A., 2001. Molecular signals vs. the Loi de Balancement. Trends Ecol. Evol. 16: 24–29. 10.1016/S0169-5347(00)02032-2 [DOI] [PubMed] [Google Scholar]
- Leroi A. M., Chen W., and Rose M. R., 1994a Long-term laboratory evolution of a genetic life-history trade-off in Drosophila melanogaster. 2. Stability of genetic correlations. Evolution 48: 1258–1268. [DOI] [PubMed] [Google Scholar]
- Leroi A. M., Chippindale A. K., and Rose M. R., 1994b Long-term laboratory evolution of a genetic life-history trade-off in Drosophila melanogaster. 2. The role of genotype-by-environment interaction. Evolution 48: 1244–1257. [DOI] [PubMed] [Google Scholar]
- Levene H., 1953. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87: 331–333. 10.1086/281792 [DOI] [Google Scholar]
- Levine M. T., Eckert M. L., and Begun D. J., 2011. Whole-genome expression plasticity across tropical and temperate Drosophila melanogaster populations from Eastern Australia. Mol. Biol. Evol. 28: 249–256. 10.1093/molbev/msq197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., 1955. The effects of population density and composition on viability in Drosophila melanogaster. Evolution 9: 27–41. 10.1111/j.1558-5646.1955.tb01511.x [DOI] [Google Scholar]
- Lewontin R. C., 1965. Selection for colonizing ability, pp. 77–94 in The Genetics of Colonizing Species, edited by Baker H. G. and Ledyard Stebbins G.. Academic Press, New York. [Google Scholar]
- Lewontin R. C., 1974. The Genetic Basis of Evolutionary Change. Columbia University Press, New York. [Google Scholar]
- R. C. Lewontin, J. A. Moore, W. B. Provine, and B. Wallace (Editors), 2003. Dobzhansky’s Genetics of Natural Populations I–XLIII. Columbia University Press, New York. [Google Scholar]
- L’Héritier P., and Teissier G., 1933. Etude d’une population de Drosophiles en équilibre. C.R. Acad. Sc. 197: 1765–1767. [Google Scholar]
- Li H., and Stephan W., 2006. Inferring the Demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2: e166 10.1371/journal.pgen.0020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-J., Seroude L., and Benzer S., 1998. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282: 943–946. 10.1126/science.282.5390.943 [DOI] [PubMed] [Google Scholar]
- Linnen C., Tatar M., and Promislow D., 2001. Cultural artifacts: a comparison of senescence in natural, laboratory-adapted and artificially selected lines of Drosophila melanogaster. Evol. Ecol. Res. 3: 877–888. [Google Scholar]
- Lints F. A., and Lints C. V., 1969. Influence of preimaginal environment on fecundity and ageing in Drosophila melanogaster hybrids. I. Preimaginal population density. Exp. Gerontol. 4: 231–244. 10.1016/0531-5565(69)90011-4 [DOI] [PubMed] [Google Scholar]
- Lirakis M., Dolezal M., and Schlötterer C., 2018. Redefining reproductive dormancy in Drosophila as a general stress response to cold temperatures. J. Insect Physiol. 107: 175–185. 10.1016/j.jinsphys.2018.04.006 [DOI] [PubMed] [Google Scholar]
- Lobell A. S., Kaspari R. R., Serrano Negron Y. L., and Harbison S. T., 2017. The genetic architecture of ovariole number in Drosophila melanogaster: genes with major, quantitative, and pleiotropic effects. G3 (Bethesda) 7: 2391–2403. 10.1534/g3.117.042390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckinbill L. S., Arking R., Clare M. J., Cirocco W. C., and Buck S. A., 1984. Selection for delayed senescence in Drosophila melanogaster. Evolution 38: 996–1003. 10.1111/j.1558-5646.1984.tb00369.x [DOI] [PubMed] [Google Scholar]
- Luckinbill L. S., Graves J. S., Tomkiw A., and Sowirka O., 1988. A qualitative analysis of some life-history correlates of longevity in Drosophila melanogaster. Evol. Ecol. 2: 85–94. 10.1007/BF02071591 [DOI] [Google Scholar]
- Lumme J., 1978. Phenology and photoperiodic diapause in northern populations of Drosophila, pp. 145–170 in Evolution of Insect Migration and Diapause, edited by Dingle H. Springer-Verlag, New York: 10.1007/978-1-4615-6941-1_7 [DOI] [Google Scholar]
- Lyman R. F., and Mackay T. F. C., 1998. Candidate quantitative trait loci and naturally occurring phenotypic variation for bristle number in Drosophila melanogaster: the delta-hairless gene region. Genetics 149: 983–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., and Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Machado H. E., Bergland A. O., O’Brien K. R., Behrman E. L., Schmidt P. S. et al. , 2016. Comparative population genomics of latitudinal variation in Drosophila simulans and Drosophila melanogaster. Mol. Ecol. 25: 723–740. 10.1111/mec.13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 1985. A quantitative genetic analysis of fitness and its components in Drosophila melanogaster. Genet. Res. 47: 59–70. 10.1017/S0016672300024526 [DOI] [Google Scholar]
- Mackay T. F. C., 2001a The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. 10.1146/annurev.genet.35.102401.090633 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., 2001b Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2: 11–20. 10.1038/35047544 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., 2004. The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14: 253–257. 10.1016/j.gde.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., 2010. Mutations and quantitative genetic variation: lessons from Drosophila. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1229–1239. 10.1098/rstb.2009.0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 2014. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., and Huang W., 2018. Charting the genotype–phenotype map: lessons from the Drosophila melanogaster genetic reference panel. WIREs. Dev. Biol. 7: e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Roshina N. V., Leips J. W., and Pasyukova E. G., 2006. Complex genetic architecture of Drosophila longevity, pp. 181–216 in Handbook of the Biology of Aging, Ed. 6, edited by Masaro E. J. and Austad S. N.. Elsevier, New York. [Google Scholar]
- Mackay T. F. C., Stone E. A., and Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. 10.1038/nrg2612 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F. et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwere T., Chapman T., and Partridge L., 2004. Sex differences in the effect of dietary restriction on life span and mortality rates in female and male Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 59: 3–9. 10.1093/gerona/59.1.B3 [DOI] [PubMed] [Google Scholar]
- Magwire M. M., Yamamoto A., Carbone M. A., Roshina N. V., Symonenko A. V. et al. , 2010. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 6: e1001037 10.1371/journal.pgen.1001037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W., and Dillin A., 2008. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77: 727–754. 10.1146/annurev.biochem.77.061206.171059 [DOI] [PubMed] [Google Scholar]
- Mair W., Goymer P., Pletcher S. D., and Partridge L., 2003. Demography of dietary restriction and death in Drosophila. Science 301: 1731–1733. 10.1126/science.1086016 [DOI] [PubMed] [Google Scholar]
- Mair W., Sgrò C. M., Johnson A. P., Chapman T., and Partridge L., 2004. Lifespan extension by dietary restriction in female Drosophila melanogaster is not caused by a reduction in vitellogenesis or ovarian activity. Exp. Gerontol. 39: 1011–1019. 10.1016/j.exger.2004.03.018 [DOI] [PubMed] [Google Scholar]
- Mair W., Piper M., and Partridge L., 2005. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 3: e223 10.1371/journal.pbio.0030223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S., Enjin A., Jirle E. V., Ramesh V., Rehermann G. et al. , 2018. Wild African Drosophila melanogaster are seasonal specialists on marula fruit. Curr. Biol. 28: 3960–3968.e3. 10.1016/j.cub.2018.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mappes T., Koivula M., Koskela E., Oksanen T. A., Savolainen T. et al. , 2008. Frequency and density-dependent selection on life-history strategies – a field experiment. PLoS One 3: e1687 10.1371/journal.pone.0001687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A., 2015. The secret lives of Drosophila flies. Elife 4: e06793. 10.7554/eLife.06793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A., and O’Grady P. M., 2005. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu. Rev. Genet. 39: 263–291. 10.1146/annurev.genet.39.073003.112454 [DOI] [PubMed] [Google Scholar]
- Markow T. A., and O’Grady P. M., 2008. Reproductive ecology of Drosophila. Funct. Ecol. 22: 747–759. 10.1111/j.1365-2435.2008.01457.x [DOI] [Google Scholar]
- Markow T. A., Beall S., and Matzkin L. M., 2009. Egg size, embryonic development time and ovoviviparity in Drosophila species. J. Evol. Biol. 22: 430–434. 10.1111/j.1420-9101.2008.01649.x [DOI] [PubMed] [Google Scholar]
- Martin L. B., Ghalambor C. K., and Woods H. A. (Editors), 2015. Integrative Organismal Biology. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Martorell C., Toro M. A., and Gallego C., 1998. Spontaneous mutation for life-history traits in Drosophila melanogaster. Genetica 102/103: 315–324. 10.1023/A:1017051312917 [DOI] [PubMed] [Google Scholar]
- Masoro E. J., and Austad S. N., 1996. The evolution of the antiaging action of dietary restriction: a hypothesis. J. Gerontol. A Biol. Sci. Med. Sci. 51: B387–B391. 10.1093/gerona/51A.6.B387 [DOI] [PubMed] [Google Scholar]
- Mathur V., and Schmidt P. S., 2017. Adaptive patterns of phenotypic plasticity in laboratory and field environments in Drosophila melanogaster. Evolution 71: 465–474. 10.1111/evo.13144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos M., Rose M. R., Rocha Pité M. T., Rego C., and Avelar T., 2000. Adaptation to the laboratory environment in Drosophila subobscura. J. Evol. Biol. 13: 9–19. 10.1046/j.1420-9101.2000.00116.x [DOI] [Google Scholar]
- May C. M., Doroszuk A., and Zwaan B. J., 2015. The effect of developmental nutrition on life span and fecundity depends on the adult reproductive environment in Drosophila melanogaster. Ecol. Evol. 5: 1156–1168. 10.1002/ece3.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C. M., van den Heuvel J., Doroszuk A., Hoedjes K. M., Flatt T. et al. , 2019. Adaptation to developmental diet influences the response to selection on age at reproduction in the fruit fly. J. Evol. Biol. 32: 425–437. 10.1111/jeb.13425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., 1958. The effects of temperature and of egg-laying on the longevity of Drosophila subobscura. J. Exp. Biol. 35: 832–842. [Google Scholar]
- Mayr E., 1963. Animal Species and Evolution. Harvard University Press, Cambridge, MA: 10.4159/harvard.9780674865327 [DOI] [Google Scholar]
- McCracken A. W., Adams G., Hartshorne L., Tatar M., and Simons M. J. P., 2019. The hidden costs of dietary restriction: implications for its evolutionary and mechanistic origins. bioRxiv. Available at: 10.1101/533711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch D., and Gems D., 2003. Body size, insulin/IGF signaling and aging in the nematode Caenorhabditis elegans. Exp. Gerontol. 38: 129–136. 10.1016/S0531-5565(02)00147-X [DOI] [PubMed] [Google Scholar]
- McMillan I., Fitz-Earle M., and Robson D. S., 1970a Quantitative genetics of fertility. I. Lifetime egg production of Drosophila melanogaster – theoretical. Genetics 65: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan I., Fitz-Earle M., Butler L., and Robson D. S., 1970b Quantitative genetics of fertility. II. Lifetime egg production of Drosophila melanogaster – experimental. Genetics 65: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar P. B., 1946. Old age and natural death. Mod. Quart. 2: 30–49. [Google Scholar]
- Medawar P. B., 1952. An Unsolved Problem of Biology. H.K. Lewis and Company, London. [Google Scholar]
- Mertz D. B., 1975. Senescent decline in flour beetle strains selected for early adult fitness. Physiol. Zool. 48: 1–23. 10.1086/physzool.48.1.30155634 [DOI] [Google Scholar]
- Messer P. W., and Petrov D. A., 2013. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28: 659–669. 10.1016/j.tree.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A., and Partridge L., 2013. Dietary restriction extends lifespan in wild-derived populations of Drosophila melanogaster. PLoS One 8: e74681 10.1371/journal.pone.0074681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf C. J., 2016. Invisible trade-offs: Van Noordwijk and de Jong and life-history evolution. Am. Nat. 187: iii–v. 10.1086/685487 [DOI] [PubMed] [Google Scholar]
- Miller R. S., and Thomas J. L., 1958. The effects of larval crowding and body size on the longevity of adult Drosophila melanogaster. Ecology 39: 118–125. 10.2307/1929973 [DOI] [Google Scholar]
- Min K.-J., and Tatar M., 2006. Restriction of amino acids extends lifespan in Drosophila melanogaster. Mech. Ageing Dev. 127: 643–646. 10.1016/j.mad.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Min K.-J., Hogan M. F., Tatar M., and O’Brien D. M., 2006. Resource allocation to reproduction and soma in Drosophila: a stable isotope analysis of carbon from dietary sugar. J. Insect Physiol. 52: 763–770. 10.1016/j.jinsphys.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Min K.-J., Flatt T., Kulaots I., and Tatar M., 2007. Counting calories in Drosophila diet restriction. Exp. Gerontol. 42: 247–251. 10.1016/j.exger.2006.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K., and Riddiford L. M., 2007. Size assessment and growth control: how adult size is determined in insects. Bioessays 29: 344–355. 10.1002/bies.20552 [DOI] [PubMed] [Google Scholar]
- Mirth C. K., and Shingleton A. W., 2012. Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front. Endocrinol. 3: 49 10.3389/fendo.2012.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R. K., and Reeve E. C. R., 1964. Clines in body dimensions in populations of Drosophila subobscura. Genet. Res. 5: 240–256. 10.1017/S0016672300001208 [DOI] [Google Scholar]
- Mitrovski P., and Hoffmann A. A., 2001. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc. Biol. Sci. 268: 2163–2168. 10.1098/rspb.2001.1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moatt J. P., Nakagawa S., Lagisz M., and Walling C. A., 2016. The effect of dietary restriction on reproduction: a meta-analytic perspective. BMC Evol. Biol. 16: 199 10.1186/s12862-016-0768-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. E., 2018. First in Fly. Drosophila Research and Biological Discovery. Harvard University Press, Cambridge, MA: 10.4159/9780674984721 [DOI] [Google Scholar]
- Moorad J. A., and Promislow D. E., 2009. What can genetic variation tell us about the evolution of senescence? Proc. Biol. Sci. 276: 2271–2278. 10.1098/rspb.2009.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. N. S., Coogan C., Chamseddin K., Fernandez-Kim S. O., Kolli S. et al. , 2012. Development of diet-induced insulin resistance in adult Drosophila melanogaster. Biochim. Biophys. Acta 1822: 1230–1237. 10.1016/j.bbadis.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau T. A., and Roff D. A., 1987. Natural selection and the heritability of fitness components. Heredity 59: 181–197. 10.1038/hdy.1987.113 [DOI] [PubMed] [Google Scholar]
- Mueller L. D., 1987. Evolution of accelerated senescence in laboratory populations of Drosophila. Proc. Natl. Acad. Sci. USA 84: 1974–1977. 10.1073/pnas.84.7.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L. D., and Joshi A., 2000. Stability in Model Populations. Princeton University Press, Princeton, NJ. [Google Scholar]
- Mueller L. D., Shahrestani P., and Rauser C. L., 2009. Predicting death in female Drosophila. Exp. Gerontol. 44: 766–772. 10.1016/j.exger.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Mukai T., 1964. The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 50: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., 1969. The genetic structure of natural populations of Drosophila melanogaster. VII. Synergistic interactions of spontaneous mutant polygenes affecting viability. Genetics 50: 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., 1988. Genotype-environment interaction in relation to the maintenance of genetic variability in populations of Drosophila melanogaster, pp. 21–31 in Proceedings of the Second International Conference on Quantitative Genetics, edited by Weir B. S., Eisen E. J., Goodman M. J., and Namkoong G.. Sinauer, Sunderland, MA. [Google Scholar]
- Mukai T., and Nagano S., 1983. The genetic structure of natural populations of Drosophila melanogaster. XVI. Excess of additive genetic variance of viability. Genetics 105: 115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Chigusa S. I., Mettler L. E., and Crow J. F., 1972. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics 72: 335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Cardellino R. A., Watanabe T. K., and Crow J. F., 1974. The genetic variance for viability and its components in a local population of Drosophila melanogaster. Genetics 78: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1928. The measurement of mutation rate in Drosophila, its high variability and its dependence on temperature. Genetics 13: 279–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1950. Our load of mutations. Am. J. Hum. Genet. 2: 111–176. [PMC free article] [PubMed] [Google Scholar]
- Murphy P. A., Giesel J. T., and Manlove M. N., 1983. Temperature effects on life history variation in Drosophila simulans. Evolution 37: 1181–1192. 10.1111/j.1558-5646.1983.tb00233.x [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Lagisz M., Hector K. L., and Spencer H. G., 2012. Comparative and meta-analytic insights into life extension via dietary restriction. Aging Cell 11: 401–409. 10.1111/j.1474-9726.2012.00798.x [DOI] [PubMed] [Google Scholar]
- Nestel D., Papadopoulos N. T., Pascacio-Villafán C., Righini N., Altuzar-Molina A. R. et al. , 2016. Resource allocation and compensation during development in holometabolous insects. J. Insect Physiol. 95: 78–88. 10.1016/j.jinsphys.2016.09.010 [DOI] [PubMed] [Google Scholar]
- Ng’oma E., Perinchery A. M., and King E. G., 2017. How to get the most bang for your buck: the evolution and physiology of nutrition-dependent resource allocation strategies. Proc. Biol. Sci. 284: 20170445. 10.1098/rspb.2017.0445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng’oma E., Fidelis W., Middleton K. M., and King E. G., 2019. The evolutionary potential of diet-dependent effects on lifespan and fecundity in a multi-parental population of Drosophila melanogaster. Heredity 122: 582–594. 10.1038/s41437-018-0154-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T. X., and Moehring A. J., 2015. Accurate alternative measurements for female lifetime reproductive success in Drosophila melanogaster. PLoS One 10: e0116679 10.1371/journal.pone.0116679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant K. T., Singh N. D., and Alani E., 2009. Genomic mutation rates: what high-throughput methods can tell us. Bioessays 31: 912–920. 10.1002/bies.200900017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop J., 1917. The role of yeast in the nutrition of an insect (Drosophila). J. Biol. Chem. 30: 181–187. [Google Scholar]
- Nunney L., and Cheung W., 1997. The effect of temperature on body size and fecundity in female Drosophila melanogaster: evidence for adaptive plasticity. Evolution 51: 1529–1535. [DOI] [PubMed] [Google Scholar]
- Nusbaum T. J., and Rose M. R., 1999. The effects of nutritional manipulation and laboratory selection on lifespan in Drosophila melanogaster. J. Gerontol. A 54: B192–B198. 10.1093/gerona/54.5.B192 [DOI] [PubMed] [Google Scholar]
- Nylin S., and Gotthard K., 1998. Plasticity in life-history traits. Annu. Rev. Entomol. 43: 63–83. 10.1146/annurev.ento.43.1.63 [DOI] [PubMed] [Google Scholar]
- O’Brien D. M., Min K. J., Larsen T., and Tatar M., 2008. Use of stable isotopes to examine how dietary restriction extends Drosophila lifespan. Curr. Biol. 18: R155–R156. 10.1016/j.cub.2008.01.021 [DOI] [PubMed] [Google Scholar]
- Ohnishi O., 1977. Spontaneous and ethyl methylsulfonate-induced mutations controlling viability in Drosophila melanogaster. II. Homozygous effects of polygenic mutations. Genetics 87: 529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S., and Hafen E., 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13: 79–85. 10.1016/S0962-8924(02)00042-9 [DOI] [PubMed] [Google Scholar]
- Orozco-terWengel P., Kapun M., Nolte V., Kofler R., Flatt T. et al. , 2012. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Mol. Ecol. 21: 4931–4941. 10.1111/j.1365-294X.2012.05673.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr W. C., and Sohal R. S., 1994. Extension of life span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science 263: 1128–1130. 10.1126/science.8108730 [DOI] [PubMed] [Google Scholar]
- Ørsted M., Hoffmann A. A., Rohde P. D., Sørensen P., and Kristensen T. N., 2019. Strong impact of thermal environment on the quantitative genetic basis of a key stress tolerance trait. Heredity 122: 315–325. 10.1038/s41437-018-0117-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., and Schmidt P. S., 2008. Functional significance of allelic variation at methuselah, an aging gene in Drosophila. PLoS One 3: e1987 10.1371/journal.pone.0001987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., and Schmidt P. S., 2009. Dissecting the genetics of longevity in Drosophila melanogaster. Fly (Austin) 3: 29–38. 10.4161/fly.3.1.7771 [DOI] [PubMed] [Google Scholar]
- Paaby A. B., Blacket M. J., Hoffmann A. A., and Schmidt P. S., 2010. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol. Ecol. 19: 760–774. 10.1111/j.1365-294X.2009.04508.x [DOI] [PubMed] [Google Scholar]
- Paaby A. B., Bergland A. O., Behrman E. L., and Schmidt P. S., 2014. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution 68: 3395–3409. 10.1111/evo.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker Musselman L., Fink J. L., Narzinski K., Ramachandran P. V., Sukumar Hathiramani S. et al. , 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Model. Mech. 4: 842–849. 10.1242/dmm.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons P. A., 1961. Fly size, emergence time and sternopleural chaeta number in Drosophila. Heredity 16: 455–473. 10.1038/hdy.1961.52 [DOI] [Google Scholar]
- Parsons P. A., 1964. Egg lengths in Drosophila melanogaster and correlated responses to selection. Genetica 35: 175–181. 10.1007/BF01804886 [DOI] [PubMed] [Google Scholar]
- Parsons P. A., 1975. The comparative evolutionary biology of the sibling species, Drosophila melanogaster and D. simulans. Q. Rev. Biol. 50: 151–169. 10.1086/408437 [DOI] [PubMed] [Google Scholar]
- Parsons, P. A., 1977 Genotype-temperature interaction for longevity in natural populations of Drosophila simulans Exp. Gerontol. 12: 241–244. 10.1016/0531-5565(77)90012-2 [DOI] [PubMed] [Google Scholar]
- Parsons P. A., 1978. The effect of genotype and temperature on longevity in natural populations of Drosophila melanogaster. Exp. Gerontol. 13: 167–169. 10.1016/0531-5565(78)90009-8 [DOI] [PubMed] [Google Scholar]
- Parsons P. A., and Hosgood S. M., 1967. Genetic heterogeneity among the founders of laboratory populations of Drosophila. I. Scutellar chaetae. Genetica 38: 328–339. 10.1007/BF01507465 [DOI] [PubMed] [Google Scholar]
- Partridge L., 1988. Lifetime reproductive success in Drosophila, pp. 11–23 in Reproductive Success – Studies of Individual Variation in Contrasting Breeding Systems, edited by Clutton-Brock T. H. University of Chicago Press, Chicago. [Google Scholar]
- Partridge L., and Barton N. H., 1993. Evolution of aging - testing the theory using Drosophila. Genetica 91: 89–98. 10.1007/BF01435990 [DOI] [PubMed] [Google Scholar]
- Partridge L., and Fowler K., 1992. Direct and correlated responses to selection on age at reproduction in Drosophila melanogaster. Evolution 46: 76–91. 10.1111/j.1558-5646.1992.tb01986.x [DOI] [PubMed] [Google Scholar]
- Partridge L., and Fowler K., 1993. Responses and correlated responses to artificial selection on thorax length in Drosophila melanogaster. Evolution 47: 213–226. 10.1111/j.1558-5646.1993.tb01211.x [DOI] [PubMed] [Google Scholar]
- Partridge L., and Gems D., 2002. Mechanisms of ageing: public or private? Nat. Rev. Genet. 3: 165–175. 10.1038/nrg753 [DOI] [PubMed] [Google Scholar]
- Partridge L., and Harvey P. H., 1988. The ecological context of life history evolution. Science 241: 1449–1455. 10.1126/science.241.4872.1449 [DOI] [PubMed] [Google Scholar]
- Partridge L., Fowler K., Trevitt S., and Sharp W., 1986. An examination of the effects of males on the survival and egg-production rates of female Drosophila melanogaster. J. Insect Physiol. 32: 925–929. 10.1016/0022-1910(86)90140-X [DOI] [Google Scholar]
- Partridge L., Green A., and Fowler K., 1987. Effects of egg-production and of exposure to males on female survival in Drosophila melanogaster. J. Insect Physiol. 33: 745–749. 10.1016/0022-1910(87)90060-6 [DOI] [Google Scholar]
- Partridge L., Barrie B., Fowler K., and French V., 1994a Thermal evolution of pre-adult life-history traits in Drosophila melanogaster. J. Evol. Biol. 7: 645–663. 10.1046/j.1420-9101.1994.7060645.x [DOI] [PubMed] [Google Scholar]
- Partridge L., Barrie B., Fowler K., and Vernon F., 1994b Evolution and development of body size and cell size in Drosophila melanogaster in response to temperature. Evolution 48: 1269–1276. 10.1111/j.1558-5646.1994.tb05311.x [DOI] [PubMed] [Google Scholar]
- Partridge L., Prowse N., and Pignatelli P., 1999a Another set of responses and correlated responses to selection on age at reproduction in Drosophila melanogaster. Proc. Biol. Sci. 266: 255–261. 10.1098/rspb.1999.0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Langelan R., Fowler K., Zwaan B., and French V., 1999b Correlated responses to selection on body size in Drosophila melanogaster. Genet. Res. 74: 43–54. 10.1017/S0016672399003778 [DOI] [PubMed] [Google Scholar]
- Partridge L., Gems D., and Withers D. J., 2005a Sex and death: what is the connection? Cell 120: 461–472. 10.1016/j.cell.2005.01.026 [DOI] [PubMed] [Google Scholar]
- Partridge L., Piper M. D., and Mair W., 2005b Dietary restriction in Drosophila. Mech. Ageing Dev. 126: 938–950. 10.1016/j.mad.2005.03.023 [DOI] [PubMed] [Google Scholar]
- Partridge L., Pletcher S. D., and Mair W., 2005c Dietary restriction, mortality trajectories, risk and damage. Mech. Ageing Dev. 126: 35–41. 10.1016/j.mad.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Pasyukova E. G., Vieira C., and Mackay T. F., 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156: 1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova E. G., Roshina N. V., and Mackay T. F., 2004. Shuttle craft: a candidate quantitative trait gene for Drosophila lifespan. Aging Cell 3: 297–307. 10.1111/j.1474-9728.2004.00114.x [DOI] [PubMed] [Google Scholar]
- Pearl R., 1932. The influence of density of population upon egg production in Drosophila melanogaster. J. Exp. Zool. 63: 57–84. 10.1002/jez.1400630103 [DOI] [Google Scholar]
- Pearl R., and Parker S. L., 1921. Experimental studies on the duration of life. I. Introductory discussion of the duration of life in Drosophila. Am. Nat. 55: 481–509. 10.1086/279836 [DOI] [Google Scholar]
- Pearl R., and Parker S. L., 1922. Experimental studies on the duration of life. IV. Data on the influence of density of population on duration of life in Drosophila. Am. Nat. 56: 312–321. 10.1086/279870 [DOI] [Google Scholar]
- Pearl R., and Parker S. L., 1924. Experimental studies on the duration of life. LX. New life tables for Drosophila. Am. Nat. 58: 71–82. 10.1086/279956 [DOI] [Google Scholar]
- Pease C. M., and Bull J. J., 1988. A critique of methods for measuring life-history trade-offs. J. Evol. Biol. 1: 293–303. 10.1046/j.1420-9101.1988.1040293.x [DOI] [Google Scholar]
- Pegoraro M., Zonato V., Tyler E. R., Fedele G., Kyriacou C. P. et al. , 2017. Geographical analysis of diapause inducibility in European Drosophila melanogaster populations. J. Insect Physiol. 98: 238–244. 10.1016/j.jinsphys.2017.01.015 [DOI] [PubMed] [Google Scholar]
- Perrin N., and Sibly R. M., 1993. Dynamic models of energy allocation and investment. Annu. Rev. Ecol. Syst. 24: 379–410. 10.1146/annurev.es.24.110193.002115 [DOI] [Google Scholar]
- Pigliucci M., 2001. Phenotypic Plasticity: Beyond Nature and Nurture. The Johns Hopkins University Press, Baltimore. [Google Scholar]
- Pigliucci M., and Preston K. (Editors), 2004. Phenotypic Integration–Studying the Ecology and Evolution of Complex Phenotypes. Oxford University Press, New York. [Google Scholar]
- Piper M. D., Skorupa D., and Partridge L., 2005. Diet, metabolism and lifespan in Drosophila. Exp. Gerontol. 40: 857–862. 10.1016/j.exger.2005.06.013 [DOI] [PubMed] [Google Scholar]
- Piper M. D. W., Partridge L., Raubenheimer D., and Simpson S. J., 2011. Dietary restriction and aging: a unifying perspective. Cell Metab. 14: 154–160. 10.1016/j.cmet.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. D. W., Blanc E., Leitao-Goncalves R., Yang M., He X. et al. , 2014. A holidic medium for Drosophila melanogaster. Nat. Methods 11: 100–105 (erratum: Nat. Methods 11: 971); [corrigenda: Nat. Methods 12: 1098 (2015)]. 10.1038/nmeth.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. D. W., Soultoukis G. A., Blanc E., Mesaros A., Herbert S. L. et al. , 2017. Matching dietary amino acid balance to the in silico-translated exome optimizes growth and reproduction without cost to lifespan. Cell Metab. 25: 610–621 (erratum: Cell Metab. 25: 1206). 10.1016/j.cmet.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchers W., Pool J. E., and Dworkin I., 2013. Altitudinal clinal variation in wing size and shape in African Drosophila melanogaster: one cline or many? Evolution 67: 438–452. 10.1111/j.1558-5646.2012.01774.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S., Markow T. A., and Spicer G. S., 1995. Delayed male maturity is a cost of producing large sperm in Drosophila. Proc. Natl. Acad. Sci. USA 92: 10614–10618. 10.1073/pnas.92.23.10614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S. D., Houle D., and Curtsinger J. W., 1998. Age-specific properties of spontaneous mutations affecting mortality in Drosophila melanogaster. Genetics 148: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S. D., Macdonald S. J., Marguerie R., Certa U., Stearns S. C. et al. , 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12: 712–723. 10.1016/S0960-9822(02)00808-4 [DOI] [PubMed] [Google Scholar]
- Pletcher S. D., Libert S., and Skorupa D., 2005. Flies and their golden apples: the effect of dietary restriction on Drosophila aging and age-dependent gene expression. Ageing Res. Rev. 4: 451–480. 10.1016/j.arr.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Porcelli D., Westram A. M., Pascual M., Gaston K. J., Butlin R. K. et al. , 2016. Gene expression clines reveal local adaptation and associated trade-offs at a continental scale. Sci. Rep. 6: 32975 10.1038/srep32975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., 1997. Progress and Prospects in Evolutionary Biology – The Drosophila Model. Oxford University Press, Oxford. [Google Scholar]
- Prasad N. G., and Joshi A., 2003. What have two decades of laboratory life-history evolution studies on Drosophila melanogaster taught us? J. Genet. 82: 45–76. 10.1007/BF02715881 [DOI] [PubMed] [Google Scholar]
- Price T., and Schluter D., 1991. On the low heritability of life-history traits. Evolution 45: 853–861. 10.1111/j.1558-5646.1991.tb04354.x [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., and Di Rienzo A., 2010. Adaptation – not by sweeps alone. Nat. Rev. Genet. 11: 665–667. 10.1038/nrg2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., and Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. 10.1016/j.cub.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow D. E., and Tatar M., 1998. Mutation and senescence: where genetics and demography meet. Genetica 102–103: 299–314. 10.1023/A:1017047212008 [DOI] [PubMed] [Google Scholar]
- Promislow D. E. L., and Bugbee M., 2000. Direct and correlated responses to selection on age at physiological maturity in Drosophila simulans. J. Evol. Biol. 13: 955–966. 10.1046/j.1420-9101.2000.00240.x [DOI] [Google Scholar]
- Prout T., 1962. The effects of stabilizing selection on the time of development in Drosophila melanogaster. Genet. Res. 3: 364–382. 10.1017/S0016672300003219 [DOI] [Google Scholar]
- Prout T., 1971a The relation between fitness components and population prediction in Drosophila. I. The estimation of fitness components. Genetics 68: 127–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout T., 1971b The relation between fitness components and population prediction in Drosophila. II. Population prediction. Genetics 68: 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit S., Gefen E., Bergland A. O., Petrov D. A., Gibbs A. G. et al. , 2018. Spatiotemporal dynamics and genome-wide association genome-wide association analysis of desiccation tolerance in Drosophila melanogaster. Mol. Ecol. 27: 3525–3540. 10.1111/mec.14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rako L., Anderson A. R., Sgrò C. M., Stocker A. J., and Hoffmann A. A., 2006. The association between inversion In(3R)Payne and clinally varying traits in Drosophila melanogaster. Genetica 128: 373–384. 10.1007/s10709-006-7375-7 [DOI] [PubMed] [Google Scholar]
- Reed S. C., and Reed E. W., 1948. Natural selection in laboratory populations of Drosophila. Evolution 2: 176–186. 10.1111/j.1558-5646.1948.tb02739.x [DOI] [PubMed] [Google Scholar]
- Reed S. C., and Reed E. W., 1950. Natural selection in laboratory populations of Drosophila. II. Competition between a white-eye gene and its wild type allele. Evolution 4: 34–42. [Google Scholar]
- Reed L. K., Williams S., Springston M., Brown J., Freeman K. et al. , 2010. Genotype-by-diet interactions drive metabolic phenotype variation in Drosophila melanogaster. Genetics 185: 1009–1019. 10.1534/genetics.109.113571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. P., and Fairbairn D. J., 1999. Change in sexual size dimorphism as a correlated response to selection on fecundity. Heredity 83: 697–706. 10.1046/j.1365-2540.1999.00616.x [DOI] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J., and Kern A. D., 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197: 361–373. 10.1534/genetics.114.161463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis T., 2016. Effects of synthetic diets enriched in specific nutrients on Drosophila development, body fat, and lifespan. PLoS One 11: e0146758 10.1371/journal.pone.0146758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remolina S. C., Chang P. L., Leips J., Nuzhdin S. V., and Hughes K. A., 2012. Genomic basis of aging and life-history evolution in Drosophila melanogaster. Evolution 66: 3390–3403. 10.1111/j.1558-5646.2012.01710.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. M., Temiyasathit S., Reedy M. M., Ruedi E. A., Drnevich J. M. et al. , 2007. Age specificity of inbreeding load in Drosophila melanogaster and implications for the evolution of late-life mortality plateaus. Genetics 177: 587–595. 10.1534/genetics.106.070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D., Bryant M., and Holmes D., 2005. The evolution of senescence and post-reproductive lifespan in guppies (Poecilia reticulata). PLoS Biol. 4: e7 10.1371/journal.pbio.0040007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rion S., and Kawecki T. J., 2007. Evolutionary biology of starvation resistance: what we have learned from Drosophila. J. Evol. Biol. 20: 1655–1664. 10.1111/j.1420-9101.2007.01405.x [DOI] [PubMed] [Google Scholar]
- Robertson A., 1955. Selection in animals: synthesis. Cold Spring Harb. Symp. Quant. Biol. 20: 225–229. 10.1101/SQB.1955.020.01.021 [DOI] [PubMed] [Google Scholar]
- Robertson A., 1966. A mathematical model of the culling process in dairy cattle. Anim. Sci. 8: 95–108. 10.1017/S0003356100037752 [DOI] [Google Scholar]
- Robertson F. W., 1957a Studies in quantitative inheritance X. Genetic variation of ovary size in Drosophila. J. Genet. 55: 410–427. 10.1007/BF02984060 [DOI] [PubMed] [Google Scholar]
- Robertson F. W., 1957b Studies in quantitative inheritance. XI. Genetic and environmental correlation between body size and egg production in Drosophila melanogaster. J. Genet. 55: 428 10.1007/BF02984061 [DOI] [PubMed] [Google Scholar]
- Robertson F. W., 1959. Studies in quantitative inheritance. XII. Cell size and number in relation to genetic and environmental variation of body size in Drosophila. Genetics 44: 869–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson F. W., 1960a The ecological genetics of growth in Drosophila. 1. Body size and developmental time on different diets 1. Body size and developmental time on different diets. Genet. Res. 1: 288–304. 10.1017/S0016672300000264 [DOI] [Google Scholar]
- Robertson F. W., 1960b The ecological genetics of growth in Drosophila. 2. Selection for large body size on different diets. Genet. Res. 1: 305–318. 10.1017/S0016672300000276 [DOI] [Google Scholar]
- Robertson F. W., 1960c The ecological genetics of growth in Drosophila. 3. Growth and competitive ability of strains selected on different diets. Genet. Res. 1: 333–350. 10.1017/S001667230000032X [DOI] [Google Scholar]
- Robertson F. W., and Reeve E., 1952. Studies in quantitative inheritance. I. The effects of selection of wing and thorax length in Drosophila melanogaster. J. Genet. 50: 414 10.1007/BF02986839 [DOI] [Google Scholar]
- Robinson S. J. W., and Partridge L., 2001. Temperature and clinal variation in larval growth efficiency in Drosophila melanogaster. J. Evol. Biol. 14: 14–21. 10.1046/j.1420-9101.2001.00259.x [DOI] [PubMed] [Google Scholar]
- Rockman M. V., 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17. 10.1111/j.1558-5646.2011.01486.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D., 1981. On being the right size. Am. Nat. 118: 405–422. 10.1086/283832 [DOI] [Google Scholar]
- Roff D. A., 1992. The Evolution of Life Histories - Theory and Analysis. Chapman and Hall, New York. [Google Scholar]
- Roff D. A., 1995. The estimation of genetic correlations from phenotypic correlations: a test of Cheverud’s conjecture. Heredity 74: 481–490. 10.1038/hdy.1995.68 [DOI] [Google Scholar]
- Roff D. A., 1997. Evolutionary Quantitative Genetics. Chapman and Hall, London, New York: 10.1007/978-1-4615-4080-9 [DOI] [Google Scholar]
- Roff D. A., 2002. Life History Evolution. Sinauer, Sunderland, MA. [Google Scholar]
- Roff D. A., 2007. Contributions of genomics to life-history theory. Nat. Rev. Genet. 8: 116–125. 10.1038/nrg2040 [DOI] [PubMed] [Google Scholar]
- Roff D. A., and Fairbairn D. J., 2007. The evolution of trade-offs: where are we? J. Evol. Biol. 20: 433–447. 10.1111/j.1420-9101.2006.01255.x [DOI] [PubMed] [Google Scholar]
- Roff D. A., and Mousseau T. A., 1987. Quantitative genetics and fitness: lessons from Drosophila. Heredity 58: 103–118. 10.1038/hdy.1987.15 [DOI] [PubMed] [Google Scholar]
- Rogina B., Reenan R. A., Nilsen S. P., and Helfand S. L., 2000. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290: 2137–2140. 10.1126/science.290.5499.2137 [DOI] [PubMed] [Google Scholar]
- Rogina B., Wolverton T., Bross T. G., Chen K., Müller H. G. et al. , 2007. Distinct biological epochs in the reproductive life of female Drosophila melanogaster. Mech. Ageing Dev. 128: 477–485. 10.1016/j.mad.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. R., 1982. Antagonistic pleiotropy, dominance, and genetic variation. Heredity 48: 63–78. 10.1038/hdy.1982.7 [DOI] [Google Scholar]
- Rose M. R., 1984a Artificial selection on a fitness-component in Drosophila melanogaster. Evolution 38: 516–526. 10.1111/j.1558-5646.1984.tb00317.x [DOI] [PubMed] [Google Scholar]
- Rose M. R., 1984b Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution 38: 1004–1010. 10.1111/j.1558-5646.1984.tb00370.x [DOI] [PubMed] [Google Scholar]
- Rose M. R., 1985. Life history evolution with antagonistic pleiotropy and overlapping generations. Theor. Popul. Biol. 28: 342–358. 10.1016/0040-5809(85)90034-6 [DOI] [Google Scholar]
- Rose M. R., 1991. Evolutionary Biology of Aging. Oxford. University Press, New York, Oxford. [Google Scholar]
- Rose M. R., and Bradley T. J., 1998. Evolutionary physiology of the cost of reproduction. Oikos 83: 443–451. 10.2307/3546672 [DOI] [Google Scholar]
- Rose M. R., and Charlesworth B., 1981a Genetics of life history in Drosophila melanogaster. I. Sib analysis of adult females. Genetics 97: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. R., and Charlesworth B., 1981b Genetics of life history in Drosophila melanogaster. II. Exploratory selection experiments. Genetics 97: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. R., Vu L. N., Park S. U., and Graves J. L. Jr, 1992. Selection on stress resistance increases longevity in Drosophila melanogaster. Exp. Gerontol. 27: 241–250. 10.1016/0531-5565(92)90048-5 [DOI] [PubMed] [Google Scholar]
- Rose M. R., Nusbaum T. J., and Chippindale A. K., 1996. Laboratory evolution: the experimental wonderland and the Cheshire cat syndrome, pp. 221–241 in Adaptation, edited by Rose M. R. and Lauder G. V.. Academic Press, San Diego. [Google Scholar]
- Rose M. R., Passananti H. B., and Matos M., 2004. Methuselah Flies - A Case Study in Aging. World Scientific, Singapore: 10.1142/5457 [DOI] [Google Scholar]
- Rose M. R., Mueller L. D., and Burke M. K., 2011. New experiments for an undivided genetics. Genetics 188: 1–10. 10.1534/genetics.111.128900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewell J., and Shorrocks B., 1987. The implication of survival rates in natural-populations of Drosophila - capture recapture experiments on domestic species. Biol. J. Linn. Soc. Lond. 32: 373–384. 10.1111/j.1095-8312.1987.tb00438.x [DOI] [Google Scholar]
- Rueffler C., Van Dooren T. J. M., Leimar O., and Abrams P. A., 2006. Disruptive selection and then what? Trends Ecol. Evol. 21: 238–245. 10.1016/j.tree.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Salmon A. B., Marx D. B., and Harshman L. G., 2001. A cost of reproduction in Drosophila melanogaster: stress susceptibility. Evolution 55: 1600–1608. 10.1111/j.0014-3820.2001.tb00679.x [DOI] [PubMed] [Google Scholar]
- Saltz J. B., Alicuben E. T., Grubman J., Harkenrider M., Megowan N. et al. , 2012. Nonadditive indirect effects of group genetic diversity on larval viability in Drosophila melanogaster imply key role of maternal decision-making. Mol. Ecol. 21: 2270–2281. 10.1111/j.1365-294X.2012.05518.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrelli F., Tauber E., Pegoraro M., Mazzotta G., Cisotto P. et al. , 2007. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science 316: 1898–1900. 10.1126/science.1138426 [DOI] [PubMed] [Google Scholar]
- Sang J. H., 1956. The quantitative nutritional requirements of Drosophila melanogaster. J. Exp. Biol. 33: 45–72. [Google Scholar]
- Sang J. H., 1978. The nutritional requirements of Drosophila, pp. 159–192 in The Genetics and Biology of Drosophila, Vol. 2, edited by Ashburner M. and Wright T. R. F.. Academic Press, New York. [Google Scholar]
- Sang J. H., and King R. C., 1961. Nutritional requirements of axenically cultured Drosophila melanogaster adults. J. Exp. Biol. 38: 793–809. [Google Scholar]
- Santos M., Fowler K., and Partridge L., 1994. Gene-environment interaction for body size and larval density in Drosophila melanogaster: an investigation of effects on development time, thorax length and adult sex ratio. Heredity 72: 515–521. 10.1038/hdy.1994.69 [DOI] [PubMed] [Google Scholar]
- Santos M., Céspedes W., Balanyà J., Trotta V., Calboli F. C. et al. , 2005. Temperature-related genetic changes in laboratory populations of Drosophila subobscura: evidence against simple climatic-based explanations for latitudinal clines. Am. Nat. 165: 258–273. 10.1086/427093 [DOI] [PubMed] [Google Scholar]
- Saunders D. S., and Bertossa R. C., 2011. Deciphering time measurement: the role of circadian ‘clock’ genes and formal experimentation in insect photoperiodism. J. Insect Physiol. 57: 557–566. 10.1016/j.jinsphys.2011.01.013 [DOI] [PubMed] [Google Scholar]
- Saunders D. S., and Gilbert L. I., 1990. Regulation of ovarian diapause in Drosophila melanogaster by photoperiod and moderately low temperature. J. Insect Physiol. 36: 195–200. 10.1016/0022-1910(90)90122-V [DOI] [Google Scholar]
- Saunders D. S., Henrich V. C., and Gilbert L. I., 1989. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc. Natl. Acad. Sci. USA 86: 3748–3752. 10.1073/pnas.86.10.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D. S., Richard D. S., Applebaum S. W., Ma M., and Gilbert L. I., 1990. Photoperiodic diapause in Drosophila melanogaster involves a block to the juvenile hormone regulation of ovarian maturation. Gen. Comp. Endocrinol. 79: 174–184. 10.1016/0016-6480(90)90102-R [DOI] [PubMed] [Google Scholar]
- Scheiner S. M., 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24: 35–68. 10.1146/annurev.es.24.110193.000343 [DOI] [Google Scholar]
- Scheiner S. M., 2002. Selection experiments and the study of phenotypic plasticity. J. Evol. Biol. 15: 889–898. 10.1046/j.1420-9101.2002.00468.x [DOI] [Google Scholar]
- Scheiner S. M., Caplan R. L., and Lyman R. F., 1989. A search for trade-offs among life history traits in Drosophila melanogaster. Evol. Ecol. 3: 51–63. 10.1007/BF02147931 [DOI] [Google Scholar]
- Schlichting C. D., and Pigliucci M., 1998. Phenotypic Evolution: A Reaction Norm Perspective. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Schlötterer C., Tobler R., Kofler R., and Nolte V., 2014. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15: 749–763. 10.1038/nrg3803 [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Kofler R., Versace E., Tobler R., and Franssen S. U., 2015. Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity (Edinb) 114: 431–440 [corrigenda: Heredity (Edinb) 116: 248 (2016)]. 10.1038/hdy.2014.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., 2011. Evolution and mechanisms of insect reproductive diapause: a plastic and pleiotropic life history syndrome, pp. 230–242 in Mechanisms of Life History Evolution: The Genetics and Physiology of Life History Traits and Trade-Offs, edited by Flatt T. and Heyland A.. Oxford University Press, Oxford: 10.1093/acprof:oso/9780199568765.003.0018 [DOI] [Google Scholar]
- Schmidt P. S., and Conde D. R., 2006. Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution 60: 1602–1611. 10.1111/j.0014-3820.2006.tb00505.x [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., and Paaby A. B., 2008. Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution 62: 1204–1215. 10.1111/j.1558-5646.2008.00351.x [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Duvernell D. D., and Eanes W. F., 2000. Adaptive evolution of a candidate gene for ageing in Drosophila. Proc. Natl. Acad. Sci. USA 97: 10861–10865. 10.1073/pnas.190338897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., Matzkin L., Ippolito M., and Eanes W., 2005a Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59: 1721–1732. [PubMed] [Google Scholar]
- Schmidt P. S., Paaby A. B., and Heschel M. S., 2005b Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution 59: 2616–2625. 10.1111/j.0014-3820.2005.tb00974.x [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Zhu C.-T., Das J., Batavia M., Yang L. et al. , 2008. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 105: 16207–16211. 10.1073/pnas.0805485105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., and Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. 10.1534/genetics.113.151670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf L., Blows M. W., and Caley M. J., 1999. Life-history consequences of divergent selection on egg size in Drosophila melanogaster. Am. Nat. 154: 333–340. 10.1086/303242 [DOI] [PubMed] [Google Scholar]
- Service P. M., 1987. Physiological mechanisms of increased stress resistance in Drosophila melanogaster selected for postponed senescence. Physiol. Zool. 60: 321–326. 10.1086/physzool.60.3.30162285 [DOI] [Google Scholar]
- Service P. M., and Rose M. R., 1985. Genetic variation among life-history components: the effect of novel environments. Evolution 39: 943–945. 10.1111/j.1558-5646.1985.tb00436.x [DOI] [PubMed] [Google Scholar]
- Service P. M., Hutchinson E. W., MacKinley M. D., and Rose M. R., 1985. Resistance to environmental stress in Drosophila melanogaster selected for postponed senescence. Physiol. Zool. 58: 380–389. 10.1086/physzool.58.4.30156013 [DOI] [Google Scholar]
- Service P. M., Hutchinson E. W., and Rose M. R., 1988. Multiple genetic mechanisms for the evolution of senescence in Drosophila melanogaster. Evolution 42: 708–716. 10.1111/j.1558-5646.1988.tb02489.x [DOI] [PubMed] [Google Scholar]
- Sevenster J. G., and Van Alphen J. J. M., 1993. A life history trade-off in Drosophila species and community structure in variable environments. J. Anim. Ecol. 62: 720–736. 10.2307/5392 [DOI] [Google Scholar]
- Sgrò C. M., and Partridge L., 2000. Evolutionary responses of the life history of wild-caught Drosophila melanogaster to two standard methods of laboratory culture. Am. Nat. 156: 341–353. 10.1086/303394 [DOI] [Google Scholar]
- Sgrò C. M., and Partridge L., 2001. Laboratory adaptation of life history in Drosophila. Am. Nat. 158: 657–658. 10.1086/323592 [DOI] [PubMed] [Google Scholar]
- Sgrò C. M., van Heerwaarden B., Kellermann V., Wee C. W., Hoffmann A. A. et al. , 2013. Complexity of the genetic basis of ageing in nature revealed by a clinal study of lifespan and methuselah, a gene for ageing, in Drosophila from eastern Australia. Mol. Ecol. 22: 3539–3551. 10.1111/mec.12353 [DOI] [PubMed] [Google Scholar]
- Shanley D., and Kirkwood T., 2000. Calorie restriction and aging: a life-history analysis. Evolution 54: 740–750. 10.1111/j.0014-3820.2000.tb00076.x [DOI] [PubMed] [Google Scholar]
- Shapiro H., 1932. The rate of oviposition in the fruit fly, Drosophila. Biol. Bull. (Woods Hole) 63: 456–471. 10.2307/1537346 [DOI] [Google Scholar]
- Shiotsugu J., Leroi A. M., Yashiro H., Rose M. R., and Mueller L. D., 1997. The symmetry of correlated selection responses in adaptive evolution: an experimental study using Drosophila. Evolution 51: 163–172. 10.1111/j.1558-5646.1997.tb02397.x [DOI] [PubMed] [Google Scholar]
- Sibly R. M., and Calow P., 1985. Physiological Ecology of Animals: An Evolutionary Approach. Blackwell, Oxford. [Google Scholar]
- Silbermann R., and Tatar M., 2000. Reproductive costs of heat shock protein in transgenic Drosophila melanogaster. Evolution 54: 2038–2045. 10.1111/j.0014-3820.2000.tb01247.x [DOI] [PubMed] [Google Scholar]
- Simões P., Rose M. R., Duarte A., Goncalves R., and Matos M., 2007. Evolutionary domestication in Drosophila subobscura. J. Evol. Biol. 20: 758–766. 10.1111/j.1420-9101.2006.01244.x [DOI] [PubMed] [Google Scholar]
- Simmons F. H., and Bradley T. J., 1997. An analysis of resource allocation in response to dietary yeast in Drosophila melanogaster. J. Insect Physiol. 43: 779–788. 10.1016/S0022-1910(97)00037-1 [DOI] [PubMed] [Google Scholar]
- Simmons M. J., and Crow J. F., 1977. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11: 49–78. 10.1146/annurev.ge.11.120177.000405 [DOI] [PubMed] [Google Scholar]
- Simpson S. J., and Raubenheimer D., 2012. The Nature of Nutrition. A Unifying Framework from Animal Adaptation to Human Obesity. Princeton University Press, Princeton, NJ. [Google Scholar]
- Skorupa D. A., Dervisefendic A., Zwiener J., and Pletcher S. D., 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7: 478–490. 10.1111/j.1474-9726.2008.00400.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1975. Gene flow and selection in a two-locus system. Genetics 81: 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth H. T., 1974. Courtship behavior in Drosophila. Annu. Rev. Entomol. 19: 385–405. 10.1146/annurev.en.19.010174.002125 [DOI] [PubMed] [Google Scholar]
- Sokal R. R., 1970. Senescence and genetic load: evidence from Tribolium. Science 167: 1733–1734. 10.1126/science.167.3926.1733 [DOI] [PubMed] [Google Scholar]
- Sokolowski M. B., Pereira H. S., and Hughes K., 1997. Evolution of foraging behavior in Drosophila by density-dependent selection. Proc. Natl. Acad. Sci. USA 94: 7373–7377. 10.1073/pnas.94.14.7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen J. G., Schou M. F., Kristensen T. N., and Loeschcke V., 2016. Thermal fluctuations affect the transcriptome through mechanisms independent of average temperature. Sci. Rep. 6: 30975 10.1038/srep30975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P. D., Ng’oma E., O’Day S., and King E. G., 2017. Genetic dissection of nutrition-induced plasticity in insulin/insulin-like growth factor signaling and median life span in a Drosophila multiparent population. Genetics 206: 587–602. 10.1534/genetics.116.197780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns S. C., 1976. Life-history tactics: a review of the ideas. Q. Rev. Biol. 51: 3–47. 10.1086/409052 [DOI] [PubMed] [Google Scholar]
- Stearns S. C., 1977. The evolution of life history traits: a critique of the theory and a review of the data. Annu. Rev. Ecol. Syst. 8: 145–171. 10.1146/annurev.es.08.110177.001045 [DOI] [Google Scholar]
- Stearns S. C., 1984. How much of the phenotype is necessary to understand evolution at the level of the gene? pp. 31–45 in Population Biology and Evolution, edited by Wöhrmann K. and Loeschcke V.. Springer, Berlin: 10.1007/978-3-642-69646-6_4 [DOI] [Google Scholar]
- Stearns S. C., 1989a Trade-offs in life-history evolution. Funct. Ecol. 3: 259–268. 10.2307/2389364 [DOI] [Google Scholar]
- Stearns S. C., 1989b Comparative and experimental approaches to the evolutionary ecology of development. Geobios 22: 349–355. 10.1016/S0016-6995(89)80035-X [DOI] [Google Scholar]
- Stearns S. C., 1989c The evolutionary significance of phenotypic plasticity. Bioscience 39: 436–445. 10.2307/1311135 [DOI] [Google Scholar]
- Stearns S. C., 1992. The Evolution of Life Histories. Oxford University Press, Oxford. [Google Scholar]
- Stearns S. C., 2000. Life history evolution: successes, limitations, and prospects. Naturwiss. 87: 476–486. 10.1007/s001140050763 [DOI] [PubMed] [Google Scholar]
- Stearns S. C., and Kaiser M., 1993. The effects of enhanced expression of elongation factor EF-1 alpha on lifespan in Drosophila melanogaster. IV. A summary of three experiments. Genetica 91: 167–182. 10.1007/BF01435996 [DOI] [PubMed] [Google Scholar]
- Stearns S. C., and Kaiser M., 1996. The effects on fitness components of P-element inserts in Drosophila melanogaster: analysis of trade-offs. Evolution 50: 795–806. [DOI] [PubMed] [Google Scholar]
- Stearns S. C., and Koella J. C., 1986. The evolution of phenotypic plasticity in life-history traits: predictions of reaction norms for age and size at maturity. Evolution 40: 893–913. 10.1111/j.1558-5646.1986.tb00560.x [DOI] [PubMed] [Google Scholar]
- Stearns S. C., and Medzhitov R., 2016. Evolutionary Medicine. Sinauer, Sunderland, MA. [Google Scholar]
- Stearns S. C., and Partridge L., 2001. The genetics of aging in Drosophila, pp. 353–368 in Handbook of the Biology of Aging, edited by Masoro E. and Austad S.. Academic Press, San Diego. [Google Scholar]
- Stearns S., de Jong G., and Newman B., 1991. The effects of phenotypic plasticity on genetic correlations. Trends Ecol. Evol. 6: 122–126. 10.1016/0169-5347(91)90090-K [DOI] [PubMed] [Google Scholar]
- Stearns S. C., Kaiser M., and Hillesheim E., 1993. Effects on fitness components of enhanced expression of elongation factor EF-1a in Drosophila melanogaster. I. The contrasting approaches of molecular and population biologists. Am. Nat. 142: 961–993. 10.1086/285584 [DOI] [PubMed] [Google Scholar]
- Stearns S. C., Ackermann M., Doebeli M., and Kaiser M., 2000. Experimental evolution of aging, growth, and reproduction in fruitflies. Proc. Natl. Acad. Sci. USA 97: 3309–3313. 10.1073/pnas.97.7.3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefana M. I., Driscoll P. C., Obata F., Pengelly A. R., Newell C. L. et al. , 2017. Developmental diet regulates Drosophila lifespan via lipid autotoxins. Nat. Commun. 8: 1384 10.1038/s41467-017-01740-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner U. K., and Tuljapurkar S., 2012. Neutral theory for life histories and individual variability in fitness components. Proc. Natl. Acad. Sci. USA 109: 4684–4689. 10.1073/pnas.1018096109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., 2016. Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol. Ecol. 25: 79–88. 10.1111/mec.13288 [DOI] [PubMed] [Google Scholar]
- Stern D. L., 2000. Perspective: evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. 10.1111/j.0014-3820.2000.tb00544.x [DOI] [PubMed] [Google Scholar]
- Stern D. L., 2014. Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet. 30: 547–554. 10.1016/j.tig.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1921. The North American Species of Drosophila (No. 301). Carnegie Inst., Washington: 10.5962/bhl.title.10480 [DOI] [Google Scholar]
- Sved J. A., 1971. An estimate of heterosis in Drosophila melanogaster. Genet. Res. 18: 97–105. 10.1017/S0016672300012453 [DOI] [PubMed] [Google Scholar]
- Sved J. A., 1975. Fitness of third chromosome homozygotes in Drosophila melanogaster. Genet. Res. 25: 197–200. 10.1017/S0016672300015603 [DOI] [PubMed] [Google Scholar]
- Sved J. A., 1989. The measurement of fitness in Drosophila, pp. 99–104 in Evolution and Animal Breeding, edited by Hill W. G. and Mackay T. F. C.. C. A. B. International, Wallingford, UK. [Google Scholar]
- Svetec N., Saelao P., Cridland J. M., Hoffmann A. A., and Begun D. J., 2019. Functional analysis of a putative target of spatially varying selection in the Menin1 gene of Drosophila melanogaster. G3 (Bethesda) 9: 73–80. 10.1534/g3.118.200818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., and Yamazaki T., 1990. Fitness and its components in Drosophila melanogaster. Jap. J. Genet. 65: 417–426. 10.1266/jjg.65.417 [DOI] [PubMed] [Google Scholar]
- Tatar M., 1999. Transgenes in the analysis of life span and fitness. Am. Nat. 154: S67–S81. 10.1086/303284 [DOI] [PubMed] [Google Scholar]
- Tatar M., 2000. Transgenic organisms in evolutionary ecology. Trends Ecol. Evol. 15: 207–211. 10.1016/S0169-5347(00)01834-6 [DOI] [PubMed] [Google Scholar]
- Tatar M., 2007. Diet restriction in Drosophila melanogaster. Design and analysis. Interdiscip. Top. Gerontol. 35: 115–136. [DOI] [PubMed] [Google Scholar]
- Tatar M., 2011. The plate half-full: status of research on the mechanisms of dietary restriction in Drosophila melanogaster. Exp. Gerontol. 46: 363–368. 10.1016/j.exger.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., and Carey J. R., 1995. Nutrition mediates reproductive trade-offs with age-specific mortality in the beetle Callosobruchus maculatus. Ecology 76: 2066–2073. 10.2307/1941681 [DOI] [Google Scholar]
- Tatar M., and Yin C.-M., 2001. Slow aging during insect reproductive diapause: why butterflies, grasshoppers and flies are like forms. Exp. Gerontol. 36: 723–738. 10.1016/S0531-5565(00)00238-2 [DOI] [PubMed] [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M.-P., Yin C.-M. et al. , 2001a A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110. 10.1126/science.1057987 [DOI] [PubMed] [Google Scholar]
- Tatar M., Chien S. A., and Priest N. K., 2001b Negligible senescence during reproductive dormancy in Drosophila melanogaster. Am. Nat. 158: 248–258. 10.1086/321320 [DOI] [PubMed] [Google Scholar]
- Tatar M., Bartke A., and Antebi A., 2003. The endocrine regulation of aging by insulin-like signals. Science 299: 1346–1351. 10.1126/science.1081447 [DOI] [PubMed] [Google Scholar]
- Tatar M., Post S., and Yu K., 2014. Nutrient control of Drosophila longevity. Trends Endocrinol. Metab. 25: 509–517. 10.1016/j.tem.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum E. L., 1939. Nutritional requirements of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 25: 490–497. 10.1073/pnas.25.9.490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber E., Zordan M., Sandrelli F., Pegoraro M., Osterwalder N. et al. , 2007. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 316: 1895–1898. 10.1126/science.1138412 [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M., McIntyre L. M., and Wayne M. L., 2005. Genetic architecture of two fitness-related traits in Drosophila melanogaster: ovariole number and thorax length. Genetica 125: 211–222. 10.1007/s10709-005-8549-4 [DOI] [PubMed] [Google Scholar]
- Tennessen J. M., and Thummel C. S., 2011. Coordinating growth and maturation insights from Drosophila. Curr. Biol. 21: R750–R757. 10.1016/j.cub.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio H., and Rose M. R., 2000. Variation in the reversibility of evolution. Nature 408: 463–466. 10.1038/35044070 [DOI] [PubMed] [Google Scholar]
- Teotónio H., Chelo I. M., Bradic M., Rose M. R., and Long A. D., 2009. Experimental evolution reveals natural selection on standing genetic variation. Nat. Genet. 41: 251–257. 10.1038/ng.289 [DOI] [PubMed] [Google Scholar]
- Tobler R., Franssen S. U., Kofler R., Orozco-terWengel P., Nolte V. et al. , 2013. Massive habitat-specific genomic response in D. melanogaster populations during experimental evolution in hot and cold environments. Mol. Biol. Evol. 31: 364–375. 10.1093/molbev/mst205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler R., Hermisson J., and Schlötterer C., 2015. Parallel trait adaptation across opposing thermal environments in experimental Drosophila melanogaster populations. Evolution 69: 1745–1759. 10.1111/evo.12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., 2000. Transgenic methods for increasing Drosophila lifespan. Mech. Ageing Dev. 118: 1–14. 10.1016/S0047-6374(00)00152-4 [DOI] [PubMed] [Google Scholar]
- Tower J., 2011. Heat shock proteins and Drosophila aging. Exp. Gerontol. 46: 355–362. 10.1016/j.exger.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis J. A., 1994. The ecological genetics of life histories: variation and its evolutionary significance, pp. 171–204 in Ecological Genetics, edited by Real L. A. Princeton University Press, Princeton, NJ. [Google Scholar]
- Trotta V., Calboli F. C., Ziosi M., Guerra D., Pezzoli M. C. et al. , 2006. Thermal plasticity in Drosophila melanogaster: a comparison of geographic populations. BMC Evol. Biol. 6: 67 10.1186/1471-2148-6-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M.-P., and Tatar M., 2003. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell 2: 327–333. 10.1046/j.1474-9728.2003.00064.x [DOI] [PubMed] [Google Scholar]
- Tucić N., Cvetković D., and Milanović D., 1988. The genetic variation and covariation among fitness components in Drosophila melanogaster females and males. Heredity 60: 55–60. 10.1038/hdy.1988.9 [DOI] [PubMed] [Google Scholar]
- Turner T. L., 2014. Fine-mapping natural alleles: quantitative complementation to the rescue. Mol. Ecol. 23: 2377–2382. 10.1111/mec.12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., and Begun D. J., 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473. 10.1534/genetics.107.083659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Stewart A. D., Fields A. T., Rice W. R., and Tarone A. M., 2011. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 7: e1001336 10.1371/journal.pgen.1001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delden W., and Kamping A., 1997. Worldwide latitudinal clines for the alcohol dehydrogenase polymorphism in Drosophila melanogaster: what is the unit of selection? pp. 97–115 in Environmental Stress, Adaptation and Evolution, edited by Bijlsma R. and Loeschcke V.. Birkhäuser Basel, Basel: 10.1007/978-3-0348-8882-0_6 [DOI] [PubMed] [Google Scholar]
- van Heerwaarden B., and Sgrò C. M., 2017. The quantitative genetic basis of clinal divergence in phenotypic plasticity. Evolution 71: 2618–2633. 10.1111/evo.13342 [DOI] [PubMed] [Google Scholar]
- van Noordwijk A., and de Jong G., 1986. Acquisition and allocation of resources: their influence on variation in life history tactics. Am. Nat. 128: 137–142. 10.1086/284547 [DOI] [Google Scholar]
- Van’t Land J., van Putten P., Zwaan B., Kamping A., and van Delden W., 1999. Latitudinal variation in wild populations of Drosophila melanogaster: heritabilities and reaction norms. J. Evol. Biol. 12: 222–232. 10.1046/j.1420-9101.1999.00029.x [DOI] [Google Scholar]
- Via S., Gomulkiewicz R., De Jong G., Scheiner S. M., Schlichting C. D. et al. , 1995. Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol. Evol. 10: 212–217. 10.1016/S0169-5347(00)89061-8 [DOI] [PubMed] [Google Scholar]
- Vieira C., Pasyukova E. G., Zeng Z.-B., Hackett J. B., Lyman R. F. et al. , 2000. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijendravarma R. K., Narasimha S., and Kawecki T. J., 2011. Plastic and evolutionary responses of cell size and number to larval malnutrition in Drosophila melanogaster. J. Evol. Biol. 24: 897–903. 10.1111/j.1420-9101.2010.02225.x [DOI] [PubMed] [Google Scholar]
- Vijendravarma R. K., Narasimha S., and Kawecki T. J., 2012. Adaptation to abundant low quality food improves the ability to compete for limited rich food in Drosophila melanogaster. PLoS One 7: e30650 10.1371/journal.pone.0030650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonesch S. C., Lamparter D., Mackay T. F. C., Bergmann S., and Hafen E., 2016. Genome-wide analysis reveals novel regulators of growth in Drosophila melanogaster. PLoS Genet. 12: e1005616 10.1371/journal.pgen.1005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P. (Editor), 2001. The Character Concept in Evolutionary Biology. Academic Press, New York. [Google Scholar]
- Walsh B., and Lynch M., 2018. Evolution and Selection of Quantitative Traits. Oxford University Press, Oxford: 10.1093/oso/9780198830870.001.0001 [DOI] [Google Scholar]
- Wang Y., Salmon A. B., and Harshman L. G., 2001. A cost of reproduction: oxidative stress susceptibility is associated with increased egg production in Drosophila melanogaster. Exp. Gerontol. 36: 1349–1359. 10.1016/S0531-5565(01)00095-X [DOI] [PubMed] [Google Scholar]
- Wang Y., Pot D., Kachman S. D., Nuzhdin S. V., and Harshman L. G., 2006. A quantitative trait locus analysis of natural genetic variation for Drosophila melanogaster oxidative stress survival. J. Hered. 97: 355–366. 10.1093/jhered/esl009 [DOI] [PubMed] [Google Scholar]
- Wayne M. L., Hackett J. B., and Mackay T. F. C., 1997. Quantitative genetics of ovariole number in Drosophila melanogaster. 1. Segregating variation and fitness. Evolution 51: 1156–1163. 10.1111/j.1558-5646.1997.tb03963.x [DOI] [PubMed] [Google Scholar]
- Weber A. L., Khan G. F., Magwire M. M., Tabor C. L., Mackay T. F. C. et al. , 2012. Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster. PLoS One 7: e34745 10.1371/journal.pone.0034745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks A. R., McKechnie S. W., and Hoffmann A. A., 2002. Dissecting adaptive clinal variation: markers, inversions and size/stress associations in Drosophila melanogaster from a central field population. Ecol. Lett. 5: 756–763. 10.1046/j.1461-0248.2002.00380.x [DOI] [Google Scholar]
- Welbergen P., and Sokolowski M. B., 1994. Development time and pupation behavior in the Drosophila melanogaster subgroup (Diptera: Drosophilidae). J. Insect Behav. 7: 263–277. 10.1007/BF01989734 [DOI] [Google Scholar]
- West-Eberhard M. J., 2003. Developmental Plasticity and Evolution. Oxford University Press, New York. [Google Scholar]
- Whitaker R., Gil M. P., Ding F., Tatar M., Helfand S. L. et al. , 2014. Dietary switch reveals fast coordinated gene expression changes in Drosophila melanogaster. Aging (Albany N.Y.) 6: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman D. W., and Ananthakrishnan T. N. (Editors), 2009. Phenotypic Plasticity of Insects: Mechanisms and Consequences. Science Publishers, Enfield: 10.1201/b10201 [DOI] [Google Scholar]
- Williams G. C., 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11: 398–411. 10.1111/j.1558-5646.1957.tb02911.x [DOI] [Google Scholar]
- Williams G. C., 1966. Natural selection, the costs of reproduction, and a refinement of Lack’s principle. Am. Nat. 100: 687–690. 10.1086/282461 [DOI] [Google Scholar]
- Williams K. D., and Sokolowski M. B., 1993. Diapause in Drosophila melanogaster females: a genetic analysis. Heredity 71: 312–317. 10.1038/hdy.1993.141 [DOI] [PubMed] [Google Scholar]
- Williams K. D., Busto M., Suster M., So A., Ben-Shahar Y. et al. , 2006. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc. Natl. Acad. Sci. USA 103: 15911–15915. 10.1073/pnas.0604592103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. H., Morgan T. J., and Mackay T. F. C., 2006. High-resolution mapping of quantitative trait loci affecting increased life span in Drosophila melanogaster. Genetics 173: 1455–1463. 10.1534/genetics.105.055111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton A. N., and Sved J. A., 1979. X-chromosomal heterosis in Drosophila melanogaster. Genet. Res. 34: 303–315. 10.1017/S0016672300019534 [DOI] [PubMed] [Google Scholar]
- Wit J., Loeschcke V., and Kellermann V., 2015. Life span variation in 13 Drosophila species: a comparative study on life span, environmental variables and stress resistance. J. Evol. Biol. 28: 1892–1900. 10.1111/jeb.12706 [DOI] [PubMed] [Google Scholar]
- Wittmann M. J., Bergland A. O., Feldman M. W., Schmidt P. S., and Petrov D. A., 2017. Seasonally fluctuating selection can maintain polymorphism at many loci via segregation lift. Proc. Natl. Acad. Sci. USA 114: E9932–E9941. 10.1073/pnas.1702994114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1984. Evolution and the Genetics of Populations. Volume 3. Experimental Results and Evolutionary Deductions. University of Chicago Press, Chicago. [Google Scholar]
- Wright S., and Dobzhansky T., 1946. Genetics of natural populations. XII. Experimental reproduction of some of the changes caused by natural selection in certain populations of Drosophila pseudoobscura. Genetics 31: 125–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Anholt R. R. H., and Mackay T. F. C., 2009. Epistatic interactions attenuate mutations affecting startle behaviour in Drosophila melanogaster. Genet. Res. 91: 373–382. 10.1017/S0016672309990279 [DOI] [PubMed] [Google Scholar]
- Yamazaki T., 1984. Measurement of fitness and its components in six laboratory strains of Drosophila melanogaster. Genetics 108: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., and Hirose Y., 1984. Genetic analysis of natural populations of Drosophila melanogaster in Japan. II. The measurement of fitness and fitness components in homozygous lines. Genetics 108: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S., 2015. Local adaptation by alleles of small effect. Am. Nat. 186: S74–S89. 10.1086/682405 [DOI] [PubMed] [Google Scholar]
- Zajitschek F., Zajitschek S. R. K., Canton C., Georgolopoulos G., Friberg U. et al. , 2016. Evolution under dietary restriction increases male reproductive performance without survival cost. Proc. Biol. Sci. 283: 20152726 10.1098/rspb.2015.2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajitschek F., Georgolopoulos G., Vourlou A., Ericsson M., Zajitschek S. R. K. et al. , 2019. Evolution under dietary restriction decouples survival from fecundity in Drosophila melanogaster females. J. Gerontol. A 74: 1542–1548. 10.1093/gerona/gly070 [DOI] [PubMed] [Google Scholar]
- Zamudio K. R., Huey R. B., and Crill W. D., 1995. Bigger isn’t always better: body size, developmental time and parental temperature and male territorial success in Drosophila melanogaster. Anim. Behav. 49: 671–677. 10.1016/0003-3472(95)80200-2 [DOI] [Google Scholar]
- Zandveld J., van den Heuvel J., Mulder M., Brakefield P. M., Kirkwood T. B. L. et al. , 2017. Pervasive gene expression responses to a fluctuating diet in Drosophila melanogaster: the importance of measuring multiple traits to decouple potential mediators of life span and reproduction. Evolution 71: 2572–2583. 10.1111/evo.13327 [DOI] [PubMed] [Google Scholar]
- Zera A. J., and Harshman L. G., 2001. The physiology of life history trade-offs in animals. Annu. Rev. Ecol. Syst. 32: 95–126. 10.1146/annurev.ecolsys.32.081501.114006 [DOI] [Google Scholar]
- Zhao X., Bergland A. O., Behrman E. L., Gregory B. D., Petrov D. A. et al. , 2016. Global transcriptional profiling of diapause and climatic adaptation in Drosophila melanogaster. Mol. Biol. Evol. 33: 707–720. 10.1093/molbev/msv263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Campbell T. G., Stone E. A., Mackay T. F. C., and Anholt R. R. H., 2012. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 8: e1002593 10.1371/journal.pgen.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. T., Chang C., Reenan R. A., and Helfand S. L., 2014. Indy gene variation in natural populations confers fitness advantage and life span extension through transposon insertion. Aging (Albany N.Y.) 6: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonato V., Collins L., Pegoraro M., Tauber E., and Kyriacou C. P., 2017. Is diapause an ancient adaptation in Drosophila? J. Insect Physiol. 98: 267–274. 10.1016/j.jinsphys.2017.01.017 [DOI] [PubMed] [Google Scholar]
- Zonato V., Vanin S., Costa R., Tauber E., and Kyriacou C. P., 2018. Inverse european latitudinal cline at the timeless locus of Drosophila melanogaster reveals selection on a clock gene: population genetics of ls-tim. J. Biol. Rhythms 33: 15–23. 10.1177/0748730417742309 [DOI] [PubMed] [Google Scholar]
- Zwaan B., Bijlsma R., and Hoekstra R. F., 1995a Artificial selection for developmental time in Drosophila melanogaster in relation to the evolution of aging: direct and correlated responses. Evolution 49: 635–648. 10.1111/j.1558-5646.1995.tb02300.x [DOI] [PubMed] [Google Scholar]
- Zwaan B., Bijlsma R., and Hoekstra R. F., 1995b Direct selection on life span in Drosophila melanogaster. Evolution 49: 649–659 [corrigenda: Evolution 50: 475 (1996)]. 10.1111/j.1558-5646.1995.tb02301.x [DOI] [PubMed] [Google Scholar]
- Zwaan, B. J., 1993 Genetical and environmental aspects of ageing in Drosophila melanogaster: an evolutionary perspective. Ph.D. Thesis, University of Groningen, Groningen. [Google Scholar]
- Zwaan B. J., Bijlsma R., and Hoekstra R. F., 1991. On the developmental theory of ageing. I. Starvation resistance and longevity in Drosophila melanogaster in relation to pre-adult breeding conditions. Heredity 66: 29–39. 10.1038/hdy.1991.4 [DOI] [PubMed] [Google Scholar]
- Zwaan B. J., Bijlsma R., and Hoekstra R. F., 1992. On the developmental theory of ageing. II. The effect of developmental temperature on longevity in relation to adult body size in D. melanogaster. Heredity 68: 123–130. 10.1038/hdy.1992.19 [DOI] [PubMed] [Google Scholar]