Abstract
Diels–Alder reactions are among the most powerful synthetic transformations to construct complex natural products. Despite that increasing of enzymatic intramolecular Diels–Alder reactions have been discovered, natural intermolecular Diels–Alderases are rarely described. Here, we report an intermolecular hetero-Diels–Alder reaction in the biosynthesis of tropolonic sesquiterpenes and functionally characterize EupfF as the first fungal intermolecular hetero-Diels–Alderase. We demonstrate that EupfF catalyzed the dehydration of a hydroxymethyl-containing tropolone (5) to generate a reactive tropolone o-quinone methide (6) and might further stereoselectively control the subsequent intermolecular hetero-Diels–Alder reaction with (1E,4E,8Z)-humulenol (8) to produce enantiomerically pure neosetophomone B (1). Our results reveal the biosynthetic pathway of 1 and expand the repertoire of activities of Diels–Alder cyclases.
Diels–Alder (DA) reactions, in which a 1,3-diene and a dienophile react to form an unsaturated cyclohexane regio- and stereoselectively are one of the most powerful synthetic strategies for the synthesis of complex natural products.1 Both intramolecular and intermolecular Diels–Alder cycloadditions are presumed to introduce structure complexity and biological activity in natural compounds. Although increasing numbers of intramolecular Diels–Alderases or [4 + 2]-cyclases2 such as LepI,3 SpnF,4 PyrE3,5 IccD,6 PvhB,7 and others,8 have been discovered in Nature, direct biochemical evidence of enzyme-catalyzed intermolecular DA reactions have rarely been described except for macrophomate synthase and riboflavin synthase.2 Notably, natural products probably biosynthesized via an intermolecular Diels–Alder reaction are frequently reported from bacteria, fungi, and plants9 (Figure S1).
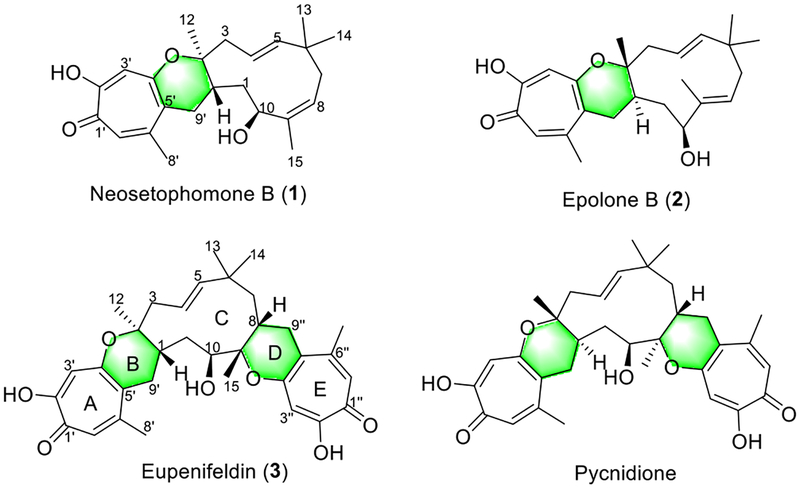
Remarkably, several members of the tropolone-sesquiterpene family of meroterpenoids (Figure S2) represented by neosetophomone B (1),10 epolone B (2),11 eupenifeldin (3),12 and pycnidione,13 isolated as optically pure compounds from a variety of fungi, were proposed to be biosynthesized via one or two tandem hetero Diels–Alder (hDA) reactions (Figure 1).14 Structurally, these meroterpenoids possess a unique central 11-membered macrocycle flanked by one or two dihydropyrans coupled to tropolones. Biologically, 1 and 3 were found as very potent antitumor agents with activity at the nanomole level in in vitro tumor cell lines,10,12 while 2 and pycnidione are stromelysin inhibitors and potential antiarthritic drugs.13
Figure 1.
Representative tropolone-sesquiterpenes probably generated from intermolecular Diels–Alder reactions.
Studies toward the biomimetic synthesis of tropolone-sesquiterpenes showed the generation of monotropolone adducts via hDA reaction under thermal conditions achieved success only for unnatural model compounds.15 In fact, the total synthesis of these tropolone-sesquiterpenes has not been completed thus far. It is therefore of interest to identify strategies and enzymes Nature uses to synthesize compounds of this family, especially the structures with observed in 1.
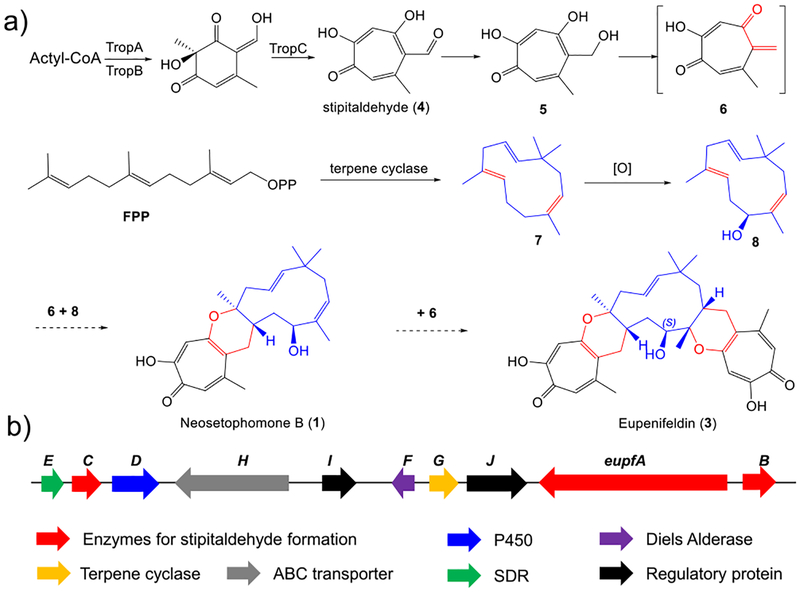
Biosynthetically, hDA reactions of a tropolone o-quinone methide with humulene were proposed for the biosynthesis of 1 and 2, which further served as an intermediate for the biosynthesis of eupenifeldin and pycnidione, respectively11,16- (Figure 2a). While biosynthesis of the tropolone core in stipitatonic acid has been identified (Figure 2a), including a nonreduced polyketide synthase (NR-PKS, TropA), a FAD-dependent monooxygenase (FMO, TropB), and an α-ketoglutarate-dependent dioxygenase (α-KGD, TropC),17 the enzymatic basis for the DA reaction for 1 biosynthesis has remained unexplored. Here, we report mapping of the cascade of enzymatic steps that furnish the structure of 1, which include a cytochrome P450 monooxygenase (P450), a short-chain dehydrogenase (SDR), and an intermolecular hetero-Diels–Alderase (hDAase) as keys. We demonstrate that the hDAase is responsible for the formation of a highly reactive tropolone o-quinone methide and may further stereoselectively control the subsequent intermolecular hDA cycloaddition to produce 1.
Figure 2.
Proposed biosynthetic pathways of 1 and 3 (a) and the eupf gene cluster in P. janthinellum (b).
Recently, we coisolated 1 and 3 from the wild-type Penicillium janthinellum, a producer of alanditrypinone.18 Analysis of the crude extract by liquid chromatography mass spectrometry (LC–MS) led to the observation of minor 1 and major 3 with characteristic UV absorption of tropolones and molecular weights of 384 and 548, respectively (Figure S3). Compounds 1 and 3 were fully characterized by NMR and circular dichroism spectra as enantiomerically pure neosetophomone B and eupenifeldin, respectively. To investigate the biosynthetic pathway of 1, the genome of P. janthinellum was queried for the presence of the trop-like gene cluster. Bioinformatics analysis led to the identification of only one trop-like NR-PKS gene cluster (named eupf cluster) in its genome (Figures 2b and S4). In addition to three enzymes (EupfA–C) which are homologues to TropA, TropB, and TropC, respectively, the eupf cluster encodes additionally a cytochrome P450 monooxygenase (EupfD), a short-chain dehydrogenase (EupfE), two hypothetic enzymes (EupfF and EupfG), as well as an ABC transporter and regulators (Figure 2b). EupfF and EupfG show high homology to AsR5 and AsR6 (64% and 62%), respectively, which were recently suggested as putative hetero-Diels–Alderase and humulene synthase, respectively, in the biosynthesis of xenovulene A.19 A biosynthetic gene cluster eup in Phoma sp. has been recently identified by gene deletion for the biosynthesis of eupenifeldin and EupF was also suggested as putative hDAase.21 However, the function of AsR5 and EupF has not been successfully characterized. It was scrutable that EupfABC were responsible for the formation of stipitaldehyde, and EupfG served as a humulene synthase to form a sesquiterpene, while the only oxygenase EupfD could catalyze the hydroxylation at the terpene moiety at an uncertain stage. The putative hDAase EupfF, homologous to AsR5 and EupF, was proposed to be involved in the cyclization between diene and dienophile.
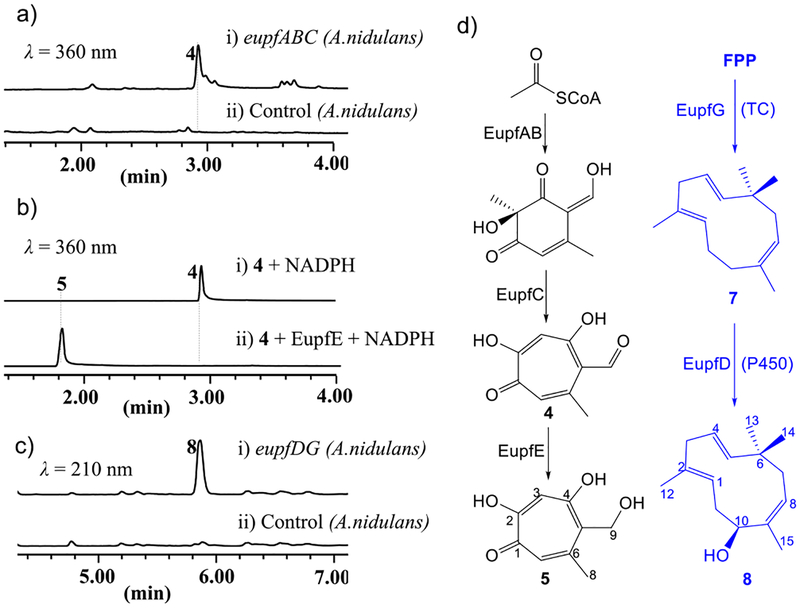
To verify the eupf gene cluster and elucidate the biosynthetic pathway of 1, heterologous expression experiments in Aspergillus nidulans were conducted. As expected, expression of eupfABC in A. nidulans led to the production of major stipitaldehyde (4, Figure 3a), which was suggested as an intermediate for biosynthesis of xenovulene A and other tropolone-sesquiterpenes. To form a diene precursor for the DA reaction, the aldehyde moiety in 4 has to be reduced first. As the only potential reductase in the eupf cluster, Eupf E is highly likely to be involved in this transformation. To verify the function of EupfE, intron-free eupf E was cloned from cDNA, heterologously expressed in Escherichia coli, and purified to homogeneity (Figure S6). Compared to control experiments where missing EupfE did not generate new products, incubation of EupfE with 4 and NADPH led to complete conversion to a new product 5 (Figure 3b) with characteristic UV absorption of tropolone and MW of 182, 2 Da more than that of 4 (Figure S7). Although 5 was not stable enough, careful isolation and quick NMR analysis led to the full identification of 5 as we expected (Figure 3d). Dehydration of 5 either spontaneously or enzymatically was proposed to generate the reactive tropolone o-quinone methide (6).
Figure 3.
Generation of potential precursors in the biosynthesis of 1. (a–c) LCMS traces from heterologous expression of eupfABC/DG in A. nidulans or from in vitro assay of EupfE. (d) Generation of 5 and 8.
To identify the dienophile precursor, EupfG, the putative humulene synthase was expressed in A. nidulans, which led to the detection of a compound 7 with the same molecular weight (m/z 204) but different retention time from that of α-humulene (9) standard indicated by GC–MS analysis (Figure S8), which suggested 7 was an isomer of α-humulene. Isolation of 7 was found to be difficult due to its high volatility. The structure of 7 was suggested by coexpression of eupf G and eupf D in A. nidulans, which led to the production of 8 (Figure 3c, trace (i)) with MW of 220, 16 Da more than that of α-humulene. Compound 8 was fully characterized by MS and NMR spectra as a hydroxylated product of 7 (Figure 3d), which also supported that 7 was (1E,4E,8Z)-humulene. Different from α-humulene in which all three olefins are in the E form, compounds 7 and 8 possess a Z form of the olefinic bond at Δ8,9, which is consistent with the Z form of alkene in 1. As a result, EupfG was identified as a new sesquiterpene synthase producing (1E,4E,8Z)-humulene other than α-humulene.
We realized that 5 could be O-methylated at C9-OH gradually and partly in methanol (Figure S9). Methylated 5 was isolated and fully characterized by NMR spectra as a tautomer mixture (2:1). The identification of methylated 5 (6a) also suggested the spontaneous conversion from 5 to 6 occurred before it underwent 1,4- or 1,6-addition by methanol (Figure S9). Due to the incomplete spontaneous conversion, we believed that enzyme was required to generate 6 from 5, and EupfF, the putative hetero-Diels–Alderase, was presumed to be involved in this process. However, repeated attempts to obtain soluble EupfF for biochemical assay failed. Fortunately, an enzyme, EupF, highly homologous to EupfF (81% similarity/67% identity), was found in the genome of Phoma sp., which was suggested but not characterized as putative hDAase for the biosynthesis of eupenifeldin.20 Attempts to obtain soluble EupF (42.0 kDa) from Escherichia coli BL21(DE3) were successful (Figure S10). To explore if EupF catalyzed the dehydration of 5, we performed in vitro EupF assay with 5 as the sole substrate. Since the expected product 6 was extremely instable, we added 1% glycerol in the in vitro system to trap 6. Consequently, 72.5% of 5 converted to two main products, 6b and 6c, with molecular weights of 256 and 346, respectively (Figure S11), by EupF after 2 h, while only 21.8% of 5 could be transformed without EupF. On the basis of the MS and UV, 6b and 6c were suggested to be 1,4-nucleophilic addition products of 6 with glycerol and 5, respectively. The results from the in vitro assay supported that dehydration of 5 could occur spontaneously, but EupF remarkably accelerated this process. Hence, the diene precursor 6 could be identified as tropolone o-quinone methide.
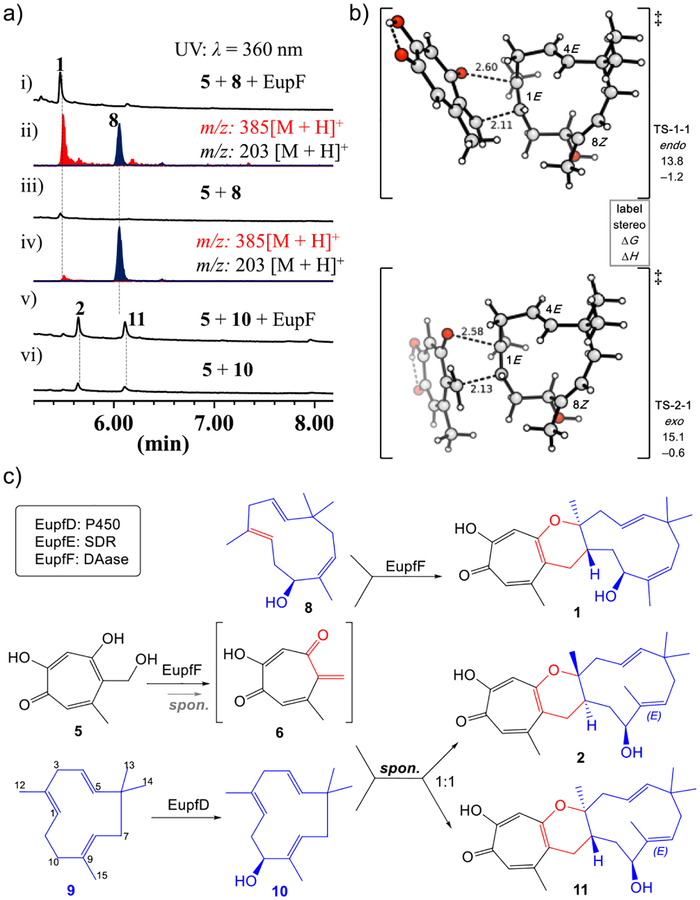
With 5 and 8 in hand, we were able to examine whether EupF was directly involved in the hDA reaction. We performed an in vitro reaction containing 30 μM EupF, 4.0 mM 5, and 1.0 mM 8 in 50 mM bicine buffer (pH 8.0). Whereas the control reaction without EupF did not lead to apparent new products, EupF catalyzed the conversion of 5 and 8 into 1 (Figures 4a and S12). Compound 1’s functionality as an enantiopure compound indicated the hDA reaction was enzymatically controlled since racemic products would be expected from the spontaneous reaction. Interestingly, when the natural substrate 8 was switched to unnatural substrate humulenol (10), which was obtained from expression of eupf D in A. nidulans by feeding humulene (9), in the in vitro EupF assay, two products 2 and 11 (1:1 ratio) were detected (Figure 4a, trace v). To obtain enough 2 and 11 for structural elucidation, 9 was fed to A. nidulans coexpressing eupfABCDEF (Figure S12). Finally, compounds 2 and 11 were fully characterized by NMR and CD as natural epolone B and its isomer, named isoepolone B (Figure 4c), respectively. Trace amounts of 2 and 11 were also detectable (Figure 4a, trace vi) in the absence of EupF with low yield compared to the enzymatic reaction. Hence, we suggested (1) the cycloaddition between the reactive tropolone o-quinone methide 6 and dienophile (8 or 10) could occur spontaneously, (2) EupF directly controlled the stereo-selectivity of hDA reaction in the case of natural substrate 8, and (3) unnatural substrate 10 was not recognized by EupF but underwent spontaneous cycloaddition with 6. Thus, EupF was fully characterized as the first hDAase, which differs from all intramolecular DAases in phylogenetic analysis (Figure S13) and has no recognizable cofactor-binding site. A model of EupfF (EupF) computed with SWISS-MODEL and template 2p4o suggest a predicted six-bladed propeller (Figure S14) that may provide suitable space for substrates. Although His, Lys, and Arg have been found as potential active sites for dehydration and DA reaction,21 mutagenesis experiments showed that conserved H37, R51, R92, and R323 were not catalytic residues for EupF (Figures S15–16), which suggested EupF was different from other DAases.
Figure 4.
Biosynthesis of 1 and 2. (a) LCMS traces from in vitro assay of EupF. (b) DFT-computed free energies for hDA reaction between 8 and 6. (c) Biosynthetic pathway of 1.
To support the proposed pathway, we performed density functional theory (DFT) studies on the Diels–Alder reaction at the PBE0-D3(BJ)/def2-TZVP level of theory.22–27 These calculations indicate that this cycloaddition is very facile (Figures 4b and S17). Transition states TS-1 and TS-2 lead to product 1 and are highly asynchronous with the two forming bonds differing by 0.49 and 0.45 Å, respectively. There is a 0.25 electron charge transfer in the transition state, in TS-1 the heterodiene has a −0.25 charge, and dienophile has a +0.25 charge. The charges are similar in TS-2. It is probable that the rate-determining step is generation of the o-quinone methide 6. After formation of 6, the reaction barriers of TS-1 and TS-2 are purely entropic (TS-1 ΔH‡ = −1.2, ΔG‡ = +13.8 kcal·mol−1, TS-2 ΔH‡ = −0.6, ΔG‡ = +15.1 kcal·mol−1). These reactions are enthalpically barrierless because the heterodiene is highly electron-deficient and a highly stable aromatic product is formed.
Unexpectedly, neither in vitro assay with 5 and 8 as substrates nor feeding experiments (Figure S18) led to production of 3 (only 1 was detected), which highly indicated that other or additional enzymes were required for the right side DA reaction in the biosynthesis of 3. Further investigation on the biosynthesis of bistropolone-sesquiterpenes is in progress.
In conclusion, we biochemically characterized EupfF (EupF) as the first fungal intermolecular hetero-Diels–Alderase and demonstrated that EupfF catalyzed the generation of a reactive tropolone o-quinone methide and further stereoselectively controlled the subsequent intermolecular hetero-Diels–Alder reaction to produce 1. Our discovery expands the collection of increasing DAases from fungi and opens a new strategy for mining and combinational biosynthesis of tropolone-sesquiterpenes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported financially by the CAMS Innovation Fund for Medical Sciences (CIFMS, No. 2017-I2M-4-004 to Y.H., and 2018-I2M-3-005 to Y.C.) and the Drug Innovation Major Project (2018ZX09711001-006 to Y.H.) as well as R01GM 124480 and NSF (CHE-1806581) to K.N.H. The UCLA Institute of Digital Research and Education (IDRE) provided computational resources for all calculations. We are grateful to Dr. Minghua Chen for ECD calculations.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b06592.
Experimental details, spectroscopic and computational data (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).(a) Dhambri S; Mohammad S; Van Buu ON; Galvani G; Meyer Y; Lannou MI; Sorin G; Ardisson J Recent advances in the synthesis of natural multifunctionalized decalins. Nat. Prod. Rep 2015, 32, 841–64. [DOI] [PubMed] [Google Scholar]; (b) Stocking EM; Williams RM Chemistry and biology of biosynthetic Diels-Alder reactions. Angew. Chem., Int. Ed 2003, 42, 3078–3115. [DOI] [PubMed] [Google Scholar]
- (2).Jeon BS; Wang SA; Ruszczycky MW; Liu HW Natural [4 + 2]-Cyclases. Chem. Rev 2017, 117, 5367–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ohashi M; Liu F; Hai Y; Chen M; Tang MC; Yang Z; Sato M; Watanabe K; Houk KN; Tang Y SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 2017, 549, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kim HJ; Ruszczycky MW; Choi SH; Liu YN; Liu HW Enzyme-catalysed [4 + 2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature 2011, 473, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tian Z; Sun P; Yan Y; Wu Z; Zheng Q; Zhou S; Zhang H; Yu F; Jia X; Chen D; Mandi A; Kurtan T; Liu W An enzymatic [4 + 2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol 2015, 11, 259–265. [DOI] [PubMed] [Google Scholar]
- (6).Zhang Z; Jamieson CS; Zhao YL; Li D; Ohashi M; Houk KN; Tang Y Enzyme-Catalyzed Inverse-Electron Demand Diels-Alder Reaction in the Biosynthesis of Antifungal Ilicicolin H. J. Am. Chem. Soc 2019, 141, 5659–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tan D; Jamieson CS; Ohashi M; Tang MC; Houk KN; Tang Y Genome-Mined Diels-Alderase Catalyzes Formation of the cis-Octahydrodecalins of Varicidin A and B. J. Am. Chem. Soc 2019, 141, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).(a) Li L; Tang MC; Tang S; Gao S; Soliman S; Hang L; Xu W; Ye T; Watanabe K; Tang Y Genome Mining and Assembly-Line Biosynthesis of the UCS1025A Pyrrolizidinone Family of Fungal Alkaloids. J. Am. Chem. Soc 2018, 140, 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li L; Yu P; Tang MC; Zou Y; Gao SS; Hung YS; Zhao M; Watanabe K; Houk KN; Tang Y Biochemical Characterization of a Eukaryotic Decalin-Forming Diels-Alderase. J. Am. Chem. Soc 2016, 138, 15837–15840. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhang B; Wang KB; Wang W; Wang X; Liu F; Zhu J; Shi J; Li LY; Han H; Xu K; Qiao HY; Zhang X; Jiao RH; Houk KN; Liang Y; Tan RX; Ge HM Enzyme-catalysed [6 + 4] cycloadditions in the biosynthesis of natural products. Nature 2019, 568, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Kusumi T; Ichikawa A; Kakisawa H; Tsunakawa M; Konishi M; Oki T The structures of quartromicins A1, A2, and A3: novel macrocyclic antiviral antibiotics possessing four tetronic acid moieties. J. Am. Chem. Soc 1991, 113, 8947–8948. [Google Scholar]; (b) Li CJ; Ma J; Sun H; Zhang D; Zhang DM Guajavadimer A, a Dimeric Caryophyllene-Derived Meroterpenoid with a New Carbon Skeleton from the Leaves of Psidium guajava. Org. Lett 2016, 18, 168–171. [DOI] [PubMed] [Google Scholar]; (c) Yang M-H; Gu M-L; Han C; Guo X-J; Yin G-P; Yu P; Kong L-Y; Aureochaeglobosins A-C Three [4 + 2] Adducts of Chaetoglobosin and Aureonitol Derivatives from Chaetomium globosum. Org. Lett 2018, 20, 3345–3348. [DOI] [PubMed] [Google Scholar]
- (10).El-Elimat T; Raja HA; Ayers S; Kurina SJ; Burdette JE; Mattes Z; Sabatelle R; Bacon JW; Colby AH; Grinstaff MW; Pearce CJ; Oberlies NH Meroterpenoids from Neosetophoma sp.: A Dioxa[4.3.3]propellane Ring System, Potent Cytotoxicity, and Prolific Expression. Org. Lett 2019, 21, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cai P; Smith D; Cunningham B; Brown-Shimer S; Katz B; Pearce C; Venables D; Houck D Epolones: novel sesquiterpene-tropolones from fungus OS-F69284 that induce erythropoietin in human cells. J. Nat. Prod 1998, 61, 791–795. [DOI] [PubMed] [Google Scholar]
- (12).Mayerl F; Gao Q; Huang S; Klohr SE; Matson JA; Gustavson DR; Pirnik DM; Berry RL; Fairchild C; Rose WC Eupenifeldin, a novel cytotoxic bistropolone from Eupenicillium brefeldianum. J. Antibiot 1993, 46, 1082–1088. [DOI] [PubMed] [Google Scholar]
- (13).Hsiao CJ; Hsiao SH; Chen WL; Guh JH; Hsiao G; Chan YJ; Lee TH; Chung CL Pycnidione, a fungus-derived agent, induces cell cycle arrest and apoptosis in A549 human lung cancer cells. Chem.-Biol. Interact 2012, 197, 23–30. [DOI] [PubMed] [Google Scholar]
- (14).Guo H; Roman D; Beemelmanns C Tropolone natural products. Nat. Prod. Rep 2019, 36, 1137–1155. [DOI] [PubMed] [Google Scholar]
- (15).(a) Adlington RM; Baldwin JE; Mayweg AV; Pritchard GJ Biomimetic cycloaddition approach to tropolone natural products via a tropolone ortho-quinone methide. Org. Lett 2002, 4, 3009–3011. [DOI] [PubMed] [Google Scholar]; (b) Baldwin JE; Mayweg AV; Neumann K; Pritchard GJ Studies toward the biomimetic synthesis of tropolone natural products via a hetero Diels-Alder reaction. Org. Lett 1999, 1, 1933–1935. [DOI] [PubMed] [Google Scholar]; (c) Adlington RM; Baldwin JE; Pritchard GJ; Williams AJ; Watkin DJ A biomimetic synthesis of lucidene. Org. Lett 1999, 1, 1937–1939. [DOI] [PubMed] [Google Scholar]
- (16).Zhang J; Liu L; Wang B; Zhang Y; Wang L; Liu X; Che Y Phomanolides A and B from the Fungus Phoma sp.: Meroterpenoids Derived from a Putative Tropolonic Sesquiterpene via Hetero-Diels-Alder Reactions. J. Nat. Prod 2015, 78, 3058–3066. [DOI] [PubMed] [Google Scholar]
- (17).al Fahad A; Abood A; Simpson TJ; Cox RJ The biosynthesis and catabolism of the maleic anhydride moiety of stipitatonic acid. Angew. Chem., Int. Ed 2014, 53, 7519–7523. [DOI] [PubMed] [Google Scholar]
- (18).Yan D; Chen Q; Gao J; Bai J; Liu B; Zhang Y; Zhang L; Zhang C; Zou Y; Hu Y Complexity and Diversity Generation in the Biosynthesis of Fumiquinazoline-Related Peptidyl Alkaloids. Org. Lett 2019, 21, 1475–1479. [DOI] [PubMed] [Google Scholar]
- (19).Schor R; Schotte C; Wibberg D; Kalinowski J; Cox RJ Three previously unrecognised classes of biosynthetic enzymes revealed during the production of xenovulene A. Nat. Commun 2018, 9, 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhai Y; Li Y; Zhang J; Zhang Y; Ren F; Zhang X; Liu G; Liu X; Che Y Identification of the gene cluster for bistropolone-humulene meroterpenoid biosynthesis in Phoma sp. Fungal Genet. Biol 2019, 129, 7–15. [DOI] [PubMed] [Google Scholar]
- (21).Cai Y; Hai Y; Ohashi M; Jamieson CS; Garcia-Borras M; Houk KN; Zhou J; Tang Y Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI. Nat. Chem 2019, 11, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Adamo C; Barone V Toward Reliable Density Functional Methods without Adjustable Parameters: The PBE0Model. J. Chem. Phys 1999, 110, 6158–6170. [Google Scholar]
- (23).Grimme S; Antony J; Ehrlich S; Krieg H A Consistent and Accurate Ab Initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J. Chem. Phys 2010, 132, 154104. [DOI] [PubMed] [Google Scholar]
- (24).Grimme S; Ehrlich S; Goerigk L Effect of the Damping Function in Dispersion Corrected Density Functional Theory. J. Comput. Chem 2011, 32, 1456–1465. [DOI] [PubMed] [Google Scholar]
- (25).Weigend F; Ahlrichs R Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys 2005, 7, 3297–3305. [DOI] [PubMed] [Google Scholar]
- (26).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Petersson GA; Nakatsuji H; Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Petersson GA; Nakatsuji H; Li X; Caricato M; Marenich AV; Bloino J; Janesko BG; Gomperts R; Mennucci B; Hratchian HP; Ortiz JV; Izmaylov AF; Sonnenberg JL; Williams-Young D; Ding F; Lipparini F; Egidi F; Goings J; Peng B; Petrone A; Henderson T; Ranasinghe D; Zakrzewski VG; Gao J; Rega N; Zheng G; Liang W; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Throssell K; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark MJ; Heyd JJ; Brothers EN; Kudin KN; Staroverov VN; Keith TA; Kobayashi R; Normand J; Raghavachari K; Rendell AP; Burant JC; Iyengar SS; Tomasi J; Cossi M; Millam JM; Klene M; Adamo C; Cammi R; Ochterski JW; Martin RL; Morokuma K; Farkas O; Foresman JB; Fox DJ; et al. Gaussian 16, revision A.03; Gaussian, Inc., 2016. [Google Scholar]
- (27).Ernzerhof M; Scuseria GE Assessment of the Perdew–Burke–Ernzerhof Exchange-Correlation Functional. J. Chem. Phys 1999, 110, 5029–5036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.