Abstract
Genetic diversity in human natural killer (NK) cell receptors is linked to resistance and susceptibility to many diseases. Here, we tested the effect of this diversity on the nanoscale organization of killer cell immunoglobulin (Ig)-like receptors (KIRs). Using superresolution microscopy, we found that inhibitory KIRs encoded by different genes and alleles were organized differently at the surface of primary human NK cells. KIRs that were found at low abundance assembled into smaller clusters than those formed by KIRs that were more highly abundant, and at low abundance there was a greater proportion of KIRs in clusters. Upon receptor triggering, a structured interface called the immune synapse assembles, which facilitates signal integration and controls NK cell responses. Here, triggering of low-abundance receptors resulted in less phosphorylation of the downstream phosphatase SHP-1 but more phosphorylation of the adaptor protein Crk than did that of high-abundance receptors. In cells with greater KIR abundance, SHP-1 dephosphorylated Crk, which potentiated NK cell spreading during activation. Thus, genetic variation modulates both the abundance and nanoscale organization of inhibitory KIRs. In other words, as well as the number of receptors at the cell surface varying with genotype, the way in which these receptors are organized in the membrane also varies. Essentially, a change in the average surface abundance of a protein at the cell surface is a coarse descriptor entwined with changes in local nanoscale clustering. Together, our data indicate that genetic diversity in inhibitory KIRs affects membrane-proximal signaling and, unexpectedly, the formation of activating immune synapses.
Introduction
Natural killer (NK) cells use various germline-encoded receptors to detect signs of disease. Inhibitory receptors that recognize human leukocyte antigen (HLA) class I prevent NK cells from killing healthy cells but enable NK cells to attack virus-infected or malignant cells that have downregulated HLA class I and thereby escaped the CD8+ T cell response. There are seven different inhibitory killer cell immunoglobulin (Ig)-like receptors (KIRs), one leukocyte Ig-like receptor (LILR), and one lectin-like receptor (NKG2A) that NK cells use to recognize HLA class KIR2DL1 recognizes the C2 epitope carried by a subset of HLA-C allotypes. All other HLA-C allotypes have the C1 epitope, which is recognized by KIR2DL2 and KIR2DL3. KIR3DL1 recognizes the Bw4 epitope of some HLA-B allotypes. NKG2A recognizes nonclassical HLA-E, whereas LILRB1 binds to all folded HLA class I. The KIR family is particularly diverse. Within human populations, there are many allotypic variants, and each individual carries only a selection of receptors. Diversity in the KIR genes is associated with disease susceptibility. For example, different inhibitory KIRs are associated with clearance of hepatitis C virus (1), AIDS-free survival for HIV-infected individuals (2), and successful pregnancy without complication (3).
To investigate how KIR variation influences disease susceptibility, research has focused so far on four aspects of how receptors recognize their ligands. First, inhibitory KIR signaling can be disrupted by certain peptides presented by HLA class I (4), and viral peptides also have this effect (5). If HLA class I proteins load viral peptides at a sufficiently high abundance, then those particular peptides might influence the immune response to the virus. Second, KIRs bind to HLA class I with variable avidities (6, 7), which could influence the threshold for NK cell activation when activating and inhibitory ligands are present simultaneously. Third, the amounts of KIRs at NK cell surfaces varies with both gene and allele (8, 9), and overall KIR genotype strongly influences the frequency of NK cells that express each KIR (10, 11). Thus, homozygosity for a KIR, which can lead to more cells expressing the receptor and at a greater abundance, can be beneficial (1). Fourth, KIR variation can influence NK cell education or licensing (12). However, as yet unexplored in the functional context of NK cell receptor diversity is the spatial organization of KIRs at the NK cell surface.
In exerting their biological functions, NK cells form a highly structured interface—the immune synapse—with target cells. On the target cell, HLA class I ligands cluster at the immune synapse (13), whereas on the NK cell, KIRs assemble in microclusters within 1 min of ligation (14) and KIR phosphorylation is sustained for several minutes (15). Although NKG2A and LILRB1 are not as well studied as is KIR, the evidence suggests that both receptors are recruited to the immune synapse upon ligation with their respective ligands (16, 17). If the immune synapse is disrupted by zinc chelation (13, 18) or by changing the sizes of the proteins present (19), inhibitory function is lost.
KIR2DL1 constitutively assembles in nanometer-scale clusters at NK cell surfaces (20). Using fluorescence correlation spectroscopy, Guia et al. showed that activating receptors differ in their nanoscale movements according to whether the NK cell is educated (21). In addition, activating ligands spaced 60 nm apart on a glass surface induce NK cell spreading, whereas ligands spaced 104 nm apart fail to induce NK cell activation (22). These results point to the functional importance of the nanoscale organization of NK cell receptors. Thus, we set out to determine whether the KIRs encoded by different genes or alleles varied in their nanoscale organization. We found that low-abundant, but highly clustered arrangements of receptors were associated with the increased phosphorylation of the adaptor protein Crk than were highly abundant receptors, and that this was associated with an impaired spreading response at activating synapses. Together, these data establish that different inhibitory KIRs organize differently at cell surfaces, which is associated with distinct receptor signaling and function.
Results
Inhibitory receptors encoded by different genes have distinct nanometer-scale arrangements
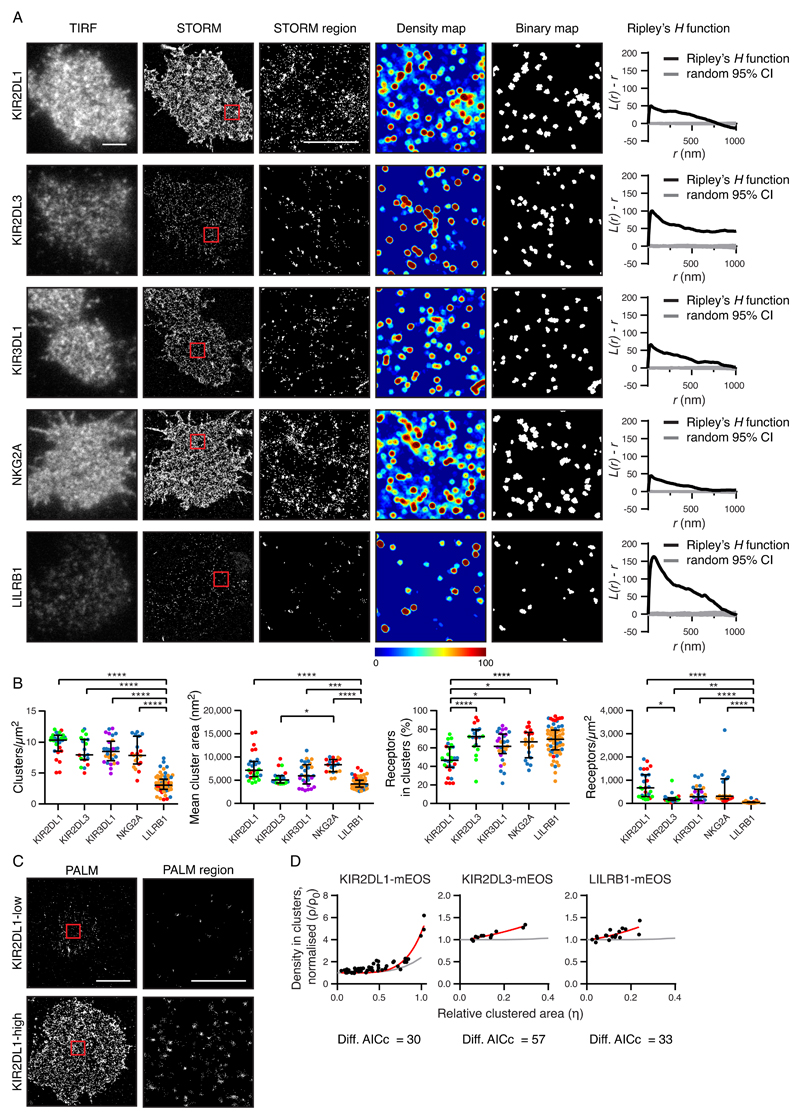
To determine whether inhibitory NK cell receptors encoded by different genes varied in their nanoscale organization, clones of primary human NK (pNK) cells were analyzed by flow cytometry and selected for their expression of five inhibitory NK cell receptors: KIR2DL1, KIR2DL3, KIR3DL1, NKG2A, and LILRB1 (fig.S1, A and B). The amount of receptor on pNK clones was of a similar order of magnitude to the range of receptor abundance on bulk NK cells (fig. S1C). Cells were imaged by total internal reflection (TIRF) microscopy and direct stochastic optical reconstruction microscopy (STORM), which produced coordinate-based data (fig. S2) with a localization precision of approximately 20 nm (23). All five receptors clustered on a nanometer-scale (Fig. 1A). This result was confirmed by Ripley’s H function analysis, where positive L(r) – r values indicate that receptors are more clustered and have more neighbors than in a random distribution. For further comparison of receptor organization, Getis and Franklin’s local point pattern analysis (G&F analysis) was used as the basis for quantitative clustering maps (Fig. 1A, fourth column). Density maps were color-coded on a spectrum from red (highest degree of clustering) to blue (lowest clustering). A suitable threshold (fig. S3A) was applied to generate a binary map containing clustered (above threshold) and nonclustered (below threshold) areas.
Fig. 1. Inhibitory receptors encoded by different genes have distinct nanometer-scale arrangements.
(A) Representative TIRF microscopy and STORM images of pNK cell clones (scale bar: 3 μm). STORM panels show the Gaussian-rendered image of coordinate data. The 2 × 2 μm regions (red boxes in the STORM images) are magnified, and the corresponding scatter plots, density maps (G&F analysis), and binary maps are shown (scale bar: 1 μm). The Ripley’s H function is plotted for a 3 × 3 μm region containing the enlarged region. CI, confidence interval of randomized control. (B) Quantitative analysis of binary maps. Each dot represents the mean value for a cell, and different colors represent different donors. Black bars show the median and interquartile range (IQR). Kruskal-Wallis test with Dunn’s multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; KIR2DL1 (27 cells, 3 donors), KIR2DL3 (19 cells, 3 donors), KIR3DL1 (29 cells, 3 donors), NKG2A (19 cells, 3 donors), and LILRB1 (65 cells, 3 donors). (C) Representative PALM images of YTS cells expressing low and high amounts of KIR2DL1 tagged with a photoactivatable protein (mEos2). PALM images (scale bar: 5 μm) contain 2 × 2 μm regions (red boxes) that are magnified and shown as scatter plots (scale bar: 1 μm). (D) The normalized density within clusters [according to (24); see Materials and Methods] was plotted against the relative clustered area for three different receptors. Akaike’s Information Criteria (AICc) were used to compare theoretical curves for randomly organized events (gray lines) with the best fit curves (red lines). KIR2DL1 (73 cells, five experiments), KIR2DL3 (11 cells, two experiments), and LILRB1 (18 cells, three experiments). See also figs. S1 to S3, for details on how NK clones were selected, various imaging control experiments, and how alternative methods for image analysis led to similar conclusions.
To ensure our results were not dependent on the way in which protein clustering was measured, two alternative analyses were applied to the STORM data. In the first analysis (fig. S3B), clustering was assessed in a ring around each receptor and compared to an estimated inhomogeneous background. This method confirmed that KIR2DL3, KIR3DL1, and LILRB1 clustered on a scale less than 100 nm, whereas KIR2DL1 and NKG2A also clustered on a larger scale. In the second analysis (fig. S3C), the sum of the distances to the ten nearest neighbors produced a ”clustering value” for each event. By this analysis, the proportion of receptors in clusters differed for KIR2DL1 and KIR2DL3 (fig. S3D).
Binary maps (Fig. 1A, fifth column) were used to quantitatively compare nanoclusters. This analysis revealed that LILRB1 had the lowest density of clusters, the smallest clusters, and the lowest number of receptors at the cell surface (Fig. 1B). Despite the structural similarity of the KIRs studied, KIR2DL1 was distinguished by being present at a smaller proportion of receptors inside clusters (Fig.1B), which is consistent with the nearest-neighbor analysis (fig. S3D). KIR and NKG2A formed clusters of a similar sizes and densities. These conclusions are independent of specific data processing parameters (fig. S2D).
We also compared the nanometer-level organization of the NK cell receptors using a complementary superresolution microscopy technique: photoactivated localization microscopy (PALM). For this approach, an NK-like cell line, YTS, was transfected to express KIR2DL1, KIR2DL3, or LILRB1 tagged at the C terminus with the photoswitchable protein, mEos2. An advantage of PALM is that it gives a closer one-to-one relationship between receptor and reported fluorescence localizations than does STORM. A density-based analysis (24) determines the clustering of receptors independently of how many events are associated with a single receptor. For all receptors studied (KIR2DL1, KIR2DL3, and LILRB1) the density in clusters scaled with receptor abundance to a modest extent (Fig. 1, C and D), which was at the threshold of what is detectable by this method (25). This further establishes that these proteins are clustered on a nanoscale in the plasma membrane.
Allelic variation affects the nanometer-scale organization of receptors at the cell surface
As well as the differences between receptors, the same KIR could exhibit donor-specific differences [indicated by the spread of differently colored data points (Fig. 1B)]. We hypothesized that this was a consequence of allelic variation. Among the NK cell clones derived from one donor, some exhibited a high abundance of KIR2DL1, whereas others exhibited low abundance. Genomic analysis revealed that this donor was heterozygous for KIR2DL1*001 and KIR2DL1*004, which were previously shown to be expressed at high and low levels, respectively (8, 9). By isolating cDNA from each NK cell clone and performing a quantitative polymerase chain reaction (qPCR) assay that distinguished between the two alleles, we established that the low-expressing NK cell clones transcribed KIR2DL1*004, whereas the high-expressing clones transcribed KIR2DL1*001 (Fig. 2A).
Fig. 2. KIR2DL1 allotypes differ in their nanoscale organization.
(A) Flow cytometry analysis of representative pNK cell clones from a single donor that expressed different KIR2DL1 allotypes, stained for KIR2DL1 (gray) or with an isotype-matched control (white). (B) Representative TIRF microscopy and Gaussian-rendered STORM images of pNK cell clones with different KIR2DL1 allotypes (scale bar: 3 μm). The 2 × 2 μm regions (red boxes in the STORM images) are magnified and the corresponding scatter plots, density maps, and binary maps are shown (scale bar: 1 μm). The Ripley’s H function is plotted for a 3 × 3 μm region containing the enlarged region. (C) Quantitative analysis of the binary maps. Each dot represents the mean value for a cell. Red bars represent the median and IQR. Mann Whitney test: KIR2DL1*004 (13 cells, 1 clone, 1 donor), KIR2DL1*001 (11 cells, 3 clones, 1 donor). **P < 0.01. See also fig. S4 for analogous data with KIR3DL1 and KIR2DL3.
To examine the receptor organization of the different KIR2DL1 allotypes, clones expressing each allotype separately were stained and imaged by STORM. KIR2DL1*001+ cells had more receptor at the cell surface, which was evenly distributed, with most receptors outside of clusters (as evidenced by more cyan and yellow in the density maps and a shallow, broad peak on the Ripley’s H curve; Fig. 2B). Quantification of clusters (Fig. 2C) showed that KIR2DL1*001+ cells formed larger nanoclusters (when stringent data-processing parameters were applied; fig. S2E), but these clusters contained a lower proportion of receptors than did those on KIR2DL1*004+ cells. This difference suggests that allotypic variation influences the nanometer-scale organization of KIR2DL1.
We performed similar analyses of NK cells expressing different cell surface amounts of KIR2DL3 and KIR3DL1 (fig. S4). For both of these KIRs, the high-expressing clones had more clusters, which were larger than those formed by the low-expressing clones. Differences were slight for KIR2DL3, but more pronounced for KIR3DL1. Thus, these data suggest that the effect of receptor abundance on the nanoscale organization of receptors is applicable to the wider KIR family.
Variation in KIR-coding sequence does not inherently affect nanometer-scale receptor organization
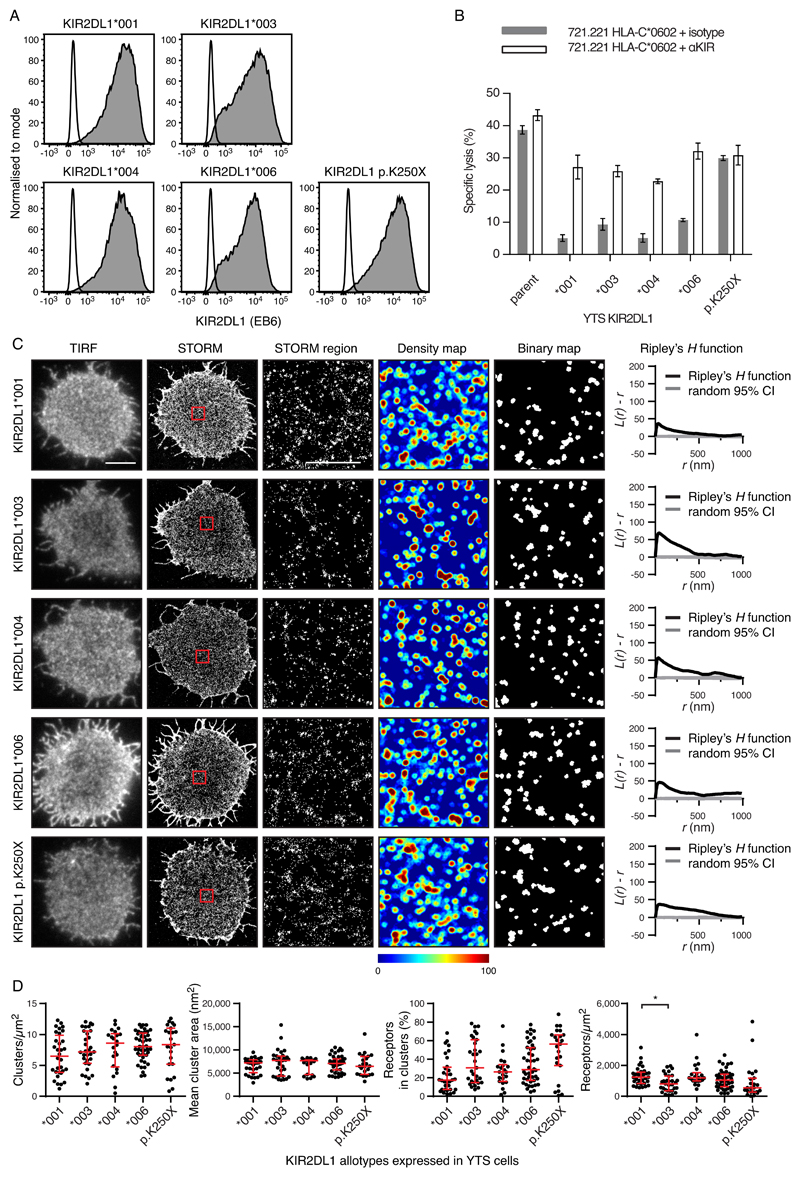
To investigate how the amino acid substitutions that distinguish between KIR allotypes affected nanometer-scale organization, we studied transfectants of the YTS cell line that expressed the coding region sequences for KIR2DL1 (Fig. 3) and KIR3DL1 allotypes (fig. S5). The YTS cell line does not endogenously express KIR, NKG2A, or LILRB1 (fig. S5A), which makes it ideal for studying the function of each inhibitory receptor independently. Three common KIR2DL1 allotypes of European populations (KIR2DL1*001, KIR2DL1*003, and KIR2DL1*004) and one common KIR2DL1 allotype of African populations (KIR2DL1*006) were compared. Four KIR3DL1 allotypes were also studied. Of these, KIR3DL1*001 and KIR3DL1*1502 are expressed at high abundance on the surface of pNK cells, whereas KIR3DL1*005 and KIR3DL1*007 are expressed at low abundance (26). Cell sorting was used to isolate transfected cell lines in which each of the KIR2DL1 or KIR3DL1 allotypes was present at a similar abundance at the cell surface (Fig. 3A and fig. S5B). KIR2DL1+ YTS cells were unable to kill target cells expressing C2+ HLA-C but killing was re-established by the addition of a monoclonal antibody (mAb) that specifically blocks the interaction of KIR2DL1 with C2+ HLA-C (Fig. 3B). Thus, cells expressing exogenous KIR2DL1 can engage C2+ HLA-C and generate an inhibitory signal (Fig. 3B). In parallel experiments, KIR3DL1+ YTS cells could not kill target cells expressing Bw4+ HLA-B, unless a mAb was used to specifically block the interaction (fig. S5C). When investigated by STORM, there were no differences in the size or density of the clusters, or in the proportion of receptors in clusters for all allotypes of the same receptor (Fig. 3, C and D and fig. S5, D and E). Thus, when the cell-surface expression of the KIR is held constant, the allotypic differences of KIR2DL1 and KIR3DL1 have no detectable effect on clustering.
Fig. 3. Allotypic differences do not affect the nanoscale organization of receptors when their abundance is controlled.
(A) Flow cytometry analysis of YTS cells expressing different KIR2DL1 allotypes or KIR2DL1*003 with a premature stop codon (KIR2DL1 p.K250X) stained for KIR2DL1 (gray) or with an isotype-matched control (white). (B) Lysis of 721.221 cells expressing a ligand for KIR2DL1 (HLA-C*0602) by YTS transfectants in the presence of a mAb that blocked KIR or an isotype-matched control. One representative experiment of two is shown, measured in triplicate, showing means ± the standard error of the mean (SEM). (C) Representative TIRF microscopy and Gaussian-rendered STORM images of YTS transfectants (scale bar: 5 μm). The 2 × 2 μm regions (red boxes) are magnified and the corresponding scatter plots, density maps, and binary maps are shown (scale bar: 1 μm). The Ripley’s H function is plotted for a 3 × 3 μm region containing the enlarged region. (D) Quantitative analysis of the binary maps. Each dot represents the mean value for a cell and the red bars show the median and IQR. Kruskal-Wallis test with Dunn’s multiple comparisons: KIR2DL1*001 (32 cells, four experiments), KIR2DL1*003 (31 cells, four experiments), KIR2DL1*004 (22 cells, four experiments), KIR2DL1*006 (44 cells, five experiments), KIR2DL1 p.K250X (23 cells, four experiments). *P < 0.05. See also fig. S5 for analogous data for KIR3DL1.
To determine whether constitutive KIR clustering depended on signaling through the immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic region, YTS cells were transfected to express a truncated form of KIR2DL1*003 that lacks these motifs (KIR2DL1-p.K250X). Although expressed to a similar extent as KIR2DL1*003 (Fig. 3A), this mutant did not prevent the killing of C2+ HLA-C–bearing target cells (Fig. 3B). In contrast, the KIR2DL1-p.K250X mutant formed cell-surface clusters such that the fraction of receptors in clusters and the size and number of clusters were comparable to those formed by KIR2DL1*003 (Fig. 3, C and D). This result demonstrates that the nanoscale clustering of KIR2DL1 does not require the cytoplasmic tail and its associated signaling function.
Receptor abundance correlates with the size and number of nanometer clusters
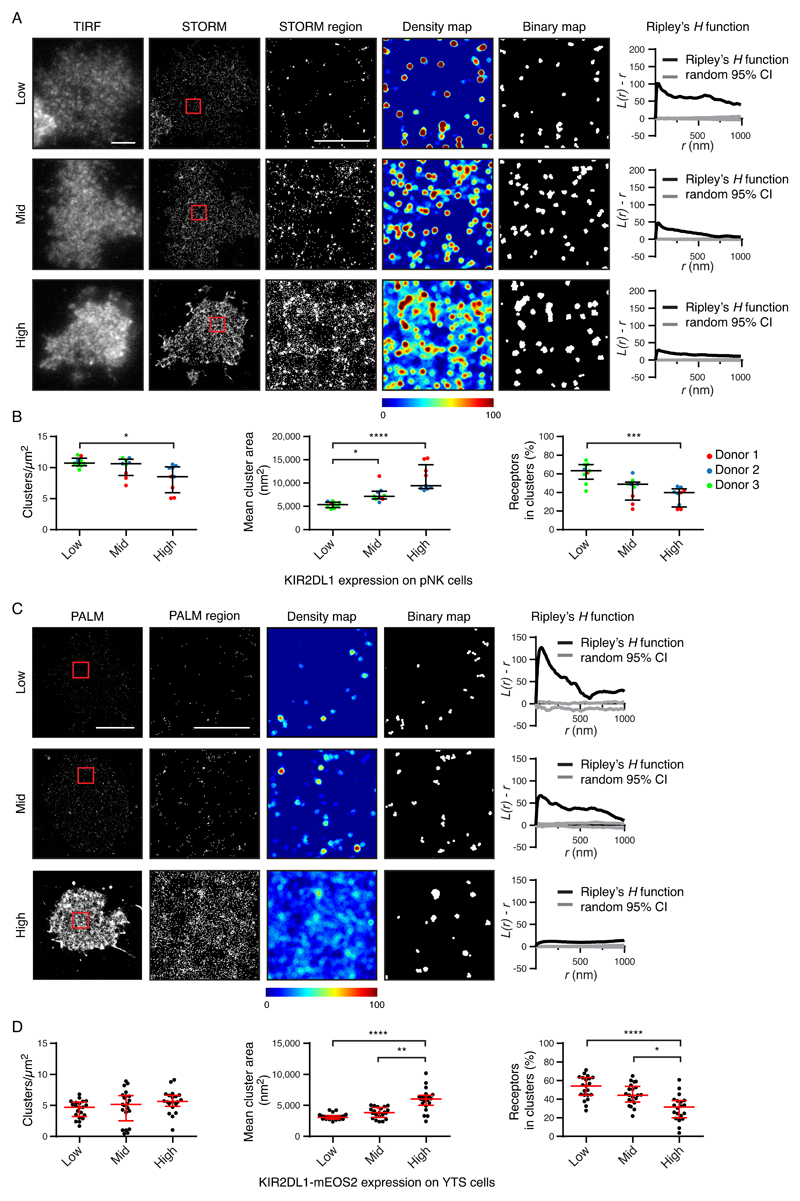
We next tested whether differences seen in nanoscale clustering on the primary cells related to cell surface abundance. According to their cell surface expression of KIR2DL1 (Fig. 4, A and B) and KIR3DL1 (fig. S6, A and B), pNK cells were stratified into three groups having low, medium, or high abundance. Increasing receptor abundance correlated with larger clusters, with a greater proportion of receptors outside clusters for both KIR2DL1 and KIR3DL1. These receptors differed in their relationship between abundance and cluster density. However, this could be accounted for by the fact that the actual abundance of protein within the three categories of low, middle, and high, varied between receptors.
Fig. 4. The abundance of KIR2DL1 determines its nanoscale organization.
(A and B) The KIR2DL1+ pNK cell clones analyzed in Fig. 1 are presented stratified according to receptor abundance. (A) Representative TIRF microscopy and Gaussian-rendered STORM images (scale bar: 3 μm) are shown. The 2 × 2 μm regions (red boxes) are magnified, and the corresponding scatter plots, density maps, and binary maps are shown (scale bar: 1 μm). The Ripley’s H function is plotted for a 3 × 3 μm region containing the enlarged region. (B) Quantitative analysis of the binary maps. Each dot represents the mean value for a cell and different colors represent different donors. Black bars show the median and IQR. Kruskal-Wallis test with Dunn’s multiple comparisons: *P < 0.05, ***P < 0.001, ****P < 0.0001; 9 cells per tertile; three experiments. (C and D) The YTS cells expressing KIR2DL1-mEos2 analyzed in Fig. 1 were stratified according to receptor abundance. (C) Representative Gaussian-rendered PALM images (scale bar: 5 μm) contain 2 × 2 μm regions (red boxes) that are magnified, and the corresponding scatter plots, density maps, and binary maps are shown (scale bar: 1 μm). The Ripley’s H function is plotted for a 3 × 3 μm region containing the enlarged region. (D) Quantitative analysis of the binary maps. Each dot represents the mean value for a cell and the red bars show the median and IQR. Kruskal-Wallis test with Dunn’s multiple comparisons: 20 to 21 cells per tertile, five experiments. *P < 0.05, **P < 0.01, ****P < 0.0001. See also figs. S6 and S7 for analogous data for KIR3DL1, and for KIR2DL1 as assessed by an alternative imaging method.
The cell surface abundance of KIRs in YTS transfectants also affected their nanoscale organization. STORM analysis (fig. S6, C and D, fig. S7, A and B) and PALM analysis (Fig. 4, C and D) gave similar results. As was observed for pNK cells, a higher abundance of KIR2DL1 and KIR3DL1 was associated with a greater proportion of receptors being outside of clusters and the receptors being distributed more evenly. The average cluster area increased with KIR2DL1 and KIR3DL1 abundance (which was statistically significantly different in the PALM analysis of KIR2DL1 and STORM analysis of KIR3DL1, but not in the STORM analysis of KIR2DL1). Thus overall, KIRs at high density are organized differently to KIRs at low density.
In a simulation of the effects of increasing receptor expression (fig. S8), the cluster size did not inevitably increase with increasing abundance (central panels). In other words, the different organizations of KIRs at low and high abundance could not be accounted for by the density of clusters being so high that they appeared to coalesce. Indeed, only if receptors were added into existing clusters (fig. S8, right panels) did the cluster size increase with increasing abundance.
KIR nanoclusters increase in size upon ligation
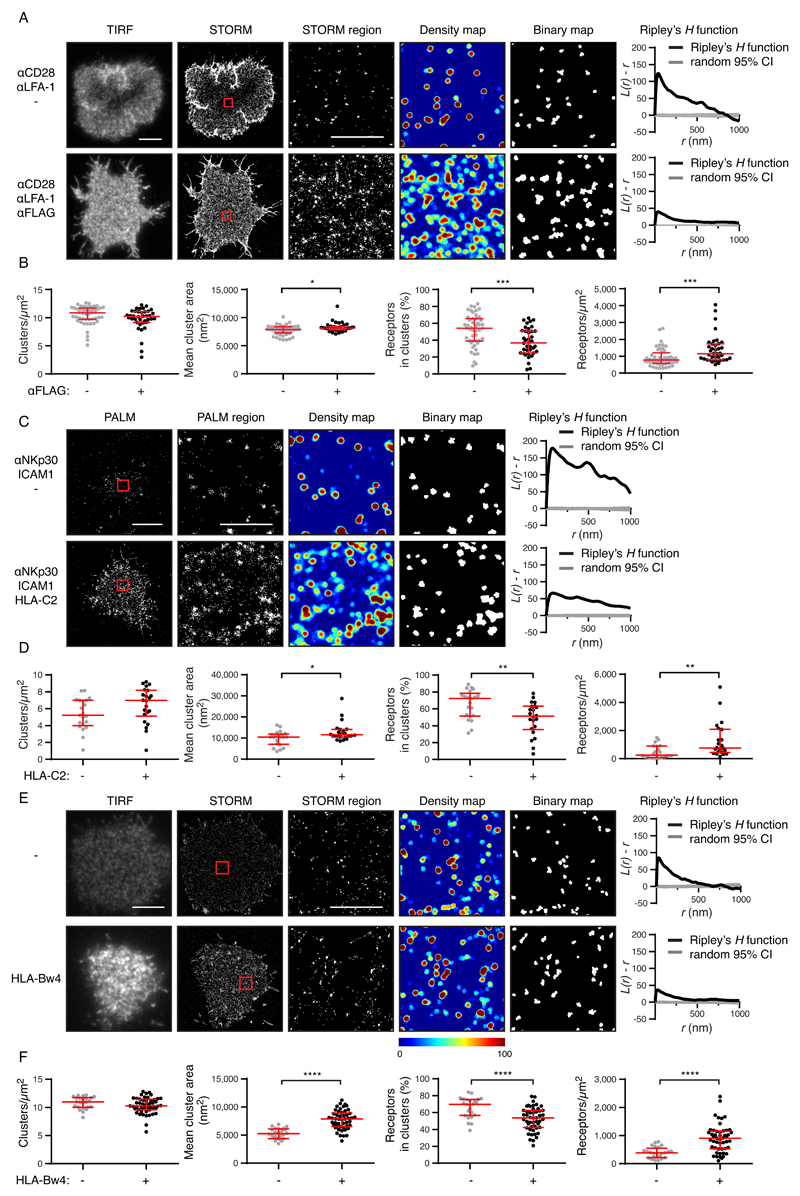
Having characterized the nanoscale distribution of KIRs in unactivated NK cells, we studied how this organization changes upon ligation. We made use of YTS cells transfected with a KIR2DL1 construct that also contained a FLAG tag at the N terminus of KIR2DL1. KIR2DL1+ YTS cells can be activated by anti-CD28 (a co-stimulatory receptor in primary NK cells) and anti–LFA-1 mAb, giving rise to the dense actin rings that are typical of lytic synapses (fig. S9A). When anti-FLAG mAb was added as well as anti-CD28 and anti-LFA-1, the KIR2DL1+ cells were not activated, as seen by their failure to form actin rings (fig. S9B). Under these conditions, KIR2DL1 crosslinked by anti-FLAG generated an inhibitory signal that overrode the activating signals from activated CD28 and LFA-1; but only for the transfectants capable of inhibitory signaling, not the KIR2DL1-p.K250X control. In the presence of anti-FLAG mAb, KIR2DL1 became gathered at the interface of the cells with the mAb-coated glass surface (Fig. 5A). This increased the density of KIR2DL1 at the contact site and the size of the KIR2DL1 nanoclusters (Fig. 5B). There was an increased number of receptors outside clusters and they were distributed more homogeneously. The same changes in receptor organization were observed by PALM when YTS-KIR2DL1-mEos2 cells interacted with surfaces coated with mAb against the natural cytotoxicity receptor NKp30, with and without a bona fide ligand for KIR2DL1, HLA-C*0401 (Fig. 5, C and D).
Fig. 5. Ligation reorganizes KIRs.
(A) Representative TIRF microscopy and Gaussian-rendered STORM images of YTS transfectants expressing KIR2DL1-FLAG cells on surfaces coated with mAbs against CD28, LFA-1, and an isotype-matched control antibody (activating conditions) or with mAbs against CD28, LFA-1, and FLAG (inhibitory conditions). (B) Quantitative analysis of the binary maps. Each dot represents the mean value for a cell and the red bars represent the median and IQR. Mann Whitney tests: *P < 0.05, ***P < 0.001. Activation conditions: 48 cells from five experiments; inhibitory conditions (FLAG ligation): 38 cells from five experiments. (C) Representative Gaussian-rendered PALM images of YTS transfectants expressing KIR2DL1-mEos2 on surfaces coated with mAbs against NKp30 and LFA-1, with HLA-C*0401 (“HLA-C2”) or a control protein, BSA. (D) Quantitative analysis of the binary maps depicted in (C). Each dot represents the mean value for a cell and the red bars represent the median and IQR. Mann Whitney tests: *P < 0.05, **P < 0.01. Activation conditions: 21 cells from three experiments; HLA-C2 ligation conditions: 22 cells from three experiments. (E) Representative TIRF microscopy and Gaussian-rendered STORM images of mAb 177407 staining of KIR3DL1+ YTS cells interacting with HLA-B*5701 (“HLA-Bw4”) or control protein, IgG2a. (F) Quantitative analysis of the binary maps of KIR3DL1. No ligation: 24 cells from three experiments; HLA-Bw4 ligation: 49 cells from six experiments. In images of whole cells (scale bars: 5 μm), 2 × 2 μm regions (red boxes) are magnified, and the corresponding scatter plots, density maps, and binary maps are shown alongside (scale bar: 1 μm). In each case, the Ripley’s H function is plotted for a 3 × 3 μm region containing the enlarged region. Each dot represents the mean value for a cell and the red bars represent the median and IQR. Mann Whitney tests: ****P < 0.0001.
KIR3DL1 was reorganized in a comparable manner when KIR3DL1+ YTS cells interacted with surfaces coated with HLA-B*5701, which bears the Bw4 epitope. Upon ligation, KIR3DL1 increased in density at the contact site, nanoclusters became larger, and there were more receptors outside of clusters (Fig.5, E and F). Inhibitory KIR organization upon ligation (whether or not in the presence of activating receptor co-ligation) is therefore similar to the organization found constitutively on cells that have higher KIR abundance. These changes in clusters are therefore likely to be a consequence of increased KIR2DL1 density at the synapse and there is no need to invoke a specific mechanism of cluster reorganization to explain these changes.
CrkII/CrkL is phosphorylated at the immune synapse upon KIR2DL1 ligation
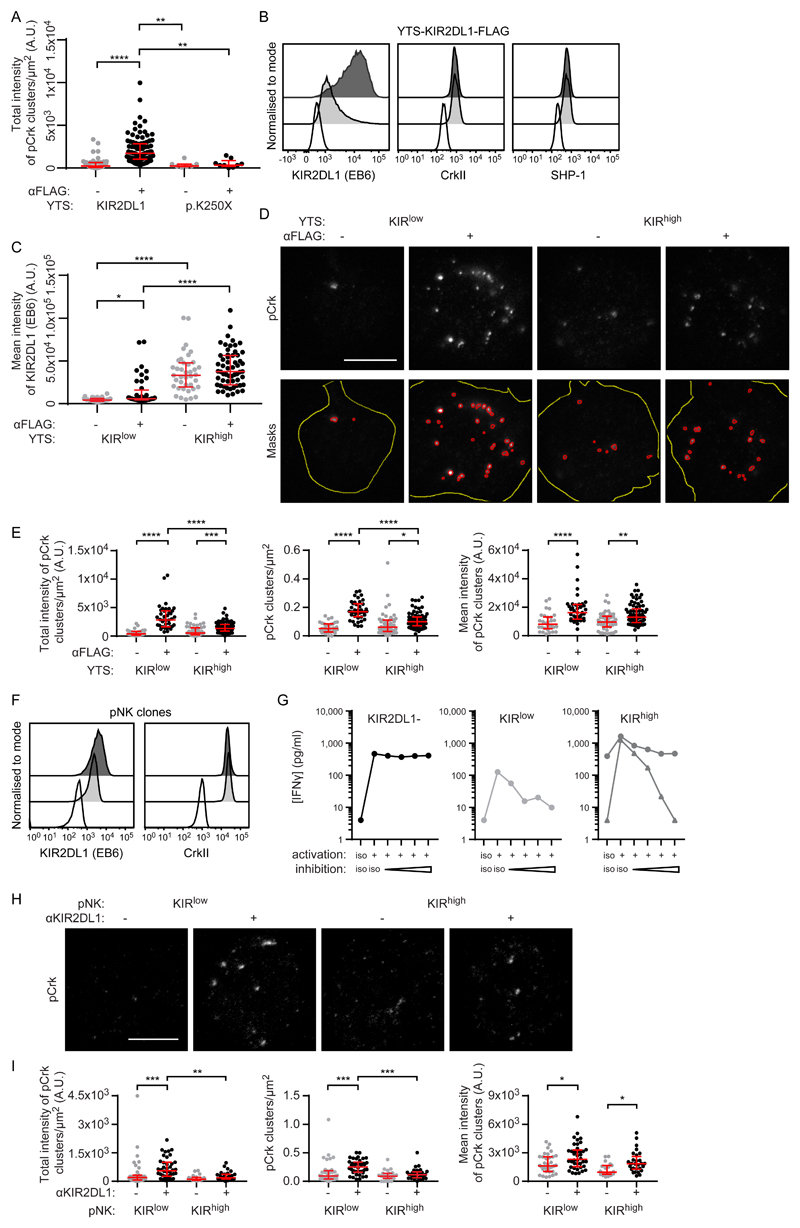
To investigate how receptor nanoclusters affect signaling, we used stimulated emission depletion (STED) microscopy, a method of superresolution microscopy readily able to detect receptors and membrane-proximal signaling molecules simultaneously. We analyzed the distribution of the adaptor protein CrkII and its close relative CrkL. Upon ligation of inhibitory NK cell receptors, c-Abl phosphorylates CrkII at tyrosine 221 (Tyr221) and CrkL at Tyr207 (27). Stimulation of KIR2DL1+ YTS with anti-FLAG mAb, but not an isotype-matched control, was sufficient to generate clusters of phosphorylated CrkII/CrkL (pCrk) at the immune synapse, as detected by an antibody that binds to both phosphorylated CrkII and phosphorylated CrkL (Fig. 6A). Although some clusters of pCrk were detected constitutively, there was a statistically significant increase in the total intensity of pCrk in KIR2DL1+ cells upon ligation (and not in cells expressing the signaling-deficient mutant KIR2DL1-p.K250X).
Fig. 6. KIRlow cells generate more pCrk than do KIRhigh cells.
(A to E) KIR2DL1+ YTS cells were ligated with surfaces coated with mAbs against FLAG or an isotype-matched control. (A) Quantification of the intensity of pCrk clusters triggered by the ligation of KIR2DL1 or KIR2DL1-p.K250X as measured by STED microscopy. Kruskal-Wallis test with Dunn’s multiple comparisons: **P < 0.01, ****P < 0.0001. KIR2DL1 no ligation (57 cells, four experiments), KIR2DL1 FLAG ligation (98 cells, four experiments), KIR2DL1 p.K250X no ligation (7 cells, three experiments), KIR2DL1 p.K250X FLAG ligation (9 cells, three experiments). (B) Representative flow cytometry analysis of unstimulated YTS clones that expressed KIR2DL1*003 at high (KIRhigh, dark gray) or low (KIRlow, light gray) amounts stained for surface KIR2DL1, or intracellular CrkII or SHP-1, compared to an isotype-matched control mAb (white). (C to E) Quantification and representative images relating to STED microscopy analysis of KIRlow and KIRhigh cells on surfaces that ligated KIR2DL1. (C) Quantification of KIR2DL1 staining. Kruskal-Wallis test with Dunn’s multiple comparisons: *P < 0.05, ****P < 0.0001. KIRlow no ligation (32 cells, four experiments), KIRlow FLAG ligation (40 cells, four experiments), KIRhigh no ligation (39 cells, three experiments), KIRhigh FLAG ligation (58 cells, three experiments). (D) Representative images of pCrk (scale bar: 5 μm) and outlines of the high-intensity regions of pCrk staining selected for analysis using a custom ImageJ script (masks). Red outlines show the high intensity areas of pCrk staining, whereas yellow outlines show the cell area (from IRM images). (E) Quantitative analysis of pCrk nanoclusters. Kruskal-Wallis test with Dunn’s multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. KIRlow no ligation (32 cells, four experiments), KIRlow FLAG ligation (40 cells, four experiments), KIRhigh no ligation (48 cells, four experiments), KIRhigh FLAG ligation (74 cells, four experiments). (F) Flow cytometry analysis of representative unstimulated pNK cell clones expressing high (KIRhigh; dark gray) or low (KIRlow; light gray) amounts of KIR2DL1, stained for surface KIR2DL1, intracellular CrkII, or with an isotype-matched control (white). (G) The concentration of IFN-γ in supernatants from pNK clones stimulated with ICAM-1 and a combination of mAbs against activating receptors (NKp30) or inhibitory receptors (KIR2DL1) at a range of concentrations (1, 2, 5, and 7.5 μg/ml), and isotype-matched controls, was measured by ELISA. Graphs are shown for representative clones without KIR2DL1 (left, 1 clone), or expressing low (central, 1 clone) or high (right, 3 clones) amounts of KIR2DL1. Each line represents a different clone. Triplicate measurements were performed. (H) Representative STED microscopy images of pCrk in pNK clones stimulated with coated surfaces (scale bar: 3 μm). (I) Quantitative analysis of pCrk nanoclusters. Each dot represents the mean value for a cell. Red bars represent the median and IQR. Kruskal-Wallis test with Dunn’s multiple comparisons: KIRlow no ligation (35 cells, four experiments), KIRlow KIR ligation (40 cells, four experiments), KIRhigh no ligation (20 cells, three experiments), KIRhigh KIR ligation (28 cells, three experiments). *P < 0.05, **P < 0.01, ***P < 0.001.
To test how receptor organization affected signaling, we exploited our observation that YTS-KIR2DL1*003 exhibits a broad range of surface expression by isolation of clones with high (KIRhigh) and low (KIRlow) abundance (Fig. 6B). Theses clones did not differ in their amounts of the signaling proteins CrkII and Src-homology region 2 domain-containing phosphatase-1 (SHP-1) (Fig. 6B). Activation of both types of cells was inhibited by anti-FLAG ligation (fig. S9C) or target cell expression of C2+ HLA-C (fig. S9D). The amount of KIR2DL1 at the contact interface increased upon ligation with anti-FLAG mAb in KIRlow cells but was not statistically significantly different for KIRhigh cells (Fig. 6C). For both KIRlow and KIRhigh cells, Crk phosphorylation was increased upon receptor ligation (Fig. 6, D and E). Unexpectedly, KIRlow cells generated more pCrk clusters and a greater total intensity of pCrk compared to KIRhigh cells. Thus, this aspect of membrane-proximal signaling was greater in KIRlow cells than in KIRhigh cells.
The abundance of KIR2DL1 decreases the extent of pCrk nanoclustering at the immune synapse
We next tested whether receptor abundance affected the generation of pCrk in primary cells. We isolated pNK cell clones expressing high and low amounts of KIR2DL1, but similar amounts of CrkII, from the same donor (Fig. 6F). The cellular activation and interferon γ (IFN-γ) secretion triggered by anti-NKp30 mAb was reduced to an extent similar to that in unactivated cells by ligation with anti-KIR2DL1 mAb for both KIRhigh and KIRlow clones (Fig. 6G). When the receptor was ligated by mAb, KIRlow pNK clones generated more pCrk clusters, which resulted in a greater intensity of pCrk clusters overall in KIRlow pNK cells compared to that in KIRhigh pNK cells (Fig. 6, H and I). Together, these data establish that pCrk is organized in nanoscale clusters and, unexpectedly, the intensity of pCrk is inversely correlated with the abundance of KIR2DL1. Thus, genetic diversity affects cell surface receptor expression and nanoscale clustering, and subsequently one or both of these factors affects membrane-proximal signaling.
Crk phosphorylation triggered by KIR2DL1 ligation is reduced by SHP-1 activity
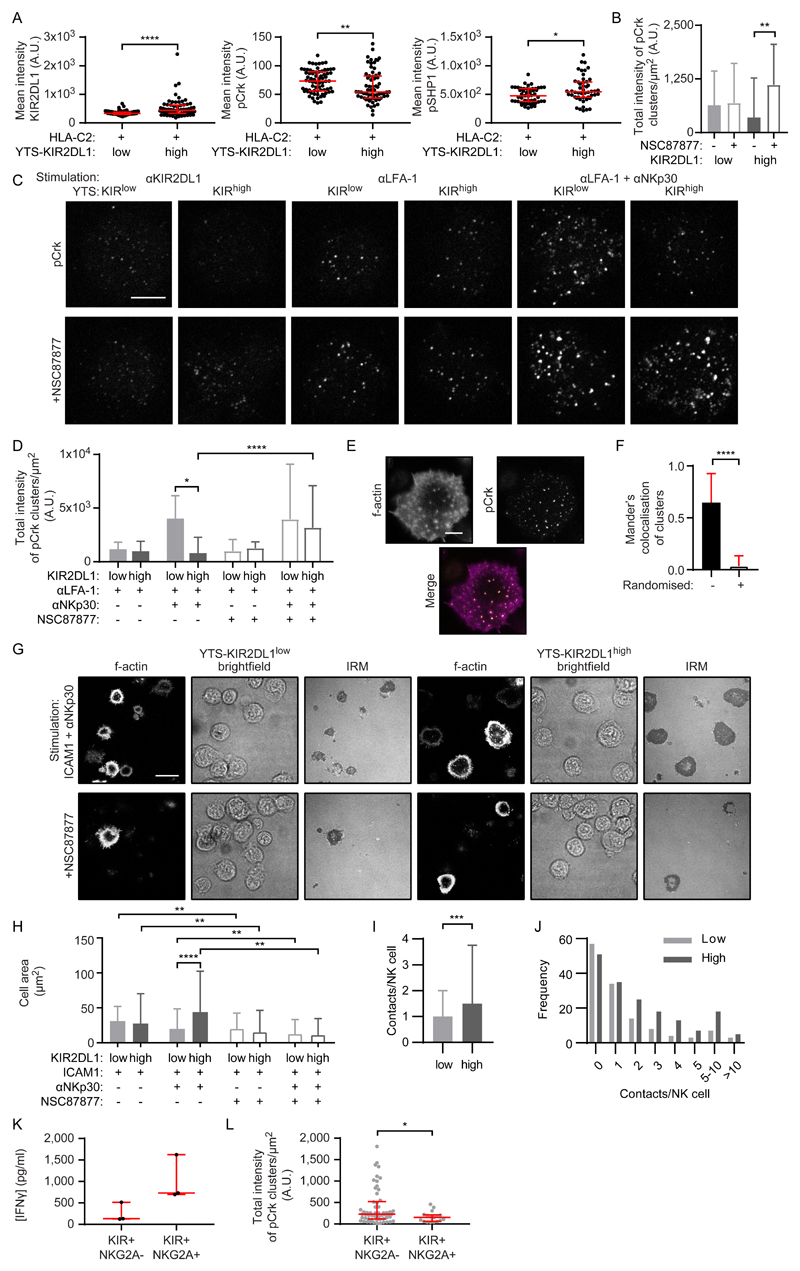
Larger nanoclusters of KIR2DL1 are associated with more phosphorylation of SHP-1 at Tyr536 (to generate pSHP1) (28), which is a marker of SHP-1 activity (29). In addition, SHP-1 can dephosphorylate CrkII in T cells (30). We therefore asked whether SHP-1 activity led to a local decrease in pCrk abundance near larger clusters of KIR2DL1. To establish whether SHP-1 activity varied with KIR2DL1 abundance, we incubated KIRlow and KIRhigh YTS cells on slides coated with HLA-C*0401. For the KIRhigh cells, more KIR2DL1 and pSHP1 were present at the contact site but they had less pCrk compared to KIRlow cells (Fig. 7A). These data lead us to suggest a model whereby c-Abl phosphorylates Crk in a manner that is independent of KIR abundance, but SHP-1 activity increases as KIR abundance and cluster size increases, which leads to the dephosphorylation of pCrk. The balance between these two processes would then determine the abundance of pCrk at the synapse.
Fig. 7. SHP-1 activity controls Crk phosphorylation and potentiates immune synapse formation.
(A) Mean intensity of KIR2DL1 (FLAG), pCrk, and pSHP-1 in YTS KIRlow and KIRhigh cells ligated with HLA-C*0401 coated onto slides, as measured by confocal microscopy. Each dot represents the mean value for a cell. Mann Whitney test: *P < 0.05, **P < 0.01, ****P < 0.0001. KIRlow KIR2DL1/pCrk (72 cells, three experiments), KIRhigh KIR2DL1/pCrk (66 cells, three experiments), KIRlow pSHP-1 (51 cells, three experiments), KIRlow pSHP-1 (50 cells, three experiments). (B to D) Quantification of pCrk clusters in confocal images of YTS KIRlow and KIRhigh cells on surfaces coated with mAbs against (B) KIR2DL1 or (D) LFA-1 ± NKp30. YTS cells were pretreated with the SHP-1/2 inhibitor NSC87877 (empty) or were untreated (filled). Kruskal-Wallis test with Dunn’s multiple comparisons: *P < 0.05, **P < 0.01, ****P < 0.0001. KIRlow anti-KIR (61 cells, three experiments), KIRlow anti-KIR + NSC87877 (52 cells, three experiments), KIRhigh anti-KIR (50 cells, three experiments), KIRhigh anti-KIR + NSC87877 (47 cells, three experiments), KIRlow anti-LFA-1 (60 cells, three experiments), KIRhigh anti-LFA-1 (60 cells, three experiments), KIRlow anti-LFA-1 anti-NKp30 (59 cells, three experiments), KIRhigh anti-LFA-1 anti-NKp30 (60 cells, three experiments), KIRlow anti-LFA-1 + NSC87877 (57 cells, three experiments), KIRhigh anti-LFA-1 + NSC87877 (60 cells, three experiments), KIRlow anti-LFA-1 anti-NKp30 + NSC87877 (59 cells, three experiments), KIRhigh anti-LFA-1 anti-NKp30 + NSC87877 (58 cells, three experiments). (C) Representative confocal images of pCrk (scale bar: 5μm) relating to (B) and (D). (E) Representative F-actin and pCrk STED microscopy images of a KIR2DL1+ YTS cell on a surface ligating LFA-1 and NKp30 (scale bar: 5μm). (F) Mander’s colocalization of pCrk clusters with the brightest spots of actin, compared to colocalization when the pCrk clusters are artificially placed at random within the cell region of interest. Mann Whitney test: ****P < 0.0001 (38 cells, six experiments). (G) Representative F-actin, brightfield, and IRM images of YTS KIRlow and KIRhigh cells on surfaces coated with ICAM-1 ± mAb against NKp30 ± NSC87877 (scale bar: 20 μm). (H) Quantification of spread cell area from IRM images of the cells represented in (G). Kruskal-Wallis test with Dunn’s multiple comparisons: **P < 0.01, ****P < 0.0001. KIRlow ICAM-1 (731 cells, six experiments), KIRhigh ICAM-1 (711 cells, six experiments), KIRlow ICAM-1 anti-NKp30 (535 cells, six experiments), KIRhigh ICAM-1 anti-NKp30 (547 cells, six experiments), KIRlow ICAM-1 + NSC87877 (246 cells, six experiments), KIRhigh ICAM-1 + NSC87877 (259 cells, six experiments), KIRlow ICAM-1 anti-NKp30 + NSC87877 (390 cells, six experiments), KIRhigh ICAM-1 anti-NKp30 + NSC87877 (459 cells, six experiments). (I and J) Quantification of the number of immune synapses (“contacts”) made by KIRlow and KIRhigh cells with 721.221 cells when suspended in a 3D extracellular matrix and imaged by confocal microscopy for 8 hours. (I) Mean number of synapses made per YTS cell and (J) the number of synapses made by each YTS cell over 8 hours (KIRlow 130 cells, three experiments; KIRhigh, 172 cells, three experiments). Mann Whitney test: **P < 0.01. (K) The amounts of IFN-γ secreted from NKG2A- and NKG2A+ pNK clones stimulated with ICAM-1 and anti-NKp30 mAb were measured by ELISA. Each dot represents a clone. NKG2A- (3 clones, 1 donor), NKG2A+ (3 clones, 1 donor). (L) Quantitative analysis of pCrk nanoclusters in STED microscopy images of NKG2A- and NKG2A+ pNK clones. Each dot represents the mean value for a cell. Mann Whitney test: *P < 0.05. NKG2A- (58 cells, 12 experiments, 1 donor), NKG2A+ (17 cells, three experiments, 1 donor). In all graphs, bars represent the median and IQR.
Towards testing this model, we treated YTS cells that had either low or high KIR2DL1 abundance with the SHP-1/2 inhibitor NSC87877 (31), and then incubated the cells on slides coated with mAbs that ligated KIR, LFA-1, or NKp30 and LFA-1 (Fig. 7, B to D). Treatment of KIRlow and KIRhigh cells with NSC87877 did not alter KIR organization (fig. S10), but the amount of pCrk detected upon KIR2DL1 ligation increased in KIRhigh cells (Fig. 7, B and C). There was no effect of treatment on KIRlow cells and indeed, the amount of pCrk was similar in high and low-expressing cells when SHP-1 was inhibited. This is consistent with cells with high KIR2DL1 abundance having less pCrk upon ligation, because high SHP-1 activity dephosphorylates pCrk. Crucially, at activating synapses, KIRhigh cells had less pCrk than had KIRlow cells (Fig. 7, C and D). Upon treatment with NSC87877, KIRhigh cells gained more pCrk, such that there was no longer a difference in pCrk abundance between the low- and high-expressing cells. This suggests that KIR2DL1 could activate SHP-1 to some extent and affect the amount of pCrk, even in absence of ligation.
SHP-1 activity potentiates NK cell spreading
The colocalization of pCrk with the actin cytoskeleton under activating conditions in YTS cells (Fig. 7, E and F) led us to test whether KIR abundance and the differential regulation of pCrk could affect the formation of activating synapses. We found that there was no difference in the spreading response of KIRhigh and KIRlow YTS cells stimulated by ICAM1 alone; however, when the slides were coated with anti-NKp30 mAb and ICAM1, KIRhigh cells had a greater spreading response (Fig. 7, G and H). When treated with an SHP-1/2 inhibitor, both KIRhigh and KIRlow cells failed to spread on activating surfaces, with most of the cells making small contacts. Thus, independently of being ligated, KIR abundance affects the ability of NK cells to adhere upon activation.
We next asked whether differences in activating synapse formation would be apparent when YTS KIRhigh and KIRlow cells interacted with HLA-deficient 721.221 cells. Both YTS cells and 721.221 cells were labelled with fluorescent dyes and suspended in an extracellular matrix at a 1:1 ratio. The cells were imaged in a heated chamber for 8 hours, and the number of prolonged contacts formed with 721.221 cells were scored for the KIRhigh and KIRlow cells. KIRhigh cells made more synapses with 721.221 cells (Fig. 7, I and J), suggesting that serial engagement of target cells is different between cells expressing different amounts of KIR2DL1.
One consequence of having either a low or high abundance of KIR2DL1 (32) and KIR3DL1 (33) is setting the education status of NK cells. Co-expression of NKG2A effectively increases the density of inhibitory receptors in the cell membrane and is another process by which NK cells become more educated (34, 35). Indeed, we found that NK cell clones expressing NKG2A and KIR responded more strongly to NKp30 activation (in terms of the amount of IFN-γ that they produced) than did clones lacking NKG2A (Fig. 7K). When we quantified the amount of pCrk in educated NKG2A+ cells, we found that in the unactivated state there was less pCrk detectable at the cell membrane than in NKG2A- cells (Fig. 7L). Thus, before activation, uneducated NK cells have more pCrk at the cell membrane but educated NK cells have less. This suggests that the baseline state of signaling molecules within the cell can be altered according to the receptors present at the cell surface and that the abundance of pCrk inversely correlates with education status in NK cells.
Discussion
Understanding the thresholds for activation and inhibition of the human immune system is an important goal, both for fundamental understanding of immunity and the design of medicines to harness immune responses. Superresolution microscopy is now revealing an additional level of complexity in the control of activation thresholds. An emerging theme is that immune cell activities are controlled by interactions between nanoscale clusters of activating and inhibitory surface receptors, membrane-proximal adaptors, and signaling molecules. For T cells and NK cells, there is evidence that receptor nanoclusters of greater densities are associated with increased signaling potential (28, 36). In addition, the relative positions of membrane nanoclusters in B cells (37) and macrophages (38) are important. TCR clusters associate with membrane protrusions (39), although more broadly, the mechanisms that underlie immune receptor clustering and how this is effected by membrane composition and dynamics is not entirely understood.
Here, we showed that the nanoscale organization of the highly related receptors KIR2DL1 and KIR2DL3 differ. These receptors are structurally very similar but are often associated with opposing risk or susceptibility in genetic studies. KIR2DL1 was more abundant than KIR2DL3 in pNK cells, and a lower proportion of KIR2DL1 than KIR2DL3 accumulated in clusters. At the level of allotypes, KIR2DL1*004 differs from other common KIR2DL1 allotypes by five to seven amino acid residues, including residue 245 in the transmembrane sequence. KIR2DL1*004+ NK cells are less educated than NK cells expressing other common KIR2DL1 allotypes (32, 40). In genetic studies, KIR2DL1*004 is not a risk factor for diseases of pregnancy, unlike other for KIR2DL1 alleles (40), but it is associated with a worse outcome in hematopoietic stem cell transplantation (41). We found that KIR2DL1*004 was less abundant on the surface of pNK cells than was KIR2DL1*001, another common KIR2DL1 allotype. This observation is consistent with those from previous studies (8, 9, 42). Using STORM, we showed that KIR2DL1*004 formed smaller nanoclusters than did KIR2DL1*001. This indicates that, as well as being expressed at different amounts, different KIR2DL1 allotypes organize differently on the cell surface.
These findings raised the question of whether the amino acid sequence inherently determines the nanoscale organization of the KIR. In testing this, we found that distinct KIR organizations were largely a consequence of different cell surface abundances and not the difference in sequence. Thus, KIR2DL1*004 had the same cell-surface organization as did KIR2DL1*001, for example, when expressed at similar amounts on the cell surface. This also holds true for KIR3DL1 allotypes. We found that when KIR3DL1 and KIR2DL3 were expressed in high amounts on the surface of pNK cells, they similarly formed larger clusters with more receptors outside of the clusters. It is well-established that cell surface expression of KIR3DL1 (and to a lesser extent that of KIR2DL3) is greatly affected by allotype (8–9,26). Therefore, in pNK cells, genetic variation influences receptor abundance, which in turn affects receptor arrangements. Other questions raised by our data that remain to be tested include whether or not KIR organization is affected by education or by the co-expression of activating or inhibitory receptors.
Ligation of inhibitory NK cell receptors increases the density of receptors at the immune synapse, which induces changes in clustering. There is evidence that a modest change in nanocluster size may affect downstream signals, because we have previously shown that larger nanoclusters of KIR2DL1 preferentially localize where phosphorylation of SHP-1 occurs (28). However, large clusters of KIR can be just as frequent in the surface membrane when the receptor density is endogenously high, such as when receptor density is increased locally by ligation at the synapse. The first study of the NK cell immune synapse determined that gathering of KIR2DL1 at the interface is not dependent on ATP or the cytoskeleton (13), suggesting that it is a passive process. Here, we found that even with differing KIR organizations, the extent of phosphorylation of Crk increases upon KIR ligation. Together, this suggests that some unknown factor beyond simply receptor clustering is required for signaling, such as a conformational change in KIR2DL1; a change in receptor organization that is beyond the resolution of our methodology; or the position of KIR2DL1 relative to that of other proteins.
Clusters of pCrk were found in discrete locations at the NK cell immune synapse. We did not distinguish between the phosphorylation of CrkII and CrkL, because antibodies previously used to investigate one or the other have since been found to cross-react. Crk is regulated by tyrosine-phosphorylation, such that when it is phosphorylated, it binds to its own Src homology 2 (SH2) domain, thereby displacing other SH2 domain–bound proteins. Here, we demonstrated, as others have shown (43), that engagement of the inhibitory receptors alone was sufficient to induce Crk phosphorylation. We also found that engagement of activating receptors resulted in the accumulation of pCrk at the synapse. This was not seen in a previous study (43); a discrepancy that might be accounted for by our targeting of NKp30 for activation and ICAM-1 for co-stimulation, whereas the previous study used CD16-mediated activation. More importantly, and unexpectedly, the number of pCrk clusters was greater in KIRlow NK cells than in KIRhigh NK cells, irrespective of whether the KIR itself or activating receptors were engaged. These data are consistent with a model in which KIR2DL1 ligation results in c-Abl activation and Crk phosphorylation; however, higher KIR abundance and larger clusters increased the local activity of SHP-1, which dephosphorylates pCrk. Functionally, cells with greater KIR abundance and larger clusters at the surface exhibited an enhanced NK cell spreading response, which was dependent on SHP-1 activity. Thus, KIR abundance can, even in the absence of ligation, potentiate this early step in contact-dependent NK cell activation. When multiple parameters change together, in this case receptor abundance, cluster size, and the proportion of receptors in clusters, it is difficult to ascertain which particular feature is important. However, our data are not inconsistent with receptor organization being a determinant of receptor signaling, possibly even a major factor. This is also supported by a study in which larger clusters of NKG2D (44) or specific engagement of CD16 by graphene-templated protein nanoclusters (45) led to enhanced functional responses.
Co-expression of KIR and NKG2A was associated with the presence of less pCrk at the cell membrane, even before inhibitory receptors were engaged. Thus, cells educated through high abundance of KIR or through the co-expression of inhibitory receptors, both had less pCrk at the membrane and more Crk available for cytotoxic processes. Other lines of evidence support the hypothesis that the availability of unphosphorylated Crk affects the ability of an NK cell to become activated and thus its education status. First, SHP-1 is essential for education (46) and, as we showed here, it is necessary for the dephosphorylation of Crk (Fig. 7). In addition, educated cells have more dynamic migration and conjugation behavior (47, 48), and Crk is a key determinant of integrin signaling and adhesion, which are important for migration and conjugation (30). Together, these data suggest a mechanism for an important aspect of NK cell education: the activation of SHP-1 determines the availability of unphosphorylated Crk which in turn, can affect the spreading response.
Overall, our data indicate that genetic variants of KIR are expressed with different nanometer-scale arrangements, such that receptor abundance and organization are covariates. One or both of these covariates in turn affects the efficiency of membrane-proximal signaling and activating synapse formation.
Materials and Methods
Primary human cell culture
Peripheral blood was acquired from the NHS blood service under ethics license REC 05/Q0401/108 (University of Manchester). Peripheral blood mononuclear cells (PBMCs) were purified by centrifugation on density gradient medium (Ficoll-Paque Plus, GE Healthcare) and pNK cells were isolated by negative selection with magnetic beads (Miltenyi Biotec). These NK cells were then cultured at a limiting dilution in clone medium (DMEM, 30% HAM’s F-12, 10% human serum, 1 mM sodium pyruvate, 1% MEM nonessential amino acids, 2 mM L-glutamine, 50 U/ml penicillin and streptomycin, 50 μM 2-mercaptoethanol) and stimulated with irradiated feeder cells (RPMI 8866 and allogeneic PBMC, irradiated at 40 Gy), 1 μg/ml phytohemagglutinin, and 400 U/ml IL-2 (Roche) on days 0 and 7. After three weeks, the expanded NK cells were assessed for receptor expression and maintained in clone medium with 200 U/ml IL-2 added every 5 to 7 days to maintain viability.
Cell lines
The NK-like cell line, YTS (RRID:CVCL_D324); the chronic myelogenous leukemia B cell line, RPMI-8866 (RRID:CVCL_1668); and the EBV-transformed B cell line, 721.221 (RRID:CVCL_6263) were all cultured at 37°C in culture medium [RPMI-1640 (Sigma), 10% fetal calf serum, 2 mM L-glutamine, 50 U/ml penicillin streptomycin (Gibco)]. All cell lines were routinely monitored for mycoplasma infection using a PCR-based kit (PromoCell).
Plasmids and transfected cell lines
Codon-optimized sequences encoding full-length KIR2DL1*001, KIR2DL1*003, KIR2DL1*004, KIR2DL1*006, and KIR2DL1*003 with a premature stop codon at p.K250X, all with N-terminal FLAG tags in the pcDNA3.1+ expression vector (7); and full-length KIR3DL1*001, KIR3DL1*005, KIR3DL1*007, and KIR3DL1*01502 in pEF/V5-His-TOPO (49) were used to transfect YTS cells by electroporation (Amaxa Nucleofector, Lonza). Cells were sorted on the basis of similar cell surface receptor expression and maintained with 1.5 μg/ml G418 (Life Technologies). Clones of KIR2DL1*003+ YTS cells with different cell surface receptor abundances were isolated from a serial dilution of the electroporated population. The sequence encoding full-length KIR2DL3*001 (a kind gift of Ashley Moffett, Cambridge University) was initially cloned into pmCherry-N1 (Clontech) using the primers and restriction sites described in table S1. The sequence encoding KIR2DL3 was then subcloned using the Bam HI and Nhe I restriction sites into the vector pmEos2-N1 (50). The sequence encoding full-length KIR2DL1*002 (20) and the sequence encoding full-length LILRB1 with the leader peptide of CD8a and an HA tag (a kind gift of Des Jones, Cambridge University) (51) were amplified with the primers described in the table S1. The KIR2DL1 (Bam HI and Eco RI) and LILRB1 (Xho I and Age I) amplicons were then digested and inserted into the vector pmEos2-N1 (50). These constructs were then used to transfect YTS cells by electroporation, as described earlier. 721.221 cells lack expression of HLA-B and HLA-C, and transfectants expressing HLA-B*5701 or HLA-C*0602 were generated as described previously (52).
Antibodies for staining
Staining of cells for microscopy analysis was performed with the following mAbs: EB6 for KIR2DL1 (mouse IgG1, 10 μg/ml), GL183 (mouse IgG1, 10 μg/ml) for KIR2DL3 and Z199 (mouse IgG2b, 10 μg/ml) for NKG2A (all Beckman Coulter); DX9 (mouse IgG1, 10 μg/ml) (R&D Systems) for KIR3DL1; and HP-F1 (mouse IgG1, 5 μg/ml) (eBioscience) for LILRB1. Because the mAbs EB6, GL183, and Z199 cross-react with KIR2DS1/L3, KIR2DL2/S2, and NKG2C, respectively, we detected inhibitory receptors of interest by using flow cytometry to screen clonal NK cells that lacked expression of the confounding receptors. To do this we used competitive staining (53) with the generic mAbs mentioned earlier and specific mAbs against KIR2DL1, 143211 (mouse IgG1, 10 μl/test); KIR2DL3, 180701 (mouse IgG2a, 10 μl/test); and NKG2C, 134591 (mouse IgG1, 5 μl/test) (all R&D Systems), antibodies that cannot be used for immunocytochemistry. For example, competitive staining with the mAb 143211 (which binds to KIR2DL1 and blocks staining by EB6) and the mAb EB6 (which binds to KIR2DL1/S1/L3*005/10) enabled the identification of clones that expressed KIR2DL1 (143211+ EB6-) and did not co-express KIR2DS1/L3*005/10 (143211+ EB6+) (fig. S1A). The anti-receptor mAbs used for microscopy were directly conjugated to Alexa Fluor 647 (Thermo Fisher). Staining was specific, because NK cell clones lacking these receptors showed no staining (fig. S1B). For all batches of antibody, the ratio of fluorophore to mAb (degree of labelling) was measured by absorption and calculated according to the manufacturer’s instructions (mean: 5.0/mAb, SD: 1.68). mAb against CD56 (HCD56, mouse IgG1, 1 μl/test, Biolegend), KIR2DL2/3 (DX27, mouse IgG2a, 5 μl/test, Biolegend), and KIR3DL1 (177407, mouse IgG2a, 10 μl/test R&D Systems) were also used for flow cytometry.
Antibodies and proteins used to stimulate and stain cells
Most, if not all, mAbs against KIR2DL1 compete with each other for binding, and compete with HLA class I ligands. Thus, in experiments with YTS-KIR2DL1 cells, we made use of a FLAG-tag at the N terminus of KIR2DL1 so that the receptor could be ligated with an anti-FLAG mAb and stained with a noncompeting mAb. Anti-FLAG (clone 9A3, mouse IgG1, 1 in 40 dilution, Cell Signaling) and EB6–Alexa Fluor 647 were used respectively to stimulate and stain these cells. Antibodies against the signaling proteins CrkII (3G11C1, mouse IgG2b, 1 in 300 dilution, Abcam), SHP-1 (255402, rat IgG2a, 10 μg/ml, R&D Systems), SHP-1 pY536 (rabbit polyclonal, 1 in 200 dilution, Acris Antibodies Gmbh), CrkL pY207 and CrkII pY221 (both rabbit polyclonal antibodies, 1 in 100 dilution, New England Biolabs) were detected using the appropriate cross-adsorbed secondary antibody (Alexa Fluor 568, 10 μg/ml for microscopy, ThermoFisher). The directly conjugated anti-pCrkL-Y207 Alexa Fluor 488 antibody (clone K30-391.50.80, mouse IgG2a, 1/10 dilution, BD Biosciences) was used in confocal experiments. DX9, an anti-KIR3DL1 antibody (5 μg/ml, R&D Systems), and HP3E4, an anti-KIR2DL1 (mouse IgM, 2.5 μg/ml, BD Pharmingen) antibody, were used to block cytotoxicity. Anti-CD28 (CD28.2) and anti-LFA-1 (HI111; both mouse IgG1, 5 μg/ml, eBioscience) were used to activate YTS cells for imaging. In other experiments, anti-NKp30/NCR3 (210847, mouse IgG2a, 5 μg/ml, R&D Systems) and anti-LFA-1 (HI111; mouse IgG1, 1.25 μg/ml, eBioscience) or ICAM-1 (1.25 μg/ml; produced in-house) was used to activate YTS and pNK cells for imaging and ELISA. HLA-C*0401 and HLA-B*5701 class I biotinylated monomers (NIH Tetramer Core Facility) were coated onto slides at 5 μg/ml and 40 μg/ml, respectively. KIR proteins interacting with HLA were detected using anti-FLAG Alexa Fluor 647 (clone L5, rat IgG2a, 10 μg/ml, Biolegend) for KIR2DL1 or anti-KIR3DL1 (177407, mouse IgG2a, 10 μl/test R&D Systems) conjugated to Alexa Fluor 647. Staining of KIR on these surfaces was compared to the staining of KIR on slides coated at the same density with control proteins (bovine serum albumin or mouse IgG2a) that did not ligate the receptor.
Flow cytometry
To assess the amounts of cell surface proteins, cells were first washed in PBS. Approximately 200,000 washed cells in 200 μl of PBS were stained with live/dead discriminator dyes (Zombie, 1 in 1000 dilution, Biolegend) for 20 min at 4°C. Cells were then washed in 1% FCS/PBS, blocked for 20 min at 4°C (1% human serum, 1% FCS/PBS) and stained with antibodies for 30 min at 4°C. Alternatively, if staining was competitive, then the first mAb was added for 10 min at 4°C, which was followed, without washing, by the second mAb for 20 min at 4°C. After washing, the cells were fixed with 2% paraformaldehyde (PFA)/PBS for 20 min at room temperature. For intracellular staining, cells were stained with live/dead dyes (Biolegend) as described earlier, washed and then fixed and permeabilized (Cytofix/Cytoperm, BD Biosciences), according to the manufacturer’s instructions. CrkII and SHP-1 were stained with primary antibodies (listed earlier) for 30 min at room temperature. The cells were then washed and stained with Alexa Fluor 647–conjugated goat anti-mouse IgG2b or goat anti-rat cross-absorbed secondary antibodies (4 μg/ml, ThermoFisher) for 30 min at room temperature.
Radioactive-release cytotoxicity assay
NK cell cytotoxicity was assessed using a standard 35S-methionine release assay, as described previously (20). The release was quantified using a scintillation counter (MicroBeta, PerkinElmer). The percentage lysis was calculated from the release of 35S-methionine from target cells as follows: (Experimental release – Spontaneous Release) / (Maximum release – Spontaneous Release). Experimental data were not included in the analysis if the spontaneous release was >25% of the maximum release.
SHP-1/2 inhibitor
Cells were resuspended in culture medium and incubated with the SHP-1/2 inhibitor (NSC-87877, Calbiochem) at 50 μM for 30 min at 37°C. After this incubation, the cells were added to coated slides for 5 min at 37°C in the presence of the inhibitor. The concentration of the drug was titrated for its effect on inhibitory immune synapses with minimal effect on viability.
Sample preparation
Chambered coverglass slides (Lab-Tek II, Nunc) were coated with 0.01% PLL (Sigma) for 20 min at room temperature, before being washed in water and dried for 1 hour at 60°C. In unstimulating conditions, cells were allowed to adhere to PLL for 20 min at 37°C. The mAbs were coated at various concentrations in PBS onto PLL-coated coverglass slides overnight at 4°C. After washing and blocking the slides in culture medium, the cells were allowed to interact with the coated surfaces for 5 min at 37°C. Cells were fixed in 4% PFA/PBS (ThermoFisher) for 30 min at room temperature and then washed in PBS. Samples for PALM analysis were imaged immediately, whereas samples for TIRF, STORM, and STED analyses were either stained immediately or stored overnight at 4°C with 0.02% azide/PBS. For surface protein imaging, after cells were fixed, they were incubated in blocking buffer 1 (1% BSA, 1% human serum/PBS) for 1 hour at room temperature and then stained in blocking buffer 1 containing antibodies against surface proteins at appropriate concentrations (listed earlier) as determined by titration for 1 hour at room temperature, and then washed in PBS. For assessment of their spreading responses, cells were fixed, permeabilized, and blocked with 0.1% saponin (Sigma) in blocking buffer 1 for 1 hour. Saponin-mediated cell permeabilization is reversible and so saponin was maintained in the washing and blocking steps thereafter. Cells were stained with phalloidin–Alexa Fluor 568 or phalloidin–Alexa Fluor 488 (Life Technologies) at 13.2 nM (1 in 500 dilution) and directly conjugated antibodies for confocal imaging for 1 hour at room temperature, and then washed in PBS. In experiments in which cells were stained with phalloidin alone, cthe ells were permeabilized with 0.1% Triton-X-100/PBS (Sigma) for 5 min at room temperature. For signaling protein imaging, after cells were fixed, they were incubated with 100 μg/ml lysophosphatidylcholine/PBS (Sigma) for 10 min at room temperature [a reversible permeabilizing agent used previously for membrane proteins; 50]. Cells were washed and then incubated with blocking buffer 2 [1% goat serum, 4% BSA/50 mM Tris-buffered saline (TBS)] for 1 hour at room temperature, followed by incubation with blocking buffer 2 containing primary antibodies for 1 hour at room temperature. After washing, the cells were incubated with goat secondary antibodies conjugated to Alexa Fluor 568 (ThermoFisher) in blocking buffer 2 for 1 hour at room temperature and then washed in TBS. After staining, cells were post-fixed with 4% PFA/PBS for 5 min at room temperature and then washed. Samples for STORM and TIRF analyses were either imaged immediately or stored overnight at 4°C in 0.02% azide/PBS. Samples for STED analysis were mounted in a hard-drying medium that preserved fluorescence and matched the refractive index of the STED microscope lens (Fluorescent Mounting Media, Dako).
Microscopy
For STORM imaging, slides were immersed in oxygen-scavenging buffer [560 μg/ml glucose oxidase, 34 μg/ml catalase, 1% β-mercaptoethanol, 25 mM glucose, 5% glycerol (all Sigma), 25 mM HEPES/PBS (Fisher Scientific, pH 8], 0.22 μm filter-sterilized], which was refreshed regularly to maintain a low oxygen environment for the fluorophores. For PALM imaging, slides were kept in PBS. STORM and PALM were performed with an inverted microscope (Nikon Ti Eclipse) fitted with focus stabilization (perfect focus system), a TIRF 100× oil immersion lens (N.A. 1.49), and an EMCCD camera (Andor iXon Ultra 897). For STORM analysis, images were acquired with a 170mW 647 nm laser (Agilent Ultra High Power Dual Output Laser set to 30%) with an 18-ms exposure. The optimal frame number was determined as the point at which reconstructed cluster maps did not change with increasing frame acquisition (for NK cell receptors: up to 30,000 frames). Cells were imaged until the optimal frame number was reached or the cells were bleached. For PALM, images were acquired using a 120mW 561 nm laser (Agilent Ultra High Power Dual Output Laser, set to 30%) and 20mW 405 nm laser (increasing from 0.1% to 10% to maintain the blink rate) with a 50-ms exposure. Frames were acquired until the rate of photoswitching was negligible. STORM analysis of KIR3DL1+ YTS cells was performed separately with an inverted microscope (Leica SR GSD microscope) fitted with a TIRF 160× oil immersion lens (NA 1.43), and an EMCCD camera (Andor iXon Ultra 897). These images were acquired with a 647-nm laser (set to 30%) with a 15-ms exposure. STED was performed with a continuous wave STED microscope (Leica TCS SP8 STED CW), a 100× oil immersion lens (N.A. 1.40), and a HyD hybrid photon detector (Leica Microsystems). Two-color STED microscopy was achieved by sequential excitation/depletion with wavelengths of 568 nm/660 nm and 495 nm/592 nm (1.5 mW/channel supercontinuum White Light Laser and 1.5 W STED depletion lasers, Leica Microsystems). Time gating of 1.0 to 6.0 ms was applied.
Single-molecule localization microscopy data
Single-molecule localization microscopy (SMLM) image reconstruction was performed with ThunderSTORM software (54). Raw images were initially filtered to remove noise (wavelet filter B-spine method, order 3, scale 2). The approximate location of molecules “events”) was determined with the local maximum localization method (peak intensity threshold = 3 × SD and 8 neighborhood connectivity for each pixel screened) for STORM data or 1.5× SD for the PALM data. The precise location (sub-pixel coordinates) of events was calculated by fitting a point spread function to the approximate location of molecules (integrated Gaussian method, using maximum likelihood fitting, with a fitting radius of 3px and an initial sigma of 1.6 px). STORM events were filtered on their intensity (> 1500 photons), sigma (> 10), and uncertainty (< 25 nm) values. These values filtered out noise, as determined by imaging uncoated glass. Any drift during acquisition was corrected using cross-correlation. Events believed to have originated from a single fluorophore in consecutive frames were removed by averaging (merging) the location of events that occurred within a maximum distance of 50 nm and with a limited break in the number of frames between consecutive events (20 off frames). These values were determined by imaging sparse antibodies on glass, where individual fluorophores could be tracked across frames. See the cluster of events originating from two individual mAbs detected on a glass surface (fig. S2C). Before merging of events is applied, the mAb appear as two dense clusters in xy (fig. S2C, left panels), but many of these events originate in bursts in time (fig. S2C, right panels), where a single fluorophore is detected over a short sequence of frames. After merging of events is applied, each fluorophore is represented by a single event, so that the average number of events per mAb is equal to the number of fluorophores per mAb (as measured by absorption). Throughout the text, we refer to detected events as ”receptors” for readability. It is important to note, however, that the detected events are indicative of receptor distributions and are not exactly equivalent to receptors, a caveat for the microscope technologies used here. STORM and PALM images were rendered using the normalized Gaussian method for localized events, but the enlarged regions show scatter plots of the events. The brightness of representative microscopy images has been adjusted to make details visible, but the scaling factor is the same for each figure section and unaltered data was used for quantitation.
Ripley’s H function analysis
In the Ripley’s K function analysis (55), each event is assigned a value, L(r), based upon the number of neighboring events within a given search radius (r). This value is adjusted in the Ripley’s H function analysis, L(r)-r, to account for the increasing number of neighbors that would be expected in a random distribution as r increases. Therefore, a random distribution would have no peak on a Ripley’s H function curve. STORM data derived from mAbs labelled with several fluorescent moieties is inherently clustered; however, this analysis and the relative position of the Ripley’s H function peak is informative for comparative study of the different receptors, because they are labelled to a similar extent.
G&F local point, pattern-clustering analysis of SMLM data
A Ripley’s K function (55) and univariate G&F analysis (56) were used as the basis of quantitative clustering maps for STORM and PALM coordinate data, as described previously (28). In this analysis, a value, L(r), is assigned to each event based on the number of neighboring events within a given radius and adjusted for the expected number of neighbors for randomly distributed events at that density: the higher the number, the more neighboring events, the greater the clustering of that receptor. Multiple regions of 3 × 3 μm were defined for each cell and events within these regions were assessed with a sampling radius, r, of 50 nm and a threshold value of 90 to consider an event to be clustered [the L(50) threshold]. The L(50) threshold was determined by randomly placing events at the same density as each receptor and selecting a value that excluded most of the randomized events from clusters across all receptors (fig. S3A). A threshold of 90 is sensitive to clusters in dense datasets (for example, KIR2DL1), but for sparse data (for example, LILRB1), a small proportion of events was assigned to clusters in the randomized data. Density maps are colored according to the L(50) value of the individual events (shown as black dots in the density maps), and the space between events is colored by interpolation. To generate binary maps, all events above an L(50) threshold of 90, which corresponds to red and orange points in the density maps, are depicted by a white circle of radius 50 nm to generate clusters. Quantitative analyses of the white area in binary maps, using an ImageJ plugin, were used to compare receptor cluster density, mean cluster area, and the proportion of receptors in clusters. It should be noted that the analysis is dependent on the search radius used and the threshold applied. Consequently, the parameters that describe the extent of clustering for each receptor are not absolute but do show the differences between the five receptors.
Inhomogeneous O-ring analysis
An inhomogeneous point pattern analysis (57) was estimated using the Kinhom function from the ‘spatstat’ package for R. It differs from the Ripley’s H analysis described earlier in that it does not assume receptors to be homogeneously distributed in the null condition, for example, if the receptors are concentrated on membrane protrusions. The O-ring (or Neighborhood Density Function) analysis (58), similarly to the G&F analysis, measures the clustering of events at different radii, but subtracts the number of events within a smaller radius from the number of events within a larger radius to generate a measure of events within a ring. Unlike the G&F analysis, this method assesses clustering independently of how dispersed the clusters are from each other.
Distance to nearest neighbors analysis of SMLM data
In addition to the G&F analysis, a nearest neighbor–based method was used to assess the distribution of the STORM data. To determine if an event was clustered, a threshold was applied that was based on the sum of the distances to the ten nearest neighboring events, and this threshold varied according to the density of the data. In this analysis, for each data point within each image, the distance to the ten nearest data points was calculated, inverted, and normalized to randomized data. The threshold was chosen as 75% of the spread of the randomized data, when it was at the same density as the experimental data. These analyses were performed in MATLAB R2014a (MathWorks) software.
STED and confocal analysis
STED and confocal images were analyzed for clusters using a custom script in Java for ImageJ. The pipeline is based on the Laplacian of Gaussian (59) filter and morphological segmentation techniques to identify and segment bright spots in the image at sites of signaling against the noisy and uneven background. First, the Laplacian of Gaussian filter standard deviation σLoG was convolved with the raw image. A binary map was then generated by applying a threshold tLoG to the filtered image; pixels that fell below the threshold were considered as background and set to zero, whereas bright spots were set to 1. A region-labelling algorithm [provided in the MorphoLibJ library for ImageJ (60)] was then applied to the binary map to identify isolated disjoint regions of nonzero valued connected pixels. The area of each region was computed, Ai, and any region that contained Ai ≤ 9 pixels was considered as noise because of nonspecific binding (when compared to the isotype-matched control) and therefore discarded. For each remaining region (labelled as ”masks” in figures), the local background intensity was estimated from the raw image by computing the median intensity of the pixels directly outside the perimeter of the region. Finally, the pixel intensity descriptors for each region, such as the mean or integrated intensity, were computed by subtracting the local background estimate from the pixels inside the region and computing the respective statistic on each set of pixels. The parameters used in the aforementioned analysis pipeline were the mean radius of the bright spots, r = σLoG = 5 pixels, and the threshold applied to the filtered image tLoG = 15 A.U. These values were chosen empirically and kept constant between conditions. Mander’s colocalization of clusters was applied to the STED clusters defined earlier with a custom ImageJ script. For a negative control, the clusters in one channel were randomly placed within the cell region of interest. Spread area was measured by drawing around the outline of cells in IRM images in ImageJ.
Determination of the KIR alleles expressed by pNK clones
Genomic DNA was isolated from pNK cells with a DNeasy Blood and Tissue Kit (Qiagen). Exons 1, 4, and 5 of KIR2DL1 were amplified using the primers described by Norman et al. (61) for KIR2DL1 (table S2). The following conditions were used for PCR using Thermopol Taq (New England Biolabs): 95°C for 30 s; 95°C for 15 s, 10 cycles of 65°C for 60s; 20 cycles of 95°C for 20 s, 61°C for 50 s, and 68°C for 30 s; 68°C for 5 min. Sanger sequencing with the same primers was used to determine the presence of specific KIR alleles (GATC Biotech). RNA was isolated from pNK clones with an RNeasy mini kit (Qiagen) with DNAse treatment (Qiagen) and converted to cDNA with the High Capacity RNA-to-cDNA kit (Applied Biosystems). The primers listed in table S3, which were adapted from Shilling et al. (62), were used on cDNA from pNK clones to discriminate between alleles identified in the genomic DNA. Quantitative PCR was analyzed by the ΔΔ cycle threshold (CT) method and normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward primer: GAAGGTGAAGGTCGGAGT; reverse primer: CATGGGTGGAATCATATTGGAA).
Enzyme-linked immunosorbent assay (ELISA)
Flat-bottomed, 96-well plates were coated with PLL and then coated with stimulating mAb in PBS at 4°C overnight. Wells were coated with the following proteins: ICAM-1 (2.5 μg/ml), activating mAb (anti-NKp30, 10 μg/ml), inhibitory mAb (anti-KIR2DL1, EB6 titrated from 1 to 7.5 μg/ml), and isotype-matched control mAb (to maintain a final concentration of 20 μg/ml). After coating, the wells were washed with 0.05% Tween-20/PBS (Sigma) and then blocked for 5 min at room temperature with culture medium. This was then removed, and 100,000 pNK cell clones were added per well in triplicate. After overnight culture at 37°C, supernatants were harvested by centrifugation at 300g for 10 min at 4°C, and the concentration of IFN-γ was measured by ELISA, as previously described (28).
Simulation of KIR2DL1 nanoscale organization
Data for simulating clusters (fig. S8) were generated with a published script in R (63). Clusters were simulated to match the parameters of KIR2DL1 in low-expressing pNK cells: 10 receptors per cluster, 60% of receptors in clusters with an SD of 20 nm, and 7.8 clusters/μm2 randomly placed into the region of interest. The number of clusters/μm2 was then increased to match the density of receptors in mid-expressing (39 clusters/μm2) and high-expressing (78 clusters/μm2) pNK cells (receptors added to new clusters), still with 60% of receptors in clusters and an SD of 20 nm. In the second simulation, clusters were simulated to match the parameters of KIR2DL1 for low-expressing pNK cells, as described earlier. The number of receptors per cluster was then increased to match the density of receptors in mid-expressing (40 receptors per cluster) and high-expressing (100 receptors per cluster) pNK cells (receptors added to existing clusters), still with 60% of receptors in clusters and an SD of 20 nm.
Time-lapse imaging of YTS in a three-dimensional matrix
Time-lapse imaging of YTS and 721.221 cells was performed as described previously (64). YTS cells were labelled with 5 μM Cell Trace Violet, whereas 721.221 cells were labelled with 0.5 μM Calcein Green (both Thermo Fisher Scientific). YTS and 721.221 cells were mixed at a 1:1 ratio and suspended 4:1 with cold growth factor–reduced matrigel membrane matrix (Corning) containing 1 μM TO-PRO-3 (Thermo Fisher Scientific) to detect dead cells. The matrigel was solidified in chambered coverglass for 30 min at 37°C before imaging. Time-lapse images were taken of cells every 3 min over 8 hours at 37°C, 5% CO2 on a confocal microscope equipped with a White Light Laser and a 20x 0.75 NA objective (TCS SP8, Leica Microsystems). Images were analyzed with the TrackMate Plugin (65) for ImageJ and visually scored for contact formation.
Quantification and statistical analysis
Statistical tests and n numbers for each figure are given in the figure legends. Datasets were tested for normality with the D’Agostino and Pearson omnibus normality test. The Mann-Whitney test, the Kruskal-Wallis with Dunn’s multiple comparisons test, the Wilcoxon signed rank test, and Akaike’s Information Criteria (AICc) were all calculated with GraphPad Prism 7.01 software. The Fisher exact test was performed with an online tool (www.openepi.com).
Supplementary Material
One-sentence summary.
The abundance and clustering patterns of inhibitory receptors on natural killer cells are genetically encoded.
Editor’s summary.
Diversity starts at the gene level
Natural killer (NK) cells are immune cells with potent antiviral and antitumor activities. These cells express several germline-encoded inhibitory receptors (KIRs) that prevent NK cells from killing healthy cells but enable the targeting of infected and transformed cells. Using various imaging techniques and functional assays, Kennedy et al. investigated the effects of genetic diversity in KIR-encoding genes on receptor organization and activity. They found that KIRs encoded by distinct genes were present at varying abundances and in clusters of varying sizes, which led to differences in downstream signaling that affected NK cell function. Together, these data suggest that genetic diversity in KIR-encoding genes affects receptor organization, signaling, and functional outcomes.
Acknowledgments
We thank the Davis lab for useful discussions; G. Howell and the MCCIR Flow Cytometry Facility; B. Telfer for irradiation of cells; P. Paszek for advice on statistics (all University of Manchester); O. Chazara (University of Cambridge) and D. J. Morgan (University of Manchester) for advice on qPCR; D. M. Owen (Kings College London) for software relating to data simulation; and A. Moffett and D. Jones (University of Cambridge) for the KIR2DL3 and LILRB1 cDNAs. ICAM1 protein was produced by E. McKenzie and M. Ball from the Protein Expression Facility, Manchester Institute of Biotechnology, University of Manchester. HLA class I tetramers were kindly provided by MR1 tetramer technology, which was developed jointly by J. McCluskey, J. Rossjohn, and D. Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
Funding: This work was supported by the Biotechnology and Biological Sciences Research Council (BB/I013407/1), the Medical Research Council (Award G1001044), a Wellcome Trust Investigator Award (110091), and the Manchester Collaborative Centre for Inflammation Research (funded by a pre-competitive open-innovation award from GSK, AstraZeneca and The University of Manchester, UK). This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
Footnotes
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: P.R.K., C.B., and D.M.D. conceived the project and designed the experiments; P.R.K., C.B., K.H., J.C., and K.B.S. performed experiments and analyzed the data; D.J.W. and W.T.E.P. developed and performed analysis; H.G.H., M.C., and P.P. provided reagents and edited the manuscript; and P.R.K. and D.D. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Analysis software is available at https://github.com/DMDavisLab (analysis of SMLM data) and https://github.com/EricPKDavisLab/CellFeatureAnalyser (analysis of clusters in STED images).
References
- 1.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 2.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, Claas FH, Walker JJ, Redman CC, Morgan L, Tower C, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J Clin Invest. 2010;120:4102–4110. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das J, Khakoo SI. NK cells: Tuned by peptide? Immunol Rev. 2015;267:214–227. doi: 10.1111/imr.12315. [DOI] [PubMed] [Google Scholar]
- 5.Hilton HG, Parham P. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics. 2017;69:567–579. doi: 10.1007/s00251-017-1001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. 2013;190:6198–208. doi: 10.4049/jimmunol.1300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, Parham P. Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes. J Immunol. 2015;195:3160–3170. doi: 10.4049/jimmunol.1501358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babor F, Manser AR, Fischer JC, Scherenschlich N, Enczmann J, Chazara O, Moffett A, Borkhardt A, Meisel R, Uhrberg M. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood. 2014;124:2248–2251. doi: 10.1182/blood-2014-05-572065. [DOI] [PubMed] [Google Scholar]
- 9.Dunphy SE, Guinan KJ, Chorcora CN, Jayaraman J, Traherne JA, Trowsdale J, Pende D, Middleton D, Gardiner CM. 2DL1, 2DL2 and 2DL3 all contribute to KIR phenotype variability on human NK cells. Genes Immun. 2015;16:301–310. doi: 10.1038/gene.2015.15. [DOI] [PubMed] [Google Scholar]
- 10.Andersson S, Malmberg J-A, Malmberg K-J. Tolerant and diverse natural killer cell repertoires in the absence of selection. Exp Cell Res. 2010;316:1309–1315. doi: 10.1016/j.yexcr.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 11.Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, Trowsdale J, Malmberg K-J. Influence of KIR gene copy number on natural killer cell education. Blood. 2013;121:4703–4707. doi: 10.1182/blood-2012-10-461442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagné F, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Davis DM, Chiu I, Fassett M, Cohen GB, Mandelboim O, Strominger JL. The human natural killer cell immune synapse. Proc Natl Acad Sci. 1999;96:15062–15067. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abeyweera TP, Merino E, Huse M. Inhibitory signaling blocks activating receptor clustering and induces cytoskeletal retraction in natural killer cells. J Cell Biol. 2011;192:675–90. doi: 10.1083/jcb.201009135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treanor B, Lanigan PMP, Kumar S, Dunsby C, Munro I, Auksorius E, Culley FJ, Purbhoo MA, Phillips D, Neil MAA, Burshtyn DN, et al. Microclusters of inhibitory killer immunoglobulin–like receptor signaling at natural killer cell immunological synapses. J Cell Biol. 2006;174:153–161. doi: 10.1083/jcb.200601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masilamani M, Nguyen C, Kabat J, Borrego F, Coligan JE. CD94/NKG2A inhibits NK cell activation by disrupting the actin network at the immunological synapse. J Immunol. 2006;177:3590–3596. doi: 10.4049/jimmunol.177.6.3590. [DOI] [PubMed] [Google Scholar]
- 17.Purbhoo MA, Liu H, Oddos S, Owen DM, Neil MAA, Pageon SV, French PMW, Rudd CE, Davis DM. Dynamics of Subsynaptic Vesicles and Surface Microclusters at the Immunological Synapse. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000645. ra36-ra36. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan S, Winter CC, Wagtmann N, Long EO. The Ig-related killer cell inhibitory receptor binds zinc and requires zinc for recognition of HLA-C on target cells. J Immunol. 1995;155:4143–6. [PubMed] [Google Scholar]
- 19.Köhler K, Xiong S, Brzostek J, Mehrabi M, Eissmann P, Harrison A, Cordoba S-P, Oddos S, Miloserdov V, Gould K, Burroughs NJ, et al. Matched Sizes of Activating and Inhibitory Receptor/Ligand Pairs Are Required for Optimal Signal Integration by Human Natural Killer Cells. PLoS One. 2010;5:e15374. doi: 10.1371/journal.pone.0015374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pageon SV, Cordoba S-P, Owen DM, Rothery SM, Oszmiana A, Davis DM. Superresolution Microscopy Reveals Nanometer-Scale Reorganization of Inhibitory Natural Killer Cell Receptors upon Activation of NKG2D. Sci Signal. 2013;6:ra62–ra62. doi: 10.1126/scisignal.2003947. [DOI] [PubMed] [Google Scholar]
- 21.Guia S, Jaeger BN, Piatek S, Mailfert S, Trombik T, Fenis A, Chevrier N, Walzer T, Kerdiles YM, Marguet D, Vivier E, et al. Confinement of activating receptors at the plasma membrane controls natural killer cell tolerance. Sci Signal. 2011;4:ra21. doi: 10.1126/scisignal.2001608. [DOI] [PubMed] [Google Scholar]
- 22.Delcassian D, Depoil D, Rudnicka D, Liu M, Davis DM, Dustin ML, Dunlop IE. Nanoscale ligand spacing influences receptor triggering in T cells and NK cells. Nano Lett. 2013;13:5608–5614. doi: 10.1021/nl403252x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumgart F, Arnold AM, Leskovar K, Staszek K, Fölser M, Weghuber J, Stockinger H, Schütz GJ. Varying label density allows artifact-free analysis of membrane-protein nanoclusters. Nat Methods. 2016;13:661–664. doi: 10.1038/nmeth.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossboth B, Arnold AM, Ta H, Platzer R, Kellner F, Huppa JB, Brameshuber M, Baumgart F, Schütz GJ. TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat Immunol. 2018;19:821–827. doi: 10.1038/s41590-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yawata M, Yawata N, Draghi M, Little A-M, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–45. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson ME, Long EO. Inhibitory receptor signaling via tyrosine phosphorylation of the adaptor Crk. Immunity. 2008;29:578–88. doi: 10.1016/j.immuni.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oszmiana A, Williamson DJ, Cordoba S-P, Morgan DJ, Kennedy PR, Stacey K, Davis DM. The Size of Activating and Inhibitory Killer Ig-like Receptor Nanoclusters Is Controlled by the Transmembrane Sequence and Affects Signaling. Cell Rep. 2016;15:1957–1972. doi: 10.1016/j.celrep.2016.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Shen K, Lu W, Cole PA. The role of C-terminal tyrosine phosphorylation in the regulation of SHP-1 explored via expressed protein ligation. J Biol Chem. 2003;278:4668–4674. doi: 10.1074/jbc.M210028200. [DOI] [PubMed] [Google Scholar]
- 30.Azoulay-alfaguter I, Strazza M, Peled M, Novak HK, Muller J, Dustin ML, Mor A. The tyrosine phosphatase SHP-1 promotes T cell adhesion by activating the adaptor protein CrkII in the immunological synapse. Sci Signal. 2017;1:1–12. doi: 10.1126/scisignal.aal2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Sung S, Yip MLR, Lawrence HR, Ren Y, Guida WC, Sebti SM, Lawrence NJ, Wu J. Discovery of a Novel Shp2 Protein Tyrosine Phosphatase Inhibitor. Mol Pharmacol. 2006;70:562–570. doi: 10.1124/mol.106.025536. [DOI] [PubMed] [Google Scholar]
- 32.Yawata M, Yawata N, Draghi M, Partheniou F, Little A-M, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–80. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boudreau JE, Mulrooney TJ, Le Luduec J-B, Barker E, Hsu KC. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J Immunol. 2016;196:3398–3410. doi: 10.4049/jimmunol.1502469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol. 2009;182:4572–80. doi: 10.4049/jimmunol.0803900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sim MJW, Stowell J, Sergeant R, Altmann DM, Long EO, Boyton RJ. KIR2DL3 and KIR2DL1 show similar impact on licensing of human NK cells. Eur J Immunol. 2016;46:185–191. doi: 10.1002/eji.201545757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.V Pageon S, Tabarin T, Yamamoto Y, Ma Y, Bridgeman JS, Cohnen A, Benzing C, Gao Y, Crowther MD, Tungatt K, Dolton G, et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc Natl Acad Sci. 2016;113:E5454–E5463. doi: 10.1073/pnas.1607436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38:461–74. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Lopes FB, Bálint Š, Valvo S, Felce JH, Hessel EM, Dustin ML. Davis DM Membrane nanoclusters of FcγRI segregate from inhibitory SIRPα upon activation of human macrophages. J Cell Biol. 2017;216:1123–1141. doi: 10.1083/jcb.201608094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, Gérard A, Liu T-L, Chen B-C, Betzig E, Bartumeus F, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science (80-. ) 2017;356 doi: 10.1126/science.aal3118. eaal3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huhn O, Chazara O, Ivarsson MA, Retière C, Venkatesan TC, Norman PJ, Hilton HG, Jayaraman J, Traherne JA, Trowsdale J, Ito M, et al. High-Resolution Genetic and Phenotypic Analysis of KIR2DL1 Alleles and Their Association with Pre-Eclampsia. J Immunol. 2018;201:2593–2601. doi: 10.4049/jimmunol.1800860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bari R, Rujkijyanont P, Sullivan E, Kang G, Turner V, Gan K, Leung W. Effect of donor KIR2DL1 allelic polymorphism on the outcome of pediatric allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. 2013;31:3782–3790. doi: 10.1200/JCO.2012.47.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, Leung W. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu D, Peterson ME, Long EO. The adaptor protein Crk controls activation and inhibition of natural killer cells. Immunity. 2012;36:600–11. doi: 10.1016/j.immuni.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bálint Š, Lopes FB, Davis DM. A nanoscale reorganization of the IL-15 receptor is triggered by NKG2D in a ligand-dependent manner. Sci Signal. 2018;11 doi: 10.1126/scisignal.aal3606. eaal3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loftus C, Saeed M, Davis DM, Dunlop IE. Activation of Human Natural Killer Cells by Graphene Oxide-Templated Antibody Nanoclusters. Nano Lett. 2018;18:3282–3289. doi: 10.1021/acs.nanolett.8b01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viant C, Fenis A, Chicanne G, Payrastre B, Ugolini S, Vivier E. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms6108. [DOI] [PubMed] [Google Scholar]
- 47.Forslund E, Sohlberg E, Enqvist M, Olofsson PE, Malmberg K-J, Önfelt B. Microchip-Based Single-Cell Imaging Reveals That CD56 dim CD57 – KIR – NKG2A + NK Cells Have More Dynamic Migration Associated with Increased Target Cell Conjugation and Probability of Killing Compared to CD56 dim CD57 – KIR – NKG2A – NK Cells. J Immunol. 2015;195:3374–3381. doi: 10.4049/jimmunol.1500171. [DOI] [PubMed] [Google Scholar]
- 48.Thomas LM, Peterson ME, Long EO. Cutting Edge: NK Cell Licensing Modulates Adhesion to Target Cells. J Immunol. 2013;191:3981–3985. doi: 10.4049/jimmunol.1301159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, et al. Cutting Edge: Allele-Specific and Peptide-Dependent Interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 50.Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat Immunol. 2011;12:655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- 51.Jones DC, Roghanian A, Brown DP, Chang C, Allen RL, Trowsdale J, Young NT. Alternative mRNA splicing creates transcripts encoding soluble proteins from most LILR genes. Eur J Immunol. 2009;39:3195–3206. doi: 10.1002/eji.200839080. [DOI] [PubMed] [Google Scholar]
- 52.Kennedy PR, Barthen C, Williamson DJ, Davis DM. HLA-B and HLA-C Differ in Their Nanoscale Organization at Cell Surfaces. Front Immunol. 2019;10:1–10. doi: 10.3389/fimmu.2019.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fauriat C, Ivarsson MA, Ljunggren H-G, Malmberg K-J, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115:1166–1174. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- 54.Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ripley BD. Modelling spatial patterns. J R Stat Soc Ser B. 1977;39:172–212. [Google Scholar]
- 56.Getis A, Franklin J. Second-Order Neighborhood Analysis of Mapped Point Patterns. Ecology. 1987;68:473–477. [Google Scholar]
- 57.Baddeley A, Turner R. Practical Maximum Pseudolikelihood for Spatial Point Patterns. Australian & New Zealand Journal of Statistics. 2000;42:283–322. [Google Scholar]
- 58.Wiegand T, Moloney KA. Handbook of Spatial Point-Pattern Analysis in Ecology. CRC Press; 2014. [Google Scholar]
- 59.Smal I, Loog M, Niessen W, Meijering E. Quantitative comparison of spot detection methods in fluorescence microscopy. IEEE Trans Med Imaging. 2010;29:282–301. doi: 10.1109/TMI.2009.2025127. [DOI] [PubMed] [Google Scholar]
- 60.Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: Integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 61.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein La, Hilton HG, Pando MJ, Koram KA, Riley EM, Abi-Rached L, Parham P. Co-evolution of Human Leukocyte Antigen (HLA) Class I Ligands with Killer-Cell Immunoglobulin-Like Receptors (KIR) in a Genetically Diverse Population of Sub-Saharan Africans. PLoS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 63.Griffié J, Shannon M, Bromley CL, Boelen L, Burn GL, Williamson DJ, Heard NA, Cope AP, Owen DM, Rubin-Delanchy P. A Bayesian cluster analysis method for single-molecule localization microscopy data. Nat Protoc. 2016;11:2499–2514. doi: 10.1038/nprot.2016.149. [DOI] [PubMed] [Google Scholar]
- 64.Srpan K, Ambrose A, Karampatzakis A, Saeed M, Cartwright ANR, Guldevall K, Dos Santos Cruz De Matos G, Önfelt B, Davis DM. Shedding of CD16 disassembles the NK cell immune synapse and boosts serial engagement of target cells. J Cell Biol. 2018;217:3267–3283. doi: 10.1083/jcb.201712085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceri KW. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2016;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.