Olduvai (formerly DUF1220) protein domains have undergone the largest human-specific increase in copy number of any coding region in the genome (∼300 copies of which 165 are human-specific) and have been implicated in human brain evolution...
Keywords: brain, evolution, gene duplication, genome, hyperamplification
Abstract
Sequences encoding Olduvai protein domains (formerly DUF1220) show the greatest human lineage-specific increase in copy number of any coding region in the genome and have been associated, in a dosage-dependent manner, with brain size, cognitive aptitude, autism, and schizophrenia. Tandem intragenic duplications of a three-domain block, termed the Olduvai triplet, in four NBPF genes in the chromosomal 1q21.1-0.2 region, are primarily responsible for the striking human-specific copy number increase. Interestingly, most of the Olduvai triplets are adjacent to, and transcriptionally coregulated with, three human-specific NOTCH2NL genes that have been shown to promote cortical neurogenesis. Until now, the underlying genomic events that drove the Olduvai hyperamplification in humans have remained unexplained. Here, we show that the presence or absence of an alternative first exon of the Olduvai triplet perfectly discriminates between amplified (58/58) and unamplified (0/12) triplets. We provide sequence and breakpoint analyses that suggest the alternative exon was produced by an nonallelic homologous recombination-based mechanism involving the duplicative transposition of an existing Olduvai exon found in the CON3 domain, which typically occurs at the C-terminal end of NBPF genes. We also provide suggestive in vitro evidence that the alternative exon may promote instability through a putative G-quadraplex (pG4)-based mechanism. Lastly, we use single-molecule optical mapping to characterize the intragenic structural variation observed in NBPF genes in 154 unrelated individuals and 52 related individuals from 16 families and show that the presence of pG4-containing Olduvai triplets is strongly correlated with high levels of Olduvai copy number variation. These results suggest that the same driver of genomic instability that allowed the evolutionarily recent, rapid, and extreme human-specific Olduvai expansion remains highly active in the human genome.
SEQUENCES encoding Olduvai protein domains (formerly DUF1220) (Sikela and van Roy 2017) have undergone a human-specific hyperamplification that represents the largest human-specific increase in copy number of any coding region in the genome (Popesco et al. 2006; O’Bleness et al. 2012). The current human reference genome (hg38) is reported to contain 302 haploid copies (Zimmer and Montgomery 2015), ∼165 of which have been added to the human genome since the Homo/Pan split (O’Bleness et al. 2012). Olduvai sequences are found almost entirely within the NBPF gene family (Vandepoele et al. 2005) and have undergone exceptional amplification exclusively within the primate order, with copy numbers generally decreasing with increasing phylogenetic distance from humans: humans have ∼300 copies, great apes 97–138, monkeys 48–75, and nonprimate mammals 1–8 (Popesco et al. 2006; O’Bleness et al. 2012; Zimmer and Montgomery 2015).
Olduvai copy number increase has been implicated in the evolutionary expansion of the human brain, showing a linear association with brain size, neuron number, and several other brain size-related phenotypes among primate species (Dumas and Sikela 2009; Dumas et al. 2012; Keeney et al. 2014, 2015; Zimmer and Montgomery 2015). Among humans, Olduvai copy number (total or subtype specific) has been linked, in a dosage-dependent manner, to brain size, gray matter volume, and cognitive aptitude in healthy populations (Dumas et al. 2012; Davis et al. 2015b), as well as with brain size pathologies (microcephaly/macrocephaly) (Dumas et al. 2012). Increasing Olduvai copy number has also been linearly associated with increasing severity of autism symptoms (Davis et al. 2014, 2015a, 2019), as well as increasing severity of negative schizophrenia symptoms, which are phenotypically similar to the social deficits associated with autism (Searles Quick et al. 2015). Conversely, decreasing copy number shows a linear relationship with the severity of positive symptoms of schizophrenia (e.g., hallucinations and delusions). Lending further support for a role in these disorders, 1q21.1-associated duplications and deletions that encompass many Olduvai copies are among the most common structural variations (SVs) associated with autism and schizophrenia, respectively (Brunetti-Pierri et al. 2008; Mefford et al. 2008; International Schizophrenia Consortium et al. 2009). Taken together, these findings suggest that variation in Olduvai copy number can produce both beneficial (e.g., increased brain size and cognitive function) and deleterious brain-related phenotypic outcomes (Dumas and Sikela 2009). Such a duality in function fits well with models that propose that the key genes that drove human brain evolution are also significant contributors to autism and schizophrenia (Crow 1995; Burns 2007; Dumas and Sikela 2009; Sikela and Searles Quick 2018). Finally, recent studies have shown that most human-specific Olduvai copies are encoded by four NBPF genes on 1q21.1–2, three of which are adjacent to and coregulated with three human-specific NOTCH2NL genes (Fiddes et al. 2019). The three NOTCH2NL genes have been shown to promote cortical neurogenesis (Fiddes et al. 2018; Suzuki et al. 2018), suggesting that the human-specific Olduvai expansions and their adjacent NOTCH2NL partners may work in a coordinated, complementary manner to drive human brain expansion in a dosage-related fashion (Fiddes et al. 2019).
Olduvai domains are, on average, 65 amino acids in length (ranging from 61 to 74) (Finn et al. 2014) and are encoded within a small exon and large exon doublet (Popesco et al. 2006). The domains have been subdivided into six primary subtypes based on sequence similarity: Conserved 1–3 (CON1-3) and Human Lineage-Specific 1–3 (HLS1-3) (O’Bleness et al. 2012). Sequences encoding the various subtypes are almost always found in the same order within NBPF genes: one or more CON1 domains; a single CON2 domain; one or more instances of a triplet composed of HLS1, HLS2, and HLS3 domains; and a single CON3 domain at the C-terminal end. A significant majority (114 copies) of the human-specific copies have been generated by intragenic domain amplification (i.e., increases in the number of tandemly arranged copies) primarily involving four of the 24 human NBPF genes (O’Bleness et al. 2012). These tandem expansions are predominantly organized as contiguous three domain blocks, named Olduvai triplets, each of which is ∼4.7 kb in length and composed of an HLS1, HLS2, and HLS3 subtype (O’Bleness et al. 2012). While Olduvai sequences show a human-specific range in copy number, the domains are highly copy-number polymorphic among humans, exhibiting a broad normal distribution (Davis et al. 2014). Despite the rapid and extreme nature of the Olduvai triplet expansion in the human species, the genomic factors that drove the process have remained unexplained. Similarly, the genomic mechanism that underlies the extensive Olduvai copy number variation observed within the human population remains unknown.
G-quadruplexes (G4s) are non-B-form DNA structures characterized by two or more stacked planar guanine tetrads. The characteristic sequence motif of these structures is four or more closely spaced runs of three or more guanines (Bochman et al. 2012). The guanines are stabilized by noncanonical H-bonding in a coplanar arrangement to form G-quartets and a stacked array of G-quartets makes up a G4 structure. G4 sequence motifs are overrepresented in meiotic and mitotic double-stranded breaks and are thought to promote genomic instability (Bochman et al. 2012; Maizels and Gray 2013). Here, we provide evidence supporting the view that a potential G4 motif specific to Olduvai sequences may be behind both their evolutionary hyperamplification and the high degree of variation associated with Olduvai triplets in existing humans.
Materials and Methods
Sequence analysis of predicted breakpoint regions
The coordinates of the relevant introns were obtained as described in Astling et al. (2017). Reference sequence data for the relevant introns were extracted from hg38 utilizing bedtools v2.26.0 (Quinlan and Hall 2010). The sequences were aligned with Clustal Ω (Rice et al. 2000) and the alignments were visualized with Geneious v10.2.3.
Analysis of the putative G-quadraplex sequence in nonhuman primates
Olduvai sequences in nonhuman primate genomes were located as described in Zimmer and Montgomery (2015). Assignment of each nonhuman Olduvai sequence to one of the previously described subtypes was accomplished by aligning the nonhuman sequences with the sequences of human Olduvai long exons with Clustal Ω (Rice et al. 2000), generating a percent identity distance matrix with Geneious 10.2.3, and identifying the human sequence that is the closest match to the nonhuman sequence. The nonhuman sequence was assigned the subtype of the closest human match. The locations of putative G-quadraplex (pG4) motif-containing exons had to be ascertained separately from this initial step because the search parameters only located the long exon of the Olduvai exon doublet. The locations of pG4-containing exon sequences were identified by generating a hidden Markov model based on all pG4-containing exon sequences in hg38 and using this hidden Markov model to search the nonhuman reference genomes with nhmmer. These steps were accomplished with HMMER version 3.1b2 (Eddy 2011). The locations of pG4-containing exon sequences were compared to the locations of Olduvai long exon sequences to determine which subtypes the G4-containing exons were associated with.
Optical mapping data generation and analysis
Cell lines for the 154 unrelated individuals were obtained from the Coriell Institute for Medical Research (Camden, NJ) and are part of the 1000 Genomes Project (1000 Genomes Project Consortium et al. 2015). Data for the 16 trios came from a research study of patients with rare genetic disorders consented for SV analysis. Raw Irys data were generated as described in Mak et al. (2016). Briefly, high-molecular-weight DNA was extracted from cells and fluorescently labeled at sites nicked by the nicking endonuclease Nt.BspQI. DNA was linearized and imaged by the Bionano Genomics Irys system.
Raw Irys data were assembled de novo and aligned to the reference genome (hg38) using Bionano’s default assembly pipeline. Molecules relevant to the Olduvai regions were selected with the following criteria: (1) molecules that aligned to the Olduvai regions on chromosome 1 and (2) molecules that aligned to Bionano-assembled contigs that aligned to the Olduvai regions on chromosome 1. These Olduvai-relevant molecules were realigned to chromosome 1 using RefAligner with the parameters -endoutlier 1e-2 -outlier 1e-4 -A 5 -M 1 -Mfast 0 -MultiMatches 5 -biaswt 0 -T 1e-8 -BestRef 1, as well as error values (-FP -FN -sf -sd -bpp -sr) that had been calculated during the de novo assembly process for each sample with respect to the reference genome.
The -MultiMatches 5 parameter meant that all possible alignments that met the other criteria were reported in the xmap file, as long as they had at least five labels in the reference or query that were not present in the next highest-scoring alignment. This prevented alternate alignments that highly overlapped one another (e.g., originating from the same gene) from being reported as separate alignments. We required all possible alignments to be reported at this stage, so that we could identify and filter out any molecules that aligned to multiple locations with similar confidence levels.
We performed a preview of the alignments prior to the quantification analysis. All assembled contigs with alignments across HLS regions of NBPF genes were extracted for the visualization. To identify different alleles within the populations, we clustered all contigs based on their alignment breakpoints and relative positions of nicking sites.
We identified some clusters that had high confidence scores but had misalignments at consistent positions surrounding the HLS regions. Analysis of these clusters led to the identification of alternative loci on the flanking regions of NBPF1, NBPF10, and NBPF12, which were confirmed with the visualization tool OMTools. In the population analyzed, NBPF10 and NBPF12 had only one alternative allele for each gene, so we integrated the alternative loci into the hg38 reference used for analysis as tentative alternative reference chromosomes (NBPF10_alt and NBPF12_alt). We then repeated the alignment process to identify the molecules that aligned well to the alternative loci.
We observed an extremely high amount of SV in and around NBPF1, suggesting several alternative alleles and potentially multiple copies of the NBPF1 gene. We have not yet identified a means to resolve this complexity in a manner that would allow us to confidently analyze the intragenic SV, so NBPF1 data are not reported in this paper.
To measure the sizes of the HLS region in NBPF genes, we utilized a suite of custom code (https://github.com/IleaHeft/olduvai-optical-map-analyzer). The code identifies the locations within NBPF genes where fluorescent labeling is expected to occur based on the predicted nicking locations of the endonuclease used. The positions of these expected labels in the reference genome are referred to as “reference labels of interest” (for the HLS region, these were the labels in the CON2 and CON3 sequences). The method then identifies all molecules that have been aligned to the reference labels of interest by the alignment software and performs several quality checks.
We applied a series of alignment filters as a quality control step. First, molecules were removed if their alignments had low confidence (score < 8) or appeared to map almost equally well to more than one location, or to both the original reference and an alternative locus, which we established as a difference between the highest alignment confidence score and the second-highest alignment confidence score of < 1. Second, we discarded any alignments that did not completely encompass the HLS region, which we defined as having at least two aligned labels on both flanking sides. Third, we identified and discarded all molecules with additional, unexpected labels at consistent positions within the HLS region of the molecule.
After filtering was complete, the distances between the labels of interest in each molecule were calculated. This distance is referred to as the “size call.” Because molecules originating from the same location can have slightly different size calls, molecules with size calls within 1 kb of one another are grouped together, and the median molecule length for the group is reported as the size call. The “support” for a size call is the number of molecules in the group. Size calls are normalized by the distance between the labels of interest in the reference genome to determine the amount of DNA inserted or deleted (e.g., a size call of 10 kb minus a reference distance of 5 kb would be reported as an insertion of 5 kb). Unless otherwise noted, the results presented are for instances in which one of the following conditions are met: the SV is supported by at least eight molecules or the SV is supported by four or more molecules, and those molecules represent ≥ 40% of the total molecules aligned to that gene. These conditions were selected based on changes in the total number of alleles with changing thresholds, and two alternative conditions were allowed to balance the need to reduce false positives with the need to analyze genes for which there are fewer total molecules aligned. Additionally, we have found that the general conclusions reported here are robust to variations in the number of molecules required to report a size call.
To determine the proportion of size calls that were concordant with an expected size, we performed the following steps. Expected sizes are the lengths expected to be observed from the amplification or deletion of repeat units observed in NBPF genes in hg38. In the HLS region, the most common repeat is of the sequence encoding an HLS triplet (HLS1, HLS2, and HLS3). In addition to repeats of HLS triplets, in hg38, there are a few HLS doublets. Therefore, HLS region sizes that could result from the amplification or deletion of repeat units observed in hg38 could include multiples of the HLS triplet length, multiples of the HLS doublet length, or a combination of the two. The various combinations of triplets and doublets all produce size changes that are integer multiples of a single HLS unit. Therefore, expected SV sizes for each gene were calculated by determining the mean length of a single HLS exon unit for each gene and generating integer multiples of that value. The proportions of SVs falling within 300 bp of an expected size were calculated for SVs that were supported by at least eight molecules or supported by four or more molecules when those molecules represent 40% or more of the total molecules aligned to that gene. Additionally, we required observed deviations from the reference to be at least 1-kb different from the reference size to be called as an SV.
Circular dichroism spectroscopy
An oligonucleotide containing the pG4 motif (5′-GGGGAAGGGGAAGAAAAGAAGGGGAAGAAGATCAAAGAAGGAAAGAAGAAGGGGAAGAAAAGAAGGGG-3′) was purchased from Integrated DNA Technologies, and all experiments were carried out in 10 mM NaPi buffer with 50 mM KCl or 50 mM NaCl. Next, 4 µM of oligonucleotide was heated at 95° for 10 min and then cooled slowly to room temperature (22°) overnight for circular dichroism (CD) analyses the following day. CD measurements were performed on a JASCO J-815 CD Spectrometer (JASCO Inc., Easton, MD) with a 1-mm path length quartz cuvette. CD spectra were collected across 340–200 nm in 1-nm increments, and the reported spectra correspond to the average of at least three scans. The conformation of the motif was determined by spectral investigation at 25°, and an appropriate buffer blank correction was made for all spectra.
Fluorescence analysis of nuclease footprinting assay
Double-stranded DNA (dsDNA) templates containing differential fluorescent labels at each terminus were generated using PCR on genomic DNA. The primers used were as follows: forward, /56-FAM/CTGTTGCCTCCAGGTGTTA and reverse, /5HEX/AGCTTAATGTGTCTGTCCATG. PCR amplification was carried out in a total reaction volume of 25 μl containing 1 × PCR reaction buffer with 1.5 mM MgCl2 (Roche Diagnostics), 0.5 μM of each primer (Integrated DNA Technologies), 0.2 μM of each dNTP, 1 M betaine, 0.5 U Fisher Taq-ti polymerase (Fisher Biotec, Wembley, Australia), and ∼20 ng of human genomic DNA. Touchdown PCR (Korbie and Mattick 2008; Roux 2009) was performed over 30 cycles and consisted of an initial denaturation step of 95° for 2 min and a final extension of 72° for 5 min. Cycling conditions consisted of denaturation at 95° for 30 sec and extension at 72° for 45 sec. The initial annealing temperature was 65°, and this was decreased by 1° per cycle for 10 cycles, followed by 20 cycles at 55°.
PCR products were then visualized by gel electrophoresis, and the target band was extracted and purified using a MEGA-Quick Spin Kit (iNtRON Biotechnology, Seongnam-si, South Korea) according to the manufacturer’s recommendations. Sanger DNA sequencing was carried out on nonfluorescent purified PCR amplicons (∼10 ng) with the appropriate primer. Primer sequences were as shown above but lacking fluorescent labels. Sequencing reaction products were generated using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), following the manufacturer’s protocol, and were run on an AB3130xl fragment analysis system equipped with a 50-cm capillary array using POP7 polymer.
To investigate the DNA conformation, 150 ng of purified fluorescent PCR products were resuspended in 10 mM Tris (pH 8.0), 1 mM EDTA, and 100 mM of relevant ion, KCl or LiCl (negative control). One of two incubation steps was then performed: incubation at 95° for 5 min followed by controlled cooling to room temperature at a rate of 0.2° per min or incubation at 37° for 36 hr (no heat denaturation). Prepared templates were then incubated with 1 unit of the single-strand-specific endonuclease Mung Bean nuclease (New England Biolabs, Beverly, MA) in appropriate buffer at 30° for 11 min in an 80-μl reaction volume. This reaction was terminated by the addition of phenol:chloroform at a 1:1 ratio. After vortexing and centrifugation at 14,000 × g for 5 min, the aqueous phase was removed and concentrated using a centrifugal evaporator. The lyophilized pellet was then resuspended in 20 μl deionized formamide, and 10 μl of the sample was assessed by automated capillary electrophoresis. To determine if there was any non-B structure in the PCR product (before the addition of ions), an additional control was performed in which the Mung Bean nuclease was added directly to purified fluorescent PCR product (resuspended in Tris and EDTA as described above). Only signals of ≥ 50 relative fluorescence units (RFUs) are shown, as peaks with an RFU value < 100 are likely to correspond to single-stranded DNA (ssDNA) generated as an artifact during template synthesis.
Fluorescent molecules were sized by automated capillary electrophoresis. Reaction products were prepared in deionized formamide containing the size standard GS500LIZ. Samples were heated at 95° for 5 min to denature the DNA, and capillary electrophoresis was then performed on an AB3130xl fragment analysis system equipped with a 50-cm capillary array using POP7 polymer. To confirm the fragment length of the PCR product before any ions or nuclease were added, the purified PCR product (without any other treatment) was also sized as described above.
Raw data were visualized with Applied Biosystems Peak Scanner v2.0 software and subsequently exported into Microsoft Excel (2013). The inferred nucleotide length of each peak was rounded to the nearest whole number, and peaks of a size less than or equal to the primer were removed. A minimum of three replicates were performed for each experiment and the RFUs were averaged at each nucleotide position.
Data availability
Cell lines used in this study as a source of genomic DNA can be obtained from the Coriell Institute for Medical Research and are part of the 1000 Genomes Project. Individual samples and their identification numbers are listed in Supplemental Material, Table S7. The 16 trio samples have been consented for research at the University of California, San Francisco, and corresponding DNAs were sent to P.-Y. Kwok anonymously under his local IRB approval. Data corresponding to the samples used in this study are being deposited in public databases at the National Center for Biotechnology Information. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10673792.
Results
A pG4 motif is perfectly correlated with the human-specific intragenic amplification of Olduvai triplets
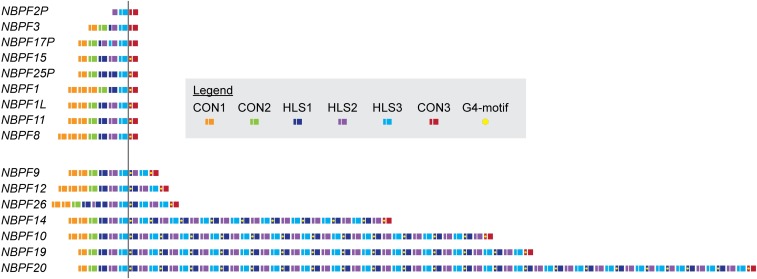
Seventeen of 24 human NBPF genes in hg38 contain at least one Olduvai triplet or doublet in hg38. The seven remaining genes encode only two to five Olduvai copies per gene (22 in total). Because doublets are relatively rare in hg38 [9 doublets out of 74 total (triplet or doublet) HLS repeats], we will here use triplets to refer to both arrangements. As depicted in Figure 1, in all NBPF genes with more than one triplet, the second-nth triplets contain a pG4 motif (Figure 2) in their first exon. This sequence is not present in the first (ancestral) triplet of any NBPF gene. The pG4 motif-containing exon is almost always 108 bp in length and is found in 70 locations in hg38: 0/12 ancestral (first) triplets, 58/58 amplified triplets, and 12/16 CON3 domains (Figure S1). Thus, the presence or absence of the pG4 motif in the first exon of Olduvai triplets perfectly discriminates between amplified and ancestral (unamplified) triplets.
Figure 1.
The location of Olduvai sequences and the pG4 motif within NBPF genes in hg38. Genes are grouped into clusters based on whether they contain intragenic amplifications of HLS sequences (bottom cluster) or not (top cluster). The vertical black line and the alignment of the genes are intended to visually highlight the occurrence of the pG4 motif in amplified HLS triplets (to the right of the vertical black line) and the absence of the motif in ancestral HLS triplets/doublets (to the left of the vertical black line). Each Olduvai-encoding exon doublet is represented by a narrow and wide rectangle to indicate the short and long exon, respectively. A wide rectangle without a corresponding narrow one represents instances where the reference contains only the long exon of an Olduvai-exon doublet. The color of each rectangle corresponds to the subtype to which that sequence has been assigned. The presence of the pG4 motif is indicated by a yellow hexagon. For clarity, exons and genes that are not Olduvai encoding that lack the CON3 sequence are not shown. CON3, Conserved 3; HLS, Human Lineage-Specific; pG4, putative G-quadraplex.
Figure 2.
The pG4 motif found in NBPF genes. The portion of Olduvai short exons that is thought to be involved in the formation of the pG4 structure. This exact sequence is found in 56/58 HLS1 pG4-containing exons. The two exceptions differ by only a single nucleotide. With respect to CON3 pG4-containing exons, this exact sequence is found in 3/13 exons, 6/13 have only a single-nucleotide difference, 1 differs by 2 nucleotides, 1 differs by 4, and 2 differ by 5. The five runs of guanines thought to contribute to formation of the G4 structure are shaded in green. CON3, Conserved 3; HLS, Human Lineage-Specific; pG4, putative G-quadraplex.
Analysis of the pG4 sequence in nonhuman primates
We examined whether the pattern observed in the human genome was also maintained in other primate genomes. None of the genes in the chimpanzee or gorilla reference genomes (panTro3-5 and gorGor3-5) contain any perfect triplet amplifications. Given this lack of triplet amplification, we would conclude that the pattern was consistent with that observed in humans if the sequence of the pG4-containing exon was only found in association with CON3 long exons. Using the two most recent chimp and gorilla assemblies (panTro5 and gorGor5), we found no amplified triplets and no pG4-containing exon sequences outside of CON3 subtypes. While this is consistent with predictions of our model, there are some ambiguities on these points in previous chimp and gorilla assemblies (Tables S1–S6). Such uncertainty is not surprising, as it is well known that complex, highly duplicated sequences such as the Olduvai/NBPF family are not well handled by most mammalian genome assemblies. Given these limitations, we have chosen to not delve into questions related to the specific organization and evolution of these sequences in nonhuman primates at this time.
Breakpoint analysis supports a nonallelic homologous recombination-mediated mechanism for the generation of pG4-containing Olduvai triplets
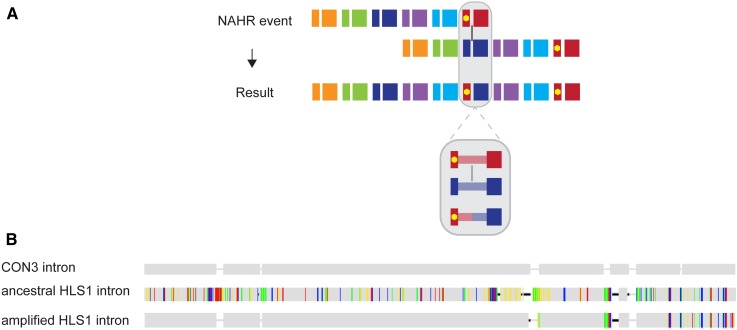
All pG4-containing exons within amplified Olduvai triplets are identical (or nearly so) to those within CON3 sequences. Because even genes with no amplified HLS sequences have the pG4 motif in their CON3 exons, the origin of the pG4-containing Olduvai triplet could be explained by a nonallelic homologous recombination event with breakpoints in the introns of the CON3 exon doublet and the ancestral HLS1 exon doublet (Figure 3A). This event would have duplicated an Olduvai triplet and replaced the original first exon with the pG4-containing first exon from a CON3, producing the organization observed in hg38.
Figure 3.
Proposed mechanism for amplification of pG4-containing HLS triplets (A) and supporting sequence data (B). (A) Two alleles of an ancestral NBPF gene are shown at the top of the figure, aligned in the manner that would produce the intragenic amplifications observed in the current human refence genome (hg38). Recombination is proposed to occur between the introns of the CON3 and HLS1 exon doublets, as indicated with the vertical black line. A zoomed-in view of the shaded regions is shown on the bottom of (A) depicting that a nonallelic homologous recombination event, as proposed here, would be expected to produce an intron that resembles the CON3 intron at its 5′ end and transitions to resembling the ancestral HLS1 intron toward its 3′ end. (B) Visual representation of sequence similarities between the introns of the CON3, ancestral HLS1, and amplified HLS1 introns in NBPF12 shows agreement with the expected pattern. Gray regions represent identical nucleotides and colored bars represent nucleotide differences from the CON3 intron. CON3, Conserved 3; HLS, Human Lineage-Specific; pG4, putative G-quadraplex.
If this were the case, we would expect to observe a certain sequence pattern in introns of amplified HLS1 exon doublets that would reflect the predicted breakpoint. Specifically, we would expect the 5′ ends of HLS1 introns to resemble the introns of pG4-containing CON3 sequences and the 3′ ends to resemble the introns of the first (i.e., ancestral) HLS1 sequences of each gene. Analysis revealed that the sequences of the relevant introns are consistent with this expectation (Figure 3B). Alignment of all relevant introns can be found in Figure S2. The same mechanism responsible for the generation of the first pG4-containing Olduvai triplet may be responsible for the continued amplification and deletion of Olduvai repeats.
The pG4 motif is strongly associated with interindividual intragenic SV due to amplification and deletion of Olduvai repeats
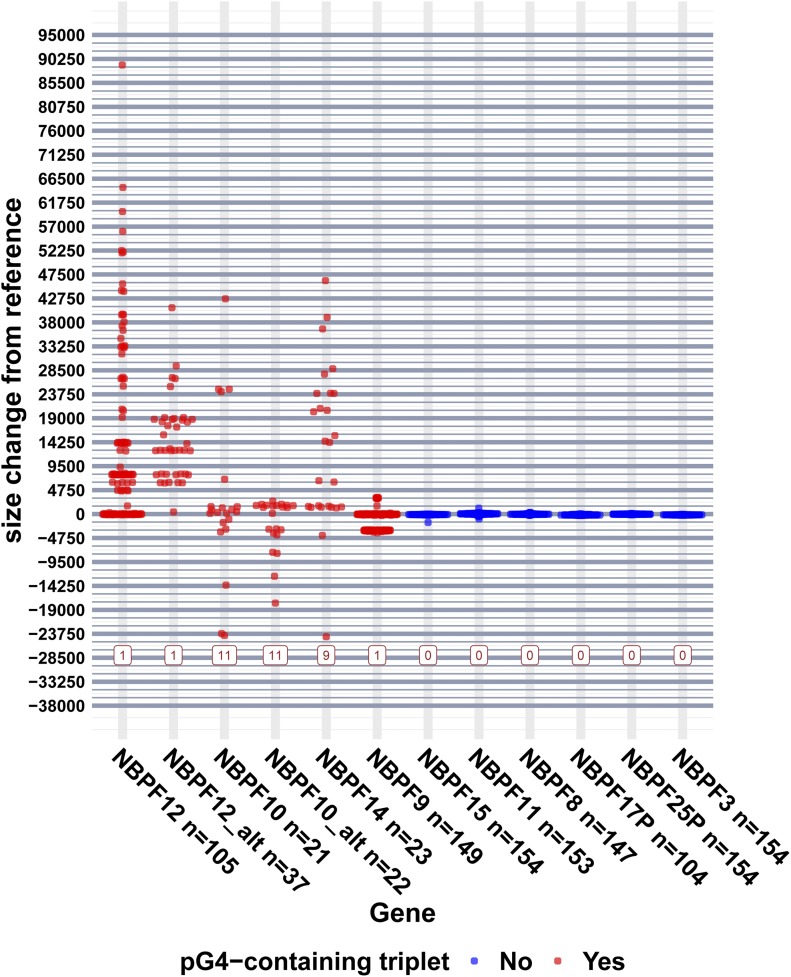
We applied Bionano Genomics optical mapping technology to analyze Olduvai-related SV in 154 unrelated individuals from 26 populations (Table S7). Single-molecule maps were used to measure the lengths of the HLS regions in different NBPF genes (Figure 4). Our results provide strong evidence of common and extensive intragenic SV involving Olduvai triplets in a subset of NBPF genes (Figure 4), and the pattern is perfectly consistent with the pG4-containing Olduvai triplets being the primary driver of variation. For the 10 NBPF genes for which sufficient data were obtained, intragenic variation was detected for four out of four genes that have one or more pG4-containing Olduvai triplets in hg38, while variation was not detected for six out of six genes that contain only a single ancestral (non-pG4) triplet.
Figure 4.
Structural variation in the HLS region of NBPF genes detected by optical mapping. Size calls are shown normalized by the reference size for each gene. Positive size changes are insertions, and negative changes are deletions relative to the reference. The interval between thick vertical lines is the median size of an HLS triplet, and the interval between thin vertical lines is the median size of a single HLS domain. The boxed numbers along the bottom of the plot denote the number of pG4-containing triplets in that gene in the current human refence genome (hg38). On the x-axis labels, the value following “n” is the number of samples for which data are shown. If only one size allele was detected for a sample, that data point was duplicated so that alleles in homozygous individuals are not underrepresented. The color of the data point denotes whether the gene has a pG4-containing triplet in hg38. HLS, Human Lineage-Specific; pG4, putative G-quadraplex.
The precise extent of variation differed slightly depending on the parameters used to decide if SV was truly present. However, the general conclusions reported below were robust to changes in the number of DNA molecules required to call an SV. The results of varying the required number of supporting molecules from 2 to 14 are shown in Figure S3. The results presented here are for instances in which one of the following conditions were met: the SV was supported by at least eight molecules or the SV was supported by four or more molecules, and those molecules represented ≥ 40% of the total molecules aligned to that gene. The 14 NBPF genes that were analyzed with the optical mapping data set contained HLS domains flanked by CON2 and CON3 domains. Four genes (NBPF9, NBPF10, NBPF12, and NBPF14) showed strong evidence of common intragenic SV in the HLS region, and six (NBPF3, NBPF8, NBPF15, NBPF11, NBPF17P, and NBPF25P) showed no evidence of intragenic SV in the HLS region for the population studied (Figure 4). No or very little data were obtained for three of the remaining genes (NBPF1L, NBPF19, and NBPF20), and NBPF1 was excluded from the analysis due to high complexity in the flanking regions that made it difficult to make accurate SV calls. It is important to note that the number of samples for which data could be obtained varied between genes. This variability was driven in part by differences in gene length, with longer genes being harder to span with single molecules. For that reason, care should be taken in comparing the level of variability observed in different genes (e.g., data points for NBPF14 could only be obtained for 23 samples, but data for NBPF9 are shown for 143 samples).
As mentioned previously, little or no data were obtained for NBPF1L, NBPF19, and NBPF20. Data were not obtained for NBPF1L because it is located on an unplaced contig and the alignment strategy used for this analysis focused only on chromosome 1. NBPF19 and NBPF20 contain extreme Olduvai triplet expansions that make them among the longest members of the NBPF family, which likely made it difficult for a sufficient number of molecules to align across them.
The SV sizes observed for the pG4-containing genes clustered at regular intervals, suggesting that the observed SVs in the population studied may be due to amplifications/deletions of similar sequences. Most measured SV sizes line up very well with the sizes predicted to result from amplifications or deletions of Olduvai triplets, doublets, or a combination of the two (e.g., an added doublet plus one or more additional triplets). The possibility of SV due to insertion or deletion of either doublets or triplets is supported by the hg38 reference sequences for the NBPF genes, which contain mostly triplets, as well as several doublets (Figure 1).
For NBPF9, 100% (60/60) of the SV calls were within 300 bp of the size predicted from amplification or deletion of an Olduvai doublet. The version of NBPF9 in hg38 contains a single ancestral Olduvai triplet plus an amplified doublet. The observed NBPF9 SVs are consistent with amplification and deletion of that doublet unit.
For NBPF12, NBPF10, and NBPF14, 94% (30/32), 69% (165/239), and 61% (127/208) of SV calls, respectively, were within 300 bp of a size predicted to result from Olduvai triplet amplifications, or a combination of triplet and doublet amplifications. A combination of triplet and doublet amplification/deletion is consistent with the reference versions of NBPF10 and NBPF14, both of which contain at least one doublet in their HLS region.
One explanation for SV sizes differing from expectation is that pG4 motifs may inhibit the ability of the molecule to fully linearize and therefore could distort the measurement of the distance between two labels. If pG4 motifs are a factor, genes with greater numbers of these motifs are likely to be affected to a greater degree. This may explain why the percentages of SV calls within 300 bp of expectation were lower for NBPF10 and NBPF14, as these genes have 12 and 10 pG4 motifs, respectively. It is also possible that other factors may be influencing SV sizes, such as the insertion or deletion of a sequence other than Olduvai (e.g., a transposable element).
De novo SV is observed for NBPF12, a gene with a pG4-containing Olduvai triplet
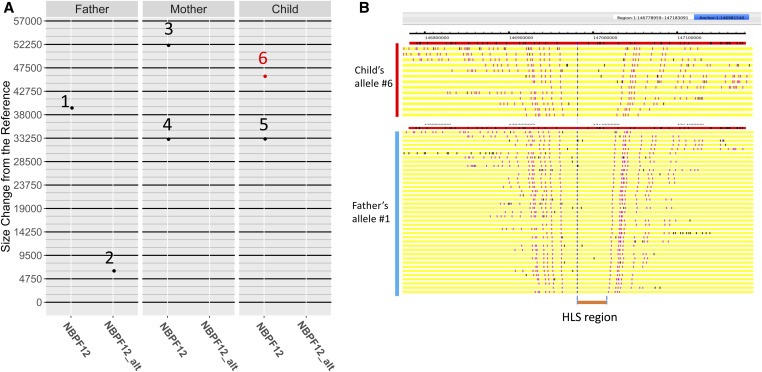
An analysis of 16 parent–child trios with the Bionano Genomics single-molecule optical mapping approach revealed evidence of de novo SV in the HLS region of NBPF12. As shown in Figure 5, a de novo insertion was found in the NBPF12 gene of the child in one trio. Specifically, the length of one of the child’s NBPF12 HLS regions was not consistent with the lengths of the HLS region observed in either parent. The child’s other allele was consistent with one of the maternal alleles, suggesting that the size of the child’s discordant allele resulted from a de novo mutation within the inherited paternal allele. The difference between the child’s discordant HLS region size and the most similar paternal allele was 6412 bp, consistent with the insertion of two Olduvai doublets (expected size is ∼6280 bp). Among the 154 unrelated individuals in this study, SVs that were multiples of ∼6280 bp were relatively common in NBPF12.
Figure 5.
Suggestive evidence of de novo variation in the HLS region of NBPF12. (A) The de novo insertion event found in a trio on NBPF12. All alleles of the trio on either the current human refence genome (hg38) NBPF12 reference or the NBPF12_alt reference are plotted and labeled with numbers 1–6. The y-axis shows size calls for the HLS region, and each column denotes the alleles on the hg38 NBPF12 reference or NBPF12_alt reference for each sample in the trio. The median size of a single HLS domain (1583 bp) is used as the minor grid, and the median size of amplified HLS triplets (4750 bp) is used as the major grid. The de novo allele is labeled as “6” and highlighted in red. One of the child’s alleles (5) appears to be inherited from the mother’s allele (4). Considering that one of the paternal alleles (2) is on the alternative locus, while neither of the child’s are, the child’s de novo allele (6) likely originates from the other paternal allele (1). (B) The optical map single-molecular alignments of the de novo allele and its original paternal allele, visualized in OMTools. As indicated on the left side of the figure, the yellow bars grouped toward the top are single molecules from the de novo allele, and the yellow bars in the lower group are from the original paternal allele. Labels colored with red denote the aligned nicking sites while the labels colored with black denote unaligned sites. The two labels used to identify the HLS region are shown. HLS, Human Lineage-Specific.
The pG4 motif forms a G4 structure in vitro
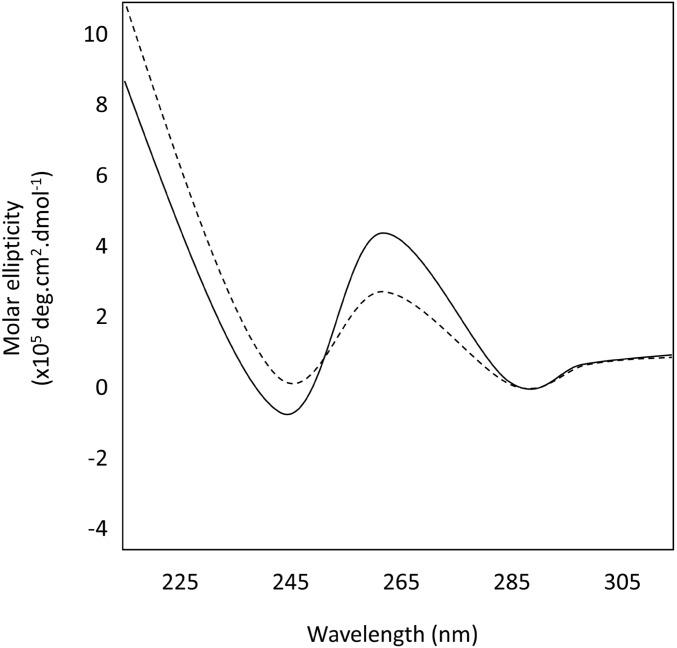
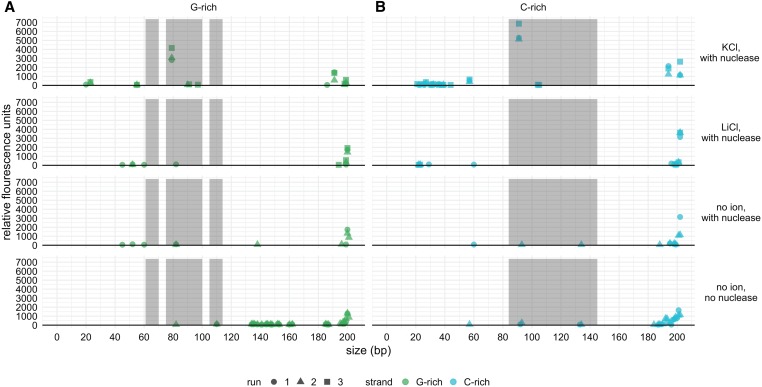
The results of both CD spectroscopy (Figure 6) and fluorescence analysis of nuclease footprinting assays (FANFA) (Figure 7) (Stevens et al. 2016) support the ability of the Olduvai pG4 motif to form a G4 structure in vitro. CD spectroscopy is a commonly used technique for assessing the G4-forming potential of single-stranded oligonucleotides in differing ionic conditions. Using this technique, G4 formation produces a characteristic spectral signature (Kypr et al. 2009; Randazzo et al. 2012), which is distinguishable from nonstructured ssDNA or dsDNA. This spectral signature originates from the specific Hoogsteen bonds formed during G4 formation, where parallel stranded G4s are identified by a trough at 245 nm and a peak at 265 nm. In the presence of both K+ and Na+, the Olduvai sequence demonstrated CD profiles representative of parallel stranded G4s (Figure 6) (Kypr et al. 2012). In the FANFA analysis, cleavage of the dsDNA template was only observed when the template DNA was incubated in KCl, which is known to promote the stability of G4 structures. The cleavage positions are consistent with the expected regions of ssDNA on both the G-rich and C-rich strands (Figure 7). Cleavage of the DNA was observed at the same positions both when the template was heat denatured (enhancing the likelihood of structure formation by separating the dsDNA) and when it was incubated with KCl at 37° for 36 hr. However, the quantity of cleaved DNA products and therefore of structure formation appeared to be less in the nonheat-denatured condition. The appearance of cleaved DNA in the nonheat-denatured condition suggests that, under the conditions of the experiment, the dsDNA was able to transition to the G4 structure. We confirmed that the cleaved DNA lengths in the KCl conditions were not present in the starting DNA template (no ions, no nuclease) and that there was no structure present in the template prior to the addition of KCl (no ions, with nuclease) (Figure 7).
Figure 6.
Results of circular dichroism spectroscopy performed on single-stranded oligonucleotides containing the pG4 motif. Solid line represents the spectra obtained in 50 mM KCl and the broken line represents the spectra obtained in 50 mM NaCl. Molar ellipticity (×105 deg.cm2.dmol−1) is on the y-axis and wavelength (nm) on the x-axis. These spectra are consistent with formation of a parallel-stranded G4 in the presence of potassium ions. pG4, putative G-quadraplex.
Figure 7.
The abundance of DNA fragment sizes resulting from the fluorescence analysis of nuclease footprinting assays under varying conditions supports G4 formation in the presence of KCl. The lengths of DNA fragments following each treatment are represented on the x-axis in nucleotides, and the abundances of each fragment are represented by relative fluorescent units on the y-axis. Full-length template DNA is represented by peaks at the far right of the x-axis, and results are shown separately for the G-rich (A) and C-rich strands (B). Shaded regions denote the expected locations of single-stranded DNA (loops) during G4 formation. The G-rich strand (A) is expected to have three single-stranded loop regions (from left to right: loop 1, loop 2, and loop 3). Data are shown for three replicates (run 1, run 2, and run 3) for the KCl and LiCl conditions, and for two replicates for the no ion control conditions. G4, G-quadraplex.
Discussion
The role of the CON3 short exon in human-specific hyperamplification of Olduvai domains
The human-specific hyperamplification of sequences encoding Olduvai protein domains represents one of the most extreme examples of evolutionarily rapid genomic change. The striking nature of this coding region increase is consistent with it serving an important evolutionary role in a key human-specific phenotype, and such an expectation is supported by the multiple reports that have implicated Olduvai copy number expansion in brain size and cognition (Dumas and Sikela 2009; Dumas et al. 2012; Keeney et al. 2014, 2015; Davis et al. 2015a, b; Zimmer and Montgomery 2015). The link with brain expansion has been given additional support by the recent finding that human-specific Olduvai expansions may function in a coordinated and complementary manner with adjacent human-specific NOTCH2NL genes to promote cortical neurogenesis (Fiddes et al. 2019). Although it has been established that the human increase was primarily due to tandem intragenic increases involving the Olduvai triplet rather than duplications of entire genes (O’Bleness et al. 2012, 2014), the genomic events responsible for this increase have until now remained unknown. Here, we report a specific mechanism through which the expansions are likely to have occurred. Specifically, we show that (1) the presence or absence of a pG4-forming sequence in the first exon of an Olduvai triplet perfectly discriminates between amplified and unamplified triplets, and (2) the results obtained by CD and FANFA support the formation of a G4 in vitro. These results point to the conclusion that the intragenic duplicative transposition of the pG4 motif-containing exon from the CON3 domain into HLS triplets in specific human NBPF genes generated a highly unstable local genomic environment within each gene that, in conjunction with strong selection pressures favoring additional copies, resulted in the repeated tandem addition of Olduvai triplets to the human genome.
Mechanism of instability
G4 structures are reported to promote instability in a number of ways. During transcription of the pG4 sequence, the formation of a G4 structure on the nontranscribed strand may promote R-loop stabilization (Aguilera and Garcia-Muse 2012; Kim and Jinks-Robertson 2012). Several mechanisms have been proposed for how R-loop formation may facilitate the formation of DNA breaks that subsequently lead to homologous recombination-based repair processes. These include prolonged exposure of ssDNA to endonucleases and DNA-damaging agents (Li and Manley 2006), the collision of transcription/replication machinery (Santos-Pereira and Aguilera 2015), and cleavage by proteins of the transcription-coupled nucleotide excision repair pathway as part of the normal process to resolve R-loops (Sollier et al. 2014; Stirling and Hieter 2017). Break-induced replication is a homology- or microhomology-dependent mechanism that can be used to repair DSBs, particularly those formed in the context of DNA replication (Sakofsky and Malkova 2017). If a directly oriented nonallelic homologous sequence is used to restart the replication fork, then deletions or duplications of the intervening DNA can be generated (Carvalho and Lupski 2016).
Potential chronology of the pG4-mediated human-specific Olduvai hyperamplification
There are a number of possible ways in which the Olduvai triplet copy number could be increased in the human lineage, e.g., pG4-mediated intragenic triplet expansion, duplication of an already expanded NBPF gene, or a combination of both. The great majority of expanded Olduvai triplets in the reference genome can be found in four human genes (NBPF10, NBPF14, NBPF19, and NBPF20) (Figure 1), while single triplet expansions are seen in NBPF9, NBPF12, and NBPF26. Given our findings, we believe that the initial intragenic expansion of pG4-containing triplets occurred in one or more of these genes. Recent studies have begun to sort out some of the likely scenarios that could have led to the human-specific Olduvai triplet expansions. These reports indicate that three of the four most highly expanded NBPF genes (NBPF10, NPBF14, and NBPF19) are adjacent to three human-specific NOTCH2NL genes (O’Bleness et al. 2014; Fiddes et al. 2018; Suzuki et al. 2018). These NBPF/NOTCH2NL gene pairs evolved jointly as two-gene units and transcriptionally are tightly coregulated (Fiddes et al. 2019). Additional analyses of these events indicate that most of the extreme human-specific Olduvai expansions (i.e., those encoded by NBPF10, NBPF14, and NPBF19) may have appeared between 0.4 and 3 MYA (Fiddes et al. 2019). This timeframe coincides with the period during which the human brain is thought to have undergone its most extreme expansion (Du et al. 2018).
The role of the pG4-containing short exon in the extreme Olduvai copy-number variability in existing human populations
As described in this manuscript, a subset of NBPF genes display common and extensive intragenic variation specifically in the regions of the genes that encode Olduvai triplets. In the population studied and with the parameters described in this manuscript, there is a perfect correlation between the presence of the pG4-containing short exon in an HLS triplet and interindividual variation in the Olduvai region of NBPF genes. Furthermore, the SV sizes measured are highly consistent with amplification or deletion of Olduvai triplet repeats. These results suggest that the pG4-containing exon may be driving SV in the extant human population. In addition to the extensive interindividual variation, we also observed evidence of de novo variation, which further supports the proposal that the pG4-containing exon continues to promote new genotypic (and possibly phenotypic) diversity in the population.
The extensive SV reported here for NBPF and Olduvai sequences is consistent with previous genome-wide studies that identified 1q21.1–2 as a complex and highly variable part of the human genome (Fortna et al. 2004; Levy-Sakin et al. 2019). The results described here extend this view, placing NBPF genes among the genes with the most variable intragenic coding regions known (Warburton et al. 2008). In the 154 unrelated individuals studied, we observed SV suggesting a range of 1–20 Olduvai triplet repeats in NBPF12, 4–19 repeats in NBPF14, 6–20 repeats in NBPF10, and 0–1 repeat in NBPF9. This striking interindividual variation in Olduvai triplet repeat copy number represents a rich source of previously unrecognized allelic diversity. Other genes known to have a large range of protein domain repeat copy numbers in nondiseased individuals include the LPA gene (1–40 copies of the Kringle IV repeat) (Kronenberg 2016), the DMBT1 gene [7–20 copies of the scavenger-receptor cysteine-rich (SRCR) domain] (Polley et al. 2015), and PRDM9 (8–18 zinc finger repeats) (Berg et al. 2010). Intragenic copy number variation in the LPA gene has been associated with lipoprotein (a) levels (Lanktree et al. 2010), and variation in PRDM9 has been associated with recombination hotspot activity (PRDM9) (Berg et al. 2010).
Relevance to human disease and evolution
By generating an unstable local genomic architecture within specific human NBPF genes, the duplicative transposition of the Olduvai pG4 sequence, as described here, is likely to have been both beneficial and harmful. Such a possibility is supported by several reports (Dumas et al. 2012; Davis et al. 2015a; Searles Quick et al. 2015), and a model has been proposed in which harmful effects of Olduvai variation have been, and continue to be, the cost evolution has placed on the emergence of the human brain (Sikela and Searles Quick 2018). At the genomic level, our ability to distinguish between harmful and beneficial Olduvai copy number variations is currently limited and, except in rare instances (Dumas et al. 2012; Davis et al. 2014, 2015a, b; Searles Quick et al. 2015), conventional genomic disease studies do not directly measure Olduvai sequences. Also, given the unusual genomic organization of the NBPF and Olduvai sequences, there are a myriad of ways in which these sequences can recombine. Thus, it may be that which, where, how, and when copies are changing determine whether the phenotypic effects are beneficial, innocuous, or harmful (Sikela and Searles Quick 2018). Sorting out the relative contributions of these variables can be expected to be an important future direction for Olduvai research.
In summary, the findings reported here describe a possible genomic mechanism, addition of Olduvai copies via pG4-mediated expansion of Olduvai triplets, that produced the hyperamplification of the Olduvai domain in the human lineage. Thus, the duplicative transposition of the pG4 motif, an instance of genomic serendipity, may have been the initial trigger behind one of the most extreme copy number expansions in the human genome. In addition, we show that this mechanism appears to remain highly active in present-day human populations. The pG4 mechanism would be expected to generate local genomic instability within NBPF genes that would lead to both gains and losses in Olduvai copy number, which we observe in our analysis of the extant population. However, the large human-lineage-specific increase in Olduvai copy number suggests that a strong selection bias favoring copy gains was in place over the 5–7 MY during which the human lineage emerged. The studies linking increased Olduvai dosage to increased measures of brain size, including the recent pairing of Olduvai expansions with adjacent NOTCH2NL genes, are consistent with the possibility that such selection pressures may have been related to improved cognitive function.
Acknowledgments
We thank Michael Dickens, Kirk Hansen, and Majesta O’Bleness for helpful discussions. Support for this work was provided by National Institutes of Health (NIH) grants R01 MH-108684 (J.M.S. and P.-Y.K.), R01 HG-005946 (P.-Y.K.), and T32 HL-007731 (Y.M.); Simons Foundation (SFARI) award 309230 (J.M.S.); The California Initiative to Advance Precision Medicine (D.I.M., D.B., and P.-Y.K.); and The Jim and Mary Carney Charitable Trust and Marsden Fund (11-UOO-175 BMS) administered by the Royal Society of New Zealand (M.A.K. and A.J.S.).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.10673792.
Communicating editor: M. Hahn
Literature Cited
- Aguilera A., and Garcia-Muse T., 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46: 115–124. 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Astling D. P., Heft I. E., Jones K. L., and Sikela J. M., 2017. High resolution measurement of DUF1220 domain copy number from whole genome sequence data. BMC Genomics 18: 614 10.1186/s12864-017-3976-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg I. L., Neumann R., Lam K. W., Sarbajna S., Odenthal-Hesse L. et al. , 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42: 859–863. 10.1038/ng.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M. L., Paeschke K., and Zakian V. A., 2012. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13: 770–780. 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N., Berg J. S., Scaglia F., Belmont J., Bacino C. A. et al. , 2008. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 40: 1466–1471. 10.1038/ng.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J., 2007. The Descent of Madness: Evolutionary Origins of Psychosis and the Social Brain. Routledge, London. [Google Scholar]
- Carvalho C. M. B., and Lupski J. R., 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17: 224–238. 10.1038/nrg.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow T. J., 1995. A continuum of psychosis, one human gene, and not much else -- the case for homogeneity. Schizophr. Res. 17: 135–145. 10.1016/0920-9964(95)00059-U [DOI] [PubMed] [Google Scholar]
- Davis J. M., Searles V. B., Anderson N., Keeney J., Dumas L. et al. , 2014. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet. 10: e1004241 (erratum: PLoS Genet. 10: e10043743). 10.1371/journal.pgen.1004241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. M., Searles Quick V. B., and Sikela J. M., 2015a Replicated linear association between DUF1220 copy number and severity of social impairment in autism. Hum. Genet. 134: 569–575. 10.1007/s00439-015-1537-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. M., Searles V. B., Anderson N., Keeney J., Raznahan A. et al. , 2015b DUF1220 copy number is linearly associated with increased cognitive function as measured by total IQ and mathematical aptitude scores. Hum. Genet. 134: 67–75. 10.1007/s00439-014-1489-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. M., Heft I., Scherer S. W., and Sikela J. M., 2019. A third linear association between Olduvai (DUF1220) copy number and severity of the classic symptoms of inherited autism. Am. J. Psychiatry 176: 643–650. 10.1176/appi.ajp.2018.18080993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A., Zipkin A. M., Hatala K. G., Renner E., Baker J. L. et al. , 2018. Pattern and process in hominin brain size evolution are scale-dependent. Proc. Biol. Sci. 285: 20172738. 10.1098/rspb.2017.2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L., and Sikela J. M., 2009. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harb. Symp. Quant. Biol. 74: 375–382. 10.1101/sqb.2009.74.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L. J., O’Bleness M. S., Davis J. M., Dickens C. M., Anderson N. et al. , 2012. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am. J. Hum. Genet. 91: 444–454. 10.1016/j.ajhg.2012.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R., 2011. Accelerated profile HMM searches. PLOS Comput. Biol. 7: e1002195 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes I. T., Lodewijk G. A., Mooring M., Bosworth C. M., Ewing A. D. et al. , 2018. Human-specific NOTCH2NL genes affect Notch signaling and cortical neurogenesis. Cell 173: 1356–1369.e22. 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes I. T., Pollen A. A., Davis J. M., and Sikela J. M., 2019. Paired involvement of human-specific Olduvai domains and NOTCH2NL genes in human brain evolution. Hum. Genet. 138: 715–721. 10.1007/s00439-019-02018-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y. et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G. et al. , 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2: e207 10.1371/journal.pbio.0020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium, Purcell S. M., Wray N. R., Stone J. L., Visscher P. M. et al. , 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460: 748–752. 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J. G., Dumas L., and Sikela J. M., 2014. The case for DUF1220 domain dosage as a primary contributor to anthropoid brain expansion. Front. Hum. Neurosci. 8: 427 10.3389/fnhum.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J. G., Davis J. M., Siegenthaler J., Post M. D., Nielsen B. S. et al. , 2015. DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Struct. Funct. 220: 3053–3060. 10.1007/s00429-014-0814-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., and Jinks-Robertson S., 2012. Transcription as a source of genome instability. Nat. Rev. Genet. 13: 204–214. 10.1038/nrg3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbie D. J., and Mattick J. S., 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 3: 1452–1456. 10.1038/nprot.2008.133 [DOI] [PubMed] [Google Scholar]
- Kronenberg F., 2016. Human genetics and the causal role of lipoprotein(a) for various diseases. Cardiovasc. Drugs Ther. 30: 87–100. 10.1007/s10557-016-6648-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypr J., Kejnovská I., Renciuk D., and Vorlickova M., 2009. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37: 1713–1725. 10.1093/nar/gkp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypr J., Kejnovská I., Bednarova K., and Vorlickova M., 2012. Circular dichroism spectroscopy of nucleic acids, pp. 575–586 in Comprehensive Chiroptical Spectroscopy, Volume 2: Applications in Stereochemical Analysis of Synthetic Compounds, Natural Products, and Biomolecules, edited by Berova N., Polavarapu P., Nakanishi K., and Wood R.. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- Lanktree M. B., Anand S. S., Yusuf S., and Hegele R. A., and SHARE Investigators , 2010. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet 3: 39–46. 10.1161/CIRCGENETICS.109.907642 [DOI] [PubMed] [Google Scholar]
- Levy-Sakin M., Pastor S., Mostovoy Y., Li L., Leung A. K. Y. et al. , 2019. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat. Commun. 10: 1025 10.1038/s41467-019-08992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., and Manley J. L., 2006. Cotranscriptional processes and their influence on genome stability. Genes Dev. 20: 1838–1847. 10.1101/gad.1438306 [DOI] [PubMed] [Google Scholar]
- Maizels N., and Gray L. T., 2013. The G4 genome. PLoS Genet. 9: e1003468 10.1371/journal.pgen.1003468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A. C., Lai Y. Y., Lam E. T., Kwok T. P., Leung A. K. et al. , 2016. Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics 202: 351–362. 10.1534/genetics.115.183483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H. C., Sharp A. J., Baker C., Itsara A., Jiang Z. et al. , 2008. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 359: 1685–1699. 10.1056/NEJMoa0805384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M., Searles V. B., Dickens C. M., Astling D., Albracht D. et al. , 2014. Finished sequence and assembly of the DUF1220-rich 1q21 region using a haploid human genome. BMC Genomics 15: 387 10.1186/1471-2164-15-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M. S., Dickens C. M., Dumas L. J., Kehrer-Sawatzki H., Wyckoff G. J. et al. , 2012. Evolutionary history and genome organization of DUF1220 protein domains. G3 (Bethesda) 2: 977–986. 10.1534/g3.112.003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley S., Louzada S., Forni D., Sironi M., Balaskas T. et al. , 2015. Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy. Proc. Natl. Acad. Sci. USA 112: 5105–5110. 10.1073/pnas.1416531112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco M. C., Maclaren E. J., Hopkins J., Dumas L., Cox M. et al. , 2006. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science 313: 1304–1307. 10.1126/science.1127980 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo A., Spada G. P., and da Silva M. W., 2012. Circular dichroism of quadruplex structures. Top. Curr. Chem. 330: 67–86. 10.1007/128_2012_331 [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., and Bleasby A., 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Roux K. H., 2009. Optimization and troubleshooting in PCR. Cold Spring Harb. Protoc. 2009: pdb.ip66. 10.1101/pdb.ip66 [DOI] [PubMed] [Google Scholar]
- Sakofsky C. J., and Malkova A., 2017. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 52: 395–413. 10.1080/10409238.2017.1314444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira J. M., and Aguilera A., 2015. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16: 583–597. 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- Searles Quick V. B., Davis J. M., Olincy A., and Sikela J. M., 2015. DUF1220 copy number is associated with schizophrenia risk and severity: implications for understanding autism and schizophrenia as related diseases. Transl. Psychiatry 5: e697 [corrigenda: Transl. Psychiatry 6: e735 (2016)]. 10.1038/tp.2015.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela J. M., and van Roy F., 2017. Changing the name of the NBPF/DUF1220 domain to the Olduvai domain. F1000Res. 6: 2185 10.12688/f1000research.13586.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela J. M., and Searles Quick V. B., 2018. Genomic trade-offs: are autism and schizophrenia the steep price of the human brain? Hum. Genet. 137: 1–13. 10.1007/s00439-017-1865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Stork C. T., Garcia-Rubio M. L., Paulsen R. D., Aguilera A. et al. , 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56: 777–785. 10.1016/j.molcel.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A. J., Kennedy H. L., and Kennedy M. A., 2016. Fluorescence methods for probing G-quadruplex structure in single- and double-stranded DNA. Biochemistry 55: 3714–3725. 10.1021/acs.biochem.6b00327 [DOI] [PubMed] [Google Scholar]
- Stirling P. C., and Hieter P., 2017. Canonical DNA repair pathways influence R-loop driven genome instability. J. Mol. Biol. 429: 3132–3138. 10.1016/j.jmb.2016.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I. K., Gacquer D., Van Heurck R., Kumar D., Wojno M. et al. , 2018. Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell 173: 1370–1384.e16. 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P. et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Van Roy N., Staes K., Speleman F., and van Roy F., 2005. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol. Biol. Evol. 22: 2265–2274. 10.1093/molbev/msi222 [DOI] [PubMed] [Google Scholar]
- Warburton P. E., Hasson D., Guillem F., Lescale C., Jin X. et al. , 2008. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics 9: 533 10.1186/1471-2164-9-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer F., and Montgomery S. H., 2015. Phylogenetic analysis supports a link between DUF1220 domain number and primate brain expansion. Genome Biol. Evol. 7: 2083–2088. 10.1093/gbe/evv122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Cell lines used in this study as a source of genomic DNA can be obtained from the Coriell Institute for Medical Research and are part of the 1000 Genomes Project. Individual samples and their identification numbers are listed in Supplemental Material, Table S7. The 16 trio samples have been consented for research at the University of California, San Francisco, and corresponding DNAs were sent to P.-Y. Kwok anonymously under his local IRB approval. Data corresponding to the samples used in this study are being deposited in public databases at the National Center for Biotechnology Information. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10673792.