Abstract
Understanding the resistance and resilience of foundation plant species to climate change is a critical issue because the loss of these species would fundamentally reshape communities and ecosystem processes. High levels of population genetic diversity may buffer foundation species against climate disruptions, but the strong selective pressures associated with climatic shifts may also rapidly reduce such diversity. We characterized genetic diversity and its responsiveness to experimental drought in the foundation plant, black grama grass (Bouteloua eriopoda), which dominates many western North American grasslands and shrublands. Previous studies suggested that in arid ecosystems, black grama reproduces largely asexually via stolons, and thus is likely to have low genetic variability, which might limit its potential to respond to climate disruptions. Using genotyping-by-sequencing, we demonstrated unexpectedly high genetic variability among black grama plants in a 1 ha site within the Sevilleta National Wildlife Refuge in central New Mexico, suggesting some level of sexual reproduction. Three years of experimental, growing season drought reduced black grama survival and biomass (the latter by 96%), with clear genetic differentiation (higher FST) between cohorts succumbing to drought and those remaining alive. Reduced genetic variability in the surviving cohorts in drought plots indicated that the experimental drought had forced black grama populations through selection bottlenecks. These results suggest that foundation grass species, such as black grama, may experience rapid evolutionary change if future climates include more severe droughts.
Keywords: climate change, genotyping-by-sequencing, natural selection, experimental evolution, Sevilleta Long Term Ecological Research
Introduction
Climate change can directly reduce plant fitness and population abundance (Parmesan 2006; Anderson 2016). Declines in population abundance under a new climate have the potential to feed back to influence the resistance or resilience of populations to future changes by reducing population genetic diversity (Pauls et al. 2013). Maintaining genetic diversity is important because it can buffer against future climate stress, disturbance, inbreeding depression, and antagonistic species interactions (e.g., Jump et al. 2009; Hart et al. 2016; Isbell et al. 2015). For example, high genetic diversity can buffer populations against unpredictable environmental conditions, such as extreme drought or flooding (e.g. Prati et al. 2016), in part through the portfolio of genotypes that vary in their sensitivity to extremes. The loss of genetic diversity can also influence population-level processes, depressing species-level productivity, population growth, and the ability to invade new habitats (Crutsinger et al. 2006; Crawford and Whitney 2010; Cook-Patton et al. 2011; Atwater and Callaway 2015). Mechanisms underlying feedback from genetic diversity to population processes include the loss of specific genotypes that can survive extreme climate events (selection or dominance effect, Fox 2005) and the loss of non-additive effects of diversity, such as niche complementarity or facilitation among genotypes, that can increase population growth and productivity (reviewed by Hughes et al. 2008; Jump et al. 2009; Whitlock 2014).
There is increasing evidence for climate-induced shifts in population-genetic structure (reviewed in Pauls et al. 2013; see also Carroll et al. 2014; Franks et al. 2014) and for the adaptive significance of these shifts in both plants and animals (reviewed by Merilä and Hendry 2014). For example, Rubidge et al. (2012) associated climate warming in Yosemite National Park with a contraction in range size and a reduction in overall genetic diversity for the alpine chipmunk (Tamias alpinus). Nevo et al. (2012) reported genetically-based changes in flowering time and a significant reduction in simple-sequence allele diversity over a 28-year period for wild populations of both wheat (Triticum dicoccoides) and barley (Hordeum spontaneum). Franks et al. (2007; 2016) described an evolutionary shift to shorter flowering time for Brassica rapa that followed a multi-year, naturally occurring drought, with corresponding shifts in allele frequencies in genes related to flowering time and drought stress. Jump et al. (2008) imposed experimental drought and warming on the early-successional Mediterranean shrub Fumana thymifolia over a seven-year period and found elevated genetic divergence between seedlings in treatment vs. control plots, signaling climate-induced selection. In contrast, other studies have found relatively little evolutionary change in response to climate (e.g., Lau and Lennon 2012; Huber et al. 2016). For example, Lau and Lennon (2012) detected only weak evolutionary responses of B. rapa populations to greenhouse-imposed drought; instead, plant fitness responded to rapid changes in the composition of soil microbes following drought.
Losses of genetic diversity could be particularly consequential in dominant or foundation species, which shape the composition and diversity of associated plant, animal, and microbial communities (Ellison et al. 2005; Bangert et al. 2008; Hughes et al. 2008). Declines in the genetic diversity of foundation plant species could cascade to community-level interactions. For example, genetic diversity in dominant species can enhance resistance to both invasion (Moreira et al. 2014; Yang et al. 2017) and herbivores (Moreira et al. 2014). In addition, high genetic diversity in foundation species can interact with species diversity to produce synergistic gains in ecosystem productivity and functioning (e.g., Crawford and Rudgers 2012; Schoeb et al. 2015). Thus, understanding the genetic consequences of climate change for foundation species could help to refine predictions of future community and ecosystem trajectories (e.g., Ikeda et al. 2017).
We know of just two studies that have examined the effects of experimental climate change on genetic diversity and differentiation in foundation plant species in the field. In a 10-year rainfall manipulation, Avolio et al. (2013) found that genotype richness of big bluestem (Andropogon gerardii) decreased under higher within-season variability in precipitation, with a few successful genotypes increasing in abundance. These successful genotypes were both genetically and phenotypically divergent from each other (Avolio et al. 2013; Avolio and Smith 2013), suggesting a role for niche differentiation in population resistance to stress. Ravenscroft et al. (2015) imposed drought, water addition, and warming on grasslands, and after 15 years found elevated genetic divergence between treatments for the forb Plantago lanceolata and the foundation grass Festuca ovina. We note that both of these studies occurred in mesic grasslands, leaving open the question of climate-change induced evolution in the plants that dominate arid ecosystems. These ecosystems are of global importance: of all the land cover classes, arid and semi-arid ecosystems contribute most to inter-annual variability in global carbon flux due to their high year-to-year variability in primary production (Ahlstrom et al. 2015; Huang et al. 2016a) and large surface area (45% of global land area, Pravalie 2016), which is rapidly expanding (Huang et al. 2016b).
In the current study, we leveraged an established drought experiment in an arid grassland to address the questions: (a) What are standing levels of genetic variation in a foundation plant species? (b) How does drought affect the genetic diversity and composition of a xeric-adapted foundation plant? We focused on black grama grass (Bouteloua eriopoda), a dominant of desert grasslands in western North America, which has been reported to reproduce mostly clonally through stolons (Peters 2002). Clonal populations may be highly sensitive to climate disruptions, such as drought, if low standing diversity provides little buffer against change (Jimenez-Alfaro et al. 2016). In our experiment, a 66% reduction of growing season precipitation caused substantial mortality of B. eriopoda plants, raising the possibility that differential survival had a genetic basis. We tested this hypothesis using a reduced representation sequencing approach, employing DNA extracted from root tissues from living vs. recently dead plants. To our knowledge, this is the first study to examine whether climate alters population genetic diversity for an arid ecosystem dominant.
Methods
Study species
Black grama (Bouteloua eriopoda (Torr.) Torr. (Poaceae) is a stoloniferous, C4 perennial grass that dominates desert grasslands of North America. It is a diploid (2n=20) (Streetman and Wright 1960; Gould 1979) with a geographic distribution ranging from Texas to southern California and from Mexico northward to Colorado, Utah, and Wyoming. Black grama is present in diverse dryland ecosystems including desert grasslands, creosote bush shrublands, mesquite scrub, and piñon- juniper woodlands, and it dominates uplands (800–1900m) with sandy loam soils (Simonin 2000). Historically, black grama covered extensive areas of western Texas, southern New Mexico, southeastern Arizona, and northern Mexico (Nichol 1952; Dick-Peddie 1993), but many desert grasslands have been replaced by encroaching shrublands over the past 150 years (e.g., van Auken 2000; Connin et al. 1997; Báez and Collins 2008; Peters et al. 2010). Prior work has documented the sensitivity of black grama to several abiotic factors, including drought (Báez et al. 2013), grazing (Gosz and Gosz 1996), and fire (Reynolds and Bohning 1956; Parmenter 2008). Monsoon precipitation (July-September) is a more important correlate of black grama production than total annual precipitation (Paulsen & Ares 1962; Thomey et al. 2011; Rudgers et al. 2018).
Study site and drought experiment
The study was conducted at the Sevilleta National Wildlife Refuge (SNWR), a Long-Term Ecological Research (LTER) site in the northern Chihuahuan Desert of central New Mexico, USA. Within the SNWR, the study site was located in a black grama-dominated grassland near a zone where creosote bush (Larrea tridentata)-dominated shrubland transitions to grassland. Black grama has been increasing in abundance at this site over the past three decades (Collins and Xia 2015). Total annual precipitation is ~250 mm, ~60% of which falls during the summer monsoon from July through early September (Notaro et al. 2010).
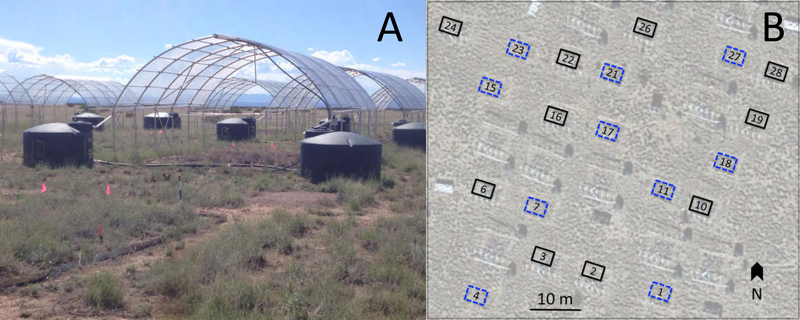
The Extreme Drought in Grassland Experiment (EDGE) was established in Spring 2013 and includes a chronic drought treatment (66% reduction in growing season rainfall) designed to mimic droughts that are predicted to occur in this region by the end of this century (Cook et al. 2015). Drought was imposed by installing roof panels constructed of plastic strips that cover 66% of the surface area (Yahdjian and Sala 2001) to reduce the size of each rain event (Fig 1A). Roof panels are in place only during the growing season (April through September). These passive drought shelters create a 1-in-100-year drought (Knapp et al. 2015) and effectively mimic rainfall patterns (size, frequency) during natural drought years (Knapp et al. 2017). The shelters were designed to allow substantial airflow, with completely open ends and sides that are open from ground level to 1.2 meters above ground (Fig. 1A). Shelters had a minimal effect (~0.4°C) on mean 2013–2017 growing season air temperatures (mean ± SE: shelter 22.84 ± 0.86 °C; control 22.42 ± 0.86 °C). The portion of the experiment used in the current study consists of 20 plots (3 m × 4 m each), with 10 drought plots and 10 controls, paired spatially into blocks (Fig. 1B). Drought treatment was assigned randomly within a block.
Fig. 1.
Extreme Drought in Grasslands Experiment (EDGE) at the Sevilleta National Wildlife refuge. (a) During summer (monsoon) months, rainout shelters are partially covered with plastic sheeting to remove approximately two-thirds of precipitation from Drought plots. (b) Black grama EDGE plot layout. Dashed (Control) and solid (Drought) rectangles superimposed on a satellite image indicate paired treatments within each block. A color version of this figure accompanies the online version of this article
Field sampling: biomass
Standing live biomass of black grama was estimated using a non-destructive volumetric method (Huenneke et al. 2001; Muldavin et al. 2008). Volume measurements (% cover x height) of all individual plants were recorded in four permanently located 1-m2 quadrats in each replicate plot during peak biomass (September) in the 2015-2017 growing seasons, the third through the fifth growing seasons under experimental drought. Volume was then converted to biomass via allometric equations derived from coupled volume measurements and destructive harvests of black grama biomass outside of the experimental plots; volume explained 70% of the variation in biomass in this equation (R2 = 0.70). We analyzed the black grama biomass response with a linear mixed effects model that included the fixed effect of drought treatment, year, and their interaction, as well the random effect of plot to account for the non-independence of replicate quadrats within a plot, using the lmer function in the lme4 package (Bates et al. 2015; R Core Team 2017). We report estimated marginal means and standard errors from the model (emmeans function).
Field sampling: genetic diversity
We quantified drought effects on the genetic diversity and composition of live (at least one green leaf) versus recently dead (no green tissue present) black grama. At the end of the third growing season (28-Oct-2015), we collected plant leaves and roots from 20 black grama individuals per plot. This sample size was determined by a goal of balanced sampling of live and dead individuals within and across plots, and the fact that some drought plots contained only 10 live individuals by the third year of treatment. Collection occurred one week after a late three-day monsoon rain period (which ended 21-Oct-2015), totaling 23 mm. In drought plots, we marked pairs of live and dead individual plants (defined as contiguous patches separated by a minimum of 5 cm, Laurenroth and Adler 2008) to meet the following criteria: (i) at least two pairs were chosen in each of four quadrants of the plot, (ii) no individuals occurred within 50 cm of the plot edge (defined by aluminum flashing), and (iii) members of a live/dead pair were closer to each other than to other marked individuals within a plot. The paired strategy was adopted to ensure that sampling of live vs. dead individuals occurred across equal spatial scales in a plot, in case of within-plot spatial population genetic structure. In control plots, we marked pairs of plants similarly, but both individuals in each control group were live due to the lack of mortality. All plots received equal levels of disturbance and vegetation removal.
From each individual, we removed 1–3 tillers and 3–5 root segments (min. ~3 cm long) using a soil knife. Tools were cleaned with bleach germicidal wipes (sodium hypochlorite 0.55%, Clorox Healthcare, Oakland, CA) between each sample. Samples were collected into sterile Whirl-Paks (Nasco, Fort Atkinson, Wisconsin) and returned to the lab where roots were washed in DI water, dried with a contaminant-free KimWipe (Kimberly-Clark, Irving, Texas; McManus and Kelley 2005), and stored at - 20C within 72 h.
We checked that plants we classified as “dead” were indeed dead (as opposed to temporarily dormant) via two methods. First, we continued monitoring of live biomass in the experimental plots over two more growing seasons to see if there was any evidence of recovery in the drought plots. Second, on 15-Sep-2018 we collected 10 apparently dead plants (one from each drought plot) and transplanted them to individual pots containing a native soil / sterilized sand mixture in the greenhouse, which was maintained between 18.3 – 23.9 °C. We placed them on a drip irrigation system which watered them three times daily. Over a four-month period, we monitored each plant for signs of new growth.
Molecular methods and genetic analyses
Root tissue from each individual was ground in liquid nitrogen and 0.25 grams of ground material was extracted using PowerSoil kits (MoBio, Carslbad, California). DNA extracts were cleaned and concentrated using ZR-96 DNA Clean & Concentrator (Zymo Research, Irvine, California). DNA concentration was determined using Nanodrop 2000c (Thermo Scientific, Waltham, Massachusetts). We obtained high-molecular-weight DNA from the roots of all samples. Because of limited financial resources for sequencing and because our questions about genetic diversity do not require tracking individual genotypes, we then pooled DNA into two pools per plot (each containing DNA from 10 individuals). For each individual plant, the amount of DNA added to the pool was normalized to the sample in the cohort with the lowest DNA concentration. Thus, there were a total of 40 DNA pools corresponding to 10 pools in each of four cohorts: “control live 1”, “control live 2”, “drought live” and “drought dead”.
Potential clonality in black grama (Peters 2002) could have limited the effective number of genotypes contributing to each pool, affecting the absolute values of estimated population genetic parameters such as allele frequency and FST. A related issue is that the small pool sizes of 10 individuals (necessitated by the small number of living individuals remaining in drought plots post-treatment) could have resulted in some imprecision in allele frequency estimates. Allele frequency estimates using pooled sequencing are cost effective and robust, but can be undermined by small pool sizes (Kofler et al. 2016; Gautier et al. 2013; Anand et al. 2016; Fracassetti et al. 2015; Schlotterer et al. 2014; Lynch et al. 2014). Nevertheless, Anand et al. (2016) found robust reproduction of allele frequencies (R2>0.98) in pools with only 12 individuals. Using the PIFs software (Gautier et al. 2013), we compared our pool-based sequencing to expectations for individual sequencing. Using our minimum pool coverage of 5X and 10 individuals (either sequenced as a pool or individually), along with an Illumina error rate of 0.24% error rate (Pfeiffer et al. 2018), we estimate that the standard deviation of the allele frequency increased only 1.265X by sequencing pools instead of individuals. Furthermore, though our pool size is small, it represents a substantial portion of the population from which it is derived.
We further note, as described above, that we applied treatments randomly to plots in space, and further balanced our sampling by making sure that each pool had the same number of individuals and that samples collected were spaced similarly within each plot (see “Field sampling: genetic diversity”). Thus any potential effects of clonality and small pool size should apply equally to parameters estimated for the drought and control treatments, and are not expected to bias the results. At worst, they should create noise, making it harder to detect significant differences between the treatments and resulting in a conservative test for the presence of a genetic response to drought. While we do not expect the data to be biased between drought and control plots, we have avoided generating locus-specific genetic parameters because they are more susceptible to the noise in the data. We expect that locus-specific genetic parameters would have lower power and sensitivity and inflated false positive rates (Luikart et al. 2003).
Sequencing
We constructed standard Illumina libraries for DNA samples digested with PstI and MspI, except that TruSeq Universal Adapters were changed to include an inline barcode, as described in Elshire et al. (2011). The 40 samples were multiplexed together and sequenced in two lanes on an Illumina HiSeq2000 machine using Illumina TruSeq v3 chemistry. Samples were filtered and base called using the Illumina RTA 1.13.48.0 and CASAVA 1.8.2 pipelines and were de-multiplexed with an in-house script. A total of 331,701,293 sequence pairs and 66,340,258,600 nts of filtered sequence were generated.
Genotyping
Genotyping of bulk samples aimed to identify the allele(s) present within the bulk rather than to obtain individual plant genotypes. We used the UNEAK program (Lu et al. 2013), which uses a network-based approach to identify alleles present in sequencing tags. This approach removes alleles that appear to be sequencing errors and focuses on loci with two alleles (Lu et al. 2013). In this way we could differentiate between loci that were fixed in a given bulk from those that had at least two alleles. We did not include putative triallelic and quadrallelic loci in our analyses, because loci with >2 alleles are expected to be quite rare, and thus these putative loci are much more likely to be false positives. UNEAK identified a total of 559,023 polymorphic loci (SNPs), with genotypes called on pooled DNA samples. Only alleles with at least 5% allele frequency were converted into genotypes in order to reduce sequencing error. With 10 samples in each pool, alleles present in only one sample should have a frequency of approximately 10% (assuming samples are equally represented) and, therefore, alleles with <5% frequency likely represented sequencing error. Sites with only one allele with a frequency ≥ 5% were called as homozygous and those with two called as heterozygous. Because these genotypes were called on pools, a heterozygous call indicates simply that both alleles are present in the pooled sample, but does not provide information on the frequencies of homozygous and heterozygous individuals within the pool. The R package SeqArray was used to import the VCF file into Genomic Data Structure (GDS) format in R (Zheng and Gogarten 2015). A genotype matrix was generated with the R package SeqVarTools (Gogarten and Zheng 2017).
Analyses of genetic similarity and FST
We first evaluated spatial heterogeneity of genetic diversity within the study area by assessing genetic versus physical distance. Because each sequenced DNA sample represented pooled DNA from a cohort of 10 individual plants, we assessed pairwise genetic distances between cohorts rather than plants. Pairwise genetic distances were calculated using genetic loci that were fixed for each cohort (i.e., only one allele was called) and that were supported by at least 5X coverage. We focused on fixed loci in order to clearly identify those that differed between cohorts, and to reduce noise arising from the fact that our limited sample size may have missed alleles in populations, especially those present at low frequency. Our focus on fixed loci avoided comparisons of a pool with two alleles to a pool with only one, which could falsely be declared different despite the fact that the second allele might be present (but unsampled) in the latter population. We focused on sites that were fixed across all samples given that differences seen between fixed sites are more likely to be real. Under our approach, false differences would arise only if both pools were missing an allele present in the population. The same set of loci was used for all samples, and we therefore expect that any potential bias that resulted from using only fixed loci was applied equally to all samples, and that the relative genetic distances between pairs of pools was preserved. Genetic similarity was defined as the percentage of loci that were identical between two cohorts, divided by the total number of loci shared by both. Physical distances were determined to the nearest 0.1 m, by measuring distances between the centers of plots in a pairwise fashion. Graphics were generated in R with the ggplot2 package (Wickham 2009) unless otherwise noted.
The FST (fixation index) was calculated in R using the snpgdsFST function in the SNPRelate package (Zheng et al. 2012) based on all genotypes from the UNEAK program, including those that were not fixed. To test for drought treatment effects on FST, we ran two analyses. First, we determined the pairwise FST for the two cohorts of 10 individuals sampled within each plot. In control plots, both cohorts consisted of randomly selected live individuals and should have low pairwise FST. In drought plots, if drought-induced mortality were selective among genotypes, it should amplify the within-plot pairwise FST relative to two randomly selected living cohorts in control plots. Thus, our a priori hypothesis was that the within-plot pairwise FST would be larger for dead-live pairs within the drought treatment than for the live-live pairs within control plots. We evaluated this hypothesis using a one-tailed, paired t-test (pairing the control and drought plots within each spatial block), given that the distribution of residuals was Gaussian. Pairing by block and separately analyzing within-plot differences from between-plot differences was motivated by the finding of effects of physical distance on genetic distance (see Results).
Second, we compared pairwise FST values among cohorts between plots. For both the live and dead drought cohorts, we expected their mean pairwise FST against live plants in control plots to be larger than the mean pairwise FST between any two control plots. Thus, we used a general linear model to compare among these three sets of between-plot pairwise FSTs: drought dead-control live, drought live-control live, and control live-control live (lm function in R). Because the analysis was of pairwise distances among all pairs of plots, it was not meaningful to include spatial block in the analysis. If the pairwise FST between drought dead-control live was larger than that between drought live-control live, it could indicate that drought selectively killed a unique set of genotypes, whereas the reverse could indicate the drought selectively favored unique genotypes. Data were plotted using SigmaPlot version 12 (Systat Software, San Jose, California).
Analyses of allelic diversity and composition
We determined a measure of within-locus allelic diversity for each bulked sample by calculating the percentage of polymorphic loci. A minimum of five sequence reads was required in order to improve the chances of sampling multiple alleles from loci where multiple alleles were present in a bulk. A minimum of 40% of the reads had to call each allele in order to minimize sequencing error. We then used a general linear mixed effects model to test the random effects of plot and block and the fixed effect of treatment cohort on allelic diversity, implemented in the lme4 package. Pairwise contrasts using FDR correction for multiple comparisons tested whether drought live plants had significantly lower allelic diversity than drought dead or control live plants; data were plotted in SigmaPlot v. 12. Next, we determined a within-sample measure of allelic richness by calculating the total number of alleles present across 3,614 loci that had genotype calls in all samples. Paired t-tests were used to compare between cohorts.
To examine drought effects on allelic composition, we used the matrix of allele frequencies across all alleles identified by UNEAK to conduct a permutational MANOVA using the adonis function in the vegan package in R (Oksanen et al. 2016). We used the Bray-Curtis distance metric in all allelic composition analyses, with 9999 permutations that were stratified by the random effect of spatial block. The global analysis compared allelic compositional differences among all four groups (control live 1, control live 2, drought live, or drought dead), and we followed with planned contrasts between drought live and drought dead, and between drought dead and all controls. To examine drought treatment effects on allelic dispersion, we used the betadisper function in the vegan package with 9999 random permutations. To visualize differences among treatment cohorts, we conducted non-metric multidimensional scaling (NMDS) analysis using default settings in metaMDS in the vegan package. In addition, heatmaps were generated in R v 3.2.3 (R Core Team 2017) using the SNPrelate package (Zheng et al. 2012). The genotypes from UNEAK were fed into the snpgdsIBS function the heatmap plotted with R’s core image function. Finally, we conducted a principal component analysis (PCA) by feeding the genotype matrix into the pca function of the SeqVarTools package. Plots used the ggrepel function to separate labels (Slowikowski 2016).
Results
Experimental drought caused black grama mortality and reduced biomass
After three growing seasons, extreme drought (66% reduction in growing season rainfall) resulted in substantial mortality, with numerous ramets of recently dead black grama grass noted in drought plots but none found in control plots. At the time of genetic sampling (September 2015), average (± s.e.) above-ground live (green) biomass of black grama was only 4.6 (±5.3) g m−2 in drought plots compared to 119.0 (±5.3) g m−2 in control plots (X2 = 245.48, P < 0.0001), a reduction of 96.2%. This represented actual mortality (vs. temporary dormancy) as live biomass continued to decline in the drought plots, to 3.0 (± 5.3) and 1.6 (± 5.3) g m−2 in September 2016 and September 2017, respectively. Further, we saw no evidence of new growth in 10 dead plants transplanted from the drought plots to the greenhouse and watered daily over a four-month period.
Substantial genetic variation in black grama populations
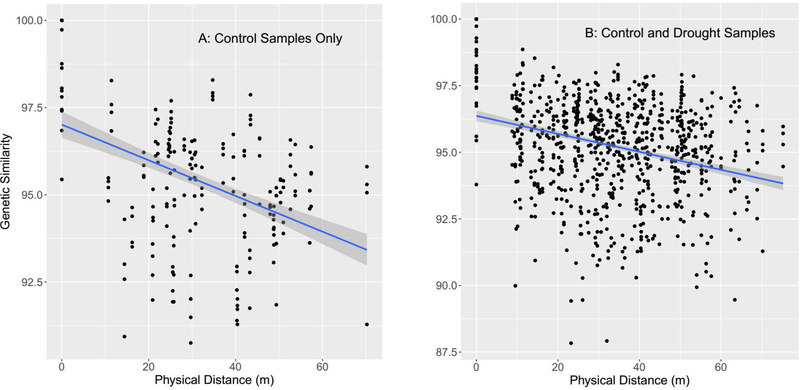
We observed substantial genetic variation among black grama plants across the study site. Sequencing analysis produced a total of 559,023 SNPs, of which 46.90% of called SNPs on average (range 43.89%−51.38%), showed variation between control samples from different plots, at a minimum rejecting the idea that the study site comprises one large black grama clone. DNA samples from the cohorts of 10 plants collected from within the same plot were more similar to each other than to cohorts from other plots (t-test on pairwise FST within-plot vs. between-plot, two-tailed with unequal variances, P < 0.0001). In addition, the physical distance between plots was negatively correlated with the genetic similarity between cohorts (r = −0.29 (control and drought treatments) and −0.43 (control treatment only), both P < 0.0001, Fig. 2), supporting an isolation-by-distance model. The apparent continuous distribution of genetic similarities along the y-axes of the similarity vs. distance plots (Fig. 2) is further evidence against widespread clonality, as a clone large enough to encompass two or more of our plots would have instead generated clumps of identical pairwise similarities. The data thus suggest a minimum of 20 distinct black grama clones in our study (one per plot).
Fig. 2.
Relationships between genetic similarity and physical distance. Points on the graphs represent pairwise comparisons of genetic similarity among cohorts of 10 black grama individuals. Points at zero physical distance represent comparisons among cohorts obtained from the same plot. Each point at 100% genetic similarity represents a cohort compared with itself. Linear regression lines with 95% confidence level shading are shown. (a) Comparisons among cohorts in the control treatment only; r = −0.43, P < 0.0001. (b) Comparisons among all cohorts in both treatments (drought and control); r = −0.29, P < 0.0001.
Drought altered the population genetic structure of black grama
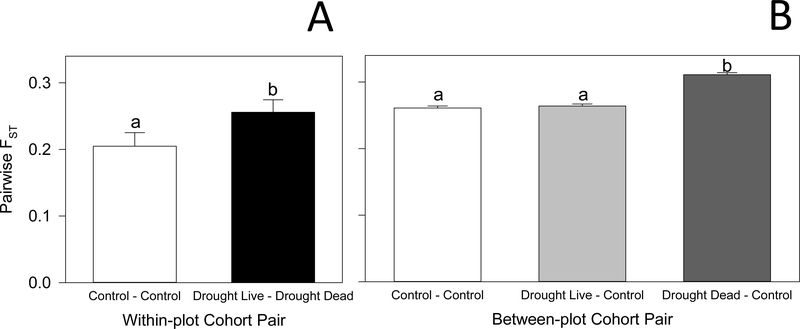
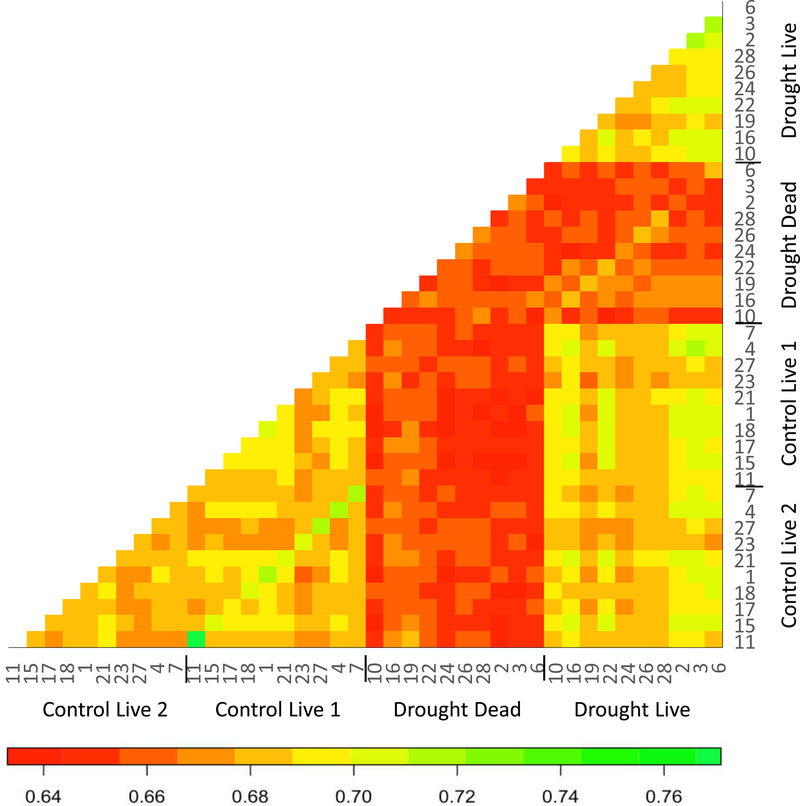
Within plots, pairwise FST values were ~25% larger between drought dead and drought live cohorts than between the two randomly selected cohorts of live plants from control plots (Fig. 3A, paired t-test, P = 0.0494). In addition, between-plot pairwise FST values for drought dead versus controls were~20% larger than any FST comparing cohorts of live plants (control live vs. control live, or drought live vs. control live; Fig. 3B, F2,577 = 75.0, P < 0.0001). Thus, the cohorts of plants that died in the drought treatment were genetically differentiated from the remaining live cohorts. This differentiation was apparent in a heat-map analysis of genetic similarity, in which the plants that died under drought were dissimilar to the control cohorts and to the drought live cohorts (Fig. 4).
Fig. 3.
Mean (± s.e.) pairwise FST values among pairs of cohorts of 10 black grama individuals. Panel A: Within-plot Cohort Pair. Panel B: Between-plot Cohort Pair. (a) Mean FST between cohorts within plots. (b) Mean FST among cohorts between different plots. Letters show statistically significant differences among the means within each graph.
Fig. 4.
Heat map of genetic similarity among black grama subpopulations. Each square represents pooled DNA from a cohort of 10 individuals collected from a single plot. Warmer (darker) colors reflect lower proportions of SNP loci that are identical by state (i.e., greater genetic distances between cohorts). Plot numbers are given at the margins (cross-reference to Fig. 1). A color version of this figure accompanies the online version of this article
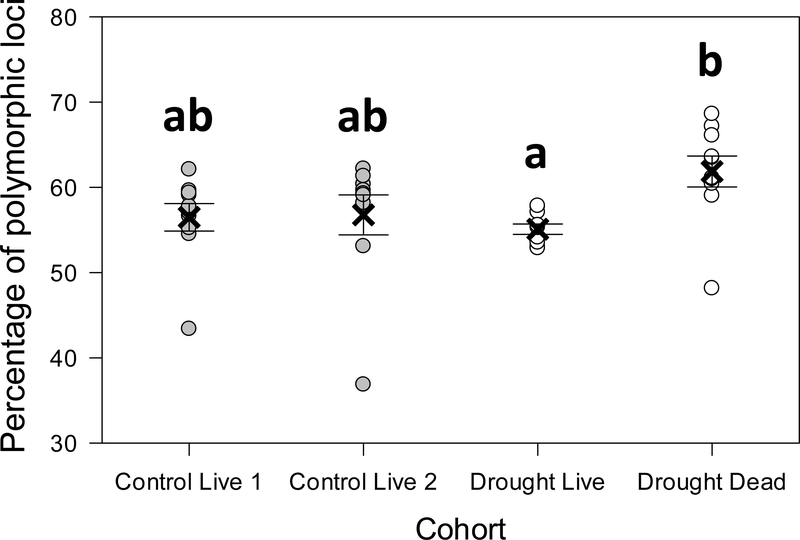
Plants that survived drought had a lower within-locus allelic diversity, as measured by an 11% drop in the percentage of polymorphic loci compared to plants that died in the drought treatment (Fig. 5, Wald X2 = 24.5, P < 0.0001). Although not statistically significant, plants that survived drought had a 3% drop in within-locus allelic diversity compared to the pre-drought population, as represented by the control plots.
Fig. 5.
Loss of genetic diversity among black grama plants that survived drought. The percentage of polymorphic loci was calculated for sample pools using data for loci for which there was 5-fold or greater sequence coverage in GBS analyses. Circles show the data from each plot. The “X” shows the treatment mean ± s.e. Letters indicate significant differences between means following FDR correction for pairwise contrasts.
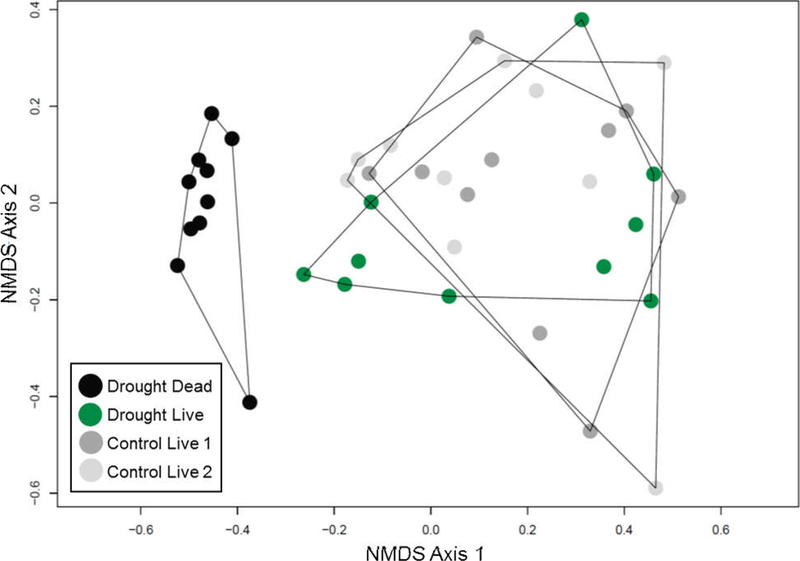
Allelic composition significantly diverged among the four cohort types (perMANOVA: R2 = 0.30, pseudo-F3,36 = 5.1, P < 0.0001; visualized with NMDS in Fig. 6). Composition differed both between the dead and living plants exposed to drought (R2 = 0.31, pseudo-F1,18 = 9.1, P < 0.0001) and between the dead plants in drought treatments and living plants in control plots (R2 = 0.30, pseudo-F1,28 = 11.8, P < 0.0001). Multivariate dispersion in allelic composition was not significantly different between live and dead cohorts within drought treatments (permutation test, F = 2.0, P = 0.18) or between cohorts of dead plants in drought treatments and controls (permutation test, F = 2.7, P = 0.11), indicating similar among-plot variability. PCA analysis employing genetic distances derived from sequence variants also supported results of these other analyses. Cohorts of plants that survived drought clustered more closely with each other than did control cohorts or drought dead cohorts. In fact, none of the outliers in this analysis were from plants that survived drought treatments. In contrast, plants that died under drought were more unique than those that survived drought, or those in adjacent control plots (Online Resource 1). These patterns are further supported by measures of within-sample allelic diversity. Drought dead samples had, on average, a higher number of alleles than any other cohort: 6206 alleles in drought dead vs 5641 in drought live (P = 0.0036), 5852 in control live 1 (P = 0.0684), and 5665 in control live 2 (P = 0.0022), although the difference was not always significant.
Fig. 6.
Nonmetric multidimensional scaling analysis showing two-dimensional plot of all cohorts examined for allelic composition, each cohort is an open circle. 2D stress = 0.15. Lines show ellipses around each type of cohort: two randomly selected cohorts of live plants from control plots (control live 1, control live 2), dead plants in drought treatments, or live plants in drought treatments. A color version of this figure accompanies the online version of this article
Discussion
Several lines of evidence support the conclusion that black grama populations experienced substantial genetic changes caused by the harsh 1-in-100-year experimental drought we imposed. Drought caused substantial mortality and reduced live black grama biomass by 96% relative to control plots. Allelic diversity of survivors, as measured by the percentage of polymorphic loci, was on average 11% lower than the plants killed by drought. In addition, cohorts of plants that died during drought were differentiated from both drought survivors and living plants in control plots (20–25% larger FST, and altered allelic composition). These results are consistent with previous studies on both plants and animals that have correlated specific climatic events with reduced genetic variability (e.g., Nevo et al. 2012; Rubidge et al. 2012). However, like Jump et al. (2008), Avolio et al. (2013), and Ravenscroft et al. (2015), our experimental approach allowed us to assign causality to drought, side-stepping the correlation-not-causation issue inherent in tracking changes in genetic structure following a natural event.
The observed genetic differentiation between drought survivors and non-survivors is strong evidence of the action of natural selection. A key future direction would be to functionally characterize the genes differing between these groups, which may provide insight into the basis of drought adaptation in this species and allow comparisons to studies on other plant species (e.g., Franks et al. 2016; Yoder et al. 2014). However, the general reduction in allelic diversity in the drought-stressed populations is also consistent with (but does not prove) a population bottleneck, which would involve genome-wide loss of diversity at neutral loci via genetic drift. Again, characterization of genes as neutral vs. non-neutral could resolve the bottleneck question by determining both the degree to which neutral loci have been affected.
Potential community-level consequences of genetic diversity loss
Our experiment simulated the effects of a short-term (three-year), but extreme, drought. A corresponding event in the wild might have varied genetic effects, depending for instance, on whether the drought encompassed only a restricted geographic area or the entire range of the species. In the former case, gene flow from unaffected populations might reduce or erase the genetic signature of the drought over time. In the latter, genetic diversity loss across the species range might limit future adaptation, reduce population-level resilience to future climatic extremes, and/or increase inbreeding depression (Potvin and Tousignant 1996).
As in most other plant species (see Pauls et al. 2013), we currently lack resolution on what the functional consequences of drought-induced losses of genetic diversity in black grama would be for the communities and ecosystems where it dominates. A recent meta-analysis demonstrated that adaptive genetic diversity within plant populations had more significant ecological consequences (e.g., for productivity, or diversity of food webs) than neutral genetic diversity (Whitlock 2014). Thus, it will be important to determine what traits, if any, are affected by the loss of allelic diversity in black grama. While trait-based studies remain dominated by interspecific trait variation, new work has been documenting significant within-species trait variability (reviewed by Siefert et al. 2015); in some cases, this variability is as ecologically important as the functional differences among species (Cook-Patton et al. 2011). A trait-based approach (e.g., McGill et al. 2006) could provide a mechanism to connect intraspecific phenotypic variation with genetic variation in dryland plant populations. Future experiments that manipulate genetic diversity in black grama populations by planting monocultures and genetic mixtures (e.g., Crawford and Whitney 2010, Atwater and Callaway 2015) could help to resolve the long-term ecological consequences of drought-induced changes to population genetic structure (Milla et al. 2009).
Unexpectedly high levels of genetic variation in black grama
To our knowledge, this is the first study to examine the population genetics of black grama. The high level of variation observed among plants on a local scale (< 1 hectare) was unexpected because field studies on levels of vegetative reproduction versus recruitment from seed have suggested that black grama reproduces primarily vegetatively (Valentine 1970; Peters and Yao 2012). Our results are more consistent with recent seed addition experiments that suggest high soil moisture levels favor episodic recruitment of black grama over relatively short time intervals (5–8 years) relative to the lifespan of this perennial grass (Moreno-de las Heras et al. 2016); episodic recruitment of sexually produced seeds could therefore underlie the relatively high standing genetic variation observed at small spatial scales. The existence of substantial local variation was critical to our ability to detect the effects of short-term abiotic stress in shaping population-genetic structure, and it suggests that black grama is an excellent candidate for future studies of evolutionary change in response to climate events in arid grasslands.
Although we captured substantial genetic variation using DNA-based methods, there may yet be additional epigenetic variation in black grama that we have missed with our approach. For example, Rico et al. (2014) compared control versus experimentally drought-stressed Holm oak (Quercus ilex) and found significant differences in methylation patterns. Such epigenetic modification could have important population- and community level effects. For example, using similar genetic lines of Arabidopsis thaliana, Latzel et al. (2013) showed that an increase in epigenetic diversity alone boosted plant productivity by 20%. Epigenetic variation may be an important component of intraspecific functional diversity, and in some plant species, epigenetic diversity exceeds genetic diversity (Medrano et al. 2014). Transcriptomic approaches applied to drought-stressed versus unstressed field plants (e.g., Travers et al. 2010) could help to resolve possible epigenetic consequences of drought as well as to identify changes in plant gene expression that may explain the drought-tolerance mechanism in surviving plants.
Implications for management and the future of dryland ecosystems
Our results have applied significance for the management of black grama-dominated grasslands, for which restoration is desirable (Cox et al. 1986; Peters et al. 2006). Higher than expected genetic diversity in natural populations suggests that restoration efforts may benefit from maintaining naturally high levels of genetic diversity in plantings, rather than using clonal transplants (Lucero et al. 2010) or seed stock derived from few individuals. Because drought-induced suppression of genetic diversity could constrain the resilience of desert grasslands to future abiotic stress, efforts to augment diversity following extreme drought events could benefit long-term grassland stability.
Understanding the evolutionary consequences of climate change may be important for predicting the future of dryland ecosystems. Drylands constitute 45% of global land area (Pravalie 2016) and are expanding globally (Huang et al. 2016b). In many drylands, primary productivity is fueled by long-lived, perennial plants; changes in their population genetic structure could thus have long-lasting effects on the resistance and resilience of primary production to future stressors and contribute to the ecology memory of response to drought (Ogle et al. 2015). Drought is increasingly likely to be a significant stressor in drylands (IPCC 2013; Williams et al. 2013; Garfin et al. 2014; Shi et al. 2014), although the consequences of more frequent, longer, or more severe droughts for the population genetics of most dryland primary producers remain unresolved.
Supplementary Material
Acknowledgements.
This work was supported by the U. S. National Science Foundation EAGER 1748133, DEB 1456955, DEB 1257965, and EF-01137363; and in part by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103451. This research was also supported by U. S. National Science Foundation grants to the University of New Mexico for Long-term Ecological Research. Thanks for field assistance from Anny Chung, Aaron Robinson, and Eva Dettweiler-Robinson.
Footnotes
Author declaration statement: This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- Ahlstrom A, Raupach MR, Schurgers G, Smith B, Arneth A, Jung M, Reichstein M, Canadell JG, Friedlingstein P, Jain AK, Kato E, Poulter B, Sitch S, Stocker BD, Viovy N, Wang YP, Wiltshire A, Zaehle S, Zeng N (2015) The dominant role of semi-arid ecosystems in the trend and variability of the land CO2 sink. Science 348:895–899. doi: 10.1126/science.aaa1668 [DOI] [PubMed] [Google Scholar]
- Anand S, Mangano E, Barizzone N, Bordoni R, Sorosina M, Clarelli F, Corrado L, Boneschi FM, D’Alfonso S, De Bellis G (2016) Next generation sequencing of pooled samples: Guideline for variants’ filtering. Scientific Reports 6. doi: 10.1038/srep33735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT (2016) Plant fitness in a rapidly changing world. New Phytol. 210:81–87. doi: 10.1111/nph.13693 [DOI] [PubMed] [Google Scholar]
- Atwater DZ, Callaway RM (2015) Testing the mechanisms of diversity-dependent overyielding in a grass species. Ecology 96:3332–3342. doi: 10.1890/15-0889.1 [DOI] [PubMed] [Google Scholar]
- Avolio ML, Beaulieu JM, Smith MD (2013) Genetic diversity of a dominant C-4 grass is altered with increased precipitation variability. Oecologia 171:571–581. doi: 10.1007/s00442-012-2427-4 [DOI] [PubMed] [Google Scholar]
- Avolio ML, Smith MD (2013) Mechanisms of selection: Phenotypic differences among genotypes explain patterns of selection in a dominant species. Ecology 94:953–965 [Google Scholar]
- Báez S, Collins SL (2008) Shrub invasion decreases diversity and alters community stability in northern Chihuahuan Desert plant communities. Plos One 3:e2332. doi: 10.1371/journal.pone.0002332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez S, Collins SL, Pockman WT, Johnson JE, Small EE (2013) Effects of experimental rainfall manipulations on Chihuahuan Desert grassland and shrubland plant communities. Oecologia 172:1117–1127. doi: 10.1007/s00442-012-2552-0 [DOI] [PubMed] [Google Scholar]
- Bangert RK, Lonsdorf EV, Wimp GM, Shuster SM, Fischer D, Schweitzer JA, Allan GJ, Bailey JK, Whitham TG (2008) Genetic structure of a foundation species: scaling community phenotypes from the individual to the region. Heredity 100:121–131. doi: 10.1038/sj.hdy.6800914 [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67:1–48 [Google Scholar]
- Carroll SP, Jorgensen PS, Kinnison MT, Bergstrom CT, Denison RF, Gluckman P, Smith TB, Strauss SY, Tabashnik BE (2014) Applying evolutionary biology to address global challenges. Science 346:313-+. doi: 10.1126/science.1245993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SL, Xia Y (2015) Long-term dynamics and hotspots of change in a desert grassland plant community. Am. Nat. 185:E30–E43. doi: 10.1086/679315 [DOI] [PubMed] [Google Scholar]
- Connin SL, Virginia RA, Chamberlain CP (1997) Carbon isotopes reveal soil organic matter dynamics following arid land shrub expansion. Oecologia 110:374–386. doi: 10.1007/s004420050172 [DOI] [PubMed] [Google Scholar]
- Cook BI, Ault TR, Smerdon JE (2015) Unprecedented 21st century drought risk in the American Southwest and Central Plains. Science Advances 1. doi: 10.1126/sciadv.1400082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook-Patton SC, McArt SH, Parachnowitsch AL, Thaler JS, Agrawal AA (2011) A direct comparison of the consequences of plant genotypic and species diversity on communities and ecosystem function. Ecology 92:915–923. doi: 10.1890/10-0999.1 [DOI] [PubMed] [Google Scholar]
- Cox JR, Martin MH, Ibarra FA, Morton HL (1986) Establishment of range grasses on various seedbeds at creosotebush Larrea tridentata sites in Arizona, USA, and Chihuahua, Mexico. J. Range Manag. 39:540–546. doi: 10.2307/3898767 [DOI] [Google Scholar]
- Crawford KM, Rudgers JA (2012) Plant species diversity and genetic diversity within a dominant species interactively affect plant community biomass. J. Ecol. 100:1512–1521. doi: 10.1111/j.1365-2745.2012.02016.x [DOI] [Google Scholar]
- Crawford KM, Whitney KD (2010) Population genetic diversity influences colonization success. Mol. Ecol. 19:1253–1263 [DOI] [PubMed] [Google Scholar]
- Crutsinger GM, Collins MD, Fordyce JA, Gompert Z, Nice CC, Sanders NJ (2006) Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313:966–968. doi: 10.1126/science.1128326 [DOI] [PubMed] [Google Scholar]
- Dick-Peddie WA (1993) New Mexico vegetation: past, present, and future. University of New Mexico Press, Albuquerque, NM [Google Scholar]
- Ellison AM, Bank MS, Clinton BD, Colburn EA, Elliott K, Ford CR, Foster DR, Kloeppel BD, Knoepp JD, Lovett GM, Mohan J, Orwig DA, Rodenhouse NL, Sobczak WV, Stinson KA, Stone JK, Swan CM, Thompson J, Von Holle B, Webster JR (2005) Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front. Ecol. Environ. 3:479–486. doi: 10.1890/1540-9295(2005)003[0479:LOFSCF]2.0.CO;2 [DOI] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6. doi: 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW (2005) Interpreting the ‘selection effect’ of biodiversity on ecosystem function. Ecol. Lett. 8:846–856. doi: 10.1111/j.1461-0248.2005.00795.x [DOI] [Google Scholar]
- Fracassetti M, Griffin PC, Willi Y (2015) Validation of pooled whole-genome re-sequencing in Arabidopsis lyrata. PLoS One 10. doi: 10.1371/journal.pone.0140462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Kane NC, O’Hara NB, Tittes S, Rest JS (2016) Rapid genome-wide evolution in Brassica rapa populations following drought revealed by sequencing of ancestral and descendant gene pools. Mol. Ecol. 25:3622–3631. doi: 10.1111/mec.13615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE (2007) Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Nat. Acad. Sci. USA 104:1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Weber JJ, Aitken SN (2014) Evolutionary and plastic responses to climate change in terrestrial plant populations. Evol. Appl. 7:123–139. doi: 10.1111/eva.12112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfin G, Franco G, Blanco H, Comrie A, Gonzalez P, Piechota T, Smyth R, Waskom R (2014) Chapter 20: Southwest In: Melillo JM, Richmond TC, Yohe GW (eds) Climate Change Impacts in the United States: The Third National Climate Assessment. U.S. Global Change Research Program, pp 462–486 [Google Scholar]
- Gautier M, Foucaud J, Gharbi K, Cezard T, Galan M, Loiseau A, Thomson M, Pudlo P, Kerdelhue C, Estoup A (2013) Estimation of population allele frequencies from next-generation sequencing data: pool-versus individual-based genotyping. Mol. Ecol. 22:3766–3779. doi: 10.1111/mec.12360 [DOI] [PubMed] [Google Scholar]
- Gogarten SM, Zheng X (2017) SeqVarTools: Tools for variant data. R package version 1.8.1, R package version 1.8.1 edn [Google Scholar]
- Gosz RJ, Gosz JR (1996) Species interactions on the biome transition zone in New Mexico: Response of blue grama (Bouteloua gracilis) and black grama (Bouteloua eripoda) to fire and herbivory. J. Arid Environ. 34:101–114. doi: 10.1006/jare.1996.0096 [DOI] [Google Scholar]
- Gould F (1979) The genus Bouteloua (Poaceae). Annals of the Missouri Botanical Garden 66:348–416. doi: 10.2307/2398834 [DOI] [Google Scholar]
- Hart SP, Schreiber SJ, Levine JM (2016) How variation between individuals affects species coexistence. Ecol. Lett. 19:825–838. doi: 10.1111/ele.12618 [DOI] [PubMed] [Google Scholar]
- Huang JP, Ji MX, Xie YK, Wang SS, He YL, Ran JJ (2016a) Global semi-arid climate change over last 60 years. Climate Dynamics 46:1131–1150. doi: 10.1007/s00382-015-2636-8 [DOI] [Google Scholar]
- Huang JP, Yu HP, Guan XD, Wang GY, Guo RX (2016b) Accelerated dryland expansion under climate change. Nature Climate Change 6:166-+. doi: 10.1038/nclimate2837 [DOI] [Google Scholar]
- Huber H, During HJ, de Bruin FB, Vermeulen PJ, Anten NPR (2016) Genotypic and phenotypic diversity does not affect productivity and drought response in competitive stands of Trifolium repens. Frontiers in Plant Science 7:364. doi: 10.3389/fpls.2016.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huenneke LF, Clason D, Muldavin E (2001) Spatial heterogeneity in Chihuahuan Desert vegetation: implications for sampling methods in semi-arid ecosystems. J. Arid Environ. 47:257–270. doi: 10.1006/jare.2000.0678 [DOI] [Google Scholar]
- Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M (2008) Ecological consequences of genetic diversity. Ecol. Lett. 11:609–623. doi: 10.1111/j.1461-0248.2008.01179.x [DOI] [PubMed] [Google Scholar]
- Ikeda DH, Max TL, Allan GJ, Lau MK, Shuster SM, Whitham TG (2017) Genetically informed ecological niche models improve climate change predictions. Global Change Biol. 23:164–176. doi: 10.1111/gcb.13470 [DOI] [PubMed] [Google Scholar]
- IPCC (2013) Climate Change 2013: The Physical Science Basis Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge Press, United Kingdom and New York, NY, USA [Google Scholar]
- Isbell F, Craven D, Connolly J, Loreau M, Schmid B, Beierkuhnlein C, Bezemer TM, Bonin C, Bruelheide H, de Luca E, Ebeling A, Griffin JN, Guo QF, Hautier Y, Hector A, Jentsch A, Kreyling J, Lanta V, Manning P, Meyer ST, Mori AS, Naeem S, Niklaus PA, Polley HW, Reich PB, Roscher C, Seabloom EW, Smith MD, Thakur MP, Tilman D, Tracy BF, van der Putten WH, van Ruijven J, Weigelt A, Weisser WW, Wilsey B, Eisenhauer N (2015) Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 526:574–U263. doi: 10.1038/nature15374 [DOI] [PubMed] [Google Scholar]
- Jimenez-Alfaro B, Garcia-Calvo L, Garcia P, Luis Acebes J (2016) Anticipating extinctions of glacial relict populations in mountain refugia. Biol. Conserv. 201:243–251. doi: 10.1016/j.biocon.2016.07.015 [DOI] [Google Scholar]
- Jump AS, Marchant R, Penuelas J (2009) Environmental change and the option value of genetic diversity. Trends Plant Sci. 14:51–58. doi: 10.1016/j.tplants.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Jump AS, Penuelas J, Rico L, Ramallo E, Estiarte M, Martinez-Izquierdo JA, Lloret F (2008) Simulated climate change provokes rapid genetic change in the Mediterranean shrub Fumana thymifolia. Global Change Biol. 14:637–643. doi: 10.1111/j.1365-2486.2007.01521.x [DOI] [Google Scholar]
- Knapp AK, Avolio ML, Beier C, Carroll CJW, Collins SL, Dukes JS, Fraser LH, Griffin-Nolan RJ, Hoover DL, Jentsch A, Loik ME, Phillips RP, Post AK, Sala OE, Slette IJ, Yahdjian L, Smith MD (2017) Pushing precipitation to the extremes in distributed experiments: recommendations for simulating wet and dry years. Global Change Biol. 23:1774–1782. doi: 10.1111/gcb.13504 [DOI] [PubMed] [Google Scholar]
- Knapp AK, Hoover DL, Wilcox KR, Avolio ML, Koerner SE, La Pierre KJ, Loik ME, Luo YQ, Sala OE, Smith MD (2015) Characterizing differences in precipitation regimes of extreme wet and dry years: implications for climate change experiments. Global Change Biol. 21:2624–2633. doi: 10.1111/gcb.12888 [DOI] [PubMed] [Google Scholar]
- Kofler R, Nolte V, Schloetterer C (2016) The impact of library preparation protocols on the consistency of allele frequency estimates in Pool-Seq data. Mol. Ecol. Res. 16:118–122. doi: 10.1111/1755-0998.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latzel V, Allan E, Silveira AB, Colot V, Fischer M, Bossdorf O (2013) Epigenetic diversity increases the productivity and stability of plant populations. Nat. Comm. 4:UNSP-2875. doi: 10.1038/ncomms3875 [DOI] [PubMed] [Google Scholar]
- Lau JA, Lennon JT (2012) Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Nat. Acad. Sci. USA 109:14058–14062. doi: 10.1073/pnas.1202319109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauenroth WK, Adler PB (2008) Demography of perennial grassland plants: survival, life expectancy and life span. J. Ecol. 96:1023–1032. doi: 10.1111/j.1365-2745.2008.01415.x [DOI] [Google Scholar]
- Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES, Costich DE (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet. 9:e1003215. doi: 10.1371/journal.pgen.1003215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero ME, Dreesen DR, VanLeeuwen DM (2010) Using hydrogel filled, embedded tubes to sustain grass transplants for arid land restoration. J. Arid Environ. 74:987–990. doi: 10.1016/j.jaridenv.2010.01.007 [DOI] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: From genotyping to genome typing. Nat. Rev.Gen. 4:981–994. doi: 10.1038/nrg1226 [DOI] [PubMed] [Google Scholar]
- Lynch M, Bost D, Wilson S, Maruki T, Harrison S (2014) Population-genetic inference from pooled-sequencing data. Genome Biol. and Evol. 6:1210–1218. doi: 10.1093/gbe/evu085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill BJ, Enquist BJ, Weiher E, Westoby M (2006) Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21:178–185. doi: 10.1016/j.tree.2006.02.002 [DOI] [PubMed] [Google Scholar]
- McManus CJ, Kelley ST (2005) Molecular survey of aeroplane bacterial contamination. J. Appl. Microbiol. 99:502–508. doi: 10.1111/j.1365-2672.2005.02651.x [DOI] [PubMed] [Google Scholar]
- Medrano M, Herrera CM, Bazaga P (2014) Epigenetic variation predicts regional and local intraspecific functional diversity in a perennial herb. Mol. Ecol. 23:4926–4938. doi: 10.1111/mec.12911 [DOI] [PubMed] [Google Scholar]
- Merilä J, Hendry AP (2014) Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol. Appl. 7:1–14. doi: 10.1111/eva.12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla R, Forero DM, Escudero A, Iriondo JM (2009) Growing with siblings: a common ground for cooperation or for fiercer competition among plants? Proc. Roy. Soc. B – Biol. Sci. 276:2531–2540. doi: 10.1098/rspb.2009.0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira X, Abdala-Roberts L, Parra-Tabla V, Mooney KA (2014) Positive effects of plant genotypic and species diversity on anti-herbivore defenses in a tropical tree species. PLoS One 9:e105438. doi: 10.1371/journal.pone.0105438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-de las Heras M, Turnbull L, Wainwright J (2016) Seed-bank structure and plant-recruitment conditions regulate the dynamics of a grassland-shrubland Chihuahuan ecotone. Ecology 97:2303–2318. doi: 10.1002/ecy.1446 [DOI] [PubMed] [Google Scholar]
- Muldavin EH, Moore DI, Collins SL, Wetherill KR, Lightfoot DC (2008) Aboveground net primary production dynamics in a northern Chihuahuan Desert ecosystem. Oecologia 155:123–132. doi: 10.1007/s00442-007-0880-2 [DOI] [PubMed] [Google Scholar]
- Nevo E, Fu Y-B, Pavlicek T, Khalifa S, Tavasi M, Beiles A (2012) Evolution of wild cereals during 28 years of global warming in Israel. Proc. Nat. Acad. Sci. USA 109:3412–3415. doi: 10.1073/pnas.1121411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol AA (1952) The natural vegetation of Arizona [revisions by Phillips, W. S.], Tucson, AZ, pp 189–230 [Google Scholar]
- Notaro M, Liu ZY, Gallimore RG, Williams JW, Gutzler DS, Collins S (2010) Complex seasonal cycle of ecohydrology in the Southwest United States. J. Geophys. Res. – Biogeosci. 115. doi: 10.1029/2010jg001382 [DOI] [Google Scholar]
- Ogle K, Barber JJ, Barron-Gafford GA, Bentley LP, Young JM, Huxman TE, Loik ME, Tissue DT (2015) Quantifying ecological memory in plant and ecosystem processes. Ecol. Lett. 18:221–235. doi: 10.1111/ele.12399 [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2016) vegan: Community Ecology Package. R package version 2.3–5. http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Parmenter RR (2008) Long-term effects of a summer fire on desert grassland plant demographics in now Mexico. Rangeland Ecol. Manag. 61:156–168. doi: 10.2111/07-010.1 [DOI] [Google Scholar]
- Parmesan C (2006) Ecological and evolutionary responses to recent climate change. Ann. Rev. Ecol. Syst. 37:637–669 [Google Scholar]
- Pauls SU, Nowak C, Balint M, Pfenninger M (2013) The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 22:925–946. doi: 10.1111/mec.12152 [DOI] [PubMed] [Google Scholar]
- Paulsen HA Jr., Ares FN (1962) Grazing values and management of black grama and tobosa grasslands and associated shrub ranges of the Southwest. U.S. Department of Agriculture, Forest Service, Washington, DC, p 56 [Google Scholar]
- Peters DPC (2002) Recruitment potential of two perennial grasses with different growth forms at a semiarid-arid transition zone. Am. J. Bot. 89:1616–1623. doi: 10.3732/ajb.89.10.1616 [DOI] [PubMed] [Google Scholar]
- Peters DPC, Herrick JE, Monger HC, Huang H (2010) Soil-vegetation-climate interactions in arid landscapes: Effects of the North American monsoon on grass recruitment. J. Arid Environ. 74:618–623. doi: 10.1016/j.jaridenv.2009.09.015 [DOI] [Google Scholar]
- Peters DPC, Mariotto I, Havstad KM, Murray LW (2006) Spatial variation in remnant grasses after a grassland-to-shrubland state change: Implications for restoration. Rangeland Ecol. Manag. 59:343–350. doi: 10.2111/05-202R1.1 [DOI] [Google Scholar]
- Peters DPC, Yao J (2012) Long-term experimental loss of foundation species: consequences for dynamics at ecotones across heterogeneous landscapes. Ecosphere 3:UNSP-27. doi: 10.1890/ES11-00273.1 [DOI] [Google Scholar]
- Pfeiffer F, Groeber C, Blank M, Haendler K, Beyer M, Schultze JL, Mayer G (2018) Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Scientific Reports 8. doi: 10.1038/s41598-018-29325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin C, Tousignant D (1996) Evolutionary consequences of simulated global change: Genetic adaptation or adaptive phenotypic plasticity. Oecologia 108:683–693. doi: 10.1007/BF00329043 [DOI] [PubMed] [Google Scholar]
- Prati D, Peintinger M, Fischer M (2016) Genetic composition, genetic diversity and small-scale environmental variation matter for the experimental reintroduction of a rare plant. J. Plant Ecol. 9:805–813. doi: 10.1093/jpe/rtv067 [DOI] [Google Scholar]
- Pravalie R (2016) Drylands extent and environmental issues. A global approach. Earth-Sci. Rev. 161:259–278. doi: 10.1016/j.earscirev.2016.08.003 [DOI] [Google Scholar]
- R Core Team (2017) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Ravenscroft CH, Whitlock R, Fridley JD (2015) Rapid genetic divergence in response to 15 years of simulated climate change. Global Change Biol. 21:4165–4176. doi: 10.1111/gcb.12966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds HG, Bohning JW (1956) Effects of burning on a desert grass-shrub range in southern Arizona. Ecology 37:769–777. doi: 10.2307/1933068 [DOI] [Google Scholar]
- Rico L, Ogaya R, Barbeta A, Penuelas J (2014) Changes in DNA methylation fingerprint of Quercus ilex trees in response to experimental field drought simulating projected climate change. Plant Biol. 16:419–427. doi: 10.1111/plb.12049 [DOI] [PubMed] [Google Scholar]
- Rubidge EM, Patton JL, Lim M, Burton AC, Brashares JS, Moritz C (2012) Climate-induced range contraction drives genetic erosion in an alpine mammal. Nature Climate Change 2:285–288. doi: 10.1038/NCLIMATE1415 [DOI] [Google Scholar]
- Rudgers JA, Chung YA, Maurer GE, Moore DI, Muldavin EH, Litvak ME, Collins SL (2018) Climate sensitivity functions and net primary production: A framework for incorporating climate mean and variability. Ecology 99:576–582. doi: 10.1002/ecy.2136 [DOI] [PubMed] [Google Scholar]
- Schloetterer C, Tobler R, Kofler R, Nolte V (2014) Sequencing pools of individuals-mining genome-wide polymorphism data without big funding. Nat. Rev. Gen. 15:749–763. doi: 10.1038/nrg3803 [DOI] [PubMed] [Google Scholar]
- Schoeb C, Kerle S, Karley AJ, Morcillo L, Pakeman RJ, Newton AC, Brooker RW (2015) Intraspecific genetic diversity and composition modify species-level diversity-productivity relationships. New Phytol. 205:720–730. doi: 10.1111/nph.13043 [DOI] [PubMed] [Google Scholar]
- Shi Z, Thomey ML, Mowll W, Litvak M, Brunsell NA, Collins SL, Pockman WT, Smith MD, Knapp AK, Luo Y (2014) Differential effects of extreme drought on production and respiration: synthesis and modeling analysis. Biogeosciences 11:621–633. doi: 10.5194/bg-11-621-2014 [DOI] [Google Scholar]
- Siefert A, Violle C, Chalmandrier L, Albert CH, Taudiere A, Fajardo A, Aarssen LW, Baraloto C, Carlucci MB, Cianciaruso MV, Dantas VdL, de Bello F, Duarte LDS, Fonseca CR, Freschet GT, Gaucherand S, Gross N, Hikosaka K, Jackson B, Jung V, Kamiyama C, Katabuchi M, Kembel SW, Kichenin E, Kraft NJB, Lagerstrom A, Le Bagousse-Pinguet Y, Li Y, Mason N, Messier J, Nakashizuka T, McC Overton J, Peltzer DA, Perez-Ramos IM, Pillar VD, Prentice HC, Richardson S, Sasaki T, Schamp BS, Schoeb C, Shipley B, Sundqvist M, Sykes MT, Vandewalle M, Wardle DA (2015) A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 18:1406–1419. doi: 10.1111/ele.12508 [DOI] [PubMed] [Google Scholar]
- Simonin KA (2000) Bouteloua eriopoda In: Fire Effects Information System, [Online]. U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station, Fire Sciences Laboratory [Google Scholar]
- Slowikowski K (2016) ggrepel: Repulsive text and label geoms for ‘ggplot2’. R package version 0.5, R package version 0.5. edn [Google Scholar]
- Streetman LJ, Wright N (1960) A cytological study of black gramagrass Bouteloua eriopoda. Am. J. Bot. 47:786–793. doi: 10.2307/2439116 [DOI] [Google Scholar]
- Thomey ML, Collins SL, Vargas R, Johnson JE, Brown RF, Natvig DO, Friggens MT (2011) Effect of precipitation variability on net primary production and soil respiration in a Chihuahuan Desert grassland. Global Change Biol. 17:1505–1515. doi: 10.1111/j.1365-2486.2010.02363.x [DOI] [Google Scholar]
- Travers SE, Tang ZW, Caragea D, Garrett KA, Hulbert SH, Leach JE, Bai JF, Saleh A, Knapp AK, Fay PA, Nippert J, Schnable PS, Smith MD (2010) Variation in gene expression of Andropogon gerardii in response to altered environmental conditions associated with climate change. J. Ecol. 98:374–383. doi: 10.1111/j.1365-2745.2009.01618.x [DOI] [Google Scholar]
- Valentine KA (1970) Influence of grazing intensity on improvement of deteriorated black grama range Bulletin 553 New Mexico State University Agricultural Experiment Station, Las Cruces, New Mexico, USA [Google Scholar]
- Van Auken OW (2000) Shrub invasions of North American semiarid grasslands. Ann. Rev. Ecol. Syst. 31:197–215. doi: 10.1146/annurev.ecolsys.31.1.197 [DOI] [Google Scholar]
- Whitlock R (2014) Relationships between adaptive and neutral genetic diversity and ecological structure and functioning: a meta-analysis. J. Ecol. 102:857–872. doi: 10.1111/1365-2745.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2009) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York, NY [Google Scholar]
- Williams AP, Allen CD, Macalady AK, Griffin D, Woodhouse CA, Meko DM, Swetnam TW, Rauscher SA, Seager R, Grissino-Mayer HD, Dean JS, Cook ER, Gangodagamage C, Cai M, McDowell NG (2013) Temperature as a potent driver of regional forest drought stress and tree mortality. Nature Climate Change 3:292–297. doi: 10.1038/nclimate1693 [DOI] [Google Scholar]
- Yahdjian L, Sala OE (2002) A rainout shelter design for intercepting different amounts of rainfall. Oecologia 133:95–101. doi: 10.1007/s00442-002-1024-3 [DOI] [PubMed] [Google Scholar]
- Yang L, Callaway RM, Atwater DZ (2017) Ecotypic diversity of a dominant grassland species resists exotic invasion. Biol. Invasions 19:1483–1493. doi: 10.1007/s10530-017-1373-9 [DOI] [Google Scholar]
- Yoder JB, Stanton-Geddes J, Zhou P, Briskine R, Young ND, Tiffin P (2014) Genomic signature of adaptation to climate in Medicago truncatula. Genetics 196:1263-+. doi: 10.1534/genetics.113.159319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Gogarten SM (2015) SeqArray: Big data management of genome-wide sequence variants. R package version 1.10.6. http://github.com/zhengxwen/SeqArray [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. doi: 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.