Abstract
Objective
To evaluate the miR-21-5p and miR-200c-3p expressions in the urine of patients with prostate cancer (PCa) and to investigate their potential as biomarkers.
Material and methods
The urine samples collected from 80 patients, including 20 patients diagnosed with benign prostate hyperplasia (BPH) and 60 patients diagnosed with PCa, were examined. The exosome isolation was performed using the miRCURY exosome isolation kit (Exiqon, Denmark), total RNA was extracted using the miRCURY RNA Isolation Kit-Biofluid kit (Exiqon, Denmark), and complementary DNA (cDNA) was synthesized using the Universal cDNA Synthesis kit (Exiqon, Denmark). A quantitative polymerase chain reaction (qPCR) analysis of gene expression was performed using the qPCR CFX 96 Thermocycler (Bio-Rad). All the procedures followed the manufacturer’s recommendations.
Results
The overexpressions of miR-21 in the non-metastatic PCa and metastatic PCa group compared to the BPH group were statistically significant with a p-value of 0.001 and 0.018, respectively. The non-metastatic PCa compared to the metastatic PCa group was also statistically significant with a p-value of 0.037. The under expressions of miR-200c in the non-metastatic PCa and metastatic PCa group compared to the BPH group are statistically significant with a p-value of 0.001 and 0.001, respectively.
Conclusion
The overexpressions of miR-21 found in this study could be a potential non-invasive diagnostic tool for patients with PCa. Despite the significant results in our study, the use of micro-RNA in urine samples may vary due to epigenetic variation. Further studies with larger populations are required to investigate the role of miR-21 and miR-200c as biomarkers in PCa.
Keywords: Biomarker, metastasis, micro-RNA, miR-21, miR-200c, prostate cancer
Introduction
Cancer prevalence among men has recently increased globally, with prostate cancer (PCa) as the second most common cancer.[1] In 2012, over 300,000 of deaths could be attributed to PCa, making it the fifth leading cause of death for men worldwide.[1] The global incidence has increased from 3 to 30 per 100,000,[2] while overall mortality rates have actually declined particularly in developed countries, mostly due to a more successful diagnosing and treatment.[2] The diagnosis of suspicious PCa is increased when an abnormality is found during a digital the rectal examination or when there is an elevated level of serum prostate specific antigen (PSA). Furthermore, an invasive procedure of prostate biopsy is required to determine the histological type. Prostate biopsy is recommended in men with the PSA level >4.0 ng/mL,[3] and this threshold has a positive predictive value of only 37%, and a negative predictive value of 91%,[3] and therefore a new potential biomarker is needed to overcome these challenges.
Used in the diagnosis of many cancers, including breast, colorectal, and lung cancer, one of the first known and the main cancer-promoting micro-RNA, miR-21, targets several tumor suppressor genes linked to proliferation, apoptosis, and invasion.[4] miR-21 was observed to be increased in patients with chronic lymphocytic leukemia.[5] Micro-RNA was revealed to be a potential biomarker in both serum and urine samples of patients with PCa.[6] Zhang et al.[7] found that the expression of miR-21 in patients with PCa is high and counteracts the tumor-suppressive target, such as the phosphatase and tensin homolog deleted on chromosome 10, and programed cell death 4. Thus, the expression of the miR-21 can provide a promising approach to diagnosing. The essential role of the miR-200 family in combating tumor invasion, metastasis, and epithelial-mesenchymal transition has been reported in numerous studies.[8] Various types of cancer have been shown to express the miR-200 family.[9] According to a previous study by Shi et al.[8] comparing the human non-transformed prostate epithelial cells, the cells in patients with PCa exhibited a significantly reduced miR-200c expression. In this study, we aimed to investigate the miR-21 and miR-200c expression in urine samples of patients with PCa and investigate their potential as biomarkers.
Material and methods
Sample collection and exosome isolation
Urine samples collected from 80 patients were examined, including those of 20 patients diagnosed with benign prostate hyperplasia (BPH) and 60 patients diagnosed with PCa. All the patients who participated in this study signed a written consent form. TThehis study received ethical approval from the Universitas Gadjah Mada Ethical Review Board (Ref. No., KE/FK/0449/EC/2019). We collected 15 ml of urine from each patient. The samples were then distributed into four vials (1.5 mL), and each vial contained 1 mL of urine sample. The urine sample was then centrifuged for 5 minutes at 10,000 × g to separate the debris. After the centrifugation, the supernatant was extracted and filled into new a vial and kept in a refrigerator at −80°C. The exosomes isolation was conducted using the miRCURY exosome isolation kit (Exiqon, Denmark), by adding 400 uL precipitation buffer B into the vial, and the mixture was then incubated in a refrigerator at 4°C for 60 minutes. After the incubation, the sample was centrifuged for 30 minutes at 10,000 × g, and the supernatant was removed from the pellet. All the above procedures followed the manufacturer’s recommendation.
RNA isolation and cDNA synthesis
The total RNA was extracted using a miRCURY RNA Isolation Kit-Biofluid kit (Exiqon, Denmark). The pellets obtained from the exosome isolation were lysed by adding 350 μL of lysis solution and mixed by using vortex for 15 seconds, then adding 200 μL ethanol 96% into the vial and mixing the mixture by using vortex for 10 seconds. The mixture was then transported into the mini spin column and centrifuged for 1 minute at 3,500 × g. 400 μL of wash solution was added into the spin column and centrifuged for 1 minute at 14,000 × g. The tube was centrifuged at 14,000 × g for 2 minutes, and then the collection tube and the liquid inside were removed and changed with a new vial to collect RNA.
Complementary DNA (cDNA) were conducted using a Universal cDNA Synthesis kit (Exiqon, Denmark). The preparation of the master mix was conducted by mixing 4 μL of 5× reaction buffer, 9 μL of nuclease free water, 2 μL of enzyme mix, and 1 μL spike in (sp6) to a total volume of 16 μL reagent and 4 μL RNA sample (20 μL/reaction). The reaction mixture was incubated at 42°C for 60 min, inactivated reverse transcriptase at 95°C for 5 min, and cooling down was conducted at 4°C.
Quantitative polymerase chain reaction (qPCR) and data analysis
cDNA was diluted with RNase-free water at a ratio of 1:80 (1 μL cDNA with 79 μL RNase-free water). Quantitative PCR was conducted using a ExiLent SYBR Green Master mix kit (Exiqon, Denmark), primers set (forward and reverse) of micro-RNA and diluted cDNA. The primers (hsa-miR-16 as the reference gene, hsa-miR-21-5p, hsa-miR-200c-3p) were diluted with the SYBR Green master mix at a ratio of 1:6 (5 μL SYBR Green master mix and 1 μL primary PCR mix). Then, 6 μL of master mix was mixed with 4 μL cDNA, the reaction mixture was incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 1 min ramp-rate 1.6°C/s optical read and analyzed the melting curve. miR-16 was used as the internal control, and the relative miR-21 and miR-200c expression were calculated using the equation.
The qPCR analysis of gene expression was performed using the qPCR CFX 96 thermocycler (Bio-Rad). All of the procedures followed the manufacturer’s recommendations, and statistical analyses were performed using the SPSS Version 23 and Graph-Pad Prism 7. In this study, statistical significance was set at a p-value <0.05.
Results
In this study, urine samples were collected from 80 patients, of who 20 were diagnosed with BPH and 60 with PCa. The median age of the patients in the BPH group was 65 years, and the median PSA level was 2.05 ng/mL. The median age of patients in the non-metastatic PCa group was 72 years, and the mean PSA level was 25.76 ng/mL. The median age of patients in the metastatic PCa group was 69.5 years old, and the mean PSA level was 95.22 ng/mL (Table 1).
Table 1.
Demographic characteristics of recruited participants
| BPH | Non-metastatic PCa | Metastatic PCa | |
|---|---|---|---|
| Subject | 20 | 30 | 30 |
| Age (minimum–maximum, median) [years] | 44–79, 65 | 52–84, 72 | 49–82, 69.5 |
| PSA (minimum–maximum, median) [ng/mL] | 0.4–8.8, 2.05 | 0.17–292, 25.76 | 22–509,95.22 |
BPH: benign prostate hyperplasia; PCa: prostate cancer; PSA: prostate specific antigen
The characteristics of patients with PCa according to the ISUP Grade Group were similar between non-metastatic and metastatic PCa, and both groups were dominated by the high-risk group/ISUP Grade Group 4–5 (Table 2).
Table 2.
Characteristics of patients with prostate cancer according to the ISUP grade group
| Risk group | ISUP grade group | Gleason Score | Non-metastasic PCa | Metastasic PCa |
|---|---|---|---|---|
| n (%) | n (%) | |||
| Low | Group 1 | ≤6 | 1 (3.33) | 3 (10) |
| Intermediate Favorable | Group 2 | 7 (3 + 4) | 1 (3.33) | 3 (10) |
| Intermediate Unfavorable | Group 3 | 7 (4 + 3) | - | 2 (6.67) |
| High | Group 4 | 8 | 8 (26.67) | 8 (26.67) |
| High | Group 5 | 9–10 | 20 (66.67) | 14 (46.67) |
ISUP: international society of urological pathology; PCa: prostate cancer
The overexpressions of miR-21 in non-metastatic PCa and metastatic PCa group compared to the BPH group were statistically significant with a p-value of 0.001 and 0.018, respectively. The non-metastatic PCa compared to the metastatic PCa group was also statistically significant with a p-value of 0.037 (Table 3). The non-metastatic PCa group had the highest expression of miR-21 compared to BPH and metastatic PCa (Figure 1).
Table 3.
P-value of Independent T-test for each micro-RNA
| Groups | miR- 21-5p | miR- 200c-3p | |
|---|---|---|---|
| BPH | Non-metastatic PCa | 0.0001 | 0.0007 |
| BPH | Metastatic PCa | 0.0182 | 0.0003 |
| Metastatic PCa | Non-metastatic PCa | 0.0369 | 0.2743 |
BPH: benign prostate hyperplasia; PCa: prostate cancer
Figure 1.
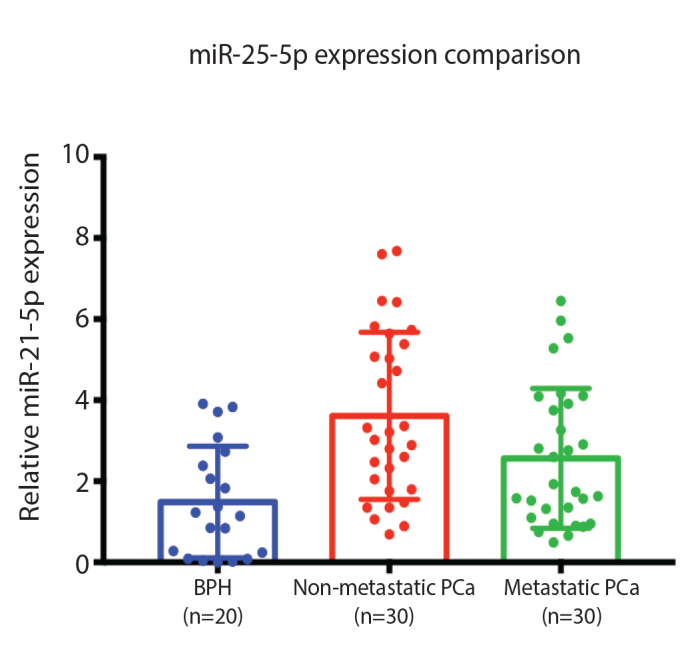
miR-21-5p expression comparison between BPH, non-metastatic, and metastatic PCa BPH: benign prostate hyperplasia; PCa: prostate cancer
The underexpressions of miR-200c in the non-metastatic PCa and metastatic PCa group compared to the BPH group were statistically significant with a p-value of 0.001 and 0.001, respectively. The non-metastatic PCa compared to the metastatic PCa group was not statistically significant, with a p-value of 0.274 (Table 3). The BPH group had the highest expression of miR-200c compared to the non-metastatic and metastatic PCa groups (Figure 2).
Figure 2.
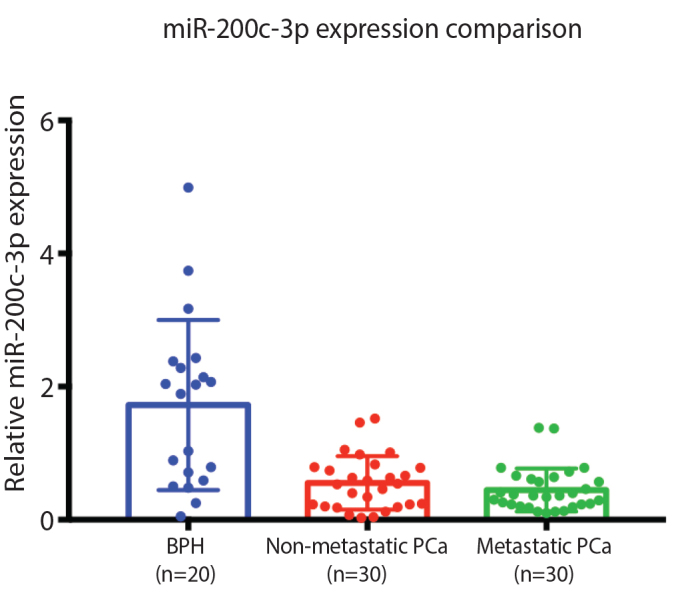
miR-200c-3p expression comparison between BPH, non-metastatic, and metastatic PCa BPH: benign prostate hyperplasia; PCa: prostate cancer
Discussion
In this study, the overexpressions of miR-21 in the non-metastatic PCa and metastatic PCa groups compared to the BPH group were statistically significant. Studies conducted by Melbø-Jørgensen et al.[10] showed the overexpression of miR-21 in patients with PCa who received radical prostatectomy. Ribas et al.[11] and Li et al.[6] also showed an increase in the miR-21 expression in PCa compared to the normal prostate tissue. Urine-based miR-21 was studied by Ghorbanmehr et al.[12] The study showed an upregulation of urine-based miR-21 in patients with PCa. These studies indicated that the overexpression of urine-based miR-21 could be a potential non-invasive biomarker for diagnostic aspects of PCa. Contrary to a previous study, this study showed that the miR-21 expression was lower in the metastatic PCa compared to the non-metastatic PCa group. Several studies found that the tissue- and blood-based miR-21 was higher in the PCa metastatic groups.[13–16] This result indicates that further studies on urinary-based miR-21 are required to clarify the role of miR-21 in metastatic PCa as a potential biomarker for early signs of metastases.
On the contrary, the tumor suppressor miR-200c was underexpressed in PCa. A decrease in the expression of miR-200c in the non-metastatic PCa and metastatic PCa group compared to the BPH group were statistically significant. The role of miR-200c has been known for tumor progressivity, cell renewal, and metastasis.[17,18] A study conducted by Shi et al.[8] showed miR-200c as an inhibitor factor of PCa proliferation, and a decrease in the expression of miR-200c on the cell line was correlated with a PCa progression. We found similar results with miR-200c underexpressed in PCa. In our study, the metastatic PCa group had a lower miR-200c expression compared to the non-metastatic PCa group but was not statistically significant, and this is likely due to the small sample population. This decrease of miR-200c expression requires further research to determine the role of miR-200c as a prognostic biomarker for PCa.
In conclusion, the overexpression of miR-21 shown in this study could be a potential non-invasive diagnostic tool for patients with PCa. Despite the significant results in our study, the usage of micro-RNA in urine samples may vary due to the epigenetic variation. Further studies with a larger population are required to investigate the role of miR-21 and miR-200c as biomarkers in PCa. Potentially, the combination of both miRNA can provide important data for an accurate and timely diagnosis for patients with PCa.
Acknowledgements
The authors thank the Faculty of Medicine, Universitas Gadjah Mada for providing the research laboratory and facilities to support this research.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the ethics committee of Faculty of Medicine, Universitas Gadjah Mada/Dr. Sardjito Hospital gave approval for this study (Ref. No.: KE/FK/0449/EC/2019).
Informed Consent: Written informed consent was obtained from all patients who participated in this study.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept – R.D., I.A., R.U., S.M.H.; Design – R.D., I.A., R.U., S.M.H.; Supervision – R.D., I.A., R.U., S.M.H.; Fundings – R.D., S.M.H.; Material – R.D., S.M.H.; Data Collection and Processing – R.D., S.M.H.; Analysis and Interpretation – R.D., I.A., R.U., S.M.H.; Literature review – R.D., I.A., R.U., S.M.H.; Writer – R.D.; Critical review – I.A., R.U., S.M.H.
Conflict of Interest: The authors have no conflicts of interest to declare.
Financial Disclosure: The authors declared that this study has received no financial support.
References
- 1.Zhou CK, Check DP, Lortet-Tieulent J, Laversanne M, Jemal A, Ferlay J, et al. Prostate cancer incidence in 43 populations worldwide: An analysis of time trends overall and by age group. Int J Cancer. 2016;138:1388–400. doi: 10.1002/ijc.29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ha Chung B, Horie S, Chiong E. The incidence, mortality, and risk factors of prostate cancer in Asian men. Prostate Int. 2019;7:1–8. doi: 10.1016/j.prnil.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nogueira L, Corradi R, Eastham JA. PSA for Prostate Cancer Detection. Int Braz J Urol. 2009;35:521–31. doi: 10.1590/S1677-55382009000500003. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 5.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–51. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 6.Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, et al. MiR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol. 2012;187:1466–72. doi: 10.1016/j.juro.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 7.Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–31. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 8.Shi R, Xiao H, Yang T, Chang L, Tian Y, Wu B, et al. Effects of miR-200c on the migration and invasion abilities of human prostate cancer Du145 cells and the corresponding mechanism. Front Med. 2014;8:456–63. doi: 10.1007/s11684-014-0353-z. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Zhang L. Members of the microRNA-200 family are promising therapeutic targets in cancer (Review) Exp Ther Med. 2017;14:10–7. doi: 10.3892/etm.2017.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melbø-Jørgensen C, Ness N, Andersen S, Valkov A, Dønnem T, Al-Saad S, et al. Stromal expression of miR-21 predicts biochemical failure in prostate cancer patients with Gleason score 6. PLoS One. 2014;9:e113039. doi: 10.1371/journal.pone.0113039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, et al. miR-21: An androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–9. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghorbanmehr N, Gharbi S, Korsching E, Tavallaei M, Einollahi B, Mowla SJ. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate. 2019;79:88–95. doi: 10.1002/pros.23714. [DOI] [PubMed] [Google Scholar]
- 13.Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, et al. miR-21: An oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selcuklu SD, Donoghue MTA, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–25. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 16.Bonci D, Coppola V, Patrizii M, Addario A, Cannistraci A, Francescangeli F, et al. A microRNA code for prostate cancer metastasis. Oncogene. 2015;35:1180–92. doi: 10.1038/onc.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Wu L, Zhao JC, Jin H, Yu J. TMPRSS2 - ERG gene fusions induce prostate tumorigenesis by modulating microRNA miR-200c. Oncogene. 2014;33:5183–92. doi: 10.1038/onc.2013.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]


